Abstract
Newly emerged SARS-CoV-2 made recent pandemic situations across the globe is accountable for countless unwanted death and insufferable panic associated with co-morbidities among mass people. The scarcity of appropriate medical treatment and no effective vaccine or medicine against SARS-CoV-2 has turned the situation worst. Therefore, in this study, we made a deep literature review to enlist plant-derived natural compounds and considered their binding mechanism with the main protease of SARS-CoV-2 through combinatorial bioinformatics approaches. Among all, a total of 14 compounds were filtered where Carinol, Albanin, Myricetin were had better binding profile than the rest of the compounds with having binding energy of –8.476, –8.036, –8.439 kcal/mol, respectively. Furthermore, MM-GBSA calculations were also considered in this selection process to support docking studies. Besides, 100 ns molecular dynamics simulation endorsed the rigid nature, less conformational variation and binding stiffness. As this study, represents a perfect model for SARS-CoV-2 main protease inhibition through bioinformatics study, these potential drug candidates may assist the researchers to find a superior and effective solution against COVID-19 after future experiments.
Communicated by Ramaswamy Sarma
Keywords: Protease inhibitors, phytochemicals, virtual screening, binding modes, MD simulation
Introduction
The world has been thundered by a sudden epidemic attack since December 2019, due to the outbreak of a novel strain of virus known as SARS-CoV-2, commonly called COVID-19 or coronavirus. The outbreak occurred in Wuhan, the capital of Hubei province in China became pandemic and killed hundreds of thousands of people globally (Wu et al., 2020; Zhang et al., 2020; Zhou et al., 2020). Coronavirus belongs to the family Coronaviridae that is single-stranded RNA virus (+ ssRNA) and has similarity with SARS coronavirus. Family Coronaviridae is classified into four genera: alpha, beta, delta and gamma among which the members of alpha and beta virus genera infect humans (de Wilde et al., 2018; Khan, Jha, et al., 2020) . The genome of COVID-19 and SARS coronavirus (SARS-CoV) is about 82% identical to each other and both viruses belong to clade b of the genus betacoronavirus (Wu et al., 2020; Zhou et al., 2020). Exact source from where this disease has spread is not clear, yet it is guessed to be a zoonotic disease and spread through wild animals as the nearest members SARS and MERS spread. In the nature, Chinese horseshoe bats act as a reservoir of SARS-CoV (Lau et al., 2005) and intermediate hosts of this virus are civet cats and raccoon dogs (Guan et al., 2003). One of these animals might spread coronavirus among humans. At the last update in 22 April 2020 WHO reported 2,402,250 coronavirus confirmed cases and 163,097 confirmed death in 213 countries (https://www.who.int/emergencies/diseases/novel-coronavirus-2019).
For the maturation of CoVs, a highly complex cascade of proteolytic processing events is involved that control viral gene expression and replication. CoV main protease also (CoV Mpro) known as 3CL protease or 3CLpro, a three domain (domains I to III) cysteine protease with a chymotrypsin-like two-domain fold at the N terminus mediate most maturation cleavage events within the precursor polyprotein of coronavirus (Gorbalenya et al., 1989; Lee et al., 1991; Ziebuhr et al., 2000). From the structure of CoV Mpros it is clear that there are two CoV Mpro molecules form an active homodimer. One of these two is a Cys-His catalytic dyad which is located in a cleft between domains I and II and the other is N-terminal residues 1 to 7 (or N finger). These two Mpro are considered to play an important role in proteolytic activity (Anand et al., 2002, 2003; Xue et al., 2007; Yang et al., 2003). The C-terminal domain III is reported to be required for dimerization (Shi et al., 2004). The Mpro in SARS-CoV2 is the key enzyme processing, assembling and multiplication of the virus. Interrupting the active of this enzyme may disrupt the replication, multiplication of the virus. Thereby, disrupting its life cycle without causing any harm to the host, makes it an ideal target as the main inhibitor of SARS-CoV2. As Mpro possess active site for inhibitor, compound can interact at the active site of the enzyme, result in interfering the replication process of the enzyme. Due to interruption occur in replication and transcription process, the virus cannot multiply (Daidone et al., 2012; Ton et al., 2020; Wang, 2020)
Still, now no effective drugs are available in the market therefore it is the supreme need to work heart and soul to develop drugs for the treatment of coronavirus. Computer-aided drug design (CADD) can contribute a great help in this regard which is frequently used to estimate the probable inhibitors that could prevent the activity of an enzyme. The valuable time and cost can be reduced significantly by CADD in order to develop a new drug. The most important and crucial factor in CADD is the determination of the ligand-binding energy (Yu & MacKerell, 2017)
Method and materials
Protein preparation
The three dimensional structure of the main protease enzyme was retrieved from protein data bank (Berman et al., 2000) and subjected to preparation using the protein preparation wizard of Schrödinger suite (Sastry et al., 2013). All the waters were removed beyond 5.0 Å from het groups and the protein was pre-processed by; assigning bond orders, adding Hydrogens and creating disulfide bonds, converting selenomethionines to methionines whereby it is applicable. H-bond networks were optimized, and protonation states were generated at pH 7.0. Finally, The energy minimization process was done by employing OPLS3e force field (Roos et al., 2019).
Ligand preparation
About 1480 compounds were enlisted form plants through literature search as Supporting Information file 1 and 2. The compounds were selected on the basis of experimental evidence of different enzymatic and antiviral assay. The abundance and availability of the plants throughout the world especially south Asian region also considered while screening. The compounds were extracted from PubChem Database (Kim et al., 2016) and prepared by using default parameter of Ligprep module of Schrödinger suite (Schrödinger, 2020). Epik was used for generating the possible states of compounds at target pH 7.0 ± 2, and the high energy ionization/tautomer states were removed for the likelihood of reliability in biological condition (Riza et al., 2019; Shelley et al., 2007). Before, undergoing virtual screening, Qikprop simulation was run (Schrödinger, 2019), to filter ligands based on Lipinski’s rules (Lipinski, 2004).
Virtual screening
To identify potent inhibitor of main protease of SARS-CoV-2 virtual screening against natural compounds were done. The virtual screening workflow (VSW) of Glide program was employed for primary screening. Generally, the VSW is combined with three different docking scoring protocols such as high throughput virtual screening (HTVS), standard precision (SP) and extra precision (XP). The HTVS protocol docks each ligand to receptor and generates one pose, where it retains all the states of scoring, but the SP protocol is bit stricter at its scoring function, instead it retains only good scoring states (Friesner et al., 2004), facilitating to filter out the false positive results. However, the XP protocol uses very sophisticated procedure, and it only retains the best scoring states (Friesner et al., 2006; Repasky et al., 2007). In order to have the accuracy of pose ranking and final selectivity, the poses generated by XP was limited to three for each ligand, and those were allowed to be post-processed through MM-GBSA. The 50% of total compounds were passed from HTVS to SP. Thereafter, the 30% of total compounds from SP, were filtered to XP. Then, only 10% of compounds were kept before going for MM-GBSA rescoring (Chen et al., 2016; Mahmud et al., 2019). Finally, the best three ligands were selected and those were considered further, to observe their stability and dynamic nature in biological condition.
ADMET prediction
The pharmacological properties of the lead compounds were explored through pkCSM (Pires et al., 2015) and admetSAR (Cheng et al., 2012) server and QikProp (Jorgensen, 2016) to evaluate drug likeness properties. To design rational drugs, Lipinski rule of five (Lipinski, 2004) need to be assessed carefully and, therefore, SwissAdme server used to check Lipinski rules. The structure data file and simplified molecular data input format were used as an input system to evaluate absorption, distribution, metabolism, excretion and toxicity (ADMET) value.
Molecular dynamics simulation
Molecular dynamics simulation was carried out through YASARA software (Krieger et al., 2004). The drug-protein complex were initially cleaned, hydrogen bonding networks were optimized, and AMBER14 force field (Case et al., 2006) was applied. The Particle Mesh Ewald method was applied to calculate long range electrostatic potential with a distance of 8 Å. The physiological condition of the system was defined as (310 K, pH 7.4, 0.9% NaCl). The TIP3P (Transferable Intermolecular Potential 3 Points) was applied with NA/Cl ions and system density was 1.012 gm/cm3(Krieger et al., 2006). The Berendsen thermostat was used to control simulation temperature, where the pressure was kept constant during the simulation. By applying a multiple time step algorithm, the simulation time step was selected as 2.0 fs. Initially, the energy minimization of each system was performed by steepest gradient approach (5000 cycles; Krieger & Vriend, 2015). Finally, 100 ns of MD simulation was run, as where the snapshot interval was set to 100ps for analyzing the trajectory data. Therefore, all of the trajectories were concatenated to calculate and plot root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg) and solvent accessible surface area (SASA) data.
Principle component analysis (PCA)
The PCA has been performed to understand variation of conformational changing upon ligand binding. We performed PCA in two different methods; one is utilizing trajectory coordinates to understand the transition sate before and after ligand binding, while another one is applied, to all the structures related variables for understating the quality of conformers and overall variation among protein-ligand complexes, compared to apo and control drug-protein complex. We used Bio3d program, implementing into R program and pc man package for PCA.
Result
Molecular docking analysis
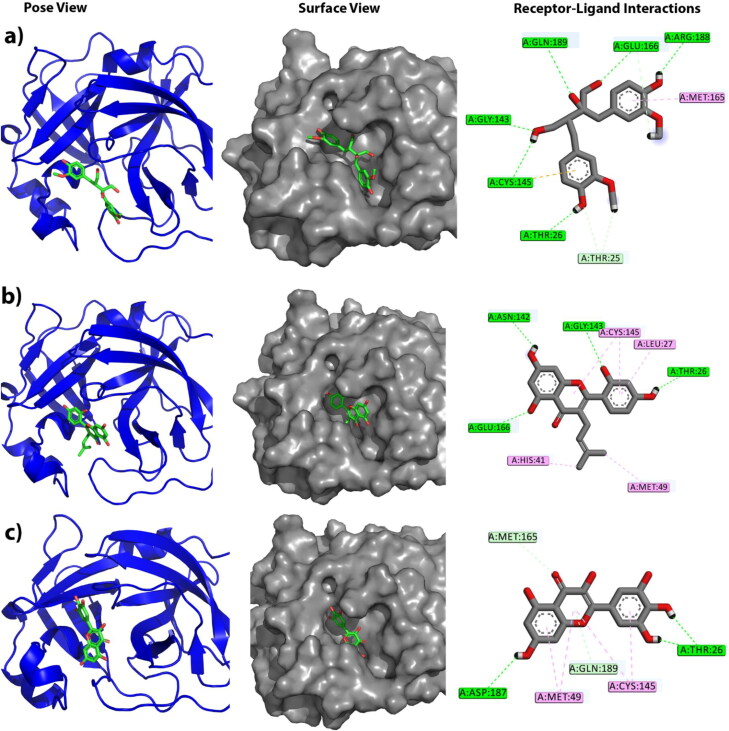
After the competition of virtual screening, about 14 compounds were found to be potential, as they were passed through rigorous screening process and found to interact with the catalytic dyad of the main protease of SARS-CoV-2. However, all the poses of compounds generated by XP was further processed with MM-GBSA, to reach more accurate approximation. Therefore, Top 3 candidates, which showed the best binding affinity (ΔG Bind) with the catalytic dyad of main protease, were chosen. The docking result and binding affinity estimation of 14 compounds are shown in the Table 1, and the interaction details of top three compounds with the active site residues are shown in Table 2. Figure 1 illustrates different bunding modes of protein-ligand complexes.
Table 1.
Docking result (kcal/mol) and binding affinity (kcal/mol) estimation of top 14 candidates.
| PubChem CID | Glide ligand efficiency | XP GScore | Glide evdw | Glide ecoul | Glide energy | Glide emodel | MMGBSA ΔG bind |
|---|---|---|---|---|---|---|---|
| 5280343 | –0.404 | –8.916 | –33.982 | –8.344 | –42.327 | –55.652 | –40.39 |
| 586373 | –0.314 | –8.476 | –33.129 | –12.172 | –45.301 | –59.218 | –46.37 |
| 5281672 | –0.365 | –8.439 | –36.917 | –7.311 | –44.228 | –54.131 | –46.96 |
| 5280681 | –0.354 | –8.178 | –34.079 | –9.163 | –43.243 | –62.916 | –42.02 |
| 5481961 | –0.308 | –8.036 | –35.007 | –10.879 | –45.886 | –60.092 | –49.66 |
| 6476139 | –0.303 | –7.891 | –31.452 | –11.422 | –42.874 | –51.915 | –45.86 |
| 57161864 | –0.464 | –7.996 | –18.869 | –10.685 | –29.555 | –36.942 | –26.61 |
| 439533 | –0.356 | –7.861 | –36.554 | –7.974 | –44.528 | –53.124 | –41.94 |
| 25243950 | –0.32 | –7.678 | –30.95 | –12.212 | –43.161 | –52.839 | –37.1 |
| 5281670 | –0.345 | –7.852 | –34.803 | –5.631 | –40.434 | –52.901 | –34.34 |
| 44258704 | –0.316 | –7.586 | –32.402 | –7.714 | –40.115 | –51.303 | –46.07 |
| 5281855 | –0.341 | –7.565 | –31.108 | –6.149 | –37.257 | –49.183 | –42.26 |
| 5280863 | –0.356 | –7.505 | –33.638 | –7.44 | –41.078 | –52.278 | –38.73 |
| 5281605 | –0.373 | –7.505 | –26.253 | –7.789 | –34.042 | –48.068 | –41.86 |
Table 2.
Non bond interaction between SARS-CoV-2 main protease and top 3 compound.
| PubChem CID | Residues in contact | Interaction type | Distance in Å |
|---|---|---|---|
| 5281672 | ASP187 | Conventional Hydrogen Bond | 2.96 |
| THR26 | Conventional Hydrogen Bond | 1.98 | |
| THR26 | Conventional Hydrogen Bond | 1.91 | |
| MET165 | Carbon Hydrogen Bond | 2.75 | |
| GLN189 | Pi-Donor Hydrogen Bond | 3.2 | |
| MET49 | Pi-alkyl | 5.2 | |
| CYS145 | Pi-alkyl | 5.22 | |
| MET49 | Pi-alkyl | 4.39 | |
| CYS145 | Pi-alkyl | 4.77 | |
| 5481961 | GLY143 | Conventional hydrogen bond | 2.12 |
| GLU166 | Conventional hydrogen bond | 2.33 | |
| ASN142 | Conventional hydrogen bond | 1.98 | |
| THR26 | Conventional hydrogen bond | 1.76 | |
| MET49 | Alkyl | 3.81 | |
| HIS41 | Pi-alkyl | 3.91 | |
| CYS145 | Pi-alkyl | 5.27 | |
| LEU27 | Pi-alkyl | 5.45 | |
| CYS145 | Pi-alkyl | 4.26 | |
| 586373 | GLY143 | Conventional hydrogen bond | 2.37 |
| GLU166 | Conventional hydrogen bond | 2.17 | |
| GLN189 | Conventional hydrogen bond | 3.02 | |
| CYS145 | Conventional hydrogen bond | 2.54 | |
| ARG188 | Conventional hydrogen bond | 2.24 | |
| THR26 | Conventional hydrogen bond | 1.86 | |
| THR25 | Carbon hydrogen bond | 2.99 | |
| GLU166 | Carbon hydrogen bond | 2.54 | |
| THR25 | Carbon hydrogen bond | 2.62 | |
| CYS145 | Pi-sulfur | 5.33 | |
| MET165 | Pi-alkyl | 4.23 |
Figure 1.
The figure illustrates different binding modes of selected compounds within the active and catalytic sites of main protease. The alphabetical orders indicate the respective complex of alpha-ketoamide, Carinol, Albanin and Myricetin, respectively. The block and line colors at receptor-ligand interactions such as green, light sky and pink define conventional hydrogen bonding, C-H bonding and hydrophobic interactions, respectively.
ADMET
To be a potent drug candidate against biological target, several parameters such as Lipinski rule of five; molecular weight, hydrogen bond donor, hydrogen bond acceptor, must be followed (Mahmud et al., 2019). The screened main protease inhibitors have better oral absorption and blood brain barrier permeability which is required for a drug candidate. All three compound have no carcinogenic profile and no toxicity found in hepatotoxicity and AMES toxicity evaluation. ATP binding cassette transporter, P-glycoprotein act as a biological barrier by extruding foreign particles from the cell. From the Table 3, it was observed that all screened compound derived from virtual screening inhibit p-glycoprotein.
Table 3.
Pharmacological properties of the screened compound assessed through admetSAR and PkCSM tools.
| Parameter | Carinol | Myricetin | Albanin |
|---|---|---|---|
| CNS | –2 | –2 | –2 |
| MW | 378.421 | 318.29 | 354.359 |
| SASA | 643.934 | 530.521 | 602.242 |
| Donor HB | 5 | 5 | 3 |
| Acceptor HB | 7.25 | 6 | 4.5 |
| QPlog Poct | 22.094 | 20.824 | 19.224 |
| LogHERG | –4.594 | –5.008 | –5.152 |
| Human Oral absorption | 77.046 | 26.816 | 75.848 |
| Caco2 permeability | –0.174 | 0.095 | –0.449 |
| P-glycoprotein inhibitor | No | No | No |
| BBB permeability | –0.1489 | –1.493 | –1.034 |
| Hepatotoxicity | No | No | No |
| Carcinogenicity | 0.8714(–) | 1.00(–) | 0.957(–) |
| AMES toxicity | No | No | No |
Here, MW: molecular weight; SASA: solvent accessible surface area; CNS: central nervous system activity; LogHERG: predicted Ic50 value for blockage of HERG K+ channels; LogPo/w: predicted octanol/water partition coefficient.
Molecular dynamics
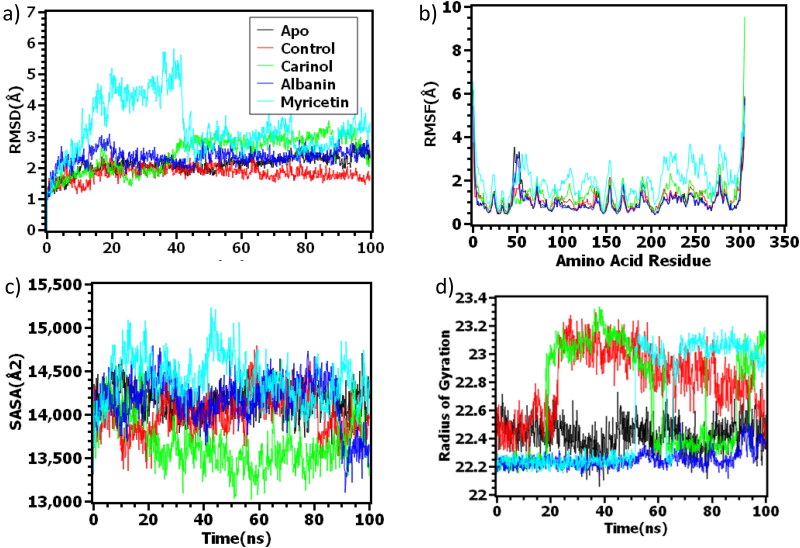
The RMSD profile of all the docked complexes were measured to understand their stability in biological environment. Furthermore, Apo and the control co-crystalized structures were simulated for better contrast, on how the newly proposed inhibitors might affect protein structure.
From Figure 2(a), it is observed that alpha ketoamide and its protein complex shows a bit lower RMSD profile than apo structure. Moreover, Carinol-Mpro complex had similar trend like apo till 10 ns and thereafter slightly flexibility observed till 25 ns and stabilized consequently.
Figure 2.
Time series analysis of all the simulated systems. The alphabetical orders from (a) to (d) indicate RMSD analysis of alpha carbon atoms (a), flexibility analysis of amino acid residue (b), protein volume with expansion analysis(c) and degree of rigidity and compactness analysis (d).
Although Albanin-Mpro complex was seen to have flexible nature at the initial phase of dynamics, it reached at stable state at 20 ns. This complex had almost similar trend as apo structure for the next phase of the simulation. In addition, Myricetin-Mpro complex had higher RMSD value from the beginning of simulation and it was reached at highest peak at 40 ns. Soon this complex reached in stable condition with very lower degree of fluctuations during the rest of the simulation time.
The RMSF value of the protein complex represents fluctuated protein region during molecular dynamics simulation. As seen from Figure 2(b), the overall residues; Ser46, Glu47, Leu50, Pro52 from beta turn region, Asn151, Ile152 from B strand and Asp153, Tyr154, Asp155 from beta hairpin region, Met276, Asn277, Gly278, Arg279, Thr280, Ser301, Gly302, Val303, Thr304, Phe305, Gln306 from beta turn region, had high level of fluctuations compared to the other amino acid residues. However, apo protein structure had less fluctuation than control which indicates that the main protease protein structure of SARS-CoV-2 protein is less flexible without ligand. The RMSFs of the Albanin-Mpro complex and Carinol-Mpro complex had similar drift as apo and control one, as where the Myricetin-Mpro complex had higher RMSF value for almost all the amino acid residues. This behavior indicates, Myricetin-Mpro complex had flexible nature compared to all other complexes.
Moreover, SASA was checked to understand the change in protein surface area. From Figure 2(c), it can be understood that, the ligand Myricetin-Mpro complex had shrunken its surface area than apo and control structure. Additionally, the surface area of apo protein, control protein and Albanin-Mpro complex did not change too much and remained at stable state during MD simulation. On the other hand, the Myricetin-Mpro complex was seen to have little increased its surface area from 0 to 40 ns, then it got down a bit. This complex had almost similar trend for rest of the simulation time and did not deviate too much.
On top of that, the degree of compactness and stability was evaluated through Rg. It can be observed from the Figure 2(d) that, apo structure had almost similar Rg value from 0 to 100 ns, which indicates that the protein structure almost stable. However, Albanin-Mpro complex had lower Rg value than apo structure, which means the higher level of rigidity. On the other hand, Myricetin-Mpro complex and Carinol-Mpro complex had similar profile like control protein complex.
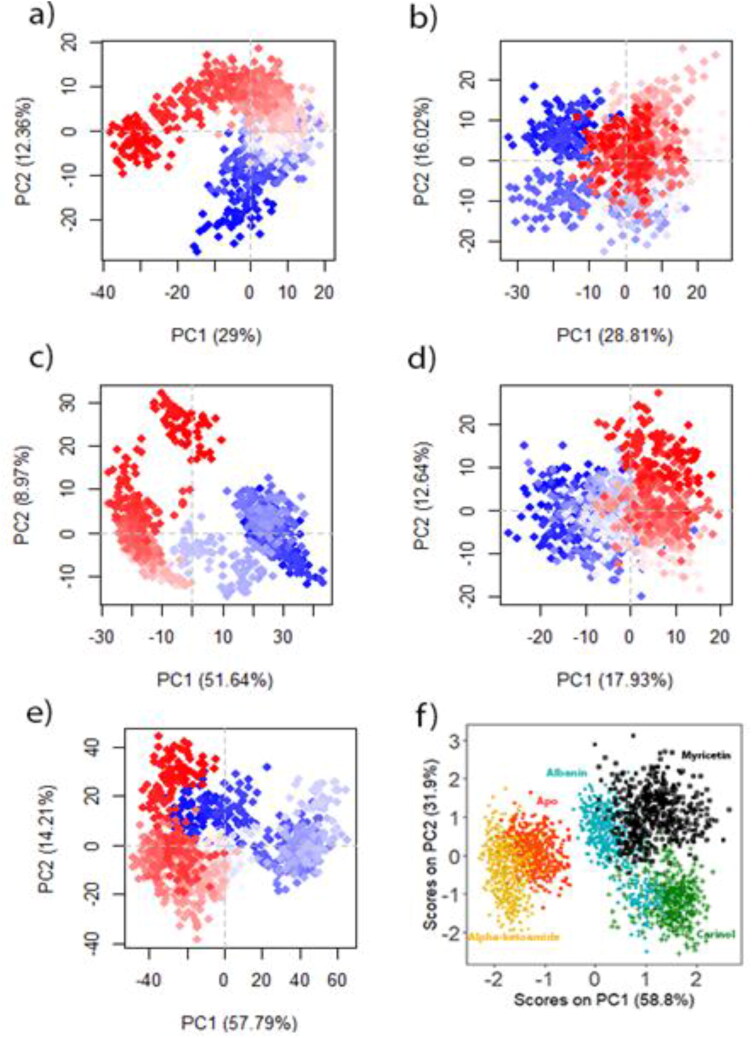
Principle component analysis
PCA is an essential approach for projecting multi-dimensional data into 2D and 3D space as well as understanding the variation of given sets of data. Figure 3 projects PCA from all trajectories including coordinates and structurally relevant data such as: RMSD, Rg and SASA. Note that, we could perform PCA for alpha carbon atoms only, throughout the trajectories, but we also considered data as aforementioned to see, if they would have any correlation among themselves.
Figure 3.
Principle component analysis of (a) Apo, (b) Alpha ketoamide-Mpro complex, (c) Carinol-Mpro complex, (d) Albanin-Mpro complex and (e) Myricetin-Mpro complex. Each dot denotes its conformation of the protein throughout the X and Y axis. The spread of blue and red color dots described the degree of conformational changes in the simulation, where the color spectrum from blue to white to red is equivalent to simulation time. The blue specifies the initial timestep, white specifies intermediate, and the final timestep is represented by red color. (f) PCA of trajectory data (RMSD, Rg and SASA) of all systems.
It is clear from PCA analysis (Figure 3) that the separation the of most conformers of Apo shift positively with reduced variance as 29%, and the separation of Alpha ketoamide-Mpro complex seems to be highly distributed around, indicating its conformation stability throughout the trajectory. Abd it achieved higher scores on PC1 (51.64%). Conversely, the conformational distribution of Carinol-Mpro complex, is seemed to shift positive, as compared to Apo structure. On the other hand, the conformational distributions of Albanin-Mpro complex is seemed to be bit similar to that of Alpha ketoamide- Mpro complex. The most PCA1 score (57.79%) was seemed to be covered by Myricetin-Mpro complex, where the conformational distribution look like more positive directions as seen for Apo and Carinol-Mpro complex.
Finally, PCA of few trajectories’ data was analyzed, to grasp structural properties of all systems. The data shows the distribution of Albanin complex is near to Apo, while the other complexes are seemed overlap a bit onto each other. The overlapping confers the similar conformational states resembling to their dynamic properties and behavior.
Discussion
Computer aided drug design and virtual screening has become essential tool to identify new lead compound. This combinatorial process allows us to reduce experimental time and cost by narrowing down biological target. Additionally, molecular dynamics, molecular docking, virtual screening have become essential part in computer aided drug design for their reliability and accurate prediction probability (Mahmud et al., 2019; Talele et al., 2010). Recently, number of virtual screening process based on plant derived compound have been successful to predict the potential blocker of the biological target (Mahmud et al., 2019, 2020).
The main protease of SARS-CoV-2 has become an attractive target for different therapeutic approaches. It is comprised of three domains; Domain I (residues 10 to 99), Domain II (residues 100 to 182) and Domain III (residues 198 to 303; Bacha et al., 2004; Shi & Song, 2006; Zhang et al., 2020 ). Among three domains, two catalytic residues such as Cys145 and His41, are reported to initiate activation through dimerization mechanism. Thus, blocking the catalytic site can be logical to inhibit the function of the main protease (Zhang et al., 2020). Beside this, the substrate-binding pocket consists of His41, Phe140, Asn142, Gly143, Ser144, Cys145, Tyr161, His163, Glu166 and His172 residues which can be alternatively targeted to inhibit the activity of main protease (Khan, Jha, et al., 2020). Alongside the catalytic center, there are two other deeply buried subsites (S1 & S2) and three shallow subsites(S3-S5), where the S1 include His163, Glu166, Cys145, Gly143, His172, Phe140 amino acid residues and S2 include Cys145, His41 and Thr25 amino acid residues; mainly involved in hydrophobic and electrostatic interactions. The Shallow subsites consist of Met49, His41, Met165, Glu166 and Gln189 amino acid residues; can endure different functionalities (Khan, Zia, et al., 2020).
Our study suggests that, the best three candidates interacted with both or at least one catalytic residue. The compound 5281672 (PubChem CID) or Myricetin was stabilized by several hydrogen bonds and hydrophobic bonds, in which it formed two hydrophobic bonds with catalytic residue Cys145, having the distance of 5.22 Å and 4.77 Å, along with two hydrophobic bonds with the shallow subsite MET49, having the distance of 5.2 Å and 4.39 Å. The compound also formed several hydrogen bonds with the shallow subsite Met165 and Gln189 at the distance of 2.75 Å, and 3.20 Å. Furthermore, it formed three hydrogen bonds with Asp187 at the distance of 2.96 Å, Thr26 at the distance of 1.98 and 1.91 Å, respectively (Khan, Zia, et al., 2020; Pant et al., 2020).
Besides, the compound 5481961 (PubChem CID) or Albanin was also stabilized by one hydrophobic bond with catalytic residue His41 at a distance of 3.91 Å and two hydrophobic bonds with catalytic residue Cys145 at the distance of 5.27 Å and 4.26 Å. More importantly, it also formed one hydrophobic bond with the shallow subsite Met49, at the distance 3.81 Å, one hydrogen bond with the buried subsite residue Gly143 at the distance of 2.12 Å, and one hydrophobic bond with both shallow and buried subsite Glu166 at the distance of 2.33 Å. Interestingly, the interaction with Glu166 could satisfy our target, since it acts as a key residue in dimerization (Zhang et al., 2020). Furthermore, it also formed two important hydrogen bonds with Asn142 at 1.98 Å, and Thr26 at 1.76 Å. Additionally, one hydrophobic bonds with Leu27 having the distance 5.45 Å (Khan, Zia, et al., 2020).
The interaction with the compound 586373 (PubChem CID) or Carinol was also studied, which was found to be stabilized by forming one hydrogen bond with catalytic residue Cys145 at 2.54 Å, and one pi-sulfur interaction with catalytic residue Cys145 at 5.33 Å. Apart from that, it forms one hydrophobic bond with the shallow subsite Met165, at 4.23 Å and one hydrogen bond with the shallow subsite Gln189, at 3.02 Å. Interestingly, it was also found to interact with the buried subsite Thr25 at 2.62 Å and 2.99 Å. Importantly, it forms two hydrogen bonds with buried subsite residues Glu166 at 2.54 Å and 2.17 Å. However, several important hydrogen bonds were seen to be formed with Thr26 at 2.62 Å, and ARG188 at 2.24 Å, respectively (Khan, Zia, et al., 2020; Pant et al., 2020).
After the development in late 1970s, molecular dynamics simulation suppress the limitation of computational prediction based on Newtonian physics (George Priya Doss et al., 2014;; McCammon et al., 1977; Priya Doss et al., 2014). Research from previous studies suggest that, combinatorial dynamics and docking approaches can be employed to get structural information as well as impact of protein structure stability upon ligand binding (Dasgupta et al., 2003; Nagasundaram & George Priya Doss, 2013). Also, different studies give indications that concordance between experimental and computational studies (Abu-Aisheh et al., 2019; Chandra Babu et al., 2017). However, we run molecular dynamics simulation for both of Apo and ligand bound complexes to understand the structural change of the protein upon ligand binding. The molecular dynamics study revealed that in Figure 2, the RMSD of the protein complex stabilized after ligand binding. However, from figure. It was also observed that RMSF, SASA, Rg changed after ligand attached to the protein structure as conformational change due to ligand attaching to the protein might happen (Gilis & Rooman, 1996; Mahmud et al., 2019; Villanueva et al., 2004). These computational calculation and data may offer necessary information to design rational drug candidates against main protease of SARS-CoV-2.
Conclusion
In summary, we tried to evaluate phytochemicals derived from plants by targeting the main protease of SARS-CoV-2. The binding information and interaction nature were found to be suitable for obstructing the target main protease. Moreover, the phytochemicals derived from virtual screening found to be non-toxic and appropriate pharmacological properties. Therefore, binding conformation and stability were confirmed with the aid of molecular dynamics simulation. Therefore, these outcomes especially came from computational based drug designing process, hence further assessment and validation is required from the wet lab to find out effective and better treatment against Covid-19.
Supplementary Material
Acknowledgment
We express our gratitude and thanks to Allah, our parents, teachers and friends.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abu-Aisheh, M. N., Al-Aboudi, A., Mustafa, M. S., El-Abadelah, M. M., Ali, S. Y., Ul-Haq, Z., & Mubarak, M. S. (2019). Coumarin derivatives as acetyl- and butyrylcholinestrase inhibitors: An in vitro, molecular docking, and molecular dynamics simulations study. Heliyon, 5(4), e01552. 10.1016/j.heliyon.2019.e01552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, K., Palm, G. J., Mesters, J. R., Siddell, S. G., Ziebuhr, J., & Hilgenfeld, R. (2002). Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. The EMBO Journal, 21(13), 3213–3224. 10.1093/emboj/cdf327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science (New York, N.Y.), 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Bacha, U., Barrila, J., Velazquez-Campoy, A., Leavitt, S. A., & Freire, E. (2004). Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry, 43(17), 4906–4912. 10.1021/bi0361766 [DOI] [PubMed] [Google Scholar]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The protein data bank. Nucleic acids research, 28(1), 235–242. [DOI] [PMC free article] [PubMed]
- Case, D. A., Darden, T. A., Cheatham III, T. E., Simmerling, C. L., Wang, J., Duke, R. E., Luo, R., Merz, K. M., Pearlman, D. A., Crowley, M., & Walker, R. (2006). AMBER 9.0 software package.
- Chandra Babu, T. M., Rajesh, S. S., Bhaskar, B. V., Devi, S., Rammohan, A., Sivaraman, T., & Rajendra, W. (2017). Molecular docking, molecular dynamics simulation, biological evaluation and 2D QSAR analysis of flavonoids from Syzygium alternifolium as potent anti-Helicobacter pylori agents. RSC Advances, 7(30), 18277–18292. 10.1039/C6RA27872H [DOI] [Google Scholar]
- Chen, F., Liu, H., Sun, H., Pan, P., Li, Y., Li, D., & Hou, T. (2016). Assessing the performance of the MM/PBSA and MM/GBSA methods. 6. Capability to predict protein-protein binding free energies and re-rank binding poses generated by protein-protein docking. Physical Chemistry Chemical Physics: PCCP, 18(32), 22129–22139. 10.1039/c6cp03670h [DOI] [PubMed] [Google Scholar]
- Cheng, F., Li, W., Zhou, Y., Shen, J., Wu, Z., Liu, G., Lee, P. W., & Tang, Y. (2012). AdmetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. Journal of Chemical Information and Modeling, 52(11), 3099–3105. 10.1021/ci300367a [DOI] [PubMed] [Google Scholar]
- Daidone, F., Montioli, R., Paiardini, A., Cellini, B., Macchiarulo, A., Giardina, G., Bossa, F., & Borri Voltattorni, C. (2012). Identification by virtual screening and in vitro testing of human DOPA decarboxylase inhibitors. PLoS One, 7(2), e31610. 10.1371/journal.pone.0031610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta, J., Sen, U., & Dattagupta, J. K. (2003). In silico mutations and molecular dynamics studies on a winged bean chymotrypsin inhibitor protein. Protein Engineering, 16(7), 489–496. 10.1093/protein/gzg070 [DOI] [PubMed] [Google Scholar]
- de Wilde, A. H., Snijder, E. J., Kikkert, M., & van Hemert, M. J. (2018). Host factors in coronavirus replication. Current Topics in Microbiology and Immunology, 419, 1–42. 10.1007/82_2017_25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner, R. A., Banks, J. L., Murphy, R. B., Halgren, T. A., Klicic, J. J., Mainz, D. T., Repasky, M. P., Knoll, E. H., Shelley, M., Perry, J. K., Shaw, D. E., Francis, P., & Shenkin, P. S. (2004). Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry, 47(7), 1739–1749. 10.1021/jm0306430 [DOI] [PubMed] [Google Scholar]
- Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A., Sanschagrin, P. C., & Mainz, D. T. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. Journal of Medicinal Chemistry, 49(21), 6177–6196. 10.1021/jm051256o [DOI] [PubMed] [Google Scholar]
- George Priya Doss, C., Rajith, B., Chakraboty, C., Balaji, V., Magesh, R., Gowthami, B., Menon, S., Swati, M., Trivedi, M., Paul, J., Vasan, R., & Das, M. (2014). In silico profiling and structural insights of missense mutations in RET protein kinase domain by molecular dynamics and docking approach. Molecular Biosystems, 10(3), 421–436. 10.1039/c3mb70427k [DOI] [PubMed] [Google Scholar]
- Gilis, D., & Rooman, M. (1996). Stability changes upon mutation of solvent-accessible residues in proteins evaluated by database-derived potentials. Journal of Molecular Biology, 257(5), 1112–1126. 10.1006/jmbi.1996.0226 [DOI] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Koonin, E. V., Donchenko, A. P., & Blinov, V. M. (1989). Coronavirus genome: Prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Research, 17(12), 4847–4861. 10.1093/nar/17.12.4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y., Zheng, B. J., He, Y. Q., Liu, X. L., Zhuang, Z. X., Cheung, C. L., Luo, S. W., Li, P. H., Zhang, L. J., Guan, Y. J., Butt, K. M., Wong, K. L., Chan, K. W., Lim, W., Shortridge, K. F., Yuen, K. Y., Peiris, J. S. M., & Poon, L. L. M. (2003). Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science (New York, N.Y.), 302(5643), 276–278. 10.1126/science.1087139 [DOI] [PubMed] [Google Scholar]
- Jorgensen, W. L. (2016). QikProp. Schrödinger LLC. [Google Scholar]
- Khan, R. J., Jha, R. , Amera, G. M., Jain, M., Singh, E., Pathak, A., & Singh, A. (2020). Targeting novel coronavirus 2019: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2’-O-ribose methyltransferase. Journal of Biomolecular Structure & Dynamics, (2), 1–40. 10.26434/chemrxiv.11888730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, S. A., Zia, K., Ashraf, S., Uddin, R., & Ul-Haq, Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure & Dynamics, 1–13. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Kim, S., Thiessen, P. A., Bolton, E. E., Chen, J., Fu, G., Gindulyte, A., Han, L., He, J., He, S., Shoemaker, B. A., Wang, J., Yu, B., Zhang, J., & Bryant, S. H. (2016). PubChem substance and compound databases. Nucleic Acids Research, 44(D1), D1202–D1213. 10.1093/nar/gkv951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, E., & Vriend, G. (2015). New ways to boost molecular dynamics simulations. Journal of Computational Chemistry, 36(13), 996–1007. 10.1002/jcc.23899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, E., Darden, T., Nabuurs, S. B., Finkelstein, A., & Vriend, G. (2004). Making optimal use of empirical energy functions: Force‐field parameterization in crystal space. Proteins: Structure, Function, and Bioinformatics, 57(4), 678–683. 10.1002/prot.20251 [DOI] [PubMed] [Google Scholar]
- Krieger, E., Nielsen, J. E., Spronk, C. A. E. M., & Vriend, G. (2006). Fast empirical pKa prediction by Ewald summation. Journal of Molecular Graphics & Modelling, 25(4), 481–486. 10.1016/j.jmgm.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Lau, S. K. P., Woo, P. C. Y., Li, K. S. M., Huang, Y., Tsoi, H.-W., Wong, B. H. L., Wong, S. S. Y., Leung, S.-Y., Chan, K.-H., & Yuen, K.-Y. (2005). Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proceedings of the National Academy of Sciences of the United States of America, 102(39), 14040–14045. 10.1073/pnas.0506735102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. J., Shieh, C. K., Gorbalenya, A. E., Koonin, E. V., La Monica, N., Tuler, J., Bagdzhadzhyan, A., & Lai, M. M. (1991). The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology, 180(2), 567–582. 10.1016/0042-6822(91)90071-I [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski, C. A. (2004). Lead- and drug-like compounds: The rule-of-five revolution. Drug Discovery Today: Technologies, 1(4), 337–341. 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Mahmud, S., Parves, M. R., Riza, Y. M., Sujon, K. M., Ray, S., Tithi, F. A., Zaoti, Z. F., Alam, S., & Absar, N. (2019). Exploring the potent inhibitors and binding modes of phospholipase A2 through in silico investigation. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2019.1680440 [DOI] [PubMed] [Google Scholar]
- Mahmud, S., Rahman, E., Nain, Z., Billah, M., Karmokar, S., Mohanto, S. C., Paul, G. K., Amin, A., Acharjee, U. K., & Saleh, M. A. (2020). Computational discovery of plant-based inhibitors against human carbonic anhydrase IX and molecular dynamics simulation. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1753579 [DOI] [PubMed] [Google Scholar]
- McCammon, J. A., Gelin, B. R., & Karplus, M. (1977). Dynamics of folded proteins. Nature, 267(5612), 585–590. 10.1038/267585a0 [DOI] [PubMed] [Google Scholar]
- Nagasundaram, N., & George Priya Doss, C. (2013). Predicting the impact of single-nucleotide polymorphisms in CDK2-flavopiridol complex by molecular dynamics analysis. Cell Biochemistry and Biophysics, 66(3), 681–695. 10.1007/s12013-012-9512-5 [DOI] [PubMed] [Google Scholar]
- Pant, S., Singh, M., Ravichandiran, V., Murty, U. S. N., & Srivastava, H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure & Dynamics, 1–15. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires, D. E. V., Blundell, T. L., & Ascher, D. B. (2015). pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of Medicinal Chemistry, 58(9), 4066–4072. 10.1021/acs.jmedchem.5b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priya Doss, C. G., Chakraborty, C., Chen, L., & Zhu, H. (2014). Integrating in silico prediction methods, molecular docking, and molecular dynamics simulation to predict the impact of ALK missense mutations in structural perspective. BioMed Research International, 2014, 895831. 10.1155/2014/895831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repasky, M. P., Shelley, M., & Friesner, R. A. (2007). Flexible ligand docking with glide. Current Protocols in Bioinformatics, 18(1), 8–12. 10.1002/0471250953.bi0812s18 [DOI] [PubMed] [Google Scholar]
- Riza, Y. M., Parves, M. R., Tithi, F. A., & Alam, S. (2019). Quantum chemical calculation and binding modes of H1R; a combined study of molecular docking and DFT for suggesting therapeutically potent H1R antagonist. In Silico Pharmacology, 7(1), 1. 10.1007/s40203-019-0050-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos, K., Wu, C., Damm, W., Reboul, M., Stevenson, J. M., Lu, C., Dahlgren, M. K., Mondal, S., Chen, W., Wang, L., Abel, R., Friesner, R. A., & Harder, E. D. (2019). OPLS3e: Extending force field coverage for drug-like small molecules. Journal of Chemical Theory and Computation, 15(3), 1863–1874. 10.1021/acs.jctc.8b01026 [DOI] [PubMed] [Google Scholar]
- Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R., & Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design, 27(3), 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Schrödinger . (2019). QikProp. Schrödinger LLC. [Google Scholar]
- Schrödinger . (2020). LigPrep. Schrödinger, LLC, 2018–4. [Google Scholar]
- Shelley, J. C., Cholleti, A., Frye, L. L., Greenwood, J. R., Timlin, M. R., & Uchimaya, M. (2007). Epik: A software program for pK( a ) prediction and protonation state generation for drug-like molecules. Journal of Computer-Aided Molecular Design, 21(12), 681–691. 10.1007/s10822-007-9133-z [DOI] [PubMed] [Google Scholar]
- Shi, J., & Song, J. (2006). The catalysis of the SARS 3C-like protease is under extensive regulation by its extra domain. The FEBS Journal, 273(5), 1035–1045. 10.1111/j.1742-4658.2006.05130.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Wei, Z., & Song, J. (2004). Dissection study on the severe acute respiratory syndrome 3C-like protease reveals the critical role of the extra domain in dimerization of the enzyme: defining the extra domain as a new target for design of highly specific protease inhibitors. The Journal of Biological Chemistry, 279(23), 24765–24773. 10.1074/jbc.M311744200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talele, T., Khedkar, S., & Rigby, A. (2010). Successful applications of computer aided drug discovery: Moving drugs from concept to the clinic. Current Topics in Medicinal Chemistry, 10(1), 127–141. 10.2174/156802610790232251 [DOI] [PubMed] [Google Scholar]
- Ton, A. T., Gentile, F., Hsing, M., Ban, F., & Cherkasov, A. (2020). Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Molecular Informatics, 39(2020), 2000028. 10.1002/minf.202000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva, J., Hoshino, M., Katou, H., Kardos, J., Hasegawa, K., Naiki, H., & Goto, Y. (2004). Increase in the conformational flexibility of beta 2-microglobulin upon copper binding: A possible role for copper in dialysis-related amyloidosis. Protein Science: A Publication of the Protein Society, 13(3), 797–809. 10.1110/ps.03445704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. (2020). Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. Journal of Chemical Information and Modeling, 60(6), 3277–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C., & Zhang, Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, X., Yang, H., Shen, W., Zhao, Q., Li, J., Yang, K., Chen, C., Jin, Y., Bartlam, M., & Rao, Z. (2007). Production of Authentic SARS-CoV M(pro) with enhanced activity: Application as a novel tag-cleavage endopeptidase for protein overproduction. Journal of Molecular Biology, 366(3), 965–975. 10.1016/j.jmb.2006.11.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Yang, M., Ding, Y., Liu, Y., Lou, Z., Zhou, Z., Sun, L., Mo, L., Ye, S., Pang, H., Gao, G. F., Anand, K., Bartlam, M., Hilgenfeld, R., & Rao, Z. (2003). The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proceedings of the National Academy of Sciences of the United States of America, 100(23), 13190–13195. 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W., & MacKerell, A. D. (2017). Computer-aided drug design methods. Methods in Molecular Biology, 1520, 85–106. 10.1007/978-1-4939-6634-9_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr, J., Snijder, E. J., & Gorbalenya, A. E. (2000). Virus-encoded proteinases and proteolytic processing in the Nidovirales. The Journal of General Virology, 81(Pt 4), 853–879. 10.1099/0022-1317-81-4-853 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.