Abstract
Computer-aided drug screening by molecular docking, molecular dynamics (MD) and structural–activity relationship (SAR) can offer an efficient approach to identify promising drug repurposing candidates for COVID-19 treatment. In this study, computational screening is performed by molecular docking of 1615 Food and Drug Administration (FDA) approved drugs against the main protease (Mpro) of SARS-CoV-2. Several promising approved drugs, including Simeprevir, Ergotamine, Bromocriptine and Tadalafil, stand out as the best candidates based on their binding energy, fitting score and noncovalent interactions at the binding sites of the receptor. All selected drugs interact with the key active site residues, including His41 and Cys145. Various noncovalent interactions including hydrogen bonding, hydrophobic interactions, pi–sulfur and pi–pi interactions appear to be dominant in drug–Mpro complexes. MD simulations are applied for the most promising drugs. Structural stability and compactness are observed for the drug–Mpro complexes. The protein shows low flexibility in both apo and holo form during MD simulations. The MM/PBSA binding free energies are also measured for the selected drugs. For pattern recognition, structural similarity and binding energy prediction, multiple linear regression (MLR) models are used for the quantitative structural–activity relationship. The binding energy predicted by MLR model shows an 82% accuracy with the binding energy determined by molecular docking. Our details results can facilitate rational drug design targeting the SARS-CoV-2 main protease.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, SARS-CoV-2, FDA approved drug, structural–activity relationship, molecular docking, molecular dynamics, principal component analysis
1. Introduction
A novel coronavirus was identified in Wuhan, China in December 2019 spreading an infectious pneumonia-like disease (Huang et al., 2020). The International Committee on Taxonomy of Viruses (ICTV) named the novel coronavirus (2019-nCoV) as SARS-CoV-2 (Gorbalenya et al., 2020). The World Health Organization (WHO) officially named the disease caused by SARS-CoV-2 as Coronavirus disease (COVID-19) on 11 February 2020. On the basis of ‘alarming levels of spread and severity, and by the alarming levels of inaction’, on 11 March 2020, the Director-General of WHO characterized the COVID-19 situation as a pandemic (Bedford et al., 2020). To date, the total numbers of reported cases have reached over 5 million and the number of death is more than 300 thousand (Worldometer, 2020). Earlier, epidemics of severe acute respiratory syndrome coronavirus (SARS-CoV) in Guangdong, China in 2003 and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012 have shown a high fatality rate (Hilgenfeld & Peiris, 2013; Ton et al., 2020). SARS-CoV-2 was found to be an enveloped positive-sense, single- strained RNA virus belonging to the genus betacoronavirus (Beta-CoV) (Chan et al., 2020; Lu et al., 2020). Phylogenetic analysis showed that SARS-CoV-2 is closely related to (with 88% identity) two bat-derived severe acute respiratory syndrome (SARS)-like coronaviruses, bat-SL-CoVZC45 and bat-SL- CoVZXC21, collected in 2018 in Zhoushan, Eastern China, but are more distant from SARS-CoV (about 79%) and MERS-CoV (about 50%) (Lu et al., 2020). CoVs have the largest known RNA virus genomes, ranging from 27 to 34 kb (Sexton et al., 2016).
Inside the host cell, coronavirus genomes are translated into two groups of proteins; structural proteins, such as Spike (S), Envelope (E), Matrix (M) and Nucleocapsid (N) and nonstructural proteins, such as 3C-Like Protease (3CL-PRO, nsp5), RNA Dependent RNA Polymerase (RdRp, nsp12) (Elfiky, 2020). Coronavirus genome replication and transcription takes place at cytoplasmic membranes and involves a coordinated process of both continuous and discontinuous RNA synthesis that is mediated by the viral replicate (Sola et al., 2015). The main protease (Mpro) of SARS-CoV-2 plays an important role in cutting down polyproteins into functional pieces (Chen et al., 2020; Shaghaghi, 2020). Thus, the main protease plays a vital role in viral replication. The main protease (also known as 3C-like protease) can be an attractive drug target to inhibit coronavirus. Inhibiting the activity of the main protease could block the replication of the virus inside infected cells.
The crystallized structure of the main protease (Mpro) or Chymotrypsin-like protease (3CL-Pro) of SARS-CoV-2 was identified and repositioned at the RCSB Protein Data bank (PDB) (Jin et al., 2020). The crystallized structure (PDB ID: 6LU7) contains two chains, A and B, which form a homodimer. Chain A was used for macromolecule preparation. The protein chain is composed of 306 residues. His41 and Cys145 form an uncharged catalytic dyad (Paasche et al., 2014). The active site of the protease is activated by a protonation reaction in the catalytic site (Paasche et al., 2014). Inhibiting the activity of the catalytic dyad will ultimately help to inhibit the activity of the main protease.
Currently, no effective drugs targeting SARS-CoV-2 are available. Drug repurposing, a strategy to identify new uses of approved drugs, could shorten the time and reduce the cost for identifying effective drugs against COVID-19 (Zhou et al., 2020). We have selected 1615 FDA approved drugs from the ZINC database (Irwin & Shoichet, 2005). The focus of this study is to identify the binding affinities and molecular interactions of these drugs against the main protease using computational and statistical tools. Molecular docking, molecular dynamics (MD), principal components analysis (PCA) and quantitative structure–activity relationships (QSAR) were executed to evaluate the performance of the drugs.
2. Computational methods
2.1. Virtual screening and molecular docking protocols
The structures of 1615 FDA approved drugs are obtained from the ZINC database. The database was then checked for redundant molecules. The three-dimensional crystal structure of SARS-CoV-2 main protease (Mpro,3CLpro) was collected from the RCSB Protein Data Bank (PDB) database (PDB-ID:6LU7, Resolution = 2.16 Å, Method: X-ray diffraction, Organism: Bat-SARS like coronavirus) (Zhang et al., 2020). The crystal structure of Mpro was prepared according to previously published methods for molecular docking (Islam et al., 2020). Potential active site residues were specified by their vicinity to the ligand, N3 (Sekhar, 2020). PyRx Virtual Screening Tools incorporated with Autodock Vina was used for virtual screening (Dallakyan & Olson, 2015). Structural optimization of the drugs was carried out with universal force field (UFF) using the steepest descent optimization algorithm, a total of 2000 minimization steps (Ahmed et al., 2020). These drugs were converted to AutoDock ligand (PDBQT format) for docking. The grid center points were set to X = −11.5693, Y = 14.9663, Z = 67.9041 and box dimensions (Angstrom) X = 34.6857, Y = 45.2264, Z = 42.8980. Grid box center points and dimensions were set to target the substrate binding-binding pocket of the protein (Chen et al., 2006). The binding affinities of the drugs were measured in kcal/mol unit and sorted according to the higher negative values, which imply the best binding affinities (de Sousa et al., 2020). Based on calculated binding affinities, the 31 top-ranked potential drugs were chosen for further analysis. The top-ranked drugs were then optimized by Gaussian 09 software using semi-empirical PM6 method (Frisch, 2009). These optimized drug structures are then docked again using AutoDock Vina software and GOLD (Jones et al., 1997) docking programs. The molecular interactions of the drugs as predicted by docking simulation were analyzed in BIOVIA Discovery Studio Visualizer (Studio, 2015).
2.2. MD simulation protocols
The MD simulations for the main protease were conducted (6LU7) in apo-form (protein without ligand) and in holo-form (protein–drug complex) to assess any probable conformational changes to and interactions with their structures over the 100 nanoseconds (ns). AMBER14 force field (Dickson et al., 2014) was used on YASARA Dynamics (Krieger et al., 2013) for the simulation. Water molecules were added and the system was neutralized by the addition of NaCl salt of 0.9% concentration at a temperature of 310 K (Krieger et al., 2006). The particle-mesh Ewald method was used for calculating long-range electrostatic interactions. A periodic boundary condition was utilized to carry out the simulation. In all cases, the cell size was larger than the protein by 20 Å. The temperature of the simulation was controlled by the Berendsen thermostat. The initial energy minimization process of each simulation was performed by the simulated annealing method, using the steepest gradient approach (5000 cycles). Normal simulation speed (1.25 fs time step) was maintained during all the simulations (Krieger & Vriend, 2015). A snapshot was saved in every 100 ps during simulations. After completion of 100 ns MD simulation for all systems, the results were analyzed in YASARA. During the analysis of the simulations, time, energy, bond distances, bond angles, dihedral angles, solvent–accessible surface area (SASA), molecular surface area (MolSA), coulombic and Van der Waals interactions, root mean square fluctuation (RMSF) and root mean square deviation (RMSD) values for backbone, alpha carbon and heavy atoms were recorded. For more accuracy, the 100 ns MD simulation was conducted twice for every complex and the average result was used for analysis.
For binding free energy calculations, molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) method was used in YASARA Dynamics software (Baker et al., 2001; Chirag et al., 2020; Massova & Kollman, 2000). The AMBER14 force field was used during the binding free energy calculation. The default macro file was modified for the calculation. The following equation was used for the binding free energy calculation:
Here, ΔGMM is the molecular mechanics interaction (sum of electrostatic and van der Waals interaction), ΔGPB and ΔGSA correspond to polar and nonpolar solvation energyies, respectively. TΔS is the entropic contribution. Due to the high computational time, the entropy contribution was not considered. The final 50 ns data of MD simulation was used for binding free energy calculations.
Principal component analysis (PCA) is applied to identify similarities and dissimilarities among the collected energy profiles of MD trajectory data (Martens & Naes, 1998). The important components of a PCA model are highlighted in the following equation: X = TkPkT + E, where the X matrix expresses a product of two new matrices, i.e. Tk, Pk, Tk is the matrix of scores that shows how samples relate to each other, Pk is the matrix of loadings, which carries information about how the variables relate to each other, k represents the number of factors involved in the model and E is the matrix of residuals.
The selected four drug–main protease complexes may have dissimilarities with the apo–Mpro during MD simulation regarding the energy profile. These dissimilarities can be identified using the PCA algorithm (Islam et al., 2019, 2020). All calculations were carried out on R platform employing in-house developed codes and plots were produced using the package ggplot2 (Kassambara, 2016). Data were preprocessed employing autoscale function before applying the PCA algorithm (Martens & Naes, 1998). The last 40 ns data of the MD trajectory was utilized for PCA.
2.3. Structure–activity relationship protocols
Structure–activity relationship (SAR) is a method for establishing computational or mathematical models that aim to detect a statistically significant correlation between structure and function employing chemometric technique (Verma et al., 2010). 31 initially selected drugs are explored in this study. Topological polar surface area (TPSA, Å2), molecular weight (MW, gmol−1), XLogP3-AA, hydrogen bond donor (HBD), hydrogen bond acceptor (HBA), number of the rotatable bonds (ROTB), number of H, C, O, Cl, F atoms, single bonds (SB) count, number of double bonds (DB) and number of benzene rings of the drug candidates were studied as variables. Along with calculated binding energies, these variables were applied to correlate with the SAR by multiple linear regression (MLR) (Fakayode et al., 2009; Mark & Workman, 2007). MLR was carried out with XLSTAT (Adinsoft, 2010).
3. Results and discussions
3.1. Virtual screening and molecular docking
The structure of 1615 FDA approved drugs is considered for virtual screening using Autodock Vina. Considering −5.1 kcal/mol as a cutoff value, the binding affinities of the drugs are distributed between −5.1 to −6.0, −6.1 to −7.0, −7.1 to −8.0, −8.1 to −9.0 and −9.1 to −10.4 kcal/mol (Figure 1). Most drugs are found to have binding affinities between −6.1 to −7.0 kcal/mol. The top 31 candidates are selected for further analysis according to their binding affinities. The binding affinity of two already reported drugs Remdesivir and Lopinavir were observed −7.5 and −7.4 kcal/mol, respectively. After optimizing by PM6 level of theory, the selected drugs are screened again in AutoDock Vina. GOLD suit was also employed to understand the binding fitness using the CHEMPLP fitness score. Here, a higher fitness score suggests better docking interaction between protein and ligand. Along with 5 previously suggested FDA approved drugs (Contini, 2020), the binding affinities and GOLD fitting scores of 31 drugs are summarized in Table 1. According to best binding affinities, Simeprevir, Ergotamine, Bromocriptine and Tadalafil are selected for further analysis (Figure 2). Remdesivir and Lopinavir are also considered as control drugs for MD simulations.
Figure 1.
Frequency distribution of FDA approved drugs over the range of docking score (Cutoff value –5.1 kcal/mol).
Table 1.
AutoDock Vina predicted binding affinity (kcal/mol) and GOLD fitting score of 31 drugs and 5 already reported drugs with SARS-CoV-2 Mpro.
| Drug | Binding affinity (kcal/mol) | GOLD fitting score |
|---|---|---|
| Simeprevir | –10.3 | 73.83 |
| ZINC000164760756 | ||
| Ergotamine | –9.8 | 81.8 |
| ZINC000052955754 | ||
| Bromocriptine | –9.6 | 70.66 |
| ZINC000053683151 | ||
| Tadalafil | –9.5 | 59.32 |
| ZINC000003993855 | ||
| Dihydroergotamine | –9.2 | 75.46 |
| ZINC000003978005 | ||
| Perampanel | –9.2 | 74.57 |
| ZINC000030691797 | ||
| Nilotinib | –9.2 | 84.77 |
| ZINC000006716957 | ||
| Rolapitant | –9.1 | 72.66 |
| ZINC000003816514 | ||
| Naldemedine | –8.9 | 70.68 |
| ZINC000100378061 | ||
| Irinotecan ZINC000001612996 | –8.9 | 72.3 |
| Raltegravir | –8.9 | 64.33 |
| ZINC000013831130 | ||
| Lumacaftor | –8.8 | 68.84 |
| ZINC000064033452 | ||
| Eltrombopag | –8.8 | 69.34 |
| ZINC000011679756 | ||
| Saquinavir | –8.7 | 87.79 |
| ZINC000003914596 | ||
| Sildenafil | –8.7 | 80.21 |
| ZINC000019796168 | ||
| Pimozide | –8.6 | 77.44 |
| ZINC000004175630 | ||
| Paliperidone | –8.5 | 62.18 |
| ZINC000004214700 | ||
| Suvorexant | –8.5 | 62.23 |
| ZINC000049036447 | ||
| Nintedanib | –8.5 | 79.48 |
| ZINC000100014909 | ||
| Maraviroc | –8.5 | 74.25 |
| ZINC000100003902 | ||
| Paliperidone | –8.4 | 58.75 |
| ZINC000001481956 | ||
| Conivaptan | –8.4 | 69.76 |
| ZINC000012503187 | ||
| Pazopanib | –8.4 | 66.93 |
| ZINC000011617039 | ||
| Ibrutinib | –8.4 | 76.69 |
| ZINC000035328014 | ||
| Tipranavir | –8.4 | 79.31 |
| ZINC000100022637 | ||
| Dabrafenib ZINC000068153186 | –8.3 | 68.28 |
| Telotristat | –8.3 | 74.89 |
| ZINC000084758235 | ||
| Teniposide | –8.2 | 58.82 |
| ZINC000004099009 | ||
| Apixaban | –8.2 | 72.74 |
| ZINC000011677837 | ||
| Rifaximin | –7.7 | 42.56 |
| ZINC000169621200 | ||
| Lifitegrast | –7.5 | 73.73 |
| ZINC000084668739 | ||
| Montelukast (Contini, 2020) | –8.3 | 93.62 |
| GHRP-2 (Contini, 2020) | –8.1 | 95.49 |
| Indinavir (Contini, 2020) | –8 | 88.35 |
| Cobicistat (Contini, 2020) | –7.5 | 86.53 |
| Angiotensin II (Contini, 2020) | –6.9 | 89.85 |
Figure 2.
Two dimensional (2D) structures of the selected drugs.
3.2. Molecular interactions of the selected drugs with the main protease
Noncovalent interactions of the selected drugs which are detected by Autodock Vina reveals that all of them interact with either both catalytic residues (His41 and Cys145) or at least one of them, as shown in Figure 3.
Figure 3.
Nonbonding interactions of top drug candidates with the main protease of SARS-CoV-2 (pose predicted by AutoDock Vina). (a) Simeprevir, (b) Ergotamine, (c) Bromocriptine, and (d) Tadalafil.
Simeprevir, Ergotamine, Bromocriptine and Tadalafil form a number of hydrogen bonds and hydrophobic interactions with the protein (noncovalent interactions detected by GOLD are reported in (Table S2, supporting information). Among them, most of the interactions with the active site residues are categorized as hydrophobic (Table 2). Both Simeprevir and Ergotamine form a hydrogen bond with at least one of the catalytic residues.
Table 2.
Noncovalent interactions of selected four drugs with main protease of SARS-CoV-2 (pose predicted by AutoDock Vina) where, H = Hydrogen bond, CH = Conventional Hydrogen bond, C = Carbon Hydrogen bond.
| Interacting residue | Distance | Bond category | Bond type |
|---|---|---|---|
| Simeprevir | |||
| CYS145 | 2.7202 | H | CH |
| GLN189 | 2.29 | H | CH |
| GLY143 | 2.19455 | H | CH |
| THR190 | 3.81766 | Hydrophobic | Amide-Pi Stacked |
| PRO168 | 4.48443 | Hydrophobic | Alkyl |
| LEU50 | 5.06348 | Hydrophobic | Alkyl |
| LEU50 | 4.94019 | Hydrophobic | Alkyl |
| PRO168 | 4.58666 | Hydrophobic | Pi-Alkyl |
| ALA191 | 4.47775 | Hydrophobic | Pi-Alkyl |
| PRO168 | 3.91598 | Hydrophobic | Pi-Alkyl |
| Ergotamine | |||
| HIS41 | 2.88425 | H | C |
| MET165 | 1.89419 | H | C |
| MET165 | 4.81695 | Hydrophobic | Pi-Alkyl |
| HIS41 | 4.81113 | Hydrophobic | Pi-Pi-T Shaped |
| MET165 | 5.65835 | Other | Pi-sulfur |
| Bromocriptine | |||
| GLY143 | 2.32271 | H | CH |
| GLY143 | 1.86614 | H | CH |
| ARG188 | 2.28318 | H | CH |
| ASN142 | 2.55112 | H | C |
| GLY143 | 2.91712 | H | C |
| GLN189 | 1.99597 | H | C |
| LEU27 | 3.98094 | Hydrophobic | Alkyl |
| MET165 | 5.33935 | Hydrophobic | Alkyl |
| LEU27 | 4.43789 | Hydrophobic | Alkyl |
| CYS145 | 3.95427 | Hydrophobic | Alkyl |
| MET49 | 4.83683 | Hydrophobic | Alkyl |
| HIS41 | 5.27854 | Hydrophobic | Pi-Alkyl |
| MET165 | 4.15194 | Hydrophobic | Pi-Alkyl |
| MET165 | 4.68951 | Hydrophobic | Pi-Alkyl |
| GLN189 | 2.38092 | Hydrophobic | Pi-Sigma |
| HIS41 | 5.13653 | Hydrophobic | Pi-Pi-T Shaped |
| Tadalafil | |||
| ASN142 | 2.16312 | H | C |
| GLY143 | 2.66564 | H | C |
| MET165 | 2.64728 | H | C |
| CYS145 | 5.22621 | Hydrophobic | Alkyl |
| MET49 | 5.50154 | Other | Pi-Sulfur |
| MET49 | 5.03037 | Hydrophobic | Pi-Alkyl |
| CYS145 | 5.47403 | Hydrophobic | Pi-Alkyl |
| MET49 | 4.6017 | Hydrophobic | Pi-Alkyl |
| HIS41 | 5.18874 | Hydrophobic | Pi-Alkyl |
Simeprevir is observed to show three hydrophobic interactions with Pro168, two with Leu50 and one with each of the Thr190 and Ala191. Ergotamine exhibits one pi–alkyl and one pi–pi T-shaped hydrophobic interaction with Met165 and His41, respectively. Among all the protein–drug complexes, Bromocriptine demonstrates the most noncovalent interactions with the main protease. It shows two hydrophobic interactions with His41, two with Leu27, three with Met165 and one with each of the Met49, Cys145 and Gln189 residues. Tadalafil exhibits hydrophobic interactions with both catalytic residues. Both Ergotamine and Tadalafil show one pi–sulfur interaction with Met165 and Met49 residues, respectively.
3.3. MD simulation
MD simulation for all complexes of the main protease (6LU7) with the top four selected drugs and apo–form is performed for 100 ns. RMSD of alpha carbon atoms, RMSF of all amino acid residues, solvent accessible surface area, radius of gyration and the number of hydrogen bonds of all the drug–protein complexes and apo–Mpro are investigated.
The RMSD of Simeprevir–Mpro and Ergotamine–Mpro are stabilized after 20 ns of simulation, with an average value of 1.81 ± 0.30 Å and 1.90 ± 0.32 Å, respectively (Table 3). Figure 4a reveals that Bromocriptine–Mpro follows a similar trend in MD trajectory like apo–Mpro. Tadalafil–Mpro complex shows fluctuation until 30 ns, while it reaches a peak of 3.01 Å at 26.8 ns. It exhibits structural stability after 30 ns of MD simulation. These drug–Mpro complexes exhibit better stability in the RMSD trajectory than the control drugs. Unstable nature is observed for the RMSD of Remdesivir–Mpro after 70 ns of MD simulation, while the Lopinavir–Mpro complex followed the similar trend like the Bromocriptine–Mpro complex. Figure 4b displays root-mean-square fluctuation as a function of the residue number of the apo–Mpro and drug–Mpro complexes. This is explored to understand how the binding of drug molecules changes the behavior of the amino acid residues of the protein. A low RMSF is observed for the apo–Mpro which suggests low flexibility of protein even without a ligand. It also shows flexibility at Ser46 to Asn53, Tyr154, Val303, Thr304, Phe305 and Gln306. The residues Arg188 to Asp197 show high flexibility in Tadalafil–Mpro complex. RMSF of Simeprevir–Mpro, Ergotamine–Mpro, Bromocriptine–Mpro, Remdesivir–Mpro and Lopinavir–Mpro complexes are almost similar in most areas.
Table 3.
Average RMSD, SASA, Rg, number of hydrogen bonds and binding free energy of the selected drugs–Mpro complexes.
| Complex | RMSD (Å) | SASA (Å2) | Radius of gyration (Å) | Number of hydrogen bonds | Binding free energy (kcal/mol) |
|---|---|---|---|---|---|
| Apo–Mpro | 2.07 ± 0.32 | 14137.41 ± 217.54 | 22.35 ± 0.14 | 510.2 ± 11.80 | – |
| Simeprevir–Mpro | 1.81 ± 0.30 | 13889.76 ± 295.20 | 22.39 ± 0.14 | 502 ± 12.72 | –77.44 ± 2.43 |
| Ergotamine–Mpro | 1.90 ± 0.32 | 13800.03 ± 243.83 | 22.32 ± 0.11 | 503.36 ± 11.36 | –26.33 ± 0.39 |
| Bromocriptine–Mpro | 2.07 ± 0.36 | 14035.29 ± 208.71 | 22.20 ± 0.13 | 512.50 ± 12.33 | –33.69 ± 0.11 |
| Tadalafil–Mpro | 2.24 ± 0.41 | 14197.53 ± 390.24 | 22.57 ± 0.24 | 510.86 ± 14.66 | –14.46 ± 0.63 |
| Remdesivir–Mpro | 1.89 ± 0.36 | 13892.51 ± 252.63 | 22.22 ± 0.12 | 507.96 ± 12.04 | –0.31 ± 0.06 |
| Lopinavir–Mpro | 2.03 ± 0.41 | 14168.63 ± 226.90 | 22.42 ± 0.11 | 508.66 ± 12.10 | –27.89 ± 0.35 |
Figure 4.
Analysis of RMSD, RMSF, SASA, Rg and total number of hydrogen bonds of apo–Mpro and selected four drug–protein complexes at 100 ns. (a) Root-mean-square deviation (RMSD, Å) of the Cα atoms over the phase of 100 ns, (b) RMSF values of the alpha carbon over the entire simulation, where the ordinate is RMSF (Å), (c) Solvent accessible surface area (SASA), (d) Radius of gyration (Rg) over the entire simulation, (e) Total H-bond count throughout the simulation and (f) Binding Free Energy during the last 50 ns of simulation.
The radius of gyration (Rg) is the root mean square distance of various atoms of a protein from the axis of rotation, and it illustrates structural compactness. A greater change in Rg value may occur due to the folding or conformational changes of protein structure, while a lower degree of fluctuation over the simulation period indicates rigidity and higher compactness of a structure. According to the observed Rg value, the apo–Mpro and Simeprevir–Mpro complex showed almost similar structural compactness over the phase of the 100 ns simulation. The apo–Mpro complex experienced a fluctuation in Rg value during 80–90 ns. Ergotamine showed structural compactness all over the simulation time with an average of 22.32 ± 0.11 Å. Bromocriptine–Mpro followed a similar trend like the Remdesivir–Mpro complex after 45 ns. A loose packing is observed for Tadalafil–Mpro during 60 ns to 90 ns. Except for this time frame, Tadalafil–Mpro and Lopinavir–Mpro followed a similar trend in MD trajectory. Based on the overall Rg, the average values are 22.35 ± 0.14 Å, 22.39 ± 0.14 Å, 22.20 ± 0.13 Å, 22.57 ± 0.24 Å, 22.22 ± 0.12 Å and 22.42 ± 0.11 Å for apo–Mpro, Simeprevir–Mpro, Bromocriptine–Mpro, Tadalafil–Mpro, Remdesivir–Mpro and Lopinavir–Mpro complexes, respectively. To anticipate the solvent accessibility of all complexes, the solvent-accessible surface area (SASA) of apo–Mpro and complexes are calculated (Figure 4(c)). Binding of a drug to the protein may impact its structural properties, thus, SASA may also change. A higher SASA value suggests the expansion of protein structure. A low fluctuation of SASA value is expected during the simulation. All the examined complexes exhibit a similar trend until 40 ns of MD trajectory. Simeprevir–Mpro and Ergotamine–Mpro showed lower SASA value than the control drug–Mpro complexes. The overall average SASA value suggests Lopinavir could induce protein expansion, and thus, increase the solvent accessible surface of the protein (Table 3).
Since intermolecular hydrogen bonds between protein–drug complexes have a significant contribution to the conformational stability of the complex, the total number of hydrogen bonds is calculated (Figure 4(e)). The average number of hydrogen bonds observed in apo–Mpro is 510.2 ± 11.80. The Bromocriptine–Mpro complex has the most hydrogen bonds over the 100 ns simulation time (Table 3).
Binding free energy calculations for each complex were conducted by employing the MM/PBSA method. The Simeprevir–Mpro shows the highest average binding free energy of −77.44 ± 2.43 kcal/mol. All our selected drug–Mpro complexes demonstrate better binding free energy than the Remdesivir–Mpro complex (Figure 4(f)). The binding free energy of the Remdesivir–Mpro complex persisted as positive most of the time, which indicates loose binding between the drug and the protein. The binding free energy of Ergotamine–Mpro, Bromocriptine–Mpro, Tadalafil–Mpro and Lopinavir–Mpro complexes remained negative most of the time, which also indicates good binding (Figure S2, supporting information). The average binding free energy was observed −26.33 ± 0.39, −33.69 ± 0.11, −14.46 ± 0.63, −0.31 ± 0.06 and −27.89 ± 0.35 kcal/mol for Ergotamine, Bromocriptine, Tadalafil, Remdesivir and Lopinavir, respectively.
The snapshots of Mpro–drug complexes are visualized at different time intervals to explore the drugs pose during MD simulation. All our selected drugs stayed at the binding pocket of the protein over the phase of 100 ns MD simulation (Figure 5). Additionally, all drugs interact with at least one of the catalytic residues during most of the time of MD simulation (Table S4, supporting information).
Figure 5.
Binding pose of drugs during 100 ns MD simulation. The crystal structure of Mpro is shown in beige color with (a) Simeprevir (cyan), (b) Ergotamine (green), (c) Bromocriptine (blue) and (d) Tadalafil (orange red).
3.4. Principal components analysis
PCA is applied to observe the structural properties and energy profiles of apo–Mpro and the selected four drug–Mpro complexes. A single PCA model consisting of five training sets (Mpro and four drug–Mpro complexes) was constructed for analysis. The first two PCs explained 91.74% of the total variance, where PC1 contributed 75.07% and PC2 contributed 16.67% of the variance. It is observed from the scores plot (Figure 6(a)) that the apo–Mpro (violet) and Simeprevir–Mpro complex (cyan) overlap with each other, thus, indicating similarity in energy and structural profile. Moreover, Ergotamine–Mpro (green) and Tadalafil–Mpro (orange–red) shifted slightly toward the right in the PC1 direction and resided very close to apo–Mpro and Simeprevir–Mpro complex. Thus, by forming a complex with Ergotamine and Tadalafil, Mpro showed a similar structural profile compared to Mpro alone. However, the Bromocriptine–Mpro complex (blue) shifted significantly toward the positive direction of PC1. This indicates greater dissimilarity of Bromocriptine–Mpro with apo–Mpro and other complexes. The loading plot (Figure 6(b)) revealed that the variables are separated in the PC1 direction, which explains different cluster formation in the PC1 direction. Therefore, it is evident from the PCA scoring plot that Simeprevir has the best binding stability with Mpro, while Ergotamine and Tadalafil also demonstrate almost similar behavior in MD simulation.
Figure 6.
(a) The score plot presented five data clusters in different color, where each dot represents one time point. The clustering is attributable as: apo–Mpro (violet), Simeprevir–Mpro complex (cyan), Ergotamine–Mpro (green), Bromocriptine–Mpro (blue), Tadalafil–Mpro (orange red) (b) loading plot from principal components analysis of the energy and structural data.
3.5. Structure–activity relationship
SAR has been used for decades by researchers as a cheaper way to develop relationships between physiochemical properties of chemicals and their bioactivities (Santos-Filho et al., 2009; Verma et al., 2010). It is a widely used tool in bioinformatics, drug discovery for clinical research, pharmaceutical industry, petrochemical and agrochemical sectors for modeling and predictive pattern analysis (Alam & Khan, 2017; Fakayode et al., 2014; Funar-Timofei et al., 2017). For further analysis, MLR has been employed. For instance, TPSA (Å2), molecular weight, XLogP3, H-bond donor count, H-bond acceptor count of the drugs are the most significant variables on QSAR contributors to the MLR model (Table S3, supporting information).
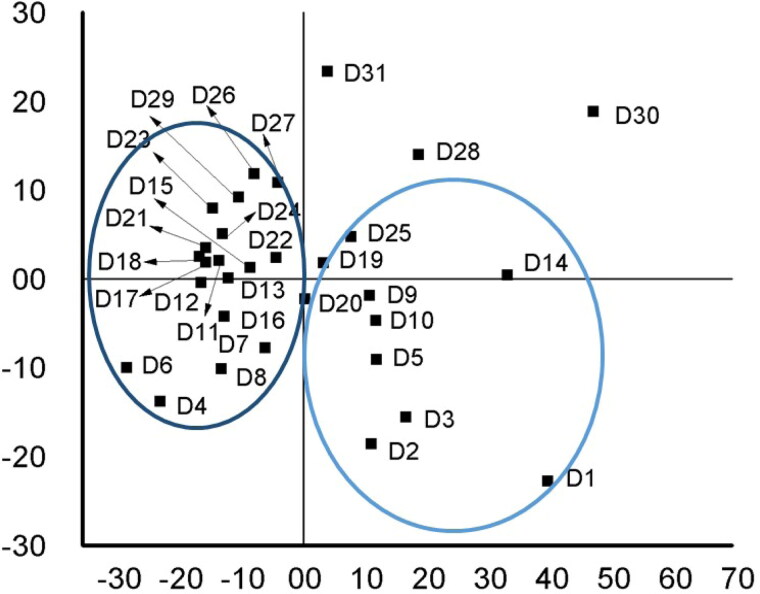
The drugs with similar functional groups are found to stay side by side on the score plot (Figure 7). For example, drugs (D2, D3 and D5) containing indole ring, amide group and alcohol group are clustered together on the fourth quadrant of the score plot. On the other hand, drugs (D23, D7, D16, D13, D22 and D24) with diazole ring attached to the benzene ring are clustered on the second and third quadrants. The grouping on the score plot was highly significant.
Figure 7.
Score plot of PCA analysis for quantitative structural–activity relationship of drugs.
The MLR model is generated and employed to predict the binding energy obtained from molecular docking. Additionally, a test set is utilized for the validation of drug candidates. The observed results of validation reveal similarity in the binding energies (Table 4).
Table 4.
Predicted binding energy by the MLR model and actual binding energy from molecular docking.
| Sample | Predicted binding energy (kcal/mol) | Actual binding energy(kcal/mol) | %RE |
|---|---|---|---|
| D2 | –8.89 | –9.80 | –2.99 |
| D5 | –8.92 | –9.20 | –0.91 |
| D8 | –8.90 | –9.10 | –0.66 |
| D12 | –8.76 | –8.80 | –0.13 |
| D15 | –8.99 | –8.70 | 0.98 |
| D20 | –9.03 | –8.50 | 1.77 |
| D24 | –8.68 | –8.40 | 0.92 |
| D26 | –8.62 | –8.30 | 1.07 |
| D28 | –9.02 | –8.20 | 2.72 |
| D29 | –8.77 | –8.20 | 1.90 |
| D31 | –8.64 | –7.50 | 3.79 |
| RMS%RE | 1.95 |
The ability of the MLR model to accurately anticipate the binding energy of the validated drugs is evaluated by the root-mean-square-relative percent-error (RMS%RE). The result obtained from MLR is 82% accurate while comparing to the binding energy. Yet, findings from this analysis can help researchers predict the drug performances against the main protease considering the binding energy, chemical behavior and pharmacological efficacy of upcoming drug candidates.
4. Conclusion
In this study, several computational and statistical tools such as molecular docking, MD simulation, SAR and PCA are used to detect the best existing drugs against the main protease of SARS-CoV-2. Among the 1615 studied drugs, Simeprevir, Ergotamine, Bromocriptine and Tadalafil are found to have the highest binding affinity. As these drugs are already certified for human use, they do not require long-term clinical trials. These drugs show a significant number of noncovalent interactions including hydrogen bond, hydrophobic interaction and electrostatic interaction with the binding site residues of Mpro. All the selected drugs interact either with both (His41 and Cys145) or at least one of the catalytic residues. Furthermore, MD simulations exhibit that Simeprevir, Ergotamine and Bromocriptine drugs with Mpro protein preserve the structural stability over the simulation period. Additionally, the MM/PBSA free energy profiles of these drugs suggest strong binding with the receptor. SAR pattern recognition reveals that drugs containing an indole ring, amide group and alcohol group are clustered together in the fourth quadrant and drugs containing an azole ring attached to benzene ring are assembled together in the second and third quadrant of the scoring plot. The predicted binding affinity values from MLR shows 82% accuracy compared to values obtained from molecular docking. It can be concluded that the selected drugs are promising and can be used to design effective drugs against the SARS-CoV-2.
Supplementary Material
Acknowledgements
We are grateful to our donors (http://grc-bd.org/donate/) who supported the building of the computational platform in Bangladesh. The authors acknowledge the World Academy of Science (TWAS) for purchasing the High-Performance computer for the MD simulations.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Adinsoft, S. ( 2010). XLSTAT-software, version 10. Addinsoft. [Google Scholar]
- Ahmed, S., Islam, N., Shahinozzaman, M., Fakayode, S. O., Afrin, N., & Halim, M. A. (2020). Virtual screening, molecular dynamics, density functional theory and quantitative structure activity relationship studies to design peroxisome proliferator-activated receptor-γ agonists as anti-diabetic drugs. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1714482 [DOI] [PubMed] [Google Scholar]
- Alam, S., & Khan, F. (2017). 3D-QSAR studies on maslinic acid analogs for anticancer activity against breast cancer cell line. Scientific Reports, 7(1), 13. 10.1038/s41598-017-06131-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, N. A., Sept, D., Joseph, S., Holst, M. J., & McCammon, J. A. (2001). Electrostatics of nanosystems: Application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America, 98(18), 10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford, J., Enria, D., Giesecke, J., Heymann, D. L., Ihekweazu, C., Kobinger, G., Lane, H. C., Memish, Z., Oh, M.-D., Sall, A. A., Schuchat, A., Ungchusak, K., & Wieler, L. H. (2020). COVID-19: Towards controlling of a pandemic. The Lancet, 395(10229), 1015–1018. 10.1016/S0140-6736(20)30673-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F.-W., Kok, K.-H., Zhu, Z., Chu, H., To, K. K.-W., Yuan, S., & Yuen, K.-Y. (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes & Infections, 9(1), 221–236. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., Wei, P., Huang, C., Tan, L., Liu, Y., & Lai, L. (2006). Only one protomer is active in the dimer of SARS 3C-like proteinase. The Journal of Biological Chemistry, 281(20), 13894–13898. 10.1074/jbc.M510745200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Liu, Q., & Guo, D. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology, 92(4), 418–423. 10.1002/jmv.25681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirag, N. P., Prasanth Kumar, S., Himanshu, A. P., & Rakesh, M. R. (2020). Identification of potential binders of the SARS-Cov-2 spike protein via molecular docking, dynamics simulation and binding free energy calculation. chemRxiv. 10.26434/chemrxiv.12278588.v1 [DOI]
- Contini, A. (2020). Virtual screening of an FDA approved drugs database on two COVID-19 coronavirus proteins. https://chemrxiv.org/articles/Virtual_Screening_of_an_FDA_Approved_Drugs_Database_on_Two_COVID-19_Coronavirus_Proteins/11847381/files/21713028.pdf
- Dallakyan, S., & Olson, A. J. (2015). Small-molecule library screening by docking with PyRx. Methods in Molecular Biology (Clifton, N.J.), 1263, 243–250. 10.1007/978-1-4939-2269-7_19 [DOI] [PubMed] [Google Scholar]
- de Sousa, A. C. C., Combrinck, J. M., Maepa, K., & Egan, T. J. (2020). Virtual screening as a tool to discover new β-haematin inhibitors with activity against malaria parasites. Scientific Reports, 10(1), 3374. 10.1038/s41598-020-60221-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, C. J., Madej, B. D., Skjevik, Å. A., Betz, R. M., Teigen, K., Gould, I. R., & Walker, R. C. (2014). Lipid14: The amber lipid force field. Journal of Chemical Theory and Computation, 10(2), 865–879. 10.1021/ct4010307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sciences, 248, 117477. 10.1016/j.lfs.2020.117477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakayode, S. O., Busch, K. W., & Busch, M. A. (2009). Chemometric approach to chiral recognition using molecular spectroscopy. Encyclopedia of Analytical Chemistry. 10.1002/9780470027318.a9082 [DOI] [Google Scholar]
- Fakayode, S. O., Mitchell, B. S., & Pollard, D. A. (2014). Determination of boiling point of petrochemicals by gas chromatography-mass spectrometry and multivariate regression analysis of structural activity relationship. Talanta, 126, 151–156. 10.1016/j.talanta.2014.03.037 [DOI] [PubMed] [Google Scholar]
- Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Mennucci, B., & Petersson, G. A. (2009). Gaussian 09, A.02. Gaussian, Inc. [Google Scholar]
- Funar-Timofei, S., Borota, A., & Crisan, L. (2017). Combined molecular docking and QSAR study of fused heterocyclic herbicide inhibitors of D1 protein in photosystem II of plants. Molecular Diversity, 21(2), 437–454. 10.1007/s11030-017-9735-x [DOI] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Baker, S. C., Baric, R. S., de Groot, R. J., Drosten, C., Gulyaeva, A. A., Haagmans, B. L., Lauber, C., Leontovich, A. M., Neuman, B. W., Penzar, D., Perlman, S., Poon, L. L. M., Samborskiy, D. V., Sidorov, I., Sola, I., & Ziebuhr, J. (2020). The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology, 5(4), 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld, R., & Peiris, M. (2013). From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Research, 100(1), 286–295. 10.1016/j.antiviral.2013.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., Liu, M., … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin, J. J., & Shoichet, B. K. (2005). ZINC-a free database of commercially available compounds for virtual screening. Journal of Chemical Information and Modeling, 45(1), 177–182. 10.1021/ci049714+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M. J., Khan, A. M., Parves, M. R., Hossain, M. N., & Halim, M. A. (2019). Prediction of deleterious non-synonymous SNPs of human STK11 gene by combining algorithms, molecular docking, and molecular dynamics simulation. Scientific Reports, 9(1), 16426. 10.1038/s41598-019-52308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, R., Parves, M. R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., Hossain, Md. N.A., Md. Ali, & Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from COVID-19 virus and discovery of its inhibitors. BioRxiv, 2020.02.26.964882. 10.1101/2020.02.26.964882 [DOI] [Google Scholar]
- Jones, G., Willett, P., Glen, R. C., Leach, A. R., & Taylor, R. (1997). Development and validation of a genetic algorithm for flexible docking 1 1Edited by F. E. Cohen. Journal of Molecular Biology, 267(3), 727–748. 10.1006/jmbi.1996.0897 [DOI] [PubMed] [Google Scholar]
- Kassambara, A. (2016). Practical guide to principal component methods in R. STHDA, 2, 1–170. CreateSpace Independent Publishing Platform. [Google Scholar]
- Krieger, E., Elmar, G. V., & Spronk, C. (2013). YASARA–Yet another scientific artificial reality application. YASARA.Org.
- Krieger, E., Nielsen, J. E., Spronk, C. A. E. M., & Vriend, G. (2006). Fast empirical pKa prediction by Ewald summation . Journal of Molecular Graphics and Modelling, 25(4), 481–486. 10.1016/j.jmgm.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Krieger, E., & Vriend, G. (2015). New ways to boost molecular dynamics simulations. Journal of Computational Chemistry, 36(13), 996–1007. 10.1002/jcc.23899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark, H., & Workman, J. (2007). Chemometrics in spectroscopy. In Chemometrics in spectroscopy. Elsevier. 10.1016/B978-0-12-374024-3.X5000-4 [DOI] [Google Scholar]
- Martens, H., & Naes, T. (1998). Multivariate calibration. In Data handling in science and technology (Vol. 20). Wiley. 10.1016/S0922-3487(98)80046-4 [DOI] [Google Scholar]
- Massova, I., & Kollman, P. A. (2000). Combined molecular mechanical and continuum solvent approach (MM- PBSA/GBSA) to predict ligand binding. Perspectives in Drug Discovery and Design, 18(1), 113–135. 10.1023/A:1008763014207 [DOI] [Google Scholar]
- Paasche, A., Zipper, A., Schäfer, S., Ziebuhr, J., Schirmeister, T., & Engels, B. (2014). Evidence for substrate binding-induced zwitterion formation in the catalytic Cys-His dyad of the SARS-CoV main protease. Biochemistry, 53(37), 5930–5946. 10.1021/bi400604t [DOI] [PubMed] [Google Scholar]
- Santos-Filho, O. A., Hopfinger, A. J., Cherkasov, A., & de Alencastro, R. B. (2009). The receptor-dependent QSAR paradigm: An overview of the current state of the art. Medicinal Chemistry (Shariqah (United Arab Emirates)), 5(4), 359–366. 10.2174/157340609788681458 [DOI] [PubMed] [Google Scholar]
- Sekhar, T. (2020). Virtual screening based prediction of potential drugs for COVID-19. 10.20944/preprints202002.0418.v2 [DOI]
- Sexton, N. R., Smith, E. C., Blanc, H., Vignuzzi, M., Peersen, O. B., & Denison, M. R. (2016). Homology-based identification of a mutation in the coronavirus RNA-dependent RNA polymerase that confers resistance to multiple mutagens. Journal of Virology, 90(16), 7415–7428. 10.1128/JVI.00080-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaghaghi, N. (2020). Molecular docking study of novel COVID-19 protease with low risk terpenoides compounds of plants. ChemRxiv, 1(1). 10.26434/chemrxiv.11935722.v1 [DOI] [Google Scholar]
- Sola, I., Almazán, F., Zúñiga, S., & Enjuanes, L. (2015). Continuous and discontinuous RNA synthesis in coronaviruses. Annual Review of Virology, 2(1), 265–288. 10.1146/annurev-virology-100114-055218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studio, D. (2015). Dassault systemes BIOVIA, Discovery studio modelling environment, Release 4.5. Accelrys Software Inc. [Google Scholar]
- Ton, A.-T., Gentile, F., Hsing, M., Ban, F., & Cherkasov, A. (2020). Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Molecular Informatics 10.1002/minf.202000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, J., Khedkar, V., & Coutinho, E. (2010). 3D-QSAR in drug design-a review. Current Topics in Medicinal Chemistry, 10(1), 95–115. 10.2174/156802610790232260 [DOI] [PubMed] [Google Scholar]
- Worldometer. (2020). Coronavirus cases. Worldometer . https://www.worldometers.info/coronavirus/coronavirus-cases/#daily-cases
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 3405(March), eabb3405. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Hou, Y., Shen, J., Huang, Y., Martin, W., & Cheng, F. (2020). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery, 6(1) 14. 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.