Abstract
The sudden outburst of Coronavirus disease (COVID-19) caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) poses a massive threat to global public health. Currently, no therapeutic drug or vaccine exists to treat COVID-19. Due to the time taking process of new drug development, drug repurposing might be the only viable solution to tackle COVID-19. RNA‐dependent RNA polymerase (RdRp) catalyzes SARS-CoV-2 RNA replication and hence, is an obvious target for antiviral drug design. Interestingly, several plant-derived polyphenols effectively inhibit the RdRp of other RNA viruses. More importantly, polyphenols have been used as dietary supplementations for a long time and played beneficial roles in immune homeostasis. We were curious to study the binding of polyphenols with SARS-CoV-2 RdRp and assess their potential to treat COVID-19. Herein, we made a library of polyphenols that have shown substantial therapeutic effects against various diseases. They were successfully docked in the catalytic pocket of RdRp. The investigation reveals that EGCG, theaflavin (TF1), theaflavin-3’-O-gallate (TF2a), theaflavin-3’-gallate (TF2b), theaflavin 3,3'-digallate (TF3), hesperidin, quercetagetin, and myricetin strongly bind to the active site of RdRp. Further, a 150-ns molecular dynamic simulation revealed that EGCG, TF2a, TF2b, TF3 result in highly stable bound conformations with RdRp. The binding free energy components calculated by the MM-PBSA also confirm the stability of the complexes. We also performed a detailed analysis of ADME prediction, toxicity prediction, and target analysis for their druggability. Overall, our results suggest that EGCG, TF2a, TF2b, TF3 can inhibit RdRp and represent an effective therapy for COVID-19.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2 and RNA‐dependent RNA polymerase (RdRp), molecular docking, molecular dynamics, MM-PBSA, natural polyphenols
1. Introduction
An outbreak of Coronavirus disease (COVID-19) has caused a pandemic situation across the globe. Although the outburst was first observed at Wuhan city in China, at present more than 200 countries and territories around the world have witnessed the COVID-19 fatalities affecting all age groups. As of July 03, 2020, more than 11 million people have been affected by the disease, with a fatality of 5,25,410 across the globe as per the WHO report. Unfortunately, there is no clinically approved drug or vaccine for COVID-19 as of now. The disease is caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), a member of the Coronaviridae family of viruses and it belongs to the same family Betacoronaviruses, like Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) (Chan et al., 2015; Elfiky & Azzam, 2020; Ibrahim et al., 2020). The symptoms of SARS-CoV-2 infection include fever, dry cough, shortness of breath, runny nose, and sore throat (Wu et al., 2020). SARS-CoV-2 is a positive-sense single-stranded RNA virus, and its genome is around 29.7 kB long with twelve putative open reading frames (ORFs) that encode different viral structural and non-structural proteins. There are four structural proteins in SARS-CoV-2, namely spike (S), envelope (E), membrane (M), and nucleocapsid (N), and all of them can potentially serve as an antigen for neutralizing antibody preparation as potential therapeutics (Boopathi et al., 2020).
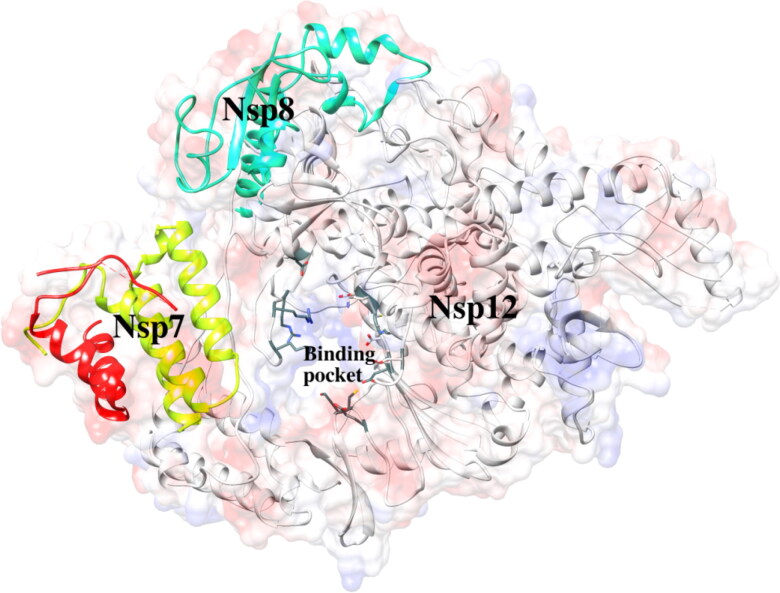
Another potential drug target for SARS-CoV-2 is RNA‐dependent RNA polymerase (RdRp) (Figure 1), which is a key component of the replication machinery of the virus to make multiple copies of the RNA genome (Elfiky 2020c). RdRp in various coronaviruses are remarkably similar. For example, the RdRp of SARS-CoV exhibits ∼97% sequence similarity with that of SARS-CoV-2. More importantly, there is no human polymerase counterpart that resembles the sequence/structural homology with RdRp from coronaviruses, and hence, the development of RdRp inhibitors could be a potential therapeutic strategy without risk of crosstalk with human polymerases (Borgio et al., 2020; Subissi et al., 2014; Zhai et al., 2005). Very recently, Yin et al. reported the crystal structure RdRp of SARS-CoV-2 complexed with an antiviral drug, Remdesivir highlighting how the template-primer RNA is recognized by the polymerase enzyme and the chain elongation is inhibited by Remdesivir providing a basis for developing a wide range of effective inhibitors to overcome from SARS-CoV-2 infection (Yin et al., 2020). RdRp has been found to be an effective drug target for several other RNA viruses, spanning from the hepatitis C virus, zika virus to coronaviruses (Elfiky, 2017, 2019; Ganesan & Barakat, 2017). The active site of RdRp is highly conserved, and the catalytic domains contain two consecutive aspartate residues in a beta-turn joining β15 and β16 (Elfiky 2020c). The general structure of RdRp consists of 7 motifs (A to G) among them inner channel of catalytic sites represented by motif A to C, and they play a crucial role during the nucleotide addition cycle (Jia & Gong, 2019; Wu & Gong, 2018).
Figure 1.
The 3-dimensional crystal structure of RNA‐dependent RNA polymerase (RdRp).
Epidemiological studies repetitively suggested that consumption of bioactive compounds (e.g. vitamins, phytochemicals, polyphenols, flavonoids, flavonols, and carotenoids etc.) has beneficial activity on human health and could minimize the risk of various diseases starting from cancers to different viral infections (Khan et al., 2020; Szajdek & Borowska, 2008). Traditional natural compounds have been consumed since ancient times as they exhibit less toxicity, low-cost availability, minimum side-effects and are rich in therapeutic resources. Some of the previous findings also suggest that naturally occurring compounds possess a wide range of antiviral properties against RNA viruses, including polio-virus type 1, parainfluenza virus type 3, and respiratory syncytial virus by inhibiting their replication (Lin et al., 2014). In that context, Ahmed-Belkacem et al. have screened more than forty potent natural flavonoids for their polymerase inhibition activity using HCV-NS5 strain and among the different flavonoids, quercetagetin showed strong HCV replication inhibitory activity in vitro (Ahmed-Belkacem et al., 2014). Previously, song et al. reported that green tea catechins, by disrupting the membrane of the influenza virus, inhibited neuraminidase in the crude system (Song et al., 2005). On a separate report, Takashi et al. reported that EGCG, a green tea polyphenol can inhibit the endonuclease activity of influenza A virus RNA polymerase. EGCG is also reported to interfere with viral replication via modulating the cellular redox environment (Ho et al., 2009; Kuzuhara et al., 2009). Therefore, the existing scientific evidence strongly suggests that natural flavonoids/polyphenol can act against SARS-CoV-2 (Aanouz et al., 2020; Elfiky 2020b; Elmezayen et al., 2020; Enmozhi et al., 2020). Because of the time-consuming process of new synthetic/semi-synthetic drug development, drug repurposing of phytomolecules is an ideal alternative in this urgent situation as the latter process is economical and scalable in a very short period of time (Adeoye et al., 2020; Islam et al., 2020; Muralidharan et al., 2020; Sinha et al., 2020). Hence, a comprehensive understanding of their binding to SARS-CoV-2 RdRp can yield interesting findings that can further be capitalized to develop COVID-19 drugs. Nevertheless, the apparent lack of cytotoxicity of polyphenols at even significantly high concentrations makes them potential antiviral drug candidates. In the present study, we selected a hundred natural polyphenols to assess their potential to act as SARS-CoV-2 RdRp inhibitors. The selected library was then explored to evaluate the binding affinity of individual polyphenols towards RdRp of the SARS-CoV-2 by molecular docking using AutoDock vina. Among the selected polyphenols, theaflavin (TF1), theaflavin-3′-O-gallate (TF2a), theaflavin-3′-gallate (TF2b), theaflavin 3,3′-digallate (TF3), hesperidin, EGCG, myricetin, and quercetagetin were found to be docked in the active site of RdRp of the SARS-CoV-2 with a highly favourable affinity for the binding pocket. Further to get a better understanding of the dynamics of the complexes, we performed a 150-nanoseconds molecular dynamic simulation with those eight polyphenols. The binding free energy components were calculated by the MM-PBSA. Remdesivir (Hendaus, 2020) and GTP (a physiological nucleotide) were taken as positive controls to validate our results.
2. Materials and methods
2.1. Molecular docking studies
2.1.1. Protein preparations
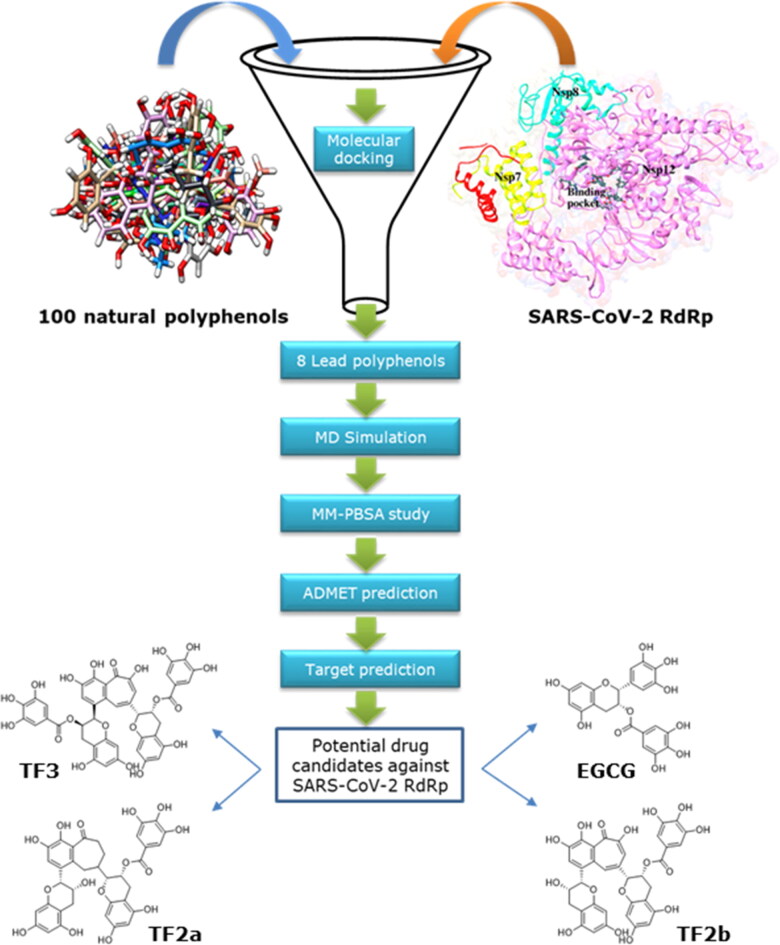
The crystal structure of SARS-CoV-2 RdRp (PDB ID: 6M71) (Yan et al., 2020) was retrieved from the protein databank (www.rcsb.org) (Berman et al., 2000). The crystal structure was prepared individually by adding hydrogen atoms and computing the Gasteiger charge using the AutoDock v4.2 program (Morris et al., 2009). Subsequently, the file was saved as .pdbqt format in preparation for molecular docking. Schematic representation of the work-flow for selecting potential natural polyphenolic inhibitors for the SARS-CoV-2 RdRp is shown in Figure 2.
Figure 2.
Flow chart of the methodology for shortlisting the best natural polyphenolic inhibitor of the SARS-CoV-2 RdRp.
2.1.2. Ligand preparations
The SDF structures of GTP, remdesivir, and selected hundred polyphenols (see Table S1 in Supplementary Information) were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (Kim et al., 2019). The compounds were converted into PDB format, and conformational energies of all the compounds were minimized by using UCSF Chimera (Pettersen et al., 2004).
Table 1.
Binding energy (kcal/mol) of the natural polyphenols along with the control compounds (GTP and remdesivir) against RdRp of the SARS-CoV-2 (PDB ID: 6M71) by molecular docking study.
| S. No. | Compound Name | Binding energy (kcal/mol) | S. No. | Compound Name | Binding energy (kcal/mol) |
|---|---|---|---|---|---|
| 1 | TF3 | −9.9 | 52 | Cyanidin | −6.3 |
| 2 | TF2b | −9.6 | 53 | Daidzein | −6.3 |
| 3 | TF1 | −9.6 | 54 | Glycitein | −6.3 |
| 4 | TF2a | −9.3 | 55 | Wogonin | −6.3 |
| 5 | Hesperidin | −8.8 | 56 | Phloretin | −6.3 |
| 6 | EGCG | −7.3 | 57 | Catechin | −6.2 |
| 7 | Myricetin | −7.2 | 58 | Urolithin B | −6.2 |
| 8 | Quercetagetin | −7.0 | 59 | Angolensin | −6.2 |
| 9 | Quercetin | −6.9 | 60 | Pinosylvin | −6.2 |
| 10 | Curcumin | −6.9 | 61 | Formononetin | −6.2 |
| 11 | Dihydrorobinetin | −6.8 | 62 | Liquiritigenin | −6.2 |
| 12 | Peonidin | −6.8 | 63 | Prunetin | −6.2 |
| 13 | Fisetin | −6.8 | 64 | Alpinetin | −6.2 |
| 14 | Robinetin | −6.7 | 65 | Biochanin A | −6.2 |
| 15 | 5-Deoxygalangin | −6.7 | 66 | Rhapontigenin | −6.1 |
| 16 | Kaempferol | −6.7 | 67 | Genistein | −6.1 |
| 17 | Scutellarein | −6.7 | 68 | Chrysin | −6.1 |
| 18 | (-)-Epicatechin | −6.7 | 69 | 6-Hydroxyflavone | −6.1 |
| 19 | Purpurin | −6.7 | 70 | Equol | −6.1 |
| 20 | Isorhamnetin | −6.7 | 71 | Piceatannol | −6.1 |
| 21 | Tricetin | −6.6 | 72 | Isorhapontigenin | −6.0 |
| 22 | Gossypetin | −6.6 | 73 | Resveratrol | −5.8 |
| 23 | Norathyriol | −6.6 | 74 | Danshensu | −5.7 |
| 24 | Coumestrol | −6.6 | 75 | Eugenin | −5.6 |
| 25 | Isosakuranetin | −6.6 | 76 | Sinapic acid | −5.5 |
| 26 | Pectolinarigenin | −6.6 | 77 | Pterostilbene | −5.5 |
| 27 | Tangeritin | −6.6 | 78 | Ferulic acid | −5.4 |
| 28 | Nobiletin | −6.6 | 79 | Caffeic acid | −5.4 |
| 29 | Pratensein | −6.6 | 80 | Isoferulic acid | −5.4 |
| 30 | Hispidulin | −6.6 | 81 | Dihydrocaffeic acid | −5.4 |
| 31 | Baicalein | −6.5 | 82 | Gentisic acid | −5.3 |
| 32 | Apigenin | −6.5 | 83 | Pyrogallol | −5.3 |
| 33 | Morin | −6.5 | 84 | 4-Hydroxycinnamic acid | −5.2 |
| 34 | Urolithin A | −6.5 | 85 | Resacetophenone | −5.2 |
| 35 | Acacetin | −6.5 | 86 | Salicyclic acid | −5.1 |
| 36 | Pelargonidin | −6.5 | 87 | Syringic acid | −5.1 |
| 37 | Irilone | −6.5 | 88 | 2-Hydroxybenzoic acid | −5.1 |
| 38 | Naringenin | −6.5 | 89 | Gallic acid | −5.0 |
| 39 | Pinocembrin | −6.5 | 90 | 3-Hydroxybenzoic acid | −5.0 |
| 40 | Kaempferide | −6.5 | 91 | 4-Hydroxybenzoic acid | −5.0 |
| 41 | Malvidin | −6.5 | 92 | Vanillin | −5.0 |
| 42 | Luteolin | −6.4 | 93 | p-Coumeric acid | −4.9 |
| 43 | Dalbergin | −6.4 | 94 | Vanillic acid | −4.8 |
| 44 | Butein | −6.4 | 95 | Paeonol | −4.8 |
| 45 | Biochanin A (1-) | −6.4 | 96 | Cinnamic acid | −4.7 |
| 46 | Fustin | −6.4 | 97 | Protocatechuic acid | −4.6 |
| 47 | 5-Hydroxyflavone | −6.4 | 98 | 4-Ethylphenol | −4.5 |
| 48 | Pinostrobin | −6.4 | 99 | Catechol | −4.5 |
| 49 | Pinobanksin | −6.4 | 100 | Tyrosol | −4.5 |
| 50 | Datiscetin | −6.3 | 101 | GTP | −7.9 |
| 51 | Galangin | −6.3 | 102 | Remdesivir | −7.7 |
2.1.3. Docking studies using AutoDock Vina
The energy-minimized structure of all the natural polyphenols, remdesivir, and GTP were docked with the receptor (RdRp of SARS-CoV-2) using AutoDock Vina 1.1.2 (Trott & Olson, 2010). The ligand files were further saved in PDBQT file format, a modified PDB format containing atomic charges, atom type definitions for ligands, and topological information (rotatable bonds). A grid box (30 Å × 30 Å × 30 Å) centered at (121, 120, 125) Å for the SARS-CoV-2 RdRp, was used in the docking experiments. After the receptor-ligand preparation, docking runs were started from the command prompt. The lowest binding energy and best-docked conformation were considered as the ligand molecule with maximum binding affinity.
2.1.4. Protein-ligand interactions
LigPlot+ was used to investigate protein-ligand interactions for a given .pdb file containing the docked conformation and also the final simulated conformation (Wallace et al., 1995). The LigPlot+ program self-generated schematic 2D representations of protein-ligand interaction. The output file represents the intermolecular interactions and their strengths, including hydrogen bonds, hydrophobic contacts, and atom accessibilities. H-bonds are shown in green dotted lines, whereas residues involved in hydrophobic interaction are represented in the red semicircle.
2.2. Molecular dynamics simulations
All-atom molecular dynamics (MD) simulations were performed on the best eight selected plant-derived natural polyphenols obtained from the molecular docking study along with remdesivir, a well-known RdRp inhibitor, for studying thermodynamic stability of the docked structure. The pmemd.cuda module in AMBER18 (Case et al. 2018) was used for conducting MD simulations, and all simulations were performed utilizing the graphics card, RTX 2080Ti. We adopted the same protocol that was used in our earlier studies (Jonniya et al., 2019; Sk, Roy, Jonniya, et al., 2020; Sk, Roy, & Kar, 2020). The receptor and small molecules were described by the Amber ff14SB (Maier et al., 2015) and GAFF2 (Wang et al., 2004) force field, respectively. Ligands were assigned the AM1-BCC (Jakalian et al., 2002) atomic charges calculated using the antechamber (Wang et al., 2001) module. The complexes were then solvated using an explicit TIP3P (Price & Brooks III, 2004) water model, and nearly 38124 water molecules were needed to solvate each system. Subsequently, all solvated systems were neutralized by adding an appropriate number of Na+ ion. All bond lengths, including hydrogen atoms, were constrained by the SHAKE algorithm (Kräutler et al., 2001). This allows the usage of a 2 fs time-step. The non-bonded cut-off was set to 8 Å and the long range electrostatic interactions were evaluated using the particle-mesh Ewald (PME) (Darden et al., 1993) method. The temperature was kept at 300 K using the Langevin thermostat (Loncharich et al., 1992) with a collision frequency of 2 ps−1. The system pressure was controlled by Berendsen’s Barostat (Berendsen et al., 1984) and fixed at 1.0 bar. We used a time-step of 2.0 fs for all simulations.
Briefly, we used two stages of minimization. Firstly, each complex was optimized by 500 steps of steepest descent followed by another 500 steps of conjugate gradient minimization, keeping all atoms of the complex restrained to their initial coordinate with a weak harmonic potential (force constant 2.0 kcal mol−1 Å−2). The second stage of minimization was carried out without any restraints by performing 100 steps of steepest descent, followed by another 900 steps of conjugate gradient minimization to remove any residual steric clashes. Subsequently, all systems were gradually heated from 0 K to 300 K at the NVT ensemble with a force constant of 2.0 kcal mol−1Å−2 acting on all solute atoms. Next, 1.0 ns equilibration MD phase was carried out without any restraint. Finally, we performed 150 ns production simulations for all four systems at the NPT ensemble. Overall, we accumulated 15000 conformations for each simulation, and we used 500 snapshots from the last 50 ns trajectories for binding affinity estimation using the MM-PBSA scheme. The trajectory analysis was done by the AmberTools19 CPPTRAJ (Roe & Cheatham III, 2013) module of Amber18.
2.3. Molecular mechanics Poisson-Boltzmann surface area calculations
The binding affinity of EGCG, TF1, TF3, TF2b, TF2a, hesperidin, myricetin, quercetagetin, and remdesivir against the SARS-CoV-2 RdRp, were calculated by the molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) methodology (Jonniya & Kar, 2020; Kar, Seel, et al., 2007; Kar, Wei, et al., 2007; Kar et al., 2011; 2013; Kar & Knecht, 2012a, 2012b, 2012c, 2012d; Kollman et al., 2000; Roy et al., 2020; Wang et al., 2006). The MM-PBSA scheme can briefly be described as follows:
| (1) |
| (2) |
| (3) |
where are the changes in molecular mechanical energy, solvation free energy, and conformational entropy, respectively. Further, molecular mechanical energy is composed of (bond, dihedral, and angle), (electrostatic) and (van der Waals) and the change in desolvation free energy is composed of polar solvation () and non-polar solvation free energy (). The polar solvation free energy, was calculated by the pbsa module of AMBER18. Due to the high computational cost, we neglect the configurational entropy calculations. Further, to understand the polyphenol-protein interaction more closely, the interaction energy was decomposed into the contributions from each residue of the protein by using the molecular mechanics generalized Born surface area (MM-GBSA) scheme (Gohlke et al., 2003).
2.4. ADMET studies
The in-silico pharmacological studies of EGCG, TF2a, TF2b, TF3, and remdesivir were predicted based on their ADMET profile. The ADMET studies (absorption, distribution, metabolism, elimination, and toxicity) were predicted using the pkCSM tool (http://biosig.unimelb.edu.au/pkcsm/prediction) (Pires et al., 2015). The canonical SMILE molecular structures of the above-mentioned compounds were retrieved from the PubChem database (www.pubchem.ncbi.nlm.nih.gov).
2.5. Molecular target prediction
Natural compounds interact with a large number of proteins, enzymes, lipids. This interaction plays a crucial role in elucidating the molecular mechanism of the small molecules. So, it is important to identify the molecular targets for new molecules (Gfeller et al., 2014). Swiss Target Prediction website (http://www.swisstargetprediction.ch/index.php) was logged on, and canonical SMILE molecular structures of remdesivir, EGCG, TF2a, TF2b, and TF3 were entered in the search bar option, and results were analyzed.
3. Results and discussion
3.1. Molecular docking analysis
3.1.1. The binding mode analysis and predicted binding affinity calculations of natural polyphenols against the SARS-CoV-2 RdRp
Herein, we investigated our natural polyphenol library against RdRp of the SARS-CoV-2 (PDB ID: 6M71) by molecular docking. The best conformation of the natural polyphenols was docked against the SARS-CoV-2 RdRp, and resulting binding energies are listed in Table 1.
Polyphenols exhibiting binding energy of −7.0 kcal/mol or lower (eight polyphenols) against RdRp of the SARS-CoV-2 are listed in Table 2 along with the ligand-amino acid interactions. Control compounds, GTP and remdesivir, exhibited the binding energy of −7.9 and −7.7 kcal/mol, respectively, against the SARS-CoV-2 RdRp. Eight polyphenols displayed significantly higher binding affinity among the selected hundred natural polyphenols docked against the SARS-CoV-2 RdRp, with binding energies of TF3, TF2b, TF1, TF2a, hesperidin, EGCG, myricetin and quercetagetin as −9.9, −9.6, −9.6, −9.3, −8.8, −7.3, −7.2 and −7.0 kcal/mol, respectively (highlighted in Table 1). Further, 2D LigPlot+ representation of RdRp and the above-mentioned eight polyphenols reveal the stable network of molecular interactions (see Table 2 and Figure S1 in the Supplementary Information).
Table 2.
Ligand-amino acid interactions of top eight scoring natural polyphenols against the SARS-CoV-2 RdRp.
| S. No. | Compound name | Binding energy (kcal/mol) | No. of non-covalent interactions | Involved amino acids |
|---|---|---|---|---|
| 1 | TF3 | −9.9 | 17 | W617, K551, S549, D623, R836, S814, E811, F812, C813, D761, D618, S759, Y619, C622, R553, K621, D760 |
| 2 | TF2b | −9.6 | 13 | K551, Y619, D760, K798, W617, W800, D761, F812, C813, E811, D618, S549, A550 |
| 3 | TF1 | −9.6 | 12 | W617, D761, D760, Y619, R553, K621, P620, F793, D164, S795, K798, D618 |
| 4 | TF2a | −9.3 | 14 | C813, F812, D761, D760, D618, K798, K551, A550, S549, K621, Y619, W800, W617, E811 |
| 5 | Hesperidin | −8.8 | 13 | Y619, D618, K798, S795, M794, P793, D164, V166, P620, K621, D623, R555, Y455 |
| 6 | EGCG | −7.3 | 9 | D623, Y619, K621, S795, C622, D618, M794, P620, K798 |
| 7 | Myricetin | −7.2 | 10 | W617, W800, D760, E811, K798, D618, Y619, C622, D761, F812 |
| 8 | Quercetagetin | −7.0 | 8 | R553, K545, K621, D623, C622, D760, P620, Y619 |
| 9 | Remdesivir (Control) | −7.7 | 13 | R553, K621, C622, D760, E811, W800, K798, P620, Y455, R624, Y619, D618, D761 |
| 10 | GTP (Control) | −7.9 | 15 | R624, T556, D623, D760, Y619, C622, K621, D452, A554, R553, Y455, R555, D761, D618, P620 |
In addition to remdesivir, here we observed that eight dietary polyphenols (TF1, TF2a, TF2b, TF3, hesperidin, EGCG, myricetin and quercetagetin) have significant potential to function as inhibitors of the SARS-CoV-2 RdRp. The top-eight scoring ligands for the SARS-CoV-2 RdRp (as highlighted in Table 1) suggest that these set of natural polyphenols can strongly bind to the catalytic site of the SARS-CoV-2 RdRp and are expected to inhibit the RdRp activity, and thus blocking the replication and preventing viral transcription. Many reports suggest polyphenols have low systemic toxicity, and they are highly beneficial for human health (Bhardwaj et al., 2020; Cory et al., 2018). TF1 and its gallate derivatives, collectively known as black tea polyphenols, previously have shown to exert antiviral activity against many viruses such as hepatitis virus and influenza A and B viruses (Chowdhury et al., 2018). Hesperidin is also known to possess antiviral activity by altering the immune system mainly via regulating interferons in the influenza A virus (Randall & Goodbourn, 2008). EGCG, a major green tea polyphenol, has several pharmacological properties, including antiviral activity (Carneiro et al., 2016; Moon & Morris, 2007). Similarly, myricetin has also been found to act as an inhibitor of the SARS coronavirus helicase (Yu et al., 2012). Quercetagetin also showed strong hepatitis C virus replication inhibitory activity in vitro (Ahmed-Belkacem et al., 2014). Thus, the set dietary polyphenols identified in the present study could be used as repurposed drugs for the treatment of the SARS-CoV-2 infection with further in-vitro and in-vivo validations.
3.2. Molecular dynamics (MD) simulation
3.2.1. Thermodynamic stability and flexibility analysis
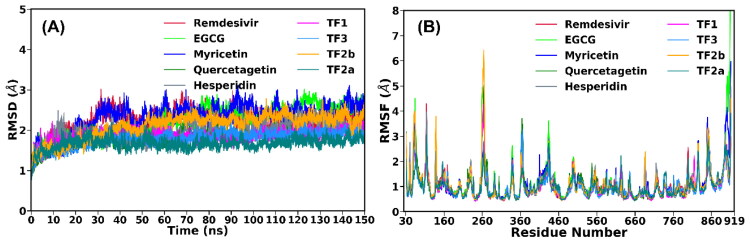
The 150 ns production simulations carried out for nine systems (complex of remdesivir, EGCG, TF1, TF2a, TF2b, TF3, hesperidin, myricetin, and quercetagetin with the SARS-CoV-2 RdRp) were stable on the basis of the potential energy and total energy (data not shown) of those complexes. Subsequently, the root-mean-square deviations (RMSDs) of backbone atoms relative to their respective initial positions were calculated for each complex and are shown in Figure 3(A). It is evident from Figure 3(A) that all the nine studied systems drifted from their initial positions during the first 50 ns, and after that, they reached equilibrium. The average RMSD values were estimated to be 2.30 Å, 2.45 Å, 1.87 Å, 2.28 Å, 1.68 Å, 2.47 Å, 1.90 Å, 2.03 Å and 1.88 Å for RdRp/remdesivir, RdRp/EGCG, RdRp/TF3, RdRp/TF2b, RdRp/TF2a, RdRp/myricetin, RdRp/quercetagetin, RdRp/hesperidin, and RdRp/TF1 complexes, respectively (Table 3). The least deviation was observed for RdRp/TF2a, while RdRp/myricetin displayed the highest deviation. We also investigated structural variations in the binding site, including all amino acids that fall within a radius of 5 Å from the inhibitor, and the same trend was observed (Figure S2A in the Supplementary Information). Overall, this suggests the convergence of our simulations.
Figure 3.
(A) Time evolution of root-mean-square deviations (RMSDs) of backbone atoms and (B) the root-mean-square fluctuations (RMSFs) of Cα atoms of nine complexes relative to their respective energy minimized structure.
Table 3.
The average backbone RMSD, radius of gyration (RoG), and solvent accessible surface area (SASA) for all nine complexes. The data are reported as average ± standard error of the mean (SEM).
| System | RMSD (Å) | RoG (Å) | SASA (Å2) |
|---|---|---|---|
| RdRp/Remdesivir | 2.30 ± 0.03 | 29.96 ± 0.02 | 34973.20 ± 91.56 |
| RdRp/EGCG | 2.45 ± 0.05 | 29.52 ± 0.06 | 35026.03 ± 63.52 |
| RdRp/TF3 | 1.87 ± 0.02 | 29.60 ± 0.01 | 34080.16 ± 53.41 |
| RdRp/TF2b | 2.28 ± 0.01 | 29.86 ± 0.02 | 35462.92 ± 50.40 |
| RdRp/TF2a | 1.68 ± 0.02 | 29.75 ± 0.02 | 34312.55 ± 112.89 |
| RdRp/Myricetin | 2.47 ± 0.03 | 29.88 ± 0.01 | 35395.35 ± 104.67 |
| RdRp/Quercetagetin | 1.90 ± 0.03 | 29.84 ± 0.01 | 34618.65 ± 51.25 |
| RdRp/Hesperidin | 2.03 ± 0.04 | 29.74 ± 0.02 | 34554.08 ± 47.25 |
| RdRp/TF1 | 1.88 ± 0.03 | 29.86 ± 0.01 | 34420.69 ± 56.90 |
Next, we investigated the structural stability of remdesivir and eight polyphenols by estimating the temporal RMSDs of heavy atoms relative to their respective initial coordinates (see Figure S2B in the Supplementary Information). It is evident from Figure S2B that EGCG, myricetin, quercetagetin, TF1, TF3 displayed a rigid behavior in the bound form with an average RMSD of < 1.0 Å. However, remdesivir, TF2a, TF2b and hesperidin showed higher fluctuations as compared to the abovementioned polyphenols, and an average RMSD of > 2.0 Å was noted.
To identify the regions which are flexible, the root-mean-square fluctuations (RMSFs) of Cα atoms of each residue are calculated and shown in Figure 3(B). From this analysis, we can get a better insight into what extent the binding of remdesivir and natural polyphenols affects the residual flexibility of RdRp (mainly Nsp12). Figure 3(B) indicates that RdRp/remdesivir and RdRp-polyphenols shared a similar RMSF pattern. Notable dynamic fluctuations were located in the non-active site domain, including both N-terminals and C-terminal. Regions around Asn150, Asp260, Arg305, Asn360, and Phe440 are found to be more flexible compared to the other area for all complexes. The binding pocket residues, such as Asp452, Lys545, Lys551, Tyr455, Arg553, Ala554, Arg555, Thr556, Asp618, Tyr619, pro620, Lys621, Cys622, Asp623 Arg624, Asn691, Asp760, Asp761, Phe793, Met794, Ser795, Lys798, Trp800, Glu811, Phe812, and Ser814 exhibited considerably low fluctuations for all the RdRp-inhibitor complexes. In the case of RdRp/TF3 and RdRp/TF2a, the binding site residues displayed lesser fluctuations compared to the other RdRp-polyphenol complexes. This suggests that TF3 and TF2a are likely to be bound to RdRp more strongly than the other polyphenols.
Since the radius of gyration (RoG) helps us to understand the protein structural compactness, RoG of each complex was monitored and represented in Figure S3A in the Supplementary Information. The average values of RoG are 29.96 Å, 29.52 Å, 29.60 Å, 29.86 Å, 29.75 Å, 29.88 Å, 29.84 Å, 29.74 Å and 29.86 Å for RdRp complexed with remdesivir, EGCG, TF3, TF2b, TF2a, myricetin, quercetagetin, hesperidin and TF1 respectively (Table 3). This suggests that the structural compactness remained unchanged during simulations. Finally, the solvent-accessible surface area (SASA) was also explored, and the time evolution of SASA for four RdRp-polyphenol complexes are shown in Figure S3B in the Supplementary Information. The average values of SASA are reported in Table 3. Binding of an inhibitor to the substrate changes SASA and sometimes could greatly affect the protein structure. Here, a relatively higher SASA value was obtained for RdRp/TF2b (35462.9 Å2) compared to the other RdRp/inhibitor complexes. On the other hand, the lowest SASA value was noted for RdRp/TF3 (34080.2 Å2). Thus, it can be suggested that the binding of TF3 could potentially reduce protein expansion.
3.2.2. Binding free energy analysis
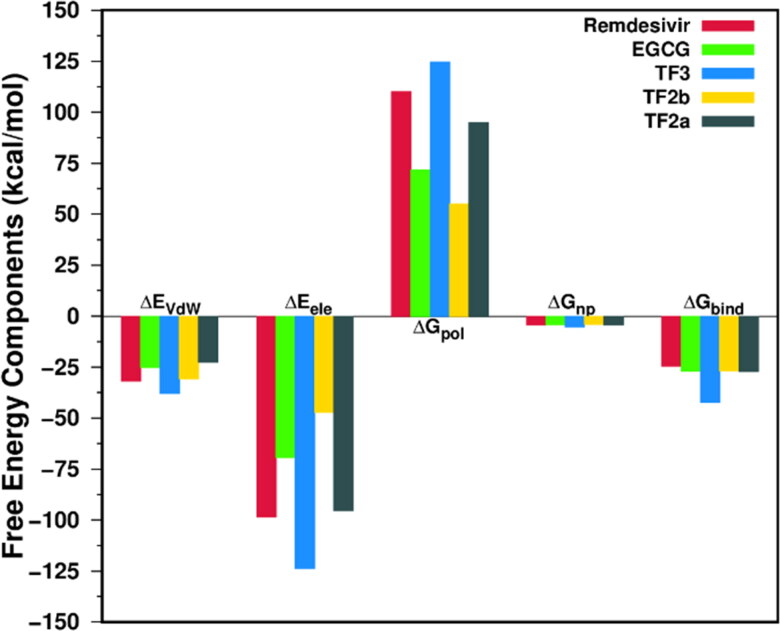
We predicted the binding free energy of all nine complexes by utilizing the MM-PBSA scheme, and four polyphenols, namely EGCG, TF3, TF2b, and TF2a, displayed a higher estimated affinity compared to remdesivir as depicted in Figure 4. Various components of the binding free energy of EGCG, TF3, TF2b, and TF2a are reported in Table 4. The remaining four polyphenols which showed lower estimated affinity compare to remdesivir are shown in Table S2 in Supplementary Information. It can be noted from Figure 4 that the intermolecular van der Waals (ΔEvdW) and electrostatic (ΔEelec) terms are favorable for the ligand binding, whereas the desolvation of polar groups (ΔGpol) opposes the complex formation. Non-polar solvation free energy (ΔGnp) is favorable to the binding for all cases. A similar trend was observed in our earlier study (Sk, Roy, Jonniya, et al., 2020).
Figure 4.
Energy components (kcal/mol) for the binding of remdesivir and four polyphenols to RdRp receptor. ΔEvdW, van der Waals interaction; ΔEele, electrostatic interaction in the gas phase; ΔGpol, polar solvation energy; ΔGnp, non-polar solvation energy, and ΔGbind, estimated binding affinity.
Table 4.
Energetic components of the binding free energy of RdRp and natural polyphenols along with remdesivir complexes in kcal/mol. Data are represented as average ± SEM.
| Components | Remdesivir | EGCG | TF3 | TF2b | TF2a |
|---|---|---|---|---|---|
| ΔEvdW | −31.85 ± 0.15 | −25.11 ± 0.18 | −37.82 ± 0.21 | −30.66 ± 0.23 | −22.55 ± 0.19 |
| ΔEelec | −98.40 ± 0.70 | −69.38 ± 0.73 | −123.63 ± 0.88 | −47.18 ± 0.64 | −95.28 ± 1.27 |
| ΔGpol | 109.97 ± 0.57 | 71.62 ± 0.48 | 124.47 ± 0.58 | 55.01 ± 0.49 | 94.94 ± 1.10 |
| ΔGnp | −4.29 ± 0.01 | −4.15 ± 0.01 | −5.29 ± 0.01 | −3.91 ± 0.01 | −4.28 ± 0.02 |
| aΔGsolv | 105.68 ± 0.57 | 67.47 ± 0.48 | 119.18 ± 0.58 | 51.1 ± 0.49 | 90.66 ± 1.10 |
| bΔGpol + elec | 11.57 ± 0.90 | 2.24 ± 0.87 | 0.84 ± 1.05 | 7.83 ± 0.80 | −0.34 ± 1.68 |
| cΔEMM | −130.25 ± 0.71 | −94.49 ± 0.75 | −161.45 ± 0.90 | −77.84 ± 0.68 | −117.83 ± 1.28 |
| ΔGbindSim | −24.57 ± 0.91 | −27.02 ± 0.89 | −42.27 ± 1.07 | −26.74 ± 0.83 | −27.17 ± 1.69 |
ΔGsolv = ΔGnp + ΔGpol,
ΔGpol + elec = ΔEelec + ΔGpol,
ΔEMM = ΔEvdW + ΔEelec.
It is evident from Table 4 that for all complexes, ΔEvdW varies between −22.55 kcal/mol and −37.82 kcal/mol while ΔEelec ranges from −47.18 kcal/mol to −123.63 kcal/mol. Furthermore, in the cases of RdRp/remdesivir, RdRp/EGCG, RdRp/TF3, and RdRp/TF2b, ΔEele is over-compensated by the desolvation energy (ΔGpol), indicating that the sum of these two components, ΔGpol + elec, is unfavorable to the binding and varies between 0.84 kcal/mol and 11.57 kcal/mol (see Table 4) and similar results are found for RdRp/myricetin, RdRp/quercetagetin, RdRp/hesperidin, and RdRp/TF1 (see Table S2 in Supplementary Information). In contrast, in the case of RdRp/TF2a, ΔGpol + elec, is favorable to the complexation (ΔGpol + elec = −0.34 kcal/mol). Overall, this suggests that the complex formation is mainly driven by the van der Waals interactions between polyphenols as well as remdesivir and RdRp. Therefore, hydrophobic residues in the binding pocket played a crucial role in the complexation process.
The estimated binding free energy (ΔGbind) of remdesivir, EGCG, TF3, TF2b, and TF2a were −24.57, −27.02, −42.27, −26.74 and −27.17 kcal/mol, respectively (Table 4) and myricetin, quercetagetin, hesperidin and TF1 show lower binding affinity compared to that of remdesivir (Table S2 in the Supplementary Information). This suggests that polyphenol TF3 binds most strongly to RdRp, followed by TF2a and EGCG. The potency of the five inhibitors decreases in the following order: TF3 > TF2a > EGCG > TF2b > remdesivir. TF3 binds most strongly to RdRp because both ΔEvdW and ΔEelec are more favorable to the binding compared to the other inhibitors. Similarly, TF2a binds more strongly to RdRp compared to EGCG or TF2b because ΔGpol + elec is favorable for TF2a (ΔGpol + elec = −0.34 kcal/mol) while it is found to be unfavorable for EGCG (ΔGpol + elec = 2.24 kcal/mol) and TF2b (ΔGpol + elec = 7.83 kcal/mol).
3.2.3. Essential residues for polyphenols binding
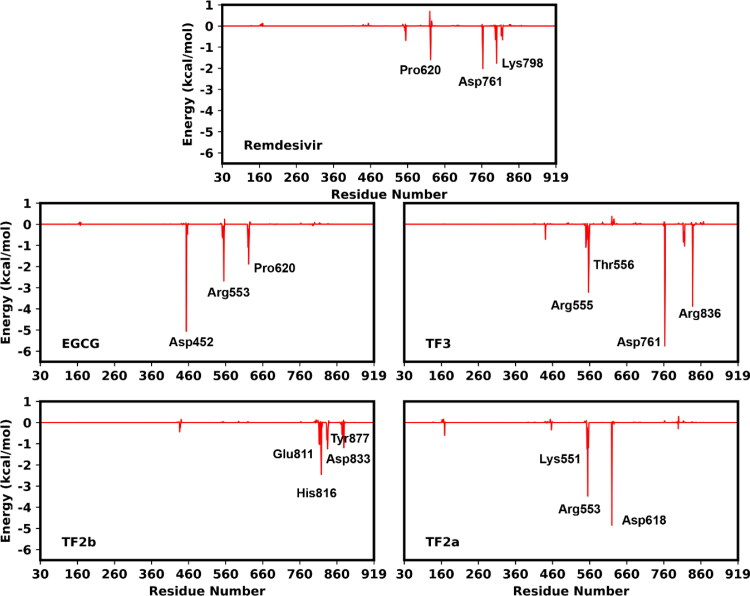
Further, to gain a deeper insight into the best four RdRp/polyphenols and remdesivir interaction pattern, the total binding free energy was decomposed into polyphenols-residue pair based on the MM-GBSA scheme. The approach of per-residue based contributions is useful to determine the binding mechanisms of an inhibitor at an atomistic level, and it also reveals the individual residue contributions. The different energy contributions from the backbone and side-chain of each residue are shown in Figure 5 and listed in Table 5.
Figure 5.
Decomposition of the binding free energy into contributions from individual residues for RdRp complexed with remdesivir, EGCG, TF3, TF2b and TF2a.
Table 5.
Per-residue based decomposition of binding free energy for the complex of remdesivir, EGCG, TF3, TF2a and TF2b with the SARS-CoV-2 RdRp.
| Residue | TvdW | Telec | Tpol | Tnp | Tside_chain | Tbackbone | Ttotal |
|---|---|---|---|---|---|---|---|
| RdRp/Remdesivir | |||||||
| Asp761 | 1.05 | −21.73 | 18.81 | −0.14 | −1.82 | −0.19 | −2.01 |
| Lys798 | −2.74 | −4.28 | 5.74 | −0.47 | −1.76 | 0.01 | −1.75 |
| Pro620 | −1.42 | −1.15 | 1.16 | −0.18 | −1.26 | −0.33 | −1.59 |
| Asp760 | −0.57 | −4.27 | 4.24 | −0.12 | −0.20 | −0.52 | −0.72 |
| Arg553 | −2.07 | −3.71 | 5.61 | −0.51 | −0.76 | 0.08 | −0.68 |
| RdRp/EGCG | |||||||
| Asp452 | 1.65 | −16.48 | 9.85 | −0.08 | −5.15 | 0.09 | −5.06 |
| Arg553 | −3.83 | −4.84 | 6.59 | −0.59 | −2.68 | 0.01 | −2.67 |
| Pro620 | −1.64 | −0.41 | 0.51 | −0.33 | −1.62 | −0.25 | −1.87 |
| Asp618 | 0.49 | −8.14 | 6.66 | −0.09 | −1.13 | −0.05 | −1.08 |
| Lys621 | −2.26 | −5.12 | 6.92 | −0.50 | −0.87 | −0.09 | −0.96 |
| RdRp/TF3 | |||||||
| Asp761 | 2.52 | −30.13 | 22.11 | −0.25 | −5.54 | −0.21 | −5.75 |
| Arg836 | −0.83 | −14.95 | 12.29 | −0.37 | −3.85 | −0.01 | −3.86 |
| Arg555 | −5.80 | −4.00 | 7.29 | −0.70 | −2.98 | −0.23 | −3.21 |
| Thr556 | −0.20 | −3.96 | 2.44 | −0.13 | −0.21 | −1.64 | −1.85 |
| Ile548 | −1.03 | −0.08 | 0.12 | −0.13 | −0.65 | −0.47 | −1.12 |
| Ser814 | −1.67 | 0.09 | 0.78 | −0.08 | −0.48 | −0.40 | −0.88 |
| Val557 | −0.65 | −0.16 | 0.17 | −0.24 | −0.65 | −0.23 | −0.88 |
| RdRp/TF2b | |||||||
| His816 | −3.00 | −0.81 | 1.69 | −0.32 | −1.63 | −0.81 | −2.44 |
| Asp833 | −1.29 | −0.23 | 0.46 | −0.18 | −0.21 | −1.03 | −1.24 |
| Tyr877 | −0.92 | −2.06 | 2.06 | −0.27 | −0.43 | −0.76 | −1.19 |
| Glu811 | 0.22 | −9.24 | 8.11 | −0.13 | −2.07 | 1.03 | −1.04 |
| His810 | −1.88 | −0.41 | 1.66 | −0.34 | −0.10 | −0.85 | −0.95 |
| Tyr831 | −1.51 | −0.36 | 1.19 | −0.13 | −0.21 | −0.60 | −0.81 |
| Asn815 | −0.60 | −0.27 | 0.09 | −0.01 | −0.25 | −0.54 | −0.79 |
| RdRp/TF2a | |||||||
| Asp618 | 2.63 | −20.71 | 13.40 | −0.17 | −4.84 | −0.01 | −4.85 |
| Arg553 | −3.80 | −6.52 | 7.55 | −0.69 | −2.68 | −0.78 | −3.46 |
| Lys551 | −2.23 | −0.80 | 2.23 | −0.45 | −0.84 | −0.41 | −1.25 |
| Arg555 | −1.33 | −0.62 | 1.05 | −0.27 | −1.12 | −0.05 | −1.17 |
| Glu167 | 0.42 | −6.82 | 5.92 | −0.12 | −0.61 | 0.01 | −0.60 |
As shown in Figure 5, it was observed that residues favoring the binding of the polyphenols with RdRp include Asp452, Arg553, Arg555, Val557, Asp618, Pro620, Lys621, Asp623, Arg624, Asp760, Asp761, and Glu811, Asp833, and Arg836. Most of these residues are located in the binding site of RdRp and can form direct contacts with polyphenols and remdesivir. Figure 5 shows that amino acids Pro620, Asp761 and Lys798 for RdRp/remdesivir; Asp452, Arg553, Pro620 and Lys621 for RdRp/EGCG; Ile548, Arg555, Thr556, Asp761 and Arg836 for RdRp/TF3; Glu811, His816, Asp833 and Tyr877 for RdRp/TF2b; Lys551, Arg553, Arg555 and Asp618 for RdRp/TF2a contributed more favorably towards the binding by contributing more than −1.0 kcal/mol in size.
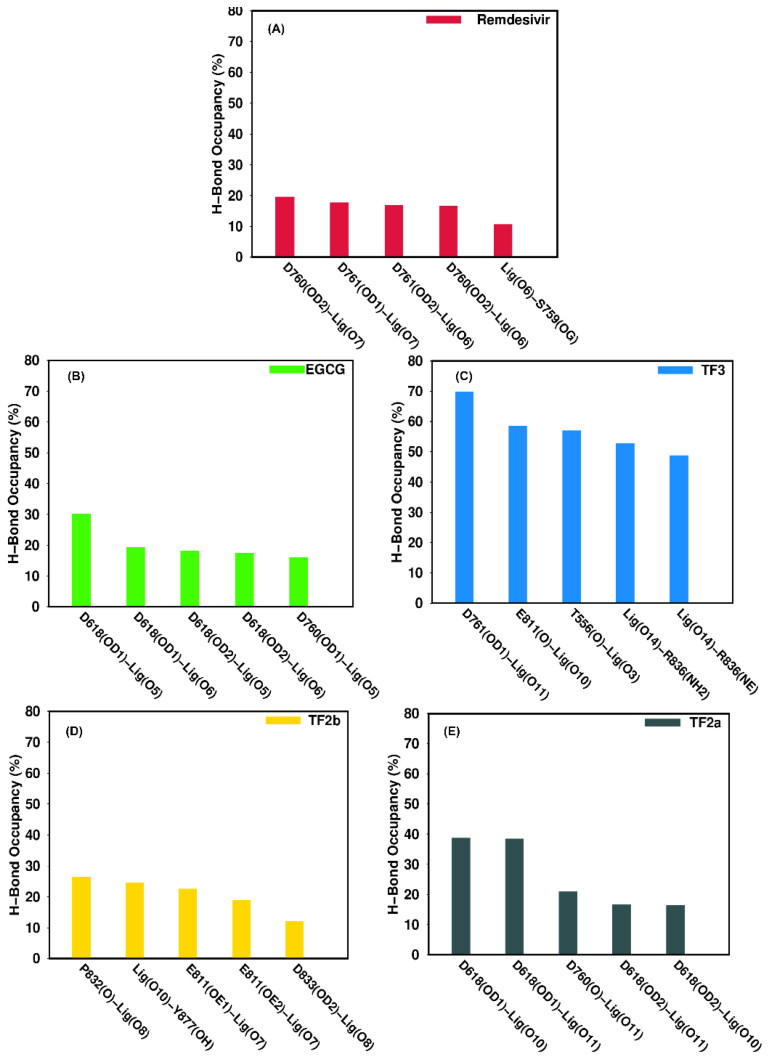
To complement the energetic analysis, we performed MD trajectory-based hydrogen bond (h-bond) analysis for all five complexes, and the h-bonds with occupancy are listed in Table 6. The h-bonds were determined by setting the acceptor-donor distance of ≤ 3.5 Å, and the angle cut off ≥ 1200. Important h-bonds between RdRp-inhibitors are shown in Figure 6. In the case of RdRp/remdesivir, key residues involved in the hydrogen bonding are Asp761, Asp760, and Ser759, respectively. Asp760 is found to form two h-bonds with remdesivir (Asp760@OD2 - Lig@O7, Asp760@OD2 - Lig@O6) with an occupancy of more than 15% (see Table 6 and Figure 6). In the case of RdRp/EGCG, both Asp618 and Asp760 form two h-bonds with the ligand with an occupancy in the range of 16.09 to 30.17%. On the other hand, Asp761 form an h-bond with TF3 (Asp761@OD1 - Lig@O11) with an occupancy of 69.84%, while Arg836 forms two h-bonds with the ligand (Arg836@NH2 - Lig@O14, Arg836@NE - Lig@O14) with an occupancy of 52.66%, and 48.70%, respectively. Glu811, Thr556 and Asp761 also formed h-bonds with the ligand during our simulations with an occupancy varying in the range of 44% to 58% (see Table 6). In the case of RdRp/TF2b, Glu811 is found to form two strong h–bonds with the ligand (Glu811@OE1 – Lig@O7, Glu811@OE2 – Lig@O7) with an occupancy of 22.45% and 18.89%, respectively. On the other hand, it can be observed from Table 6 that Pro832 and Tyr877 form strong h-bonds (Pro832@O -Lig@O8 and Lig@O10 -Tyr877@OH) with increased occupancy (> 24%). Finally, in the case of RdRp/TF2a, Asp618 is found to form two strong h-bonds with the TF2a (Asp618@OD1 – Lig@O10, Asp618@OD1 – Lig@O11) with an occupancy of 38.68% and 38.38%, respectively. Asp760 also forms a h-bond (Asp760@O – Lig@O11) with an occupancy of 20.83%.
Table 6.
Main hydrogen bond interactions formed by RdRp with remdesivir and polyphenols along with the corresponding average distance and percentage of occupancy determined using the trajectories of production simulations.
| Acceptor | Donor | Avg. Distance (Å) | Occupancy (%) |
|---|---|---|---|
| RdRp/Remdesivir | |||
| Asp760@OD2 | Lig@O7 | 2.66 | 19.46 |
| Asp761@OD1 | Lig@O7 | 2.65 | 17.70 |
| Asp761@OD2 | Lig@O6 | 2.63 | 16.86 |
| Asp760@OD2 | Lig@O6 | 2.65 | 16.65 |
| Lig@O6 | Ser759@OG | 2.80 | 11.63 |
| Asp760@OD1 | Lig@O7 | 2.66 | 10.59 |
| RdRp/EGCG | |||
| Asp618@OD1 | Lig@O5 | 2.61 | 30.13 |
| Asp618@OD1 | Lig@O6 | 2.61 | 29.28 |
| Asp618@OD2 | Lig@O5 | 2.61 | 18.25 |
| Asp618@OD2 | Lig@O6 | 2.61 | 17.38 |
| Asp760@OD1 | Lig@O5 | 2.63 | 16.09 |
| Tyr455@OH | Lig@O11 | 2.83 | 10.97 |
| RdRp/TF3 | |||
| Asp761@OD1 | Lig@O11 | 2.61 | 69.84 |
| Glu811@O | Lig@O10 | 2.76 | 58.43 |
| Thr556@O | Lig@O3 | 2.72 | 56.95 |
| Lig@O14 | Arg836@NH2 | 2.83 | 52.66 |
| Lig@O14 | Arg836@NE | 2.86 | 48.70 |
| Asp761@OD2 | Lig@O20 | 2.62 | 44.51 |
| RdRp/TF2b | |||
| Pro832@O | Lig@O8 | 2.77 | 26.31 |
| Lig@O11 | Tyr877@OH | 2.75 | 24.39 |
| Glu811@OE1 | Lig@O7 | 2.65 | 22.45 |
| Glu811@OE2 | Lig@O7 | 2.65 | 18.89 |
| Asp833@OD2 | Lig@O8 | 2.65 | 12.01 |
| Asn874@OD1 | Lig@O11 | 2.68 | 8.58 |
| RdRp/TF2a | |||
| Asp618@OD1 | Lig@O10 | 2.59 | 38.68 |
| Asp618@OD1 | Lig@O11 | 2.62 | 38.38 |
| Asp760@O | Lig@O11 | 2.70 | 20.83 |
| Asp618@OD2 | Lig@O11 | 2.62 | 16.59 |
| Asp618@OD2 | Lig@O10 | 2.58 | 16.49 |
| Asp618@OD1 | Lig@O15 | 2.67 | 16.37 |
Figure 6.
Five main hydrogen bond interactions between ligands and RdRp.
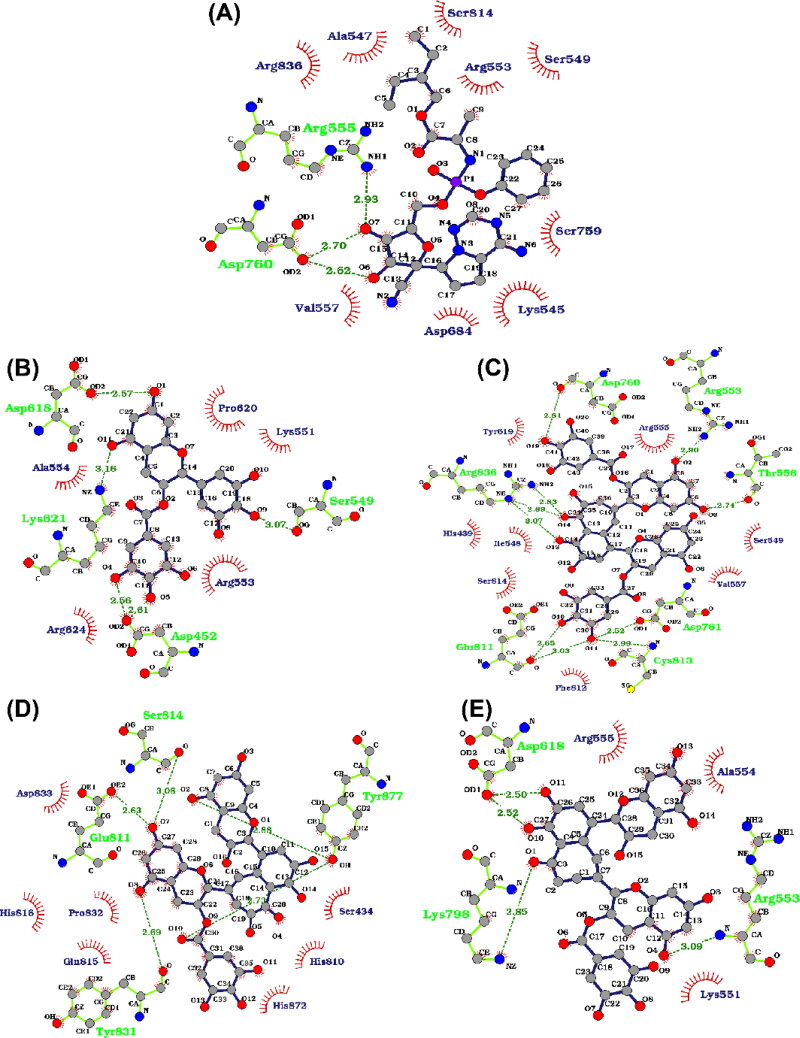
Finally, we supplemented the above results by analyzing the final conformation of each production simulation with the help of 2D LigPlot+ software, and different h-bonds and hydrophobic interactions were shown in Figure 7. Hydrogen bonds are depicted in green dotted lines, while red semicircle residues are involved in hydrophobic interactions. For the RdRp/remdesivir complex, Figure 7(A) displayed nine hydrophobic interactions with Lys545, Ala547, Ser549, Arg553, Val557, Asp684, Ser759, Ser814, and Arg836. This large number of interactions account for the high stability and good binding affinity of remdesivir to RdRp. EGCG formed hydrophobic interactions with Lys551, Ala554, Arg553, Arg624, Pro620, (Figure 7(B)). In the case of TF3, eight hydrophobic interactions with His439, Ile548, Ser814, Phe812, Val557, Ser549, Tyr619 and Arg555 were formed as revealed by Figure 7(C). Figure 7(D) shows that seven hydrophobic interactions with Asp833, His816, Pro832, Gln815, His872, His810 and Ser434 were formed for RdRp/TF2b. Finally, Figure 7(E) shows that RdRp/TF2a formed hydrophobic interactions with Arg555, Ala554 and Lys551. Overall, TF3 has a higher binding affinity toward RdRp compared to the other polyphenols due to a larger number of stable hydrogen bonds and hydrophobic interactions.
Figure 7.
The RdRp-ligands interaction profile for (A) RdRp/remdesivir, (B) RdRp/EGCG, (C) RdRp/TF3, (D) RdRp/TF2b and (E) RdRp/TF2a. The polyphenols and remdesivir are shown in balls and sticks. Hydrogen bonds are depicted in green dotted lines, and red semicircles residues are involved in hydrophobic interactions.
3.3. Prediction of the absorption, distribution, metabolism, excretion, and toxicity (ADMET) profile
In addition to testing the physiochemical efficiency of a given molecule to inhibit the target protein, other parameters such as absorption, distribution, metabolism, excretion, and toxicity (ADMET) of the inhibitor play a critical role on demonstrating the likelihood of success of a drug. Utilization of in-silico ADMET profiling, in combination with in vivo and in vitro predictions in the initial stage of the screening process, can significantly fasten the drug discovery process by minimizing the number of potential safety problems. Hence, we performed a detailed ADMET profiling to evaluate the drug likeliness of the four polyphenols: EGCG, TF2a, TF2b, TF3 that exhibited the highest score from the MD simulation and MM-PBSA study along with the positive control remdesivir.
Human colon adenocarcinoma-2 cell line (Caco2) permeability and human intestinal absorption (HIA) are key parameters to decide the total bioavailability of a drug. All the five compounds (EGCG, TF2a, TF2b, TF3, and remdesivir) showed comparatively low Caco2 permeability potential (<8 × 10−6 cm/s) and could be absorbed via the human intestine (Larregieu & Benet, 2013). EGCG, TF2a, TF2b, TF3, and remdesivir were predicted to be substrates of permeability glycoprotein (P-glycoprotein), which is an efflux membrane protein. However, remdesivir was predicted as a P-glycoprotein I inhibitor, and EGCG and TF3 as a P-glycoprotein II inhibitor, whereas TF2a and TF2b as both P-glycoprotein I and II inhibitor. Hence, above mentioned five compounds could regulate the physiological functions of P-glycoprotein (see Table S3 in the Supplementary Information).
The distribution of a drug is regulated by many parameters such as lipid-solubility, concentration in plasma and binding ability to plasma proteins, transport proteins, etc. The volume of distribution at steady-state (VDss) suggests that EGCG, TF2a, TF2b, TF3, and remdesivir had a lower theoretical dose required for uniform distribution in the plasma. Further, the degree of diffusion across the plasma membrane increases in the following order remdesivir < EGCG < TF2a < TF2b < TF3 (Table S4 in the Supplementary Information) as measured by the fraction that is in the unbound state. The predictions through the distribution of the drugs via the central nervous system and blood-brain barrier suggest that these five compounds are poorly distributed to the brain and unable to penetrate the central nervous system. However, the medium level of the lipophilicity of the drugs suggests that they would have no negative impact on nervous system exposure.
Cytochromes P450 (CYP) isozymes play crucial roles in drug metabolism. It has been observed that TF2a, TF3, and remdesivir are a substrate of CYP3A4 and hence, can be efficiently metabolized by CYP3A4. On the other hand, EGCG is a CYP3A4 inhibitor (Table S5 in the Supplementary Information). On a separate note, EGCG is predominantly metabolized in the small intestine and liver by the conjugate formation of glucuronide, methyl sulfates in the urine and plasma (Chow et al. 2005).
Among the five compounds, TF2b and TF3 were predicted as the substrate of renal organic cation transporter-2 (Renal OCT2), as shown in Table S6 in the Supplementary Information. While EGCG, TF2a, and remdesivir are possibly cleared through other available routes such as bile, breath, faces, and sweat. EGCG remains intact in the plasma and later excreted via bile and metabolized by colon microflora. It is also expected that all the compounds are absorbable via oral prescription.
We have also analyzed the toxicity profiles for EGCG, TF2a, TF2b, TF3 as well as remdesivir (see Table 7). The toxicity prediction from the AMES test (Salmonella typhimurium reverse mutation assay) exhibited that all the compounds could be considered as non-mutagenic agents. High toxicity was observed for all the compounds in Tetrahymena pyriformis. EGCG, TF2a, TF2b, TF3, and remdesivir were shown to inhibit the human ether-a-go-go-related gene II (hERG II). However, Remdesivir has been shown to induce hepatotoxicity, whilst EGCG, TF2a, TF2b, TF3 are not likely to be associated with hepatotoxicity. The maximum recommended tolerated dose (MRTD) in human prediction shows that remdesivir violate MRTD whereas natural polyphenol EGCG, TF2a, TF2b, TF3 do not fall into this category. Remdesivir does not possess high acute toxicity whereas EGCG, TF2a, TF2b, and TF3 regarded as high acute toxic compound as it falls under minnow toxicity. Additionally, none of the compounds predicted to be associated with skin sensitization.
Table 7.
Predicted toxicity profile of EGCG, TF3, TF2b, TF2a, and remdesivir.
| S. No. | Compounds name | Toxicity prediction | |
|---|---|---|---|
| Properties | Predicted values | ||
| 1 | EGCG | AMES toxicity | No |
| Maximum tolerated dose (Human) | 0.441 (log mg/kg/day) | ||
| hERG I inhibitor | No | ||
| hERG II inhibitor | Yes | ||
| Oral rat acute toxicity (LD50) | 2.522 (mol/kg) | ||
| Oral rat chronic toxicity (LOAEL) | 3.065 (log mg/kg_bw/day) | ||
| Hepatotoxicity | No | ||
| Skin sensitivity | No | ||
| T. pyriformis toxicity | 0.285 (µg/L) | ||
| Minnow toxicity | 7.713 log mM | ||
| 2 | TF3 | AMES toxicity | No |
| Maximum tolerated dose (Human) | 0.438 (log mg/kg/day) | ||
| hERG I inhibitor | No | ||
| hERG II inhibitor | Yes | ||
| Oral rat acute toxicity (LD50) | 2.482 (mol/kg) | ||
| Oral rat chronic toxicity (LOAEL) | 7.443 (log mg/kg_bw/day) | ||
| Hepatotoxicity | No | ||
| Skin sensitivity | No | ||
| T. pyriformis toxicity | 0.285 (µg/L) | ||
| Minnow toxicity | 9.738 log mM | ||
| 3 | TF2b | AMES toxicity | No |
| Maximum tolerated dose (Human) | 0.438 (log mg/kg/day) | ||
| hERG I inhibitor | No | ||
| hERG II inhibitor | Yes | ||
| Oral rat acute toxicity (LD50) | 2.482 (mol/kg) | ||
| Oral rat chronic toxicity (LOAEL) | 5.322 (log mg/kg_bw/day) | ||
| Hepatotoxicity | No | ||
| Skin sensitivity | No | ||
| T. pyriformis toxicity | 0.285 (µg/L) | ||
| Minnow toxicity | 8.685 log mM | ||
| 4 | TF2a | AMES toxicity | No |
| Maximum tolerated dose (Human) | 0.439 (log mg/kg/day) | ||
| hERG I inhibitor | No | ||
| hERG II inhibitor | Yes | ||
| Oral rat acute toxicity (LD50) | 2.484 (mol/kg) | ||
| Oral rat chronic toxicity (LOAEL) | 5.035 (log mg/kg_bw/day) | ||
| Hepatotoxicity | No | ||
| Skin sensitivity | No | ||
| T. pyriformis toxicity | 0.285 (µg/L) | ||
| Minnow toxicity | 4.898 log mM | ||
| 5 | Remdesivir | AMES toxicity | No |
| Maximum tolerated dose (Human) | 0.15 (log mg/kg/day) | ||
| hERG I inhibitor | No | ||
| hERG II inhibitor | Yes | ||
| Oral rat acute toxicity (LD50) | 2.043 (mol/kg) | ||
| Oral rat chronic toxicity (LOAEL) | 1.639 (log mg/kg_bw/day) | ||
| Hepatotoxicity | Yes | ||
| Skin sensitivity | No | ||
| T. pyriformis toxicity | 0.285 (µg/L) | ||
| Minnow toxicity | 0.291 log mM |
3.4. Identification of target class for natural polyphenol via target prediction studies
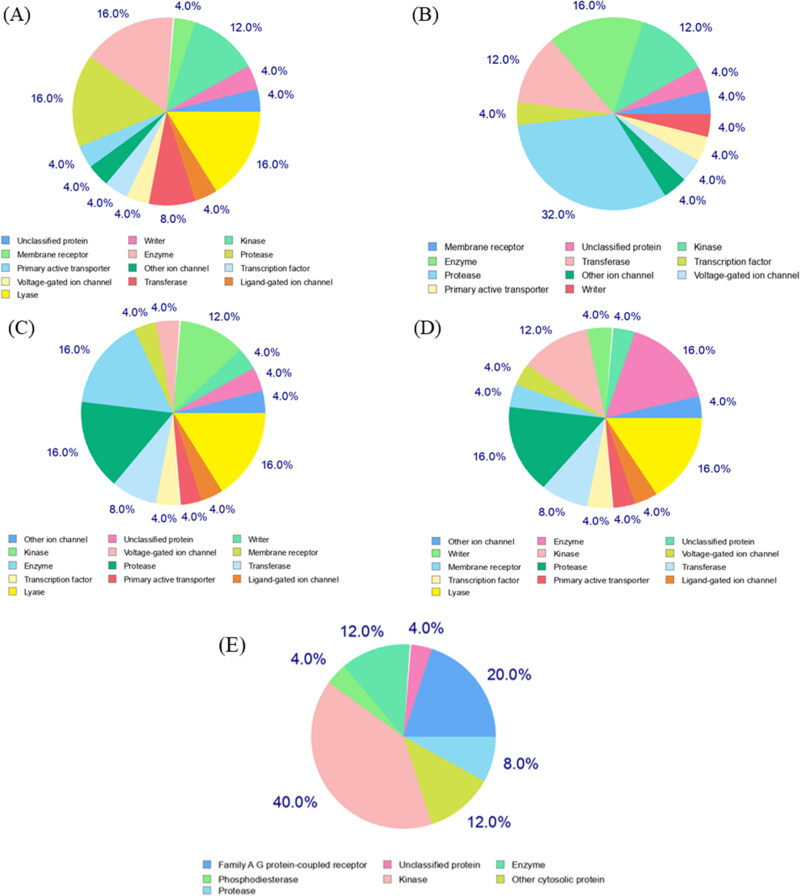
The polyphenolic structural motif of dietary polyphenols allows them to serve as excellent hydrogen bond donors which in turn help them to strongly interact with various biomacromolecules such as proteins. This interaction is a critical step in the regulatory role of polyphenols on various key proteins involved in cellular physiology. The majority, if not all, of the beneficial effect of polyphenols, can be explained via the functional consequence of proteins it interacts with. Molecular target studies help us to predict therapeutic protein targets for a given small molecule. Herein, we analyzed the predicted interacting proteins/enzymes for EGCG, TF2a, TF2b, TF3, and remdesivir. This study is particularly important in the current context as we think that these polyphenols could target RdRp, an important enzyme that catalyzes the RNA replication in the SARS-CoV-2. Notably, the molecular target analysis suggests that all four polyphenols possess excellent properties of druggability, and they interact with a diverse class of proteins/enzymes. The top 25 target classes of EGCG, TF2a, TF2b, TF3, and remdesivir are represented in the pie-chart, as shown in Figure 8. The detailed output table with the target, common name, UniProt ID, ChEMBL ID, target class, probability, and known actives in 2D/3D are listed in Tables S7–S11 in the Supplementary Information.
Figure 8.
Molecular target predictions for (A) EGCG, (B) TF2a, (C) TF2b (D) TF3, and (E) remdesivir obtained from swiss target prediction report. The frequency of the target classes (top 25) is depicted in the pie chart.
4. Conclusions
Keeping in mind that RdRp inhibitors play a crucial role to combat the SARS-CoV-2 infection, in this work, we performed a comprehensive molecular docking study with a library of hundred natural polyphenols with potential antiviral properties that may inhibit the SARS-CoV-2 RdRp and prevent the RNA replication. We shortlisted eight natural polyphenols having binding energy −7.0 kcal/mol or less for molecular dynamics simulation. Further, we performed 150 ns molecular dynamics simulation of RdRp/EGCG, RdRp/TF1, RdRp/TF2a, RdRp/TF2b, RdRp/TF3, RdRp/hesperidin, RdRp/myricetin, RdRp/quercetagetin, along with RdRp/remdesivir complex and computed the binding energies by the molecular mechanics-Poisson-Boltzmann surface area (MM-PBSA) scheme from last 50 ns trajectories. Our study suggests that the complex formation of the SARS-CoV-2 RdRp and eight natural polyphenols is favoured by the intermolecular van der Waals and electrostatic interactions as well as nonpolar solvation free energy. We have also investigated the hotspot residues controlling the receptor-ligand binding. Finally, molecular dynamics simulation and MM-PBSA study reveals that EGCG, TF2a, TF2b, and TF3 possess a better binding affinity than the control drug remdesivir against the SARS-CoV-2 RdRp. Further, we also looked at the ADME prediction, toxicity prediction, and target analysis to assess their druggability of the five compounds. The obtained results strongly suggest that EGCG, TF2a, TF2b, and TF3 have a stable binding affinity towards RdRp of the SARS-CoV-2 with favourable pharmacokinetic properties. These bioactive compounds exhibit broad ranges of therapeutic properties. Therefore, we believe that these four natural polyphenols can act as potential inhibitors for the SARS-CoV-2 RdRp. However, further in vitro and in vivo studies need to be carried out to validate their efficacy against SARS-CoV-2 infection.
Supplementary Material
Funding Statement
The authors gratefully acknowledge the financial support from the Indian Institute of Technology Indore and Indian Institute of Technology Palakkad for the provision of conducting research and the scholarship provided by the Ministry of Human Resource Development, Govt. of India to S. Singh. This work was also supported by the Department of Science and Technology-Science & Engineering Research Board (DST-SERB), Govt. of India (ECR/2017/002082). PK is supported by the Department of Biotechnology, Govt. of India (grant number BT/RLF/Re-entry/40/2014, DBT-Ramalingaswami Re-entry Fellowship), and DST-SERB, Govt. of India (grant number ECR/2017/000010). MFS would like to thank DST, Govt. of India, for providing fellowship under the INSPIRE Fellowship Scheme (DST/INSPIRE Fellowship/2017/IF170145).
Disclosure statement
The authors declare that there is no conflict of interest.
References
- Aanouz, I., Belhassan, A., El Khatabi, K., Lakhlifi, T., El Idrissi, M., & Bouachrine, M. (2020). Moroccan medicinal plants as inhibitors of COVID-19: Computational investigations. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeoye, A. O., Oso, B. J., Olaoye, I. F., Tijjani, H., & Adebayo, A. I. (2020). Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1765876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed-Belkacem, A., Guichou, J.-F., Brillet, R., Ahnou, N., Hernandez, E., Pallier, C., & Pawlotsky, J.-M. (2014). Inhibition of RNA binding to hepatitis C virus RNA-dependent RNA polymerase: A new mechanism for antiviral intervention. Nucleic Acids Research, 42(14), 9399–9409. 10.1093/nar/gku632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J., Postma, J. v., van Gunsteren, W. F., DiNola, A., & Haak, J. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Berman, H. M., Bhat, T. N., Bourne, P. E., Feng, Z., Gilliland, G., Weissig, H., & Westbrook, J. (2000). The Protein Data Bank and the challenge of structural genomics. Nucleic Acids Research, 7, 957–959. [DOI] [PubMed] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Rajendran, V., Purohit, R., & Kumar, S. (2020). Identification of bioactive molecules from Tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure Dynamics. 10.1080/07391102.2020.1766572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 Coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgio, J. F., Alsuwat, H. S., Al Otaibi, W. M., Ibrahim, A. M., Almandil, N. B., Al Asoom, L. I., Salahuddin, M., Kamaraj, B., & AbdulAzeez, S. (2020). State-of-the-art tools unveil potent drug targets amongst clinically approved drugs to inhibit helicase in SARS-CoV-2. Archives of Medical Science: AMS, 16(3), 508–518. 10.5114/aoms.2020.94567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro, B. M., Batista, M. N., Braga, A. C. S., Nogueira, M. L., & Rahal, P. (2016). The green tea molecule EGCG inhibits Zika virus entry. Virology, 496, 215–218. 10.1016/j.virol.2016.06.012 [DOI] [PubMed] [Google Scholar]
- Case, D. A., Ben-Shalom, I. Y., Brozell, S. R., Cerutti, D. S., Cheatham, T. E., III, Cruzeiro, V. W. D., Darden, T. A., Duke, R. E., Ghoreishi, D., Gilson, M. K., Gohlke, H., Goetz, A. W., Greene, D., Harris, R., Homeyer, N., Izadi, S., Kovalenko, A., Kurtzman, T., Lee, T. S., … Kollman, P. A. (2018). AMBER 2018. University of California. [Google Scholar]
- Chan, J. F., Lau, S. K., To, K. K., Cheng, V. C., Woo, P. C., & Yuen, K.-Y. (2015). Middle East respiratory syndrome coronavirus: Another zoonotic betacoronavirus causing SARS-like disease. Clinical Microbiology Reviews, 28(2), 465–522. 10.1128/CMR.00102-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, H. S., Hakim, I. A., Vining, D. R., Crowell, J. A., Ranger-Moore, J., Chew, W. M., Celaya, C. A., Rodney, S. R., Hara, Y., & Alberts, D. S. (2005). Effects of dosing condition on the oral bioavailability of green tea catechins after single-dose administration of Polyphenon E in healthy individuals. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 11(12), 4627–4633. 10.1158/1078-0432.CCR-04-2549 [DOI] [PubMed] [Google Scholar]
- Chowdhury, P., Sahuc, M.-E., Rouillé, Y., Rivière, C., Bonneau, N., Vandeputte, A., Brodin, P., Goswami, M., Bandyopadhyay, T., Dubuisson, J., & Séron, K. (2018). Theaflavins, polyphenols of black tea, inhibit entry of hepatitis C virus in cell culture. PloS One, 13(11), e0198226. 10.1371/journal.pone.0198226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory, H., Passarelli, S., Szeto, J., Tamez, M., & Mattei, J. (2018). The role of polyphenols in human health and food systems: A mini-review. Frontiers in Nutrition, 5, 87. 10.3389/fnut.2018.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98(12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Elfiky, A. A. (2017). Zika virus: Novel guanosine derivatives revealed strong binding and possible inhibition of the polymerase. Future Virology, 12(12), 721–728. 10.2217/fvl-2017-0081 [DOI] [Google Scholar]
- Elfiky, A. A. (2019). Novel guanosine derivatives as anti-HCV NS5b polymerase: A QSAR and molecular docking Study. Medicinal Chemistry, 15(2), 130–137. 10.2174/1573406414666181015152511 [DOI] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020a). Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sciences, 248, 117477. 10.1016/j.lfs.2020.117477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020b). Natural products may interfere with SARS-CoV-2 attachment to the host cell. Journal of Biomolecular Structure Dynamics. 10.1080/07391102.2020.1761881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020c). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A., & Azzam, E. B. (2020). Novel Guanosine Derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., Şahin, A. T., & Yelekçi, K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure Dynamics. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan, A., & Barakat, K. (2017). Applications of computer-aided approaches in the development of hepatitis C antiviral agents. Expert Opinion on Drug Discovery, 12(4), 407–425. 10.1080/17460441.2017.1291628 [DOI] [PubMed] [Google Scholar]
- Gfeller, D., Grosdidier, A., Wirth, M., Daina, A., Michielin, O., & Zoete, V. (2014). SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids Research, 42(Web Server issue), W32–W38. 10.1093/nar/gku293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke, H., Kiel, C., & Case, D. A. (2003). Insights into protein–protein binding by binding free energy calculation and free energy decomposition for the Ras–Raf and Ras–RalGDS complexes. Journal of Molecular Biology, 330(4), 891–913. 10.1016/S0022-2836(03)00610-7 [DOI] [PubMed] [Google Scholar]
- Hendaus, M. A. (2020). Remdesivir in the treatment of Coronavirus Disease 2019 (COVID-19): A simplified summary. Journal of Biomolecular Structure Dynamics. 10.1080/07391102.2020.1767691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, H.-Y., Cheng, M.-L., Weng, S.-F., Leu, Y.-L., & Chiu, D. T.-Y. (2009). Antiviral effect of epigallocatechin gallate on enterovirus 71. Journal of Agricultural and Food Chemistry, 57(14), 6140–6147. 10.1021/jf901128u [DOI] [PubMed] [Google Scholar]
- Ibrahim, I. M., Abdelmalek, D. H., Elshahat, M. E., & Elfiky, A. A. (2020). COVID-19 spike-host cell receptor GRP78 binding site prediction. The Journal of Infection, 80(5), 554–562. 10.1016/j.jinf.2020.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, R., Parves, R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., Hossain, M. N., Ali, M. A., & Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure Dynamics. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakalian, A., Jack, D. B., & Bayly, C. I. (2002). Fast, efficient generation of high‐quality atomic charges. AM1‐BCC model: II. Parameterization and validation. Journal of Computational Chemistry, 23(16), 1623–1641. 10.1002/jcc.10128 [DOI] [PubMed] [Google Scholar]
- Jia, H., & Gong, P. (2019). A structure-function diversity survey of the RNA-dependent RNA polymerases from the positive-strand RNA viruses. Frontiers in Microbiology, 10, 1945. 10.3389/fmicb.2019.01945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonniya, N. A., & Kar, P. (2020). Investigating specificity of the anti-hypertensive inhibitor WNK463 against With-No-Lysine kinase family isoforms via multiscale simulations. Journal of Biomolecular Structure & Dynamics, 38(5), 1306–1321. 10.1080/07391102.2019.1602079 [DOI] [PubMed] [Google Scholar]
- Jonniya, N. A., Sk, M. F., & Kar, P. (2019). Investigating phosphorylation-induced conformational changes in WNK1 kinase by molecular dynamics simulations. ACS Omega, 4(17), 17404–17416. 10.1021/acsomega.9b02187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar, P., & Knecht, V. (2012. a). Energetic basis for drug resistance of HIV-1 protease mutants against amprenavir. Journal of Computer-Aided Molecular Design, 26(2), 215–232. 10.1007/s10822-012-9550-5 [DOI] [PubMed] [Google Scholar]
- Kar, P., & Knecht, V. (2012. b). Energetics of mutation-induced changes in potency of lersivirine against HIV-1 reverse transcriptase. The Journal of Physical Chemistry B, 116(22), 6269–6278. 10.1021/jp300818c [DOI] [PubMed] [Google Scholar]
- Kar, P., & Knecht, V. (2012. c). Mutation-induced loop opening and energetics for binding of tamiflu to influenza N8 neuraminidase. The Journal of Physical Chemistry B, 116(21), 6137–6149. 10.1021/jp3022612 [DOI] [PubMed] [Google Scholar]
- Kar, P., & Knecht, V. (2012. d). Origin of decrease in potency of darunavir and two related antiviral inhibitors against HIV-2 compared to HIV-1 protease. The Journal of Physical Chemistry B, 116(8), 2605–2614. 10.1021/jp211768n [DOI] [PubMed] [Google Scholar]
- Kar, P., Lipowsky, R., & Knecht, V. (2011). Importance of polar solvation for cross-reactivity of antibody and its variants with steroids. The Journal of Physical Chemistry B, 115(23), 7661–7669. 10.1021/jp201538t [DOI] [PubMed] [Google Scholar]
- Kar, P., Lipowsky, R., & Knecht, V. (2013). Importance of polar solvation and configurational entropy for design of antiretroviral drugs targeting HIV-1 protease. The Journal of Physical Chemistry B, 117(19), 5793–5805. 10.1021/jp3085292 [DOI] [PubMed] [Google Scholar]
- Kar, P., Seel, M., Hansmann, U. H., & Höfinger, S. (2007). Dispersion terms and analysis of size- and charge dependence in an enhanced Poisson-Boltzmann approach. The Journal of Physical Chemistry B, 111(30), 8910–8918. 10.1021/jp072302u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar, P., Wei, Y., Hansmann, U. H., & Höfinger, S. (2007). Systematic study of the boundary composition in Poisson Boltzmann calculations. Journal of Computational Chemistry, 28(16), 2538–2544. 10.1002/jcc.20698 [DOI] [PubMed] [Google Scholar]
- Khan, M. T., Ali, A., Wang, Q., Irfan, M., Khan, A., Zeb, M. T., Zhang, Y.-J., Chinnasamy, S., & Wei, D.-Q. (2020). Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2. A molecular dynamic study. Journal of Biomolecular Structure and Dynamics. 10.1080/07391102.2020.1769733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., Li, Q., Shoemaker, B. A., Thiessen, P. A., Yu, B., Zaslavsky, L., Zhang, J., & Bolton, E. E. (2019). PubChem 2019 update: Improved access to chemical data. Nucleic Acids Research, 47(D1), D1102–D1109. 10.1093/nar/gky1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman, P. A., Massova, I., Reyes, C., Kuhn, B., Huo, S., Chong, L., Lee, M., Lee, T., Duan, Y., Wang, W., Donini, O., Cieplak, P., Srinivasan, J., Case, D. A., & Cheatham, T. E. (2000). Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Accounts of Chemical Research, 33(12), 889–897. 10.1021/ar000033j [DOI] [PubMed] [Google Scholar]
- Kräutler, V., van Gunsteren, W. F., & Hünenberger, P. H. (2001). A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. Journal of Computational Chemistry, 22(5), 501–508. [DOI] [Google Scholar]
- Kuzuhara, T., Iwai, Y., Takahashi, H., Hatakeyama, D., & Echigo, N. (2009). Green tea catechins inhibit the endonuclease activity of influenza A virus RNA polymerase. PLoS Currents, 1, RRN1052. 10.1371/currents.RRN1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larregieu, C. A., & Benet, L. Z. (2013). Drug discovery and regulatory considerations for improving in silico and in vitro predictions that use Caco-2 as a surrogate for human intestinal permeability measurements. The AAPS Journal, 15(2), 483–497. 10.1208/s12248-013-9456-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, L.-T., Hsu, W.-C., & Lin, C.-C. (2014). Antiviral natural products and herbal medicines. Journal of Traditional and Complementary Medicine, 4(1), 24–35. 10.4103/2225-4110.124335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncharich, R. J., Brooks, B. R., & Pastor, R. W. (1992). Langevin dynamics of peptides: The frictional dependence of isomerization rates of N-acetylalanyl-N'-methylamide. Biopolymers, 32(5), 523–535. 10.1002/bip.360320508 [DOI] [PubMed] [Google Scholar]
- Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., & Simmerling, C. (2015). ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. Journal of Chemical Theory and Computation, 11(8), 3696–3713. 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, Y. J., & Morris, M. E. (2007). Pharmacokinetics and bioavailability of the bioflavonoid biochanin A: Effects of quercetin and EGCG on biochanin A disposition in rats. Molecular Pharmaceutics, 4(6), 865–872. 10.1021/mp7000928 [DOI] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure Dynamics. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pires, D. E., Blundell, T. L., & Ascher, D. B. (2015). pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of Medicinal Chemistry, 58(9), 4066–4072. 10.1021/acs.jmedchem.5b00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, D. J., & Brooks III, C. L. (2004). A modified TIP3P water potential for simulation with Ewald summation. The Journal of Chemical Physics, 121(20), 10096–10103. 10.1063/1.1808117 [DOI] [PubMed] [Google Scholar]
- Randall, R. E., & Goodbourn, S. (2008). Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. The Journal of General Virology, 89(Pt 1), 1–47. 10.1099/vir.0.83391-0 [DOI] [PubMed] [Google Scholar]
- Roe, D. R., & Cheatham III, T. E. (2013). PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Roy, R., Ghosh, B., & Kar, P. (2020). Investigating conformational dynamics of Lewis Y oligosaccharides and elucidating blood group dependency of cholera using molecular dynamics. ACS Omega, 5(8), 3932–3942. 10.1021/acsomega.9b03398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, S. K., Shakya, A., Prasad, S. K., Singh, S., Gurav, N. S., Prasad, R. S., & Gurav, S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure Dynamics. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sk, M. F., Roy, R., Jonniya, N. A., Poddar, S., & Kar, P. (2020). Elucidating biophysical basis of binding of inhibitors to SARS-CoV-2 main protease by using molecular dynamics simulations and free energy calculations. Journal of Biomolecular Structure Dynamics. 10.1080/07391102.07392020.01768149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sk, M. F., Roy, R., & Kar, P. (2020). Exploring the potency of currently used drugs against HIV-1 protease of subtype D variant by using multiscale simulations. Journal of Biomolecular Structure Dynamics. 10.1080/07391102.07392020.01724196 [DOI] [PubMed] [Google Scholar]
- Song, J.-M., Lee, K.-H., & Seong, B.-L. (2005). Antiviral effect of catechins in green tea on influenza virus. Antiviral Research, 68(2), 66–74. 10.1016/j.antiviral.2005.06.010 [DOI] [PubMed] [Google Scholar]
- Subissi, L., Posthuma, C. C., Collet, A., Zevenhoven-Dobbe, J. C., Gorbalenya, A. E., Decroly, E., Snijder, E. J., Canard, B., & Imbert, I. (2014). One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proceedings of the National Academy of Sciences of the United States of America, 111(37), E3900–E3909. 10.1073/pnas.1323705111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajdek, A., & Borowska, E. (2008). Bioactive compounds and health-promoting properties of berry fruits: A review. Plant Foods for Human Nutrition (Dordrecht, Netherlands), 63(4), 147–156. 10.1007/s11130-008-0097-5 [DOI] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computaional Chemistry, 31, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, A. C., Laskowski, R. A., & Thornton, J. M. (1995). LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Engineering, 8(2), 127–134. 10.1093/protein/8.2.127 [DOI] [PubMed] [Google Scholar]
- Wang, J., Wang, W., Kollman, P. A., & Case, D. A. (2001). Antechamber: An accessory software package for molecular mechanical calculations. Journal of the American Chemical Society, 123(17), 3986–3994. 10.1021/ja003164o [DOI] [PubMed] [Google Scholar]
- Wang, J., Wang, W., Kollman, P. A., & Case, D. A. (2006). Automatic atom type and bond type perception in molecular mechanical calculations. Journal of Molecular Graphics & Modelling, 25(2), 247–260. 10.1016/j.jmgm.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., & Case, D. A. (2004). Development and testing of a general amber force field. Journal of Computational Chemistry, 25(9), 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C., & Zhang, Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., & Gong, P. (2018). Visualizing the nucleotide addition cycle of viral RNA-dependent RNA polymerase. Viruses, 10(1), 24. 10.3390/v10010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (New York, N.Y.), 367(6485), 1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W., Mao, C., Luan, X., Shen, D.-D., Shen, Q., Su, H., Wang, X., Zhou, F., Zhao, W., Gao, M., Chang, S., Xie, Y.-C., Tian, G., Jiang, H.-W., Tao, S.-C., Shen, J., Jiang, Y., Jiang, H., Xu, Y., … Xu, H. E. (2020). Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science (New York, N.Y.), 368(6498), 1499–1504. 10.1126/science.abc1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M.-S., Lee, J., Lee, J. M., Kim, Y., Chin, Y.-W., Jee, J.-G., Keum, Y.-S., & Jeong, Y.-J. (2012). Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorganic & Medicinal Chemistry Letters, 22(12), 4049–4054. 10.1016/j.bmcl.2012.04.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, Y., Sun, F., Li, X., Pang, H., Xu, X., Bartlam, M., & Rao, Z. (2005). Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nature Structural & Molecular Biology, 12(11), 980–986. 10.1038/nsmb999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.