Abstract
Background
T cell activation is associated with increase in glycolysis and glutaminolysis. T cell immunoglobulin and mucin domain containing protein-3 (TIM-3), a T cell surface molecule, downregulates T cell activation and leads to insufficient immunity in cancer and chronic infection. TIM-3 regulates T cell activation possibly through alterations in metabolism; however, the relationship between TIM-3 expression and T cell metabolic changes has not been well studied.
Results
We investigated the association between TIM-3 expression and metabolic changes by analyzing glucose metabolism, glutamine metabolism, and mitochondrial function in TIM-3 overexpressing or knockout Jurkat T cell lines relative to their control cell lines. Glucose uptake and consumption, and lactate release were downregulated by TIM-3 expression but upregulated by TIM-3 knockout. Concomitantly, the expression of the glucose transporter, Glut1, but not Glut2, 3, or 4 was altered by TIM-3 expression. However, TIM-3 expression alone could not account for the change in glutamine consumption, glutamate release, and mitochondrial mass, ROS production or membrane potential in these cell lines.
Conclusion
Our results show the association of TIM-3 expression with T cell glucose metabolism. These results are significant in chronic infections and cancers where it is necessary to control TIM-3 expressing T cells.
Keywords: HAVCR2, Glycolysis, CD4+ T cell, Glutaminolysis, Glucose transporter
Background
T cell activation alters cellular metabolism [1]. Shifting of a T cell from a resting to an active state is accompanied by metabolic alterations to meet the increased demand for energy and macromolecule synthesis for cell proliferation and effector functions [2]. The metabolic alterations in activated T cells include increase in glucose and glutamine consumption [3, 4]. Glucose and glutamine are catabolized through glycolysis and glutaminolysis, respectively, to supply metabolic intermediates such as lactate and glutamate [3–5]. Glucose and glutamine are also directed to the hexosamine biosynthesis pathway to produce uridine diphosphate N-acetylglucosamine (UDP-GlcNAc), which is used for protein glycosylation. Blocking glucose and glutamine metabolism interferes with T cell activation [6, 7]. Downregulation of glycolysis inhibits the production of interferon- γ by activated CD4+ T cells as well as memory CD8+ T cells [8, 9]. Restriction of glucose and glutamine supply suppresses T cell proliferation through perturbation of protein glycosylation [6, 7]. Thus, T cell activation is closely associated with and regulated by metabolism.
Mitochondria play a critical role in T cell activation and differentiation [10]. Upon activation, the mitochondrial mass and DNA level increase in the T cell [10]. Antigenic stimulation leads to the relocation of mitochondria toward the immune synapse where they modulate calcium signaling and generate ATP for T cell activation [11, 12]. Mitochondria also produce reactive oxygen species (ROS) needed for the activation of transcription factors such as NFAT and AP-1 [13, 14]. Further, mitochondria are the organelles that provide the building blocks for macromolecule synthesis through the TCA cycle and oxidative phosphorylation [10]. Mitochondrial dysfunction impairs activation-induced T cell proliferation and differentiation [15].
TIM-3 is a transmembrane glycoprotein encoded by the HAVCR2 gene and it regulates T cell activation [16]. TIM-3 molecules were initially identified on terminally differentiated TH1 cells and are also found on TH17 cells, regulatory T cells, exhausted CD4+ T cells, as well as CD8+ T cells. TIM-3 expression is inversely correlated with IL-2 or IFN-γ production [16–18]. Following the interaction of TIM-3 with its ligands galectin-9 or CEACAM1, inhibitory signals are generated, leading to suppression of T cell activation [16, 19–21]. The prevalence of TIM-3 expressing T cells increases in various diseases such as chronic hepatitis C viral infection and cancers [22–25]. In vivo, TIM-3-blocking antibodies increase T cell immunity against chronic viral infections and cancers [22, 26–28]. Understanding how TIM-3 regulates T cell activation has been an important research focus on improving the protective immune response in chronic infections and cancers.
Given that blocking glucose or glutamine metabolism leads to T cell dysregulation and that TIM-3 negatively impacts T cell activation, it is possible that TIM-3 may downregulate glycolysis or glutaminolysis; however, to the best of our knowledge there are no studies that report the effect of TIM-3 on the T cell metabolism. In this study, we examined the effect of TIM-3 expression on glucose and glutamine metabolism as well as on mitochondrial content and function because mitochondria are involved in the glucose and glutamine metabolism. We found an association between TIM-3 expression and glucose consumption and lactate release but not with glutamine consumption, glutamate release, mitochondrial ROS production or mitochondrial membrane potential in Jurkat T cells. Also, we found that TIM-3 expression was linked to glucose uptake and GLUT1 expression levels.
Results
Glucose consumption and lactate release except glutamine consumption and glutamate release, decrease in Jurkat T cells according to TIM-3 overexpression
To examine whether TIM-3 expression was associated with glucose metabolism in T cells, we analyzed glucose consumption and the release of its metabolite, lactate, by two Jurkat T cell-derived cell lines that overexpress TIM-3; JLT3 expresses TIM-3 in the context of lentiviral vector and T7 expresses TIM-3 as well as GFP in the context of pIRES2-EGFP plasmid [29, 30]. JLT3 and T7 express TIM-3 in the absence of stimulation, and are significantly upregulated in the presence of PMA stimulation [30] (Suppl. Fig. 1). Glucose consumption and lactate release decreased significantly in the TIM-3 overexpressing cells compared to the control cells stably transfected with a corresponding empty vector both in the presence and absence of stimulation with PMA and ionomycin (Fig. 1a, b). We next analyzed glutamine metabolism in these cell lines. Glutamine consumption and glutamate release were similar between JLT3 and its control cell line, JLV, but increased significantly in T7 compared to its control cell line G2 (Fig. 1c, d). This discrepancy in glutamine consumption and glutamate release in JLT3 and T7 could be due to the differences in TIM-3 expression levels (Suppl. Fig. 1) or cellular properties attributed to the process of establishing stable cell lines such as the change in certain gene expression caused by the insertion of the vector into the host chromosomal DNA. If the latter is the case, increase in glutamine consumption and glutamate release in T7 could not be linked to TIM-3 expression. Taken together, these results are supportive of the association between TIM-3 expression and glucose metabolism but inconclusive for the association between TIM-3 expression and glutamine metabolism.
Fig. 1.
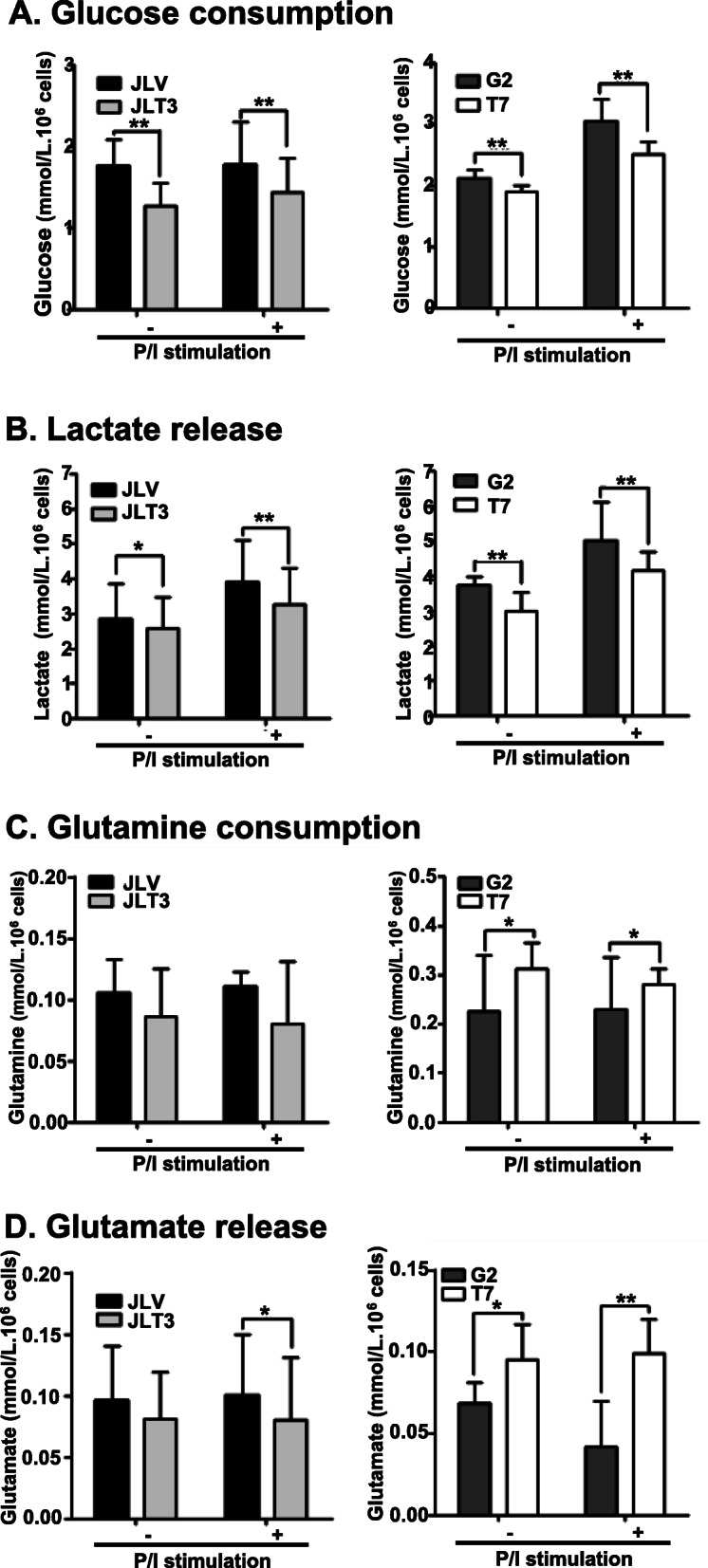
Glucose and glutamine metabolism in TIM-3 overexpressing cell lines. Glucose consumption (a), lactate release (b), glutamine consumption (c), and glutamate release (d), in control cells (JLV and G2) and TIM-3 overexpressing cells (JLT3 and T7) cultured in the absence (−) or presence (+) of PMA (25 ng/ml) and Iono (10 μM) for 6 h. A, B; Data of more than three independent experiments in quintuplicate or sextuplicate. C, D; Data represent two independent experiments performed in sextuplicate. Data are mean ± standard deviation (SD). P/I: PMA and Iono. *: P < 0.05
Glucose consumption and lactate release increase in Jurkat T cells with TIM-3 knockout
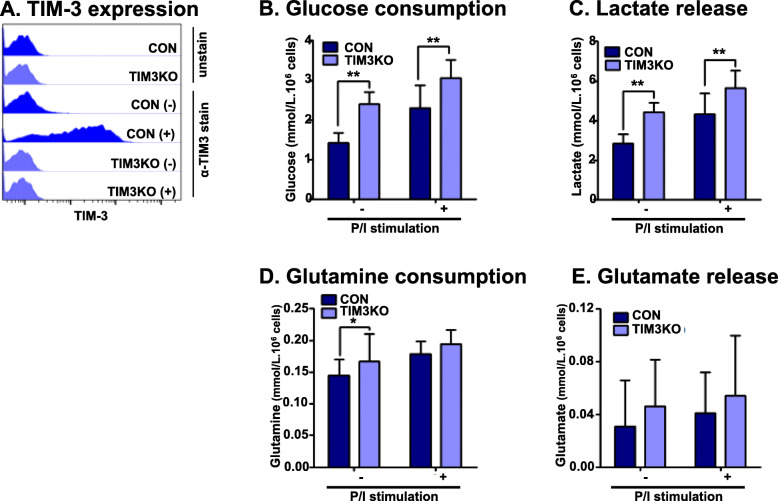
To further examine the association of TIM-3 expression with glucose and glutamine metabolism, a TIM-3 knockout cell line was established (Fig. 2). Figure 2a shows no expression of TIM-3 in TIM3KO cells even after stimulation, while the basal and stimulation-induced TIM-3 expression was observed in the CON cells transfected with an empty vector. The association could be established if glucose consumption and lactate release increased but glutamine consumption and glutamate release decreased in the TIM-3 knockout cells. As expected, glucose consumption and lactate release increased significantly in TIM3KO cells compared to CON cells in the presence and absence of stimulation (Fig. 2b, c). However, glutamine consumption and glutamate release tended to be enhanced in TIM3KO cells compared to CON cells although statistical significance was observed only in glutamine consumption in the absence of stimulation (Fig. 2d, e). Considering the results of JLT3 and T7, these data were not supportive of the association between TIM-3 expression and glutamine consumption and glutamate release; however, these results strengthened the evidence for the involvement of TIM-3 expression in glucose metabolism of Jurkat T cells.
Fig. 2.
Glucose and glutamine metabolism in TIM-3 knockout Jurkat T cells. The surface expression of TIM-3 (a), glucose consumption (b), lactate release (c), glutamine consumption (d), and glutamate release (e) in control cells (CON) and TIM-3 knockout cells (TIM3KO) in the absence (−) or presence (+) of PMA (25 ng/ml) and Iono (10 μM) for 6 h. Data represent two or three independent experiments performed in sextuplicate. Data are mean ± SD. P/I: PMA and Iono. *: P < 0.05
TIM-3 cytoplasmic tail is required to decrease glucose consumption and lactate release in Jurkat T cells by TIM-3 overexpression
We hypothesized that if TIM-3 was involved in glucose consumption and lactate release, TIM-3 signaling should be important and the cytoplasmic tail of TIM-3 (the signal-delivering part) was required for TIM-3-mediated effects on glucose consumption and lactate release. Therefore, glucose consumption and lactate release were assessed in JLct36, JLct43, JLct54, and JLct64 cell lines that express TIM-3 with a truncated cytoplasmic tail [30]. In contrast to JLT3 that expresses full-length TIM-3, these cell lines with truncated TIM-3 consumed and secreted similar amount of glucose and lactate, respectively, compared to JLV, the control cell line (Fig. 3a and b). These results indicate that the cytoplasmic tail of TIM-3 is critical to the regulation of glucose consumption and lactate release by TIM-3 in Jurkat T cells.
Fig. 3.
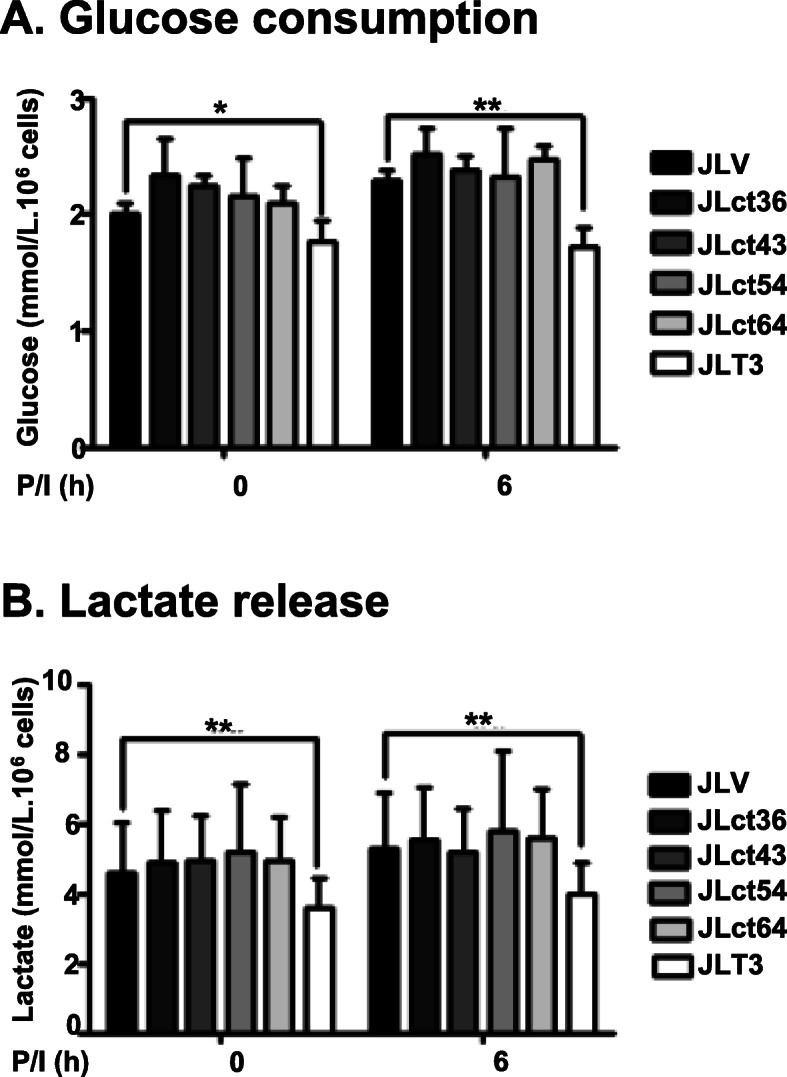
Glucose consumption and lactate release in cells expressing TIM-3 with deletion of cytoplasmic tail. Glucose consumption (a) and lactate release (b) in control cells (JLV), TIM-3 cytoplasmic tail deletional mutant cells (JLct36, JLct43, JLct54, and JLct64) and cells expressing full-length TIM-3 (JLT3) in the absence (−) or presence (+) of PMA (25 ng/ml) and Iono (10 μM) for 6 h. Data represent two independent experiments performed in sextuplicate. Data are mean ± SD. P/I: PMA and Iono. *: P < 0.05
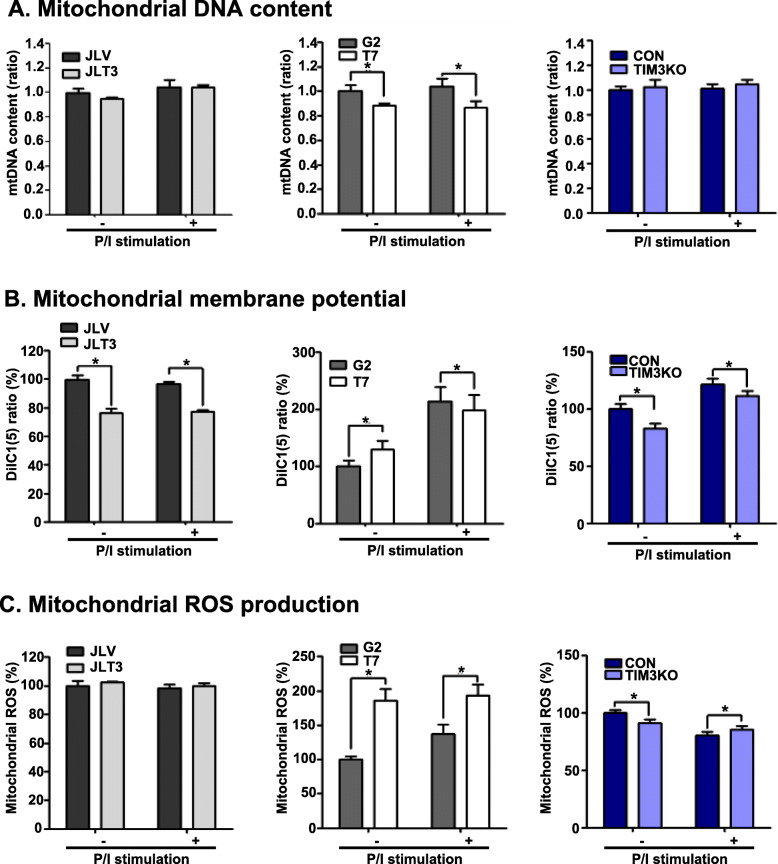
TIM3 expression in Jurkat T cells is not associated with changes in mitochondrial mass, membrane potential, or ROS production
Mitochondria contribute to cellular metabolism through the generation of ATP and regulation of the redox status [14]. To find out whether TIM-3 expression influences mitochondrial mass or function, we analyzed mitochondrial DNA content, membrane potential, and ROS production in JLT3, T7 and TIM3KO compared to their control cells. The reason was that parallel changes in mitochondrial mass or function of JLT3 and T7 and opposite changes in mitochondrial mass or function of TIM3KO compared to their control cell lines would support the possible association between TIM-3 and mitochondrial function. Mitochondrial DNA content was similar between JLT3 and JLV as well as between TIM3KO and CON but significantly less in T7 compared to G2 (Fig. 4a). Mitochondrial membrane potential decreased in JLT3 compared to JLV, whereas it increased in T7 compared to G2 in the absence of stimulation but decreased in the presence of stimulation. In the case of TIM3KO, the mitochondrial membrane potential decreased compared to that of CON cells regardless of stimulation (Fig. 4b). Mitochondrial ROS production was similar between JLT3 and JLV, but was significantly increased in T7 compared to G2; however, it slightly decreased in TIM3KO compared to CON in the absence of stimulation but not in the presence of stimulation (Fig. 4c). These results showed that TIM-3 expression might not be associated with the mitochondrial content, membrane potential, and ROS production in Jurkat T cells.
Fig. 4.
Mitochondrial DNA content, membrane potential, and ROS production in TIM-3 overexpressing or knockout cells. Mitochondrial DNA content (a), membrane potential (b), and ROS production (c), in control cells (JLV cell line, G2 clone, and CON clone), TIM-3 overexpressing cells (JLT3 cell line and T7 clone), and TIM-3 knockout cells (TIM3KO clone) cultured in the absence (−) or presence (+) of PMA (25 ng/ml) and Iono (10 μM) for 6 h. Mitochondrial DNA content was measured using qPCR. Mitochondrial membrane potential and ROS production were assessed by using flow cytometry with DilC(5) and MitoSOX™, respectively. Data represent two independent experiments performed in triplicates or sextuplets (JLT3 cell line and T7 clone). Data represent an experiment performed in sextuplets (TIM3KO clone). Data are mean ± SD. Mitochondrial DNA ratio: mitochondrial DNA to the nuclear 18S DNA. P/I: PMA and Iono. *: P < 0.05
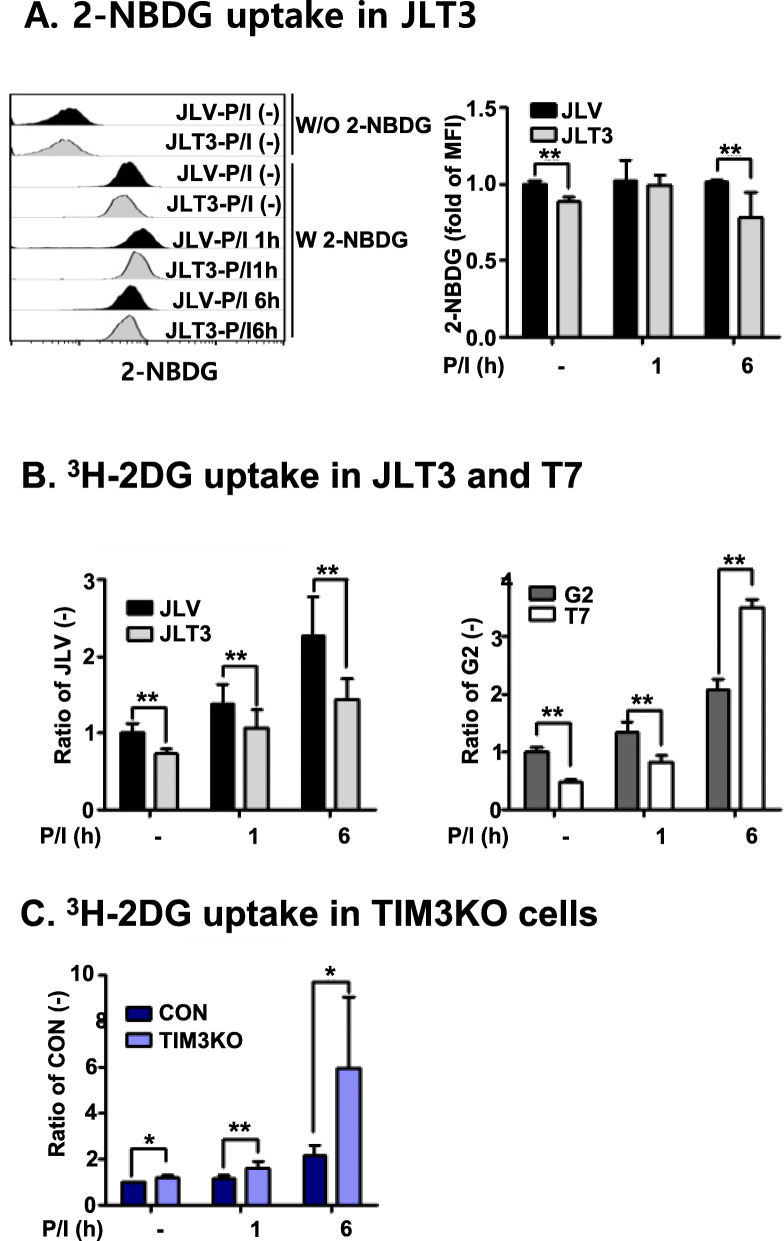
TIM-3 expression decreases glucose uptake of Jurkat T cells at resting state and at an early time point of activation
We next examined how TIM-3 expression influenced glucose consumption and lactate release. Given that glucose consumption starts with glucose uptake, the association of TIM-3 expression with glucose uptake was investigated. The glucose uptake of JLT3 cells was measured using a 2-NBDG fluorescence-labeled glucose analog (Fig. 5a). The amount of 2-NBDG uptake by JLT3 was significantly less than that by control JLV before and after stimulation. Similar results were obtained when we assessed glucose uptake by JLT3 using 3H-labeled deoxy glucose (2DG, Fig. 5b). Also, glucose uptake by T7 cells was significantly reduced compared to the control G2 cells in the absence of stimulation or 1 h post-stimulation although it was significantly increased 6 h post-stimulation. Concomitantly, glucose uptake by TIM3KO was significantly higher than that by control cells in the absence and presence of stimulation (Fig. 5c). These results indicate that TIM-3 expression may be involved in modulating glucose uptake in Jurkat T cells.
Fig. 5.
Glucose uptake by TIM-3 overexpressing or knockout cells. Glucose uptake by control cells (JLV, G2, and CON), TIM-3 overexpressing cells (JLT3 and T7), and TIM-3 knockout cells (TIM3KO) stimulated with PMA (25 ng/ml) and Iono (10 μM) for the indicated times was determined using a 2-NBDG (a), or 3H-2-DG uptake (b and c). Data represent two independent experiments performed in triplicate or sextuplicate. Data are mean ± SD. *: P < 0.005
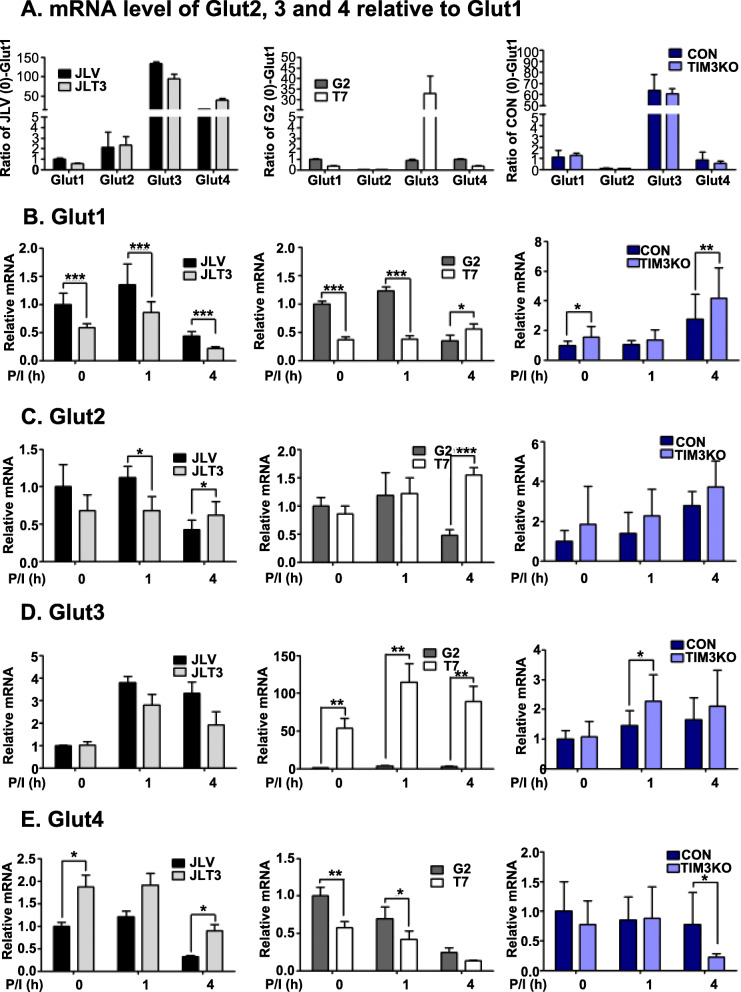
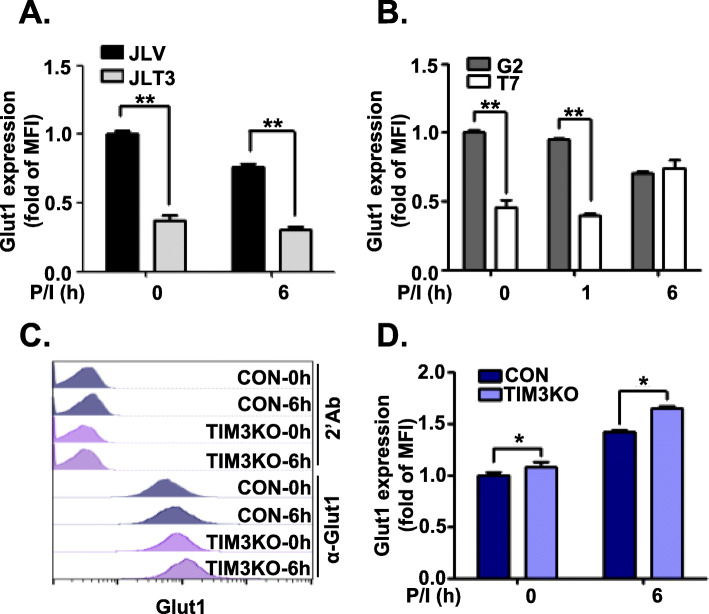
TIM-3 expression decreases Glut1 expression in Jurkat T cells at resting state and at an early time point of activation
Human T cells take up glucose through glucose transporters such as Glut1 [31]. Therefore, it would be interesting to examine whether TIM-3 expression could affect the expression of Glut protein family members. We first examined the relative mRNA expression of Glut1, Glut2, Glut3, and Glut4 in TIM3 overexpressing cell lines, TIM3KO cell line, and their respective control cell lines in the resting state by using quantitative RT-PCR (Fig. 6a). In all cell lines except the G2, Glut3 transcripts were most abundant. In JLT3 and JLV cells, Glut4 mRNA levels were higher than Glut1 mRNA levels, but not in T7, G2, TIM3KO, and CON cells. We then examined the association between the transcript level of the Glut isotypes with TIM-3 expression before and after stimulation of the cells. Glut1 transcript were less abundant in JLT3 and T7 than in JLV and G2, in the absence of stimulation and 1 h post-stimulation, whereas Glut1 transcript were more abundant in TIM3KO than in CON (Fig. 6b). Thus, Glut1 transcript level was associated with TIM-3 expression in the absence of stimulation and 1 h post-stimulation. In the case of Glut2, Glut3, and Glut4 mRNAs, there was no correlation between their mRNA levels and TIM-3 expression (Fig. 6c, d, and e). These results indicate that TIM-3 expression may affect the expression of Glut1 but not that of Glut2, Glut3 or Glut4. Therefore, Glut1 protein expression was assessed in these cells (Fig. 7). Similar to Glut1 mRNA levels, Glut1 protein expression was significantly lower in JLT3 than in JLV before and after stimulation (Fig. 7a). Also, Glut1 protein level was significantly lower in T7 than in G2 in the absence of stimulation and 1 h post-stimulation, but higher 6 h post-stimulation (Fig. 7b). In TIM3KO cells, Glut1 protein level was significantly increased compared to the CON cells both before and after stimulation (Fig. 7c and d). Taken together, these results suggest an association between TIM-3 and Glut1 expression, which may account for the link between TIM-3 expression and glucose uptake as well as glucose consumption.
Fig. 6.
Transcript levels of Glut1, 2, 3 and 4 in TIM-3 overexpressing or knockout cells. Transcript levels of Glut2, 3, and 4 relative to Glut1 in each cell line in the absence of stimulation (a). mRNA levels of Glut1 (b), Glut2 (c), Glut3 (d), and Glut4 (e) in control cells (JLV, G2, and CON), TIM-3 overexpressing cells (JLT3 and T7), and TIM-3 knockout cells (TIM3KO) stimulated with PMA (25 ng/ml) and Iono (10 μM) for the indicated time was determined using qRT-PCR. Relative transcript: mRNA level of each cell line at the indicated time point relative to mRNA level in the corresponding control cells at the 0 time point. Data represent two independent experiments performed in triplicate or sextuplicate. Data are mean ± SD. *: P < 0.05
Fig. 7.
Glut1 protein level in TIM-3 overexpressing or knockout cells. Glut1 protein levels analyzed by using flow cytometry in TIM-3 overexpressing cells (a and b), and TIM-3 knockout cells (TIM3KO) (c and d) along with the corresponding control cells (JLV, G2, and CON, respectively). Cells stimulated for the indicated time with PMA (25 ng/ml) and Iono (10 μM) were labeled with PE-conjugated anti-mouse Ig Ab alone or together with mouse anti-Glut1 Ab and then the mean fluorescence intensity (MFI) was assessed. Representative histogram (c). Data represent two independent experiments performed in triplicate or sextuplicate. Data are mean ± SD. Fold of MFI: MFI relative to MFI of corresponding control cells at the 0 time point. P/I: PMA and Iono. *: P < 0.005
Discussion
TIM-3-mediated T cell regulation is a critical research agenda because manipulation of TIM-3 pathway may lead to enhancement of immunity against tumor cells or certain pathogens [22–27]. In this study, the possibility that TIM-3 may alter T cell activation-associated metabolism was evaluated, and three novel findings were demonstrated. Firstly, TIM-3 is involved in glucose metabolism but not glutamine consumption and release, mitochondrial DNA content and membrane potential, or ROS production. Secondly, there is a link between TIM-3 expression and glucose uptake as well as Glut1 expression. Thirdly, the cytoplasmic tail of TIM-3 is required for its effect on glucose metabolism.
TIM-3 involvement in glucose consumption and lactate release was revealed using CD4+ Jurkat T cell-derived cell lines namely, two TIM-3 overexpressing JLT3 and T7, and a TIM3KO. Contrary to no apparent consistent results for glutamine consumption and glutamate release, mitochondrial DNA content and membrane potential, or ROS production, glucose consumption and lactate release were decreased in JLT3 and T7, but increased in TIM3KO, indicating that TIM-3 downregulates glucose consumption and lactate release. In line with our results, reduced glucose consumption was also reported in the exhausted T cells, of which approximately 30% expressed TIM-3, although 80% of these T cells also expressed other inhibitory molecules such as PD-1 and the role of TIM-3 in glucose consumption was not investigated [32]. The role of PD-1 in reduced glucose consumption was revealed in CD8+ T cells but not in CD4+ T cells [32]. Notably, PD-1 transcription was not detected in Jurkat T cells (data not shown). Thus, the novel finding of our study is the association between TIM-3 and glucose metabolism in CD4+ human T cell lines. Due to the limitation of Jurkat T cells that were derived from acute T cell leukemia, further study is needed to prove this finding by using primary T cells.
The decrease in glucose uptake and Glut1 expression according to TIM-3 expression was demonstrated in Jurkat T cells. Glucose uptake and Glut1 expression levels were lower in JLT3 and T7, but higher in TIM3KO. Thus, we propose that TIM-3 expression may affect glucose uptake and consumption partly through regulation of Glut1 expression in a human T cell line. Glut1 is a principal glucose transporter required for the activation of human primary T cells which express Glut1, 3, 6, and 8 [31, 33]. The cell lines used in this study express Glut1, 2, 3, 4, 6, and 8 mRNAs and the relative amount of each mRNA vary between the cell lines (Fig. 6 and Suppl. Fig. 2). Our results show that Glut1 mRNA and protein levels, but not Glut2, 3, 4, 6 and 8 mRNA levels, are associated with TIM-3 expression (Fig. 6 and Suppl. Fig. 2A, B). Our results are similar to the previous report that Glut1 expression increased following T cell activation and decreased in exhausted T cells expressing PD-1 [32]. We ruled out the role of sodium-glucose linked transporter 1 (SGLT1), which are efficient at low extracellular glucose concentrations [34], in alteration of glucose uptake in a TIM-3 expression-dependent way by showing no association of SGLT1 transcript level with TIM-3 expression (Suppl. Fig. 2C). Thus, low Glut1 expression probably accounts for the reduced glucose uptake and consumption, and lactate release in TIM-3 expressing cells.
We should however mention that in TIM-3 expressing T7 cells, the reduced glucose uptake and Glut1 expression were transient, and increased 6 h post-stimulation compared to the control G2 cells. These finding suggest that additional mechanisms leading to reduced glucose consumption exist in the TIM-3 expressing cells. Given that hexokinase 2 (HK2) is the rate-limiting enzyme in glycolysis and that 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) directs glucose metabolism to either the pentose phosphate pathway or glycolysis-dependent pathway depending on its expression level and modification [35–38], we examined HK2 and PFKFB3 expression. However, any association between their expression and that of TIM-3 was not found (Suppl. Fig. 3 and and Suppl. methods). Thus, the additional mechanisms by which TIM-3 reduces glucose consumption remain inconclusive.
Activation-induced glycolysis is required for cytokine production [8, 9], hence, TIM-3-associated reduction in glucose consumption and lactated release may contribute to decrease in the production of IL-2 and IFN-γ by TIM-3. Downregulation of IL-2 expression has been reported in JLT3 and T7 cells [30]. Therefore, we tested this hypothesis by assessing IL-2 production in JLT3 or T7 cells overexpressing Glut1. If the hypothesis is true, then IL-2 production should increase in these cells after Glut1 expression. Unfortunately, Glut1 overexpression was not achieved using the transfection of Glut1 expression vector into these cells. Further study is required to prove this hypothesis.
Since the cytoplasmic tail of TIM-3 mediates the delivery of signals and is essential for reduced glucose consumption and lactate release in TIM-3 overexpressing Jurkat T cells, expression of TIM-3 with a deletion in its cytoplasmic tail did not reduce glucose consumption and lactate release. It interacts with Bat3 or the long non-coding RNA, Lnc-Tim3, to activate distinct signaling pathways [39–41]. TIM-3 is also reported to interact with the interleukin-inducible T cell kinases (ITKs), Fyn and Lck, and p85 phosphatidylinositol 3-kinase (PI3K) and to regulate their activation [42]. PI3K/AKT/mTOR activation promotes Glut1 expression, glycolysis, and IL-2 production by T cells [32, 33, 36]. It is plausible that Glut1 expression and glucose consumption are modulated by signaling transmitted through the TIM-3 cytoplasmic tail in TIM-3 expressing Jurkat T cells.
Conclusions
Our results show an association between TIM-3 expression and Jurkat T cell glucose metabolism. This association is a novel and significant finding in the alteration of T cell function by TIM-3 and raises the possibility of regulating TIM-3 expressing T cells through controlling glycolysis and/or glucose transporter expression. This is relevant to chronic infection and cancer in which increased frequency of hypofunctional TIM-3-expressing effector T cells have been observed.
Methods
Cell and culture
Cell lines over-expressing TIM-3 (JLT3 and JT7), cell lines expressing TIM-3 with a truncated cytoplasmic tail (JLct36, JLct43, JLct54, and JLct64), and control cells (JLV and JG2) were generated as described previously [29, 30]. Cells were grown in complete RPMI medium (Invitrogen, Life technologies, Gaithersburg, MD, USA) with 10% heat-inactivated FBS (Invitrogen), and penicillin/ streptomycin (Invitrogen). For activation, cells (1 × 106 cells/well) were stimulated with phorbol 12-myristate 13-acetate (PMA; 25 ng/ml; Sigma-Aldrich, St Louis, MO, USA) and Ionomycin (Iono; 1 μM; Sigma-Aldrich) for an appropriate time.
Establishment of TIM-3 knockout cell line
The pSpCas9(BB)-2A-GFP (PX458) plasmid (Addgene plasmid #48138) was used to generate TIM-3 knockouts [43]. Oligonucleotides for single guide RNA (sgRNA) for TIM-3 gene were cloned into the sgRNA scaffold of pSpCas9(BB)-2A-GFP (PX458) according to the Feng Zhang Lab CRISPR plasmid instructions [43]. The guide RNA sequences used for targeting TIM-3 gene were; 5′-CAC CGT TAT GCC TGG GAT TTG GAT CCG C-3′ and 5′-AAA CCC GGA TCC AAA TCC CAG GCA TAA C-3′. The construct was transfected into Jurkat T cells using the Neon Transfection System (Invitrogen). Transfected cells expressing green fluorescence were sorted 24 h post-transfection using the FACSAriaII (Becton–Dickinson, Franklin Lakes, NJ). The cells were further sorted according to the expression of TIM-3 after stimulation with PMA (25 ng) and Iono (1 μM) for 6 h by staining cells with a PE-conjugated anti-TIM-3 antibody (R&D Systems,Inc., Minneapolis, MN). After sorting four times, the cells were cloned by limiting dilution. The mutation in the TIM-3 genomic DNA sequence of the selected clone was confirmation by DNA sequencing.
Quantitative PCR for glut transcription levels
Total RNA was isolated using RNAiso (TAKARA Bio Inc., Shiga, Japan) and subjected to reverse transcription using oligo (dT) primer and then real-time PCR using SYBR green Ex Taq Premix (TAKARA Bio, Inc.) with the appropriate primer sets and an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Foster, CA). The primers used were: Glucose transporter 1, Glut1-F; 5′-ACT GCA ACG GCT TAG ACT TCG AC-3′, Glut1-R; 5′-TCT CTG GGT AAC AGG GAT CAA ACA-3′, Glut2-F; 5′-TCC TGA GTG GCT TCT GA-3′, Glut2-R; 5′-GGC CAT AGA ATA GTT TGG CTA GAA T-3′, Glut3-F; 5′-CCT ATG CCG AAT GCC CTC A-3′, Glut3-R; 5′-TGA CAG TGC ACA TAC ATT CAAT CCT C-3′, Glut4-F; 5′-GGG CTG AGA CAG GGA CCA TAA C-3′, Glut4-R; 5′-CAT GAG CAA TGG CAT CCA GAA-3′, β-actin-F; 5′-TGG CAC CCA GCA CAA TGA A-3′, β-actin-R; 5′-CTA AGT CAT AGT CCG CCT AG-3′. All PCR results were normalized to β-actin mRNA.
Assessment of Glut1 protein expression
The cells were fixed in 2% paraformaldehyde solution for 20 min at 24 °C, and subsequently washed with PBS. The fixed cells were permeabilized in PBS containing 0.3% saponin buffer for 15 min at room temperature. For blocking the Fcγ receptors (FcRs), the cells were incubated with human FcR binding inhibitor (ebioscience, San Diego, CA) and then, incubated with anti-Glut1 antibody (Abcam, Cambridge, UK) for 1 h followed by PE- anti-mouse Ig antibody (ebioscience) for 1 h. Then, the cells were washed twice in PBS containing 0.1% saponin buffer, and resuspended in 0.25 mL 0.5% paraformaldehyde in PBS and analyzed using FACS CantoII (Becton–Dickinson).
Assessment of glucose consumption, lactate release, glutamine consumption, and glutamate release
Glucose consumption, lactate release, glutamine consumption, and glutamate release were determined by measuring their concentrations in the final medium with a YSI biochemical analyzer (YSI Life Sciences; Simpsonville, SC). Cells (1 × 106/ml) were either stimulated or not with PMA (25 ng) and Iono (1 μM) at 37 °C for 6 h and cell supernatant was harvested. Glucose or glutamine consumption and lactate or glutamate release were calculated as follows: consumption = mmol/L of ingredient in fresh complete RPMI media – mmol/L of ingredient in cultured media, release = mmol/L of ingredient in cultured media – mmol/L of ingredient in fresh complete RPMI. The rates were reported as mmol/L per 106 cells.
Assessment of glucose uptake
Glucose uptake was analyzed using the fluorescent glucose analog 2-(N-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl) amino)-2-deoxyglucose (2-NBDG; Invitrogen) or Tritium (3H)-labeled 2-Deoxyglucose (2-DG; Perkin Elmer, Boston, MA). In the assessment using 2-NBDG, cells were unstimulated or stimulated with PMA (25 ng) and Iono (1 μM) at 37 °C for 1 h or 6 h, and then washed with N buffer (5 nM KCl, 2.5 mM CaCl2, 1 mM MgSO4, 1 mM KH2PO4, 10 mM HEPES pH 7.2) to label with 2-NBDG by incubating them in N buffer containing 400 nM 2-NBDG and 140 mM NaCl at 37 °C for 10 min. Subsequently, cells were washed with Na+-free buffer twice and analyzed by FACS CantoII (Becton–Dickinson). In the assay using 3H-labeled 2-DG, unstimulated or stimulated cells as above were incubated in the presence of 3H-labeled 2-DG (5 μCi/mL) for 30 min. The uptake of 3H-labeled 2-DG was stopped by the addition of cold PBS containing the glucose transporter inhibitor cytochalasin B (2.5 μg/mL, Sigma-Aldrich). The cells were washed once with ice-cold PBS and lysed in 400 μL RIPA buffer with 1% SDS. Radioactive counts were determined using a scintillation counter.
Assessment of mitochondrial function
We assessed mitochondrial membrane potential by labeling the cells with 25 nM 1,1′,3,3,3′,3′-hexamethylindodicarbo-cyanine iodide (DIlC(5), Invitrogen) for 30 min at 37 °C and then performed flow cytometry (Becton–Dickinson). Mitochondrial ROS was measured by flow cytometry following the labeling of cells with 5 μM MitoSOX™Red (Invitrogen) for 10 min at 37 °C prior to harvest.
Assessment of mitochondrial DNA content
Cells (1 × 106) were lysed with 500 μL of lysis buffer (100 mM NaCl, 10 mM Tris pH 8.0, 25 mM EDTA pH 8.0, 0.5% SDS, and 0.8 mg/ml protease K) and incubated at 56 °C overnight. Total DNA was isolated using the phenol/chloroform method and subjected to qPCR using primer sets specific to human mitochondrial DNA mt3212 to mt3319 (5′-CAC CCA AGA ACA GGG TTT GT-3′, human mt3319-R; 5′-TGG CCA TGG GTA TGT TGT TAA-3′) and nuclear 18S DNA (18S DNA-F; 5′-TAG AGG GAC AAG TGG CGT TC-3′, 18S DNA-R; 5′-CGC TGA GCC AGT CAG TGT-3′) [37]. A ratio of mitochondrial DNA to the nuclear 18S DNA was generated for each sample.
Statistics
Student’s t-test was performed and p-values were determined to identify significant differences as indicated in the figures.
Supplementary information
Additional file 1. Supplemental Figure 1. TIM-3 expression in TIM-3 overexpressing and control cells. Supplemental Figure 2. Transcript levels of Glut6, 8 and SGLT1 in TIM-3 overexpressing or knockout cells. Supplemental Figure 3. HK2 and PFKFB3 expression in TIM-3 overexpressing or knockout cells.
Acknowledgements
We would like to thank Editage (www.editage.co.kr) for English language editing.
Abbreviations
- DilC(5)
1,1′,3,3,3′,3′-hexamethylindodicarbo-cyanine iodide
- FcRs
Fcγ receptors
- Glut
Glucose transporter
- HK2
Hexokinase 2
- Iono
Ionomycin
- ITKs
Interleukin-inducible T cell kinases
- mTOR
Mammalian target of rapamycin
- PFKFB3
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
- PI3K
Phosphatidylinositol 3-kinase
- PMA
Phorbol 12-myristate 13-acetate
- ROS
Reactive oxygen species
- SGLT1
Sodium-glucose linked transporter 1
- TIM-3
T cell immunoglobulin and mucin domain containing protein-3
- UDP-GlcNAc
Uridine diphosphate N-acetylglucosamine
- 2-NBDG
2-(N-(7-nitrobenz-2-oxa-1, 3-diazol-4-yl) amino)-2-deoxyglucose
- 2-DG
2-Deoxyglucose
Authors’ contributions
ML performed and designed most experiments, analyzed the data, and produced Figs. SY established TIM3KO and CON cell lines. BL and EJ performed qPCR. GY and KK contributed to analysis and interpretation of the data. SP conceived the idea, interpreted the results, and wrote the manuscripts. The authors read and approved the final manuscript.
Funding
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2017R1A6A3A11033467, 2018R1D1A1B07049040).
Availability of data and materials
The datasets analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12865-020-00377-6.
References
- 1.Johnson MO, Siska PJ, Contreras DC, Rathmell JC. Nutrients and the microenvironment to feed a T cell army. Semin Immunol. 2016;28(5):505–513. doi: 10.1016/j.smim.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maciolek JA, Pasternak JA, Wilson HL. Metabolism of activated T lymphocytes. Curr Opin Immunol. 2014;27:60–74. doi: 10.1016/j.coi.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010;185(2):1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol. 2013;14(5):500–508. doi: 10.1038/ni.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck MD, O'Sullivan D, Pearce EL. T cell metabolism drives immunity. J Exp Med. 2015;212(9):1345–1360. doi: 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, van Aalten DM, et al. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat Immunol. 2016;17(6):712–720. doi: 10.1038/ni.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araujo L, Khim P, Mkhikian H, Mortales CL, Demetriou M. Glycolysis and glutaminolysis cooperatively control T cell function by limiting metabolite supply to N-glycosylation. Elife. 2017;6:e21330. [DOI] [PMC free article] [PubMed]
- 8.Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O'Sullivan D, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153(6):1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gubser PM, Bantug GR, Razik L, Fischer M, Dimeloe S, Hoenger G, et al. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat Immunol. 2013;14(10):1064–1072. doi: 10.1038/ni.2687. [DOI] [PubMed] [Google Scholar]
- 10.Bantug GR, Galluzzi L, Kroemer G, Hess C. The spectrum of T cell metabolism in health and disease. Nat Rev Immunol. 2018;18(1):19–34. doi: 10.1038/nri.2017.99. [DOI] [PubMed] [Google Scholar]
- 11.Dimeloe S, Mehling M, Frick C, Loeliger J, Bantug GR, Sauder U, et al. The immune-metabolic basis of effector memory CD4+ T cell function under hypoxic conditions. J Immunol. 2016;196(1):106–114. doi: 10.4049/jimmunol.1501766. [DOI] [PubMed] [Google Scholar]
- 12.Baixauli F, Acin-Perez R, Villarroya-Beltri C, Mazzeo C, Nunez-Andrade N, Gabande-Rodriguez E, et al. Mitochondrial respiration controls lysosomal function during inflammatory T cell responses. Cell Metab. 2015;22(3):485–498. doi: 10.1016/j.cmet.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaminski MM, Sauer SW, Kaminski M, Opp S, Ruppert T, Grigaravicius P, et al. T cell activation is driven by an ADP-dependent glucokinase linking enhanced glycolysis with mitochondrial reactive oxygen species generation. Cell Rep. 2012;2(5):1300–1315. doi: 10.1016/j.celrep.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38(2):225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bengsch B, Johnson AL, Kurachi M, Odorizzi PM, Pauken KE, Attanasio J, et al. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8(+) T cell exhaustion. Immunity. 2016;45(2):358–373. doi: 10.1016/j.immuni.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf Y, Anderson AC, Kuchroo VK. TIM3 comes of age as an inhibitory receptor. Nat Rev Immunol. 2019;20(3):173–185. doi: 10.1038/s41577-019-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koguchi K, Anderson DE, Yang L, O'Connor KC, Kuchroo VK, Hafler DA. Dysregulated T cell expression of TIM3 in multiple sclerosis. J Exp Med. 2006;203(6):1413–1418. doi: 10.1084/jem.20060210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomkowicz B, Walsh E, Cotty A, Verona R, Sabins N, Kaplan F, et al. TIM-3 suppresses anti-CD3/CD28-induced TCR activation and IL-2 expression through the NFAT signaling pathway. PLoS One. 2015;10(10):e0140694. doi: 10.1371/journal.pone.0140694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang YH, Zhu C, Kondo Y, Anderson AC, Gandhi A, Russell A, et al. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature. 2015;517(7534):386–390. doi: 10.1038/nature13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van de Weyer PS, Muehlfeit M, Klose C, Bonventre JV, Walz G, Kuehn EW. A highly conserved tyrosine of Tim-3 is phosphorylated upon stimulation by its ligand galectin-9. Biochem Biophys Res Commun. 2006;351(2):571–576. doi: 10.1016/j.bbrc.2006.10.079. [DOI] [PubMed] [Google Scholar]
- 21.Clayton KL, Haaland MS, Douglas-Vail MB, Mujib S, Chew GM, Ndhlovu LC, et al. T cell Ig and mucin domain-containing protein 3 is recruited to the immune synapse, disrupts stable synapse formation, and associates with receptor phosphatases. J Immunol. 2014;192(2):782–791. doi: 10.4049/jimmunol.1302663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMahan RH, Golden-Mason L, Nishimura MI, McMahon BJ, Kemper M, Allen TM, et al. Tim-3 expression on PD-1+ HCV-specific human CTLs is associated with viral persistence, and its blockade restores hepatocyte-directed in vitro cytotoxicity. J Clin Invest. 2010;120(12):4546–4557. doi: 10.1172/JCI43127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Wu K, Tao K, Chen L, Zheng Q, Lu X, et al. Tim-3/galectin-9 signaling pathway mediates T-cell dysfunction and predicts poor prognosis in patients with hepatitis B virus-associated hepatocellular carcinoma. Hepatology. 2012;56(4):1342–1351. doi: 10.1002/hep.25777. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Han H, He X, Li S, Wu C, Yu C, et al. Expression of the galectin-9-Tim-3 pathway in glioma tissues is associated with the clinical manifestations of glioma. Oncol Lett. 2016;11(3):1829–1834. doi: 10.3892/ol.2016.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu W, Shi Y, Li J, Chen F, Chen Z, Zheng M. Tim-3 expression on peripheral T cell subsets correlates with disease progression in hepatitis B infection. Virol J. 2011;8:113. doi: 10.1186/1743-422X-8-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ju Y, Hou N, Zhang XN, Zhao D, Liu Y, Wang JJ, et al. Blockade of Tim-3 pathway ameliorates interferon-gamma production from hepatic CD8+ T cells in a mouse model of hepatitis B virus infection. Cell Mol Immunol. 2009;6(1):35–43. doi: 10.1038/cmi.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JE, Patel MA, Mangraviti A, Kim ES, Theodros D, Velarde E, et al. Combination therapy with anti-PD-1, anti-TIM-3, and focal radiation results in regression of murine gliomas. Clin Cancer Res. 2017;23(1):124–136. doi: 10.1158/1078-0432.CCR-15-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das M, Zhu C, Kuchroo VK. Tim-3 and its role in regulating anti-tumor immunity. Immunol Rev. 2017;276(1):97–111. doi: 10.1111/imr.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jun KJ, Lee MJ, Shin DC, Woo MY, Kim K, Park S. Identification of CCL1 as a gene differentially expressed in CD4 T cells expressing TIM-3. Immune Netw. 2011;11(4):203–209. doi: 10.4110/in.2011.11.4.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee MJ, Woo MY, Chwae YJ, Kwon MH, Kim K, Park S. Down-regulation of interleukin-2 production by CD4(+) T cells expressing TIM-3 through suppression of NFAT dephosphorylation and AP-1 transcription. Immunobiology. 2012;217(10):986–995. doi: 10.1016/j.imbio.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Thorens B, Mueckler M. Glucose transporters in the 21st century. Am J Physiol Endocrinol Metab. 2010;298(2):E141–E145. doi: 10.1152/ajpendo.00712.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siska PJ, van der Windt GJ, Kishton RJ, Cohen S, Eisner W, MacIver NJ, et al. Suppression of Glut1 and glucose metabolism by decreased Akt/mTORC1 signaling drives T cell impairment in B cell leukemia. J Immunol. 2016;197(6):2532–2540. doi: 10.4049/jimmunol.1502464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macintyre AN, Gerriets VA, Nichols AG, Michalek RD, Rudolph MC, Deoliveira D, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20(1):61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhavsar SK, Singh Y, Sharma P, Khairnar V, Hosseinzadeh Z, Zhang S, et al. Expression of JAK3 sensitive Na+ coupled glucose carrier SGLT1 in activated cytotoxic T lymphocytes. Cell Physiol Biochem. 2016;39(3):1209–1228. doi: 10.1159/000447827. [DOI] [PubMed] [Google Scholar]
- 35.Le Bourgeois T, Strauss L, Aksoylar HI, Daneshmandi S, Seth P, Patsoukis N, et al. Targeting T cell metabolism for improvement of cancer immunotherapy. Front Oncol. 2018;8:237. doi: 10.3389/fonc.2018.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16(6):769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 37.Bai RK, Perng CL, Hsu CH, Wong LJ. Quantitative PCR analysis of mitochondrial DNA content in patients with mitochondrial disease. Ann N Y Acad Sci. 2004;1011:304–309. doi: 10.1007/978-3-662-41088-2_29. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto T, Takano N, Ishiwata K, Ohmura M, Nagahata Y, Matsuura T, et al. Reduced methylation of PFKFB3 in cancer cells shunts glucose towards the pentose phosphate pathway. Nat Commun. 2014;5:3480. doi: 10.1038/ncomms4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q, et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. 2018;9(5):478. doi: 10.1038/s41419-018-0528-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rangachari M, Zhu C, Sakuishi K, Xiao S, Karman J, Chen A, et al. Bat3 promotes T cell responses and autoimmunity by repressing Tim-3-mediated cell death and exhaustion. Nat Med. 2012;18(9):1394–1400. doi: 10.1038/nm.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saresella M, Piancone F, Marventano I, La Rosa F, Tortorella P, Caputo D, et al. A role for the TIM-3/GAL-9/BAT3 pathway in determining the clinical phenotype of multiple sclerosis. FASEB J. 2014;28(11):5000–5009. doi: 10.1096/fj.14-258194. [DOI] [PubMed] [Google Scholar]
- 42.Lee J, Su EW, Zhu C, Hainline S, Phuah J, Moroco JA, et al. Phosphotyrosine-dependent coupling of Tim-3 to T-cell receptor signaling pathways. Mol Cell Biol. 2011;31(19):3963–3974. doi: 10.1128/MCB.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplemental Figure 1. TIM-3 expression in TIM-3 overexpressing and control cells. Supplemental Figure 2. Transcript levels of Glut6, 8 and SGLT1 in TIM-3 overexpressing or knockout cells. Supplemental Figure 3. HK2 and PFKFB3 expression in TIM-3 overexpressing or knockout cells.
Data Availability Statement
The datasets analysed during the current study are available from the corresponding author on reasonable request.