Abstract
Purpose
Huashi Baidu formula (HSBDF) was developed to treat the patients with severe COVID-19 in China. The purpose of this study was to explore its active compounds and demonstrate its mechanisms against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) through network pharmacology and molecular docking.
Methods
All the components of HSBDF were retrieved from the pharmacology database of TCM system. The genes corresponding to the targets were retrieved using UniProt and GeneCards database. The herb–compound–target network was constructed by Cytoscape. The target protein–protein interaction network was built using STRING database. The core targets of HSBDF were analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). The main active compounds of HSBDF were docked with SARS-CoV-2 and angiotensin converting enzyme II (ACE2).
Results
Compound–target network mainly contained 178 compounds and 272 corresponding targets. Key targets contained MAPK3, MAPK8, TP53, CASP3, IL6, TNF, MAPK1, CCL2, PTGS2, etc. There were 522 GO items in GO enrichment analysis (p < .05) and 168 signaling pathways (p < .05) in KEGG, mainly including TNF signaling pathway, PI3K–Akt signaling pathway, NOD-like receptor signaling pathway, MAPK signaling pathway, and HIF-1 signaling pathway. The results of molecular docking showed that baicalein and quercetin were the top two compounds of HSBDF, which had high affinity with ACE2.
Conclusion
Baicalein and quercetin in HSBDF may regulate multiple signaling pathways through ACE2, which might play a therapeutic role on COVID-19.
Keywords: Huashi Baidu formula, coronavirus, SARS-CoV-2, network pharmacology, molecular docking
Introduction
Coronavirus disease (COVID-19) was caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a newly discovered coronavirus with a size of 60–140 nm, located in the SARB subgene of the Betacoronavirus family [1]. Moreover, the population's susceptibility to this highly pathogenic coronavirus led to massive global outbreak which turned into an international public health event [2]. There is a late surge of the confirmed cases in European region and region of the Americas. As on 2:00am CEST, 18 April 2020, there have been 2,160,207 confirmed cases of COVID-19, including 146,088 deaths globally [3]. A total of 83,160 cases have been confirmed in China, with 3342 deaths and a case fatality rate of 4% by now. According to an analysis of 44,672 confirmed cases out of 72,314 reported cases by the Chinese Center for Disease Control and Prevention (CDC), the crude case fatality rate of the patients with severe COVID-19 reached 49%. The fatality rate in severe patients with acute respiratory distress syndrome (ARDS) was close to 50%, and even as high as 70% if ARDS reached moderate to severe [4]. The older people or the crowd of underlying medical problems were more likely to infect with severe symptoms. The symptoms of patients were common with fever, shortness of breath, upper airway congestion, dry cough, fatigue, sputum production, myalgia/arthralgia, and breathing difficulties [5]. Critical patients can lead to pneumonia, kidney failure, and even death [5].
No vaccine for SARS-CoV-2 has been published publicly, and there was no medication specific for the treatment of COVID-19 so far [6]. It was proved that Traditional Chinese Medicine (TCM) could obviously shorten fever duration and symptomatic relief of the patients with severe COVID-19 [7–9]. Up to now, ‘three medicines and three formulas’ have been proven effective in treating the COVID-19. Among them, Huashi Baidu formula (HSBDF) was used as an auxiliary medicine for the treatment of patients with severe COVID-19. HSBDF was developed by Chinese Academy of Traditional Chinese Medicine [10]. HSBDF is consisted of glycyrrhiza, apricot kernel, agastache rugosus, magnolia officinalis, atractylodes, amomum tsao-ko, pinellia ternate, poria cocos, rhubarb, astragalus, lepidium seed, red peony root, and ephedra. According to the theory of TCM, the core pathogenesis of COVID-19 was the wet epidemic caused by the cold and humidity outside the lung and spleen, which was transformed into heat and lead to heat stagnation due to the imbalance of qi mechanism and endogenous stagnated heat [11]. Agastache rugosus and ephedra in HSBDF have the functions of detoxifying dampness, clearing heat, and relieving asthma [10]. HSBDF was applied to the treatment of severe patients, and proved to be effective in resisting SARS-CoV-2, eliminating inflammation, and improving immunity [10,12].
However, the mechanism of HSBDF for the treatment of COVID-19 was not clear. HSBDF contained 14 Chinese herbs, and the components of each herb were complex. The therapeutic effect of each herb or each component was not clear.
Network pharmacology was proposed as a promising method to understand herbal formulas [13] and predict potential new drugs or targets for the specific diseases [14–16]. Molecular docking is a significant pathway of structural molecular biology and the computer aided drug design in new medicines [17,18]. Study showed that SARS-CoV-2 binded to angiotensin converting enzyme II (ACE2) receptor with nearly 10–20 times higher affinity than SARS-CoV [17]. The combination of SARS-CoV-2 and ACE2 was the main cause of COVID-19. Therefore, ACE2 and SARS-CoV-2 3CL were regarded as receptors in molecular docking. In this study, we aim to utilize network pharmacology and molecular docking to understand the active compounds of HSBDF, predict their potential targets and signal pathways, and explore the association between the active compounds of HSBDF with ACE2.
Materials and methods
Identification and screening of active compounds
Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://tcmspw.com/) was used to authenticate all compounds of the fourteen Chinese medicinal herbs in HSBDF. TCMSP had the ability to identify ingredient–target networks and the large number of herbal items. It was composed of 499 Chinese herbs recorded in the Chinese pharmacopeia with 29,384 compounds and 3311 targets [19]. The names of herbs were used as the key words to retrieve all components. We screened compounds of HSBDF based on absorption, distribution, metabolism, and excretion (ADME), which were strategic processes in drug discovery and development [20]. Oral bioavailability (OB) and drug-likeness (DL) were the most significant pharmacokinetic parameters. In ADME system [21], OB was defined as ‘the degree to which active ingredients are used by the body, including the dose and rate at which they enter the bloodstream’ by Food and Drug Administration (FDA) [22]. The degree of OB largely determined the effect of the compound on disease. DL was used to screen out first-rank compounds and improve candidate compounds in the early period of drug development [14]. In this study, the active compounds in HSBDF were selected according to the criterion of OB ≥30% and DL ≥0.18 [23].
Identification of protein targets
The protein targets associated with active compounds were retrieved from the TCMSP database (http://lsp.nwsuaf.edu.cn/tcmsp.php), which provided information of 6511 drug molecules and 3987 targets as well as the interactions between them [19]. As an authoritative database of protein sequences, UniProt Knowledgebase (UniProtKB) comprised 54,247,468 sequence items [24]. The targets, including the gene names and gene ID, were further extracted using UniProtKB (http://www.uniprot.org).
Construction of component–target gene network
Visual component–target network was established based on aforementioned data sets through Cytoscape 3.7.2 (http://www.cytoscape.org/) to reflect the complex relationships between active compounds and their potential targets [25]. Cytoscape 3.7.2 is an open-source software platform which is used to visualize complicated networks and integrate different types of attribute data [26]. In the network, nodes represent the screened active ingredients and targets, while the connections between the nodes represent the interactions between these biological analyses. The top eight compounds were screened as ligands for molecular docking based on the degree value. The degree value of the molecular represents the number of connections between the molecular and target in the core architecture of the network [27]. The larger the value is, the more likely the component is to become the key ingredient of HSBDF.
Predicting the targets of COVID-19
GeneCards database (https://www.genecards.org/) and Therapeutic Target Database (TTD, https://db.idrblab.org/ttd/) were used to gather the information on COVID-19-associated target genes [28]. GeneCards is a comprehensive database of functions involving proteomics, genomics, and transcriptomics [29]. The keywords ‘novel coronavirus pneumonia,’ ‘cough,’ and ‘fever’ were utilized to screen the COVID-19-associated targets. The names of targets were collected from TTD, which provided information about the therapeutic protein targets, the corresponding ID of each targets and the targeted disease. The common targets of HSBDF and COVID-19 were then gathered as the core targets of HSBDF for COVID-19.
Construction of protein–protein interactions (PPI) network
The core targets of HSBDF were put into STRING (https://string-db.org/cgi/input.pl) to build the PPI network interaction [30]. Cytoscape V3.7.2 [26] was used to construct and visualize the PPI network. CytoNCA, a network topology analysis plug-in in Cytoscape, was used for topological analysis of PPI networks. ‘Degree’ referred to the number of connections of the node in the whole network, which reflected the interaction information between nodes. The value of ‘Degree’ was used as a reference for the importance of the core target.
GO and KEGG pathway enrichment analysis
Webgestalt (www.webgestalt.org) was used to process data and visualize the enrichment results of Gene Ontology (GO) enrichment analysis. GO enrichment analysis included biological processes (BP), molecular functions (MF), and cellular components (CC). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using the Rpackage ‘ClusterProfifiler’ [31]. Statistical significance threshold of enrichment analysis was set at p ≤ .05.
Component–target molecular docking
The three-dimensional (3D) structure of the SARS-CoV-2 3CL (PDB ID: 6lu7) was downloaded from the RCSB PDB database (https://www.rcsb.org/). The 3D structure of ACE2 (PDB ID: 1r42) was from Qingdao Ocean Science and Technology National Laboratory 2019-nCoV drug target system structure of information sharing platform (http://ncovtarget.qnlm.ac/web/mg/hm). AutoDock Tools 1.5.6 software was used to remove the water molecules, isolate proteins, add the nonpolar hydrogen and calculate Gasteiger charges for the structure and save it as a PDBQT file. PubChem database (https://pubchem.ncbi.nlm.nih.gov/) was used to download the two-dimensional (2D) structures of the top eight compounds. The 2D structure was processed and transformed into PDB format through Chem3D, and they were saved in PDBQT format as docking ligands in AutoDock Tools 1.5.6 software. SARS-CoV-2 3CL protein and ACE2 were used as receptors, and the active compounds were used as ligands. The active site of molecular docking was determined by the ligand coordinate in the target protein complex. The ligand was set to be flexible and the receptor was rigid. Autodock Vina 1.1.2 was used to dock small molecules with ACE2 protein and SARS-CoV-2 3CL protein, respectively. For results of docking, a total of 20 conformations were generated each. The conformation with the best affinity was selected as the final docking conformation and visualized in Pymol 2.3. These active ingredients were compared with lopinavir, ritonavir, and remdesivir.
Results
Active compounds of HSBDG
A total of 222 active compounds were retrieved in TCMSP database. These active compounds were primarily originated from glycyrrhiza (92 compounds), red peony root (29 compounds), ephedra (23 compounds), astragalus (20 compounds), apricot kernel (19 compounds), rhubarb (16 compounds), poria cocos (15 compounds), pinellia ternate (13 compounds), lepidium seed (12 compounds), agastache rugosus (11 compounds), atractylodes (nine compounds), amomum tsao-ko (eight compounds), etc. The main active components of HSBDG are shown in Table 1.
Table 1.
Basic information of some active components of HSBDG.
| Mol ID | Chemical component | OB/% | DL | Herb |
|---|---|---|---|---|
| MOL004990 | 7,2′,4′-Trihydroxy-5-methoxy-3-arylcoumarin | 83.71 | 0.27 | Glycyrrhiza |
| MOL002311 | Glycyrol | 90.78 | 0.67 | |
| MOL001925 | Paeoniflorin_qt | 68.18 | 0.40 | Red peony root |
| MOL001918 | Paeoniflorgenone | 87.59 | 0.37 | |
| MOL004328 | Naringenin | 59.29 | 0.21 | Ephedra |
| MOL005190 | Eriodictyol | 71.79 | 0.24 | |
| MOL000378 | 7-O-methylisomucronulatol | 74.69 | 0.30 | Astragalus |
| MOL000398 | Isoflavanone | 109.99 | 0.30 | |
| MOL010922 | Diisooctyl succinate | 31.62 | 0.23 | Apricot kernel |
| MOL005030 | Gondoic acid | 30.70 | 0.20 | |
| MOL002293 | Sennoside D_qt | 61.06 | 0.61 | Rhubarb |
| MOL000471 | Aloe-emodin | 83.38 | 0.24 | |
| MOL000282 | Ergosta-7,22E-dien-3beta-ol | 43.51 | 0.72 | Poria cocos |
| MOL000300 | Dehydroeburicoic acid | 44.17 | 0.83 | |
| MOL006967 | Beta-D-Ribofuranoside, xanthine-9 | 44.72 | 0.21 | Pinellia ternate |
| MOL006957 | (3S,6S)-3-(benzyl)-6-(4-hydroxybenzyl)piperazine-2,5-quinone | 46.89 | 0.27 | |
| MOL000422 | Kaempferol | 41.88 | 0.24 | Lepidium seed |
| MOL000098 | Quercetin | 46.43 | 0.28 | |
| MOL005911 | 5-Hydroxy-7,4′-dimethoxyflavanon | 51.54 | 0.27 | Agastache rugosus |
| MOL005890 | Pachypodol | 75.06 | 0.40 | |
| MOL000186 | Stigmasterol 3-O-beta-D-glucopyranoside_qt | 43.83 | 0.76 | Atractylodes |
| MOL000179 | 2-Hydroxyisoxypropyl-3-hydroxy-7-isopentene-2,3-dihydrobenzofuran-5-carboxylic | 45.20 | 0.20 | |
| MOL000096 | (−)-Catechin | 49.68 | 0.24 | Amomum tsao-ko |
| MOL000074 | (4E,6E)-1,7-bis(4-hydroxyphenyl)hepta-4,6-dien-3-one | 67.92 | 0.24 | |
| MOL005980 | Neohesperidin | 57.44 | 0.27 | Magnolia officinalis |
| MOL005970 | Eucalyptol | 60.62 | 0.32 |
The construction of herb–compound–target network
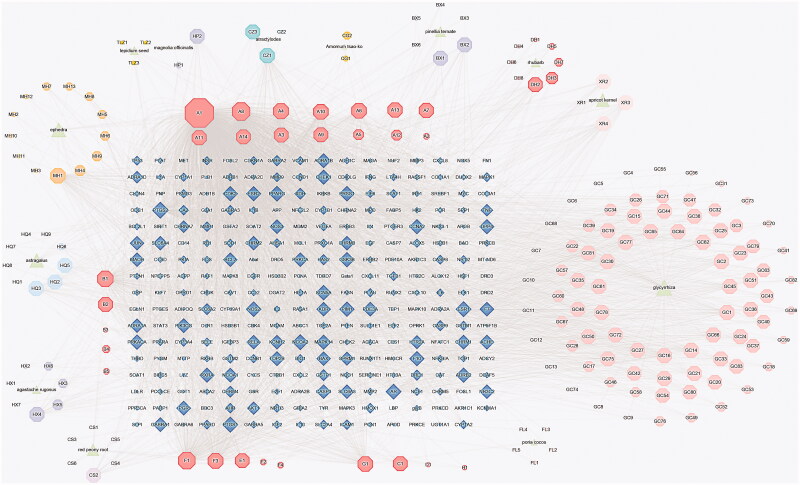
After 44 compounds without target were removed, herb–compound–target network contained 463 nodes (including 13 herbs, 178 compounds and 272 genes) and 4081 edges (Figure 1). The larger node meant more importance. According to the degree analysis, the top eight compounds were MOL000098 (quercetin), MOL000422 (kaempferol), MOL000358 (beta-sitosterol), MOL000449 (stigmasterol), MOL000354 (isorhamnetin), MOL002714 (baicalein), MOL004328 (naringenin), and MOL000392 (formononetin), with 886°, 240°, 185°, 120°, 99°, 76°, 72°, and 72°, respectively. More details of these top eight compounds are shown in Table 2.
Figure 1.
Herb-compound-target network of HSBDF (The triangle nodes are composed of all the herbs of HSBDF, which are surrounded with their particular compounds. The octagon nodes represent the compounds of HSBDF. The rhombus nodes, arranged into a rectangular matrix, represent the relative gene targets of HSBDF).
Table 2.
Basic information of the top eight degree of the compounds.
| MOL ID | compound | CAS | Molecular formula |
|---|---|---|---|
| MOL000098 | Quercetin | 117-39-5 | 2-(3,4-Dihydroxyphenyl)-3,5,7-trihydroxychromen-4-one |
| MOL000422 | Kaempferol | 520-18-3 | 3,5,7-Trihydroxy-2-(4-hydroxyphenyl)chromen-4-one |
| MOL000358 | Beta-sitosterol | 474-58-8 | Beta-sitosterol 3-O-glucoside_qt |
| MOL000449 | Stigmasterol | 83-48-7 | (3S,8S,9S,10R,13R,14S,17R)-17-[(2R,5S)-5-ethyl-6-methylhept-3-en-2-yl]-10,13-dimethyl-2,3,4,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-3-ol |
| MOL000354 | Isorhamnetin | 480-19-3 | 3,5,7-Trihydroxy-2-(4-hydroxy-3-methoxyphenyl)chromen-4-one |
| MOL002714 | Baicalein | 491-67-8 | 5,6,7-Trihydroxy-2-phenylchromen-4-one |
| MOL004328 | Naringenin | 480-41-1 | (2S)-5,7-dihydroxy-2-(4-hydroxyphenyl)chroman-4-one |
| MOL000392 | Formononetin | 485-72-3 | 7-Hydroxy-3-(4-methoxyphenyl)-4H-benzopyran-4-one |
Prediction results of virus targets and the construction of PPI network
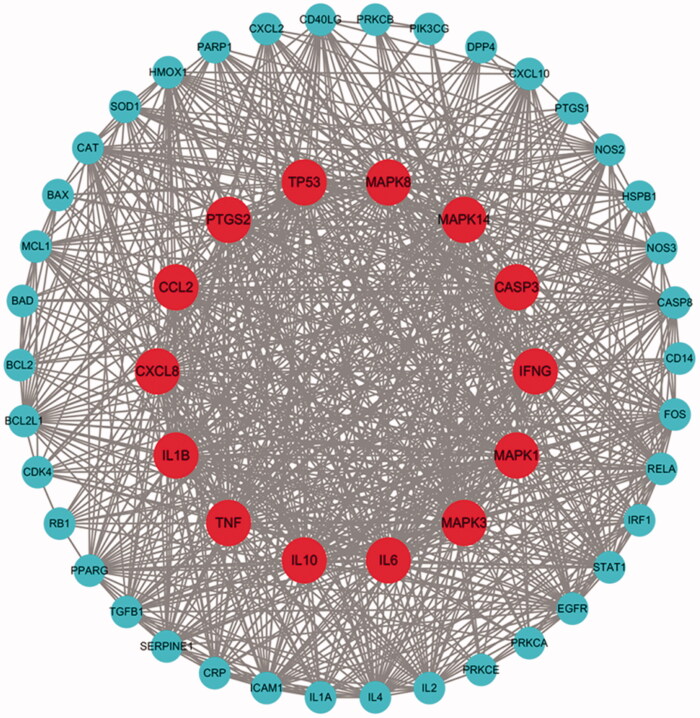
A total of 216 possible targets of COVID-19 were screened through ‘novel coronavirus pneumonia,’ ‘cough,’ and ‘fever.’ These 216 targets were combined with 272 targets of HSBDF to obtain a total of 53 core targets (Figure 2, Table 3). The PPI network showed that there were strong correlations among targets and a complex interlaced network. The network contained a total of 53 nodes, which were the core targets of HSBDF in the treatment of COVID-19. Among them, MAPK3 (45), MAPK8 (44), TNF (44), IL6 (44), and TP53 (44) were considered to be hub genes (Table 3).
Figure 2.
The PPI network of 53 nodes (genes). The larger nodes in the inner ring represent more important hub nodes. The smaller nodes in the outer ring represent the other nodes.
Table 3.
A partial information table for the core targets.
| Targets | Degree | Targets | Degree |
|---|---|---|---|
| MAPK3 | 45.0 | MAPK1 | 42.0 |
| MAPK8 | 44.0 | IL10 | 41.0 |
| TNF | 44.0 | CXCL8 | 39.0 |
| IL6 | 44.0 | PTGS2 | 38.0 |
| TP53 | 44.0 | CCL2 | 38.0 |
| CASP3 | 43.0 | IL1B | 38.0 |
GO and KEGG enrichment analyze
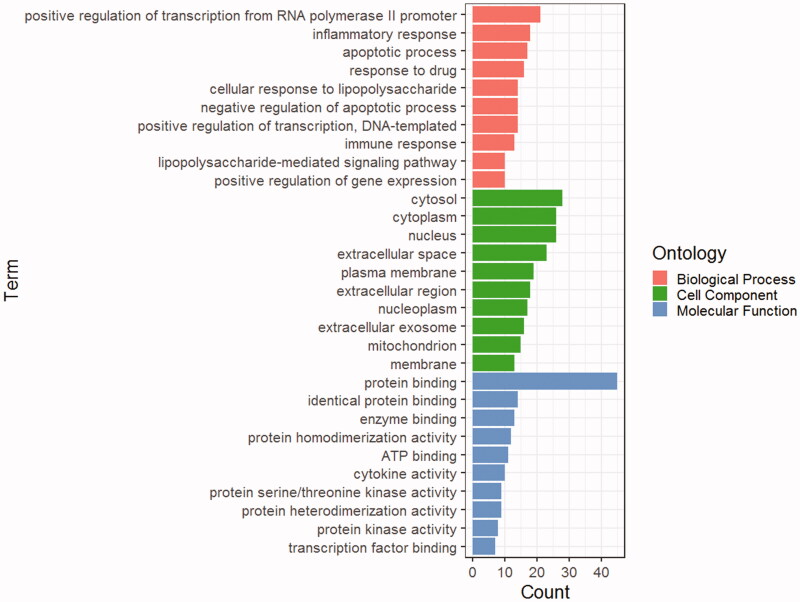
To further understand the intersection genes, GO enrichment analysis was conducted. (1) A total 328 top-ranking terms about biological processes were selected, mainly including inflammatory response, response to drug, and cellular response to lipopolysaccharide (Figure 3). The active targets essentially connected to positive regulation of transcription from RNA polymerase II promoter or apoptotic process, including IL6, TNF, and TP53. (2) According to enrichment analysis of cellular components, the targets mainly contained extracellular space, Cytosol, extracellular region, Cytoplasm, Nucleus, and so on. (3) Simultaneously, molecular function terms mainly contained protein binding, identical protein binding, enzyme binding, cytokine activity, protein homodimerization activity, etc.
Figure 3.
The GO enrichment analysis of 53 nodes (BP, MF, CC).
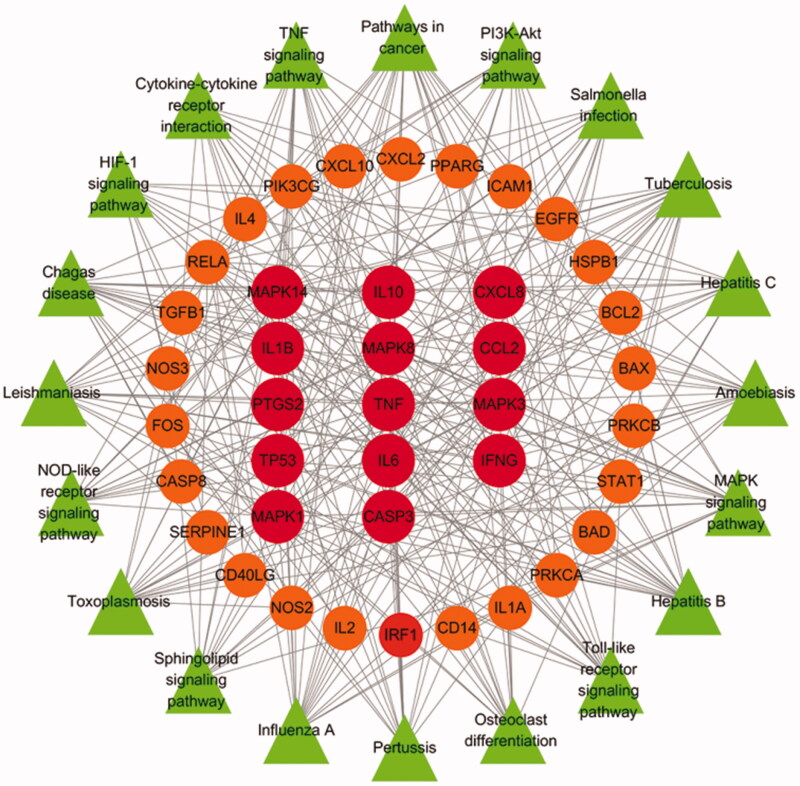
KEGG pathway enrichment was conducted to cluster the major effects that associated to the HSBDF. A total of 20 top-ranking pathway (Figure 4) screened out (p < .05). The main pathway included TNF signaling pathway, PI3K–Akt signaling pathway, pathways in cancer, MAPK signaling pathway as well as some related to cancer.
Figure 4.
Target–pathway network of HSBDF. (The round nodes in the center represent the important hub nodes, and the smaller round nodes in the middle ring represent the other nodes. The outermost triangles represent the related pathways).
Results of molecular docking
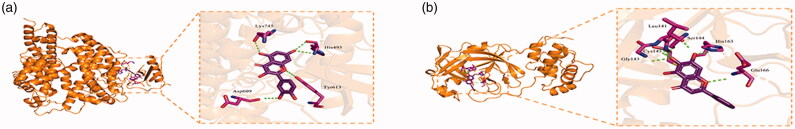
Molecular docking was used to verify if the top eight compounds had a significant role in regulation of ACE2. The result showed that all the key compounds in the network had strong affinity with ACE2 protein and SARS-CoV-2 3CL protein. Baicalein (MOL002714) was the most stable active ingredient in SARS-CoV-2 3CL binding. Meanwhile, quercetin (MOL000098) showed strong association with ACE2. The small molecule quercetin showed a compact binding pattern with ACE2 protein active pocket. Quercetin formed four hydrogen bonds with the amino acid residues Lys745, Tyr613, His493, and Asp609, making quercetin and ACE2 form a stable complex (Figure 5). The binding energies of the various compounds are shown in Table 4.
Figure 5.
(a) ACE2 protein-quercetin. (b) SARS-CoV-2 3CL protein-baicalein.
Table 4.
The binding energy of top eight compounds.
| CAS | Molecule name | Molecular formula | MW | SARS-CoV-2 3CL docking score (kcal/mol) | ACE2 docking score (kcal/mol) |
|---|---|---|---|---|---|
| 117-39-5 | Quercetin | C15H10O7 | 302.25 | −7.5 | −8.4 |
| 520-18-3 | Kaempferol | C15H10O6 | 286.25 | −7.6 | −8.2 |
| 83-46-5 | Beta-sitosterol | C29H50O | 414.79 | −7.7 | −8.3 |
| 83-48-7 | Stigmasterol | C29H48O | 412.77 | −7.0 | −8.2 |
| 480-19-3 | Isorhamnetin | C16H1207 | 316.28 | −7.2 | −8.2 |
| 491-67-8 | Baicalein | C15H10O5 | 270.25 | −7.8 | −8.2 |
| 480-41-1 | Naringenin | C15H12O5 | 272.27 | −7.7 | −7.9 |
| 485-72-3 | Formononetin | C16H12O4 | 268.28 | −7.2 | −7.5 |
| 192725-17-0 | Lopinavir | C37H48N4O5 | 628.80 | −8.0 | −8.2 |
| 1809249-37-3 | Remdesivir | C27H35N6O8P | 602.58 | −7.6 | −7.9 |
| 155213-67-5 | Ritonavir | C37H48N6O5S2 | 720.96 | −7.8 | −8.5 |
Discussion
In this study, we screened out eight main active components of HSBDF: quercetin, kaempferol, beta-sitosterol, stigmasterol, isorhamnetin, baicalein, naringenin, and formononetin. In molecular docking, these active ingredients were compared with lopinavir, ritonavir, and remdesivir, which were currently recommended for clinical therapeutics [32–34]. Results showed that baicalein showed strong affinity for SARS-CoV-2, and quercetin had strong affinity for ACE2 3CL. It indicated that baicalein and quercetin might play an important role in the treatment of SARS-CoV-2. Baicalein and quercetin were both flavonoids. Flavonoids reduced the barrier dysfunction induced by influenza A virus by inhibiting the NOX4/NF-κB/MLCK pathway, which might be a potential drug for the prevention and treatment of influenza A virus and pulmonary endothelial barrier dysfunction [35]. A study showed that baicalein inhibited the overactivation of the complement system in vivo and improved the acute lung injury induced by influenza a virus [36]. Since both SARS and COVID-19 were caused via binding S-protein to ACE2 [37,38], quercetin acted as a competitive antagonist to inhibit infection of SARS-CoV-2. Other research also reported that quercetin had the functions of reducing capillary brittleness, angiogenesis, detoxification, apoptosis, cell cycle, and antioxidant replication [39].
KEGG enrichment analysis showed that the key targets were mainly concentrated in TNF signaling pathway, PI3K–Akt signaling pathway, MAPK signaling pathway, HIF-1 signaling pathway, NOD-like receptor signaling pathway. The results of PPI network showed that, MAPK3, MAPK8, TNF, IL6, and TP53 were considered to be hub genes. According to these results, we think HSBDF had effect to treat SARS-CoV-2 through the following pathways. (1) TNF signaling pathway: GO enrichment analysis also showed that the major biological processes included inflammatory response in our study. TNF signaling pathway was an important pathway in inflammatory response [40], in which related factor receptors can also induce apoptosis. Quercetin acted as an anti-inflammatory by regulating the TNF signaling pathway [41]. Among the hub genes, IL6 and TNF were involved in TNF signaling pathway. IL6 and TNF were important inflammatory cytokines and commonly involved in the process of inflammation [42]. TNF induced the production of IL-6 and other cytokines, participated in the process of inflammation and oxidative stress [43]. (2) MAPK signaling pathway: Previous studies showed that MAPK signaling pathway participated in the progression of ARDS [44,45]. Many inflammatory factors such as IL-1β, TNF-α, and IL-6 were produced via MAPK signaling pathway [46]. Some anti-inflammatory medications work by targeting MAPK signaling pathway [47,48]. Quercetin was found to regulate the activation of MAPK signaling pathway in retinoblastoma, cardiomyocytes, and chorionic carcinoma cells [49]. Other studies have found that baicalin may inhibit the expression and invasion of cancer cells by inhibiting the p38 MAPK signaling pathway [50]. (3) PI3K–Akt signaling pathway: The PI3K/AKT signaling pathway regulated the activation of inflammatory response cells and the release of inflammatory transmitters to play a role in chronic inflammatory response in the lungs and airways [51]. Quercetin inhibited the PI3K–Akt signaling pathway by inhibiting the expression of AKT1 to silence the anti-apoptotic effect of lung fibroblast, thus realize the treatment of pulmonary fibrosis [52]. Baicalin also played an anti-pulmonary fibrosisrole by the PI3K–Akt pathway [53–55].
There are several limitations in our study. First, our results need to be further verified by experiments. Second, more comprehensive TCM target genes database was needed, which made the results of network pharmacology analysis more reliable. Third, even if the results of network pharmacology and molecular docking were combined, we still could not completely understand the accurate therapeutic mechanism of HSBDF. A comprehensive understanding of HSBDF and COVID-19 depended on the common development of multi-disciplines.
Conclusions
In summary, network pharmacology showed that the main active components of HSBDF, particularly baicalein, and quercetin could act on multiple targets. HSBDF had effect to treat SARS-CoV-2 mainly through the following pathways: TNF signaling pathway, PI3K–Akt signaling pathway, MAPK signaling pathway. Molecular docking showed baicalein and quercetin were the top two compounds which indicated that they might play an important role in the treatment of SARS-CoV-2.
Acknowledgments
All the authors of the manuscript are immensely grateful to their respective universities and institutes for their technical assistance and valuable support in the completion of this research project.
Funding Statement
This study was funded by the National Natural Science Foundation of China [Grant no. 81403296], the Natural Science Foundation of Guangdong Province [Grant no. 2017A030313827], and Science Program for Overseas Scholar (Xinhuo plan) of Guangzhou University of Chinese Medicine.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Lipsitch M, Swerdlow DL, Finelli L.. Defining the epidemiology of Covid-19 – studies needed. N Engl J Med. 2020;382(13):1194–1196. [DOI] [PubMed] [Google Scholar]
- 2.Nkengasong J. China's response to a novel coronavirus stands in stark contrast to the 2002 SARS outbreak response. Nat Med. 2020;26(3):310–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Novel Coronavirus (COVID-19) Situation dashboard. [cited 2020 Jul 1]. Available from: https://covid19.who.int/.
- 4.Liu Y, Sun W, Li J. Clinical features and progression of acute respiratory distress syndrome in coronavirus disease. medRxiv. 2019;02:2020.DOI: 10.1101/2020.02.17.20024166 [DOI] [Google Scholar]
- 5.World Health Organization . Coronavirus Overview. [cited 2020 Jul 1]. Available from: https://www.who.int/health-topics/coronavirus/coronavirus#tab=tab_1.
- 6.Heymann DL, Shindo N.. COVID-19: what is next for public health? Lancet (London, England). 2020;395(10224):542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren JL, Zhang AH, Wang XJ.. Corrigendum to ‘Traditional Chinese medicine for COVID-19 treatment’. Pharmacol Res. 2020;155:104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Liu X, Guo L, et al. Traditional Chinese herbal medicine for treating novel coronavirus (COVID-19) pneumonia: protocol for a systematic review and meta-analysis. Syst Rev. 2020;9(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Islam MS, Wang J, et al. Traditional Chinese medicine in the treatment of patients infected with 2019-new coronavirus (SARS-CoV-2): a review and perspective. Int J Biol Sci. 2020;16(10):1708–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. NHC:Diagnosis and treatment of pneumonia caused by new coronavirus (trial version 7). China: National Health Commission; 2020. Available from: http://www.nhc.gov.cn/xcs/zhengcwj/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml
- 11.Ma J, Huo XQ, Chen X, et al. Study on screening potential traditional Chinese medicines against 2019-nCoV based on Mpro and PLP], Zhongguo Zhong yao za zhi = Zhongguo zhongyao zazhi. China J Chinese Materia Medica. 2020;45:1219–1224. [ [DOI] [PubMed] [Google Scholar]
- 12.Ye H, Wei J, Tang K, et al. Drug repositioning through network pharmacology. Curr Top Med Chem. 2016;16(30):3646–3656. [DOI] [PubMed] [Google Scholar]
- 13.Zhang GB, Li QY, Chen QL, et al. Network pharmacology: a new approach for Chinese herbal medicine research. Evid. Based Complement. Altern. Med.: eCAM. 2013;2013:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao W, Xu X, Wang X, et al. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J Ethnopharmacol. 2013;145(1):1–10. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Yu H, Qi J, et al. Natural formulas and the nature of formulas: exploring potential therapeutic targets based on traditional Chinese herbal formulas. PloS One. 2017;12(2):e0171628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan JF, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris GM, Lim-Wilby M.. Molecular docking. Methods Mol Biol (Clifton, NJ). 2008;443:365–382. [DOI] [PubMed] [Google Scholar]
- 18.Saikia S, Bordoloi M.. Molecular docking: challenges, advances and its use in drug discovery perspective. Curr Drug Targets. 2019;20(5):501–521. [DOI] [PubMed] [Google Scholar]
- 19.Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su X, Kong L, Lei X, et al. Biological fingerprinting analysis of traditional Chinese medicines with targeting ADME/Tox property for screening of bioactive compounds by chromatographic and MS methods. Mini Rev Med Chem. 2007;7(1):87–98. [DOI] [PubMed] [Google Scholar]
- 21.Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 2012;13(6):6964–6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen ML, Shah V, Patnaik R, et al. Bioavailability and bioequivalence: an FDA regulatory overview. Pharm Res. 2001;18(12):1645–1650. [DOI] [PubMed] [Google Scholar]
- 23.Ning K, Zhao X, Poetsch A, et al. Computational molecular networks and network pharmacology. Biomed Res Int. 2017;2017:7573904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, Yuan Y, Zhou W, et al. Network pharmacology analysis of Chaihu Lizhong Tang treating non-alcoholic fatty liver disease. Comput Biol Chem. 2020;86:107248. [DOI] [PubMed] [Google Scholar]
- 25.Qin T, Wu L, Hua Q, et al. Prediction of the mechanisms of action of Shenkang in chronic kidney disease: a network pharmacology study and experimental validation. J Ethnopharmacol. 2020;246:112128. [DOI] [PubMed] [Google Scholar]
- 26.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang S, Jing H, Huang Z, et al. Identification of key candidate genes in neuropathic pain by integrated bioinformatic analysis. J Cell Biochem. 2020;121(2):1635–1648. [DOI] [PubMed] [Google Scholar]
- 28.Li YH, Yu CY, Li XX, et al. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018;46(D1):D1121–D1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paolacci S, Precone V, Acquaviva F, et al. Genetics of lipedema: new perspectives on genetic research and molecular diagnoses. Eur Rev Med Pharmacol Sci 2019;23:5581–5594. [DOI] [PubMed] [Google Scholar]
- 30.UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Zhang YH, Wang S, et al. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One. 2017;12(9):e0184129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu CM, Cheng VC, Hung IF, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends. 2020;14(1):69–71. [DOI] [PubMed] [Google Scholar]
- 35.Yu WY, Li L, Wu F, et al. Moslea Herba flavonoids alleviated influenza A virus-induced pulmonary endothelial barrier disruption via suppressing NOX4/NF-κB/MLCK pathway. J Ethnopharmacol. 2020;253:112641. [DOI] [PubMed] [Google Scholar]
- 36.Zhi H, Jin X, Zhu H, et al. Exploring the effective materials of flavonoids-enriched extract from Scutellaria baicalensis roots based on the metabolic activation in influenza A virus induced acute lung injury. J Pharm Biomed Anal. 2020;177:112876. [DOI] [PubMed] [Google Scholar]
- 37.Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song W, Gui M, Wang X, et al. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14(8):e1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q, Zhao XH, Wang ZJ.. Cytotoxicity of flavones and flavonols to a human esophageal squamous cell carcinoma cell line (KYSE-510) by induction of G2/M arrest and apoptosis. Toxicol In Vitro: Int J Published Assoc BIBRA. 2009;23(5):797–807. [DOI] [PubMed] [Google Scholar]
- 40.Noack M, Miossec P.. Selected cytokine pathways in rheumatoid arthritis. Semin Immunopathol. 2017;39(4):365–383. [DOI] [PubMed] [Google Scholar]
- 41.Kashyap D, Mittal S, Sak K, et al. Molecular mechanisms of action of quercetin in cancer: recent advances. Tumour Biol. 2016;37(10):12927–12939. [DOI] [PubMed] [Google Scholar]
- 42.Shivappa N, Hébert JR, Rosato V, et al. Inflammatory potential of diet and risk of oral and pharyngeal cancer in a large case-control study from Italy. Int J Cancer. 2017;141(3):471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ge Q, Chen L, Tang M, et al. Analysis of mulberry leaf components in the treatment of diabetes using network pharmacology. Eur J Pharmacol. 2018;833:50–62. [DOI] [PubMed] [Google Scholar]
- 44.Xiong LL, Tan Y, Ma HY, et al. Administration of SB239063, a potent p38 MAPK inhibitor, alleviates acute lung injury induced by intestinal ischemia reperfusion in rats associated with AQP4 downregulation. Int Immunopharmacol. 2016;38:54–60. [DOI] [PubMed] [Google Scholar]
- 45.Ma L, Zhao Y, Wang R, et al. 3,5,4′-Tri-O-acetylresveratrol attenuates lipopolysaccharide-induced acute respiratory distress syndrome via MAPK/SIRT1 pathway. Mediators Inflamm. 2015;2015:143074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bode JG, Ehlting C, Häussinger D.. The macrophage response towards LPS and its control through the p38(MAPK)-STAT3 axis. Cell Signal. 2012;24(6):1185–1194. [DOI] [PubMed] [Google Scholar]
- 47.Liu W, Jiang HL, Cai LL, et al. Tanreqing injection attenuates lipopolysaccharide-induced airway inflammation through MAPK/NF-κB signaling pathways in rats model. Evid Based Complement Alternat Med. 2016;2016:5292346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen CC, Lin MW, Liang CJ, et al. The anti-inflammatory effects and mechanisms of eupafolin in lipopolysaccharide-induced inflammatory responses in RAW264.7 macrophages. PloS One. 2016;11(7):e0158662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C, Wang T, Zhang C, et al. Quercetin attenuates cardiomyocyte apoptosis via inhibition of JNK and p38 mitogen-activated protein kinase signaling pathways. Gene. 2016;577(2):275–280. [DOI] [PubMed] [Google Scholar]
- 50.Yan H, Xin S, Wang H, et al. Baicalein inhibits MMP-2 expression in human ovarian cancer cells by suppressing the p38 MAPK-dependent NF-κB signaling pathway. Anticancer Drugs. 2015;26(6):649–656. [DOI] [PubMed] [Google Scholar]
- 51.Jiang H, Abel PW, Toews ML, et al. Phosphoinositide 3-kinase gamma regulates airway smooth muscle contraction by modulating calcium oscillations. J Pharmacol Exp Ther. 2010;334(3):703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wan Y, Xu L, Liu Z, et al. Utilising network pharmacology to explore the underlying mechanism of Wumei Pill in treating pancreatic neoplasms. BMC Complement Altern Med. 2019;19(1):158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao A, Zeng Q, Xie X, et al. MicroRNA-125b induces cancer cell apoptosis through suppression of Bcl-2 expression. J Genet Genomics. 2012;39(1):29–35. [DOI] [PubMed] [Google Scholar]
- 54.Duronio V. The life of a cell: apoptosis regulation by the PI3K/PKB pathway. Biochem J. 2008;415(3):333–344. [DOI] [PubMed] [Google Scholar]
- 55.Wang W, Zeng C, Feng Y, et al. The size-dependent effects of silica nanoparticles on endothelial cell apoptosis through activating the p53-caspase pathway. Environ Pollut (Barking, Essex: 1987). 2018;233:218–225. [DOI] [PubMed] [Google Scholar]