Abstract
SARS-CoV-2 virus outbreak poses a major threat to humans worldwide due to its highly contagious nature. In this study, molecular docking, molecular dynamics, and structure-activity relationship are employed to assess the binding affinity and interaction of 76 prescription drugs against RNA dependent RNA polymerase (RdRp) and Main Protease (Mpro) of SARS-CoV-2. The RNA-dependent RNA polymerase is a vital enzyme of coronavirus replication/transcription complex whereas the main protease acts on the proteolysis of replicase polyproteins. Among 76 prescription antiviral drugs, four drugs (Raltegravir, Simeprevir, Cobicistat, and Daclatasvir) that are previously used for human immunodeficiency virus (HIV), hepatitis C virus (HCV), Ebola, and Marburg virus show higher binding energy and strong interaction with active sites of the receptor proteins. To explore the dynamic nature of the interaction, 100 ns molecular dynamics (MD) simulation is performed on the selected protein-drug complexes and apo-protein. Binding free energy of the selected drugs is performed by MM/PBSA. Besides docking and dynamics, partial least square (PLS) regression method is applied for the quantitative structure activity relationship to generate and predict the binding energy for drugs. PLS regression satisfactorily predicts the binding energy of the effective antiviral drugs compared to binding energy achieved from molecular docking with a precision of 85%. This study highly recommends researchers to screen these potential drugs in vitro and in vivo against SARS-CoV-2 for further validation of utility.
Keywords: Antiviral drugs, SARS-CoV-2, molecular docking, molecular dynamics, quantitative structure-activity relationship, principal component analysis
Graphical Abstract
Communicated by Ramaswamy H. Sarma
1. Introduction
SARS-CoV-2, the new seventh coronavirus, known to infect humans, is responsible for COVID-19 (Andersen et al., 2020). The World Health Organization (WHO) declared the virus as a global pandemic on March 11, 2020 (Aanouz et al., 2020). The COVID-19 pandemic is the biggest calamity since the Second World War as no specific drug or vaccine has been approved globally for the treatment of human coronaviruses to date. Therefore, the outbreak of SARS-CoV-2 is considered to pose a devastating threat to human life. This virus family circulates in animals and some can be passed on between animals and humans (Boopathi et al., 2020). The common signs of infection include fever, coughing, breathing difficulties, and in severe cases, it can cause pneumonia, multiple organ failure, and death.
SARS-CoV-2 consists of crown-shaped peplomers and a single-stranded positive RNA genome with 5′ cap and 3′ Poly-A tail, belongs to family Coronaviridae of the order Nidovirales (Pant et al., 2020; Woo et al., 2010). This RNA genome consists of at least six open reading frames (ORFs) of which the first ORF (ORF1a/b) makes up the 5’ two-third and encodes two polypeptides (pp1a and pp1ab). These two polypeptides conduct the production of 16 nonstructural proteins (NSPs) (R. J. Khan et al., 2020). Another five ORFs compose the remaining one-third of the RNA genome which generates four main structural factors of the virion known as spike protein (S), envelope protein (E), membrane protein (M) and nucleocapsid protein (N) (Gordon et al., 2020; Hussain et al., 2005). The virus uses the spike (S) protein on its surface to interact with the ACE2 (Angiotensin-converting enzyme 2) cellular receptor of human tissues (Wong et al., 2004). During interaction with the cell, RNA is used as a template for direct translation of two polypeptides (pp1a and pp1ab) that encodes a number of vital nonstructural proteins (NSPs) including NSP5 (a cysteine 3C-like protease) known as main protease (Mpro) and NSP3 (a papain-like protease) (Sarma et al., 2020). These two proteins process pp1a and pp1ab to produce 16 different NSPs (Hilgenfeld, 2014; Zhou et al., 2020). The NSP3 processes the polypeptide to produce NSP1-4 while the Mpro operates at 11 cleavage sites to generate the rest of the important NSPs that include helicase, methyltransferase, and RNA-dependent RNA polymerase all of which play a critical role in the viral infection cycle (Cui et al., 2019). Hence, the main protease creates major and attractive drug target to impede the assembly of nonstructural viral components and to halt the replication event in the virus life cycle. In addition, RNA-dependent RNA polymerase is one of the most versatile enzymes for coronavirus that is indispensable for facilitating the replication of RNA from RNA templates (Romano et al., 2020). Subsequently, these phenomena led us to target two crucial proteins to fight against SARS-CoV-2.
As the coronavirus outbreak is an on-going pandemic and new drug development is very time consuming, therefore, the fastest way to defeat COVID-19 is to apply effective treatment strategies considering the available drugs in the market. This raises a high and urgent need to screen for potential drugs through either drug-repurposing or combination therapy. Drug repurposing is an efficient approach to combat novel diseases that spread rapidly (Ashburn & Thor, 2004; Ciliberto & Cardone, 2020; Xu et al., 2020) as many diseases share overlapping molecular pathways (Hodos et al., 2016). The approved drugs are safe and reported to be used in humans for countering certain viral infections (H. Li et al., 2020). The screening of drugs to repurpose has arisen as a resourceful alternative to accelerate the drug development process against rapidly spreading diseases like COVID-19 in recent years (Elmezayen et al., 2020; Sun et al., 2016). This approach has successfully led to the discoveries of potential drugs against several diseases such as Ebola, hepatitis C, and zika virus infection (Barrows et al., 2016; Johansen et al., 2015). Remdesivir which has been reported as the most promising drug against COVID-19 is an antiviral originally developed to target the Ebola virus (Zhang & Zhou, 2020). This provided the rationalization for drug repurposing with the hope to discover antiviral drugs to treat COVID-19.
2. Methodologies
2.1. Protein preparation
The crystal structure of the SARS-COV-2 RNA dependent RNA polymerase was not available when we started our study; therefore, the polymerase structure was obtained via homology modeling (Figure S1). The template search for SARS-COV-2 RdRp has been carried out with BLAST and HHBlits against the SWISS-MODEL template library (SMTL) (Waterhouse et al., 2018). The RdRp sequence was sought with BLAST against the primary amino acid sequence contained in the SMTL and the SARS coronavirus polymerase (PDB ID: 6NUR) was found as a template (Remmert et al., 2011). Considering the target-template alignment, the RdRp Model was built using ProMod3. QMEAN scoring function (Bertoni, Kiefer, Biasini, Bordoli, & Schwede, 2017) had been applied for the assessment of global and per-residue model quality. Two Zn2+ ligands existing in the template structure were transferred to the model structure. GMQE score estimated the accuracy of the tertiary structure of the resultant RdRp model. The best drugs are also docked against the new crystal structure of RdRp (PDB ID: 6M71)(Burley et al., 2019) which was newly released on April 1st, 2020. For main protease (Mpro) of SARS-CoV-2, the crystal structure of PDB ID: 6LU7 (Y. Li et al., 2020) was used. The RdRp and Mpro PDB structures were energy minimized via Swiss PDB Viewer (Guex & Peitsch, 1997).
Figure 1.
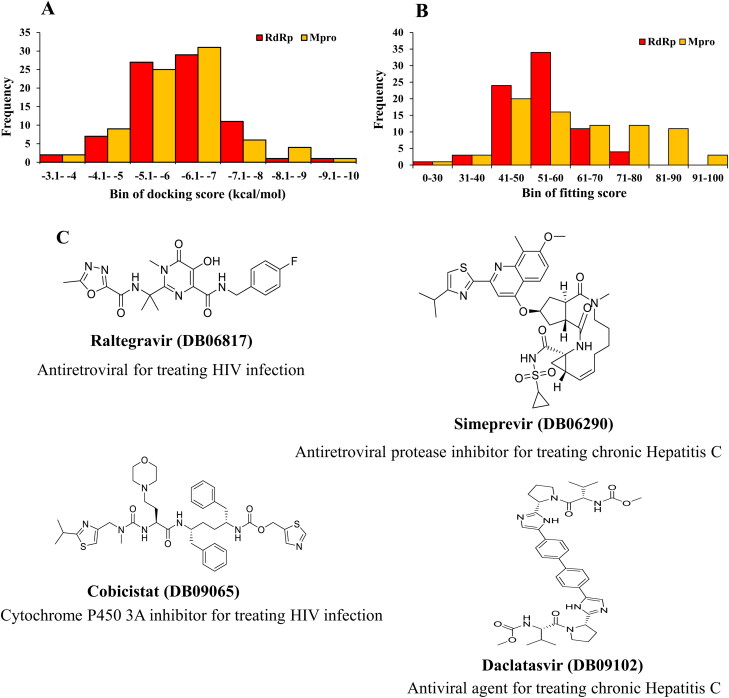
Frequency distribution of 76 drugs over range of docking results and structure of promising drugs. (A) Docking score by AutoDock Vina, (B) Fitting score by GOLD and (C) Two-dimensional structures of the four promising drugs.
2.2. Ligand preparation
Initial geometries of the selected 76 approved antiviral drugs are collected from PubChem and ChemSpider databases. Name, Drug ID, 2 D structure and reference of these drugs are summarized in Table S1. Quantum mechanics (QM) calculations were conducted to optimize antiviral drugs. Gaussian 09 program package was used for all quantum calculations (Frisch et al., 2009). To optimize the antiviral drugs, semi-empirical PM6 method (Bikadi & Hazai, 2009) was used. Subsequently the vibrational frequencies (all values are in harmonic approximation) were calculated and the absence of imaginary frequencies confirmed the stationary points correspond to minima on the Potential Energy Surface.
Table 1.
Binding affinity of the five drugs with different crystal structures of RdRp and Mpro (AutoDock Vina scores are in kcal/mol).
| Drug | RNA dependent RNA polymerase |
Main protease |
|||||
|---|---|---|---|---|---|---|---|
| AutoDock Vina |
GOLD | AutoDock Vina | GOLD | ||||
| Model | 6NUR | 6M71 | 7BTF | Model | 6LU7 | ||
| Raltegravir | −9.7 | −9.1 | −8.4 | −8.5 | 53.29 | −9.1 | 65.25 |
| Daclatasvir | −9.5 | −9.4 | −8.5 | −7.9 | 67.6 | −7.2 | 74.11 |
| Simeprevir | −9.3 | −8.9 | −9 | −9.5 | 66.65 | −8.7 | 76.18 |
| Cobicistat | −8.4 | −8.2 | −6.7 | −7.2 | 73.86 | −7.4 | 89.31 |
| Remdesivir | −7.2 | −7.6 | −7.9 | −6.5 | 54.79 | −7.8 | 81.16 |
2.3. Molecular docking protocols
RdRp and Mpro proteins were docked with 76 approved antiviral drugs by AutoDock Vina protocol (Trott & Olson, 2010) and GOLD 5.7 (Genetic Optimisation for Ligand Docking) (Jones et al., 1997). The molecular docking approach using AutoDock Vina protocol predicted the binding affinity and the interaction of the selected antiviral drugs with RdRp and Mpro. The docking grid box was set around the polymerase active site considering the (A-G) conserved motifs, around center X = 145.85, Y = 141.66, and Z = 156.30 and the dimensions were X: 31.08, Y: 29.76, and Z: 23.81. For Mpro, the grid box value remained around center X = −9.83, Y = 14.38, and Z = 68.48 and where, the dimensions were X: 24.69, Y: 31.64, and Z: 29.97 covering desired binding site residues. Besides docking by AutoDock Vina, GOLD docking Suite was employed to estimate the fitness score and binding interaction of these drugs against the receptor proteins. Hermes visualizer in the GOLD Suite was used to prepare the drugs as well as SARS-CoV-2 RdRp and Mpro proteins for docking. All other parameters were set as default and CHEMPLP was chosen for the fitness function. For visualization and detecting the non-covalent interaction in the docked drug-protein complex, PyMol (version 2.3.2) and BIOVIA Discovery Studio version 4.5 were utilized.
2.4. Molecular dynamics (MD) simulations
MD simulation was employed to validate the docking results obtained for the best antiviral drugs. 100 ns MD simulation was performed for RdRp and Mpro in apo-form (without drug) and holo-form (drug-protein). YASARA dynamics (Krieger et al., 2004) program was used for all simulation applying AMBER14 force field (Dickson et al., 2014). In the presence of a water solvent, the total system was equilibrated with 0.9% NaCl at 298 K temperature. During simulation, the particle Mesh Ewald algorithm was considered for long-range electrostatic interactions. Berendsen thermostat process was used to regulate the simulation temperature. A cubic cell was generated within 20 Å on each side of system and periodic boundary condition was preferred during the simulation. A time step of 1.25 fs was maintained to carry out 100 ns MD simulation and the snapshots were taken at every 100 ps. Based on our previous MD data analysis (Ahmed et al., 2020; M. J. Islam et al., 2019; R. Islam et al., 2020; Junaid et al., 2019; A. M. Khan, Shawon, & Halim, 2017; Shahinozzaman et al., 2019), various data including bond distances, bond angles, dihedral angles, free energy, columbic and van der Waals interactions, solvent-accessible surface area (SASA), molecular surface area (MolSA), root mean square fluctuation (RMSF) and root mean square deviation (RMSD) values for alpha carbon, backbone, and heavy atoms are retrieved from MD simulations using modified macro files.
To understand the structural and energy changes during MD simulation of RdRp and Mpro proteins in presence of the drug, the principal component analysis (PCA) method was employed for analysing the different multivariate energy factors (De Jong, 1990; Wold et al., 1987). In this analysis, bond distances, bond angles, dihedral angles, planarity, van der Waals energies, and electrostatic energies represent structural and energy information about the two studied PCA models. Typically, PCA analysis helps to visualize any dissimilarity present among different groups which are added in the model (M. J. Islam et al., 2019; R. Islam et al., 2020). The last 35 ns trajectory data were considered for PCA analysis for both the two PCA models. Using centering and scaling, data of two models were pre-processed for this analysis. The six multivariate factors were organized in the X matrix and decomposed them into a product of two new matrices by the following equation:
Here, the Tk matrix represents the relation of sample to each other, Pk is the loading matrix which correlates the variables with each other, number of factors represented by k, and E be the representative of unmolded variance. In this analysis, R (R Core Team, 2014), RStudio (Rstudio Team, 2019) and in-house developed codes were employed for performing all the calculations and plots were generated by using the package ggplot2 (Wickham, 2009).
2.5. Mm/PBSA binding free energy calculations
For binding free energy calculation, Molecular Mechanics/Poisson-Boltzmann Surface Area method was used. YASARA dynamics was used for all calculations. AMBER14 force field with the ‘single trajectory approach’ was considered (Kollman et al., 2000). Selected snapshots from the last 50 ns MD simulation were used for all 6 protein-drug complexes. Protein and drug binding free energy was calculated using the following equations: (Gilson & Honig, 1988)
Here, ΔGcomplex, ΔGligand and ΔGprotein the total free energy of the protein-ligand complex, total energy of separated ligand and protein in solvent, respectively.
Where, ΔGMM= molecular mechanics interaction energy (ΔGelec and ΔGVdWare electrostatic and van der waals interaction energy respectively). ΔGPB is denoted the polar solvation energy ΔGSA is the nonpolar solvation energy. TΔS is the contribution of entropy to the free energy (T = temperature and S = entropy) (Razzaghi-Asl et al., 2018).
2.6. Quantitative structural activity relationships (QSAR) for antiviral drugs
29 antiviral drugs were selected based on high rank in docking and distributed randomly into two data arrays. 19 drugs were considered as the “training set” or “calibration samples”. The other 10 drugs were treated as the “test set” or “validation samples”. For the structural activity relationship, various parameters such as TPSA (topological polar surface area, Å2), molecular weight, XLogPs-AA, HBD (hydrogen bond donor), HBA (hydrogen bond acceptor) and number of rotatable bond (ROTB), benzene ring, single bond, double bond, hydrogen atom, carbon atom, nitrogen atom, oxygen atom, fluorine atoms were considered for the drug candidates relating with the drug binding affinity observed from molecular docking via Partial-least-square (PLS) regression applying a standard multivariate regression analysis procedure (Ahmed et al., 2020). PLS regression data explored by XLSTAT (2020XLSTAT | Statistical Software for Excel, 2017). The PLS regression was optimized cautiously and refined properly as required.
3. Results
3.1. Results obtained from molecular docking
Molecular docking is performed for 76 approved antiviral drugs to obtain their binding affinities against the RdRp and Mpro (Table S2) by AutoDock Vina. For the second validation of molecular docking data, the GOLD docking suite is also used. The results which are obtained from the docking of the selected drugs with RdRp complex and drugs with Mpro complex using the GOLD suite are reported in ChemPLP fitness scores which is the default scoring function. The drugs with the higher fitness value, the better the docked interaction of the complexes. The frequency analysis shows that the Autodock Vina binding affinity of these drugs is varied from −3.1 kcal/mol to −10.0 kcal/mol (Figure 1A) and GOLD fitting score is ranged from 30 to100 (Figure 1B). Around 30 drugs show the binding affinity from −6.1 to −7.0 kcal/mol. A similar binding affinity pattern is observed for both proteins, RdRp and Mpro. However, Mpro demonstrated higher fitting score than RdRp.
Table 2.
Non-covalent interactions of the top ranked drugs with RdRp model protein (Pose predicted by AutoDock Vina).
| Drug | Binding affinity | Hydrogen bond (AA…Ligand) | Halogen bond (AA…Ligand) | Hydrophobic interaction (AA…Ligand) | Electrostatic interaction (AA…Ligand) |
|---|---|---|---|---|---|
| Raltegravir | −9.7 | TYR455 (2.686) O-H…F-C | TYR455 (2.686) O-H…F-C | TYR455 (3.860) Pi-Pi Stacked | ARG553 (4.308) Pi-cation |
| SER682 (2.623) O-H…O-C | ARG553 (4.940)Pi-Alkyl | ASP623 (3.474) Pi-Anion | |||
| THR680 (2.824) C-O…H-N | |||||
| THR680 (2.623) C-O…H-N | |||||
| ASP452 (2.570)) C-O…H-C | |||||
| Daclatasvir | −9.5 | ARG553 (2.713) N-H…O-C | ALA685 (4.025) Alkyl | ASP760 (3.933) Pi- Anion | |
| ARG569 (2.225) N-H…O-C | ARG555 (3.814) Alkyl | ||||
| ARG569 (2.668) N-H…O-C | ARG569 (4.112) Alkyl | ||||
| LYS577 (2.889) N-H…O-C | LEU576 (4.810) Alkyl | ||||
| ASP623 (2.519) C-O…H-N | LEU576 (3.913) Alkyl | ||||
| ASP623 (2.688) C-O…H-N | ILE494 (5.246) Alkyl | ||||
| ASP623 (2.990) C-O…H-C | TYR689 (5.106) Pi-Alkyl | ||||
| ILE494 (2.865) C-O…H-C | ALA688 (5.077) Pi-Alkyl | ||||
| ALA688 (2.714) Pi-Sigma | |||||
| Simeprevir | −9.3 | ARG553 (2.958) N-H…O-C | LYS621 (4.472) Alkyl | ASP760 (4.534) Pi-Anion | |
| ASN691 (2.541) N-H…O-C | GLU811 (3.928) Pi-Anion | ||||
| SER814 (2.996) O-H…O-C | |||||
| ASP760 (2.087) C-O…H-N | |||||
| ASP618 (2.339) C-O…H-N | |||||
| ASP618 (2.794) C-O…H-N | |||||
| SER682 (2.629) C-H…O-S | |||||
| Cobicistat | −8.4 | ARG553 (2.188) N-H…O-C | TYR455(3.958) Pi-Pi Stacked | ARG553 (4.395) Pi-Cation | |
| THR556 (2.827) N-H…O-C | CYS622 (5.167) Alkyl | ARG553 (4.357) Pi-Cation | |||
| THR556 (2.703) O-H…O-C | LYS798 (5.267) Alkyl | ASP618 (3.458) Pi- Anion | |||
| LYS621( 2.869) N-H…O-C | ARG553(4.377) Pi-Alkyl | ||||
| CYS622 ( 1.942) N-H…O-C | |||||
| ARG624 (3.059) N-H…O-C | |||||
| ARG624 (2.085) N-H…O-C | |||||
| ASP623 (2.408) C-O…H-N | |||||
| ARG555 (2.527) C-H…O-C | |||||
| ASP760 (2.590) C-O…H-C | |||||
| THR556 (2.335) C-O…H-C | |||||
| ASP760 (2.467) C-O…H-C | |||||
| Remdesivir | −7.2 | LYS545 (3.052)) N-H…N-C | TYR455 (4.616) Pi-Pi T shaped | ||
| LYS545 (2.917) N-H…N-C | LYS621 (5.080) Pi-Alkyl | ||||
| ARG553 (2.854)) N-H…O-P | ARG624 (5.445) Pi-Alkyl | ||||
| ARG553 (2.855) N-H…O-P | |||||
| ASP623 (2.129) C-O…H-N | |||||
| THR556 (2.265) C-O…H-N | |||||
| ASP760 (3.046) C-O…H-C | |||||
| THR556 (2.858) C-O…H-C | |||||
| ASP623 (3.092) C-O…H-C |
In addition, the selected drugs are also docked with newly released PDB structures (6M71 and 7BTF) to make a comparison with the model structure of RdRp. The docking results are comparable (Table 1). The selected drugs are predicted to have promising results as potential inhibitors against both proteins of the virus, clearly indicating that they may show better inhibitory effects than Remdesivir (Figures 1C and 2). The better binding affinity, as well as fitness score, is obtained for Cobicistat, Daclatasvir, Raltegravir, Simeprevir than Remdesivir in most of the cases (Table 1).
3.2. Molecular interaction of the best drugs with RdRp and Mpro
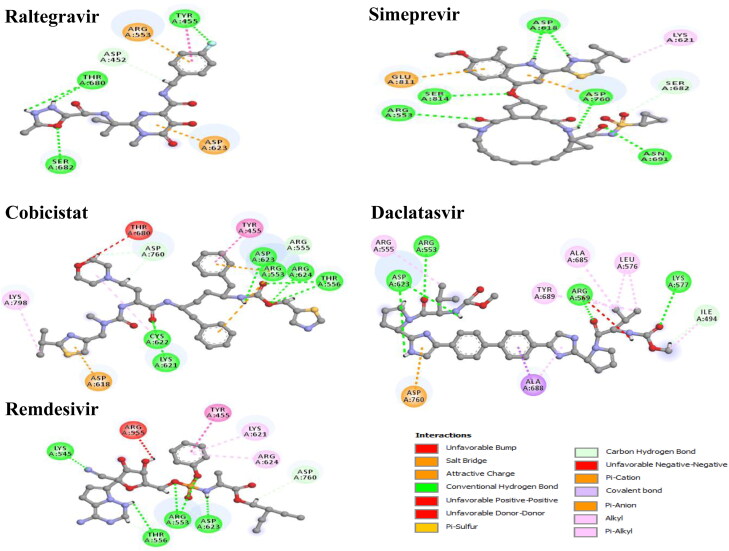
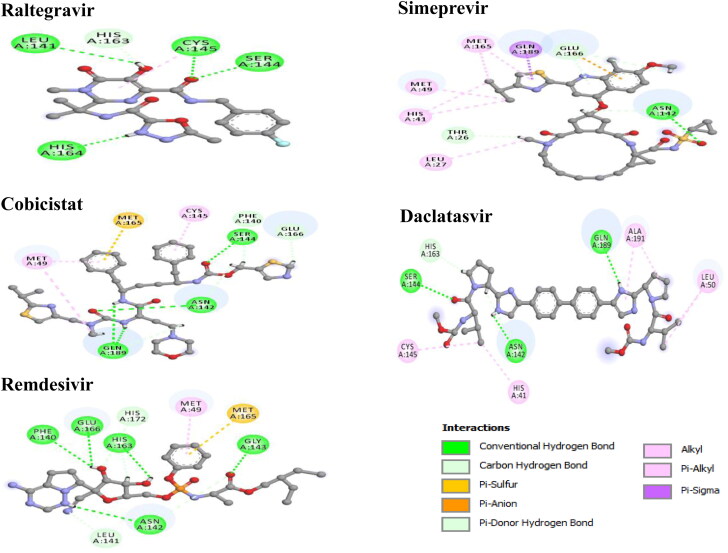
Various types of non-covalent interactions such as hydrogen bond, halogen bond, and hydrophobic interactions are found to be involved in the binding of drugs with RdRp and Mpro when the poses were predicted with AutoDock Vina (Figures 3 and 4). Among the selected drugs, the highest negative binding affinity (RdRp −9.7 kcal/mol; Mpro −9.1 kcal/mol) and fitting score (RdRp 53.29; Mpro 65.25) are noticed for Raltegravir. The Raltegravir is found to form five hydrogen bonds, one halogen bond, two hydrophobic bonds, and two electrostatic bonds with RdRp whereas six hydrogen interactions, and one hydrophobic interaction with Mpro. In Simeprevir-RdRp complex the drug is stabilized by seven hydrogen bonds, one hydrophobic bond, and two electrostatic bonds and with Mpro, forms five hydrogen bonds, seven hydrophobic bonds, and one electrostatic bond. In the Cobicistat-RdRp complex, the drug is interacted with receptor protein by twelve hydrogen bonds, four hydrophobic, and three electrostatic interactions whereas for the Cobicistat-Mpro complex, nine hydrogen interactions, three hydrophobic interaction, and one Pi-Sulphur bond. The Daclatasvir-RdRp complex is formed a stable network by eight hydrogen bonds, nine hydrophobic interactions, and one electrostatic interaction while the Daclatasvir-Mpro complex is showed five hydrogen bonds, six hydrophobic interactions. In Remdesivir-RdRp complex, nine hydrogen bonds and three hydrophobic bonds are detected whereas nine hydrogen bonds, one hydrophobic bond and one Pi-Sulfur bond are observed in Remdesivir-Mpro complex. Non-covalent interactions are predicted by the GOLD suite are presented in Figures S2 and S3. Moreover, the residues of 6M71 interacted with Remdesivir are similar compared to the model RdRp structure (Gao et al., 2020).
Figure 3.
Non-covalent interactions of selected drugs with RdRp model protein (Pose predicted by AutoDock Vina).
Figure 4.
Non-covalent interactions of selected drugs with Mpro (PDB ID:6LU7) (Pose predicted by AutoDock Vina).
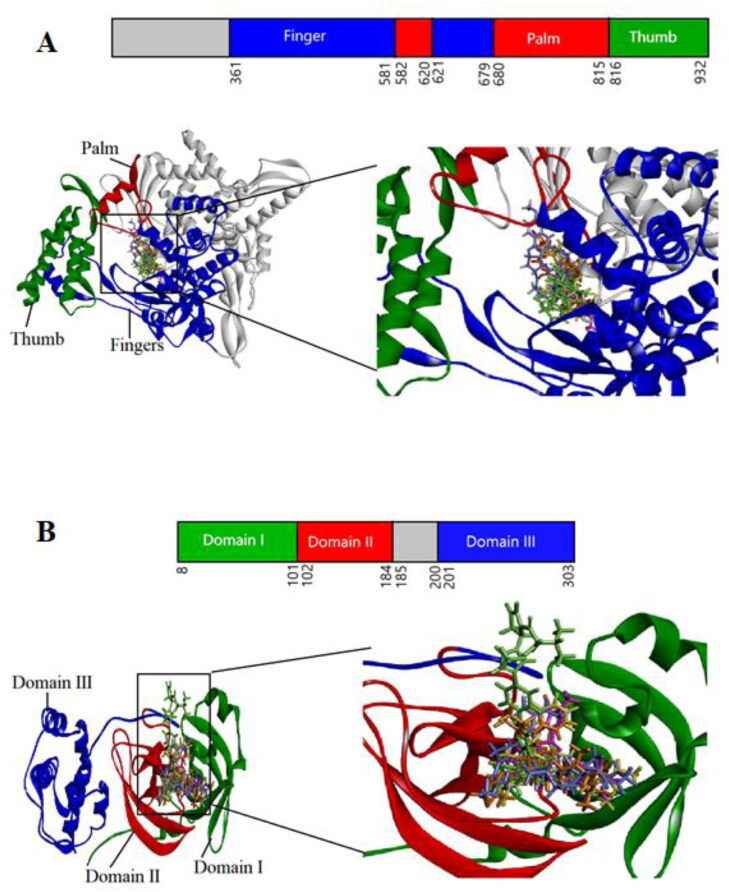
Figure 2.
Domain organization and interaction of both proteins with drugs. The inter-domain borders are labeled with amino acid residue numbers. (A) 3 D structure of RdRp (Fingers-Blue; Palm-Red; Thumb-Green) interacting with drugs (B) 3 D structure of Mpro (Domain I-Green; Domain II-Red; Domain III-Blue) interacting with drugs. Cobicistat is shown in Orange; Daclatasvir in Green; Raltegravir in Pink; Simeprevir in Violet; Remdesivir in Yellow.
3.3. Result obtained from molecular dynamics simulation
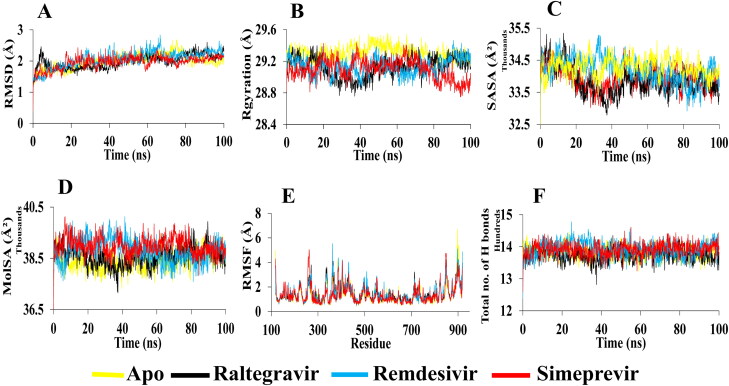
MD simulation for each complex of RdRp and Mpro with two selected drugs (Raltegravir and Simeprevir) and apo-form is performed for 100 ns. The Remdesivir-RdRp complex and the Remdesivir-Mpro complex are also subjected to MD simulation as a control. In case of RdRp, the RMSDs (0.5–2.7 Å) for α-carbon atoms in apo-RdRp is higher than Raltegravir and Simeprevir, thus suggesting that the unbound form of RdRp may be unstable in physiological conditions. In Figure 5A, RMSD values of Raltegravir are slightly increased to 2.4 Å after 4 ns and after 90 ns, indicating that the trajectories generated during the whole run are stable. Higher structural stability in the Raltegravir-RdRp complex is detected. For Remdesivir, the RMSD exhibits more fluctuations during the whole run and is increased to 2.8 Å at 67 ns. The average RMSD for Raltegravir is 1.9 Å, whereas the RMSD for Remdesivir is detected around 2.1 Å. A repeated fluctuation is found in Simeprevir from 18 to 65 ns with an average maximum deviation of 1.9 Å. It is indicated that the selected best drugs can be a stable and better drug than the control drug, Remdesivir. On the other hand, the RMSDs (0.4–2.7 Å) for α-carbon atoms in apo-Mpro is higher than Remdesivir and Simeprevir, conferring that the bound form of Mpro may be more stable in physiological conditions. In Figure 6A, RMSD values of Raltegravir are less in the initial run but significantly increased after 18 ns till 38 ns with the highest value of 6.3 Å, then again a slight increase from 58 to 62 ns, indicating that the trajectories generated during these times are unstable. However, structural stability in the Raltegravir-Mpro complex is detected in the later run (average 2.7 Å). The average RMSD for Remdesivir is 1.9 Å, whereas the RMSD for Simeprevir is detected around 1.8 Å, indicates higher structural stability in both complexes. It reflects that the selected both drugs can be a stable and better drug than the control drug, Remdesivir.
Figure 5.
Analysis of 100 ns MD simulation of RdRp in complex with selected drugs.(A) Root mean square deviation values of C-α atom of RdRp in the complexes with selected drugs along the 100 ns simulations. The structural changes of RdRp by means of (B) radius of gyration, (C) solvent accessible surface area, (D) molecular surface area, (E) root means square fluctuations, and (F) total number of hydrogen bonds formed during the simulation.
Figure 6.
Analysis of 100 ns MD simulation of Mpro in complex with selected drugs. (A) Root mean square deviation values of C-α atom of Mpro in the complexes with selected drugs along the 100 ns MD simulations. The structural changes of Mpro by means of (B) radius of gyration, (C) solvent accessible surface area, (D) molecular surface area, (E) root means square fluctuations, and (F) total number of hydrogen bonds formed during the simulation.
The radius of gyration (Rg) and solvent accessible surface area (SASA) are also observed to investigate the structural compactness and solvent accessibility of all complexes. The radius of gyration is described as the distance where the entire mass is supposed to act. It is the root mean square distance of a collection of atoms from a given axis of rotation. It depicts as an indicator of protein structural compactness. A bigger radius of gyration is detected for apo-form than all complexes which suggests the loose packing of the apo-protein structure during the whole run. A loose packing from 30 to 65 ns is obtained for apo-RdRp (Figure 5B). The average radius of gyration of Raltegravir, Simeprevir, and Remdesivir is remained nearly the same (29.1 Å). Besides, a slightly higher radius of gyration is observed for the Raltegravir-Mpro complex (average 22.4 Å) than apo-Mpro and other complexes. The Rg graph shows loose packing of the apo-protein structure during the initial 50 ns but compactness in the later phase. The average radius of gyration of the Simeprevir-Mpro complex and apo-Mpro remains the same (22.3 Å) whereas the Remdesivir-Mpro complex exhibits more compactness than others (average 22.1 Å) (Figure 6B). Similarly, the solvent-accessible surface area (SASA) is assessed which shows the highest value for Simeprevir complex (RdRp-average 34287 Å2; Mpro-average 13982 Å2) compared to the control drug (RdRp-average 34187 Å2; Mpro-13847 Å2) (Figure 5C). A slightly lower value is found for the Raltegravir-RdRp complex (average 33846 Å2) than Remdesivir and the apo-RdRp (average 33926 Å2). On the contrary, the Raltegravir-Mpro complex showed a higher average of 14376 Å2 than Remdesivir-Mpro (average 13847 Å2) and the apo-Mpro (14160 Å2) (Figure 6C). Based on Rg and SASA analysis, it is revealed that the best candidate drugs are stable.
Molecular surface area (MolSA) is also determined for the selected drug-protein complexes. In this analysis, Simeprevir-RdRp shows the highest MolSA (average 38976 Å2) whereas Raltegravir and Remdesivir demonstrate a slightly lower average value of 38580 and 38802 Å2, respectively (Figure 5D). In the case of Mpro complexes, Raltegravir reveals the highest MolSA (average 14243 Å2) than the others (Remdesivir, 13952 Å2; Simeprevir, 14103 Å2) (Figure 6D).
RMSF values are also calculated for determining the dynamic behavior of both protein residues. In the case of RdRp, it can be seen from Figure 5E that the RMSF of residues (261–431) and (850–919) fluctuates significantly during protein and ligands interaction. Although a low RMSF (within the limit of 1.3 Å) in the RdRp-Simeprevir complex is detected for most of the residues, a high fluctuation of 4.4 Å is noticed in the thumb domain close to the residue 848. A maximum fluctuation of 5 Å is detected in the N terminal extension of residues 262. Remdesivir-RdRp complex is predicted to have fluctuation with around 5.1 Å in the residue 898 of the thumb domain. The RMSF of residues Arg553, Arg555, and Val557 (finger domain), and Asp618 (palm domain) in the Raltegravir-RdRp complex are lower than other complexes, reflecting that Raltegravir can stimulate more strong interactions with these key residues than other drugs. Since the less fluctuation indicates higher protein structural stability, the highest degree of flexibility is observed for apo-Mpro, and Remdesivir-Mpro, whereas a similar fluctuation (average 1.4 Å) is noticed for Raltegravir-Mpro and Simeprevir-Mpro. Furthermore, the visual analysis of MD simulation trajectories suggests that all the drugs engaged in significant binding interactions with the hotspot residues (His163, Glu166, Cys145, Gly143, His172, Phe140, Cys145, His41, Thr25, Met49, His41, Met165, Glu166, and Gln189) of the Mpro protein (Figure 6E).
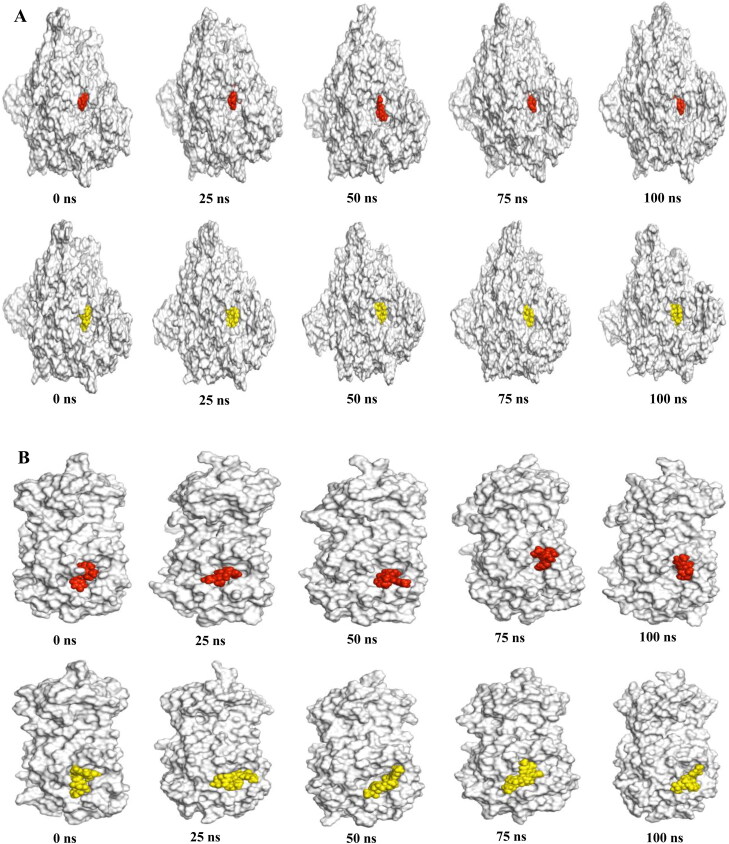
Hydrogen bonds play a significant role in the case of molecular recognition and the overall stability of the protein structure. The intermolecular hydrogen bonds formed during the whole 100 ns, is collected from the trajectories. In Figure 5F, the maximum hydrogen bonds, which maintain the rigidity of the complex, is predicted for Remdesivir and Simeprevir (average 1392) with RdRp. On the contrary, the highest number of hydrogen bonds with Mpro is observed for Raltegravir (average 515) (Figure 6F). Moreover, our snapshot conformers exhibit that both drugs most of the time stayed in the binding sites of both proteins during the whole simulation run (Figure 7A and B).
Figure 7.
Binding pose of Raltegravir and Simeprevir over the course of 100 ns simulation. The crystal structures of RdRp and Mpro are shown as white surface with Raltegravir (red) and Simeprevir (yellow) as spheres in (A) and (B) respectively.
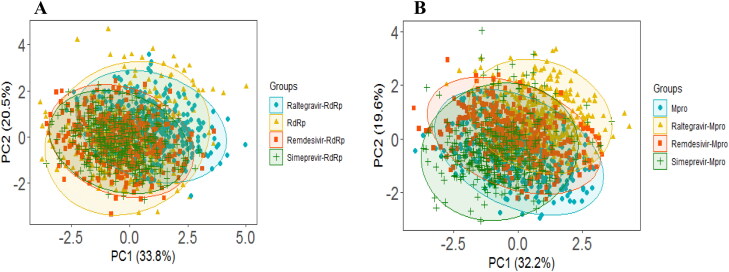
Two different PCA models are developed for two different proteins; the first PCA model is developed considering RdRp, and the second model is explored for Mpro. In each PCA model, four training sets (Raltegravir-Protein, Remdesivir-Protein, Simeprevir-Protein complexes, and apo-protein structure) are included in the model to understand the structural change of protein during MD simulation. For the RdRp PCA model (Figure 8A), 54.3% of the variance is explained by PC1 and PC2, where PC1 explains 33.8% and PC2 explains 20.5% of the variance. Similarly, two PCs explain 51.8% of the variance in the Mpro PCA Model (PC1 = 32.2%, PC2 = 19.6%) (Figure 8B). From the scores plots of PC1 and PC2 (Figure 8A and B) for both models, it is observed that different groups are overlapped with each other without any significant separation. Therefore, the structural and energy profiles are almost unchanged for all drug-protein complexes compared to both corresponding apo-protein during the simulation. However, here in this study, all three drugs (Raltegravir, Remdesivir, Simeprevir) show similar types of binding behavior with both the RdRp and Mpro.
Figure 8.
PCA analysis of RdRp and Mpro. (A) RdRp PCA model: The scores plot presented four data clusters in different colors, where each dot represented one time point. The clustering is attributable to: apo-RdRp (yellow), Raltegravir-RdRp complex (cyan), Remdesivir-RdRp complex (orange), Simeprevir-RdRp complex (green). (B) Mpro PCA model: The scores plot presented four data clusters in different colors, where each dot represented one time point. The clustering is attributable to: apo-Mpro (cyan), Raltegravir-Mpro complex (yellow), Remdesivir-Mpro complex (orange), Simeprevir-Mpro complex (green).
3.4. Mm/PBSA binding free energy analysis
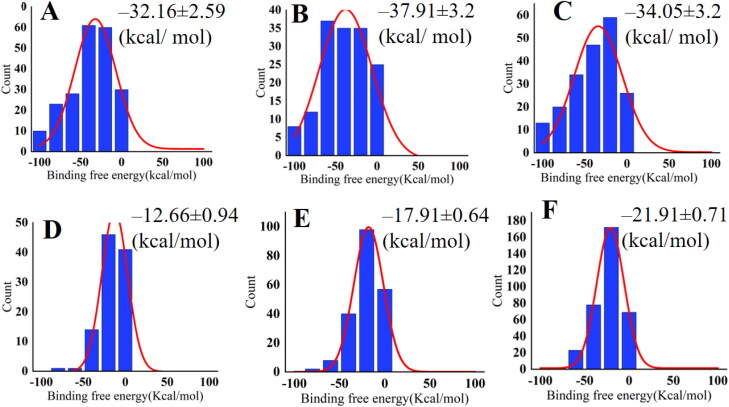
The binding free energy is calculated for six protein-drug complexes. The binding free energy values for Remdesivir-RdRp, Raltegravir-RdRp, Simeprevir-RdRp are ‒32.16 ± 2.59 (kcal/mol), ‒37.91 ± 3.2 (kcal/mol), and ‒34.05 ± 3.2 (kcal/mol), respectively (Figure 9A, B & C). Raltegravir has the most negative binding free energy out of this three complexes. On the other hand, binding free energy of Remdesivir, Raltegravir and Simeprevir against Mpro is ‒12.66 ± 0.94 (kcal/mol), ‒17.91 ± 0.64 (kcal/mol), ‒21.91 ± 0.71 (kcal/mol) (Figure 9D, E & F). Simeprevir shows the highest negative value for Mpro.
Figure 9.
Histogram of the binding free energy for (A) Remdesivir-RdRp, (B) Raltegravir-RdRp (C) Simeprevir-RdRp and (D) Remdesivir-Mpro (E) Raltegravir-Mpro (F) Simeprevir-Mpro. Here, the red color curve represents Gaussian fit.
3.5. Insights from quantitative structural activity relationships (QSAR)
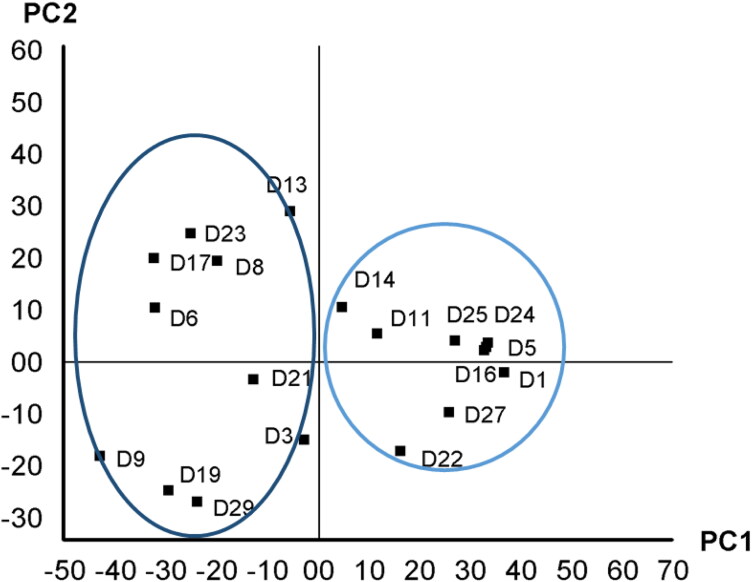
Quantitative structure activity relationship (QSAR) analysis is the most powerful and widely used method that predicts the biological property of novel compounds. In the exchange of experimental testing, QSAR methodology is highly recommended for an ultimate validation of desired models due to its fast throughput good hit rate (Neves et al., 2018). To develop PLS regression, all QSARs of drugs are vital, yet, the contribution of them may vary considerably. The most important QSAR contributors to the PLS regression are topological polar surface area (Å2), number of hydrogen bond donor (HBD), hydrogen atom, and single bond of the drugs (Table S5). Beside QSAR analysis, principal component analysis (PCA) is employed for pattern recognition of drugs’ structure-activity relationship. The scores-plot of the PCA modes is displayed in Figure 10. The PC1 elucidates 51.48% of the variability in drug QSAR. Moreover, the PC2 describes 19.09% QSAR variability of drugs.
Figure 10.
Score plot of PLS regression of quantitative structural activity relationship of drugs.
The scores plot illustrated a remarkable combination of drugs. The similar drugs QSAR were clustered together on the scores-plot. The drugs (D1, D5 (Daclatasvir), D11, D14 (Nelfinavir), D16 (Norvir), D24, D25, and D27) comprising thiazoyl ring and H2NCOO-, –CH2-, OH, -CONH- functional groups attached to benzene ring are assembled on the first and second quadrant of the scores-plot. Further, the drugs consisting –CONH–, –OH and C = O functional groups (D6, D13, D8, D17, D21 (Raltegravir), and D23) are located on the third and fourth quadrant of the score-plots. In D21 (Raltegravir), oxadiazole and pyrimidine-4(3H)-one groups are connected to a benzene ring. Moreover a fluorine atom is also present in the benzene ring.
The principal objective of any PLS model is to predict upcoming drug candidates, mainly from its SAR. Hence, the binding energies of the drug candidates are supposed to predict via developed PLS regression. The binding energy obtained from molecular docking with RdRp model protein and the predicted binding energy by PLS regression for each drug of validation drugs (test set) are exhibited in Table 4. The capability of PLS regression to predict accurately, the binding energy of the validated drugs is estimated by root-mean-square-relative percent-errors (RMS%RE). The PLS regression revealed a high accuracy of 85% for the prediction of the binding energy of drugs compared with the binding energy obtained from molecular docking.
4. Discussion
In the present study, we have screened 76 antiviral drugs to assess their binding affinity and non-covalent interaction with RdRp and Mpro proteins. Molecular docking studies, employing Autodock Vina and GOLD, have been used. Molecular docking results show that most of the screened drugs interact with the active pocket residues of the RdRp and Mpro proteins. Moreover, the selected four drugs Cobicistat, Daclatasvir, Raltegravir, and Simeprevir also show higher binding affinity and interaction with both proteins in comparison to recently approved drug, Remdesivir. Raltegravir, an inhibitor of the HIV integrase protein, is exhibited the effective antiretroviral activity (Hicks & Gulick, 2009). Whereas Simeprevir, HCV protease inhibitor, restrains or inhibits protein synthesis and has been accepted for the treatment of chronic hepatitis C infection, genotype 1 (Talwani et al., 2013). Simpeprevir can be used as a repurposing treatment for COVID-19 (Hosseini & Amanlou, 2020). Another anti-HIV drug Cobicistat, mainly inhibits human CYP3A proteins (Mathias et al., 2010). Thus, it increases the plasma concentration of other co-administered anti-HIV drugs without the risk of resistance and can be selected as a potential against COVID-19 (Harrison, 2020). Moreover, Daclatasvir is applied against Hepatitis C Virus, which directly inhibits HCV protein NS5A to resist viral RNA replication and protein translation (Hu et al., 2020). Although Dolutegravir an HIV-1 integrase inhibitor) shows good binding affinity (-8.9 kcal/mol) in our study (Table S2), we do not further investigate it as a similar study already is reported for Dolutegravir (R. J. Khan et al., 2020).
RdRp, also known as nsp12, is the key element of viral replication or transcription machinery and one of the main drug targets for SARS-CoV-2. It exists as a complex, with the polymerase (nsp12) bound with smaller proteins named nsp7 and nsp8. It has a shape of the right hand (residues 367-920), with fingers subdomain (residues 366-581 and 621-679), a palm subdomain (residues 582-620 and 680-815), and a thumb subdomain (residues 816-920). The active site of the RdRp is consists of the conserved polymerase motifs A-G in the palm domain. A major benefit of targeting the SARS-CoV-2 RdRp for drug development is that scientists already have experience to design polymerase inhibitors for HIV therapies. The binding affinities of the selected four drugs with RdRp are compared with Remdesivir. The highest binding affinity of −9.7 kcal/mol is detected for Raltegravir while the binding affinity of −7.2 kcal/mol is obtained for Remdesivir. A similar fitting score is observed with the GOLD docking for both drugs. When docking results of both methods are compared with the Remdesivir, the selected drugs show very strong binding interactions including hydrogen bond, hydrophobic, halogen bond, and electrostatic interactions with the important amino acids of RdRp. Compared to the Remdesivir, all four drugs show better and more appropriate interactions with many important residues through hydrogen bond, Pi-Alkyl and Pi-Pi interactions. The selected drugs can promote more interactions than Remdesivir, contributing to a higher binding affinity and fitting score. Amino acid residues, Met542, Lys545, Ser549, Lys551, Arg553, Arg555, Val557, Asp618, Cys622, Asp623, Ser682, Asp760, and Arg836 in the binding pocket of RdRp are found to participate in non-covalent interactions with four drugs (Figure 2A, Table 2 and Table S3).
Table 3.
Non-covalent interactions of top ranked drugs with Mpro (PDB ID-6LU7) (Pose predicted by AutoDock Vina).
| Drug candidate | Binding affinity | Hydrogen bond (AA…ligand) | Hydrophobic interaction (AA…ligand) | Electrostatic interaction (AA…ligand) | Other |
|---|---|---|---|---|---|
| Raltegravir | −9.1 | HIS164(2.245) C-O…H-N | CYS145(5.309) Pi-Alkyl | ||
| LEU141(2.803) C-O…H-O | |||||
| SER144(2.123) N-H…O-C | |||||
| SER144(2.463) O-H…O-C | |||||
| CYS145(2.244) N-H…O-C | |||||
| HIS163(3.0656) C-H…O-H | |||||
| Simeprevir | −8.7 | ASN142(1.875) N-H…O-S | MET165(4.338) Alkyl | GLU166(3.950) Pi-Anion | |
| ASN142(2.648) C-O…H-C | GLN189(2.557) Pi-Sigma | ||||
| THR26(2.926) C-O…H-C | MET49(3.910) Alkyl | ||||
| GLU166(3.042) C-O…H-C | LEU27(5.136) Alkyl | ||||
| GLU166(2.902) Pi-Donor | MET165(4.224) Pi-Alkyl | ||||
| HIS41(4.325) Pi-Alkyl | |||||
| HIS41 (4.662) Pi-Alkyl | |||||
| Remdesivir | −7.8 | ASN142(2.202) C-O…H-N | MET49(4.617) Pi-Alkyl | MET165(5.618) Pi-Sulfur | |
| PHE140(2.838) C-O…H-O | |||||
| GLU166(2.456) C-O…H-O | |||||
| HIS163(2.246) C-N…H-O | |||||
| GLY143(2.102) N-H…O-C | |||||
| HIS163(2.637) C-N…H-C | |||||
| LEU141(2.640) C-H…N-C | |||||
| ASN142(3.051) C-H…O-C | |||||
| HIS172(3.021) C-H…O-H | |||||
| Cobicistat | −7.4 | GLN189(2.571) C-O…H-N | MET49 (4.863) Alkyl | MET165(5.563) Pi-Sulfur | |
| GLN189(2.288) C-O…H-N | CYS145(4.230) Pi-Alkyl | ||||
| ASN142(2.650) N-H…O-C | MET49(4.644) Pi-Alkyl | ||||
| SER144(2.948) O-H…O-C | |||||
| GLU166(2.803) C-O…H-C | |||||
| GLN189(2.691) C-O…H-C | |||||
| ASN142(2.434) C-O…H-C | |||||
| PHE140(2.693) C-O…H-C | |||||
| GLN189(2.345) C-O…H-C | |||||
| Daclatasvir | −7.2 | ASN142(2.319) C-O…H-N | LEU50(4.214) Alkyl | ||
| GLN189(2.677) C-O…H-N | LEU50(4.224) Alkyl | ||||
| SER144(2.537) O-H…O-C | CYS145(4.379) Alkyl | ||||
| HIS163(2.527) C-N…H-C | ALA191(4.919) Alkyl | ||||
| ASN142(2.437) C-O…H-C | ALA191(4.252) Pi-Alkyl | ||||
| HIS41(4.213) Pi-Alkyl |
Mpro is highly conserved among Coronaviridae members and is a homodimer with three structural domains (domain I, II and, III). The substrate-binding site is situated in the Cys-His catalytic dyad located in a cleft between domain I and II where Cys act as nucleophile while His acts as a proton acceptor. Though dimerization of the two protomers is crucial, only one protomer is active at a time (Muralidharan et al., 2020). Moreover, there are two other deeply buried subsites known as S1, S2 and three shallow subsites (S3, S4, and S5) in addition to the catalytic site. The S1 subsite consists of His163, Glu166, Cys145, Gly143, His172, and Phe140 while S2 comprises Cys145, His41, and Thr25 amino acid residues. These residues are principally associated in hydrophobic and electrostatic interactions. Theses shallow subsites S3-S5 consists of Met49, His41, Met165, Glu166 and Gln189 amino acid residues and can endure various functionalities (S. A. Khan et al., 2020). Theses residues of the catalytic site of Mpro are found to interact with our top-ranked four drugs (Figure 2B, Table 3, and Table S4). Overall, protein-drug interactions reveal a strong inhibition of virus polymerase and protease.
Table 4.
Predicted binding energy by PLS regression and actual binding energy of RdRp model protein from molecular docking.
| Sample | Predicted binding energy | Actual binding energy | %RE |
|---|---|---|---|
| D4 | −8.42 | −8.4 | 1.27 |
| D26 | −8.23 | −9.3 | −2.18 |
| D15 | −8.04 | −8.1 | −0.1 |
| D10 | −7.96 | −7.3 | 1.36 |
| D12 | −7.88 | −7.6 | 0.58 |
| D2 | −7.86 | −8.2 | −0.67 |
| D28 | −7.82 | −8.1 | −0.57 |
| D7 | −7.81 | −8.9 | −2.22 |
| D18 | −7.8 | −7.4 | 0.82 |
| D20 | −7.57 | −7.3 | 0.56 |
| RMS%RE | 1.54 |
Based on molecular docking, four drugs show better binding affinity than Remdesivir. The best two drugs (Raltegravir and Simeprevir) are chosen for analysis with molecular dynamics. It is shown from MD simulation that the average RMSD values of Raltegravir and Simeprevir (1.9 Å) is lower compared to Remdesivir (2.1 Å), indicating more structural stability with RdRp in bound states. Although Raltegravir-Mpro complex shows much fluctuation in the primary state, the stability increases in the later phase. Simeprevir also shows lower RMSD value, marks its greater stability. For RdRp, it is revealed from the Rg and SASA that Raltegravir and Simeprevir complex are stable similar to Remdesivir (Figure 5B and C) whereas, for Mpro, the Raltegravir-Mpro complex shows some instability in the initial run. Furthermore, a similar average MolSA is detected for the Raltegravir-RdRp and the Remdesivir-RdRp whereas it is highest for the Simeprevir-RdRp (Figure 5D). For Mpro, the Raltegravir-Mpro complex exhibits the highest value whereas Simeprevir and Remdesivir have similar MolSA value. It is observed from the RMSF values that Simeprevir has more reasonable binding stability with RdRp (Figure 5E). Moreover, a nearly similar RMSF value of residues is predicted for the Raltegravir-RdRp complex when compared with the Remdesivir-RdRp complex. Although Raltegravir exhibits more fluctuations than the other complexes, all the drugs demonstrate significant interaction with important residues indicates their compactness in a complex form with Mpro (Figure 6E). Furthermore, the binding free energy analysis supports the MD results (Figure 9). PCA analysis confirms that the selected drugs can be a good candidate to stabilize both proteins as supported by the RMSD analysis (Figure 8). Overall, all analyses from the MD simulations support the docking results indicating that the selected drugs can actively interact with RdRp and Mpro.
Quantitative structure activity relationships (QSAR) analysis shows that molecular weight, TPSA (Å2), number of hydrogen donor, hydrogen atom, and C-C single bonds contribute the high impact on the binding affinity and non-covalent interaction against the protein receptor. The molecular docking result and QSAR reveal that D21 (Raltegravir), D5 (Daclatasvir), D26 (Simeprevir), and D4 (Cobicistat from test set) could be highly effective to target SARS-COV-2 RdRp and Mpro.
5. Conclusion
In this study, we employ drug repurposing approach to identify potential candidates which can bind and interact with RdRp and Mpro proteins of SARS-CoV-2. Based on binding affinity and interaction results, Raltegravir, Simeprevir, Cobicistat, and Daclatasvir are found to be very promising and therefore these four drugs can further optimize to design effective inhibitors against SARS-CoV-2. All four drugs are stable and they form a greater number of interactions through hydrogen bonds with both proteins. MD results show that the selected two drugs (Raltegravir and Simeprevir) remain in the binding pocket of the RdRp and Mpro protein without altering protein native structures. Moreover, MD simulation reveals that RMSD values of the selected drugs are relatively lower compared to control Remdesivir drug. Binding free energy calculated by MM/PBSA demonstrates the higher binding affinity of Raltegravir and Simeprevir compared to Remdesivir. Principal component analysis (PCA) illustrates “QSAR pattern recognition” and discloses that thiazoyl ring and H2NCOO-, –CH2-, OH, -CONH- functional groups attached to benzene ring are assembled together on the first and second quadrant of the scores-plot and drugs consisting –CONH–, –OH and C = O functional group located on the third and fourth quadrant of the score-plots. The predicted values of the binding energy from PLS regression is very close (accuracy 85%) compared with the values obtained from molecular docking. Thus, QSAR explores the biological properties and mechanism of drug action against targeted protein to ensure drug efficacy. These finding will help experimental medicinal chemist to test the performance of these drugs against SARS-CoV-2 in in vitro and in vivo setting.
Associated content
Supporting information
Figure S1, Superimposed structure of SARS-CoV-2 RdRp model protein with 6NUR and 6M71 in two different views; Table S1, Name, Drug ID and 2Dstructures of the 76 approved antiviral drugs; Table S2, Docking results of 76 FDA drugs with RdRp and Mpro of SARS-CoV-2; Figure S2, Non-covalent interactions of selected drugs with RdRp model protein (Pose predicted by GOLD); Figure S3, Non-covalent interactions of selected drugs with Mpro (PDB ID-6LU7) (Pose predicted by GOLD); Table S3, Non-covalent interactions of selected drugs with RdRp model protein (Pose predicted by GOLD); Table S4, Non-covalent interactions of selected drugs with Mpro (PDB ID-6LU7) (Pose predicted by GOLD); Table S5, Drug-likeness properties of 29 drugs showing high binding affinity
Supplementary Material
Acknowledgements
We are grateful to our donors (http://grc-bd.org/donate/) who supported to build a computational platform. The authors like to acknowledge The World Academy of Science (TWAS) to purchase the High-Performance Computers for molecular dynamics simulation.
Disclosure statement
The authors declare no competing financial interest.
References
- Aanouz, I., Belhassan, A., El Khatabi, K., Lakhlifi, T., El Idrissi, M., & Bouachrine, M. (2020). Moroccan Medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure & Dynamics, 1–12. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, S., Islam, N., Shahinozzaman, M., Fakayode, S. O., Afrin, N., & Halim, M. A. (2020). Virtual screening, molecular dynamics, density functional theory and quantitative structure activity relationship studies to design peroxisome proliferator-activated receptor-γ agonists as anti-diabetic drugs. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1714482 [DOI] [PubMed] [Google Scholar]
- Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., & Garry, R. F. (2020). The proximal origin of SARS-CoV-2. Nature Medicine, 26(4), 450–452. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburn, T. T., & Thor, K. B. (2004, August). Drug repositioning: Identifying and developing new uses for existing drugs. Nature Reviews. Drug Discovery, 3(8), 673–683. . Nature Publishing Group. 10.1038/nrd1468 [DOI] [PubMed] [Google Scholar]
- Barrows, N. J., Campos, R. K., Powell, S. T., Prasanth, K. R., Schott-Lerner, G., Soto-Acosta, R., Galarza-Muñoz, G., McGrath, E. L., Urrabaz-Garza, R., Gao, J., Wu, P., Menon, R., Saade, G., Fernandez-Salas, I., Rossi, S. L., Vasilakis, N., Routh, A., Bradrick, S. S., & Garcia-Blanco, M. A. (2016). A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host & Microbe, 20(2), 259–270. 10.1016/j.chom.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni, M., Kiefer, F., Biasini, M., Bordoli, L., & Schwede, T. (2017). Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Scientific Reports, 7(1), 10480. 10.1038/s41598-017-09654-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikadi, Z., & Hazai, E. (2009). Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. Journal of Cheminformatics, 1(1), 15. 10.1186/1758-2946-1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 Coronavirus Structure, Mechanism of Action, Antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure & Dynamics, 1–14. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley, S. K., Berman, H. M., Bhikadiya, C., Bi, C., Chen, L., Di Costanzo, L., Christie, C., Dalenberg, K., Duarte, J. M., Dutta, S., Feng, Z., Ghosh, S., Goodsell, D. S., Green, R. K., Guranovic, V., Guzenko, D., Hudson, B. P., Kalro, T., Liang, Y., … Zardecki, C. (2019). RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res, 47(D1), D464–D474. 10.1093/nar/gky1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciliberto, G., & Cardone, L. (2020). Boosting the arsenal against COVID-19 through computational drug repurposing. Drug Discovery Today, 25(6), 946–948. 10.1016/j.drudis.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J., Li, F., & Shi, Z. L. (2019). Origin and evolution of pathogenic coronaviruses. Nature Reviews. Microbiology, 17(3), 181–192. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong, S. (1990). Multivariate calibration, H. Martens and T. Naes, Wiley, New York, 1989. Journal of Chemometrics, 4(6), 441–441. ISBN 0 471 90979 3. Price: £75.00, US$138.00. No. of pages: 504. 10.1002/cem.1180040607 [DOI] [Google Scholar]
- Dickson, C. J., Madej, B. D., Skjevik, Å. A., Betz, R. M., Teigen, K., Gould, I. R., & Walker, R. C. (2014). Lipid14: The amber lipid force field. Journal of Chemical Theory and Computation, 10(2), 865–879. 10.1021/ct4010307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., Şahin, A. T., & Yelekçi, K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Petersson, G. A., Nakatsuji, H., Li, X., Caricato, M., Marenich, A. V., Bloino, J., Janesko, B. G., Gomperts, R., Mennucci, B., Hratchian, H. P., Ortiz, J. V., & Fox, D. J. (2009). Gaussian 09, Revision D.01. Wallingford CT: Gaussian, Inc. [Google Scholar]
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., … Rao, Z. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (New York, N.Y.). 368(6492), 779–782. 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson, M. K., & Honig, B. (1988). Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins, 4(1), 7–18. Reprint, (061/4), 10.1002/prot.340040104 [DOI] [PubMed] [Google Scholar]
- Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., O’Meara, M. J., Guo, J. Z., Swaney, D. L., Tummino, T. A., Hüttenhain, R., Kaake, R. M., Richards, A. L., Tutuncuoglu, B., Foussard, H., Batra, J., Haas, K., Modak, M., Kim, M., Haas, P., & Krogan, N. J. ( 2020). A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. Nature, 583, 459–468. 10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed]
- Guex, N., & Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis, 18(15), 2714–2723. 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- Harrison, C. (2020). Coronavirus puts drug repurposing on the fast track. Nature Biotechnology, 38(4), 379–381. 10.1038/d41587-020-00003-1 [DOI] [PubMed] [Google Scholar]
- Hicks, C., & Gulick, R. M. (2009). Raltegravir: The first HIV Type 1 Integrase Inhibitor. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America, 48(7), 931–939. 10.1086/597290 [DOI] [PubMed] [Google Scholar]
- Hilgenfeld, R. (2014). From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. The FEBS Journal, 281(18), 4085–4096. 10.1111/febs.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodos, R. A., Kidd, B. A., Shameer, K., Readhead, B. P., & Dudley, J. T. (2016). In silico methods for drug repurposing and pharmacology. Wiley Interdisciplinary Reviews. Systems Biology and Medicine, 8(3), 186–210. 10.1002/wsbm.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini, F. S., Amanlou, M. (2020). Simeprevir, potential candidate to repurpose for coronavirus infection: Virtual screening and molecular docking study. Preprints 2020, 2020020438. 10.20944/Preprints202002.0438.V1, (February), 0–2. [DOI] [PMC free article] [PubMed]
- Hu, F., Jiang, J., & Yin, P. (2020). Prediction of potential commercially inhibitors against SARS-CoV-2 by multi-task deep model, 1–8. [DOI] [PMC free article] [PubMed]
- Hussain, S., Pan, J., Chen, Y., Yang, Y., Xu, J., Peng, Y., Wu, Y., Li, Z., Zhu, Y., Tien, P., & Guo, D. (2005). Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. Journal of Virology, 79(9), 5288–5295. 10.1128/JVI.79.9.5288-5295.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M. J., Khan, A. M., Parves, M. R., Hossain, M. N., & Halim, M. A. (2019). Prediction of Deleterious Non-synonymous SNPs of Human STK11 Gene by Combining Algorithms, Molecular Docking, and Molecular Dynamics Simulation. Scientific Reports, 9(1), 16426. 10.1038/s41598-019-52308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, R., Parves, R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A., Al, … Halim, M. A. (2020). A Molecular Modeling Approach to Identify Effective Antiviral Phytochemicals against the Main Protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen, L. M., DeWald, L. E., Shoemaker, C. J., Hoffstrom, B. G., Lear-Rooney, C. M., Stossel, A., Nelson, E., Delos, S. E., Simmons, J. A., Grenier, J. M., Pierce, L. T., Pajouhesh, H., Lehár, J., Hensley, L. E., Glass, P. J., White, J. M., & Olinger, G. G. (2015). A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci Transl Med, 7(290), 290ra89. 10.1126/scitranslmed.aaa5597 [DOI] [PubMed] [Google Scholar]
- Jones, G., Willett, P., Glen, R. C., Leach, A. R., & Taylor, R. (1997). Development and validation of a genetic algorithm for flexible docking . J. Mol. Biol, 267(3), 727–748. 10.1006/jmbi.1996.0897 [DOI] [PubMed] [Google Scholar]
- Junaid, M., Islam, N., Hossain, M. K., Ullah, M. O., & Halim, M. A. (2019). Metal based donepezil analogues designed to inhibit human acetylcholinesterase for Alzheimer’s disease. Plos One, 14(2), e0211935. 10.1371/journal.pone.0211935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, R. J., Jha, R. K., Amera, G., Jain, M., Singh, E., Pathak, A., Singh, R. P., Muthukumaran, J., & Singh, A. K. (2020). Targeting SARS-CoV-2: A Systematic Drug Repurposing Approach to Identify Promising Inhibitors Against 3C-like Proteinase and 2’-O-RiboseMethyltransferase. Journal of Biomolecular Structure & Dynamics, 1–40. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, A. M., Shawon, J., & Halim, M. A. (2017). Multiple receptor conformers based molecular docking study of fluorine enhanced ethionamide with mycobacterium enoyl ACP reductase (InhA). Journal of Molecular Graphics & Modelling, 77, 386–398. 10.1016/j.jmgm.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Khan, S. A., Zia, K., Ashraf, S., Uddin, R., & Ul-Haq, Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kollman, P. A., Massova, I., Reyes, C., Kuhn, B., Huo, S., Chong, L., Lee, M., Lee, T., Duan, Y., Wang, W., Donini, O., Cieplak, P., Srinivasan, J., Case, D. A., & Cheatham, T. E. (2000). Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Accounts of Chemical Research, 33(12), 889–897. 10.1021/ar000033j [DOI] [PubMed] [Google Scholar]
- Krieger, E., Darden, T., Nabuurs, S. B., Finkelstein, A., & Vriend, G. (2004). Making optimal use of empirical energy functions: Force-field parameterization in crystal space. Proteins, 57(4), 678–683. 10.1002/prot.20251 [DOI] [PubMed] [Google Scholar]
- Li, H., Wang, Y. M., Xu, J. Y., & Cao, B. (2020). [Potential antiviral therapeutics for 2019 Novel Coronavirus]. Zhonghua Jie he he hu xi za Zhi = Zhonghua Jiehe he Huxi Zazhi = Chinese Journal of Tuberculosis and Respiratory Diseases, 43(0), E002. 10.3760/cma.j.issn.1001-0939.2020.0002 [DOI] [PubMed] [Google Scholar]
- Li, Y., Zhang, J., Wang, N., Li, H., Shi, Y., Guo, G., Liu K., Zeng H., Zou, Q. (2020). Therapeutic Drugs Targeting 2019-nCoV Main Protease by High-Throughput Screening. BioRxiv, 2020.01.28.922922. 10.1101/2020.01.28.922922 [DOI]
- Mathias, A. A., German, P., Murray, B. P., Wei, L., Jain, A., West, S., Warren, D., Hui, J., & Kearney, B. P. (2010). Pharmacokinetics and Pharmacodynamics of GS-9350: A Novel Pharmacokinetic Enhancer Without Anti-HIV Activity . Clin. Pharmacol. Ther, 87(3), 322–329. 10.1038/clpt.2009.228 [DOI] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Neves, B. J., Braga, R. C., Melo-Filho, C. C., Moreira-Filho, J. T., Muratov, E. N., & Andrade, C. H. (2018). QSAR-based virtual screening: Advances and applications in drug discovery. Frontiers in Pharmacology, 9(NOV), 1275. 10.3389/fphar.2018.01275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant, S., Singh, M., Ravichandiran, V., Murty, U. S. N., & Srivastava, H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure & Dynamics, 1–15. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2014). R Core Team (2014). R: A language and environment for statistical computing. R Foundation for Statistical Computing. URL http://www.R-Project.Org/. [Google Scholar]
- Razzaghi-Asl, N., Mirzayi, S., Mahnam, K., & Sepehri, S. (2018). Identification of COX-2 inhibitors via structure-based virtual screening and molecular dynamics simulation . J. Mol. Graph. Model, 83, 138–152. 10.1016/j.jmgm.2018.05.010 [DOI] [PubMed] [Google Scholar]
- Remmert, M., Biegert, A., Hauser, A., & Söding, J. (2011). HHblits: Lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nature Methods, 9(2), 173–175. 10.1038/nmeth.1818 [DOI] [PubMed] [Google Scholar]
- Romano, M., Ruggiero, A., Squeglia, F., Maga, G., & Berisio, R. (2020). A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells, 9(5), 1267. 10.3390/cells9051267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rstudio Team (2019). RStudio: Integrated development for R. RStudio, Inc., Boston MA. RStudio. 10.1007/978-3-642-20966-6 [DOI]
- Sarma, P., Sekhar, N., Prajapat, M., Avti, P., Kaur, H., Kumar, S., … Medhi, B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure & Dynamics, 1–11. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahinozzaman, M., Ishii, T., Ahmed, S., Halim, M. A., & Tawata, S. (2019). A computational approach to explore and identify potential herbal inhibitors for the p21-activated kinase 1 (PAK1). . Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2019.1659855 [DOI] [PubMed] [Google Scholar]
- Sun, W., Sanderson, P. E., & Zheng, W. (2016, July). Drug combination therapy increases successful drug repositioning. Drug Discovery Today, 21(7), 1189–1195. 10.1016/j.drudis.2016.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talwani, R., Heil, E. L., Gilliam, B. L., & Temesgen, Z. (2013). Simeprevir: A macrocyclic hcv protease inhibitor. Drugs of Today (Barcelona, Spain : 1998), 49(12), 769. December). 10.1358/dot.2013.49.12.2067249 [DOI] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F. T., de Beer, T. A. P., Rempfer, C., Bordoli, L., Lepore, R., & Schwede, T. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research, 46(W1), W296–W303. 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag. 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- Wold, S., Esbensen, K., & Geladi, P. (1987). Principal component analysis. Chemometrics and Intelligent Laboratory Systems, 2(1-3), 37–52. 10.1016/0169-7439(87)80084-9 [DOI] [Google Scholar]
- Wong, S. K., Li, W., Moore, M. J., Choe, H., & Farzan, M. (2004). A 193-Amino Acid Fragment of the SARS Coronavirus S Protein Efficiently Binds Angiotensin-converting Enzyme 2. The Journal of Biological Chemistry, 279(5), 3197–3201. 10.1074/jbc.C300520200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, P. C. Y., Huang, Y., Lau, S. K. P., & Yuen, K. Y. (2010). Coronavirus genomics and bioinformatics analysis. Viruses, 2(8), 1804–1820. MDPI AG. 10.3390/v2081803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- XLSTAT | Statistical Software for Excel. (2017). XLSTAT | Statistical Software for Excel. Retrieved April 26, 2020, from https://www.xlstat.com/en/
- Xu, J., Shi, P.-Y., Li, H., & Zhou, J. (2020). Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential. ACS Infectious Diseases, 6(5), 909–915. 10.1021/acsinfecdis.0c00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Zhou, R. (2020). Binding mechanism of remdesivir to SARS-CoV-2 RNA dependent RNA polymerase. Preprints, (2020030267). 10.20944/preprints202003.0267.v1 [DOI] [PubMed]
- Zhou, Y., Hou, Y., Shen, J., Huang, Y., Martin, W., & Cheng, F. (2020). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery, 6(1), 14–18. 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.