Abstract
Recent outbreak of novel coronavirus and its rapid pandemic escalation in all over the world has drawn the attention to urgent need for effective drug development. However, due to prolonged vaccine and drug development procedure against a newly emerged devastating SARS-CoV-2 virus pathogen, repurposing of existing potential pertinent drug molecules would be preferable strategy to reduce mortality immediately and further development of new drugs to combat overall global Covid-19 crisis in all over the world. Herein, we have filtered 23 prospective drug candidates through literature review. Assessing evidences from molecular docking studies, it was clearly seen that, Epirubicin, Vapreotida, and Saquinavir exhibited better binding affinity against SARS-CoV-2 Main Protease than other drug molecules among the 23 potential inhibitors. However, 50 ns molecular dynamics simulation indicated the less mobile nature of the docked complex maintaining structural integrity. Our overall prediction findings indicate that Epirubicin, Vapreotida, and Saquinavir may inhibit COVID-19 by synergistic interactions in the active cavity and those results can pave the way in drug discovery although it has to be further validated by in-vitro and in-vivo investigations.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, SARS-CoV-2, anti-viral drugs, drug discovery, epirubicin, vapreotida, saquinavir
Introduction
Following detection of SARS-CoV-2 in late December 2019 in Wuhan, capital of Central China's Hubei Province, it has quickly spread across the globe (Tang et al., 2020) and this highly spreading and devastating infection, COVID-19 has killed 3,70,120 people worldwide (https://www.worldometers.info/coronavirus/#countries, n.d.). Named after the related structured severe acute respiratory syndrome-related coronaviruses, SARS-CoV-2 has a genome sized∼30kb with the specific pattern, ORF1ab encoding the large replicase polyprotein pp1ab and forming the non-structural proteins (nsp1-16) as well as the structural proteins (S, E, M, and N) (Khailany et al., 2020). Moreover, the SARS-CoV-2 produced 6 accessory proteins, encoded by ORF3a, ORF6, ORF7a, ORF7b, ORF8, and ORF10 genes (Khailany et al., 2020). Two cysteine proteases, namely the main protease (Mpro) or the 3C-like protease (3CLpro) and the secondary papain-like protease 2 (PL2pro) mainly process the replicase complex (RNA-dependent RNA polymerase and the helicase) that ultimately facilitate the viral replication, where Mpro and PL2pro cleaves at 11 and three sites of the polyprotein, respectively. Wu et al. (2020) reported that among the non-structural proteins of SARS-CoV-2, 3CLpro plays a key role in the replication and maturation step.
Based on these facts, the Mpro is a more promising drug target against SARS-CoV-2 than the PL2pro, as well as for potential drug targets; scientists generally choose proteins and enzymes of CoVs vital for the replication process. In addition, previous studies have also shown that the main proteases of different coronaviruses are highly conserved in terms of both sequences and 3D structures (Liu & Wang, 2020). These features, together with its functional importance, have made 3CLpro an attractive target for the design of anti-corona viral drugs (Liu & Wang, 2020) than other corona viral proteins.
Until now, no clinically approved drug is available for coronavirus (SARS-CoV-2) treatment (Battegay et al., 2020). Scientists are trying to solve the puzzle through drug repurposing techniques, which is a promising option considering the global pandemic caused by the virus (Pushpakom et al., 2019). Drug repurposing, an alternative option for approved or investigational drugs outside their defined indication, has a lower risk of failure and drastically reduces the time frame for development, facilitating prompt clinical decisions at lower costs development (Cherian et al., 2020).
In the recent reports, we have come across some of the scientific endeavors, most of which are related to drug repurposing, to find effective drugs, nucleotide analogs (Chien et al., 2020; Kirchdoerfer, 2020), and peptide binder (Zhang et al., 2020). For example, Ritonavir and Lopinavir, which were under phase II/III clinical trials for MERS-CoV (Chan et al., 2015; Kim, Won, et al., 2016), have activity against HCoV-229E, HCoV-NL63 and animal CoVs (Kim, Won, et al., 2016), and can be an excellent repurposed drug for SARS-CoV-2 because the drug binding sites and 96% protein sequences were similar in SARS-CoV and SARS-CoV-2 main protease (Liu & Wang, 2020). Also, researchers have currently reported using the drug repurposing approach based on the molecular docking and dynamics study where the key target proteins are 3CL protease, RNA dependent RNA polymerase (RdRp), and spike proteins (Elfiky, 2020b; Muralidharan et al., 2020; Smith & Smith, 2020; Tahir Ul Qamar et al., 2020; Yu et al., 2020).
In this study, we explore potential drug candidate(s) against SARS-CoV-2 by performing the virtual screening based on drug repurposing concept, using available commercial medicine (23 potential drugs against other viruses) that can eventually, be predicted to act against the main protease (Mpro), or 3-C like protease (3CLpro). Due to the lack of confidence in giving the accurate binding energy, binding mode, solvation effect, constant motion effect, and entropy energy during molecular docking (Sethi et al., 2019), a molecular dynamics study was preferred.
Materials and method
Molecular docking preparation
In this study, the crystal structure of the main protease of SARS-CoV-2 was extracted (PDB ID: 6LU7) which has similar binding sites of SARS-CoV (Rose et al., 2016). The protein structure was cleaned and prepared by Discovery Studio (San Diego: Accelrys Software Inc, 2012) and Pymol (DeLano, 2002) software package. The cleaned protein structure was prepared and minimized with GROMOS 43B1 force field with the aid of SWISS PDB Viewer (Kaplan & Littlejohn, 2001). The ligand molecules were retrieved from PubChem database (Kim, Thiessen, et al., 2016) after a deep literature review process. The ligand structure was optimized and cleaned with the help of mmff94 force field (Halgren, 1996) with the steepest gradient algorithm.
Molecular docking study
Molecular docking was performed to understand the binding affinity with the ligand and drug compounds. The ligand were converted in PDBQT format to make them acceptable format for employing docking in AutoDock Vina (Trott & Olson, 2010). The center of the grid box was X:-26.289, Y: 12.606, Z: 58.9496 and dimensions were (X: 50.8171, Y: 67.5389, Z: 59.6721) Ǻ. Finally the docked pose were analyzed with the aid of Discovery studio and Pymol.
Molecular dynamics simulation
The molecular dynamics simulation was run in YASARA application (Krieger et al., 2004). The AMBER14 force field (Case et al., 2007) was employed for the minimization of the drug-protein complex and transferable intermolecular interaction potential 3 points (TIP3P) were used for the addition of Na and Cl ions. The box size of the cell was set as (96.9654 × 96.9654 × 96.9654) Ǻ with a periodic boundary condition. A cut off the radius of 8 angstroms was used for the calculation of short-range Van der Waals and Coulomb interaction. The Particle Mesh Ewalds was used to calculate long-range electrostatic interaction with a physiological condition at 300 K, pH 7.4, 0.9% NaCl (Krieger et al., 2006; Krieger & Vriend, 2015). The simulation was run for 50 ns and root mean square deviation (RMSD), root mean square fluctuation (RMSF), the radius of gyration (Rg), solvent-accessible surface area (SASA). Moreover, all the trajectory further used to calculate MM-Posson-Boltzman surface area or MM-PBSA binding energy for binding free energy calculation where positive energy denotes favorable binding (Dash et al., 2019; Krieger & Vriend, 2015).
Result
Among 23 drug molecules only three showed better binding energy while interacting with Main Protease (Table 1).
Table 1.
List of potential drug compounds and their molecular docking score energy.
| Compound ID | Compound Name | Docking Energy (kcal·mol−1) |
|---|---|---|
| CID-4748 | Perphenazine | −6.1 |
| CID-37542 | Ribavirin | −6.3 |
| CID-41867 | Epirubicin | −9.5 |
| CID-64143 | Nelfinavir | −7.1 |
| CID-65016 | Amprenavir | −7.1 |
| CID-71306 | Vapreotida | −9.1 |
| CID-92727 | Lopinavir | −9.4 |
| CID-131536 | Fosamprenavir | −6.6 |
| CID-148192 | Atazanavir | −7 |
| CID-164522 | Bepotastine | −7.1 |
| CID-213029 | Darunavir | −7.1 |
| CID-392622 | Ritonavir | −7.1 |
| CID-441243 | Saquinavir | −9.5 |
| CID-454216 | Valrubicin | −7.8 |
| CID-492405 | Favipiravir | −5.1 |
| CID-2826718 | Caspofungin | −7.1 |
| CID-5311054 | Collistin | −6.6 |
| CID-5362440 | Indinavir | −7.8 |
| CID-6918173 | Icatibant | −7.1 |
| CID-10445549 | Galidesivir | −5.9 |
| CID-54682461 | Tipranavir | −7.6 |
| CID-121304016 | Remdesivir | −7.2 |
| CID-135413536 | Aprepitant | −7.3 |
Epirubicin, formed five hydrogen bond at Phe140, Glu166, Gln189, Asn142, Pro168 and one hydrophobic bond at Pro168 position. On the other hand, Vapreotida and main protease complex stabilized by several hydrogen bond at Gln110, Asp245, Arg105, Asp245, Ile249 and one pi-pi-stacked bond Phe294, one pi-pi-T shaped at His246, one alkyl Pro252 and two pi-alkyl bond at Val202, Ile249. Besides, main protease and Saquinavir complex stabilized by six hydrogen bond at Ile249, Phe294, Gln110, Thr292, Pro293, Phe294 and six hydrophobic bond at Asn245, Phe294, Ile249, Val202, Pro293, Ile249 (Table 2 and Figure 1).
Table 2.
Docking simulation results with docking score energy and interaction with amino acids.
| Compound Name | Docking energy (kcal·mol−1) | Hydrogen Bond | Hydrophobic Bond | MM-PBSA (kj/mol) |
|---|---|---|---|---|
| Ligand atom-Amino Acid Distance(Å) | Interaction-Amino Acid Distance(Å) | |||
| Epirubicin | −9.5 | H-Phe140:O(1.88), H-Glu166:OE2(2.68), HE21-Gln189:O(2.12), HA-Asn142:O(2.65), HA-Pro168:O(2.68), |
Pi-Sigma-Pro168(4.70) | 93.405 ± 1.1 |
| Vapreotida | −9.1 | H-Gln110:OE(2.08), H-Asp245:OD1(2.98), H-Arg105:O(2.54), H54-Asp245:OD1(2.96), HA-ILE249:O(2.52), |
Pi-Pi-Stacked
Phe294(3.79), Pi-Pi-Tshaped-His246(4.79), Alkyl Pro252(4.47), Pi-Alkyl Val202(5.02), Pi-Alkyl Ile249(4.58) |
53.042 ± 0.9 |
| Saquinavir | −9.5 | H-Ile249:O(2.44), HN-Phe294:O(2.45), H21-Gln110:OE(2.38), HB-Thr292:O(2.49), HD1-Pro293:O(2.52), HA-Phe294:O(3.01), |
Pi-anion Asn245(4.56), Pi-Pi-Stacked Phe294 (3.87), Alkyl Ile249(4.45), Alkyl Val202(4.87), Alkyl Pro293(5.28), Pi-Alkyl Ile249(5.01) |
73.807 ± 1.0 |
Figure 1.
Molecular docking interactions between the SARS-CoV-1 main protease and drug compounds. A denoted Epirubicin, B denoted Vapreotida, C denoted Saquinavir.
Molecular dynamics simulation study
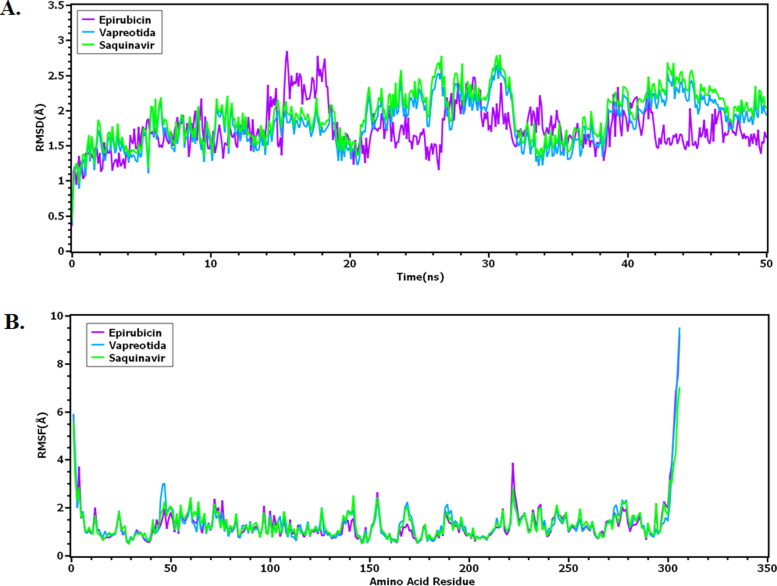
The RMSD value of docked complex analyzed from simulated trajectory to find out whether the complex remain stable or complex rigidity. From the Figure 2, it can be observed that, all three chemical drugs had lower pic than 2.5 angstrom during whole simulation time
Figure 2.
Root Mean Square Deviation (RMSD) of the C-alpha atom, (B) Root Mean Square Fluctuation (RMSF) of the drug-protein complex.
However, Epirubicin showed less flexibility till 10 ns and thereafter, fluctuated for the next 5 ns. The Epirubicin and main protease protein from SARS-CoV-2 were stabilized and then reached in a stabilized state. On the other hand, Vapreotida and Saquinavir had similar profile till 10 ns and after then, it exhibited lower RMSD value. It can be denoted from Figure 2, that all the complex stabilized despite have some minor fluctuation but they were way below 2.5 angstrom. On the other hand, root means square fluctuation of the drug-protein complex can help to understand the flexibility in the protein region. From Figure 2 it can be understood that all three drug molecules exhibit a similar level of flexibility over the protein region. Most of the residue did not have a higher peak in RMSF therefore responsible for the complex overall stability. However, Figure 2 indicates that the amino acid; Ser1 (helix), Gly2 (helix), Arg4 (helix), Glu47 (beta turn), Leu50 (beta turn), Glu55 (helix), Arg60 (helix), His64 (helix), Asn72 (beta hairpin), Arg76 (helix), Lys97 (beta turn), Tyr154 (beta hairpin), Pro168 (beta hairpin), Thr169 (beta hairpin), Arg222 (beta turn), Phe223 (beta turn), Thr224 (beta turn), Met235 (helix), Lys236 (helix), Gln244 (helix), Asn277 (beta turn), Gly278 (beta turn), Arg279 (beta turn), Phe294 (helix), Arg298 (helix), Gln299 (helix), Ser301(beta turn), Gly302 (beta turn), Val303 (beta turn), Thr304 (beta turn), Phe305 (beta turn), Gln306 (beta turn) from all the three complex were most flexible than other residue in the system.
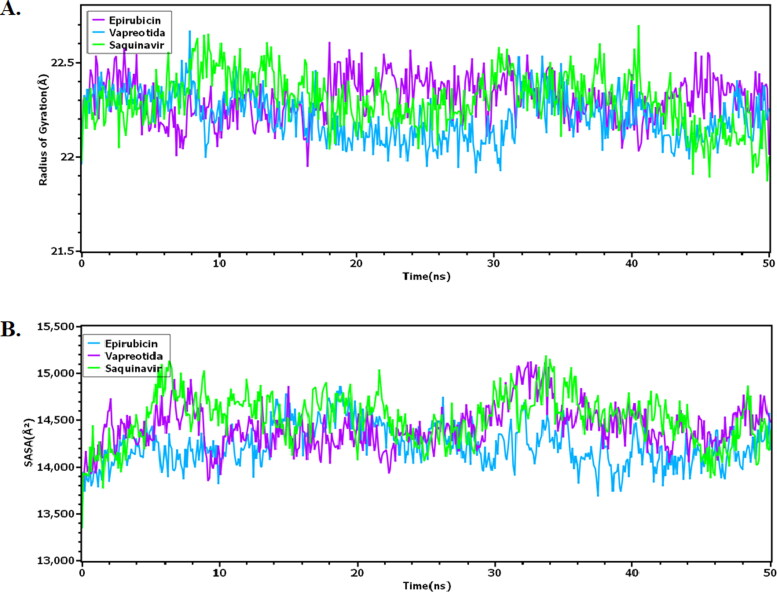
The complex compactness and rigidity can be assessed through a radius of gyration. Figure 3 denoted that, Vapreotida and main protease has lower Rg value than Saquinavir and Epirubicin which indicates the degree of compactness of the Vapreotida.
Figure 3.
(A) Radius of Gyration of the complexes was explored to find out the compact nature of the system, (B) Solvent Accessible Surface Area (SASA) was determined to understand the changes in surface area.
Moreover, Epirubicin and protein complex is less compact than the other two docked complex as they had higher Rg value among three complex.
On the other hand, Higher SASA value indicates the high degree of expansion in the protein surface area. The Saquinavir had a slightly higher peak than the other two drug molecules till 10 ns. Subsequently, Saquinavir, Vapreotida, Epirubicin had a similar trend to 30 ns which indicates protein surface area remain same during this simulation time. However, Epirubicin had a lower SASA peak than two complexes after 30 ns which may be the result of complex area expansion. Both Saquinavir and Vapreotida had similar SASA profiles for the rest of the simulation time.
Furthermore, we have calculated MM-PBSA binding energy by taking each trajectory from the simulated protein complex to understand the relation between ligand binding and structural changes (Figure 4). The main protease complex and Epirubicin exhibited lower binding energy than other two complexes. The rest two complex had higher binding energy which indicates better binding with main protease of SARS-CoV-2.
Figure 4.
Molecular mechanics Poisson-Boltzmann surface area calculation of the screened drug complex system.
Discussion
Molecular modelling especially docking and dynamic simulation study has become essential part in modern drug discovery pipeline(Bappy et al., 2020; Elfiky, 2020a; Islam et al., 2019). Moreover, time and lab cost can significantly reduce by narrowing the drug molecule choices in in silico based drug discovery process (Talele et al., 2010). Besides, this combinatorial computational approaches allow the researcher to get the binding insights of the drug molecules to the target protein and rationally design the therapeutic targets (Mullard, 2019).
Among the selected drugs from this study, Epirubicin can have a negative impact on chronic HBV patient through reactivation of the viral genome (Xu et al., 2014). The Epirubicin drug molecules showed −9.5 kCal/mol energy while binding with the main protease of SARS-CoV-2 which is the highest binding energy among the drug molecules. This might be the result of several hydrogen bonding that was found such as Phe140, Glu166, Gln189, Asn142, and Pro168 at the active cavity of the main protease enzyme. Researcher suggests that non-covalent binding at the active site of the target protein may lead to possible inhibition and blockage of the protein (Hasan et al., 2015). On the other hand, the somatostatin analog, Vapreotide (VAP) was used to control tumor growth, progression, and metastasis. Saquinavir is a protease inhibitor of HIV has shown in-vitro effect against SARS and MERS coronaviruses (Feng et al., 2014). Multiple hydrogens and hydrophobic bonds for the Vapreotida and Saquinavir may be responsible for the tight binding and better affinity among the 23 drug molecules (Hossain, Khan, et al., 2016; Hossain, Oany, et al., 2016).
Molecular dynamics study of screened drug molecules were checked to evaluate their stability at atomic scale. The average root mean square deviation (RMSD) of Epirubicin, Vapreotida, Saquinavir are 1.756, 2.008, 1.76 Å which indicates complex stability throughout the entire simulation (Islam et al., 2019). Additionally, the average radius of gyration for these three complexes were found as 22.307, 22.211, 22.301 Å and did not deviate much. Besides, the mean of the surface area of the three complexes was 14005.961, 14419.772, 14514.697 Å2. It was cleared from Figure 3 that simulation analysis of top three therapeutic drug molecules consistent with overall stable nature and complex rigidity (Mahmud et al., 2019).
In this ground, it is clearly seen that Epirubicin, Vapreotida, and Saquinavir may inhibit COVID-19 by synergistic interactions among the 23 potential inhibitors against SARS-CoV-19 main protease and those results pave the way in drug discovery although it has to be further validated by in vitro and in vivo investigations.
Conclusion
Scientists have chosen drug repurposing techniques as alternative options for approved or investigational drugs outside their defined indication for ease of utilization in an emergency with minor associated risk and clue for prompt target drug development and application with minimum cost involvement. Bioinformatics approaches, such as molecular docking and dynamics study with probable drug candidates binding to key target proteins of pathogens for neutralization and inhibition are widely acceptable effective techniques to validate the drug repurposing approach. We have evaluated potential drug candidates against the main protease of the SARS-CoV-2 to understand their binding sites and structural stability. The nature of the binding mode of drug compound found as stable and molecular dynamics simulation study supports their conformational rigidity. As these outcomes solely came from combining computational algorithms, it requires further validation of these findings conducting further in-vitro wet-lab experimentation followed by in-vivo rapid trials for further use of the drugs individually or in combination to reduce mortality during Covid-19 pandemics globally. Moreover, based on these data, further, the experimental study can aid in getting suitable new drug molecules for combating COVID-19.
Disclosure statement
The authors declares that they have no conflict of interest.
Author contributions
Md. Arif Khan: Conceptualization, Performed Bioinformatics experiment, Critical Data analysis, Manuscript writing, and Reviewing
Shafi Mahmud: Docking Study, Molecular dynamics analysis, Manuscript writing.
A. S. M. Rubayet Ul Alam: Helped in drafting the manuscript and Reviewing.
Md. Ekhtiar Rahman: Helped in Molecular dynamics study.
Firoz Ahmed: Critical Data analysis, Reviewing and performed critical revision,
Mohammed Rahmatullah: Conceptualization, Performed critical revision, and overall supervision of this study.
All authors provided critical feedback and helped shape the research, analysis, and manuscript.
References
- Accelrys Software Inc . (2012). Discovery studio modeling environment. Release 3.5. Accelrys Software Inc. [Google Scholar]
- Bappy, S. S., Sultana, S., Adhikari, J., Mahmud, S., Khan, M. A., Kibria, K. M. K., Rahman, M. M., & Shibly, A. Z. (2020). Extensive immunoinformatics study for the prediction of novel peptide-based epitope vaccine with docking confirmation against Envelope protein of Chikungunya virus: A computational biology approach. Journal of Biomolecular Structure and Dynamics, 1–16. https://doi.org/10.1080/07391102.2020.1726815 [DOI] [PubMed] [Google Scholar]
- Battegay, M., Kuehl, R., Tschudin-Sutter, S., Hirsch, H. H., Widmer, A. F., & Neher, R. A. (2020). 2019-novel Coronavirus (2019-nCoV): estimating the case fatality rate–a word of caution. Swiss Medical Weekly, 150(0506), 1–3. [DOI] [PubMed] [Google Scholar]
- Case, D. A., Cheatham III, T. E., Darden, T. O. M., Gohlke, H., Luo, R., Merz Jr, K. M., Onufriev, A., Simmerling, C., Wang, B., & Woods, R. J. (2007). The Amber biomolecular simulation programs. Journal of Combinatorial Chemistry, 26(16), 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F.-W., Yao, Y., Yeung, M.-L., Deng, W., Bao, L., Jia, L., Li, F., Xiao, C., Gao, H., Yu, P., Cai, J.-P., Chu, H., Zhou, J., Chen, H., Qin, C., & Yuen, K.-Y. (2015). Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERSCoV infection in a nonhuman primate model of common marmoset. Journal of Infectious Diseases, 212(12), 1904–1913. 10.1093/infdis/jiv392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian, S. S., Agrawal, M., Basu, A., Abraham, P., Gangakhedkar, R. R., & Bhargava, B. (2020). Perspectives for repurposing drugs for the coronavirus disease 2019. The Indian Journal of Medical Research, 151(2 & 3), 160–171. 10.4103/ijmr.IJMR_585_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, M., Anderson, T. K., Jockusch, S., Tao, C., Kumar, S., Li, X., Russo, J. J., Kirchdoerfer, R. N., & Ju, J. (2020). Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase. BioRxiv, 1–7. 10.1101/2020.03.18.997585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash, R., Junaid, M., Mitra, S., Arifuzzaman, M., & Hosen, S. M. Z. (2019). Structure-based identification of potent VEGFR-2 inhibitors from in vivo metabolites of a herbal ingredient. Journal of Molecular Modeling, 25(4), 98. 10.1007/s00894-019-3979-6 [DOI] [PubMed] [Google Scholar]
- DeLano, W. L. (2002). The PyMOL molecular graphics system, Version 1.1. Schrödinger LLC. 10.1038/hr.2014.17 [DOI] [Google Scholar]
- Elfiky, A. A. (2020. a). Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure & Dynamics, 1–16. 10.1080/07391102.2020.1761881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020. b). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Q., Yu, M.-Z., Wang, J.-C., Hou, W.-J., Gao, L.-Y., Ma, X.-F., Pei, X.-W., Niu, Y.-J., Liu, X.-Y., Qiu, C., Pang, W.-H., Du, L.-L., & Zhang, Q. (2014). Synergistic inhibition of breast cancer by co-delivery of VEGF siRNA and paclitaxel via vapreotide-modified core-shell nanoparticles. Biomaterials, 35(18), 5028–5038. 10.1016/j.biomaterials.2014.03.012 [DOI] [PubMed] [Google Scholar]
- Halgren, T. A. (1996). Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. Journal of Computational Chemistry, 17(5–6), 490–519. [DOI] [Google Scholar]
- Hasan, M. A., Mazumder, M. H. H., Chowdhury, A. S., Datta, A., & Khan, M. A. (2015). Molecular-docking study of malaria drug target enzyme transketolase in Plasmodium falciparum 3D7 portends the novel approach to its treatment. Source Code for Biology and Medicine, 10(1), 7. 10.1186/s13029-015-0037-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, M. U., Khan, M. A., Rakib-Uz-Zaman, S. M., Ali, M. T., Islam, M. S., Keya, C. A., & Salimullah, M. (2016). Treating diabetes mellitus: Pharmacophore based designing of potential drugs from gymnema sylvestre against insulin receptor protein. BioMed Research International, 2016, 3187647. 10.1155/2016/3187647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, M. U., Oany, A. R., Ahmad, S. A. I., Hasan, M. A., Khan, M. A., & Siddikey, M. A. A. (2016). Identification of potential inhibitor and enzyme-inhibitor complex on trypanothione reductase to control Chagas disease. Computational Biology and Chemistry, 65, 29–36. 10.1016/j.compbiolchem.2016.10.002 [DOI] [PubMed] [Google Scholar]
- https://www.worldometers.info/coronavirus/#countries. (n.d.).
- Islam, M. J., Parves, M. R., Mahmud, S., Tithi, F. A., & Reza, M. A. (2019). Assessment of structurally and functionally high-risk nsSNPs impacts on human bone morphogenetic protein receptor type IA (BMPR1A) by computational approach. Computational Biology and Chemistry, 80, 31–45. 10.1016/j.compbiolchem.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Kaplan, W., & Littlejohn, T. G. (2001). Swiss-PDB viewer (deep view). Briefings in Bioinformatics, 2(2), 195–197. 10.1093/bib/2.2.195 [DOI] [PubMed] [Google Scholar]
- Khailany, R. A., Safdar, M., & Ozaslan, M. (2020). Genomic characterization of a novel SARS-CoV-2. Gene Reports, 19, 100682. 10.1016/j.genrep.2020.100682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Thiessen, P. A., Bolton, E. E., Chen, J., Fu, G., Gindulyte, A., Han, L., He, J., He, S., Shoemaker, B. A., Wang, J., Yu, B., Zhang, J., & Bryant, S. H. (2016). PubChem substance and compound databases. Nucleic Acids Research, 44(D1), D1202–D1213. 10.1093/nar/gkv951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, U. J., Won, E. J., Kee, S. J., Jung, S. I., & Jang, H. C. (2016). Combination therapy with lopinavir/ritonavir, ribavirin and interferon-α for Middle East respiratory syndrome. Antiviral Therapy, 21(5), 455–459. 10.3851/IMP3002 [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer, R. N. (2020). Halting coronavirus polymerase. The Journal of Biological Chemistry, 295(15), 4780–4781. 10.1074/jbc.H120.013397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, E., Nielsen, J. E., Spronk, C. A. E. M., & Vriend, G. (2006). Fast empirical pKa prediction by Ewald summation. Journal of Computational Chemistry, 25(4), 481–486. [DOI] [PubMed] [Google Scholar]
- Krieger, E., Darden, T., Nabuurs, S. B., Finkelstein, A., & Vriend, G. (2004). Making optimal use of empirical energy functions: Force-field parameterization in crystal space. Proteins, 57(4), 678–683. 10.1002/prot.20251 [DOI] [PubMed] [Google Scholar]
- Krieger, E., & Vriend, G. (2015). New ways to boost molecular dynamics simulations. Journal of Computational Chemistry, 36(13), 996–1007. 10.1002/jcc.23899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., & Wang, X.-J. (2020). Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. Journal of Genetics and Genomics = Yi Chuan Xue Bao, 47(2), 119–121. 10.1016/j.jgg.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud, S., Parves, M. R., Riza, Y. M., Sujon, K. M., Ray, S., Tithi, F. A., Zaoti, Z. F., Alam, S., & Absar, N. (2019). Exploring the potent inhibitors and binding modes of phospholipase A2 through in silico investigation. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2019.1680440 [DOI] [PubMed] [Google Scholar]
- Mullard, A. (2019). Supersized virtual screening offers potent leads. Nature Reviews Drug Discovery, 18, 243. 10.1038/d41573-019-00037-4 [DOI] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Pushpakom, S., Iorio, F., Eyers, P. A., Escott, K. J., Hopper, S., Wells, A., Doig, A., Guilliams, T., Latimer, J., McNamee, C., Norris, A., Sanseau, P., Cavalla, D., & Pirmohamed, M. (2019). Drug repurposing: Progress, challenges and recommendations. Nature Reviews. Drug Discovery, 18(1), 41–58. 10.1038/nrd.2018.168 [DOI] [PubMed] [Google Scholar]
- Rose, P. W., Prlić, A., Altunkaya, A., Bi, C., Bradley, A. R., Christie, C. H., Costanzo, L. D., Duarte, J. M., Dutta, S., & Feng, Z. (2016). The RCSB protein data bank: Integrative view of protein, gene and 3D structural information. Nucleic Acids Research, 45(D1), D271–D281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi, A., Joshi, K., Sasikala, K., & Alvala, M. (2019). Molecular docking in modern drug discovery: Principles and recent applications. In Drug Discovery and Development-New Advances. IntechOpen. https://doi.org/10.5772/intechopen.85991 [Google Scholar]
- Smith, M., & Smith, J. C. (2020). Repurposing therapeutics for COVID-19: Supercomputer-based docking to the SARS-CoV-2 viral spike protein and viral spike protein-human ACE2 interface. ChemRxiv, 1–28. 10.26434/chemrxiv.11871402.v3 [DOI] [Google Scholar]
- Tahir Ul Qamar, M., Alqahtani, S. M., Alamri, M. A., & Chen, L. L. (2020). Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. Journal of Pharmaceutical Analysis, 1–7. 10.1016/j.jpha.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talele, T., Khedkar, S., & Rigby, A. (2010). Successful applications of computer aided drug discovery: Moving drugs from concept to the clinic. Current Topics in Medicinal Chemistry, 10(1), 127–141. 10.2174/156802610790232251 [DOI] [PubMed] [Google Scholar]
- Tang, X., Wu, C., Li, X., Song, Y., Yao, X., Wu, X., Duan, Y., Zhang, H., Wang, Y., Qian, Z., Cui, J., & Lu, J. (2020). On the origin and continuing evolution of SARS-CoV-2. National Science Review, 7(6), 1012–1023. 10.1093/nsr/nwaa036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766–788. https://doi.org/ 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Tu, Z., Xu, G., Wang, Y., Pan, W., Zhan, X., Luo, Q., Huang, Y., Chen, J., & Huang, A. (2014). Epirubicin directly promotes hepatitis B virus (HBV) replication in stable HBV-expressing cell lines: A novel mechanism of HBV reactivation following anticancer chemotherapy. Molecular Medicine Reports, 9(4), 1345–1350. 10.3892/mmr.2014.1973 [DOI] [PubMed] [Google Scholar]
- Yu, R., Chen, L., Lan, R., Shen, R., & Li, P. (2020). Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. International Journal of Antimicrobial Agents, 106012. 10.1016/j.ijantimicag.2020.106012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G., Pomplun, S., Loftis, A. R., Loas, A., & Pentelute, B. L. (2020). The first-in-class peptide binder to the SARS-CoV-2 spike protein. BioRxiv, 1–15. 10.1101/2020.03.19.999318 [DOI] [Google Scholar]