Abstract
Spike glycoprotein, a class I fusion protein harboring the surface of SARS-CoV-2 (SARS-CoV-2S), plays a seminal role in the viral infection starting from recognition of the host cell surface receptor, attachment to the fusion of the viral envelope with the host cells. Spike glycoprotein engages host Angiotensin-converting enzyme 2 (ACE2) receptors for entry into host cells, where the receptor recognition and attachment of spike glycoprotein to the ACE2 receptors is a prerequisite step and key determinant of the host cell and tissue tropism. Binding of spike glycoprotein to the ACE2 receptor triggers a cascade of structural transitions, including transition from a metastable pre-fusion to a post-fusion form, thereby allowing membrane fusion and internalization of the virus. From ancient times people have relied on naturally occurring substances like phytochemicals to fight against diseases and infection. Among these phytochemicals, flavonoids and non-flavonoids have been the active sources of different anti-microbial agents. We performed molecular docking studies using 10 potential naturally occurring compounds (flavonoids/non-flavonoids) against the SARS-CoV-2 spike protein and compared their affinity with an FDA approved repurposed drug hydroxychloroquine (HCQ). Further, our molecular dynamics (MD) simulation and energy landscape studies with fisetin, quercetin, and kamferol revealed that these molecules bind with the hACE2-S complex with low binding free energy. The study provided an indication that these molecules might have the potential to perturb the binding of hACE2-S complex. In addition, ADME analysis also suggested that these molecules consist of drug-likeness property, which may be further explored as anti-SARS-CoV-2 agents.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, molecular docking, phytochemicals, flavonoids and non-flavonoids
1. Introduction
The world population is facing a severe mass annihilation due to the rise of a global pandemic named Coronavirus Disease 2019 (COVID-19) (Boopathi et al., 2020; Chatterjee et al., 2020; Joshi et al., 2020; Kirchdoerfer et al., 2016). This pandemic is caused by a novel single-stranded RNA virus belonging to the β-coronavirus genera of the coronaviridae family (Elfiky & Azzam, 2020; Enmozhi et al., 2020; Khan et al., 2020; Rajarshi, Chatterjee & Ray 2020; Sarma et al., 2020; Sinha et al., 2020). As this virus shares significant phylogenetic similarity and structural familiarity (about 80% nucleotide identity and 89.10% nucleotide similarity) with the severe acute respiratory syndrome coronavirus (SARS-CoV), it has been named as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and placed in the same lineage (Subgenus Sarbecovirus) (2020; Boopathi et al., 2020; Das et al., 2020; Khan et al., 2020; Ou et al., 2020; Rajarshi, Chatterjee & Ray 2020). To date, no effective regime of antivirals or vaccines is available for the use of the general public to combat the effect of COVID infections, which has put the population at a more vulnerable position (Aanouz et al., 2020; Elfiky, 2020; Elmezayen et al., 2020; Muralidharan et al., 2020; Pant et al., 2020). The transmission of the virus is taking place at a massive rate worldwide, and hence the scientists all over the world are desperately looking for effective compounds to use it as anti-CoV therapeutic agents. Natural compounds with high bioavailability and low cytotoxicity seem to be the most efficient candidates in this regard. Since ancient times, humans have resorted to the use of natural compounds, especially phytochemicals, for the treatment of different diseases and disorders (Forni et al., 2019; Islam et al., 2020). Flavonoids are secondary metabolites produced by plants, play vital roles in plant physiology, possessing a variety of potential biological benefits such as antioxidant, anti-inflammatory, anticancer, antibacterial, antifungal and antiviral activities (Islam et al., 2020; Kumar & Pandey, 2013; Panche et al., 2016; Wang et al., 2018). Different flavonoids, including flavones and flavonols, have been thoroughly investigated for their potential antiviral properties. Many of them showed a significant antiviral response in both in vitro and in vivo studies (Gupta et al., 2020; Nijveldt et al., 2001). Curcumin, a component of turmeric, has been described to exhibit enhanced antiviral activity against diverse viruses such as dengue virus (serotype 2), herpes simplex virus, human immunodeficiency virus, Zika, and chikungunya viruses among others (Mounce et al., 2017; Praditya et al., 2019). Apigenin isolated from the sweet basil (Ocimum basilicum) plant has shown potent antiviral activity against hepatitis B virus, adenoviruses, african swine fever virus, and some RNA viruses in vitro (Chiang et al., 2005; Wu et al., 2017). Luteolin, another acclaimed flavone, has shown significant antiviral effects on both HIV-1 reactivation and inhibition of Epstein-Barr virus (EBV) reactivation in cells (Wu et al., 2016). Besides these antiviral activities, luteolin also showed antiviral effects against Chikungunya virus, Japanese encephalitis virus (Fan et al., 2016), severe acute respiratory syndrome coronavirus (SARS-CoV) and rhesus rotavirus. Among flavonols, the antiviral effect of quercetin has been most extensively investigated. Quercetin has been found to demonstrate an in vitro dose-dependent antiviral activity against respiratory syncytial virus, poliovirus type 1, HSV-1, and HSV-2. It was also reported that quercetin has the potential ability as a prophylactic against Ebola virus infection (Hasan et al., 2020; Kaul et al., 1985). Kaempferol, another flavonol extracted from Ficus benjamina leaves, has shown inhibitory activity against HCMV, HSV-1, HSV-2, and influenza A virus (Lyu et al., 2005; Zakaryan et al., 2017). Fisetin, a modified flavonol, has shown to inhibit CHIKV infection as well as the HIV-1 infection by blocking viral entry and virus-cell fusion (Zakaryan et al., 2017). Resveratrol and pterostibenes have been found to exhibit antiviral activities against a wide range of viruses, including HIV-1 (Abba et al., 2015; Chan et al., 2017). Resveratrol has been shown to inhibit the replication of pseudorabies virus (PVR) and Middle East Respiratory Syndrome coronavirus (MERS-CoV) and also reduced the expression of MERS-CoV nucleocapsid (N) protein (Campagna & Rivas, 2010; Docherty et al., 2005; Lin et al., 2017; Zhao et al., 2017). Pterostilbene, a natural dimethylated analog of resveratrol found in berries and grapes, has been found to inhibit the replication of several viruses, including herpes simplex viruses (HSVs) 1 and 2, varicella-zoster virus, influenza virus, and human papillomaviruses. Considering the contagiousness of the COVID-19 and its consequences, there is an imperative need to develop an effective therapy to curtail the spread of this deadly virus and safely treat the infected individuals. In that direction, the repurposing of the FDA approved existing drugs like chloroquine (CQ) and hydroxychloroquine (HCQ) either alone or in combination with other known drugs are currently being attempted (Al-Bari, 2017; Yao et al., 2020). Preliminary in vitro studies and clinical trials carried out by scientists on COVID-19 patients disclosed the effectiveness of HCQ, an anti-malarial drug in combination with azithromycin, a broad spectrum anti-bacterial drug in reducing the viral load (Khan et al., 2020). Although some of these initial studies disclosed promising results, a lot still remains to be done to analyze their compatibility, cost, accessibility, side effects, dosages, etc. Currently, scientists are indulging themselves in identifying ideal natural compounds that can target and modulate unique or novel sites like the spike glycoprotein (S) on the surface of SARS-CoV-2 (Adedeji et al., 2013; Elfiky & Azzam, 2020; Sarma et al., 2020; Walls et al., 2020). In this in silico study, we have performed molecular docking experiments to ascertain the most potent natural compounds (flavonoids) that can bind to the functional domains of the SARS-CoV-2 spike protein (SARS-CoV-2S), a viral surface glycoprotein required for initial attachment and internalisation within host cells (Abdelli et al., 2020; Adedeji et al., 2013; Khan et al., 2020; Kirchdoerfer et al., 2016). Here we found that about 10 of these compounds effectively bind to the C-terminal region of either the S1 domain or the S2 domain of SARS-CoV-2S, and their binding interaction is more stable than with that of HCQ. These natural compounds are capable of binding to either the S1 or S2 domains of the SARS-CoV-2S protein and most probably prevent it from binding to the hACE2 receptor or internalization during fusion (Hasan et al., 2020; Khan et al., 2020; Zhou & Simmons, 2012). Further, our MD simulation studies reveal that phytochemicals fisetin, quercetin, and kamferol has the propensity to bind at the junction of the hACE2-S protein complex with lesser binding energy. In addition, in-depth in vitro and in vivo with these lead molecules may help to explore as novel anti-COVID-19 agents.
2. Materials and methods
2.1. Preparation of ligands and receptor
The 3-dimensional structure of all ligands Kaempferol (CID: 5280863), Curcumin (CID: 969516), Pterostilbine (PMID: 26472352), Hydroxychloroquine (CID:3652), Fisetin (CID: 5281614), Quercetin (CID:5280343), Isorhamnetin (CID: 5281654), Genistein (CID: 5280961), Luteolin (CID: 5280445), Resveratrol (CID: 445154), and Apigenin (CID: 5280443)] was downloaded from the PubChem database, and then these structures were converted in pdb format by using PyMol (DeLano, 2002). The structure of SARS-CoV-2S protein was downloaded from the RCSB protein data bank (PDB-ID: 6VYB) (DeLano, 2002; Walls et al., 2020). The structure of all the ligands has been provided in Table S1 (Supplementary material).
Table 1.
The Grid box dimension used for different ligands.
| S. No. | Ligand | Docking
Parameters |
|||
|---|---|---|---|---|---|
| Blind Docking | Grid center | Direct Docking | Grid center | ||
| 1 | Kamferol | 96Å × 96 Å × 160 Å | 197.591 Å × 222.868 Å × 194.892 | 58 Å × 56 Å × 72 Å | 183.962 Å × 197.012 Å × 243.341 Å |
| 2 | Curcumin | 76 Å × 92 Å × 160 Å | 185.514 Å × 186.919 Å × 238.126 | 60 Å × 54 Å × 66 Å | 185.514 Å × 186.919 Å × 238.126 |
| 3 | Pterostilbene | 96Å × 96 Å × 160 Å | 197.591 Å × 222.868 Å × 194.892 | 58 Å × 56 Å × 72 Å | 197.591 Å × 222.868 Å × 194.892 |
| 4 | Hydroxychloroquine | 76 Å × 92 Å × 160 Å | 182.007 Å × 189.89 Å × 245.569 | 58 Å × 56 Å × 72 Å | 182.007 Å × 189.89 Å × 245.569 |
| 5 | Fisetin | 84 Å × 126 Å × 160 Å | 197.079 Å × 222.868 Å × 193.866 | 40 Å × 44 Å × 118 Å | 217.762 Å × 223.316 Å × 177.599 Å |
| 6 | Quercetin | 96 Å × 96 Å × 160 Å | 197.591 Å × 222.868 Å × 194.892 Å | 40 Å × 44 Å × 118 Å | 217.762 Å × 223.316 Å × 183.088 Å |
| 7 | Isorhamnetin | 96 Å × 96 Å × 160 Å | 197.079 Å × 222.868 Å × 205.563 Å | 40 Å × 44 Å × 118 Å | 197.079 Å × 222.868 Å × 205.563 Å |
| 8 | Genistein | 146 Å × 126 Å × 160 Å | 197.079 Å × 222.868 Å × 193.632 Å | 40 Å × 44 Å × 118 Å | 197.079 Å × 222.868 Å × 193.632 Å |
| 9 | Luteolin | 96Å × 96 Å × 160 Å | 197.591 Å × 222.868 Å × 194.892 Å | 40 Å × 44 Å × 118 Å | 197.591 Å × 222.868 Å × 194.892 Å |
| 10 | Resveratrol | 96Å × 96 Å × 160 Å | 197.079 Å × 222.868 Å × 195.651 Å × | 40 Å × 44 Å × 118 Å | 197.079 Å × 222.868 Å × 195.651 Å × |
| 11 | Apigenin | 126 Å × 102 Å × 160 Å | 197.079 Å × 222.868 Å × 194.797 | 44 Å × 96 Å × 98 | 224.192 Å × 224.074 Å × 194.102 Å × |
First, the SARS-CoV-2S, and ligand were converted into pdbqt format using AutoDock tools. Polar hydrogens and gasteiger charges were added to SARS-CoV-2S and other ligand structure before docking.
2.2. Molecular docking of phytochemicals on SARS-CoV-2 S (spike protein)
The cryo-electron microscopic structure of SARS-CoV-2S was used for the molecular docking analysis. SARS-CoV-2S protein is a heterotrimer consisting of chains A, B, and C (Walls et al., 2020). For the docking experiment, chain A of the spike protein was used. First, the SARS-CoV-2S, kamferol, and HCQ were converted into pdbqt format using AutoDock tools (Morris et al., 2009). Polar hydrogens and gasteiger charges were added to SARS-CoV-2S, kamferol, and HCQ structure before docking. The molecular docking tool AutoDock Vina (Trott & Olson, 2010) was used to study the binding of curcumin and hydroxychloroquine on the SARS-CoV-2S. Further, blind docking of kamferol was performed to ascertain the probable binding sites. For this, the entire SARS-CoV-2S molecule was covered with a grid box of dimension 76 Å × 92 Å × 160 Å with grid spacing 1 Å. The SARS-CoV-2S was kept rigid while the kamferol was kept flexible. The four sets of docking were performed with exhaustiveness 100. Each set of AutoDock Vina produced 9 conformations (Trott & Olson, 2010). Among them, 6 conformations were docked at one domain of SARS-CoV-2S, that domain is used for the direct docking. For local docking, SARS-CoV-2S was covered with the grid box of dimension 60 Å × 54 Å × 66 Å with grid spacing 1 Å. The exhaustiveness was kept at 100. Four sets of local docking were performed for each ligand and the site, and the site where maximum number of conformations bind was considered the binding site. The conformations with high negative binding energy are represented in the figures. The docking results were analyzed using MGL Tools 1.5.6 (Morris et al., 2009), and the hydrophilic and hydrophobic interactions were determined using PyMol (DeLano, 2002).
Similar to kamferol, the docking of other ligand was performed on the SARS-CoV-2S protein. The docking parameters of all the ligand have been provided in Table 1.
2.3. Simulation set-up and analysis
The biomolecular software package GROMACS-2018.1 (Kutzner et al., 2019) was used for the molecular dynamics simulations of the protein–ligand complex with the protein interactions approximated by the CHARMM36 force field and ligand parameters generated using CgenFF (Huang et al., 2017). For each protein–ligand complex, we place the respective complex in the center of a cubic box with 10 Å distance to the edges and fill the box with TIP3P water and counter ions (Na+ Cl−) 0.15 M added to neutralize the system (Joung & Cheatham, 2008). The simulation box of protein complexed with fistein, kamferol and quercontain contain water molecules, 178467, 178474 and 136669, respectively. Because of periodic boundary conditions, electrostatic interactions were evaluated using particle-Ewald summation, and a cut-off of 10 Å was used for the calculation of vdW-interactions. The resulting systems were energy-minimized by steepest descent and conjugate gradient methods, followed by a short (500 ps) equilibration in NVT ensemble and subsequent 500 ps in an NPT ensemble. Temperature and pressure were set T = 310 K and 1 bar, which was controlled by a Parrinello-Danadio-Bussi thermostat (Huang et al., 2017) and Parrinello-Rahman barostat (Bussi et al., 2007; Parrinello & Rahman, 1980). The integration step of 2 fs was used. Each system was simulated for 100 ns, and the snapshots were saved every 10 ps for further analysis. We analyze the resulting trajectories using the tools provided by the GROMACS package (Abraham et al., 2015). Several structural parameters that we measure and analyze include the root-mean-square deviation (RMSD), the radius of gyration (Rg), solvent-accessible-surface-area (SASA), root-mean-square-fluctuation (RMSF), hydrogen bonds, hydrophobic interactions and binding free energy (Islam et al., 2020; Jain et al., 2020; Mishra et al., 2018; Sarma et al., 2020). Hydrogen bonds were defined by a distance cut-off of 3.5 Å between the donor & acceptor atom and an angle cut-off of 30°.
2.4. Binding free energy estimation
Utilizing the MD trajectory, the binding free energies for ligands with S protein were calculated using MM-PBSA (Molecular Mechanics-Poisson-Boltzmann Surface Area). Considering the convergence issues, the snapshots at an interval of 10 ps were extracted from the last 20 ns of each trajectory and used for the MM-PBSA calculation (Batt et al., 2012; Prakash & Luthra, 2012). The solute dielectric constant value of 2 and salt concentration of 0.15 M was used for the calculation (Kumar et al., 2019; Pandey et al., 2018). Binding free energy of protein–ligand complexes were calculated as,
where, Gcomplex represents the free energy of the protein–ligand complex, Greceptor, the free energy of protein, Gligand as the free energy of ligand, and < > represent the ensemble average.
2.5. Prediction of ADME by computational analysis
ADME profiling of fisetin, quercetin, and kamferol at pH 7 were determined using online software tools (Jayaram et al., 2012). The important parameters allied with ADME properties such as Lipinski’s rule of five, solubility of drug, pharmacokinetic properties, molar refractivity, and drug likeliness were deliberated (Lipinski, 2004).
3. Results and discussion
3.1. Molecular docking study
In the field of computer-aided drug designing, particularly for the identification of a lead compound (Hughes et al., 2011; Raj et al., 2019), molecular docking is immensely employed to explore the various types of binding interaction of the prospective drug with different domains or active sites on the target molecules. Among all different types of interactions such as H-bond, π–π, amide-π interactions, etc., the binding efficacy of a ligand molecule with the active sites of a target has widely been explained by evaluating its hydrogen bonding pattern (Chen et al., 2016; Raj et al., 2019) and the nature of residues present at the active site. The binding energy (kcal/mol) data allows us to study and compare the binding affinity of different ligands/compounds with their corresponding target receptor molecule. The lower binding energy indicates a higher affinity of the ligand for the receptor. The ligand with the highest affinity can be chosen as the potential drug for further studies. The SARS-CoV-2S is a surface glycoprotein of 2019 novel coronavirus (Ou et al., 2020; Wahedi et al., 2020). This protein plays important roles during viral attachment, fusion, and entry into the host cells (Adedeji et al., 2013; Boopathi et al., 2020; Khan et al., 2020). Biologically this protein exists in a heterotrimeric form with three separate polypeptide chains: chain A, B, and C, forming each monomer (Boopathi et al., 2020; Li et al., 2006). For the present study, ten phytochemicals of flavonoids and non-flavonoids class with a broad range of biological activities along with one FDA-approved anti-malarial drug HCQ which exhibited efficacy against SARS-CoV-2 have been selected as ligands to investigate their binding affinity with SARS-CoV-2S chain A as the receptor target protein. The docking study revealed that out of the three different domains, namely S1-N terminal, S1-C terminal, and S2, these phytochemicals and HQC exhibited binding affinity mainly for two domains: S1-C terminal and S2 of the spike protein of SARS-CoV-2. Among all the ligands, 2-(3,4-dihydroxyphenyl)-3,7-dihydroxy-4H-chromen-4-one (fisetin) (Table 2) 2-(3,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-one (quercetin) and (Table 2) displayed lowest and identical binding energy of −8.5 kcal/mol as well as similar binding preferences for S2 domain of the spike protein. Despite having a similar preference for the S2 domain, the presence of an additional 5-OH group on the chromone ring of quercetin affected its hydrogen bonding interactions compared to fisetin, which was also reflected in their interaction with different residues of S2 domain.
Table 2.
Molecular docking analysis to determine the putative binding sites of selected inhibitors on SARS-CoV-2S (spike protein).
| Compound | Binding Affinity (kcal/mol) | Interacting Domain of Spike Protein | Interacting amino acid Residue Of Spike Protein |
|---|---|---|---|
| Kamferol | −7.4 | C-Terminal of S1 Domain | THR 393, ALA 522, LEU 527, CYS 391, LEU 390, ASN 544, PHE 782, ALA 1056 |
| Curcumin | −7.1 | C-Terminal of S1 Domain | THR 430, LEU 517, ALA 520, GLN 564, PHE 565, ASN 544, LEU 546, LEU 390, VAL 382 |
| Pterostilbene | −6.7 | C-Terminal of S1 Domain | THR 393, ALA 522, ALA 520, HIS 519, ASN 544, GLN 564, LEU 390, GLY 545, PHE 543, LEU 546,PHE 565 |
| Hydroxychloroquine | −5.6 | C-Terminal of S1 Domain | GLN564, PHE 565, ALA 520, ARG 567, HIS 519, LEU 518, LEU 517, CYS 391, LEU 546, ALA 522 |
| Fisetin | −8.5 | S2 Domain | ILE 870, PRO 880, SER 730, HIS 1058, THR 732, THR 778 |
| Quercetin | −8.5 | S2 Domain | ILE 870, ASP 867, ALA 1056, PRO 1057, GLY 1059, HIS 1058, SER 730, MET 730 MET 731, LYS 733, VAL 860, LEU 861, PRO 863 |
| Isorhamnetin | −8.3 | S2 Domain | HIS 1058, VAL 729, SER 730, MET 731, THR 732, LYS 733, VAL 860, LEU 861, PRO 863, THR 778, ILE, 870, PHE 782, ALA 1056 |
| Genistein | −8.2 | S2 Domain | GLY 1059. SER 730, HIS 1058, THR 732, LYS 733, PRO 863, ASN 867, THR 870, PHE 782, ALA 1056 |
| Luteolin | −8.2 | S2 Domain | ALA 1056, GLY 1059, HIS 1058, ASP 867, ILE 870, THR 871, PRO 863, LEU 861, LYS 733, MET 731 |
| Resveratrol | −7.9 | S2 Domain | THR 732, HIS 1058, LYS 733, LEU 861, PRO 863, ASP 867, ILE 870, PHE 782 |
| Apigenin | −7.7 | S2 Domain | SER 730, HIS 1058, ALA 1056, ILE 870, PRO 863, LEU 861 |
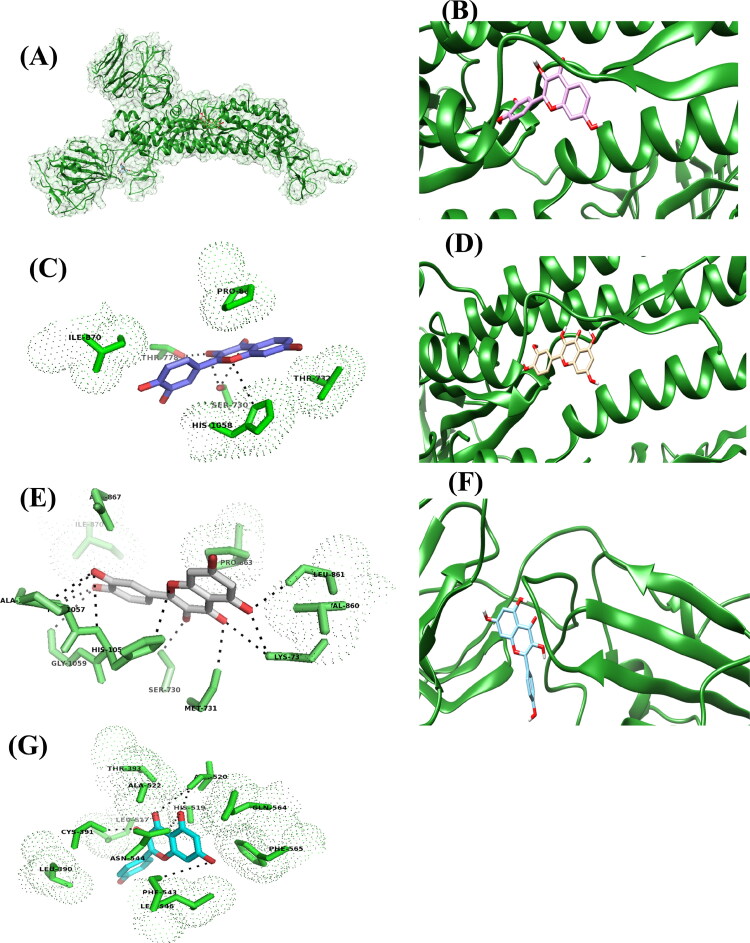
As shown in Figure 1C and Table 2, the fisetin interacts with SER 730, THR 778 and HIS 1058 residues through H-bonding and exhibits hydrophobic interaction (based on 3 D views different OH forming H-bonds and their bond length could have also been specified) with ILE 870, PRO 880 and THR 732 residues of S2 domain of the spike protein. Whereas, quercetin forms hydrogen bonds with LYS 733, LEU 861, MET 731, SER 730, PRO 1057, GLY 1059, HIS 1058 and ALA 1056 residues and displayed hydrophobic interaction with ILE 870, ASP 867, MET 730, VAL 860 and PRO 863 (Figure 1E and Table 2).
Figure 1.
The putative binding site of fisetin, quercetin and kamferol on SARS-CoV-2S protein. A) The cartoon showing the structure and surface of SARS-CoV-2 S protein, chain A and the binding site of fisetin, quercetin and kamferol, on it. Fisetin, quercetin and kamferol are shown in ball and stick model in purple, pale orange and cyan colour respectively. B) Binding site of fisetin on SARS-CoV-2 S protein. C) The residues interacting with the fisetin. D) Binding site of quercetin on SARS-CoV-2 S protein. E) The residues interacting with the quercetin. F) Binding site of kamferol on SARS-CoV-2 S protein. G) The residues interacting with the kamferol.
Similarly, lack of one hydroxyl (-OH) group at 3′-C of phenyl ring in case of 3,5,7-trihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one (kamferol) (Figure 1F, G and Table 2) not only lead to the reduction in binding affinity (−7.4 kcal/mol) but it also leads to binding at S1 domain rather than S2 domain which were observed in case of other three flavonols.
Like-wise the binding studies performed with hydroxychloroquine (HCQ), which has emerged as a potent therapeutic option against COVID-19 and received much attention in the last few days, revealed the lowest binding affinity (−5.6 kcal/mol) among all others (Table 2).
It interacts with ALA 520, GLN564, PHE 565, ARG 567, and HIS 519 residues through hydrogen bond formation. It forms the hydrophobic interaction with LEU 518, LEU 517, CYS 391, LEU 546, and ALA 522. The lowest binding affinity observed with HCQ could be attributed to the presence of a conformationally labile and sterically bulkier carbon chain, which probably disrupts the interaction of the HCQ with the binding site resulting in reduced binding affinity. Overall the docking study discloses two different sets of ligands which bind at S1 and S2 domains of SARS-CoV-2S. We thought that if these molecules interact with either S1 or S2 domain strongly, it may perturb the hACE2-S protein complex interaction. So, based on the binding energy, we selected these top phytochemicals for our further in silico study.
3.2. Structural dynamics of protein–ligand complex
The molecular docking analyses resulted in the selection of three phytochemicals, fistein, kamferol, and quercetin which show promising activity against hACE2-S protein complex. However, during the biological functions, proteins undergo substantial conformational changes, and aqueous environment around the protein plays a functional role in protein–ligand interactions (Luthra et al., 2009; Mongre et al., 2019; Yu and Dalby, 2018). Thus, to better understand the conformational stability of ligand binding and the contribution of the active site in terms of binding free energy, we performed MD simulation in solvated condition at physiological temperature for the period of 100 ns. The mediated interactions and binding energy contribution of amino acids were calculated with MM-PBSA (Wang et al., 2019).
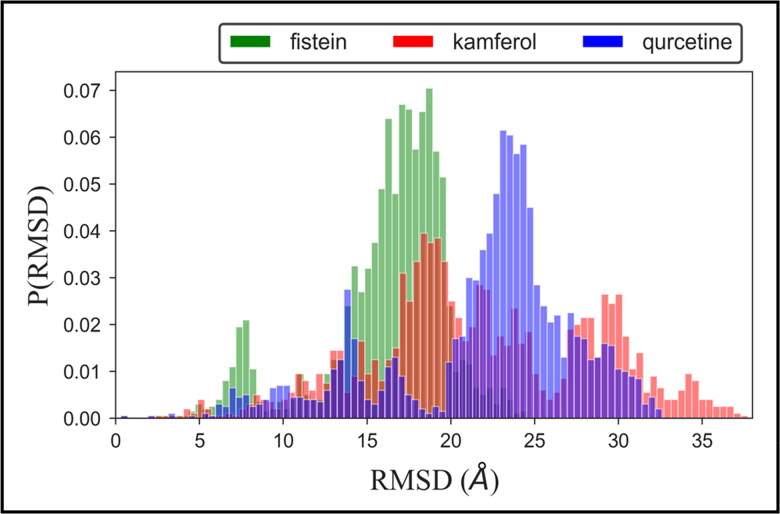
To investigate the overall conformational dynamics and stability of the protein–ligand complex, we calculated the RMSD values relative to the starting structure for the backbones of all residues (Figure 2). The probability distribution plot of the docked structure of hACE2-S protein with fistein shows that the population of stable conformational dynamics is settled around ∼18–20 Å. We find a small population density between 5 and 10 Å, which indicates the initial perturbation in the structure fistein is well-adjusted at the binding pocket; thus, the dynamic equilibrium shifted to a stable population of the complex. The time evolution plot of RMSD shown in Supplementary Figure 2 shows that the complex structure of fistein achieves equilibrium at ∼40 ns and remains stable till the simulation ends at 100 ns. Whereas, the population of complex structure with kamferol is largely stabilized around ∼17–20 Å. The splitting in the RMSD distribution plot, which can be seen near 15 Å, 20 Å, and 30 Å, respectively, suggested different drifts in the trajectory during the dynamic progression. The detailed view of the time evolution plot suggested that the structure of hACE2-S protein complexed with kamferol attains equilibrium at ∼10 ns, which is continued up to 40 ns. A consistent increase in RMSD is observed up to 70 ns; after that, the RMSD dropped quickly and attainded stable equilibrium, which is maintained until 100 ns.
Figure 2.
Probability distribution of Cα-RMSD of hACE2-S protein complex with ligands, fistein, kamferol and quercetin.
The RMSD distribution plot of complex structure with quercetin shows the higher population density >60% near 23–24 Å, which indicates the stable conformational dynamics of protein–ligand complex. The time evolution view of hACE2-S protein complexed with quercetin also provides a clear view that the complex structure attains equilibrium at ∼10 ns and remains stable for the remaining period of simulation. Thus, the RMSD results denote that all three ligands remain occupied at active site stabilized with potential interactions.
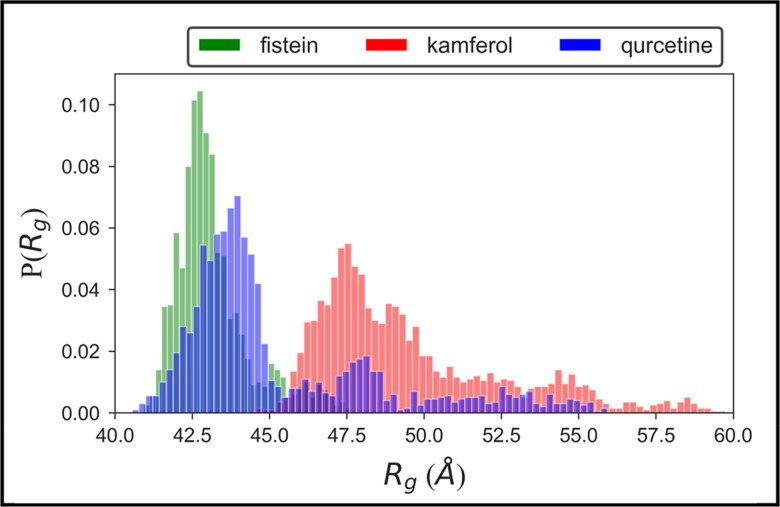
To evaluate the dynamic adaptability and overall compactness of the ligand-bound complex structure of hACE2-S protein, we calculated the probability distribution plot of radius of gyration (Rg) (Rocco et al., 2008; Singh et al., 2019). Results show that the Rg values of fistein ranged between 42.0 and 43.0 Å, whereas, the higher population density of Rg values ranged between ∼42.0–44.0 Å and ∼47.0–48.0 Å for kamferol and quercetin, respectively (Figure 3). The narrow Rg distribution ranges of highly populated conformational dynamics suggested the minimal variations in the structural compactness of protein. Thus, the overall protein–ligand complexes remain stable during the simulation, which is observed consistent with RMSD analyses.
Figure 3.
Probability distribution of radius of gyration (Rg) for the complex structure of hACE2-S protein with fistein, kamferol and quercetin.
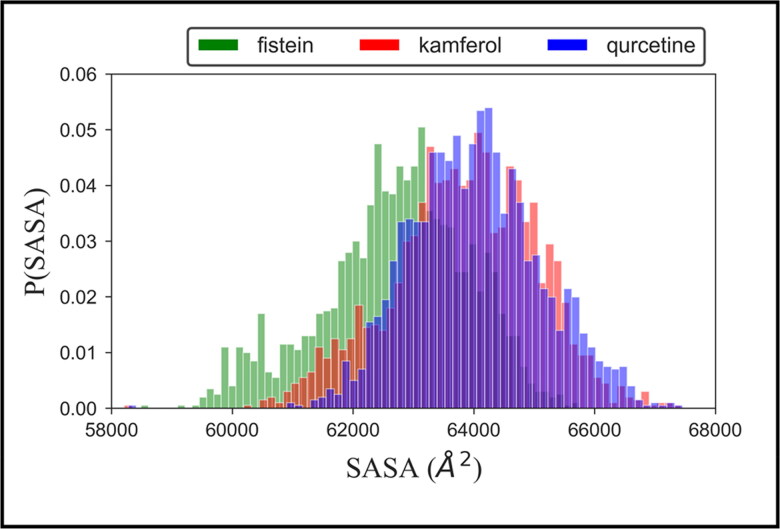
To monitor the structural stability of the ligand-bound complex of hACE2-S protein, we also computed the probability distribution plot of the solvent-accessible surface area (SASA), shown in Figure 4. This plot shows that the distribution of highly populated structure of all three complex restricted around ∼64,000 Å2, which provide elegant evidence of stable conformational dynamics of the protein–ligand complex in an aqueous environment. Thus, the results collectively obtained from probability distribution plots of RMSD, Rg, and SASA suggested that the complexation of ligands at the active site enhances the conformational stability hACE2-S protein.
Figure 4.
Probability distribution plot of solvent accessible surface area (SASA) of hACE2-S protein complex with fistein, kamferol and quercetin.
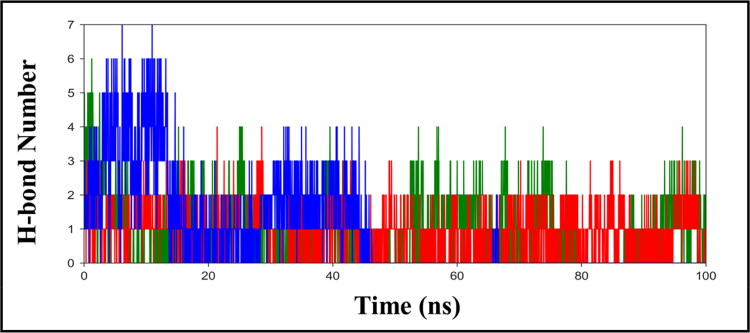
In addition, to evaluate the molecular interaction, we also examine the time evolution plot of hydrogen bond (H-bond) formed between the protein and ligands (Figure 5). H-bond acts as a key interaction in determining the binding affinity, selectivity, and stabilization effect of ligands with protein. From Figure 5, we find that the initial trajectory of fistein shows the appearance of five H-bonds with hACE2-S protein, out of which three H-bonds were observed consistently during the simulation. The H-bond plot of kamferol shows the occurrence of three-four H-bonds with protein; however, two H-bond interactions were found more stable, which can be seen retained throughout the simulation. Whereas, the H-bond plot of quercetin shows a maximum of six H-bond interactions with hACE2-S protein, during the initial 0–15 ns. During the progression of simulation, three H-bonds were observed up to 40 ns, and only two remained long-lived, which were maintained during the entire simulation time.
Figure 5.
The hydrogen bond (H-bond) plot, representing the total number of Hbond formed between hACE2- S protein and ligands, fisetin (green), kamferol (red) and quercetin (blue) during the simulation in water, at 300 K.
3.2.1. Free energy landscape (FEL)
To probe the conformational dynamics of protein–ligand complexes during the simulation, we applied principal component analysis (PCA). The PCA is an efficient tool which quantitatively defines the collective motion and measures the movement directions (David & Jacobs, 2014; Kumar et al., 2020; Sarma et al., 2020). The principal components, PC1 and PC2, were obtained from diagonalization of the covariance matrix of the Cα atomic fluctuation and were used as reaction coordinates to calculate the Gibbs free energy landscape (FEL).
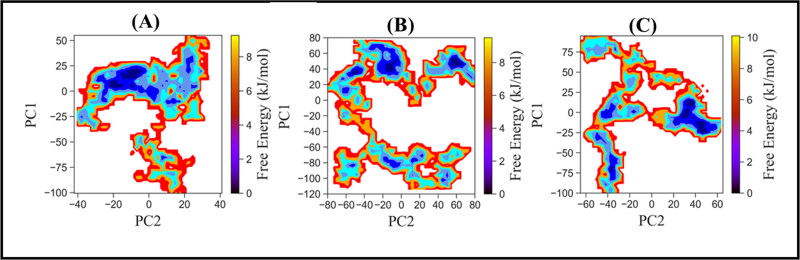
FEL plot (Figure 6) displayed that the complex of hACE2-S protein with three compounds, fisetin, kamferol, and quercetin, spans different subspace of structures during the period of 100 ns simulation. It is observed that the collective motion of fistein is confined to a small conformational space. The population of complex structures occupying the small energy basins represents the initial conformation of the optimization phase, the ligand well settled at the binding pocket which is subsequentially visited is broad energy basins, having the population of stable protein–ligand complex. The structure of hACE2-S protein complexed with kamferol navigated a broad conformational space, the collective motion of protein in the three different energy wells represents the different conformational stability of protein–ligand complex. The small energy basin represents the initial equilibration phase; then, the complex structure shifted to another energy basin, which is relatability more stable. Finally, the structure transverse through the small energy barrier to a broader energy basin, having population of a more stable complex. It may be the reason we observed two drifts at ∼20 ns and ∼75 ns in RMSD trajectory, representing the two different but stable conformational states between ∼25–60 ns and ∼80–100 ns. Quercetin experiences wide conformational space, visiting through the three energy basins. The structure crosses smoothly from one subspace to another subspace through the small energy barrier; thus, we observed that stable conformation of protein–ligand during the initial 0–40 ns, due to excursion protein jump out from the stable energy basin, navigated the broad conformational, representing the rugged energy basin. However, during the dynamics progression, protein acquired equilibrium with the dropdown in RMSD and entered to deep energy basin, suggesting the population of stable protein–ligand structures. Collectively, the structural dynamics analyses and FEL suggested the stable conformation dynamics of the docked complex of hACE2-S protein complex with ligands, fistein, kamferol, and quercetin.
Figure 6.
Free energy landscape of hACE2-S protein complex with ligands (A) Fisetin (B) Kamferol and (C) Quercetin, using PC1 and PC2 as reaction coordinates.
3.4. Binding free energy
In order to understand the contribution of active site residues which influence the binding free energy and structural stability of ligands during the dynamic progression, we applied MM-PBSA(Kumar et al., 2020; Pandey et al., 2018). MM-PBSA determines the strength of binding by calculating the absolute binding free energies (ΔGbind) which is sum of the electrostatic, Van der Waals interaction, polar and non-polar solvation free energies (Supplementary Figure 3). In addition, the contribution of each residue to the binding free energy was also calculated for the aforesaid energy terms. The estimated energy components for binding of fistein, kamferol, and quercetin with hACE2-S protein are summarized in Table 2. The compound quercetin shows the lowest binding free energy ΔGbind = −22.17 ± 3.04 kcal mol−1 which indicates the highest binding affinity for hACE2-S protein. The lowest binding affinity was observed with kamferol which shows ΔGbind = −15.07 ± 2.42 kcal mol−1. Whereas, the binding of fistein with hACE2-S protein shows ΔGbind = −21.11 ± 3.49 kcal mol−1. This suggests that the binding affinity of quercetin and fistein to hACE2-S protein remains almost with equal potency, which is consistent with molecular docking results, showing similar docked score ΔG = −8.50 kcal mol−1. Thus, MM-PBSA results strengthen the molecular docking and conformational dynamics observation, which suggested the stable binding of fistein, kamferol, and quercetin with hACE2-S protein. These results may contribute to the understanding of structure-based designing in the development of COVID-19 therapy.
3.5. Lipinski rule of five
It is generally used to (Das et al., 2020; Gupta et al., 2020; Jayaram et al., 2012; Lipinski, 2004) evaluate potential interactions between the drug and other target non-drug molecules. It evaluates the propensity of a compound with a certain pharmacological or biological activity to be used as a potential drug. It acts as a filter to screen potential therapeutic agents/drugs just at the initiation of the program, thereby minimizing the labour and cost of exercises involving clinical drug development and to a large extent preventing late-stage clinical failures. The rule mainly determines the molecular properties of a compound, which are its prime requisition to be a potential drug, like absorption, distribution, metabolism, and excretion (ADME). Lipinski’s rule states that for any compound to be selected as a potential drug it should have (a) Molecular mass < 500 Dalton (b) high lipophilicity (expressed as LogP ≤5) (c) less than 5 hydrogen bond donors (d) Less than 10 hydrogen bond acceptors (e) molar refractivity between 40 and 130. If a compound of interest possesses more than two of the aforementioned criteria, then the compound is likely to be a potential candidate for drug development. All three selected phytochemicals used in this study were found to pass all the five criteria mentioned above in Lipinski’s rule (Table 3). Thus we suggest that all of these phytochemicals have the potential ability to work effectively as novel drugs.
Table 3.
ADME Properties of selected inhibitors against SARS-CoV-2S (spike protein).
| S.No. | Ligand/Phytochemicals | ADME
Properties (Lipinski’s Rule of Five |
Drug Likeliness | |
|---|---|---|---|---|
| Properties | Values | |||
| 1 | Fisetin | Molecular weight (<500 Da) | 286 | Yes |
| LogP (<5) | 2.3 | |||
| H-bond donor (5) | 4 | |||
| H-bond acceptor (<10) | 6 | |||
| Molar Refractivity (40-130) | 72.4 | |||
| Violations | NO | |||
| 2 | Quercetin | Molecular weight (<500 Da) | 302 | Yes |
| LogP (<5) | 2 | |||
| H-bond donor (5) | 5 | |||
| H-bond acceptor (<10) | 7 | |||
| Molar Refractivity (40-130) | 74 | |||
| Violations | NO | |||
| 3 | Kamferol | Molecular weight (<500 Da) | 286 | Yes |
| LogP (<5) | 2.3 | |||
| H-bond donor (5) | 4 | |||
| H-bond acceptor (<10) | 6 | |||
| Molar Refractivity (40-130) | 72.4 | |||
| Violations | NO | |||
In vitro and in vivo experiments with our selected group of potential anti-viral compounds will strengthen our perspectives that natural products based therapeutic interventions is the need of the hour. Herbal medicines formulated from the phytochemicals we predicted, will be extremely essential for the identification of anti-COVID-19 inhibitors.
4. Conclusion
Molecular docking and MD simulation is the best platform to screen potential drug candidates. In this study, we described the binding ability of naturally occurring phytochemicals with the SARS-CoV2-S protein. Further, using MD simulation, we showed that kamferol, quercetin, and fisetin bind to the hACE2- S-protein complex, near the interface of hACE2 and S-protein binding. Our results were supported by other analyses like the free energy estimation of binding using MM-PBSA and free energy landscape. Further, we found that these compounds satisfy all criteria of Lipinski’s rule of five. Therefore, we believe that these compounds could be potential anti-COVID drugs; hence the researcher in this field should verify this using in vitro experimental techniques.
Supplementary Material
Acknowledgements
We thank the Indian Institute of Technology Delhi for providing online facilities of Supercomputing facility for Bioinformatics and Computational Biology. AK and SR thanks Mahatma Gandhi Central University Motihari, Bihar.
Glossary
Abbreviations
- COVID-19
Coronavirus Disease 2019
- SARS-CoV-2S
Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein
- hACE2
Human Angiotensin Converting Enzyme-2
- hACE2-S protein complex
Human Angiotensin Converting Enzyme-2 receptor and Severe Acute Respiratory Syndrome Coronavirus 2 Spike protein complex
- HCQ
Hydroxychloroquine
- CQ
Chloroquine
- ACE2
Angiotensin-Converting Enzyme-2
- MERS-CoV
Middle East Respiratory Syndrome coronavirus
- PDB
protein data bank
- ADME
absorption, distribution, metabolism and excretion
Disclosure statement
The authors report no conflicts of interest.
References
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses . (2020). The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology, 5, 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aanouz, I., Belhassan, A., El-Khatabi, K., Lakhlifi, T., El-Ldrissi, M., & Bouachrine, M. (2020). Moroccan Medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abba, Y., Hassim, H., Hamzah, H., & Noordin, M. M. (2015). Antiviral Activity of Resveratrol against Human and Animal Viruses. Advances in Virology, 2015, 184241. 10.1155/2015/184241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelli, I., Hassani, F., Bekkel Brikci, S., & Ghalem, S. (2020). In silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from Western Algeria. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1763199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1-2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Adedeji, A. O., Severson, W., Jonsson, C., Singh, K., Weiss, S. R., & Sarafianos, S. G. (2013). Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. Journal of Virology, 87(14), 8017–8028. 10.1128/JVI.00998-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bari, M. A. A. (2017). Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacology Research & Perspectives, 5(1), e00293. 10.1002/prp2.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batt, S. M., Jabeen, T., Bhowruth, V., Quill, L., Lund, P. A., Eggeling, L., Alderwick, L. J., Futterer, K., & Besra, G. S. (2012). Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors. Proceedings of the National Academy of Sciences of the United States of America, 109(28), 11354–11359. 10.1073/pnas.1205735109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi, G., Donadio, D., & Parrinello, M. (2007). Canonical sampling through velocity rescaling. J Chem Phys, 126(1), 014101 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- Campagna, M., & Rivas, C. (2010). Antiviral activity of resveratrol. Biochemical Society Transactions, 38(Pt 1), 50–53. 10.1042/BST0380050 [DOI] [PubMed] [Google Scholar]
- Chan, C. N., Trinite, B., & Levy, D. N. (2017). Potent Inhibition of HIV-1 replication in resting CD4 T cells by resveratrol and pterostilbene. Antimicrobial Agents and Chemotherapy, 61(9), e00408–17. 10.1128/AAC.00408-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, P., Nagi, N., Agarwal, A., Das, B., Banerjee, S., Sarkar, S., Gupta, N., & Gangakhedkar, R. R. (2020). The 2019 novel coronavirus disease (COVID-19) pandemic: A review of the current evidence. Indian Journal of Medical Research, 0(0), 0. 10.4103/ijmr.IJMR_519_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Oezguen, N., Urvil, P., Ferguson, C., Dann, S. M., & Savidge, T. C. (2016). Regulation of protein–ligand binding affinity by hydrogen bond pairing. Science Advances, 2(3), e1501240. 10.1126/sciadv.1501240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, L. C., Ng, L. T., Cheng, P. W., Chiang, W., & Lin, C. C. (2005). Antiviral activities of extracts and selected pure constituents of Ocimum basilicum . Clinical and experimental pharmacology & physiology, 32(10), 811–816. 10.1111/j.1440-1681.2005.04270.x [DOI] [PubMed] [Google Scholar]
- Das, S., Sarmah, S., Lyndem, S., & Singha Roy, A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, C. C., & Jacobs, D. J. (2014). Principal component analysis: A method for determining the essential dynamics of proteins. Methods in Molecular Biology (Clifton, N.J.).), 1084, 193–226. 10.1007/978-1-62703-658-0_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W. L. (2002). The PyMOL Molecular Graphics System. Schrödinger LLC. [Google Scholar]
- Docherty, J. J., Fu, M. M., Hah, J. M., Sweet, T. J., Faith, S. A., & Booth, T. (2005). Effect of resveratrol on herpes simplex virus vaginal infection in the mouse. Antiviral Research, 67(3), 155–162. 10.1016/j.antiviral.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A., & Azzam, E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., Sahin, A. T., & Yelekci, K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, W., Qian, S., Qian, P., & Li, X. (2016). Antiviral activity of luteolin against Japanese encephalitis virus. Virus Research, 220, 112–116. 10.1016/j.virusres.2016.04.021 [DOI] [PubMed] [Google Scholar]
- Forni, C., Facchiano, F., Bartoli, M., Pieretti, S., Facchiano, A., D'Arcangelo, D., Norelli, S., Valle, G., Nisini, R., Beninati, S., Tabolacci, C., & Jadeja, R. N. (2019). Beneficial Role of Phytochemicals on Oxidative Stress and Age-Related Diseases. BioMed Research International, 2019, 8748253. 10.1155/2019/8748253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, M. K., Vemula, S., Donde, R., Gouda, G., Behera, L., & Vadde, R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, A., Paray, B. A., Hussain, A., Qadir, F. A., Attar, F., Aziz, F. M., Sharifi, M., Derakhshankhah, H., Rasti, B., Mehrabi, M., Shahpasand, K., Saboury, A. A., & Falahati, M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., Rauscher, S., Nawrocki, G., Ran, T., Feig, M., de Groot, B. L., Grubmuller, H., & MacKerell, A. D. Jr. (2017). CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nature Methods, 14(1), 71–73. 10.1038/nmeth.4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J. P., Rees, S., Kalindjian, S. B., & Philpott, K. L. (2011). Principles of early drug discovery. British Journal of Pharmacology, 162(6), 1239–1249. 10.1111/j.1476-5381.2010.01127.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, R., Parves, M. R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., Hossain, M. N., Ali, M. A., & Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, M., Muthukumaran, J., & Singh, A. K. (2020). Comparative structural and functional analysis of STL and SLL, chitin-binding lectins from Solanum spp. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1781693 [DOI] [PubMed] [Google Scholar]
- Jayaram, B., Singh, T., Mukherjee, G., Mathur, A., Shekhar, S., & Shekhar, V. (2012). Sanjeevini: A freely accessible web-server for target directed lead molecule discovery. BMC Bioinformatics, 13 (Suppl 17), S7. 10.1186/1471-2105-13-S17-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, R. S., Jagdale, S. S., Bansode, S. B., Shankar, S. S., Tellis, M. B., Pandya, V. K., Chugh, A., Giri, A. P., & Kulkarni, M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung, I. S., & Cheatham, T. E. (2008). Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. The journal of Physical Chemistry. B, 112(30), 9020–9041. 10.1021/jp8001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul, T. N., Middleton, E., Jr., & Ogra, P. L. (1985). Antiviral effect of flavonoids on human viruses. Journal of Medical Virology, 15(1), 71–79. 10.1002/jmv.1890150110 [DOI] [PubMed] [Google Scholar]
- Khan, R. J., Jha, R. K., Amera, G. M., Jain, M., Singh, E., Pathak, A., Singh, R. P., Muthukumaran, J., & Singh, A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, S. A., Zia, K., Ashraf, S., Uddin, R., & Ul-Haq, Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kirchdoerfer, R. N., Cottrell, C. A., Wang, N., Pallesen, J., Yassine, H. M., Turner, H. L., Corbett, K. S., Graham, B. S., McLellan, J. S., & Ward, A. B. (2016). Pre-fusion structure of a human coronavirus spike protein. Nature, 531(7592), 118–121. 10.1038/nature17200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N., Srivastava, R., Prakash, A., & Lynn, A. M. (2020). Structure-based virtual screening, molecular dynamics simulation and MM-PBSA toward identifying the inhibitors for two-component regulatory system protein NarL of Mycobacterium tuberculosis. Journal of Biomolecular Structure & Dynamics, 38(11), 3396–3410. 10.1080/07391102.2019.1657499 [DOI] [PubMed] [Google Scholar]
- Kumar, S., & Pandey, A. K. (2013). Chemistry and biological activities of flavonoids: An overview. TheScientificWorldJournal, 2013, 162750. 10.1155/2013/162750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, N., Srivastava, R., Prakash, A. & Lynn, A. M. (2019). Structure-based virtual screening, molecular dynamics simulation and MM-PBSA toward identifying the inhibitors for two component regulatory system protein NarL of Mycobacterium Tuberculosis. Journal of Biomolecular Structure and Dynamics, 38(11), 3396–3410. 10.1080/07391102.2019.1657499 [DOI] [PubMed] [Google Scholar]
- Kutzner, C., Pall, S., Fechner, M., Esztermann, A., de Groot, B. L., & Grubmuller, H. (2019). More bang for your buck: Improved use of GPU nodes for GROMACS 2018. Journal of Computational Chemistry, 40(27), 2418–2431. 10.1002/jcc.26011 [DOI] [PubMed] [Google Scholar]
- Li, F., Berardi, M., Li, W., Farzan, M., Dormitzer, P. R., & Harrison, S. C. (2006). Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. Journal of Virology, 80(14), 6794–6800. 10.1128/JVI.02744-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. C., Ho, C. T., Chuo, W. H., Li, S., Wang, T. T., & Lin, C. C. (2017). Effective inhibition of MERS-CoV infection by resveratrol. BMC Infectious Diseases, 17(1), 144. 10.1186/s12879-017-2253-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski, C. A. (2004). Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov Today Technol, 1(4), 337–341. 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Luthra, P. M., Prakash, A., Barodia, S. K., Kumari, R., Mishra, C. B. & Kumar, J. B. (2009). In silico study of naphtha [1, 2-d] thiazol-2-amine with adenosine A 2A receptor and its role in antagonism of haloperidol-induced motor impairments in mice. Neuroscience letters, 463(3), 215–218. 10.1016/j.neulet.2009.07.085 [DOI] [PubMed] [Google Scholar]
- Lyu, S. Y., Rhim, J. Y., & Park, W. B. (2005). Antiherpetic activities of flavonoids against herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2) in vitro. Archives of Pharmacal Research, 28(11), 1293–1301. 10.1007/BF02978215 [DOI] [PubMed] [Google Scholar]
- Mishra, C. B., Kumari, S., Prakash, A., Yadav, R., Tiwari, A. K., Pandey, P., & Tiwari, M. (2018). Discovery of novel Methylsulfonyl phenyl derivatives as potent human Cyclooxygenase-2 inhibitors with effective anticonvulsant action: Design, synthesis, in-silico, in-vitro and in-vivo evaluation. European Journal of Medicinal Chemistry, 151, 520–532. 10.1016/j.ejmech.2018.04.007 [DOI] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongre, R. K., Mishra, C. B., Prakash, A., Jung, S., Lee, B. S., Kumari, S., Hong, J. T. & Lee, M. S. (2019). Novel Carbazole-Piperazine Hybrid Small Molecule Induces Apoptosis by Targeting BCL-2 and Inhibits Tumor Progression in Lung Adenocarcinoma in Vitro and Xenograft Mice Model. Cancers (Basel), 11(9), 1245. https://doi.org/ 10.3390/cancers11091245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounce, B. C., Cesaro, T., Carrau, L., Vallet, T., & Vignuzzi, M. (2017). Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antiviral Research, 142, 148–157. 10.1016/j.antiviral.2017.03.014 [DOI] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Nijveldt, R. J., van Nood, E., van Hoorn, D. E., Boelens, P. G., van Norren, K., & van Leeuwen, P. A. (2001). Flavonoids: A review of probable mechanisms of action and potential applications. The American Journal of Clinical Nutrition, 74(4), 418–425. 10.1093/ajcn/74.4.418 [DOI] [PubMed] [Google Scholar]
- Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., Guo, L., Guo, R., Chen, T., Hu, J., Xiang, Z., Mu, Z., Chen, X., Chen, J., Hu, K., Jin, Q., Wang, J., & Qian, Z. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Communications, 11(1), 1620. 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panche, A. N., Diwan, A. D., & Chandra, S. R. (2016). Flavonoids: An overview. Journal of Nutritional Science, 5, e47. 10.1017/jns.2016.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, P., Srivastava, R., & Bandyopadhyay, P. (2018). Comparison of molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA) and molecular mechanics-three-dimensional reference interaction site model (MM-3D-RISM) method to calculate the binding free energy of protein–ligand complexes: Effect of metal ion and advance statistical test. Chemical Physics Letters, 695, 69–78. 10.1016/j.cplett.2018.01.059 [DOI] [Google Scholar]
- Pant, S., Singh, M., Ravichandiran, V., Murty, U. S. N., & Srivastava, H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello, M., & Rahman, A. (1980). Crystal structure and pair potentials: A Molecular-dynamics study. Physical Review Letters, 45(14), 1196–1199. 10.1103/PhysRevLett.45.1196 [DOI] [Google Scholar]
- Praditya, D., Kirchhoff, L., Bruning, J., Rachmawati, H., Steinmann, J., & Steinmann, E. (2019). Anti-infective properties of the golden spice curcumin. Frontiers in Microbiology, 10, 912. 10.3389/fmicb.2019.00912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, A., & Luthra, P. M. (2012). Insilico study of the A(2A)R-D (2)R kinetics and interfacial contact surface for face for heteromerization. Amino Acids, 43(4), 1451–1464. 10.1007/s00726-012-1218-x [DOI] [PubMed] [Google Scholar]
- Raj, S., Sasidharan, S., Dubey, V. K., & Saudagar, P. (2019). Identification of lead molecules against potential drug target protein MAPK4 from L. donovani: An in-silico approach using docking, molecular dynamics and binding free energy calculation. PLoS One, 14(8), e0221331. 10.1371/journal.pone.0221331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarshi, K., Chatterjee, A., & Ray, S. (2020). BCG vaccination strategy implemented to reduce the impact of COVID-19: Hype or Hope?. Medicine in Drug Discovery, 7, 100049. 10.1016/j.medidd.2020.100049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarshi, K., Chatterjee, A., & Ray, S. (2020). Combating COVID-19 with mesenchymal stem cell therapy. Biotechnology Reports, 26, e00467. 10.1016/j.btre.2020.e00467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco, A. G., Mollica, L., Ricchiuto, P., Baptista, A. M., Gianazza, E., & Eberini, I. (2008). Characterization of the protein unfolding processes induced by urea and temperature. Biophysical Journal, 94(6), 2241–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma, P., Shekhar, N., Prajapat, M., Avti, P., Kaur, H., Kumar, S., Singh, S., Kumar, H., Prakash, A., Dhibar, D. P., & Medhi, B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Meena, N. K., Das, T., Sharma, R. D., Prakash, A. & Lynn, A. M. (2019). Delineating the conformational dynamics of intermediate structures on the unfolding pathway of beta-lactoglobulin in aqueous urea and dimethyl sulfoxide. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2019.1695669 [DOI] [PubMed] [Google Scholar]
- Sinha, S. K., Shakya, A., Prasad, S. K., Singh, S., Gurav, N. S., Prasad, R. S., & Gurav, S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi, H. M., Ahmad, S., & Abbasi, S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell , 181(2), 281–292.e6. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T. Y., Li, Q., & Bi, K. S. (2018). Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian Journal of Pharmaceutical Sciences, 13(1), 12–23. 10.1016/j.ajps.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, E., Sun, H., Wang, J., Wang, Z., Liu, H., Zhang, J. Z. H. & Hou, T. (2019) End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chemical Reviews, 119(16), 9478–9508. 10.1021/acs.chemrev.9b00055 [DOI] [PubMed] [Google Scholar]
- Wu, C. C., Fang, C. Y., Cheng, Y. J., Hsu, H. Y., Chou, S. P., Huang, S. Y., Tsai, C. H., & Chen, J. Y. (2017). Inhibition of Epstein-Barr virus reactivation by the flavonoid apigenin. Journal of Biomedical Science, 24(1), 2. 10.1186/s12929-016-0313-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. C., Fang, C. Y., Hsu, H. Y., Chen, Y. J., Chou, S. P., Huang, S. Y., Cheng, Y. J., Lin, S. F., Chang, Y., Tsai, C. H., & Chen, J. Y. (2016). Luteolin inhibits Epstein-Barr virus lytic reactivation by repressing the promoter activities of immediate-early genes. Antiviral Research, 132, 99–110. 10.1016/j.antiviral.2016.05.007 [DOI] [PubMed] [Google Scholar]
- Yao, X., Ye, F., Zhang, M., Cui, C., Huang, B., Niu, P., Liu, X., Zhao, L., Dong, E., Song, C., Zhan, S., Lu, R., Li, H., Tan, W., & Liu, D. (2020). In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. Oxford Academic. 10.1093/cid/ciaa237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. and P. A. Dalby. (2018). Exploiting correlated molecular-dynamics networks to counteract enzyme activity–stability trade-off. Proceedings of the National Academy of Sciences, 115(52), E12192–E12200. 10.1073/pnas.1812204115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakaryan, H., Arabyan, E., Oo, A., & Zandi, K. (2017). Flavonoids: Promising natural compounds against viral infections. Archives of Virology, 162(9), 2539–2551. 10.1007/s00705-017-3417-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Cui, Q., Fu, Q., Song, X., Jia, R., Yang, Y., Zou, Y., Li, L., He, C., Liang, X., Yin, L., Lin, J., Ye, G., Shu, G., Zhao, L., Shi, F., Lv, C., & Yin, Z. (2017). Antiviral properties of resveratrol against pseudorabies virus are associated with the inhibition of IκB kinase activation. Scientific Reports, 7(1), 8782. 10.1038/s41598-017-09365-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., & Simmons, G. (2012). Development of novel entry inhibitors targeting emerging viruses. Expert Rev Anti Infect Ther, 10(10), 1129–1138. 10.1586/eri.12.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.