Abstract
We aimed to analyze the interactions of both hydroxychloroquine and chloroquine with SARS-CoV-2 and identify their possible role for the prevention/treatment of COVID-19 by molecular docking studies. Protein crystal structures of SARS-CoV-2 and ACE2, the compounds hydroxychloroquine and chloroquine, and other ligand structures were minimized by OPLS3 force field. Glide Standard Precision and Extra Precision docking are performed and MM-GBSA values are calculated. Molecular docking studies showed that hydroxychloroquine and chloroquine do not interact with SARS-CoV-2 proteins, but bind to the amino acids ASP350, ASP382, ALA348, PHE40 and PHE390 on the ACE2 allosteric site rather than the ACE2 active site. Our results showed that neither hydroxychloroquine and chloroquine bind to the active site of ACE2. However, both molecules prevent the binding of SARS-CoV-2 spike protein to ACE2 by interacting with the allosteric site. This result can help ACE2 inhibitor drug development studies to prevent viruses entering the cell by attaching spike protein to ACE2.
Communicated by Ramaswamy H. Sarma
Keywords: Hydroxychloroquine, chloroquine, SARS-CoV-2, ACE2, molecular docking
Introduction
Based on the data of World Health Organization, COVID-19 affected nearly 6.200.000 people and 216 countries as of today (World Health Organization (WHO), 2020, June 3). Worldwide, many people had to live in isolation for days in addition to dramatic changes in marketing, finances and overall national/international activities. Drug repurposing studies have been the main practice ever since the outbreak of the virus, mainly to find a fast treatment for the disease. Unfortunately, no specific drug has currently been identified (Gorbalenya et al., 2020). Hydroxychloroquine/chloroquine (HCQ/CQ) have repeatedly been reported to cause a regression in the disease (Gao et al., 2020; Liu, Cao, et al., 2020; Yao et al., 2020; Zhou et al., 2020). HCQ received a renewed attention when World leaders declared their approach of prophylaxis by using daily doses of this drug (Radcliffe, 2020; Tatiana Arias, 2020). Both CQ and its less toxic derivative, HCQ have long been used for their anti-malarial and anti-inflammatory actions (Furst et al., 2009; Rosenthal, 2009). Their anti-malarial action is a result of the heme-chloroquine complex through inhibition of polymerization of heme (Chong & Sullivan, 2003). Their anti-inflammatory properties result from their interference with ‘antigen’ processing in macrophages and other antigen-presenting cells due to their weakly basic character (Fox, 1993). In fact, by increasing the pH of lysosomal and trans-Golgi network (TGN) vesicles, weak bases are known to disrupt many enzymes including acid hydrolases and inhibit the post-translational modifications of newly synthesized proteins. The chloroquine-mediated rise in endosomal pH has been shown to interfere with iron metabolism, decrease the intracellular concentration of iron and affect the function of many enzymes involved in pathways leading to replication of cellular DNA and to expression of different genes (Byrd & Horwitz, 1991). As for anti-viral mechanism of action, CQ has been shown to inhibit pH-dependent entry of some viruses such as Borna, choriomeningitis and rabies (Gonzalez-Dunia et al., 1998). In addition, Wincent et al. showed that SARS-CoV- an enveloped virus and the etiological cause of SARS disease- may be inhibited by CQ through increasing endosomal pH and interfering with terminal glycosylation of the cellular receptor, angiotensin-converting enzyme 2 (ACE2) (Liu, Xiao, et al., 2020; Vincent et al., 2005; Yan et al., 2020).
Other studies reported different mechanisms of actions. For example, to inhibit autophagy as a possible anti-tumor drug, CQ is reported to impair autophagosome fusion with lysosomes and not alter the pH of this organelle (Mauthe et al., 2018). Moreover, CQ was shown to inhibit quinone reductase 2 and thereby sialic acid synthesis (Kwiek et al., 2004). The possible interference of CQ with sialic acid biosynthesis could account for the broad antiviral spectrum of that drug since human coronavirus HCoV-O43 has been shown to use sialic acid moieties as receptors (Tortorici et al., 2019).
SARS-CoV-2 RNA polymerase, main protease, papain-like protease, endoribonuclease, ADP ribose phosphatase, 2′-O-methyl transferase, RNA-replicase, helicase, phosphoprotein, ORF7A encoded accessory protein and spike proteins play an important role in the structure, survival, reproduction, attachment and survival of the SARS-CoV-2 virus (Satarker & Nampoothiri, 2020).
In this study, a comprehensive molecular docking study of HCQ/CQ on the proteins of SARS-CoV-2; NSP12 RNA polymerase, NSP5 main protease, NSP3 papain-like protease, spike glycoproteins in their opened and closed states, post fusion core of SARS-CoV-2 subunit, C-terminal dimerization domain and N-terminal RNA-binding domain of nucleocapsid phosphoprotein, NSP15 endoribonuclease, ADP ribose phosphatase, NSP16 2′-O-methyl transferase, Nsp9 RNA-replicase, Nsp13 helicase, ORF7A encoded accessory protein, spike proteins, and human ACE2, transmembrane protease serine 2 (TMPRSS2), V-ATPase subunit C/A and 2-epimerase was performed to understand how they affect this virus.
Materials and methods
Molecular docking
All computerized calculations and visualization of the results were done with Maestro Schrodinger software (Schrödinger, 2018). Molecular docking study was carried out through ligand and protein preparation, determination of the binding site, ligand docking and development of scores.
HCQ/CQ and other ligands were downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/) in 3 D sdf format. Possible ionization and optical isomers of the ligands at physiological pH were prepared with OPLS3 field forces by using the ‘LigPrep’ module.
SARS-Cov-2 main protease, RNA polymerase, papain like protease, human ACE2 and other protein crystal structures were imported from the Protein Data Bank (PDB) (https://www.rcsb.org/) to the ‘Protein Preparation Wizard’ module. Using the default settings, pre-process, optimization and minimization processes and protein structures were prepared. Firstly, hydrogens were added, non-bonding command with metals, the formation of disulfide bonds, deletion of water at 5 Å distance from het groups and preprocess by creating pH: 7.00 +/− 2.00 het states using Epik. In the next step, the appropriate chain was selected. Hydrogen bond determination was optimized using PROKA pH:7.00 with water sample orientation. Finally, proteins were prepared by minimizing OPLS3 field forces.
While the protein crystal structures that carry ligand on the region to be docked are determined according to the coordinates of the ligand, the possible binding sites of the protein structures that do not carry ligand or unknown active site are determined by the ‘Binding Site Detection’ module and the grid box was created with the ‘Receptor Grid Generation’ module in the size of 20x20x20 Å.
Molecular docking was performed with Glide Standard Precision (SP), Glide Extra Precision (XP), and Prime Molecular Mechanics Generalized Born Surface Area (MM-GBSA) values were calculated in high protein-ligand interactions. In addition, the interaction between HCQ and ACE2 binding site was calculated by Induced Fit Docking, where protein and ligand were flexible. 2 D and 3 D interactions between the protein-ligand were determined and some important interactions selected were shown.
Results and discussion
First, the SARS-CoV-2 RNA polymerase interaction with HCQ and CQ was investigated. SARS-CoV-2 RNA dependent polymerase (PDB: 7BV2) (Yin et al., 2020) protein structure was prepared. SP Molecular docking was performed to the active site remdesivir and HCQ/CQ compound on the crystal structure of the RNA polymerase (x: 95.12, y: 92.31, z: 109.75)and the results were shown in Table 1.
Table 1.
Interactions between RNA polymerase, remdesivir and HCQ/CQ.
| Molecule | Docking score | Glide score | Glide emodel |
|---|---|---|---|
| Remdesivir | −8.072 | −8.07 | −75.425 |
| HCQ | −2.974 | −4.677 | −47.425 |
| CQ | −2.558 | −4.261 | −42.983 |
These results suggested that neither HCQ nor CQ not enough interacts with RNA polymerase.
The active site of the SARS-CoV-2 main protease (PDB: 6LU7) is already known (x: −13.67, y: 8.5, z: 65.34). Table 2 shows the XP re-docking results of the ligand of the crystal structure, HCQ and CQ.
Table 2.
Interactions between main polymerase, its ligand and HCQ/CQ.
| Molecule | Docking score | Glide score | Glide emodel |
|---|---|---|---|
| Ligand of 6LU7 | −8.110 | −8.110 | −99.643 |
| HCQ | −4.228 | −6.469 | −52.728 |
| CQ | −3.332 | −5.008 | −44.476 |
Ligand of 6LU7: (phenylmethyl) (4 ∼ {S})-4-[[(2 ∼ {S})-4-methyl-2-[[(2 ∼ {S})-3-methyl-2-[[(2 ∼ {S})-2-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]propanoyl]amino]butanoyl]amino]pentanoyl]amino]-5-[(3 ∼ {S})-2-oxidanylidenepyrrolidin-3-yl]pent-2-enoate.
These results showed that neither HCQ nor CQ not enough interacts with the main protease.
Five possible binding sites were identified in the SARS-CoV-2 open state ectodomain (PDB: 6VYB) crystal structure. In SP molecular docking study, HCQ scored −6.841 in binding site 1, −4.494 in binding site 2, −4.682 in binding site 3, −5.400 in binding site 4 and −3.752 in binding site 5. CQ gave lower scores in all binding sites. Binding site 1 showed a high interaction with SP docking results. However, further calculations with XP resulted in a docking score of −4.108 and excluded the SARS-CoV-2 ectodomain structure as a target for HCQ and CQ.
SP docking results of identified binding sites and SARS-CoV-2 proteins were shown in Table 3.
Table 3.
Interactions between HCQ/CQ and SARS-CoV-2 proteins or proteins related to human enzymes.
| Molecule | PDB Code | Name of SARS-CoV-2 protein | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 | Result |
|---|---|---|---|---|---|---|---|---|
| HCQ | 6W9C | papain-like protease | −4.268 | −5.04 | −4.985 | −4.701 | −5.555 | NI |
| CQ | 6W9C | papain-like protease | −3.154 | −4.528 | −4.550 | −3.828 | −4.340 | NI |
| HCQ | 6VXX | closed state spike glycoprotein | −3.074 | −3.818 | −4.089 | NI | ||
| CQ | 6VXX | closed state spike glycoprotein | −2.806 |
−5.351 |
−4.977 |
NI | ||
| HCQ | 6LXT | post fusion core | −3.940 | NI | ||||
| CQ | 6LXT | post fusion core | −4.655 | NI | ||||
| HCQ | 6WJI | C-terminal dimerization domain of nucleocapsid phosphoprotein | −5.184 | −4.574 | 3.846 | NI | ||
| CQ | 6WJI | C-terminal dimerization domain of nucleocapsid phosphoprotein | −4.984 |
−3.696 |
−3.304 |
NI | ||
| HCQ | 6YI3 | N-terminal RNA-binding domain | −4.412 | NI | ||||
| CQ | 6YI3 | N-terminal RNA-binding domain | −4.457 | NI | ||||
| HCQ | 6VWW | NSP15 endoribonuclease | −6.251# | −4.309 | 4.741 | −5.449 | −5.150 | NI |
| CQ | 6VWW | NSP15 endoribonuclease | −5.119 |
−3.899 |
−3.125 |
−4.484 |
−4.700 |
NI |
| HCQ | 6W6Y | ADP ribose phosphatase of NSP3 | −2.111 | −3.681 | NI | |||
| CQ | 6W6Y | ADP ribose phosphatase of NSP3 | −1.556 |
−1.264 |
NI | |||
| HCQ | 6W75 | NSP10-NSP16 2′-O-methyl transferase |
−4.140 | −5.134 | −4.153 | −5.238 | −3.998 | NI |
| CQ | 6W75 | NSP10-NSP16 2′-O-methyl transferase |
−3.555 |
−4.401 |
−3.912 |
−4.286 |
−3.150 |
NI |
| HCQ | 6W9Q | Nsp9 RNA-replicase | −4.934 | −4.487 | NI | |||
| CQ | 6W9Q | Nsp9 RNA-replicase | −3.943 |
−4.487 |
NI | |||
| HCQ | 6VW1 | S1 unit of ACE2-Spike complex | −4.618 | −5.773# | NI | |||
| CQ | 6VW1 | S1 unit of ACE2-Spike complex | −3.997 | −5.224# | NI | |||
| HCQ | 6W37 | ORF7A encoded accessory protein | −4.099 | NI | ||||
| CQ | 6W37 | ORF7A encoded accessory protein | −2.889 | NI | ||||
| HCQ | 6JYT | Nsp13 Helicase |
−5.801 | −4.998 | −5.830 | −5.766 | −4.755 | NI |
| CQ | 6JYT | Nsp13 Helicase |
−4.593 | −4.782 | −4.192 | −5.033 | −5.111 | NI |
| HCQ | 1U7L | Vacuolar ATP synthase subunit C | −5.740 |
−5.271 |
−5.879 |
−5.551 |
NI | |
| CQ | 1U7L | Vacuolar ATP synthase subunit C | −4.731 |
−4.940 |
−5.228 |
−4.661 |
NI | |
| HCQ | 6HH0 | Vacuolar ATP synthase subunit A | −2.218 |
NI | ||||
| CQ | 6HH0 | Vacuolar ATP synthase subunit A | −1.701 |
NI | ||||
| HCQ | 2OQ5 | transmembrane serine protease | −6.517# | NI | ||||
| CQ | 2OQ5 | transmembrane serine protease | −4.376 | NI | ||||
| HCQ | 2YHW | UDP-GlcNAc 2-epimerase | −2.821 | NI | ||||
| CQ | 2YHW | UDP-GlcNAc 2-epimerase | −2.119 | NI |
Please note: Shaded boxes indicate the sites that are not involved in the activity of the particular enzyme.
further calculations through XP docking did not warrant an interaction of the molecule and the protein in question.
NI: No Interactions.
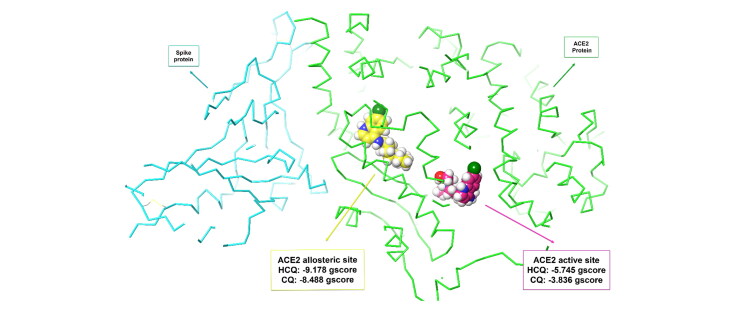
Five possible binding sites were identified on ACE2 protein crystal structure (PDB: 6VW1). Interestingly, the highest interactions were identified between HCQ and binding site 1 (x: 89.67, y: −3.1, z: 171.95) of the chimeric receptor-binding domain complexed with human ACE2 with a docking score of −7.255, a gscore of −7.305, and a glide emodel score of −79.025.
To support this process at an advanced stage, XP docking was performed with HCQ on binding site 1. A docking score of −8.157, a gscore of −8.227, and a glide emodel score of −66.104 were obtained. In addition, the calculated MM-GBSA value was −84.29.
To confirm the interaction between HCQ and ACE2, 5 possible binding sites have been identified in the ACE2 chain of another ACE2 spike complex crystal structure (PDB: 6LZG) and the results were shown in Table 4.
Table 4.
SP and XP docking values of HCQ/CQ in the potential binding sites and active site on ACE2 (PDB: 6LZG).
| PDB Code | Name of SARS-CoV-2 protein | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 allosteric site | ACE2 active site | |
|---|---|---|---|---|---|---|---|---|
| SP docking scores of HCQ | 6LZG | ACE2 unit of ACE2-Spike complex | −6.227 | −6.605 | −4.971 | −5.545 | −7.340 | −5.062 |
| SP docking scores of CQ | 6LZG | ACE2 unit of ACE2-Spike complex | −5.662 | −6.079 | −4.680 | −3.836 | −6.568 | −4.232 |
| XP docking scores of HCQ | 6LZG | ACE2 unit of ACE2-Spike complex | – | – | – | – | −9.178 | −5.745 |
| XP docking scores of CQ | 6LZG | ACE2 unit of ACE2-Spike complex | – | – | – | – | −8.488 | −3.836 |
In addition, HCQ had a gscore of −7.394, a glide emodel score of −84.594 and a MM-GBSA value of −90.49 in binding site 5.
Further analysis with XP docking was applied to the binding site 5 (allosteric site). HCQ gave a docking score of −9.178, a gscore of −9.228, glide emodel score of −63.291, and MM-GBSA value of −93.69 in allosteric site (x: −30.76, y: 21.97, z: −6.21), while it gave a docking score of −5.745, a gscore of −5.795, glide emodel score of −40.276, and MM-GBSA value of −45.98 in the active site (x: −26.3, y: 12.54, z: −32.83).
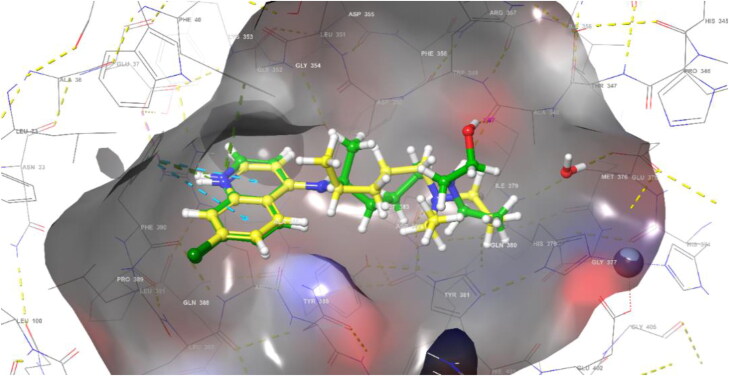
In ACE2 (PDB: 6LZG) allosteric site, ‘Induced Fit Docking’ procedure has been performed in which protein and ligand are flexible in order to better understand the interactions of HCQ/QC and ACE2 (Figure 1 and 2). HCQ formed a hydrogen bond with ASP350 (2.10 Å), ALA348 (2.02 Å) (OH) and ALA348 (1.99 Å) (N+H), two hydrogen bonds and a salt bridge with ASP382 (4.93 Å), two pi-cation interactions with HIS378 (5.03 Å) and PHE390 (4.73 Å) and hydrophobic interactions with TRP69, TYR385, PHE40, TRP349. According to the induced fit docking result, HCQ gave −11.013 docking score, −11.63 gscore and −60.299 glide emodel score.
Figure 1.
Binding mode of HCQ (green) and CQ (yellow) in the ACE2 allosteric site (PDB: 6LZG).
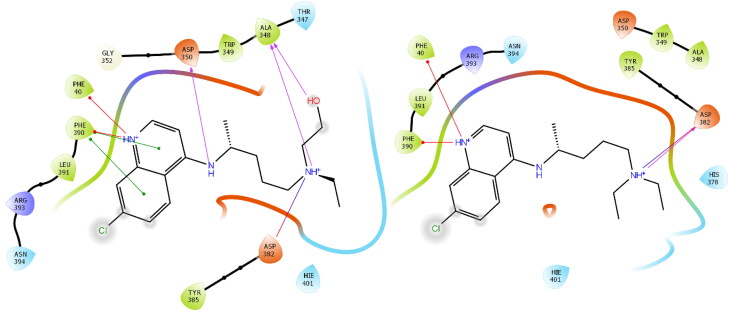
Figure 2.
2 D interaction diagrams of HCQ and CQ in ACE2 allosteric site (PDB: 6LZG).
It is known that HCQ shows a stronger anti SARS-CoV-2 activity in vitro compared to CQ (Yao et al., 2020). Our molecular docking findings support this situation. HCQ generated higher interaction on ACE2's allosteric site (-9.178) than CQ (-8.488). In the ACE2 allosteric site of HCQ and CQ, hydrogen bonding with ASP382 and pi-cation interaction with PHE390 and PHE40, and in addition to these interactions, HCQ formed two hydrogen bonds with ASP350 and ALA348. Comparison of the ligand protein interaction between HCQ and CQ in the ACE2 allosteric site was shown in Figure 2.
XP molecular docking was applied to the active area of the crystal structure with the inhibitor of ACE2 (PDB: 1R4L) (Towler et al., 2004). HCQ gave −7.598 docking score, −7.647 gscore and −64.619 glide emodel score in active site (x: 41.4, y: 5.85, z: 28.08), while it gave −8.927 docking score, −8.977 gscore and −62.108 glide emodel score in allosteric site (x: 29.61, y: 16.0, z: 24.08). These data suggested that HCQ or CQ do not bind to the active site as do ACE2 inhibitors. However, HCQ seem to prevent the binding of SARS-CoV-2 spike protein by interacting with amino acids ASP350, ASP382, ALA348, PHE40, PHE390 in its adjacent region, causing a change in the conformation of ACE2 (Figure 3).
Figure 3.
HCQ (yellow) in ACE2 allosteric site and inhibitor ORE-1001 (orange) in ACE2 active site (PDB: 1R4L).
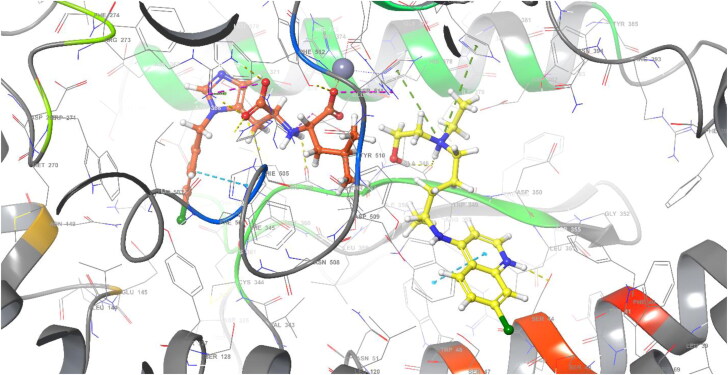
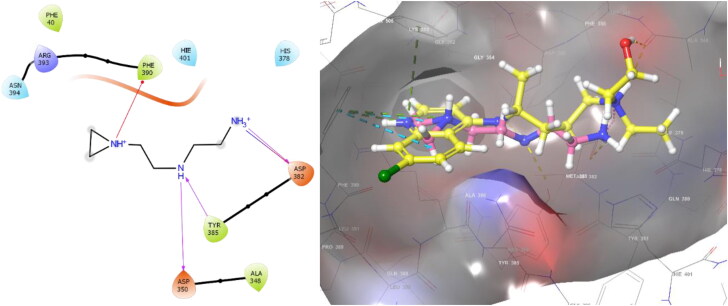
Similarly, the N-(2-aminoethyl)-1 aziridine-ethanamine (NAAE) compound was reported as an ACE2 inhibitor for SARS virus in 2004 (Huentelman et al., 2004). With XP molecular docking, NAAE gave a −6.090 gscore in the active site of ACE2 (PDB 1R4L), while it gave a −8.024 gscore in the docking process in the allosteric site (Figure 4). NAAE and HCQ exhibited similar conformation and interactions on the ACE2 allosteric site, and both NAAE and HCQ formed hydrogen bonds with ASP350 and ASP382 and formed a pi-cation interaction with PHE390 (Figure 4). In this case, it can be concluded that the inhibition of binding the spike protein to ACE2 does not occur by interacting with the active site, whereas it interacts with the allosteric site, disrupting the binding of spike protein by changing the conformation of ACE2.
Figure 4.
2 D interaction diagram of NAAE, and HCQ (yellow)/NAAE (pink) similar binding mode in ACE2 allosteric site (PDB: 6LZG).
Recent publications reported possible beneficial effects of CQ in the treatment of patients infected by SARS-CoV-2 (Colson et al., 2020; Gao et al., 2020). However, in their very recent commentary, Touret and Lamballerie pointed out that the excitement evoked by the results of initial clinical studies should be approached with caution based on the fact that they involved some issues related to design and interpretation (Touret & de Lamballerie, 2020). Not the inconclusive results of clinical trials per se, but the proposed efficiency in preventing the infection led some communities to seek for food and drink items known to contain quinine, a molecule that is chemically related to both HCQ and CQ (Valsler, 2020). In fact, a quinine-containing soda drink was sold out over several weeks in Turkey.
Thus, we evaluated possible interactions of nearly all proteins on SARS-Cov-2 with ACE2, the receptor that the virus is utilizing to infect human cells. Our results indicated no significant interactions between the proteins of the virus and HCQ or CQ. Interestingly, however, HCQ seems to be binding to an allosteric region of the receptor. This unexpected interaction may be preventing the binding of the virus indicating a preventive action of these compounds for the virus to reach the chimeric binding domain. On the other hand, an allosteric hindering of the active region by the compounds does not completely exclude a possible competitive activation of the receptor by the virus. Although highly speculative at this time, our results indicate that inhibition of further viral invasion of human cells may be the rationale of using these drugs as a preventive measure. However, based on the well-documented adverse events related to their use should still be strictly limited (FDA cautions against use of hydroxychloroquine or chloroquine for COVID-19 outside of the hospital setting or a clinical trial due to risk of heart rhythm problems).
Conclusion
In this study, the effect of HCQ/CQ on SARS-CoV-2 was investigated extensively by molecular docking studies. It was concluded that HCQ/CQ do not act on SARS-CoV-2 RNA polymerase, main protease, papain like protease, spike proteins, endoribonuclease, helicase and other NSPs. HCQ/CQ does not show its activity by binding to the active site of ACE2, whereas HCQ/CQ is thought to inhibit glycosylation of the spike protein by disrupting the conformation of ACE2 by interacting with ASP350, ASP382, ALA348, HIS378, PHE40 and PHE390 amino acids in ACE2 allosteric site. In addition, as with clinical and in vitro findings, HCQ on the ACE2 allosteric site showed higher affinity than CQ.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Byrd, T. F., & Horwitz, M. A. (1991). Chloroquine inhibits the intracellular multiplication of Legionella pneumophila by limiting the availability of iron. A potential new mechanism for the therapeutic effect of chloroquine against intracellular pathogens. The Journal of Clinical Investigation, 88(1), 351–357. 10.1172/JCI115301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, C. R., & Sullivan, D. J. Jr. (2003). Inhibition of heme crystal growth by antimalarials and other compounds: Implications for drug discovery. Biochemical Pharmacology, 66(11), 2201–2212. 10.1016/j.bcp.2003.08.009 [DOI] [PubMed] [Google Scholar]
- Colson, P., Rolain, J. M., & Raoult, D. (2020). Chloroquine for the 2019 novel coronavirus SARS-CoV-2. International Journal of Antimicrobial Agents, 55(3), 105923. 10.1016/j.ijantimicag.2020.105923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA cautions against use of hydroxychloroquine or chloroquine for COVID-19 outside of the hospital setting or a clinical trial due to risk of heart rhythm problems . https://www.fda.gov/drugs/drug-safety-and-availability/fda-cautions-against-use-hydroxychloroquine-or-chloroquine-covid-19-outside-hospital-setting-or.
- Fox, R. I. (1993). Mechanism of action of hydroxychloroquine as an antirheumatic drug. Seminars in Arthritis and Rheumatism, 23(2 Suppl 1), 82–91. 10.1016/S0049-0172(10)80012-5 [DOI] [PubMed] [Google Scholar]
- Furst, D. E., Ulrich, R. W., & Varkey-Altamirano, C. (2009). Nonsteroidal anti-inflammatory drugs, disease-modifying antirhematic drugs, nonopiod analgesics, & drugs used in gout. In Katzung B. G., Masters S. B., & Trevor A. J. (Eds.), Basic and clinical pharmacology (Vol. 11, pp. 621–642). The McGraw Hill Companies Inc. [Google Scholar]
- Gao, J., Tian, Z., & Yang, X. (2020). Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Bioscience Trends, 14(1), 72–73. 10.5582/bst.2020.01047 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Dunia, D., Cubitt, B., & de la Torre, J. C. (1998). Mechanism of Borna disease virus entry into cells. Journal of Virology, 72(1), 783–788. 10.1128/JVI.72.1.783-788.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Baker, S. C., Baric, R. S., de Groot, R. J., Drosten, C., Gulyaeva, A. A., Haagmans, B. L., Lauber, C., Leontovich, A. M., Neuman, B. W., Penzar, D., Perlman, S., Poon, L., Samborskiy, D., Sidorov, I. A., Sola, I., Ziebuhr, J. (2020). Severe acute respiratory syndrome-related coronavirus–The species and its viruses, a statement of the Coronavirus Study Group. bioRxiv. 10.1101/2020.02.07.937862 [DOI] [Google Scholar]
- Huentelman, M. J., Zubcevic, J., Hernandez Prada, J. A., Xiao, X., Dimitrov, D. S., Raizada, M. K., & Ostrov, D. A. J. H. (2004). Structure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension (Dallas, Tex.: 1979), 44(6), 903–906. 10.1161/01.HYP.0000146120.29648.36 [DOI] [PubMed] [Google Scholar]
- Kwiek, J. J., Haystead, T. A., & Rudolph, J. (2004). Kinetic mechanism of quinone oxidoreductase 2 and its inhibition by the antimalarial quinolines. Biochemistry, 43(15), 4538–4547. 10.1021/bi035923w [DOI] [PubMed] [Google Scholar]
- Liu, J., Cao, R., Xu, M., Wang, X., Zhang, H., Hu, H., Li, Y., Hu, Z., Zhong, W., Wang, M. (2020). Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 Infection in vitro. Cell Discovery, 6(1), 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Xiao, X., Wei, X., Li, J., Yang, J., Tan, H., Zhu, J., Zhang, Q., Wu, J., & Liu, L. (2020). Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. Journal of Medical Virology, 92(6), 595–601. 10.1002/jmv.25726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauthe, M., Orhon, I., Rocchi, C., Zhou, X., Luhr, M., Hijlkema, K.-J., Coppes, R. P., Engedal, N., Mari, M., & Reggiori, F. (2018). Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy, 14(8), 1435–1455. 10.1080/15548627.2018.1474314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe, S. (Producer). (2020, May 26). Trump is taking hydroxychloroquine: Why experts say you shouldn’t.
- Rosenthal, P. J. (2009). Antiprotozoal drugs. In Katzung B. G., Masters S. B., & Trevor A. J. (Eds.), Basic and clinical pharmacology (pp. 899–921). The McGraw-Hill Companies, Inc. [Google Scholar]
- Satarker, S., & Nampoothiri, M. (2020). Structural proteins in severe acute respiratory syndrome coronavirus-2. [DOI] [PMC free article] [PubMed]
- Schrödinger, L. J. S. (2018). Schrödinger release 2018-1: Maestro. Schrödinger, LLC. [Google Scholar]
- Tatiana Arias, C. (Producer). (2020). Salvadoran leader says he takes hydroxychloroquine. https://edition.cnn.com/2020/05/27/americas/salvador-president-coronavirus-hydroxychloroquine-intl/index.html
- Tortorici, M. A., Walls, A. C., Lang, Y., Wang, C., Li, Z., Koerhuis, D., Boons, G.-J., Bosch, B.-J., Rey, F. A., de Groot, R. J., & Veesler, D. (2019). Structural basis for human coronavirus attachment to sialic acid receptors. Nature Structural & Molecular Biology, 26(6), 481–489. 10.1038/s41594-019-0233-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touret, F., & de Lamballerie, X. (2020). Of chloroquine and COVID-19. Antiviral Research, 177, 104762. 10.1016/j.antiviral.2020.104762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler, P., Staker, B., Prasad, S. G., Menon, S., Tang, J., Parsons, T., Ryan, D., Fisher, M., Williams, D., Dales, N. A., Patane, M. A., & Pantoliano, M. W. (2004). ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. The Journal of Biological Chemistry, 279(17), 17996–18007. 10.1074/jbc.M311191200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsler, Ben. (2020, March 27). Popcast: Chloroquine and hydroxychloroquine. https://www.chemistryworld.com/podcasts/chloroquine-and-hydroxychloroquine/4011414.article
- Vincent, M. J., Bergeron, E., Benjannet, S., Erickson, B. R., Rollin, P. E., Ksiazek, T. G., Seidah, N. G., & Nichol, S. T. (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virology Journal, 2, 69. 10.1186/1743-422X-2-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (WHO) . (2020, June 3). Coronavirus disease (COVID-19) outbreak situation. https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. J. S. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.), 367(6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., Ye, F., Zhang, M., Cui, C., Huang, B., Niu, P., Liu, X., Zhao, L., Dong, E., Song, C., Zhan, S., Lu, R., Li, H., Tan, W., & Liu, D. (2020). In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clinical Infectious Diseases. 10.1093/cid/ciaa237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W., Mao, C., Luan, X., Shen, D.-D., Shen, Q., Su, H., Wang, X., Zhou, F., Zhao, W., Gao, M., Chang, S., Xie, Y.-C., Tian, G., Jiang, H.-W., Tao, S.-C., Shen, J., Jiang, Y., Jiang, H., Xu, Y., … Eric Xu, H. (2020). Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science, 368(6498), 1499–1504. 10.1126/science.abc1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D., Dai, S.-M., & Tong, Q. (2020). COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. Journal of Antimicrobial Chemotherapy, 75(7), 1667–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]