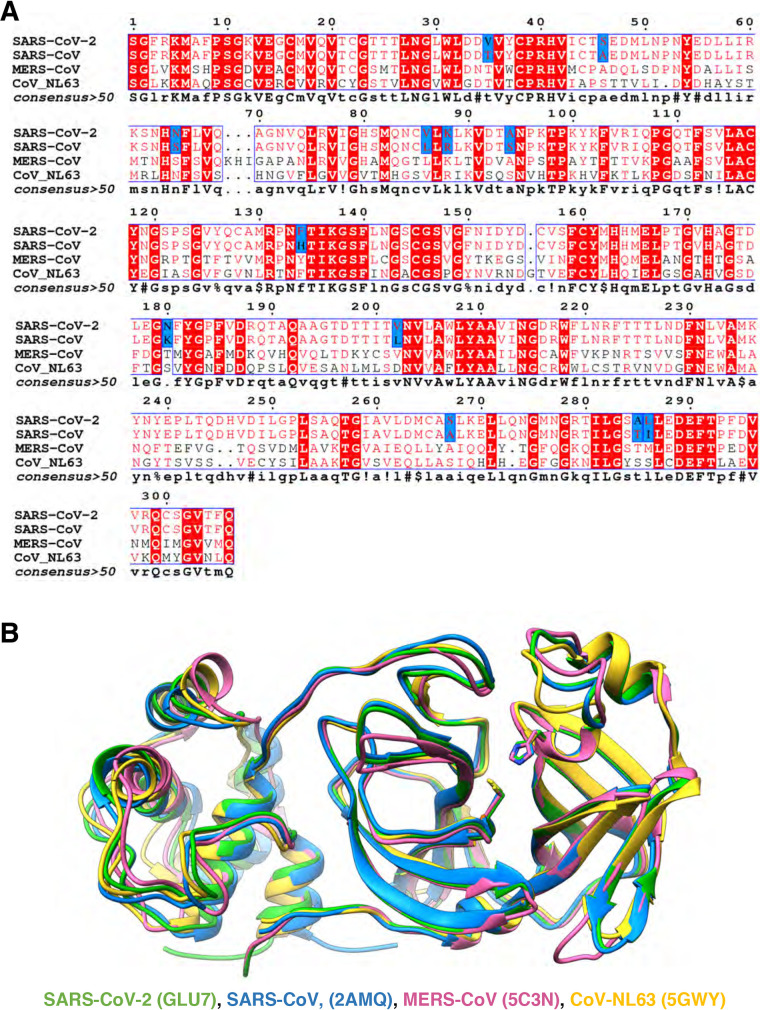
FIG 1.
Sequence conservation and structural comparison of four of coronaviruses’ main proteases. (A) The amino acid sequences of SARS-CoV-2 Mpro and SARS-CoV Mpro have 96% identity (12 amino acids [highlighted in blue] of 306 amino acids differ between the two). Conserved amino acids among the four Mpro sequences are shown in red. Distant coronavirus variants MERS-CoV and CoV-NL63 exhibit much less sequence conservation. Sequences were aligned using ENDscript server. (B) Structures of Mpro of SARS-CoV-2 (PDB entry 6LU7; in green) superimposed on Mpro of SARS-CoV (2AMQ; in blue), MERS-CoV (5C3N; in salmon), and CoV-NL63 (5GWY; in yellow) are shown. Catalytic dyad residues Cys145 and His41 are indicated in stick mode. For clarity, only a monomer structure is shown and the covalent protease inhibitor N3 was omitted. The picture was generated using UCSF Chimera.