Abstract
The entire human population over the globe is currently facing appalling conditions due to the spread of infection from coronavirus disease-2019 (COVID-19). The spike glycoprotein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) present on the surface of the virion mediates the virus entry into the host cells and therefore is targeted by several scientific groups as a novel drug target site. The spike glycoprotein binds to the human angiotensin-converting enzyme-2 (hACE2) cell surface receptor abundantly expressed in lung tissues, and this binding phenomenon is a primary determinant of cell tropism and pathogenesis. The binding and internalization of the virus is the primary and most crucial step in the process of infection, and therefore the molecules targeting the inhibition of this process certainly hold a significant therapeutic value. Thus, we systematically applied the computational techniques to identify the plausible inhibitor from a chosen set of well characterized diaryl pyrimidine analogues which may disrupt interfacial interaction of spike glycoprotein (S) at the surface of hACE2. Using molecular docking, molecular dynamics (MD) simulation and binding free energy calculation, we have identified AP-NP (2-(2-amino-5-(naphthalen-2-yl)pyrimidin-4-yl)phenol), AP-3-OMe-Ph (2-(2-amino-5-(3-methoxyphenyl)pyrimidin-4-yl)phenol) and AP-4-Me-Ph (2-(2-amino-5-(p-tolyl) pyrimidin-4-yl)phenol) from a group of diaryl pyrimidine derivatives which appears to bind at the interface of the hACE2-S complex with low binding free energy. Thus, pyrimidine derivative AP-NP may be explored as an effective inhibitor for hACE2-S complex. Furthermore, in vitro and in vivo studies will strengthen the use of these inhibitors as suitable drug candidates against SARS-COV-2.
Communicated by Ramaswamy H. Sarma
Keywords: hACE2, receptor, coronavirus, pyrimidine derivatives, binding site
1. Introduction
The world is currently going through a debilitating phase of acute health disaster attributed to the global pandemic brought about by the novel coronavirus disease-2019 (COVID-19) (Enayatkhani et al., 2020; Joshi et al., 2020; Kirchdoerfer et al., 2016; Muralidharan et al., 2020). Sequencing and simultaneous phylogenetic identification of the virus responsible for COVID-19 confirmed that it as a novel β-coronavirus that shared 88% sequence identity with two bat-derived SARS-like coronaviruses (Lu et al., 2020;; Pant et al., 2020; Wang et al., 2020). Additionally, it was shown that this coronavirus (CoV), termed as 2019-nCoV (Elfiky, 2020; Gorbalenya et al., 2020), shared 79.5% sequence identity with SARS-CoV (Elfiky & Azzam, 2020; Lu et al., 2020;; Wang et al., 2020; Xia et al., 2020) which caused the severe acute respiratory syndrome pandemic in 2012. Therefore, this newly identified virus was called as SARS-CoV-2 (Aanouz et al., 2020). The virus gains entry into the host cells via the transmembrane spike (S) glycoprotein that forms a homotrimeric structure extending outwards from the viral surface (Belouzard et al., 2012; White et al., 2008). The monomeric S protein is comprised of two distinct functional domains S1 and S2. The S1 domain is responsible for binding to the host cell receptor, while S2 domain helps in the fusion of the viral and cellular membranes (Jitendra Subhash et al., 2020; Kirchdoerfer et al., 2016). The S protein is cleaved at the boundary between the S1 and S2 subunits thereby adopting a prefusion conformation that remains bound non-covalently to the surface (Walls et al., 2020). The S1 protein subunit contains a unique receptor-binding domain (RBD) in its C-terminal region, which contributes towards the stabilization of the prefusion state of the membrane-anchored S2 subunit containing the fusion machinery (Shang et al., 2020; Walls et al., 2020). Transmembrane protease serine 2 (TMPRSS-2) further cleaves the S protein at a specific site located immediately upstream of the fusion peptide (Elmezayen et al., 2020; Shen et al., 2017). This site specific cleavage induces a set of irreversible conformational changes that essentially activates the protein for membrane fusion (Shen et al., 2017). After binding of the RBD to the hACE2 receptor on target cells, the heptad repeat 1 (HR1) and 2 (HR2) domains in the S2 subunit of S protein interact with each other to form a six-helix bundle (6-HB) fusion core, bringing viral and cellular membranes into close proximity for fusion and infection (Du et al., 2009; Li et al., 2019; Xia et al., 2020; Zhu et al., 2013).
Scientists throughout the world are currently looking for effective and immediate therapeutic strategies to intervene and prevent the infectivity or transmission of SARS-CoV-2. Till date, no suitable vaccines or anti-viral agents have been put forward.
Several types of pyrimidine bases like thymine, cytosine and uracil are found in nature, which forms the framework or building blocks of genetic materials, i.e. DNA and RNA (Sharma et al., 2014; Sohrabi et al., 2018). This prime importance makes pyrimidine moiety suitable for broad therapeutic applications (Shakibapour et al., 2019; Sharma et al., 2014). Pyrimidine moieties have gained considerable attention in the chemotherapy of various diseases like cancer (Xie et al., 2011), HIV, diabetes, cardiovascular, bacterial, fungal diseases (Sharma et al., 2014), and several viral diseases (Balzarini & McGuigan, 2002). It is also used as anti-inflammatory drugs (Amir et al., 2007), analgesics (Vega et al., 1990), antipyretic drugs, anti-leishmanial drugs, herbicidal agents and anti-oxidants (Abu-Hashem et al., 2011). As pyrimidine base is a constituent of both DNA and RNA; hence, it is useful in the chemotherapy of both DNA and RNA viruses (Sharma et al., 2014). Compounds with N-heterocyclic scaffolds are highly effective against a diverse range of diseases, and it has already been studied for their potential pharmacological activities. In particular, pyrimidine derivatives have demonstrated outstanding biological activities. For example, Dayvigo (Lemborexant), Inrebic (Fedratinib) are a few examples of drugs containing pyrimidine scaffolds which received FDA-approval in 2019. Because of the enormous therapeutic importance, we selected some well-characterized pyrimidine substituted phenols (Kumar & Rao, 2018; Table S1, supplementary material) to investigate their efficiency in binding to the interface of the hACE2-S protein complex and modulate the pattern of infectivity of the SARS-CoV-2 virus. In this study, we aim to identify diaryl pyrimidine derivatives as a potential lead molecule which may bind at the interface of the hACE2-S protein complex with high affinity. Our computational analyses show that AP-NP, AP-3-OMe-Ph and AP-4-Me-Ph have the potential to bind at the interface of the hACE2-S protein complex with high affinity. Thus, the further experimental studies with these lead molecules may facilitate the anti-COVID drug development processes.
Table 1.
The binding energy and the amino acids involved within specific distance of diaryl pyrimidine derivatives against hACE2-S protein complex.
| Compound | Binding affinity (kcal/mol) | Interacting amino acid residue of hACE2 |
|---|---|---|
| AP-NP | −8.95 | LEU 391, PHE 390, LEU 73, TRP 69, PHE 40, ASP 350, ASN 394, ARG 393 |
| AP-Ph-4-I | −7.9 | PHE 40, ASP 350, TRP 349, ALA 348, ASP 382, TYR 385, PHE 390 |
| AP-3-OMe-Ph | −8.1 | PHE 40, TRP 349, ALA 348, ASP 382, TYR 385, ARG 393, PHE 390, ASP 350, GLY 352 |
| AP-4-Me-Ph | −8.1 | ALA 348, TRP 349, SER 47, SER 44, PHE 40, ASP 350, TYR 385, HIS 401, ASP 382 |
| AP-Ph-4-OMe | −7.8 | TYR 385, ASP 382, HIS 401, ALA 348, TRP 349, PHE 40, PHE 390, GLY 352, ARG 393 |
| AP-Ph-4-Br | −7.9 | PHE 40, TRP 349, ALA 348, ASP 350, ASP 382, TYR 385, PHE 390 |
| Chloroquine | −5.7 | TRP 69, PHE 40, ASN 394, ARG 393, PHE 390, LEU 391, ALA 99, LEU 100, LEU 73 |
2. Materials and methods
2.1. Preparation of ligands and receptor
The three-dimensional structure of the hACE2-S protein complex (human angiotensin-converting enzyme 2 and spike glycoprotein complex, PDB ID: 6VW1) was downloaded from the RCSB protein data bank (Shang et al., 2020). The structure of all pyrimidine derivatives was prepared in Discovery studio 2020. The conformation of all ligands was refined using ‘Clean geometry’ command and the structure with minimum clean energy was saved in pdb format.
2.2. Molecular docking of diaryl pyrimidine derivatives on hACE2- S protein complex
The molecular docking analysis of the AP-NP compound was performed on the X-ray crystal structure of the hACE2-S protein complex (PDB ID: 6VW1). First, polar hydrogen and gasteiger charges were added on the spike glycoprotein and AP-NP compound using MGL tool (Morris et al., 2009) and saved in pdbqt format. The molecular docking tool AutoDock Vina (Trott & Olson, 2010) was used for the docking of AP-NP on the hACE2-S protein complex. To determine the probable binding site, blind docking was performed. For this, the hACE2-S protein complex was kept rigid, while AP-NP was kept flexible to allow exploration of probable binding sites. Both molecules were covered by grid box with a dimension of 116 Å × 82 Å × 86 Å with grid spacing 1 Å. Five sets of docking were performed with exhaustiveness of 100. The AutoDock Vina produced 9 docked conformations with binding energies in the range of −7.95 to −8.95 kcal/mol. Among these conformations, the conformation, where AP-NP was observed to bind at the proximal helix site present at the junction of the interacting hACE2 and S1 domains, possessed the lowest binding free energy. Therefore, the helical region was chosen for local docking. For this, the helical region was covered by the grid box with a dimension of 58 Å × 82 Å × 120 Å with grid spacing 1 Å and docking was done with exhaustiveness of 100. Five sets of local docking were performed, and the conformation with the lowest binding energy was chosen for analysis. The analyses of the results/outputs from AutoDock Vina were done using MGL tools 1.5.6 (Morris et al., 2009), and intermolecular interactions were determined using PyMoL (DeLano, 2002). The molecular docking experiments of the other diaryl pyrimidine derivatives were performed in a similar fashion as done with the AP-NP.
2.3. Molecular Dynamics (MD) Simulation study
Using GROMACS v5.1.4 biomolecular simulation package, with CHARMM27 force field and TIP3P water model, all-atoms molecular dynamics (MD) simulations were carried out on the coordinates of hACE2-S protein complex docked with compounds: AP-NP, AP-3-OMe-Ph and AP-4-Me-Ph (Abraham et al., 2015; Mokaberi et al., 2019; Sharifi-Rad et al., 2020). The topology and parameter files for ligands parameters were repaired as define by Zoete et al. (2011), Mongre et al. (2019), and Sharifi-Rad et al. (2020). The simulation box was prepared with 10 Å buffer distance, and the protein–ligand complex was placed at the centre of box, padding around with water molecules. Along with counter ions for neutralizing the system, the salt concentration of 0.15 M (0.15 M NaCl) was used for the MD simulation (Joung & Cheatham, 2008). PBC condition was defined for all three x, y and z directions (Darden et al., 1993). All energy minimization and simulations were performed at physiological temperature, 300 K (Kumar et al., 2019; Mishra et al., 2018). Energy minimization was done estimating the steepest descent, followed by the conjugant gradients involving 50,000 steps for each. SHAKE algorithm was applied to constrain all the bonds involving hydrogen atom. Particle Mesh Ewald (PME) was applied for long-range electrostatic forces. The equilibration of system was done in two steps, NVT and NPT for the period of 500 ps. Pressure and temperature during the simulation were maintained by Berendsen thermostat (Berendsen et al., 1987) and Parrinello-Rahman pressure (Parrinello & Rahman, 1980), respectively. LINC algorithm was applied to constrain the bonds and angles (Hess et al., 1997). LJ potential with a cut off of 0.10 nm was defined for Van der Waals interactions. Finally, the production run was performed on NPT ensemble for a period of 100 ns, and the trajectory was updated at the time interval of 10 ps to record the energy and velocity. All production runs were performed on CUDA enabled Tesla GPU machine (DELL T640 with V100 GPU), and OS Centos 7 (Prakash et al., 2018; Singh et al., 2019) and the GROMACS utilities were used for the analyses of obtained MD trajectories.
2.4. Binding free energy estimation
The binding free energy of the protein–ligand complexes was calculated using MM-PBSA (Molecular Mechanics-Poisson-Boltzmann Surface Area), which describes the structural stability, spatial orientation and molecular interactions of ligands in the active site of the protein (Batt et al., 2012; Prakash & Luthra, 2012; Sastry et al., 2013). The binding free energy abbreviated as ΔGbinding can be defined as,
where Gcomplex represents the total free energy of protein-ligand complex, Gprotein as the free energy of protein, Gligand used as the free energy of ligand and < > represents the ensemble average.
Neglecting the entropy term (TS), the binding free energy can be approximately written as
where ΔEMM is the change in the molecular mechanics interaction energy (gas-phase) upon ligand binding calculated as the sum of the changes in the bonded energy, electrostatics and Van der Waals interactions upon ligand binding. ΔGsolv is the change in solvation free energy upon ligand binding.
Furthermore, ΔGsolv can be written as
where is the change in the polar part of solvation free energy and is the change in nonpolar part of solvation free energy as a result of ligand binding to the proteins. In this work, Poisson–Boltzmann (PB) method was used for the estimation of the polar part of the solvation free energy.
In the current work, binding free energy () was calculated utilizing the MMPBSA.py script of the AMBER package (Wang et al., 2016). Single trajectory protocol approach was utilized for the computation, which involves only the simulation of complex forms. An ionic strength of 0.15 M was used for the PBSA calculations. Considering the convergence issues associated with the MM-PBSA calculation, only last 20 ns data was used.
2.5. ADME analysis for drug likeliness
The drug likeliness property of all diaryl pyrimidine derivatives was monitored as described earlier (Jayaram et al., 2012; Lipinski, 2004). The pdb structure of all diaryl pyrimidine derivate compounds was constructed, and ADME properties, as explained in Lipinski’s rule of five, i.e. molecular weight, solubility, H-bond donor, H-bond acceptor and molar refractivity were calculated by online software tool.
3. Results and discussion
3.1. Molecular docking analysis
Designing the targeted drug molecules by using cheminformatics tools is the initial step in drug discovery (Xu & Hagler, 2002). In this study, our focus was to investigate the efficacy of diaryl pyrimidine derivative compounds in obstructing the interaction between spike glycoprotein of COVID-19 and the hACE2 receptor, which is considered to be an essential step towards the progress of infection. The interaction of spike protein and hACE2 facilitates the attachment and internalization of the virus in the host cell (Belouzard et al., 2012). The COVID-19 virus attaches itself to the host cell using S1 domain spike glycoprotein, whereas the S2 domain of spike protein facilitates the internalization of the virus into the host membrane to begin infection (Belouzard et al., 2012; Du et al., 2009). So both domains play a crucial role in the initialization of the infection in the host cell (Enmozhi et al., 2020). The study presented here, using molecular docking tool, revealed significant binding interaction of diaryl pyrimidines with the interface of the hACE2-S receptor complex (hACE2-spike protein complex: PDB ID 6VW1) (Table 1).
In this study, we have analyzed the binding energies of all diaryl pyrimidine derivatives on the hACE2-S complex. Here we found that all diaryl pyrimidine derivatives bind at the interface of the hACE2-S protein complex. Interestingly, these molecules share the same binding site, which lies near the binding junction of hACE2 and C-terminal S1 domain of spike glycoprotein. Among these diaryl pyrimidine derivatives, AP-NP, AP-3-OMe-Ph and AP-4-Me-Ph displayed high binding affinity towards the hACE2 receptor (Figure 1) with the binding energies of −8.95, −8.1 and −8.1 kcal/mol, respectively (Table 1). These pyrimidine containing phenols binds with the active site through hydrogen bonding using –OH group of phenol part and an –NH2 group of the amino pyrimidine ring. In addition to that, the non-covalent interactions such as π–π interaction are also involved in the stabilization of the ligand within the active site.
Figure 1.
Binding site of AP-NP, AP-3-OMe-Ph and AP-4-Me-Ph on hACE2-S protein complex. (a) The putative binding site of AP-NP, AP-4-Me-Ph and AP-3-OMe-Ph on hACE2-S protein complex. The C-terminal S1 domain of spike protein is shown in orange colour, hACE2 is shown in dark green colour. (b) The zoomed view of binding site of AP-NP on hACE2-S protein complex. (c) The interacting amino acid residues of hACE2 with AP-NP. The AP-NP is shown in cream colour. (d) The zoomed view of binding site of AP-4-Me-Ph on hACE2-S protein complex. (e) The interacting amino acid residues of hACE2 with AP-4-Me-Ph. The AP-4-Me-Ph is shown in purple colour. (f) The zoomed view of binding site of AP-3OMe-Ph on hACE2-S protein complex. (g) The interacting amino acid residues of hACE2 with AP-3-OMe-Ph. The AP-3-OMe-Ph is shown in light pink colour.
In particular, the significant binding energy observed in case of AP-NP can be attributed to such non-covalent hydrophobic interactions of phenyl as well as napthyl ring with non-polar amino acid residues such as LEU 391, PHE 390, LEU 73, TRP 69 and PHE 40 along with the interaction through hydrogen bonding with polar amino acid residues such as ASP 350, ASN 394 and ARG 393 (Figure 1(b, c)). In case of AP-3-OMe-Ph, PHE 40, PHE 390, TRP 349 of hACE2 forms hydrophobic interaction with both the aryl rings attached with pyrimidine scaffold (Figure 1(d, e)). In addition to that, hydrogen bonds were also observed with ALA 348, ASP 382, ARG 393, ASP 350, and GLY 352. In case of AP-4-Me-Ph, ALA 348, TRP 349, SER 47, SER 44, PHE 40 of hACE2 forms hydrophobic interaction with the toluene ring. However, the phenol-pyrimidine base forms hydrogen bonding with ASP 350, TYR 385, HIS 401 and ASP 382 (Figure 1(f, g)).
The similarity in the binding energy of AP-3-OMe-Ph with AP-4-Me-Ph could be explained due to the presence of tetrahedral –OMe group instead of the planar aromatic group such as naphthyl. It also has the same hydrophobic interaction as AP-4-Me-Ph. This might be due to their structural similarity. Compared to the AP-4-Me-Ph, additional hydrogen bonding interactions were observed with ARG 393 residue due to the involvement of oxygen atom of –OMe group.
CQ, a well-known drug that is suggested for use in the COVID-19 therapy, was reported to interfere with the terminal glycosylation of spike protein of SARS-CoV-2S and thereby reducing the initial infection (Hu et al., 2020; Vincent et al., 2005). Hence, we also used CQ as a positive control and performed molecular docking experiment. Similar to all diaryl pyrimidine derivatives, we found that CQ also binds at the interface of the hACE2-S protein complex (Supplementary Figure 1). We observed that CQ has lesser binding affinity (−5.7 kcal/mol) with hACE2-S complex, which may be due to its high affinity towards its primary target, i.e. endosome (Al-Bari, 2017).
These selected inhibitors generated through the molecular docking studies were projected onto further analyses for molecular simulation dynamics and estimation of the free energy of binding using MM-PBSA.
3.2. Molecular Dynamics (MD) Simulation study
To examine the spatial stability and mechanistic aspects of conformational dynamics underlying the molecular interaction of diaryl pyrimidine derivatives, AP-NP, AP-3-OMe-Ph and AP-4-Me-Ph with hACE2-S protein complex, we performed MD simulation in an aqueous environment for the period of 100 ns, at physiological temperature (300 K). We analyzed the MD trajectory by employing structural parameters, RMSD (root mean square deviation) and Rg (radius of gyration), H-bond (hydrogen bonding), the minimum distance between protein and ligands, principal component analysis (PCA) and free-energy landscape (FEL) which revealed the dynamic stability of ligand-bound complex (Kumar et al., 2019; Mishra et al., 2018; Sarma et al., 2020). MM-PBSA was used for the estimation of binding free energy, which describes the conformational stability of ligand at the binding site, molecular interactions and the binding affinity with protein.
To determine the global conformational stability of ligands docked with hACE2-S protein, we computed all atoms Cα-backbone RMSD with respect to the initial structure, as shown in Figure 2. The result shows that the RMSD plot of hACE2-S protein complex docked with AP-NP quickly achieves the stable equilibrium during the initial few nanoseconds and stable conformational dynamics is observed till the end of the simulation.
Figure 2.
The evolution plot of Cα RMSD in water at 300 K of hACE2-S protein docked complex with compounds, AP-NP (black), AP-3-OMe-Ph (red) and AP-4-Me-Ph (green).
The RMSD trajectory of AP-3-OMe-Ph shows a continuous rise in RMSD during the initial 0–25 ns. The initial rise in RMSD suggested the spatial arrangement of the active site to accommodate the ligand. The ligand adjusted well at the binding groove; i.e. the protein–ligand complex achieved equilibrium at ∼25 ns and it remained stable for the period of 100 ns. Similarly, we observed consecutive two drifts in the RMSD trajectory of hACE2-S protein docked complex with AP-4-Me-Ph, during the initial 0–20 ns. The system attains equilibrium only after 20 ns, and a stable conformation of the protein–ligand complex propagated for the remaining simulation time. However, the consequences of small drifts during ∼40–70 ns indicate relatively lower conformational stability of AP-4-Me-Ph as compared to AP-NP and AP-3-OMe-Ph. Another structural order parameter, Rg, is commonly used to determine the structural compactness and stability of protein during the simulation. Thus, the time evolution plot of Rg is calculated, as shown in Figure 3.
Figure 3.
Time evolution plot of radius of gyration (Rg) in water at 300 K of hACE2-S protein docked complex with compounds, AP-NP (black), AP-3-OMe-Ph (red) and AP-4-Me-Ph (green).
In this figure, we can see that the conformation dynamics of the hACE2-S protein-AP-NP complex remain compact and stable with an average Rg value of 2.7 ± 0.01 nm, throughout the simulation period. We also did not observe any significant structural changes in the Rg trajectory of AP-3-OMe-Ph, and the protein–ligand complex remains stable with Rg value 4.4 ± 0.01 nm. However, the Rg plot of AP-4-Me-Ph shows two small drifts of ∼0.02 nm. Although, it is well settled around 35 ns and the progression of stable complex structure for the remaining period of simulation. Thus, RMSD and Rg suggest that all three compounds remain bound to the active site during the simulation period with minimal perturbation; however, AP-NP appeared more stable complex as compared to AP-3-OMe-Ph and AP-4-Me-Ph.
To further confirm the molecular interaction stability of ligands with protein, we also measured the occupancy of H-bond interactions between protein-ligands during the simulation. Besides maintaining the conformational architecture and stability of protein structure, H-bonds play a critical role in the stability of the protein–ligand complex. Molecular docking results showed the presence of two H-bonds between AP-NP and hACE2-S protein, which is consistently observed during the simulation time (Figure 4). However, during 0–20 ns and 60–100 ns, only one H-bond remains consistent, and the appearance of two H-bond is seen only for the period of 20–60 ns.
Figure 4.
Time evolution plot of hydrogen bonds (H-bond) between hACE2-S protein and ligands, AP-NP, AP-3-OMe-Ph and AP-4-Me-Ph. The representation of plots color as shown in Figure 1.
The molecular interaction of AP-3-OMe-Ph shows the occupancy of maximum three H-bonds, but only two remain stable throughout the simulation period. Appearance of three H-bond seen only between 40 and 60 ns. The maximum occupancy of four-five H-bonds is seen with AP-4-Me-Ph as observed in molecular docking; however, only two remains consistent during the simulation. But, considering the occurrence of appearance and disappearance of H-bond interactions with ligands, substantially only two H-bonds with AP-NP and AP-3-OMe-Ph observed persistent for the maximum period of simulation.
To maintain the dynamic stability of ligand at the binding pocket of protein, consistency of the spatial orientation of ligand and the optimal distance from the binding cavity of protein plays a significant role in the interaction stability of the protein-ligand complex. Thus, the time evolution plot of the mean distance of all three ligands monitored during the simulation (Figure 5).
Figure 5.
Time evolution plot of minimum (min) distances from the center of binding grooves and ligands. Color scheme as shown in Figure 1.
From Figure 5, it can be clearly depicted that the mean distance of all three ligands remains in the range of 3.7–4.2 Å, which is observed consistent almost throughout simulation time. Only, AP-4-Me-Ph moved out the cavity approximately at ∼15 ns and ∼70 ns, however, it appeared transiently, and all three ligands continue as well occupied to the binding pocket of hACE2- S protein. Furthermore, the lower flexibility of average distance with AP-NP again provides the elegance evidence of stable molecular interaction with the target protein.
3.3. Essential dynamics
Essential dynamics (ED) govern the coherent changes in the conformational dynamics of protein–ligand complex while the simulation, which can be calculated by projecting the first two principal components (PCs), PC1, and PC2 along the native structure. By diagonalizing the covariance matrix of eigenvectors, we computed PCs which determine the overall combined motion of the Cα-atoms, and localization of PCs in conformational space correlated to the protein function. ED plot of all three complexes is shown in Figure 6.
Figure 6.
Essential dynamics of hACE2-S protein with AP-NP (black), AP-3-OMe-Ph (red) and AP-4-Me-Ph (green). Conformational ensembles sampled with the projection of principal components PC1 and PC2.
From Figure 6, it is observed that the collective motion of AP-NP confined to small conformation space, reflecting stable collective atomic fluctuations. Differently, AP-4-Me-Ph explores broader conformational space during the simulation, depicting the larger collective motion of atoms; thus, it indicates a relatively less stable structure. We can see that the collective motion of AP-3-OMe-Ph shows broader conformation space than AP-NP; however, it is significantly restricted to less occupied space as compared to AP-4-Me-Ph. Thus, ED analysis indicates a better affinity of AP-NP towards ACE2-S-protein than AP-3-OMe-Ph and AP-4-Me-Ph, respectively.
3.4. Estimation of the FEL
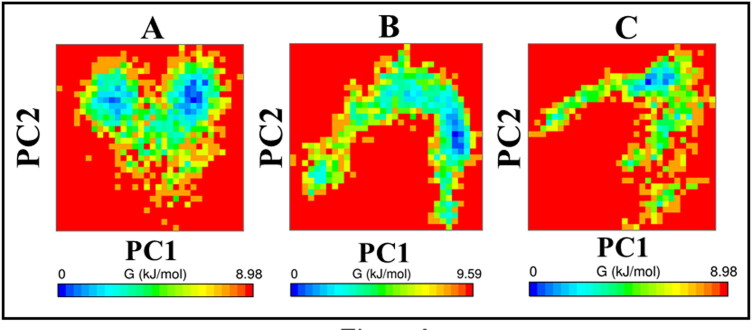
We also performed FEL analysis to examine the minimum energy conformational ensembles of all three protein–ligand complexes, using the first two PCs (PC1 and PC2) of the protein backbone atoms as reaction coordinates (Kumar et al., 2018; Sarma et al., 2020). This allows an estimation of the free energy difference of conformational stability of the hACE2-S protein bound complex with AP-NP, AP-3-OMe-Ph, and AP-4-Me-Ph with high accuracy (Figure 7).
Figure 7.
Free energy landscape calculated using the principal components PC1 and PC2 as reaction coordinates of hACE2-S protein with AP-NP, AP-3-OMe-Ph and AP-4-Me-Ph.
From Figure 7, we can see that AP-NP displays less conformational mobility, restricting to a more confined conformational space within single energy minima. This result displayed that molecular interactions of AP-NP contributed to the stabilization of the ligand-bound complex of hACE2-S protein; thus, the complex structure remains connected through the low energy barriers. The broader and sallow energy basin of AP-3-OMe-Ph characterized that ligand-bound complex navigated through the wide range of conformations; however, with small excursions, conformational ensembles shifted the dynamic equilibrium which stayed stable and confined to the low energy basin. Whereas, the sallow and fragmented energy basin of ligand AP-4-Me-Ph suggests that the conformational ensemble of complex structure broadly experiences the two different conformational spaces. The complex structure readily moves out the native basin, highly flexible structure navigated the wide conformational space, before entering to the stable energy basin. This result signifies that the proper positioning of AP-4-Me-Ph orientation in the binding grooves may require conformational adjustment, hence, unstable distributions of the binding configuration ensemble of complex transverse to stable energy basin through the higher energy barrier. Thus, the FEL plot suggests that the compound AP-NP almost stayed constant at the bonding grooves of hACE2-S protein and remains well stabilized through the molecular interactions.
3.5. Estimation of the binding free energy
The binding free energy of the protein–ligand complex provides a robust approximation of molecular interactions, which includes bonded and non-bonded interactions and the effect of solvent underlying the protein–ligand binding affinity (Kumar et al., 2019; Pandey et al., 2017). Bonded and non-bonded terms are defined by the van der Waals and electrostatic interactions. Whereas, the solvation free energies in terms of polar and non-polar solvation free energy. This analysis was performed on the fully converged trajectory of protein–ligand for the last 20 ns data (Pandey et al., 2018; Wang et al., 2016). The computed binding free energy (ΔGbinding) and the contribution of other energy parameters on the molecular interaction stabilities of AP-NP, AP-3-OMe-Ph and AP-4-Me-Ph are summarized in Table 1. Results show that the lowest binding energy, ΔG = −30.89 ± 2.24 kcal mol−1, was obtained for ligand AP-NP, suggesting the highest binding affinity with hACE2-S protein. Out of the three compounds, AP-4-Me-Ph possess the highest binding free energy −25.29 ± 2.14 kcal mol−1, which indicates the lowest binding affinity towards the target protein complex. Whereas, the binding free energy of AP-3-OMe-Ph (−28.39 ± 2.69 kcal mol−1) was observed slightly higher than AP-NP, thus, considered as the ligand with a moderate range of binding affinity. Furthermore, the results show a larger contribution of the Van der Waals energies as compared to others like electrostatic energies suggesting that ligands at the binding pocket are predominantly stabilized by hydrophobic interactions. Thus, our analyses provided substantial evidence that hydrophobic amino acid residues at the groove of the hACE2-S-protein complex is crucial for the structure-based drug development in the therapy of COVID-19.
3.6. Lipinski’s rule of five
To determine whether any compound with a particular biological activity has the potential to serve as a pharmacological agent/drug, the Lipinski rule of five is generally used. This rule acts as a filter in in-silico analyses and helps to screen potential drugs during the very beginning of the drug designing process, thus minimizing the cost of exercises, time, and labour in case of clinical drug development (Gombar et al., 2003; Hughes et al., 2011). The rule judges some of the basic molecular properties a compound possesses, like absorption, distribution, metabolism and excretion (ADME) for a selected range (Hughes et al., 2011). If a compound possesses at least two of the properties, it’s good to go (Jayaram et al., 2012; Lipinski, 2004). The potential diaryl pyrimidine derivatives used in this study were found to pass all the five criteria mentioned in Lipinski’s rule (Table 2). So it also supported the therapeutic importance of the selected diaryl pyrimidine moieties.
Table 2.
ADME values for selected diaryl pyrimidine derivatives derivatives.
| S. No. | Compound/ Ligand | ADME
properties (Lipinki’s rule of five) |
Drug likeliness | |
|---|---|---|---|---|
| Properties | Values | |||
| 1. | AP-NP | Molecular weight (<500 Da) | 313 | Yes |
| LogP (<5) | 4.3 | |||
| H-bond donor (5) | 3 | |||
| H-bond acceptor (<10) | 4 | |||
| Molar refractivity (40–130) | 96.5 | |||
| Violations | NO | |||
| 2. | AP-3-OMe-Ph | Molecular weight (<500 Da) | 293 | Yes |
| LogP (<5) | 3.1 | |||
| H-bond donor (5) | 3 | |||
| H-bond acceptor (<10) | 5 | |||
| Molar refractivity (40–130) | 85.5 | |||
| Violations | NO | |||
| 3. | AP-4-Me-Ph | Molecular weight (<500 Da) | 277 | Yes |
| LogP (<5) | 3.4 | |||
| H-bond donor (5) | 3 | |||
| H-bond acceptor (<10) | 4 | |||
| Molar refractivity (40–130) | 83.7 | |||
| Violations | NO | |||
4. Conclusion
In-silico studies provide the best alternatives to screen potential drug candidates. In this study, we have investigated the ability of some diaryl pyrimidine derivative compounds for binding to hACE2-S-protein complex and obstructing its receptor binding ability using molecular docking. Our docking results were supported by other analyses estimating the free energy of binding using MM-PBSA, free energy landscape, and dynamics simulation. We found that all the chosen compounds were capable of binding to the interface of human cell receptor ACE2 and spike protein and with high affinity and fulfill all the criteria of Lipinski’s rule of five. We believe that all of these compounds can be effective anti-COVID drugs and, therefore, should be experimentally verified by researchers working on this scheme.
Supplementary Material
Acknowledgements
We thank the Supercomputing Facility for Bioinformatics and Computational Biology, Indian Institute of Technology Delhi, for online facilities. AK and SR thanks Mahatma Gandhi Central University Motihari, Bihar.
Glossary
Abbreviations
- 6-HB
six-helix bundle
- ADME
absorption, distribution, metabolism and excretion
- AP-NP
2-(2-amino-5-(naphthalen-2-yl)pyrimidin-4-yl) phenol
- AP-4-Me-Ph
2-(2-amino-5-(p-tolyl)pyrimidin-4-yl) phenol
- AP-3-OMe-Ph
2-(2-amino-5-(3-methoxyphenyl)pyrimidin-4-yl) phenol
- COVID-19
coronavirus disease 2019
- CQ
chloroquine
- hACE2
human angiotensin converting enzyme-2
- hACE2-S protein complex
human angiotensin converting enzyme-2 receptor and severe acute respiratory syndrome coronavirus 2 spike protein complex
- HR1
heptad repeat 1
- HR2
heptad repeat 2
- PDB
protein data bank
- RBD
receptor-binding domain
- SARS-CoV-2S
severe acute respiratory syndrome coronavirus 2 spike protein
- TMPRSS-2
transmembrane protease serine 2
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Aanouz, I., Belhassan, A., El-Khatabi, K., Lakhlifi, T., El-Ldrissi, M., & Bouachrine, M. (2020). Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1-2. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., & Lindahl, E. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1-2, 19–25. 10.1016/j.softx.2015.06.001 [DOI] [Google Scholar]
- Abu-Hashem, A. A., Youssef, M. M., & Hussein, H. A. R. (2011). Synthesis, antioxidant, antituomer activities of some new thiazolopyrimidines, pyrrolothiazolopyrimidines and triazolopyrrolothiazolopyrimidines derivatives. Journal of the Chinese Chemical Society, 58(1), 41–48. 10.1002/jccs.201190056 [DOI] [Google Scholar]
- Al-Bari, M. A. A. (2017). Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacology Research & Perspectives, 5(1), e00293. 10.1002/prp2.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir, M., Javed, S., & Kumar, H. (2007). Pyrimidine as antiinflammatory agent: A review. Indian Journal of Pharmaceutical Sciences, 69(3), 337-343. 10.4103/0250-474X.34540 [DOI] [Google Scholar]
- Balzarini, J., & McGuigan, C. (2002). Bicyclic pyrimidine nucleoside analogues (BCNAs) as highly selective and potent inhibitors of varicella-zoster virus replication . The Journal of Antimicrobial Chemotherapy, 50(1), 5–9. 10.1093/jac/dkf037 [DOI] [PubMed] [Google Scholar]
- Batt, S. M., Jabeen, T., Bhowruth, V., Quill, L., Lund, P. A., Eggeling, L., Alderwick, L. J., Futterer, K., & Besra, G. S. (2012). Structural basis of inhibition of Mycobacterium tuberculosis DprE1 by benzothiazinone inhibitors. Proceedings of the National Academy of Sciences of the United States of America, 109(28), 11354–11359. 10.1073/pnas.1205735109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard, S., Millet, J., Licitra, B., & Whittaker, G. (2012). Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses, 4(6), 1011–1033. 10.3390/v4061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Grigera, J. R., & Straatsma, T. P. (1987). The missing term in effective pair potentials. The Journal of Physical Chemistry, 91(24), 6269–6271. 10.1021/j100308a038 [DOI] [Google Scholar]
- Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98(12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- DeLano, W. L. (2002). PyMOL. DeLano Scientific. [Google Scholar]
- Du, L., He, Y., Zhou, Y., Liu, S., Zheng, B. J., & Jiang, S. (2009). The spike protein of SARS-CoV-a target for vaccine and therapeutic development. Nature Reviews. Microbiology, 7(3), 226–236. 10.1038/nrmicro2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1-9. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A., & Azzam, E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1-9. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., Şahin, A. T., & Yelekçi, K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani, M., Hasaniazad, M., Faezi, S., Guklani, H., Davoodian, P., Ahmadi, N., Einakian, M. A., Karmostaji, A., & Ahmadi, K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 1–7. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombar, V. K., Silver, I. S., & Zhao, Z. (2003). Role of ADME characteristics in drug discovery and their in silico evaluation: In silico screening of chemicals for their metabolic stability. Current Topics in Medicinal Chemistry, 3(11), 1205–1225. 10.2174/1568026033452014 [DOI] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Baker, S. C., Baric, R. S., de Groot, R. J., Drosten, C., Gulyaeva, A. A., Haagmans, B. L., Lauber, C., Leontovich, A. M., Neuman, B. W., Penzar, D., Perlman, S., Poon, L. L. M., Samborskiy, D. V., Sidorov, I. A., Sola, I., Ziebuhr, J., & Coronaviridae Study Group Of The International Committee On Taxonomy Of, V. (2020). The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology, 5, 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, B., Bekker, H., Berendsen, H. J. C., & Fraaije, J. G. E. M. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18, 1463–1472. [DOI] [Google Scholar]
- Hu, T. Y., Frieman, M., & Wolfram, J. (2020). Insights from nanomedicine into chloroquine efficacy against COVID-19. Nature Nanotechnology, 15(4), 247–249. 10.1038/s41565-020-0674-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J. P., Rees, S., Kalindjian, S. B., & Philpott, K. L. (2011). Principles of early drug discovery. British Journal of Pharmacology, 162(6), 1239–1249. 10.1111/j.1476-5381.2010.01127.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram, B., Singh, T., Mukherjee, G., Mathur, A., Shekhar, S., & Shekhar, V. (2012). Sanjeevini: A freely accessible web-server for target directed lead molecule discovery. BMC Bioinformatics, 13 Suppl 17, S7. 10.1186/1471-2105-13-S17-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitendra Subhash, R., Aroni, C., Abhijeet, K., & Shashikant, R. (2020). Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: An in silco study for drug development, 10.26434/chemrxiv.12094203.v1 [DOI] [PMC free article] [PubMed]
- Joshi, R. S., Jagdale, S. S., Bansode, S. B., Shankar, S. S., Tellis, M. B., Pandya, V. K., Chugh, A., Giri, A. P., & Kulkarni, M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung, I. S., & Cheatham, T. E. 3rd (2008). Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. The Journal of Physical Chemistry. B, 112(30), 9020–9041. 10.1021/jp8001614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer, R., Cottrell, C., Nianshuang, W., Pallesen, J., Yassine, H., Turner, H., Corbett, K., Graham, B., McLellan, J., & Ward, A. (2016). Pre-fusion structure of a human coronavirus spike protein. Nature, 531(7592), 118–121. 10.1038/nature17200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V., Prakash, A., Pandey, P., Lynn, A. M., & Hassan, M. I. (2018). TFE-induced local unfolding and fibrillation of SOD1: Bridging the experiment and simulation studies. The Biochemical Journal, 475(10), 1701–1719. 10.1042/BCJ20180085 [DOI] [PubMed] [Google Scholar]
- Kumar, A., & Rao, M. L. N. (2018). Pot-economic synthesis of diarylpyrazoles and pyrimidines involving Pd-catalyzed cross-coupling of 3-trifloxychromone and triarylbismuth. Journal of Chemical Sciences, 130(12), 1–11. 10.1007/s12039-018-1565-6 [DOI] [Google Scholar]
- Kumar, N., Srivastava, R., Prakash, A., & Lynn, A. M. (2019). Structure-based virtual screening, molecular dynamics simulation and MM-PBSA toward identifying the inhibitors for two-component regulatory system protein NarL of Mycobacterium Tuberculosis. Journal of Biomolecular Structure and Dynamics, 38(11), 3396-3410. 10.1080/07391102.2019.1657499 [DOI] [PubMed] [Google Scholar]
- Li, Z., Tomlinson, A. C., Wong, A. H., Zhou, D., Desforges, M., Talbot, P. J., Benlekbir, S., Rubinstein, J. L., & Rini, J. M. (2019). The human coronavirus HCoV-229E S-protein structure and receptor binding. eLife, 8, e51230. 10.7554/eLife.51230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski, C. A. (2004). Lead- and drug-like compounds: The rule-of-five revolution. Drug Discovery Today. Technologies, 1(4), 337–341. 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet (London, England), 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, C. B., Kumari, S., Prakash, A., Yadav, R., Tiwari, A. K., Pandey, P., & Tiwari, M. (2018). Discovery of novel Methylsulfonyl phenyl derivatives as potent human cyclooxygenase-2 inhibitors with effective anticonvulsant action: Design, synthesis, in-silico, in-vitro and in-vivo evaluation. European Journal of Medicinal Chemistry, 151, 520–532. 10.1016/j.ejmech.2018.04.007 [DOI] [PubMed] [Google Scholar]
- Mokaberi, P., Reyhani, V., Amiri-Tehranizadeh, Z., Saberi, M. R., Beigoli, S., Samandar, F., & Chamani, J. (2019). New insights into the binding behavior of lomefloxacin and human hemoglobin using biophysical techniques: Binary and ternary approaches. New Journal of Chemistry, 43(21), 8132–8145. 10.1039/C9NJ01048C [DOI] [Google Scholar]
- Mongre, R. K., Mishra, C. B., Prakash, A., Jung, S., Lee, B. S., Kumari, S., Hong, J. T., & Lee, M. S. (2019). Novel carbazole-piperazine hybrid small molecule induces apoptosis by targeting BCL-2 and inhibits tumor progression in lung adenocarcinoma in vitro and xenograft mice model. Cancers (Basel), 11(9). 10.3390/cancers11091245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Pandey, P., Lynn, A. M., & Bandyopadhyay, P. (2017). Identification of inhibitors against α-Isopropylmalate Synthase of Mycobacterium tuberculosis using docking-MM/PBSA hybrid approach. Bioinformation, 13(5), 144–148. 10.6026/97320630013144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, P., Srivastava, R., & Bandyopadhyay, P. (2018). Comparison of molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA) and molecular mechanics-three-dimensional reference interaction site model (MM-3D-RISM) method to calculate the binding free energy of protein-ligand complexes: Effect of metal ion and advance statistical test. Chemical Physics Letters, 695, 69–78. 10.1016/j.cplett.2018.01.059 [DOI] [Google Scholar]
- Pant, S., Singh, M., Ravichandiran, V., Murty, U. S. N., & Srivastava, H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello, M., & Rahman, A. (1980). Crystal structure and pair potentials: A molecular-dynamics study. Physical Review Letters, 45(14), 1196–1199. 10.1103/PhysRevLett.45.1196 [DOI] [Google Scholar]
- Prakash, A., Dixit, G., Meena, N. K., Singh, R., Vishwakarma, P., Mishra, S., & Lynn, A. M. (2018). Elucidation of stable intermediates in urea-induced unfolding pathway of human carbonic anhydrase IX. Journal of Biomolecular Structure & Dynamics, 36(9), 2391–2406. 10.1080/07391102.2017.1355847 [DOI] [PubMed] [Google Scholar]
- Prakash, A., & Luthra, P. M. (2012). Insilico study of the A(2A)R-D (2)R kinetics and interfacial contact surface for face for heteromerization. Amino Acids, 43(4), 1451–1464. 10.1007/s00726-012-1218-x [DOI] [PubMed] [Google Scholar]
- Sarma, P., Shekhar, N., Prajapat, M., Avti, P., Kaur, H., Kumar, S., Singh, S., Kumar, H., Prakash, A., Dhibar, D. P., & Medhi, B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R., & Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design, 27(3), 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Shakibapour, N., Dehghani Sani, F., Beigoli, S., Sadeghian, H., & Chamani, J. (2019). Multi-spectroscopic and molecular modeling studies to reveal the interaction between propyl acridone and calf thymus DNA in the presence of histone H1: Binary and ternary approaches. Journal of Biomolecular Structure and Dynamics, 37(2), 359–371. 10.1080/07391102.2018.1427629 [DOI] [PubMed] [Google Scholar]
- Shang, J., Wan, Y., Liu, C., Yount, B., Gully, K., Yang, Y., Auerbach, A., Peng, G., Baric, R., & Li, F. (2020). Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathogens, 16(3), e1008392. 10.1371/journal.ppat.1008392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, J., Ye, G., Shi, K., Wan, Y., Luo, C., Aihara, H., Geng, Q., Auerbach, A., & Li, F. (2020). Structural basis of receptor recognition by SARS-CoV-2. Nature, 581(7807), 221–224. 10.1038/s41586-020-2179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi-Rad, A., Mehrzad, J., Darroudi, M., Saberi, M. R., & Chamani, J. (2020). Oil-in-water nanoemulsions comprising Berberine in olive oil: Biological activities, binding mechanisms to human serum albumin or holo-transferrin and QMMD simulations. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1724568 [DOI] [PubMed] [Google Scholar]
- Sharma, V., Chitranshi, N., & Agarwal, A. K. (2014). Significance and biological importance of pyrimidine in the microbial world. International Journal of Medicinal Chemistry, 2014, 202784. 10.1155/2014/202784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L. W., Mao, H. J., Wu, Y. L., Tanaka, Y., & Zhang, W. (2017). TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie, 142, 1–10. 10.1016/j.biochi.2017.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Meena, N. K., Das, T., Sharma, R. D., Prakash, A., & Lynn, A. M. (2019). Delineating the conformational dynamics of intermediate structures on the unfolding pathway of beta-lactoglobulin in aqueous urea and dimethyl sulfoxide. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2019.1695669 [DOI] [PubMed] [Google Scholar]
- Sohrabi, T., Hosseinzadeh, M., Beigoli, S., Saberi, M. R., & Chamani, J. (2018). Probing the binding of lomefloxacin to a calf thymus DNA-histone H1 complex by multi-spectroscopic and molecular modeling techniques. Journal of Molecular Liquids, 256, 127–138. 10.1016/j.molliq.2018.02.031 [DOI] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega, S., Alonso, J., Diaz, J. A., & Junquera, F. (1990). Synthesis of 3-substituted-4-phenyl-2-thioxo-1,2,3,4,5,6, 7,8-octahydrobenzo[4,5]thieno[2,3-d]pyrimidines [1]. Journal of Heterocyclic Chemistry, 27(2), 269–273. 10.1002/chin.199032196 [DOI] [Google Scholar]
- Vincent, M. J., Bergeron, E., Benjannet, S., Erickson, B. R., Rollin, P. E., Ksiazek, T. G., Seidah, N. G., & Nichol, S. T. (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virology Journal, 2, 69. 10.1186/1743-422X-2-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y. J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell, 181(2), 281–292. e286. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Nguyen, P. H., Pham, K., Huynh, D., Le, T. B., Wang, H., Ren, P., & Luo, R. (2016). Calculating protein-ligand binding affinities with MMPBSA: Method and error analysis. Journal of Computational Chemistry, 37(27), 2436–2446. 10.1002/jcc.24467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Wang, Y., Chen, Y., & Qin, Q. (2020). Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. Journal of Medical Virology, 92(6), 568–576. 10.1002/jmv.25748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. M., Delos, S. E., Brecher, M., & Schornberg, K. (2008). Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Critical Reviews in Biochemistry and Molecular Biology, 43(3), 189–219. 10.1080/10409230802058320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, S., Liu, M., Wang, C., Xu, W., Lan, Q., Feng, S., Qi, F., Bao, L., Du, L., Liu, S., Qin, C., Sun, F., Shi, Z., Zhu, Y., Jiang, S., & Lu, L. (2020). Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Research, 30(4), 343–355. 10.1038/s41422-020-0305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, F., Zhao, H., Li, D., Chen, H., Quan, H., Shi, X., Lou, L., & Hu, Y. (2011). Synthesis and biological evaluation of 2,4,5-substituted pyrimidines as a new class of tubulin polymerization inhibitors. Journal of Medicinal Chemistry, 54(9), 3200–3205. 10.1021/jm101388d [DOI] [PubMed] [Google Scholar]
- Xu, J., & Hagler, A. (2002). Chemoinformatics and drug discovery. Molecules: A Journal of Synthetic Chemistry and Natural Product Chemistry, 7(8), 566–600. 10.3390/70800566 [DOI] [Google Scholar]
- Zhu, X., Liu, Q., Du, L., Lu, L., & Jiang, S. (2013). Receptor-binding domain as a target for developing SARS vaccines. Journal of Thoracic Disease, 5 (Suppl 2), S142–S148. 10.3978/j.issn.2072-1439.2013.06.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete, V., Cuendet, M. A., Grosdidier, A., & Michielin, O. (2011). SwissParam: A fast force field generation tool for small organic molecules. Journal of Computational Chemistry, 32(11), 2359–2368. 10.1002/jcc.21816 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.