Abstract
A computational approach to in silico drug discovery was carried out to identify small drug-like compounds able to show structural and functional mimicry of the high affinity ligand X77, potent non-covalent inhibitor of SARS-COV-2 main protease (MPro). In doing so, the X77-mimetic candidates were predicted based on the crystal X77-MPro structure by a public web-oriented virtual screening platform Pharmit. Models of these candidates bound to SARS-COV-2 MPro were generated by molecular docking, quantum chemical calculations and molecular dynamics simulations. At the final point, analysis of the interaction modes of the identified compounds with MPro and prediction of their binding affinity were carried out. Calculation revealed 5 top-ranking compounds that exhibited a high affinity to the active site of SARS-CoV-2 MPro. Insights into the ligand − MPro models indicate that all identified compounds may effectively block the binding pocket of SARS-CoV-2 MPro, in line with the low values of binding free energy and dissociation constant. Mechanism of binding of these compounds to MPro is mainly provided by van der Waals interactions with the functionally important residues of the enzyme, such as His-41, Met-49, Cys-145, Met-165, and Gln-189 that play a role of the binding hot spots assisting the predicted molecules to effectively interact with the MPro active site. The data obtained show that the identified X77-mimetic candidates may serve as good scaffolds for the design of novel antiviral agents able to target the active site of SARS-CoV-2 MPro.
Communicated by Ramaswamy H. Sarma
Keywords: Coronavirus SARS-CoV-2, COVID-19, main protease, SARS-CoV-2 inhibitors, virtual screening, molecular docking, quantum chemical calculations, antiviral drugs
Introduction
The recent outbreak of coronavirus infection in China caused by the SARS-CoV-2 virus associated with COVID-19 has become a matter of serious concern to the world community, as the number of infected people is constantly increasing with significant geographical spread. As of the beginning of June 2020, the World Health Organization reports over 6.9 million confirmed cases of infection and over 400 thousand deaths. Numerous attempts are being made to develop an effective antiviral vaccine and find new therapeutic agents against COVID-19. Studies of various aspects of SARS-CoV-2, including structure, mechanism of action, epidemiology and genome sequencing, have provided important information about the new virus (Boopathi et al., 2020; Chan et al., 2020; Lu et al., 2020). According to the data obtained (Chan et al., 2020; Lu et al., 2020), SARS-CoV-2 belongs to a large family of coronaviruses that infect humans and other animal species, causing many widespread and serious diseases, such as Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) (De Wit et al., 2016). The SARS-CoV-2 coronavirus genome is positive-sense, single-stranded RNA and consists of ∼ 30,000 nucleotides, and its replicase gene encodes two overlapping polyproteins, pp1a and pp1ab, required for virus replication and transcription (Chen et al., 2020). These polyproteins undergo extensive proteolytic processing by two cysteine proteases, namely papain-like protease PLpro and 3-chymotrypsin-like protease 3CLpro (also known as the main protease MPro) which is essential for mediating viral replication and transcription (Anand et al., 2002; Yang et al., 2003). The main protease digests polyprotein at no less than 11 conserved sites, starting with the autolytic cleavage of this enzyme itself from pp1a and pp1ab (Hegyi & Ziebuhr, 2002). This indicates the extremely important role of MPro in the life virus cycle and makes this enzyme one of the most attractive targets for the development of effective antiviral drugs (Pillaiyar et al., 2016).
In the newest studies, SARS-CoV-2 MPro has been used as a target for screening clinically approved drugs as potential virus inhibitors in the hope of identifying drugs that are effective against COVID-19 (e.g. Adeoye et al., 2020; Babadaei et al., 2020; Enmozhi et al., 2020; Hendaus, 2020; Khan et al., 2020; Liu & Wang, 2020; Muralidharan et al., 2020; Rismanbaf, 2020; Yan et al., 2020; Zhijian et al., 2020; Zhou et al., 2020). Since the safety profiles of these drugs are well-documented, such an approach combining the structural design of drugs with virtual screening and molecular modeling methods can significantly facilitate and accelerate the detection of antiviral compounds with clinical potential in order to re-profile them for the treatment of patients infected with a new type of coronavirus. However, taking into account SARS-CoV-2 mutations (Forster et al., 2020; Khailany et al., 2020; Pachetti et al., 2020; Yao et al., 2020), studies on the development of novel antiviral compounds capable of blocking the functionally important sites of the viral proteins are also extremely significant.
Determination of the high-resolution X-ray structures of SARS-CoV-2 main protease (MPro) in ligand-bound and unbound states (Berman et al., 2000; http://www.rcsb.org/pdb/) laid the foundation not only for understanding the function and molecular mechanism of the enzyme action, but also for developing novel effective SARS-CoV-2 inhibitors by direct methods of computer-aided drug design (e.g. Bhardwaj et al., 2020; Fischer et al., 2020; Gupta et al., 2020; Islam et al., 2020; Jin et al., 2020; Joshi et al., 2020; Khan et al., 2020; Olubiyi et al., 2020; Pant et al., 2020; Sarma et al., 2020; Ton et al., 2020; Vega-Valdez et al., 2020; Wahedi et al., 2020; Zhang et al., 2020). In particular, SARS-CoV-2 MPro structure in the complex with the high affinity ligand X77 that is potent non-covalent inhibitor of SARS-COV-2 MPro was recently deposited in the Protein Data Bank (PDB ID: 6W63, http://www.rcsb.org/pdb/).
In this study, an integrated computational approach to in silico drug discovery was carried out to discover small drug-like compounds able to show structural and functional mimicry of the inhibitor X77. This computer-based approach included i) generation of pharmacophore model representing 3 D-arrangements of chemical functionalities that make X77 active towards the active site of SARS-CoV-2 MPro, (ii) shape/pharmacophore-based identification of the X77-mimetic candidates by a web-oriented virtual screening platform Pharmit (http://pharmit.csb.pitt.edu) allowing one to search for small molecules based on their structural and chemical similarity to another small molecule (Sunseri & Koes, 2016), iii) identification of compounds satisfying the Lipinski's “rule of five” (Lipinski et al., 2001) that recognizes molecules with drug-like properties, iv) molecular docking of these drug-like compounds with the enzyme active site, v) prediction of the interaction modes dominating the binding; vi) calculation of the values of binding free energy and dissociation constant (Kd) for the docking ligand − MPro models, vii) optimization of these models using the semiempirical quantum chemical method PM7 (Stewart, 2013), viii) molecular dynamics (MD) simulations of the identified compounds bound to MPro, ix) calculation of the values of binding energy for the PM7-based and dynamic ligand − MPro models, and x) selection of molecules most promising for biochemical assays.
As a result, an ensemble of hit compounds that bind to the active site of SARS-CoV-2 MPro and specifically interact with the functionally important residues of the enzyme was identified. These compounds are suggested to form good scaffolds for the development of novel, potent and broad drugs against COVID-19.
Methods and materials
Virtual screening
The Pharmit server software (Sunseri & Koes, 2016; http://pharmit.csb.pitt.edu) was used to generate the X77 pharmacophore model based on the X77 − MPro complex in crystal (PDB ID: 6w63; https://www.rcsb. org). This model (Table 1) was applied for virtual screening of small-molecule compounds able to block the X77-binding site of SARS-CoV-2 MPro. Virtual screening was performed in the 9 Pharmit molecular libraries containing over 213.5 million chemical structures (Sunseri & Koes, 2016; http://pharmit.csb.pitt.edu), resulting in a set of compounds that satisfied the X77 pharmacophore model (Table 1) and Lipinski's “rule of five” (Lipinski et al., 2001). These molecules were further screened by molecular docking and quantum chemical calculations to evaluate the affinity of their binding to SARS-CoV-2 MPro and identify molecules most promising for biochemical assays.
Table 1.
Pharmacophore model of X77 used for virtual screening of the Pharmit chemical databases.
| Pharmacophore type | Pharmacophore
coordinates X, Y, Z (Å) |
Pharmacophore radius (Å) | ||
|---|---|---|---|---|
| X | Y | Z | ||
| Aromatic group | −20.75 | 17.39 | −28.53 | R = 1.1 |
| Aromatic group | −20.55 | 20.33 | −31.86 | R = 1.1 |
| H-bond acceptor | −16.19 | 21.86 | −26.88 | R = 0.5 |
| H-bond acceptor | −20.84 | 19.52 | −32.66 | R = 0.5 |
| H-bond acceptor | −19.75 | 22.16 | −29.14 | R = 0.5 |
| H-bond acceptor | −18.66 | 18.65 | −25.94 | R = 0.5 |
| Hydrophobic group | −20.55 | 20.33 | −31.86 | R = 1.0 |
Molecular docking
Molecular docking of the predicted compounds with SARS-CoV-2 MPro was carried out by the QuickVina 2 program (Alhossary et al., 2015) in the approximation of rigid receptor and flexible ligands. The X77 inhibitor (Figure 1) was used in the calculations as a positive control. The 3 D structure of this compound was isolated from the crystal X77 − MPro complex (the PDB ID: 6W63; http://www.rcsb.org/pdb/). The SARS-CoV-2 MPro and ligand structures were prepared by adding hydrogen atoms with the Open Babel software (http://openbabel.org/wiki/Main_Page) followed by their optimization in the UFF force field (Rappe et al., 1992). The ligands were docked to the crystal SARS-CoV-2 MPro structure using QuickVina 2 (Alhossary et al., 2015). The grid box included the X77-binding site of SARS-CoV-2 MPro and had the following parameters: ΔX = 19 Å, ΔY = 21 Å, ΔZ = 23 Å centered at X = −20 Å, Y = 19 Å, Z = −26 Å; that is, the box volume was 19 × 21 × 23 = 9177 Å3. The value of “exhaustiveness” parameter defining number of individual sampling “runs” was set to 1000 (Alhossary et al., 2015).
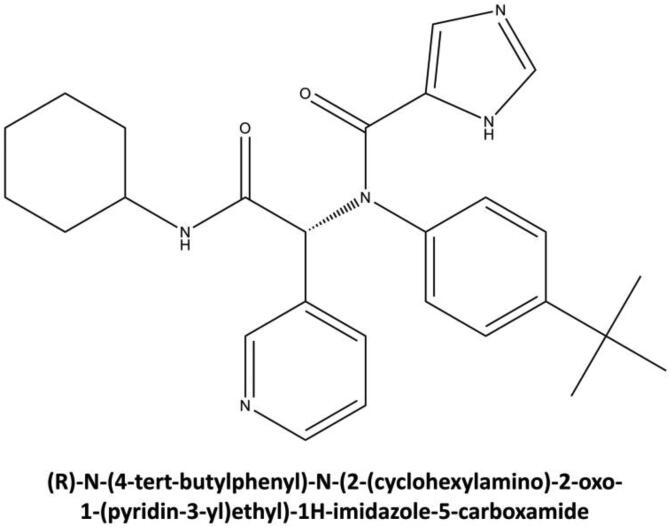
Figure 1.
Chemical structure of X77, potent non-covalent inhibitor of SARS-CoV-2 MPro (PDB ID: 6W63, http://www.rcsb.org/pdb/). The systematic name of this compound is given.
Quantum chemical calculations
The quantum chemical optimization of the docked ligand − MPro structures was carried out using the semiempirical quantum chemical method PM7 (Stewart, 2013) associated with the MOPAC2016 software package (http://OpenMOPAC.net). Before the calculations, the ligand − MPro complexes were supplemented with hydrogen atoms and optimized in the UFF force field (Rappe et al., 1992). For this purpose, the Open Babel program (http://openbabel.org/wiki/Main_Page) was used. The calculations were performed in the COSMO solvation model (COnductor-like Screening MOdel) approximation (Klamt, 2005; Klamt et al., 2015; Klamt & Schüürmann, 1993) in an implicit solvent with water's dielectric constant of 78.4 (http://OpenMOPAC.net). To speed up the calculations, the Localized Molecular Orbitals method (Høyvik et al., 2012; Lehtola & Jónsson, 2013) available in MOPAC in the form of the linear scaling SCF MOZYME algorithm (Stewart, 2013) was applied. The value of RMS gradient was set to 10 kcal/mol/Ǻ.
Molecular dynamics simulations
The classical dynamics of the ligand − MPro complexes in water was made with the implementation of Amber18 using the Amber ff14SB force field (Case et al., 2020). The Antechamber module was employed to set the Gasteiger atomic partial charges (Case et al., 2020). To prepare the force field parameters, the general Amber GAFF force field (Wang et al., 2004) was used. Hydrogen atoms were added to MPro by the tleap program of the AmberTools18 package (Case et al., 2020). Initially, the ligand − MPro complexes were each placed in a cubical box with periodic boundary conditions. In addition to the ligand − MPro complex, the box for the MD simulations included TIP3P water (Jorgensen et al., 1983) as an explicit solvent, Na+ and Cl− ions providing overall salt concentration of 0.10 M. After setting up the system, an energy minimization was performed using 500 steps of the steepest descent algorithm followed by 500 steps of the conjugate-gradient method. The backbone atoms of the complex assembly were then fixed by an additional harmonic potential with the force constant of 2.0 kcal/mol and the system was subject to the equilibration phase. The system equilibration was carried out in three consecutive stages: 1) the system was gradually heated from 0 K to 310 K for 1 ns in NVT ensemble using a Langevin thermostat with a collision frequency of 2.0 ps−1 (Case et al., 2020); 2) pressure equilibration was made for 1 ns at 1.0 bar in NPT ensemble using Berendsen barostat with a 2.0 ps characteristic time (Case et al., 2020); 3) the constraints on the complex assembly were removed and the system was equilibrated again at 310 K over 0.5 ns under constant volume conditions. After equilibration was achieved, the MD simulations were carried out for 50 ns in NPT ensemble at temperature T = 310 K and p = 1 bar. Bonds involving hydrogen atoms were constrained using SHAKE algorithm (Ryckaert et al., 1977) to achieve the integration time-step of 2 ps. Long-range electrostatic interactions were calculated using Particle Mesh Ewald (PME) algorithm (Essmann et al., 1995). Coulomb interactions and van der Waals interactions were truncated at 8 Å.
Analysis of interaction modes and binding affinity profile
The binding modes of the predicted compounds to SARS-CoV-2 MPro, namely hydrogen bonds, salt bridges, van der Waals contacts, and π-π interactions were identified by the BINANA program (Durrant & McCammon, 2011). The ligand poses in the docking ligand − MPro models were visualized with the program UCSF Chimera (Pettersen et al., 2004). To visualize van der Waals contacts, the program LigPlot (McDonald & Thornton, 1994) was employed. The values of Kd for the ligand − MPro structures were calculated using a neural-network-based scoring function NNScore 2.0 (Durrant & McCammon, 2011). The values of binding free energy were estimated from those of Kd by the formula ΔG = R × T × ln(Kd) (where ΔG is the binding free energy, R is the universal gas constant, T is the absolute temperature equal to 310 K) (Sharma & First, 2009).
For the PM7-based complexes, the ligand-binding affinity was estimated in terms of the values of binding enthalpy ΔH calculated as the differences between the heats of formation of the ligand − MPro complexes and heats of formation of the ligand and MPro in the unbound states (Stewart, 2013; http://OpenMOPAC.net). Quantum chemical calculations of the ligand and MPro structures in the unbound states were performed using the computational protocol described above for the docking ligand − MPro models.
In the case of dynamic ligand − MPro models, the values of binding energy were calculated with Amber18 (Case et al., 2020) using the MM/GBSA method (Genheden & Ryde, 2015; Sun et al., 2014; Xu et al., 2013). The calculations were made for 200 snapshots extracted from the final 40 ns of the MD trajectories, by keeping the snapshots every 0.2 ns. The polar solvation energies were computed in continuum solvent using Poisson-Boltzmann continuum-solvation model with ionic strength of 0.10. The non-polar terms were estimated using solvent accessible surface areas (Case et al., 2020). Analysis of the MD trajectories was performed by the CPPTRAJ module of AmberTools 18 (Case et al., 2020).
Results and discussion
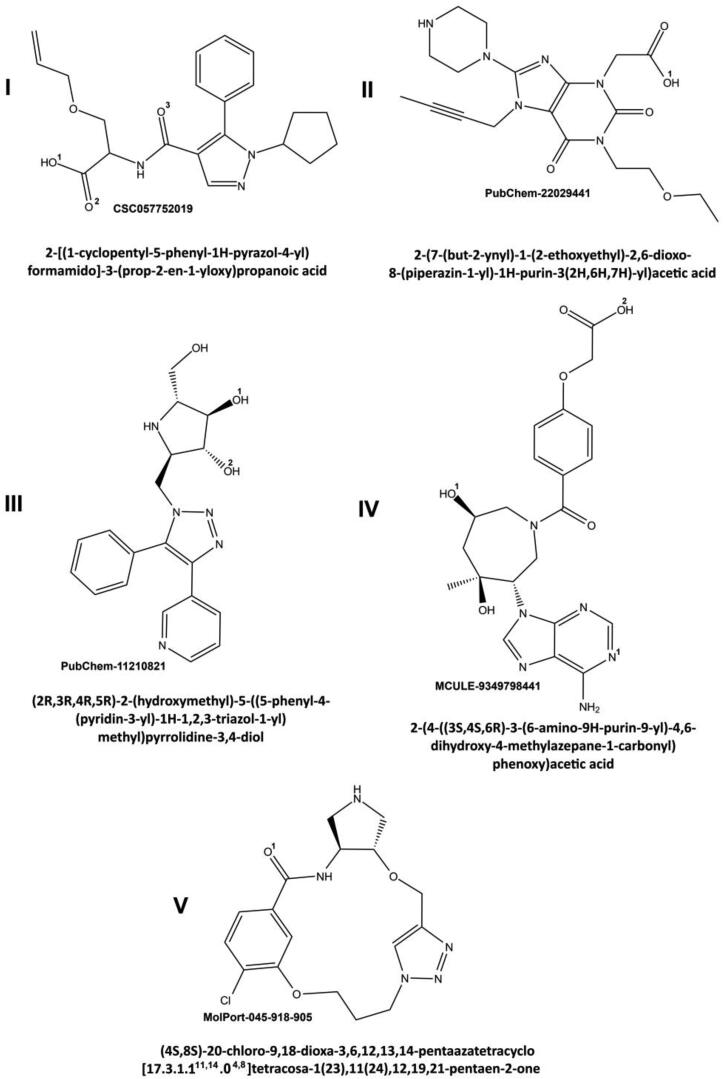
Shape/Pharmacophore-based virtual screening of the Pharmit databases resulted in 24 molecules that exhibited favorable binding energies (< −6 kcal/mol) and the values of root-mean-square deviations between the query features and the hit compound features less than 2 Å (Sunseri & Koes, 2016). Molecular docking of these molecules with the active site of MPro followed by quantum chemical calculations and MD simulations identified 5 top-ranking compounds (Figure 2) that showed a high-affinity binding in terms of Kd, binding free energy, and binding enthalpy (Table 2). This allowed one to consider these compounds as the most probable X77-mimetic candidates. Inspection of the physicochemical parameters of the predicted compounds (Table 3) providing such important characteristics for a potential drug as absorption, distribution, metabolism and excretion indicates that these molecules fully satisfy the requirements of the Lipinski's “rule of five” (Lipinski et al., 2001).
Figure 2.
Chemical structures of the potential SARS-CoV-2 MPro inhibitors. The systematic names of the compounds, as well as the corresponding databases with codes for these molecules are given. The ligand functional groups participating in the formation of intermolecular hydrogen bonds are marked by superscript numbers (see the text and Table 4).
Table 2.
Values of dissociation constant and binding energy calculated for the identified compounds and X77 bound to SARS-CoV-2 MPro.
| Ligand | I | II | III | IV | V | X77 |
|---|---|---|---|---|---|---|
| Kd1, (µM) | 0.006 | 0.039 | 0.157 | 2. 0 | 2.65 | 0.057 |
| ΔGDOC2, (kcal/mol) | −11.65 | −10.50 | −9.64 | −8.07 | −7.90 | −10.21 |
| ΔHPM73, (kcal/mol) | −80.1 | −96.6 | −90.7 | −71.4 | −53.78 | −62.8 |
| ΔGMM/GBSA4, (kcal/mol) | −27.18 ± 6.53 | −47.66 ± 4.33 | −33.56 ± 5.88 | −39.21 ± 3.83 | −35.00 ± 3.71 | −41.70 ± 4.28 |
Footnotes: 1 The values of Kd calculated for the docking ligand − MPro models; 2 The ΔG values estimated from those of Kd; 3 The values of binding enthalpy calculated for the PM7-based complexes; 4 The values of binding energy calculated for the dynamic ligand − MPro models. The averages and standard deviations corresponding to these mean values are given.
Table 3.
Physicochemical parameters of the X77-mimetic candidates associated with the Lipinski's “rule of five”.
| Ligand | Chemical formula | Molecular mass (Da) | LogP | Number of H-bond donors | Number of H-bond acceptors |
|---|---|---|---|---|---|
| I | C21H25N3O4 | 384.,00 | 2.225 | 3 | 6 |
| II | C19H26N6O5 | 419.00 | −1.206 | 3 | 9 |
| III | C19H21N5O3 | 368.00 | 0.146 | 5 | 6 |
| IV | C21H24N6O6 | 456.00 | 0.071 | 5 | 11 |
| V | C17H20ClN5O3 | 378.00 | −1.090 | 3 | 6 |
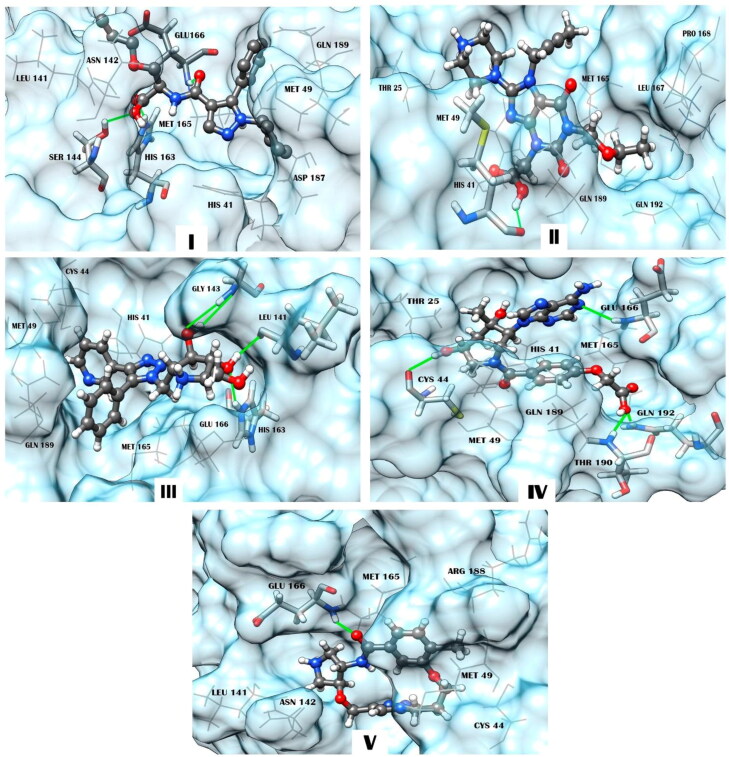
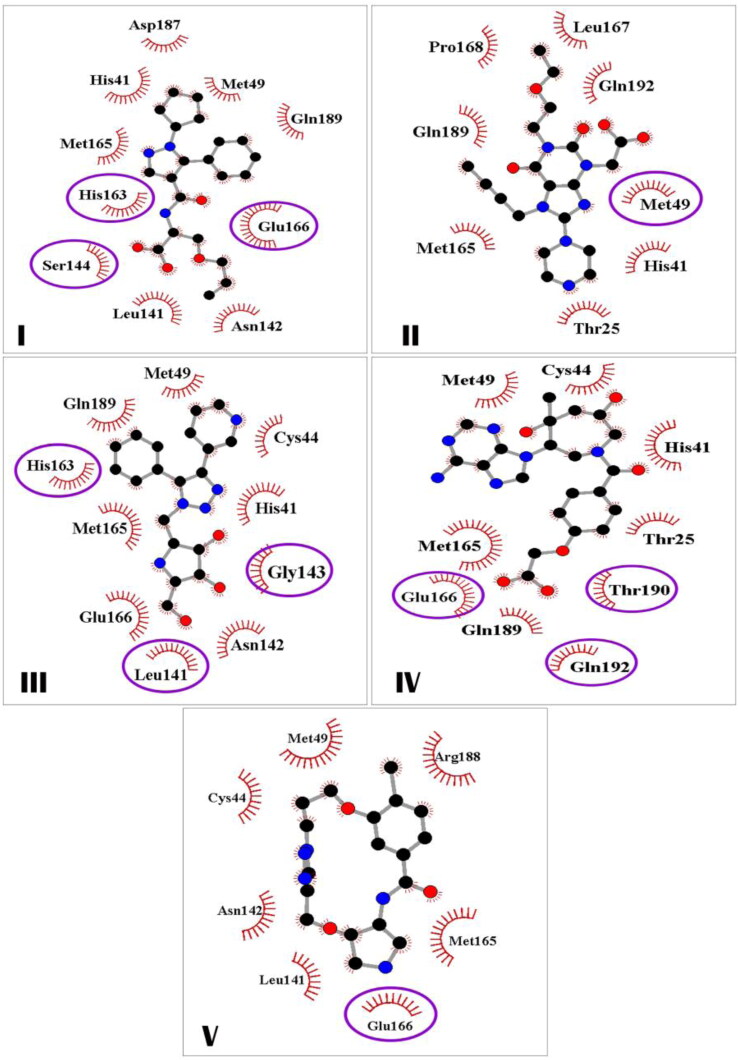
Insights into the docking ligand − MPro models (Figure 3) show that all identified X77-mimetic candidates form a wide network of intermolecular interactions involving amino acid residues of the binding pocket of MPro. In particular, compound I exhibiting the lower values of Kd and binding free energy compared to the other predicted molecules and X77 (Table 2) forms 3 hydrogen bonds with the MPro residues Ser-144, His-163 and Glu-166, a salt bridge with His-163, and 20 van der Waals contacts with the active site residues His-41, Met-49, Leu-141, Asn-142, Met-165, Glu-166, Asp-187, and Gln-189 (Table 4). In addition to these direct interatomic contacts, compound I is also involved in specific π-π interaction with His-41 which is a part of the catalytic dyad of MPro formed by this residue and Cys-145 (Chang, 2010; Qamar et al., 2020). Examination of the intermolecular interaction profile calculated for the other identified compounds indicates (Table 4) that these molecules exhibit the modes of binding to SARS-CoV-2 MPro similar to those predicted for compound I. According to the data obtained, these binding modes are provided by hydrogen bonds, van der Waals contacts, salt bridges (compounds I and III) and π-π interactions between π-conjugated systems of the ligands and the side chain of His-41 (compounds I, III and IV) (Table 4, Figure 4). Among these binding modes, intermolecular van der Waals interactions are the major contributors to the ligand − MPro interface including significant residues of the enzyme active pocket (Table 4, Figure 4).
Figure 3.
Structural complexes of compounds I, II, III, IV, and V with SARS-CoV-2 MPro generated by molecular docking. The compounds are represented by a ball-stick-ball model. The enzyme residues forming interatomic contacts with the ligands are indicated (Table 4). Residues of MPro involved in hydrogen bonding are noted using a stick model. Hydrogen bonds are shown by solid lines. A wire model is used to designate residues forming van der Waals contacts, salt bridges, and π- or T-stacking.
Table 4.
Intermolecular interactions appearing in the structural complexes of the identified compounds with SARS-CoV-2 MPro.
| Ligand | Hydrogen bond1 | Van der Waals contacts2 | Salt bridges and π-π interactions3 |
|---|---|---|---|
| I | O1…**HN[S144] O2…**HN[H163] O3…*HN[E166] | H41(3), M49(1), L141(3), N142(1), M165(6), E166(3), D187(2), Q189(1) | COO…H163 H41 (Т-stacking) |
| II | O1H…*O[M49] | T25(1), H41(2), M165(3), L167(1), P168(1), Q189(1), Q192(1) | − |
| III | O1H…*N[L141] O2H…*N[G143] O2…*NH[G143] O1…**HN[H163] | H41(4), C44(1), M49(1), M165(4), L141(2), N142(1), E166(2), Q189(5) | NCHC…E166 H41 (Т- и π-stacking) |
| IV | O1H…*O[C44] N1…*HN[E166] O2…*HN[T190] O2…**HN[Q192] | T25(2), H41(1), C44(1), M49(1), M165(3), Q189(4) | H41 (Т-stacking) |
| V | O1…*HN[E166] | C44(2), M49(1), L141(1), N142(1), M165(4), E166(1), R188(1) | − |
Footnotes: 1Atoms of the ligands are shown first, followed by the corresponding atoms of SARS-CoV-2 MPro (MPro residues are in brackets in one-letter code). Symbol * denotes the atoms of the residue main chain, and symbol ** marks the atoms of the residue side chain. 2Amino acids of SARS-CoV-2 MPro forming van der Waals contacts with the ligands. The number of contacts is given in brackets. 3For salt bridges, the functional groups of ligands are shown first, followed by the residues of SARS-CoV-2 MPro. For π- or T-stacking, residue of SARS-CoV-2 MPro involved in these interactions is shown.
Figure 4.
The SARS-CoV-2 MPro residues making direct interatomic contacts with compounds I, II, III, IV, and V. Residues involved in hydrogen bonding are marked by ellipses and highlighted in darker color.
The efficiency of the intermolecular interactions of the X77-mimetic candidates with SARS-CoV-2 MPro is supported by the low values of Kd (0.006 μM − 2.56 μM) and binding free energies (ΔG ≤ −7.9 kcal/mol), indicating their high affinity with the catalytic site of the enzyme (Table 2). Analysis of the values of Kd and binding free energy calculated for the identified compounds shows that, given the calculation errors, they are comparable with those obtained for X77 using the identical computational protocol (Table 2).
So, the data of molecular docking show that all identified compounds (Figure 2) may effectively block the key residues of the MPro catalytic site, which is confirmed by the low values of binding free energy and Kd calculated for the docking ligand − MPro models (Table 2). This is also supported by the data of quantum chemical calculations which show that, excluding compound V, the values of binding enthalpy of the analyzed molecules to MPro are lower than that predicted for the control inhibitor X77 by the same computational parameters (Table 2).
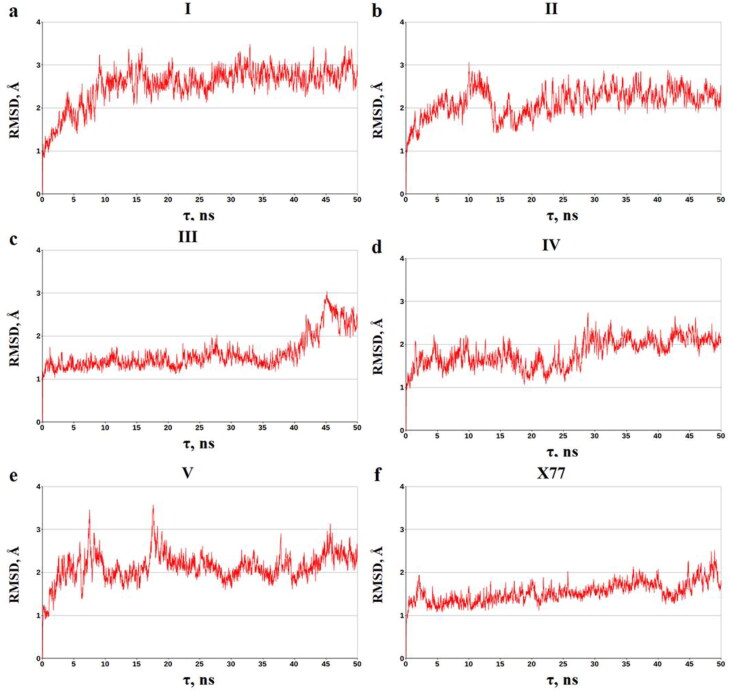
In general, the data of MD simulations are in agreement with the principal conclusions made from the analysis of the static ligand − MPro complexes. These complexes are relatively stable within the MD simulations, as evidenced by the averages of binding energies and corresponding standard deviations (Table 2). Given the MM/GBSA method errors (Genheden & Ryde, 2015; Sun et al., 2014; Xu et al., 2013), one can suggest that the dynamic ligand − MPro structures exhibit the averages of binding energy comparable with the value calculated for X77 by the identical computational protocol (Table 2). In favor of the relative stability of the dynamic ligand − MPro structures is also evidence of the data on the time dependence of the root-mean square deviations (RMSD) of the atomic positions for the dynamic and static models of the predicted compounds bound to MPro (Figure 5). Analysis of Figure 5 indicates that these complexes do not undergo significant structural rearrangements on the MD trajectories, which is confirmed by the averages of the RMSD calculated for the dynamic models of the identified molecules in the complexes with MPro. The mean values of RMSD and standard deviations, which are 2.54 ± 0.45 Å (compound I), 2.15 ± 0.33 Å (compound II), 1.59 ± 0.39 Å (compound III), 1.81 ± 0.32 Å (compound IV), and 2.13 ± 0.34 Å (compound V), are close to those of 1.54 ± 0.23 Å obtained for the SARS-CoV-2 inhibitor X77 (Figure 5).
Figure 5.
The time dependence of the RMSD (Å) calculated between all of the MD structures and the stating models of the ligand − MPro complexes. The backbone atoms of MPro were used in the calculations.
Examination of the data on the contributions of individual MPro amino-acid residues into the binding energy reveals the residues dominating the ligand − MPro interaction profile. Table 5 shows that these residues are His-41, Met-49 (excluding compound I), Cys-145, Met-165, and Gln-189. Importantly, it is those residues that are the major contributors to the X77 − MPro interaction (Table 5). Among these residues, it should first be noted the highly important His-41 and Cys-145 forming the catalytic dyad of SARS-CoV-2 Mpro (Chang, 2010; Qamar et al., 2020). These data indicate that, despite the unbound SARS-CoV-2 Mpro shows the higher mobility than a highly similar SARS-CoV Mpro (Bzówka et al., 2020), there are a number of the key anchoring residues assisting the identified compounds and X77 to effectively interact with the active site of the enzyme.
Table 5.
Averages of the binding energy for the amino-acid residues of MPro bound to the identified compounds and X77.
| Compound | ||||||
|---|---|---|---|---|---|---|
| Residue of MPro | I | II | III | IV | V | X77 |
| Residue Contribution to the Binding Energy (kcal/mol)1, 2, 3 | ||||||
| Thr-25 | − | −0.40 ± 0.18 | −0.52 ± 0.45 | −0.81 ± 0.28 | −0.47 ± 0.32 | − |
| Leu-27 | − | −0.73 ± 0.20 | −0.62 ± 0.46 | −1.04 ± 0.36 | −0.96 ± 0.29 | −0.67 ± 0.28 |
| His-41 | -0.50 ± 0.38 | -1.55 ± 0.39 | -1.50 ± 0.61 | -2.17 ± 0.68 | -2.77 ± 0.66 | -1.40 ± 0.36 |
| Val-42 | − | − | − | − | −0.47 ± 0.27 | − |
| Cys-44 | − | − | −0.59 ± 0.53 | −0.59 ± 0.33 | −0.78 ± 0.35 | − |
| Thr-45 | − | − | −0.41 ± 0.38 | − | − | − |
| Ser-46 | − | − | −0.58 ± 0.50 | −0.75 ± 0.61 | − | − |
| Met-49 | − | -2.09 ± 0.68 | -1.99 ± 0.65 | -2.24 ± 0.48 | -1.41 ± 0.63 | -0.96 ± 0.64 |
| Pro-52 | − | − | − | − | −0.52 ± 0.28 | − |
| Leu-141 | − | − | − | − | − | −0.52 ± 0.27 |
| Asn-142 | − | − | − | − | − | −0.55 ± 0.36 |
| Ser-144 | − | − | − | − | − | −0.41 ± 0.17 |
| Cys-145 | -0.48 ± 0.34 | -1.29 ± 0.31 | -0.55 ± 0.39 | -0.77 ± 0.30 | -0.74 ± 0.28 | -1.21 ± 0.34 |
| Met-165 | −1.73 ± 0.72 | -2.64 ± 0.53 | -1.01 ± 0.64 | -1.24 ± 0.44 | -0.87 ± 0.32 | -1.97 ± 0.42 |
| Glu-166 | − | −0.57 ± 0.37 | − | − | − | −0.59 ± 0.56 |
| Leu-167 | − | −0.65 ± 0.34 | − | − | − | − |
| Pro-168 | −0.67 ± 0.77 | −0.70 ± 0.35 | − | − | − | −0.87 ± 0.40 |
| Asp-187 | −0.40 ± 0.38 | −0.69 ± 0.32 | −0.60 ± 0.49 | −0.57 ± 0.46 | −0.42 ± 0.43 | − |
| Arg-188 | − | −0.60 ± 0.36 | − | − | − | − |
| Gln-189 | -1.25 ± 0.72 | -1.24 ± 0.55 | -0.74 ± 0.64 | -0.82 ± 0.50 | -0.73 ± 0.57 | -0.71 ± 0.43 |
| Ala-191 | − | −0.44 ± 0.29 | − | − | − | − |
| Ligand 4 | −19.94 ± 4.08 | −31.66 ± 2.60 | −22.83 ± 3.65 | −26.25 ± 2.26 | −23.56 ± 2.14 | −29.39 ± 2.50 |
Footnotes: 1 Data for the SARS-CoV-2 MPro residues with the binding energy ≤ -0.4 kcal/mol are presented. 2 The averages of the residue contributions to the binding energy and corresponding standard deviations are given. 3 The MPro residues dominating the ligand − MPro interaction are highlighted by bold. 4 The ligand contributions to the binding energy are presented.
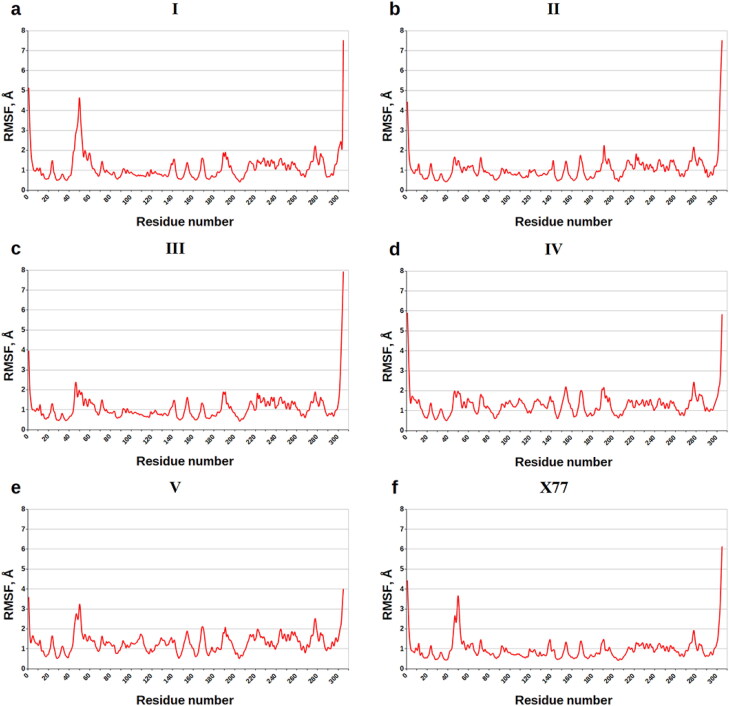
Figure 6 casts shed on the values of root-mean-square fluctuations (RMSF) of the individual residues of MPro indicating the flexibility of each amino acid during the MD simulations. Analysis of Figure 6 reveals that the majority of the MPro residues are positionally restrained on the MD trajectories, including the key anchoring residues His-41, Cys-145, Met-165, and Gln-189. The values of RMSF given for these binding hot spots in Table 6 testify to the quite small internal motions of these residues, in line with the data on their contributions into the binding energy (Table 5). The exception is Met-49 exhibiting the higher atomic fluctuations in the complexes with compound V and X77 compared with the other residues of the binding pocket of MPro (Figure 6, Table 6).
Figure 6.
Values of RMSF (Å) for each residue along the amino-acid sequence of SARS-CoV-2 MPro.
Table 6.
Values of RMSF for the MPro residues contributing to the binding energy.
| Residue of MPro | Compound |
|||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V | X77 | |
| Values of RMSF (Å) for the individual residues of MPro | ||||||
| Thr-25 | – | 0.90 | 0.98 | 0.96 | 1.20 | – |
| Leu-27 | – | 0.60 | 0.66 | 0.64 | 0.70 | 0.58 |
| His-41 | 0.72 | 0.58 | 0.69 | 0.68 | 0.85 | 0.69 |
| Val-42 | – | – | – | – | 0.94 | – |
| Cys-44 | – | – | 0.96 | 0.98 | 1.42 | – |
| Thr-45 | – | – | 1.51 | – | – | – |
| Ser-46 | – | – | 2.26 | 1.87 | – | – |
| Met-49 | – | 1.30 | 1.83 | 1.84 | 2.58 | 2.97 |
| Pro-52 | – | – | – | – | 2.00 | – |
| Leu-141 | – | – | – | – | – | 0.92 |
| Asn-142 | – | – | – | – | – | 0.95 |
| Ser-144 | – | – | – | – | – | 0.61 |
| Cys-145 | 0.62 | 0.55 | 0.58 | 0.72 | 0.66 | 0.50 |
| Met-165 | 0.70 | 0.72 | 0.64 | 0.91 | 0.90 | 0.60 |
| Glu-166 | – | 0.99 | – | – | – | 0.72 |
| Leu-167 | – | 1.29 | – | – | – | – |
| Pro-168 | 1.52 | 1.74 | – | – | – | 1.30 |
| Asp-187 | 1.00 | 0.95 | 1.06 | 1.09 | 1.02 | - |
| Arg-188 | – | 1.11 | – | – | – | – |
| Gln-189 | 1.88 | 1.35 | 1.89 | 2.05 | 1.82 | 1.30 |
| Ala-191 | – | 2.25 | – | – | – | – |
So, the findings of molecular docking, quantum chemical calculations and MD simulations suggest that the analyzed anti-SARS-CoV-2 (COVID-19) drug candidates expose the interaction modes and binding affinity profiles similar to those calculated for X77, potent, broad-spectrum inhibitor of coronavirus main protease including SARS-CoV-2 (PDB ID: 6W63, http://www.rcsb.org/pdb/).
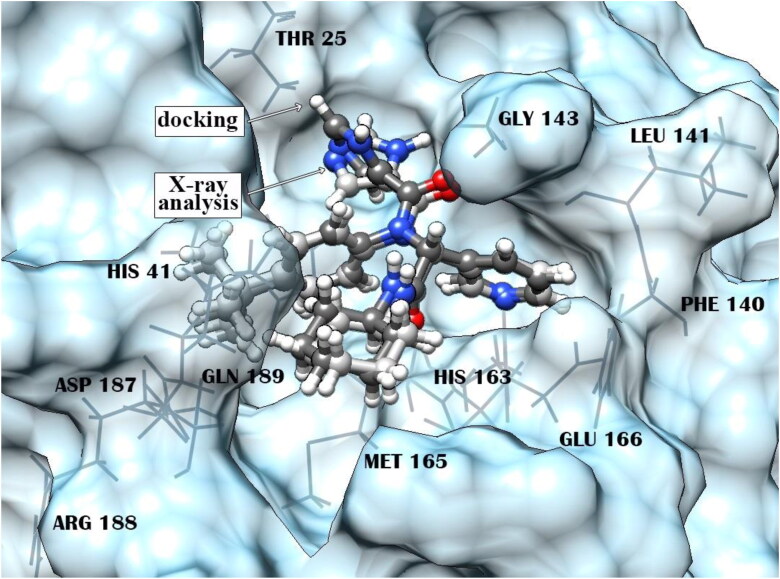
Certainly, when analyzing the obtained data, it is necessary to keep in mind that all computational approaches for modeling ligand − protein complexes and estimating the binding affinity involve various approximations. They vary from simplified forms of the first-principles equations that are easier or faster to solve, to approximations limiting the size of the system, to fundamental approximations to the underlying equations that are required to achieve any solution to them at all. Nevertheless, the findings of comparative analysis of the X77 − MPro complexes constructed by the X-ray crystallography and molecular docking (Figure 7) indicate good prediction accuracy of the computational algorithm used in the calculations, suggesting that the data obtained for the identified compounds by molecular docking, quantum chemical calculations and MD simulations adequately describe the principal geometric and energy characteristics of their complexes with SARS-CoV-2 MPro.
Figure 7.
Superposition of the X77 − MPro complexes constructed using X-ray crystallography and molecular docking. The root-mean-square deviation between the atomic coordinates of the X77 inhibitor in the calculated and experimental structures is 0.63 Å. Residues of MPro forming direct interatomic contacts with X77 are marked by a wire model.
Thus, the data on the intermolecular interaction network (Tables 4 and 5) are in line with the results of binding affinity prediction obtained for the ligand − MPro complexes using molecular docking, quantum chemical calculations and MD simulations (Table 2). From the data of Table 2, the analyzed complexes show the low values of Kd and binding energies, suggesting strong attachment of the identified X77-mimetic candidates to SARS-CoV-2 MPro. These small drug-like molecules are therefore promising candidates for further detailed experimental evaluation. However, it is clear that, despite promising in silico profile, the analyzed compounds are only starting points for the development of new highly potent drug candidates. In this connection, before biochemical assays, these compounds should go through a lead optimization, iterative process of altering the molecule structure to identify their chemical modifications with improved antiviral potency and ADMET parameters. For this purpose, the modern QSAR methods commonly used as a lead optimization approach in drug discovery may be applied (Golbraikh et al., 2017; Kuseva et al., 2019; Schultz et al., 2018).
Conclusions
Shape/Pharmacophore-based virtual screening combined with molecular docking, quantum chemical calculation and MD simulations revealed 5 top-ranking compounds that exhibited a high affinity to the catalytic site of SARS-CoV-2 MPro, allowing one to consider these small drug-like molecules as the most promising X77-mimetic candidates. Insights into the ligand − MPro models indicate that all identified compounds may specifically and effectively block the active site of SARS-CoV-2 MPro, in line with the low values of dissociation constants and binding energies. Mechanism of binding of these compounds to MPro is mainly provided by van der Waals interactions with the functionally important residues of the enzyme, such as His-41, Met-49, Cys-145, Met-165, and Gln-189 that play a role of the binding hot spots assisting the predicted molecules to effectively interact with the MPro active site.
Taken together, the data obtained show that the identified X77-mimetic candidates may serve as good scaffolds for the design of novel antiviral agents able to target the active pocket of SARS-CoV-2 MPro.
Funding Statement
This study was funded by grants from the State Program of Scientific Research “Convergence 2020” (subprogram “Consolidation”, project 3.08) and the Belarusian Republican Foundation for Fundamental Research (project X20МС-006).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Adeoye, A. O., Oso, B. J., Olaoye, I. F., Tijjani, H., & Adebayo, A. I. (2020). Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1765876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhossary, A., Handoko, S. D., Mu, Y., & Kwoh, C. K. (2015). Fast, accurate, and reliable molecular docking with QuickVina 2. Bioinformatics (Oxford, England)), 31(13), 2214–2216. 10.1093/bioinformatics/btv082 [DOI] [PubMed] [Google Scholar]
- Anand, K., Palm, G. J., Mesters, J. R., Siddell, S. G., Ziebuhr, J., & Hilgenfeld, R. (2002). Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. The EMBO Journal, 21(13), 3213–3224. 10.1093/emboj/cdf327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babadaei, M. M. N., Hasan, A., Vahdani, Y., Bloukh, S. H., Sharifi, M., Kachooei, E., Haghighat, S., & Falahati, M. (2020). Development of remdesivir repositioning as a nucleotide analog against COVID-19 RNA dependent RNA polymerase. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1767210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The protein data bank. Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Rajendran, V., Purohit, R., & Kumar, S. (2020). Identification of bioactive molecules from Tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1766572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzowka, M., Mitusińska, K., Raczyńska, A., Samol, A., Tuszyński, J. A., & Gora, A. (2020). Structural and evolutionary analysis indicate that the SARS-CoV-2 Mpro is a challenging target for small-molecule inhibitors design. International Journal of Molecular Sciences, 21(9), 3099. 10.3390/ijms21093099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D. A.,Belfon, K., Ben-Shalom, I. Y., Brozell, S. R., Cerutti, D. S., Cheatham, T. E., Cruzeiro, III, V. W. D., Darden, T. A., Duke, R.E., Giambasi, G., Gilson, M. K., Gohlke, H., Goetz, A. W., Harris, R, Izadi, S., Izmailov, S. A., Kasavajhala, K., Kovalenko, A., Krasny, R., … Kollman, P. A. (2020). AMBER 2020. University of California. [Google Scholar]
- Chan, J. F.-W., Yuan, S., Kok, K.-H., To, K. K.-W., Chu, H., Yang, J., Xing, F., Liu, J., Yip, C. C.-Y., Poon, R. W.-S., Tsoi, H.-W., Lo, S. K.-F., Chan, K.-H., Poon, V. K.-M., Chan, W.-M., Ip, J. D., Cai, J.-P., Cheng, V. C.-C., Chen, H., Hui, C. K.-M., & Yuen, K.-Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet (London, England), 395(10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, G. (2010). Quaternary structure of the SARS coronavirus main protease. In Lal, S. (Ed.), Molecular Biology of the SARS-Coronavirus, 115–128. Heidelberg, Berlin: Springer. 10.1007/978-3-642-03683-5_8 [DOI] [Google Scholar]
- Chen, Y., Liu, Q., & Guo, D. (2020). Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology, 92(4), 418–423. 10.1002/jmv.25681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit, E., van Doremalen, N., Falzarano, D., & Munster, V. J. (2016). SARS and MERS: Recent insights into emerging coronaviruses. Nature Reviews. Microbiology, 14(8), 523–534. 10.1038/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, J. D., & McCammon, J. A. (2011). BINANA: A novel algorithm for ligand-binding characterization. Journal of Molecular Graphics & Modelling, 29(6), 888–893. 10.1016/j.jmgm.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, J. D., & McCammon, J. A. (2011). NNScore 2.0: A neural-network receptor-ligand scoring function. Journal of Chemical Information and Modeling, 51(11), 2897–2903. 10.1021/ci2003889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi, S. K., Raja, K., Sebastine, I., & Joseph, J. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M. L., Darden, T., Lee, H., & Pedersen, L. G. (1995). A smooth particle mesh Ewald method. The Journal of Chemical Physics, 103(19), 8577–8593. 10.1063/1.470117 [DOI] [Google Scholar]
- Fischer, A., Sellner, M., Neranjan, S., Lill, M. A., & Smiesko, M. (2020). Inhibitors for novel coronavirus protease identified by virtual screening of 687 million compounds. ChemRxiv. 10.26434/chemrxiv.11923239.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, P., Forster, L., Renfrew, C., & Forster, M. (2020). Phylogenetic network analysis of SARS-CoV-2 genomes. Proceedings of the National Academy of Sciences, 117(17), 9241–9243. 10.1073/pnas.2004999117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden, S., & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery, 10(5), 449–461. 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbraikh, A., Wang, X. S., Zhu, H., & Tropsha, A. (2017). Predictive QSAR modeling: Methods and applications in drug discovery and chemical risk assessment. In Leszczynski J., Kaczmarek-Kedziera A., Puzyn T., Papadopoulos G. M., Reis H., Shukla K. M. (Eds.), Handbook of computational chemistry, 2303–2340. Springer. [Google Scholar]
- Gupta, M. K., Vemula, S., Donde, R., Gouda, G., Behera, L., & Vadde, R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi, A., & Ziebuhr, J. (2002). Conservation of substrate specificities among coronavirus main proteases. The Journal of General Virology, 83(Pt 3), 595–599. 10.1099/0022-1317-83-3-595 [DOI] [PubMed] [Google Scholar]
- Hendaus, M. A. (2020). Remdesivir in the treatment of Coronavirus Disease 2019 (COVID-19): A simplified summary. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1767691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyvik, I.-M., Jansik, B., & Jørgensen, P. (2012). Trust region minimization of orbital localization functions. Journal of Chemical Theory and Computation, 8(9), 3137–3146. 10.1021/ct300473g [DOI] [PubMed] [Google Scholar]
- Islam, R., Parves, R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., Hossain, M. N., Ali, M. A., & Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of MPro from 1 COVID-19 virus and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Joshi, R. S., Jagdale, S. S., Bansode, S. B., Shankar, S. S., Tellis, M. B., Pandya, V. P., Chugh, A., Giri, A. P., & Kulkarni, M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khailany, R. A., Safdar, M., & Ozaslan, M. (2020). Genomic characterization of a novel SARS-CoV-2. Gene Reports, 19, 100682. 10.1016/j.genrep.2020.100682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, S. A., Zia, K., Ashraf, S., Uddin, R., & Ul-Haq, Z. (2020). a. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Khan, R. J., Jha, R. K., Amera, G. M., Jain, M., Singh, E., Pathak, A., Singh, R. P., Muthukumaran, J., & Singh, A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klamt, A. (2005). COSMO-RS: From quantum chemistry to fluid phase thermodynamics and drug design (1st ed., p. 246.). Elsevier. [Google Scholar]
- Klamt, A., Moya, C., & Palomar, J. (2015). A comprehensive comparison of the IEFPCM and SS(V)PE continuum solvation methods with the COSMO approach. Journal of Chemical Theory and Computation, 11(9), 4220–4225. 10.1021/acs.jctc.5b00601 [DOI] [PubMed] [Google Scholar]
- Klamt, A., & Schüürmann, G. (1993). COSMO: A new approach to dielectric screening in solvents with explicit expressions for the screening energy and its gradient. Journal of the Chemical Society, Perkin Transactions, 2, 799–805. [Google Scholar]
- Kuseva, C., Schultz, T. W., Yordanova, D., Tankova, K., Kutsarova, S., Pavlov, T., Chapkanov, A., Georgiev, M., Gissi, A., Sobanski, T., & Mekenyan, O. G. (2019). The implementation of RAAF in the OECD QSAR Toolbox. Regular Toxicology & Pharmacology, 105, 51–61. 10.1016/j.yrtph.2019.03.018 [DOI] [PubMed] [Google Scholar]
- Lehtola, S., & Jónsson, H. (2013). Unitary optimization of localized molecular orbitals. Journal of Chemical Theory and Computation, 9(12), 5365–5372. 10.1021/ct400793q [DOI] [PubMed] [Google Scholar]
- Lipinski, C. A., Lombardo, F., Dominy, B. W., & Feeney, P. J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, 46(1–3), 3–26. PMID: 11259830 10.1016/s0169-409x(00)00129-0 [DOI] [PubMed] [Google Scholar]
- Liu, X., & Wang, X.-J. (2020). Potential inhibitors for 2019-nCoV coronavirus M protease from clinically approved medicines. BioRxiv. 10.1101/2020.01.29.924100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet (London, England)), 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, I. K., & Thornton, J. M. (1994). Satisfying hydrogen bonding potential in proteins. Journal of Molecular Biology, 238(5), 777–793. 10.1006/jmbi.1994.1334 [DOI] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Olubiyi, O. O., Olagunju, M., Keutmann, M., Loschwitz, J., & Strodel, B. (2020). High throughput virtual screening to discover inhibitors of the main protease of the coronavirus SARS-CoV-2. Preprint from Preprints.org. 10.20944/preprints202004.0161.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachetti, M., Marini, B., Benedetti, F., Giudici, F., Mauro, E., Storici, P., Masciovecchio, C., Angeletti, S., Ciccozzi, M., Gallo, R. C., Zella, D., & Ippodrino, R. (2020). Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. Journal of Translational Medicine, 18(1), 179. 10.1186/s12967-020-02344-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant, S., Singh, M., Ravichandiran, V., Murty, U. S. N., & Srivastava, H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure &Dynamics. https://doi.org/doi: 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pillaiyar, T., Manickam, M., Namasivayam, V., Hayashi, Y., & Jung, S. H. (2016). An overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule chemotherapy. Journal of Medicinal Chemistry, 59(14), 6595–6628. 10.1021/acs.jmedchem.5b01461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar, M. T., Alqahtani, S. M., Alamri, M. A., & Chen, L.-L. (2020). Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. Journal of Pharmaceutical Analysis. 10.1016/j.jpha.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappe, A. K., Casewit, C. J., Colwell, K. S., Goddard, W. A., III., & Skiff, W. M. (1992). UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. Journal of the American Chemical Society, 114(25), 10024–10035. 10.1021/ja00051a040 [DOI] [Google Scholar]
- Rismanbaf, A. (2020). Potential treatments for COVID-19; a narrative literature review. Archives of Academic Emergency Medicine, 8(1), e29. [PMC free article] [PubMed] [Google Scholar]
- Ryckaert, J. P., Ciccotti, G., & Berendsen, H. J. C. (1977). Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. Journal of Computational Physics, 23(3), 327–341. 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- Sarma, P., Sekhar, N., Prajapat, M., Avti, P., Kaur, H., Kumar, S., Singh, S., Kumar, H., Prakash, A., Dhibar, D. P., & Medhi, B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, T. W., Diderich, R., Kuseva, C. D., & Mekenyan, O. G. (2018). The OECD QSAR Toolbox starts its second decade. Methods in Molecular Biology (Clifton, N.J.).), 1800, 55–77. 10.1007/978-1-4939-7899-1_2 [DOI] [PubMed] [Google Scholar]
- Sharma, G., & First, E. A. (2009). Thermodynamic analysis reveals a temperature-dependent change in the catalytic mechanism of bacillus stearothermophilus tyrosyl-tRNA synthetase. The Journal of Biological Chemistry, 284(7), 4179–4190. 10.1074/jbc.M808500200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, J. J. P. (2013). Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters. Journal of Molecular Modeling, 19(1), 1–32. 10.1007/s00894-012-1667-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., Li, Y., Tian, S., Xu, L., & Hou, T. (2014). Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Physical Chemistry Chemical Physics : Pccp, 16(31), 16719–16729. − 10.1039/c4cp01388c [DOI] [PubMed] [Google Scholar]
- Sunseri, J., & Koes, D. R. (2016). Pharmit: Interactive exploration of chemical space. Nucleic Acids Research, 44(W1), W442–W448. 10.1093/nar/gkw287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, A.-T., Gentile, F., Hsing, M., Ban, F., & Cherkasov, A. (2020). Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Molecular Informatics. 10.1002/minf.202000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega-Valdez, I. R., Santiago-Quintana, J. M., Rosalez, M. N., Farfan-Garcia, E. D., & Soriano-Ursua, M. A. (2020). Theoretical evaluation of bortezomib and other boron-containing compounds as inhibitors of SARS-CoV-2 main protease. ChemRxiv. https://doi.org/ 10.26434/chemrxiv.12047346.v1 [DOI] [Google Scholar]
- Wahedi, H. M., Ahmad, S., & Abbasi, S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure & Dynamics. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., & Case, D. A. (2004). Development and testing of a general Amber force field. Journal of Computational Chemistry, 25(9), 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- Xu, L., Sun, H., Li, Y., Wang, J., & Hou, T. (2013). Assessing the performance of MM/PBSA and MM/GBSA methods. 3. The impact of force fields and ligand charge models. The Journal of Physical Chemistry. B, 117(28), 8408–8421. 10.1021/jp404160y [DOI] [PubMed] [Google Scholar]
- Yan, L., Zhang, J., Wang, N., Li, H., Shi, Y., Guo, … Zou, Q. (2020). Therapeutic drugs targeting 2019-nCoV main protease by high-throughput screening. BioRxiv. https://doi.org/ 10.1101/2020.01.28.922922 [DOI] [Google Scholar]
- Yang, H., Yang, M., Ding, Y., Liu, Y., Lou, Z., Zhou, Z., Sun, L., Mo, L., Ye, S., Pang, H., Gao, G. F., Anand, K., Bartlam, M., Hilgenfeld, R., & Rao, Z. (2003). The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proceedings of the National Academy of Sciences of the United States of America, 100(23), 13190–13195. 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H., Lu, X., Chen, O., Xu, K., Chen, Y., Cheng, L., & Li, L. (2020). Patient-derived mutations impact pathogenicity of SARS-CoV-2. MedRxiv. https://doi.org/ 10.1101/2020.04.14.20060160 [DOI] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.).), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhijian, X., Peng, C., Shi, Y., Zhu, Z., Mu, K., Wang, X., & Zhu, W. (2020). Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv. https://doi.org/ 10.1101/2020.01.27.921627 [DOI] [Google Scholar]
- Zhou, Y., Hou, Y., Shen, J., Huang, Y., Martin, W., & Cheng, F. (2020). Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery, 6(1), 14. 10.1038/s41421-020-0153-3 [DOI] [PMC free article] [PubMed] [Google Scholar]