Abstract
The recent outbreak of the coronavirus disease COVID-19 is putting the world towards a great threat. A recent study revealed COVID-19 main protease (Mpro) is responsible for the proteolytic mutation of this virus and is essential for its life cycle. Thus inhibition of this protease will eventually lead to the destruction of this virus. In-Silico Molecular docking was performed with the Native ligand and the 15 flavonoid based phytochemicals of Calendula officinals to check their binding affinity towards the COVID-19 main protease. Finally, the top 3 compounds with the highest affinity have been chosen for molecular dynamics simulation to analyses their dynamic properties and conformational flexibility or stability. In-Silico Docking showed that major phytochemicals of Calendula officinals i.e. rutin, isorhamnetin-3-O-β-D, calendoflaside, narcissin, calendulaglycoside B, calenduloside, calendoflavoside have better binding energy than the native ligand (inhibitor N3). MD simulation of 100 ns revealed that all the protease-ligand docked complexes are overall stable as compare to Mpro-native ligand (inhibitor N3) complex. Overall, rutin and caledoflaside showed better stability, compactness, and flexibility. Our in silico (Virtual molecular docking and Molecular dynamics simulation) studies pointed out that flavonoid based phytochemicals of calendula (rutin, isorhamnetin-3-O-β-D, calendoflaside) may be highly effective for inhibiting Mpro which is the main protease for SARS-CoV-2 causing the deadly disease COVID-19. Rutin is already used as a drug and the other two compounds can be made available for future use. Thus the study points a way to combat COVID-19 by the use of major flavonoid based phytochemicals of Calendula officinals.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19 Mpro, SARS-CoV-2, calendula officinals, molecular docking, molecular simulation
Introduction
The earliest history of human Corona Viruses can be traced back to 1965 when two scientists Tyrrell and Bynoe discovered that they could passage virus stain named B814 (Tyrrell & Bynoe, 1966). With the onset of 2003, at least five new types of human coronaviruses came into identification, causing huge harm to mankind. Among all of them, the severe acute respiratory syndrome (SARS) coronavirus could be identified as the most harmful till the end of 2019 causing significant morbidity and mortality (Khan et al., 2020). Some new groups of coronaviruses like the New Haven coronavirus (HCoV-NH*) and NL & NL63 were also identified globally during the year 2004–2005(Kahn & McIntosh, 2005).
The end of 2019 marked the emergence of a deadly and highly contagious new coronavirus, the 2019-nCoV which now officially termed as the SARS-CoV-2 or the “Novel Coronavirus” (Mittal et al., 2020). There has been a rapid outbreak of a pneumonia-like illness mainly originating from Wuhan City, capital of Hubei province, China which was later coined as COVID-19(Al-Khafaji et al., 2020). Community transmission was soon confirmed in Guangdong Province in China (Chen et al., 2020). The World Health Organisation (WHO) has declared “Public Health Emergency of International Concern” on 30 January 2020 and with the outspread of this disease in over 100 countries they declared this as pandemic for the first time in 21st century and urged all countries around the globe to consider it as a global public health emergency. With Lockdown in most of the countries, the most feasible and possible way to carry out research and serve the purpose of helping mankind is the in silico approach which gives us an overview and hope for new drug discovery effective against such deadly virus.
The Genome or genetic material of the virus composed of a single RNA strand which is identified as the largest among all the RNA viruses reported so far (Elfiky, 2020). Once the virus infects the cell, its genome act as a messenger RNA (mRNA) and initiates the synthesis of two long polyproteins (pp1a, pp1ab). These synthesized long polyproteins helps the virus to replicate new viruses (Boopathi et al., 2020). These polyproteins in terms make several RNAs, many more structural proteins that together contribute to the construction of a new virus and two proteases(Sarma et al., 2020). These protease hence formed are further used for cutting the polyproteins into all essential functional slices (Cui et al., 2019; St. John et al., 2015). The SARS-CoV-2 is recognized as β-coronavirus and it contains about 29.9 kb genome sequence (Wu et al., 2020). The angiotensin-converting enzyme 2 (ACE2) receptors are the major human receptors that are used by this virus to infect the cells (Lu et al., 2020; Nejadi Babadaei et al., 2020). It is on January 7, 2020, that Chinese scientists attempted to isolate SARS-CoV-2 and finally sequenced the genome SARS-CoV2. He also pointed out a potential drug target protein for the inhibition of SARS-CoV-2 replication (Figure 1). The COVID-19 Main protease (Mpro) is the major protease that plays a key role in the proteolytic mutation of the virus (Figure S1) (Jin et al., 2020a). Thus blocking the viral polypeptide cleavage would provide us a new direction towards effective treatment of this deadly disease (Gupta et al., 2020). It is urgent to identify effective and active antiviral compounds that can block the infection at the individual stage and stop its spread worldwide.
Figure 1.
The crystal structure of COVID-19 main protease in complex with an inhibitor N3.
India is always considered as the global hub for herbal medicines. More than 5000 natural plants and herbs are used for medicinal purposes (Agarwal & Raju, 2006) worldwide and the majority of them are from India. The Calendula officinals is a noted medicinal plant and used all over the world including Europe, Africa, and India (Bhatnagar & Sastri, 1960). Calendula officinals or commonly known as marigold contain diverse phytochemicals or bio-active components hence they are used in various therapies starting wound healing to ulcers (Muley et al., 2009). It is also considered to be a highly antiseptic and anti-bacterial agent and often used as antiseptic cream (Chakraborthy, 2010). Calendula officinals is also used in homeopathy to treat wounds and pain. Different in vitro studies have also pointed out that Calendula extracts have antiviral, antimicrobial, and anti-genotoxic properties (AshwlayanVD & Verma, 2018). Considering all the biological activities of Calendula officinals we have tried to screen the bioactive compounds from Calendula officinals against COVID-19 Mpro.
The main goal of our in silico studies (Virtual molecular docking and Molecular dynamics simulation) is to identify bioactive compounds from Calendula officinals which can inhibit COVID-19 Mpro effectively and hence providing a way-out to combat such deadly disease. This study perhaps first time giving an idea of how calendula can be used in blocking the actions of a deadly virus which is putting the existence of human beings towards a big threat. Further study is indeed required to validate its action on SARS-CoV-2 which is responsible for the spread of the deadly disease “COVID-19”, but an initial footstep is what this work would like to provide. A medicine can be made easily available with the continuation of the work.
Methodology
Receptor and ligand preparation
The crystal structure of a COVID-19 main protease (Mpro), vital for virus replication has been download from the protein data bank (PDB) website (http://www.rcsb.org/pdb) (PDB ID:6LU7: Resolution 2.16 Å) (Jin et al., 2020b). Major Phytochemicals (majorly flavonoids) of Calendula officinals identified earlier (Muley et al., 2009) were acquired from https://pubchem.ncbi.nlm.nih.gov/ as SDF format. Later on, the SDF format of ligand was converted into mol2 format (Figure S2) using Discovery Studio Visualizer 2017 R2. Prior to the docking, receptor (Covid-19 Mpro) and ligands have been prepared by the Dock Prep tool of UCSF Chimera. Briefly, the receptor and ligands have been optimized firstly by adding hydrogen to the system. Next charges have been assigned, to associate atoms with partial charges and other force field parameters. AMBER ff14SB force field had been selected for all standard residue and Gasteiger force field for all other residues.
Figure 2.
Molecular Re-docking of COVID-19 main protease in complex with an inhibitor N3.
Active site selection
The active site of the Covid-19 Mpro Protein (PDB ID: 6LU7) has been predicted from different receptor cavities available. Site 1 with coordinates (X= −13.669, Y = 15.134, Z = 73.529) has been chosen as the active site as the native inhibitor ligand binds to that site.
Virtual molecular docking
Virtual molecular docking and analysis have been performed according to the method described previously (Majumder et al., 2019). All the small molecules have been docked through autodock vina to active site 1 (X=-13.669, Y = 15.134, Z = 73.529) of Covid-19 Mpro. Re-docking has been also performed to validate our virtual molecular docking protocol.
Molecular dynamics simulation
MD simulation has been performed to analyze the dynamic properties and conformational flexibility or stability of apo-Mpro (COVID-19 Mpro) and selected docked ligand into the target site. GROMACS. 2019.4 package has been used to run and analyze 100 ns MD simulation. GROMOS96 43a1 in single point charge (SPC) water models and PRODRG server have been used to generate protease and ligand force field and parameter files respectively (Berendsen et al., 1995), (van Aalten et al., 1996). Each system has been solvated with water molecule (apo-Mpro: 19388, Mpro-rutin: 19341, Mpro-isorhamnetin-3-O-ß-D: 19374, Mpro-calendoflaside: 19356) in separated dodecahedron boxes (x = 9.631, y = 9.631, z = 6.810) with at least 10 Å distance from the edge of the box. Each system has been neutralized by adding 4 NA+ counter ions. The energy minimization of all four systems has been performed through running the steepest descent minimization algorithm with 5000 steps to achieve stable system with maximum force < 1000 kJ mol−1 nm−1. Prior to the running of real dynamics, the solvent and ions of all the systems have been equilibrated by NVT and NPT ensemble. First, all the systems have been run under an NVT ensemble (constant under number of particles, volume, and temperature) where the systems have been gradually heated from 0 to 300 K and equilibrated for 100 ps. Then all the systems have been run under NPT ensemble (constant number of particles, pressure, and temperature) and equilibrated at 300 K for 100 ps. Here, the backbone Cα atoms have been restrained and all the solvent molecules (water and ions) have been allowed to move freely to achieve the solvent equilibrium in the system. The h-bonds of the system have been constrained. The long range electrostatic interactions have been achieved through the particle mesh Ewald method. The systems temperature (300 K) has been controlled through the V-rescale method. Parrinello-Rahman method has been applied to equilibrate and set the pressure (1 bar), density, and total energy of systems. After that, four well-equilibrate systems have been carried out for production run without any restrain for 100 ns with a time step of 2 fs, and after every 10 ps coordinates of the structure have been saved. After the completion of the production run, MD simulation trajectories have been used for various dynamics analysis such as root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), number of hydrogen bonds, columbic interaction energy, principle component analysis (PCA), and secondary structural analysis by different inbuilt scripts of GROMACS. Also, the MM-PBSA binding free energy of receptor-ligand docked complexes have been estimated by using the Molecular Mechanics/Poisson–Boltzmann Surface Area (MMPBSA) method which utilized the MD simulation trajectories and is one of the best-used methods in this kind of analysis (Kumari et al., 2014).
Drug likeness property
The drug-likeness property of the finally selected phytochemicals has been predicted to check their pharmacological property and their likeness to be a standard drug. Drug Likness tool (DruLiTo) is an open-access software by the Department of Pharmacoinformatics, NIPER (INDIA) was used for the analysis.
Result and discussion
Virtual molecular docking
Initially, re-docking of the endogenous or native co-crystal ligand has been performed for the validation of the whole docking procedure ensuring its reproducibility. The re-docking of the native ligand has shown that it binds to the same site of the Covid-19 main protease (Mpro) as the co-crystal ligand binds (Figure 3) and the root mean square deviation (RMSD) value between the co-crystal ligand and re-docked native ligand position is 1.162 Å. RMSD where RMSD values below 2 Å is considered to be successful. Re-docking results depicted that all the major interaction between amino acid residues and co-crystal ligand resembles to the interaction between amino acid residues and the re-docked ligand. The binding energy of the re-docked ligand at the active site of Mpro is −7.7 Kcal/mol. The major amino acid interaction includes: Phe140, His164, Asn142, Gly143, Cys145, His163, Met165, Glu, 166, Gln189, Arg188, His41, Thr26, Thr25, Ser144, Leu141, Met49, Ser146, Asp187, Leu27 (Figure 2). Among this 19 amino acid residue 15 matches with that of the co-crystal ligand. Thus we can consider the re-docking to be successful.
Figure 3.
(a) Rutin in Interaction with COVID -19 Mpro; (b) Isorhamnetin-3-O-β-D, Interaction with COVID -19 Mpro; (c) Calendoflaside in Interaction with COVID -19 Mpro (d) Narcissin in Interaction with COVID -19 Mpro (e) Calendulaglycoside B in Interaction with COVID -19 Mpro (f) Calenduloside in Interaction with COVID -19 Mpro.
A total of 15 major phytochemicals of Calendula officinals have been docked to Mpro (COVID-19 main protease). The receptor-ligand interactions have indicated that all the compounds used for molecular docking have a substantial binding affinity towards the receptor as compared to the native ligand. The detailed list is depicted in Table 1.
Table 1.
Results of the docking of all 15 compounds and native ligand on the crystal structure of COVID-19 main protease.
| Sl. No. | Rank | NAME | Score | Kd(μM) | H Bond All | H Bond Ligand Atom | H Bond Receptor Atom |
|---|---|---|---|---|---|---|---|
| 1. | 1 | Rutin | −8.8 | 0.336 | 4 | 4 | 4 |
| 2. | 2 | Isorhamnetin-3-O-β-D | −8.7 | 0.398 | 2 | 2 | 2 |
| 3. | 3 | Calendoflaside | −8.5 | 0.558 | 3 | 3 | 3 |
| 4. | 4 | Narcissin | −8.4 | 0.661 | 2 | 2 | 2 |
| 5. | 5 | Calendulaglycoside B | −8.2 | 0.928 | 4 | 4 | 4 |
| 6. | 6 | Calenduloside | −7.9 | 1.542 | 6 | 6 | 5 |
| 7. | 7 | Calendoflavoside | −7.7 | 2.164 | 4 | 4 | 4 |
| 8. | - | Inhibitor N3 | −7.7 | 2.164 | 6 | 6 | 6 |
| 9. | 8 | Calenduloside A | −7.6 | 2.564 | 2 | 2 | 2 |
| 10 | 9 | Quercitrin | −7.4 | 3.598 | 1 | 1 | 1 |
| 11 | 10 | Isoquercitrin | −7.4 | 3.598 | 1 | 1 | 1 |
| 12 | 11 | Isorhamnetin | −7.3 | 4.262 | 2 | 2 | 2 |
| 13 | 12 | Calendulaglycoside A | −7.0 | 7.084 | 8 | 8 | 8 |
| 14 | 13 | Lupeol | −6.2 | 27.468 | 1 | 1 | 1 |
| 15 | 14 | Phylloquinone | −5.5 | 89.909 | 1 | 1 | 1 |
| 16 | 15 | Plastoquinone | −5.4 | 106.505 | 1 | 1 | 1 |
The compounds having binding energy greater than the native ligand (-7.7 Kcal/mol) have been taken for further analysis.
Six compounds have been taken into consideration for further analysis as reflected in Table 1. Rutin, Isorhamnetin-3-O-β-D, Calendoflaside, Narcissin, Calendulaglycoside B, Calenduloside have shown better binding energy than that of the native ligand (Table 1). The detailed interactions with the different amino acid residues of Mpro have been depicted in Table 2 and also the common interactions of native ligand observed with other ligands are highlighted in Bold front.
Table 2.
Ligands interaction with different amino acid residues of the target site.
| NAME | SCORE | AMINO ACID INTERACTION |
|---|---|---|
| Native Ligand (Inhibitor N3) | −7.7 | Phe140, His164, Asn142, Gly143, Cys145, His163, Met165, Glu166, Gln189, Arg188, His41, Thr26, Thr25, Ser144, Leu141, Met49, Ser146, Asp187, Leu27 |
| Rutin | −8.8 | Ser144, His163, Asn142, Cys145, Gly143, Leu27, His41, Phe140, Thr25, Thr26, Thr190, Arg188, Met165, Glu166, His164, Leu141, Gln189 |
| Isorhamnetin-3-O-β-D | −8.7 | Cys145, Gly143, Asn142, Ser144, His163, His172, Phe140, Glu186, Gln189, Asp187, Arg188, Tyr54, Met165, His41, Thr26, Met49 |
| Calendoflaside | −8.5 | Arg188, Asp187, Met165, His163, Ser144, Glu166, Phe140, Leu141, Cys145, Gly143, Asn142, Leu27, Met49, Gln189, His41, Tyr54 |
| Narcissin | −8.4 | Glu186, Leu141, Asn142, Cys145, Gly143, Leu27, Met49, Tyr54, Asp187, Met165, His41, Gln189, Arg188 |
| Calendulaglycoside B | −8.2 | Phe140, Ser144, His163, Glu166, Ala191, Thr190, Gln189, Arg188, Asp187, Leu141, His41, Met165, Gly143, Cys145, Asn142, His164 |
| Calenduloside | −7.9 | Cys44,Thr45,Thr25,Asn142,Cys145,His164,Gln189,Glu166, Met165, Pro168, Gly143, Leu27, Thr26, Met49, Thr24 |
Rutin has shown the best binding affinity towards the receptor with a binding energy of −8.8 Kcal/mol and Kd value of 0.336 μM. The interaction is depicted in Figure 3(a). Docking study has exhibited that Rutin interacts with 16 amino acid residue among which 15 residues (Ser144, His163, Asn142, Cys145, Gly143, His41, Phe140, Thr25, Thr26, Thr190, Arg188, Met165, Glu166, His164, Leu141, Gln189) matches with that to the inhibitor N3. Thus it can be predicted that Rutin binds to the entire amino acid residue needed for proper inhibition of receptor protein (Mpro). Rutin is not only one of the phytochemicals for marigold but many natural products including oranges, black tea, asparagus, buckwheat, onions, green tea, figs, and most citrus fruit. Thus sources of rutin are quite easily available.
Isorhamnetin-3-O-β-D another important phytochemical of Calendula officinals has shown a binding energy score of −8.7 Kcal/mol and Kd value of 0.398 μM which in terms of the native ligand is quite high. The interaction has been depicted in Figure 3(b). Docking study has been exhibited that Isorhamnetin-3-O-β-D also interacted with 16 amino acid residue among which 13 residues (Cys145, Gly143, Asn142, Ser144, His163, Phe140, Gln189, Asp187, Arg188, Met165, His41, Thr26, Met49) matches with that to the inhibitor N3. Hence we can predict that Isorhamnetin-3-O-β-D also binds to all major amino acid residue which helps in inhibiting the receptor protein.
Calendoflaside has shown a significant binding affinity towards the target receptor with a binding energy of −8.5 Kcal/mol and Kd value of 0.558 μM. The detailed interaction has been depicted in Figure 3(c). Docking study has shown that Calendoflaside also interacted with 16 amino acid residue among which 15 (Arg188, Asp187, Met165, His163, Ser144, Glu166, Phe140, Leu141, Cys145, Gly143, Asn142, Leu27, Met49, Gln189, His41) coincides with that of the native ligand which gives us a clear idea that Calendoflaside also binds to major amino acid residue responsible for inhibition of COVID-19 main protease (Mpro).
Narcissin a major flavonoid present in many natural products. Docking study with Narcissin has shown that it also exhibits a comparatively high binding affinity towards the receptor with a binding energy of −8.4 Kcal/mol and Kd value of 0.661 μM. A thorough interaction has been depicted in Figure 3(d). Narcissin has interacted with 13 major amino acid residues among which 11 (Leu141, Asn142, Cys145, Gly143, Leu27, Met49, Asp187, Met165, His41, Gln189, Arg188) amino acid matches with that of the native ligand. Thus it is very clear from the result that Narcissin can also be considered to be a good choice for inhibitor as it also interacts with major amino acid residues which the native ligand interacts with.
Calendulaglycoside B has also exhibited a considerably good binding efficiency towards the receptor with a binding energy of-8.2 Kcal/mol and Kd value of 0.928 μM. The detailed interaction has been depicted in Figure 3(e). This phytochemical interacts with 16 amino acid residues and among them, 14 amino acid residues interaction (Phe140, Ser144, His163, Glu166, Gln189, Arg188, Asp187, Leu141, His41, Met165, Gly143, Cys145, Asn142, His164) are similar to that of the native ligand. Thus we can infer that Calendulaglycoside B also interacts with major amino acid residues which are similar to the native ligand interaction.
Calenduloside on the other has a good binding affinity towards the target receptor with a binding energy of-7.9 Kcal/mol and Kd value of 1.542 μM which is greater than that of native ligand but not as good as the other ligands. The detailed interaction has been exhibited in Figure 3(f). Calenduloside interacts with 15 amino acids among which 11 amino acid residues (Thr25, Asn142, Cys145, His164, Gln189, Glu166, Met165, Gly143, Leu27, Thr26, Met49) match that of the native ligand. Thus we can infer that although calenduloside interacts with major amino acid residues similar to that of the native ligand but among the 6 chosen ligand calenduloside is the lowest in terms of efficiency.
Molecular dynamics (MD) simulation
Apo-form of COVID-19 main protease (apo-Mpro), top three (Rutin, Isorhamnetin-3-O-β-D, Calendoflaside) docked ligands with higher binding affinity and the crystal structure of COVID-19 Mpro with inhibitor N3 have been selected to find out their system stability, flexibility, and other dynamic properties through 100 ns MD simulation.
Root mean square deviation (RMSD) analysis
The Root mean square deviation (RMSD) designates the dynamic stability of all the systems. It also calculates the conformational perturbations which occur in the backbone of the protein during the simulation time scale. The RMSD values have been calculated for the apo-Mpro, and the Mpro (COVID-19 main protease) complex with Inhibitor N3, Rutin, isorhamnetin-3-O-β-D, and Calendoflaside respectively. All the trajectories have got the equilibration state after 85 ns shown in Figure 4(a). The average RMSD values have calculated for apo-Mpro, Mpro-inhibitor N3, Mpro-rutin, Mpro-isorhamnetin-3-O-β-D, and Mpro-calendoflaside are 0.354, 0.391, 0.351, 0.514, and 0.378 nm respectively. The isorhamnetin-3-O-β-D has showed a comparatively higher RMSD value from 20 ns onwards. The other selected ligands i.e. rutin and calendoflaside showed a relatively less RMSD than that of the native ligand inhibitor N3 and they are almost the same as that of the apo-Mpro and all have got almost stable after 75 ns. The Mpro-calendoflaside complex has showed a RMSD of slightly 0.4 nm. The crystal ligand i.e. the inhibitor N3 and the protease complex have showed the RMSD above 0.4 nm. The RMSD of isorhamnetin-3-O-β-D has been stable initially around 10 ns and maintained a constant RMSD of around 0.22 nm. Suddenly, it has risen to the RMSD of 0.4 nm during 10-20 ns. After 20 ns, it has shown a stable RMSD of 0.58 nm which indicates that the system got equilibrated after 20 ns concerning the initial structure of the protease-ligand complex. All the complexes are seen to be stable during the 100 ns simulation. But two complexes (Mpro-rutin, Mpro-calendoflaside) are found to be more stable than the original native co-crystal ligand: inhibitor N3.
Figure 4.
(a) RMSD analysis for the apo-Mpro and COVID-19 MPRO -ligand complexes. (b) Radius of gyration (Rg) analysis for the apo-Mpro and COVID-19 MPRO -ligand complexes (c) RMSF analysis for the apo-Mpro and COVID-19 MPRO -ligand complexes.
Radius of gyration (Rg) analysis
The compactness of the receptor-ligand complexes during the molecular dynamic simulation is best described by the Rg factor. The measurement of the distance between the center of mass of the receptor atoms with its terminal in a specified time frame is the major significance of this study. A compact receptor structure indicates less variation in the gyration value while an expanded or unstable form shows higher Rg value. The study has depicted that the variation in Rg values concerning different time points for the Apo–form of COVID-19 main protease (apo-Mpro) and all the protease-ligands complexes (Figure 4(b)). The average Rg values for apo-Mpro and protease complexes with Inhibitor N3, Rutin, isorhamnetin-3-O-β-D, and Calendoflaside are 2.12, 2.11, 2.15, 2.12, and 2.15 nm respectively. All the complexes including the native ligand inhibitor N3 have shown almost the same Rg value as compared to apo-Mpro. The native co-crystal ligand inhibitor N3 has diverged from 2.1 nm to 2.2 nm of Rg value during the 100 ns simulation. It is overlapping with the other three natural products until 18 ns. All the selected ligands from natural sources maintained a constant Rg around 2.2 nm till the end of the 100 ns simulation which indicates the compactness of all the ligand as compared to the Mpro-inhibitor N3.
Root mean square fluctuation (RMSF) analysis
The root mean square fluctuation (RMSF) value represents the mobility and flexibility of a structure. The RMSF of the amino acid residues of COVID-19 main protease apo-form (apo-Mpro) and protease (Mpro) complexes with rutin, isorhamnetin-3-O-β-D, calendoflaside, and inhibitor N3 have been analyzed (Figure 4(c)). The average RMSF value for apo-Mpro, Mpro-inhibitor, Mpro-rutin, Mpro-isorhamnetin-3-O-β-D, and Mpro-calendoflaside are 0.15, 0.19, 0.15, 0.19, and 0.13 nm respectively. It has been observed that apo-Mpro, Mpro-rutin, and Mpro-calendoflaside structure have similar kind of low RMSF value. Whereas crystal structure (Mpro-inhibitor) and Mpro-isorhamnetin-3-O-β-D have shown higher RMSF value as compared to others. Among all the predicted ligand calendoflaside and rutin have shown very good RMSF value when they bind with Mpro. All the ligands exhibited a higher RMSF peak around 0.7 nm at the C-terminal protease region. From the RMSF value analysis, we can conclude that all of our predicted ligands showed less conformational changes during binding and can act as a stable complex.
Hydrogen bond analysis
The hydrogen bond analysis is a very important factor while considering receptor-ligand stability as these bonds are transient interaction and are responsible for providing stability to the receptor-ligand complex. These are highly specific bonds. In this study, we have calculated all the hydrogen bonds for all the complexes including that of the native ligand inhibitor N3. The number of hydrogen bonds at different time points has been calculated and depicted in Figure 5(a). The average number of hydrogen bonds calculated for MPRO-inhibitor N3, MPRO-rutin, MPRO-isorhamnetin-3-O-β-D, MPRO-calendoflaside are 0-4, 0-7, 0-9, 2-12 respectively. All the predicated ligands have shown more number of hydrogen bonds compared to the native ligand inhibitor N3. The MPRO-calendoflaside complex showed the maximum number of hydrogen bonds and thus it could be concluded that this complex is the most stable among all the others. Thus calendoflaside is the best choice for ligand considering hydrogen bond into account while the other ligands including the inhibitor N3 also showed good stability towards the complex.
Figure 5.
(a) Estimation of the hydrogen bond number during the 100 ns MD simulations of Mpro-ligand complexes (b) Short-range Coulombic interaction energy of MPRO-ligand complexes. (c) Gibbs free energy calculation (MMPBSA) of MPRO-ligand complexes. (d) The quantification of the individual amino acid residue of MPRO to the total binding energies toward ligands.
Interaction energy analysis
We have analyzed interaction energy to quantify the strength of the interaction of protease-ligand complexes. It can be useful to calculate non-bonded interaction energy between protease and ligands. In Figure 5(b), the short-range columbic interaction energy of all the ligands with protease has been depicted. In this 100 ns MD simulation study, we can observe that the average interaction energy between COVID-19 MPRO receptor and co-crystal ligand N3, rutin, isorhamnetin-3-O-β-D, and calendoflaside are −48.42 ± 16.11, −107.06 ± 28.7235, −105.31 ± 50.69, and −181.99 ± 38.55 kJ/mol. Such data suggests that all the three ligands rutin, isorhamnetin-3-O-β-D, and calendoflaside have docked more strongly toward the target protease as compare to the native co-crystal ligand inhibitor N3.
MM-PBSA binding free energy calculations
The binding capacity of the ligand towards the receptor is quantitatively estimated by the binding free energy analysis. Binding free energy is the summation of all non-bonded interaction energies. The binding free energy of the interaction between MPRO (COVID-19 main protease) and docked compounds/ligands has been estimated using the G_MMPBSA tool (Kumari et al., 2014). Our binding energy analysis from 100 ns MD simulation trajectories have shown that calendoflaside, isorhamnetin-3-O-β-D, and rutin have a higher binding affinity towards COVID-19 Mpro inhibition site as compared to the native co-crystal ligand inhibitor N3 as reflected in Figure 5(c) and Table 3. The total binding energies of inhibitor N3, rutin, isorhamnetin-3-O-β-D, calendoflaside with MPRO are −195.76, −195.80, −211.08, −294.28 kJ/mol respectively. The biding energies have confirmed that the ligands isorhamnetin-3-O-β-D and calendoflaside have shown a better affinity than compared to that of the native co-crystal ligand inhibitor N3. Rutin and native ligand inhibitor N3 have shown almost similar binding energies when they bind with MPRO. Thus overall all the complexes are stable but among all the complexes MPRO- isorhamnetin-3-O-β-D, and MPRO- calendoflaside are energetically more favourable and representing the stable complex. Including binding energy, other different kinds of interaction energies like van der Waals energy, electrostatic energy, polar solvation energy, SASA energy have been also calculated for all the complexes (Figure 6, Table 3). Results indicate, van der Waals, electrostatic, and SASA energy negatively contribute to the total interaction energy while only polar solvation energy positively contributes to the total free binding energy. Considering the negative contribution, the contribution of van der Waals interaction is much more than that of the electrostatic interactions for all the cases. The contribution of SASA energy is relatively less as compared to the total binding energy. The high negative value of van der Waals energy also points to the massive hydrophobic interaction between the ligands and the COVID-19 main protease (Verma et al., 2016).
Table 3.
Van der Waals, electrostatic, polar solvation, SASA and binding energy for the docked compounds into Mpro inhibition site.
| Ligands | Van der Waals Energy | Electrostatic Energy | Polar Solvation Energy | SASA Energy | Binding Energy |
|---|---|---|---|---|---|
| Inhibitor (N3) | −222.93 ± 36.32 | −52.06 ± 22.37 | 97.02 ± 22.63 | −17.77 ± 2.16 | −195.76 ± 40.39 |
| Rutin | −189.59 ± 30.25 | −119.70 ± 33.95 | 128.01 ± 32.44 | −14.53 ± 2.62 | −195.80 ± 38.93 |
| Isorhamnetin-3-O-β-D | −232.74 ± 8.83 | −125.11 ± 55.11 | 165.85 ± 35.54 | −19.08 ± 1.42 | −211.08 ± 32.84 |
| Caledoflaside | −275.57 ± 9.24 | −209.83 ± 38.56 | 213.17 ± 23.88 | −22.05 ± 1.21 | −294.28 ± 37.47 |
Figure 6.
Representation of the Van der Waals, Electrostatic, Polar solvation, SASA and Binding energy for the docked compounds into Mpro inhibition site.
The ΔG binding energy of all the ligands binding to the COVID −19 main protease with respect to different time points has been calculated (Figure 5(c)). During the 100 ns MD simulations, there are few fluctuations in the binding energies at different time points. Close analysis shows that inhibitor N3 has more binding energies than rutin and isorhamnetin-3-O-β-D at some time points like 20 ns, 42 ns, and 78 ns but shows lower binding energy than calendoflaside throughout the 100 ns of MD simulation. MPRO-calendoflaside complex has shown very higher energy initially during 10 ns but with time it decreases but kept an average of −250 to −350 kJ/mol. The overall energies although have been found to higher for all the predicted ligands.
When considering structure-based drug designing, the residue-based binding energy is considered to be of utmost importance as based on the residue we can improve the lead compound. Thus the key residues were predicted played an important role in ligand binding. We have analysed the contribution of individual amino acid residues in the binding energy of MPRO-ligand complexes (Figure 5(c)). From the graph, we have observed that HIS41, MET49, GLN127, SER139, GLU166, GLN189, HIS172 amino acid residues at the inhibition site play a key role in the Mpro-ligand binding. Most of these amino acid residues are hydrophilic which indicates that in the receptor-ligand stable interaction non-bonded hydrogen bonds play a major role.
Principal component analysis (PCA)
To analyse the collective motion of all complexes as well as apo-form of COVID-19 main protease (apo-Mpro), the PCA analysis based on C-α atoms have been performed. It has been observed that the first few eigenvectors or the principal components (PCs) of the structures play an important role and describe overall motions of the entire system. Therefore, the first 50 eigenvectors of apo-Mpro and its complexes have been considered and plotted (Figure 7(a)). The first five eigenvectors contribute for 59.24%, 70.93%, 64.32%, 75.10%, 59.78% of the total motion found for the 100 ns MD simulation trajectory of apo-Mpro, MPRO-inhibitor N3, MPRO-rutin, MPRO-isorhamnetin-3-0-β-D, and MPRO-calendoflaside respectively. It has been found that MPRO-isorhamnetin-3-0-β-D and MPRO-inhibitor N3 have shown very high correlated motions as compared to other complexes as well as apo-Mpro. The MPRO-rutin and MPRO-calendoflaside complexes have shown almost similar kind of correlated motions as compared to the apo-Mpro. Such data suggest that rutin and calendoflaside have formed very stable complexes with MPRO (COVID-19 main protease) and can be considered as a lead compound.
Figure 7.
Principal component analysis. (a) The plot of the eigenvalues vs. eigenvector index. The first 50 eigenvectors have been considered (b) Projection of the motion of the protein in phase space along the PC1 and PC2.
We have found earlier that the first five eigenvectors contribute the majority portion of the total dynamics of the whole system. Therefore, we have plotted only the first two eigenvectors against each other where each dot represents correlated motions (Figure 7(b)). The well stable clustered dots signify the more stable structure and low clustered dots represent the weaker stable structure. It has been observed that apo-MPRO, MPRO-rutin, and MPRO-calendoflaside complexes have shown very stable clustered dots as compared to others.
Covariance plots have been generated to understand how residue atoms move relative to each other (Figure 8). Motions can be positively correlated (red region), negatively correlated (blue region), or uncorrelated (white region). The deeper color signifies a higher positive correlation or negative correlation. We have found that negatively correlated motions are dominant in all the dynamic systems except MPRO-isorhamnetin-3-0-β-D complex (Figure 8(d)). It has been observed that correlated residual motions are slightly higher in apo-MPRO as compared to other dynamic systems like MPRO-rutin, MPRO-inhibitor N3, and MPRO-calendoflaside. It has revealed that the binding of ligands induces a slightly different type of motion in MPRO-ligand complexes as compared to apo-MPRO. Here, MPRO-calendoflaside complex (Figure 8(e)) has shown lesser and MPRO-rutin complex (Figure 8(c)) has shown a similar pattern of correlated motions as compared to MPRO-inhibitor N3 (crystal structure) (Figure 8(b)).
Figure 8.
The diagonalized covariance matrix of (a) apo–form of COVID-19 main protease (apo- MPRO) and MPRO complexes docked with (b) inhibitor N3, (c) rutin, (d) isorhamnetin-3-0-β-D, and (e) calendoflaside during 100 ns MD simulation.
Free energy analysis
The Gibbs free energy landscape study provides substantial information about the receptor-ligand complex. The conformational state changes during ligand binding can be most effectively predicted by calculating the Gibbs free energy analysis for PC1 and PC2. The Gibbs free energy landscape (Figure 9) shows that the energy value ranging from 0 to 14.8, 0 to 15.7, 0 to 14.5, 0 to 15.1, 0 to 15.2 kJ/mol for apo-MPRO, MPRO-inhibitor N3, MPRO-rutin, MPRO-isorhamnetin-3-O-β-D, MPRO- calendoflaside respectively. The blue, cyan, and green regions in the plot signify a lower energy state with extremely stable structural conformation while the red region indicates higher energy conformation. The apo-MPRO showed a bluer region as compared to all other ligands. While considering MPRO-ligand complex, Mpro-rutin complexes showed the energy range from 0 to 14.5 and is the minimum value as compared to all other ligand and also control ligand and the apo- MPRO. A smaller and more concentrated blue minimal energy area in MPRO–rutin and MPRO–calendoflaside complexes compared to the crystal structure of MPRO–inhibitor N3; suggests that rutin and calendoflaside have formed highly stable complexes with COVID-19 MPRO.
Figure 9.
The Gibbs free energy landscape plot of (a) apo-form of COVID-19 main protease (apo-Mpro) and MPRO complexes docked with (b) inhibitor N3, (c) rutin, (d) isorhamnetin-3-O-β-D, and (e) calendoflaside.
Structural changes during simulation
The exclusive study is done on the 3 D structural evolution of the protease-ligand complex over 10 ns interval of time scale by comparing 5 snapshots of the protein-ligand complexes (Figure 10). The snapshots are observed to show very minimal changes in the protease structure during the 100 ns simulation. The secondary structure of the protease is conserved throughout the simulation when they are bound with the natural products, which shows the stability of the protease-ligand complex. Interestingly, the secondary structures are conserved during the MD simulations in both states. Additionally, we attribute the large RMSF peak in the residues in the ligand-binding process that signifies to the dynamical change of secondary structure in the bound form.
Figure 10.
Snapshot of (a) apo-form of COVID-19 main protease (apo-Mpro) and MPRO complexes docked with (b) inhibitor N3, (c) rutin, (d) isorhamnetin-3-O-β-D, and (e) calendoflaside over 10 ns interval of the 100 ns MD simulation trajectory.
The overall secondary structural analysis of ligand-bound Mpro (COVID-19 main protease) complexes and the apo-Mpro has been performed using gmx_do_dssp option in GROMACS (Figure 11, Table 4). The secondary structural rigidity depends on the percentage of A-Helices/B-Sheets and flexibility depends on the percentage of coils/turns present in the structure. It has been observed that there are slight structural changes in Mpro when it binds with calendoflaside, isorhamnetin-3-O-β-D, rutin, and inhibitor N3. We have found that apo-Mpro and Mpro-inhibitor N3 have a slightly higher percentage of coils as compared to other docked complexes (Table 4). Such data indicate that Mpro structure is becoming more rigid and stable after docked with compounds like calendoflaside, isorhamnetin-3-O-β-D, and rutin.
Figure 11.
The secondary structure content of (a) apo-form of COVID-19 main protease (apo-Mpro) and MPRO complexes docked with (b) inhibitor N3, (c) rutin, (d) isorhamnetin-3-O-β-D, and (e) calendoflaside.
Table 4.
Overall percentage of secondary structure elements in COVID-19 Mpro-ligand complexes.
| Complex | Structure% (A-Helix + B-Sheet + B-Bridge + Turn) | Coil % | B-Sheet % | B–Bridge % | Bend % | Turn % | A-Helix % | 5-Helix % | 3-Helix % |
|---|---|---|---|---|---|---|---|---|---|
| Apo-Protein | 57 | 29 | 24 | 1 | 12 | 9 | 23 | 0 | 1 |
| Inhibitor N3 | 57 | 28 | 25 | 2 | 13 | 9 | 22 | 0 | 2 |
| Rutin | 61 | 26 | 26 | 2 | 11 | 10 | 23 | 0 | 3 |
| Isorhamnetin-3-O-β-D | 59 | 27 | 25 | 2 | 12 | 8 | 23 | 0 | 2 |
| Calendoflaside | 59 | 28 | 25 | 1 | 12 | 9 | 24 | 0 | 1 |
Drug likeness property
Drug-likeness is a crucial consideration while choosing compounds during the initial stages of drug discovery. The pharmacological properties of the final three phytochemicals have been calculated to test their drug-likeness (Table 5). The chemical structures of all these final phytochemicals are depicted in Figure 12. Although there are few violations but all the phytochemicals can be used as a drug for proper application. Rutin on analysis has passed the Modern drug data report (MDDR Like Rule). MDDR comprises around 100000 drugs that are under development or are already launched in the market. These compounds are referenced in the various sources of literature (Kadam & Roy, 2007). Rutin (DRUGBANK No.- DB01698), in general, is also used in some commercially available drugs (Temerk et al., 2006). Like Rutin, Calendoflaside also passed the MDDR Like Rule. Thus Calendoflaside can also be considered as an important candidate for drug development. On the other hand, Isorhamnetin-3-O-β-D passed the Ghose_Filter, Weighted QED test for drug-likeness. The Ghose rule is a valid rule for selecting drugs based on qualitative and quantitative characterization of known drug databases (Ghose et al., 1999). A quantitative estimate of drug-likeness or QED depicts the fundamental distribution of molecular properties. Weighted QED is used majorly for molecular target druggability screening by arranging a large set of available bioactive compounds that are already published (Bickerton et al., 2012). Thus we can consider Isorhamnetin-3-O-β-D as a lucrative candidate for drug design. It is also reported earlier that Isorhamnetin-3-O-β-D is used as a drug for the prevention and/or treatment of diabetes and its complications in the rat model (Lee et al., 2005). Apart from this, we must consider PAINS Alert while drug design. Pan-assay interference compounds (PAINS) are chemical compounds that frequently give false positive consequences in high-throughput screening (Dahlin et al., 2015). Rutin although shows 1 alert but the other two compound that is Isorhamnetin-3-O-β-D and Calendoflaside have 0 alert, indicating low or negligible chances for false-positive results.
Table 5.
Drug Likeness Property of Rutin, Isorhamnetin-3-O-β-D & Calendoflaside.
| Pharmacological Properties | Rutin | Isorhamnetin-3-O-β-D | Calendoflaside |
|---|---|---|---|
| MW | 610.15 | 478.11 | 608.17 |
| logp | −0.735 | −0.009 | −0.14 |
| Alogp | −4.581 | −3.27 | −4.017 |
| HBA | 16 | 12 | 15 |
| HBD | 10 | 7 | 8 |
| TPSA | 265.52 | 195.6 | 234.29 |
| AMR | 147.17 | 121.18 | 150.83 |
| nRB | 6 | 5 | 6 |
| nAtom | 73 | 56 | 75 |
| nAcidic Group | 0 | 0 | 0 |
| RC | 5 | 4 | 5 |
| nRigidB | 41 | 32 | 32 |
| nAromRing | 2 | 2 | 2 |
| nHB | 26 | 19 | 23 |
| SAlerts | 2 | 1 | 1 |
| PAINS | 1 | 0 | 0 |
| Satisfying Drug Likeliness | MDDR Like Rule | Ghose_Filter, Weighted QED | MDDR Like Rule |
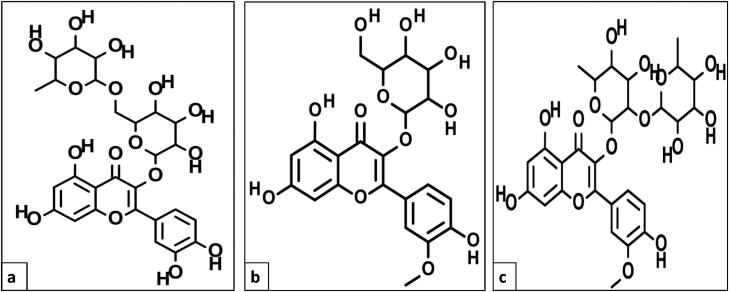
Figure 12.
Chemical 2 D structures of (a) rutin, (b) isorhamnetin-3-O-β-D, and (c) calendoflaside.
Probable mechanism of action
The Coronaviruses are positive-strand RNA viruses. The process of viral replication and transcription is controlled by two overlapping polyproteins, the pp1a (replicase 1a, 450 kD) and pp1ab (replicase 1ab, 750 kD). Ribosomal frame shifting is a necessary process for the expression of the C-proximal portion of pp1ab polyprotein. Extensive proteolytic processing finally releases functional polypeptides from both the polyproteins which are in term controlled by the main protease (Mpro). Along with the papain-like protease(s) the main protease (Mpro) is essential for proteolytic processing of the polyproteins that are translated from the viral RNA (Xue et al., 2008). The main protease slices the polyprotein at around 11 conserved sites which involve the Leu-Gln2 (↓)(Ser, Ala, Gly) sequences. This slicing process is initiated by the enzyme’s autolytic cleavage from pp1a and pp1ab. Thus we can come to the point that inhibiting the activity of the main protease would lead to blockage viral replication (Anand et al., 2003). Herein, our computational study predicts that flavonoid based inhibitors of Calendula officinals (Rutin, Isorhamnetin-3-O-β-D, Calendoflaside) may block the active site of this protease and hence blocking its action. All the three phytochemicals bind with the amino acid residues Phe140, His164, Asn142, Gly143, Cys145, His163, Met165, Glu166, Gln189, Arg188, His41, Thr26, Thr25, Ser144, Leu141, Met49, Ser146, Asp187, Leu27 and thus blocking the major amino acid residue to initiate further proteolytic activity (Figure 13). The major aim is to stop the viral replication which could be achieved by blocking the action of this main protease. As of now no human proteases possess similar cleavage specificity. Beside that inhibitors are likely to be non-toxic towards human body as they are from a well-established natural source and is used as homeopathy medicine for ages.
Figure 13.
Probable Mechanism of Action of Phytochemicals of Calendula officinals (Rutin, Isorhamnetin-3-O-β-D, Calendoflaside) blocking the action of Main Protease of COVID-19 (MPro) needed for Viral Replication.
Conclusion
Our in silico study gives a clear idea that flavonoid based phytochemicals of calendula (rutin, isorhamnetin-3-O-β-D, calendoflaside) may be highly effective for inhibiting Mpro which is the main protease for SARS-CoV-2 causing the deadly disease COVID-19. The virtual molecular docking study reveals that seven bioactive compounds from Calendula officinals have a higher binding affinity toward COVID-19 main protease (MPro) as compare to co-crystal ligand inhibitor N3. Molecular dynamics simulation (100 ns) studies of the top three docked compounds (rutin, isorhamnetin-3-O-β-D, and calendoflaside according to the docking score) and co-crystal ligand inhibitor N3 suggest that the top three docked compounds have a good amount of stability, flexibility and binding affinity toward COVID-19 main protease (Mpro) as compare to co-crystal ligand inhibitor N3. We have also observed the dynamics properties of the apo-form of COVID-19 main protease (apo-Mpro) and found that there is no significant amount of difference in terms of dynamic properties between apo-Mpro and its docked complexes Mpro-rutin, Mpro-isorhamnetin-3-O-β-D, and Mpro-calendoflaside. We have also evaluated the predicted ligands through ADMET descriptors. From the results, it can be concluded that the predicted ligands can be used for medicinal purposes and are also non-toxic.
Overall, this in silico study predicts that these three compounds from Calendula officinals, especially calendoflaside and rutin compounds due to their ability to form a stable complex with Mpro, may have the potency to be evolved as an anti-COVID-19 main protease drug to fight against the novel coronavirus but before that, it must go through under the proper preclinical and clinical trials for further experimental and/or clinical scientific validation.
Supplementary Material
Acknowledgements
We would like to acknowledge the “Centre of Excellence in Phase Transformation and Product Characterization, TEQIP-III”, Jadavpur University, Kolkata, MHRD, Indian Institute of Technology, Kharagpur, India, Indian Council of Medical Research, Council of Scientific and Industrial Research and J C Bose Fellowship (DST, India) for providing fellowship and financial support during this work. We would also like to acknowledge every lab member of Biomaterial, Cell Culture, and Microbiology lab of School of Bioscience and Engineering, Jadavpur University. All the authors would like to thank all the medical workers and emergency service providers around the globe for their incomparable contribution towards the society and mankind while combating COVID-19.
Glossary
Abbreviation
- MPRO
COVID-19 Main Protease
- MW
Molecular Weight
- HBA
Hydrogen Bond Acceptor
- HBD
Hydrogen Bond Donor
- TPSA
Topological Polar Surface Area
- AMR
Atom Molar Refractivity
- nRB
Number of Rotable Bond
- nAtom
Number of Atom
- nAcidic Group
Number of Acidic group
- RC
Rotatable bond Count
- nRigidB
Number of Rigid Bond
- nAromRing
Number of Aromatic ring
- nHB
Number of hydrogen bonds
- SAlerts
Number of Structural Alerts
- PAINS
Pan-assay interference Structures
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Agarwal, O. P., & Raju, P. S. (2006). Opportunities for India. Traditional Systems of Medicine, 5, 5–10. [Google Scholar]
- Al-Khafaji, K., Al-DuhaidahawiL, D., & Taskin Tok, T. (2020). Using integrated computational approaches to identify safe and rapid treatment for SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science (New York, N.Y.).), 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- AshwlayanVD, K. A., & Verma, M. (2018). Therapeutic potential of Calendula officinalis. Pharmacy and Pharmacology International Journal, 6(2), 149–155. [Google Scholar]
- Berendsen, H. J. C., van der Spoel, D., & van Drunen, R. (1995). GROMACS: A message-passing parallel molecular dynamics implementation. Computer Physics Communications, 91(1–3), 43–56. 10.1016/0010-4655(95)00042-E [DOI] [Google Scholar]
- Bhatnagar, S. S., & Sastri, B. N. (1960). The wealth of India raw materials (A Dictionary of Indian Raw Materials and Industrial Products). New Delhi, India, 10, 64–68. [Google Scholar]
- Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S., & Hopkins, A. L. (2012). Quantifying the chemical beauty of drugs. Nature Chemistry, 4(2), 90–98. 10.1038/nchem.1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborthy, G. S. (2010). Phytochemical screening of Calendula officinalis Linn leaf extract by TLC. International Journal of Research in Ayurveda and Pharmacy (IJRAP), 1(1), 131–134. [Google Scholar]
- Chen, Y. W., Yiu, C.-P B., & Wong, K.-Y. (2020). Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CLpro) structure: Virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research, 9, 129. 10.12688/f1000research.22457.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J., Li, F., & Shi, Z. L. (2019). Origin and evolution of pathogenic coronaviruses. Nature Reviews. Microbiology, 17(3), 181–192. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin, J. L., Nissink, J. W. M., Strasser, J. M., Francis, S., Higgins, L., Zhou, H., Zhang, Z., & Walters, M. A. (2015). PAINS in the assay: Chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. Journal of Medicinal Chemistry, 58(5), 2091–2113. 10.1021/jm5019093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose, A. K., Viswanadhan, V. N., & Wendoloski, J. J. (1999). A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. Journal of Combinatorial Chemistry, 1(1), 55–68. 10.1021/cc9800071 [DOI] [PubMed] [Google Scholar]
- Gupta, M. K., Vemula, S., Donde, R., Gouda, G., Behera, L., & Vadde, R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., & Duan, Y. (2020. a). Structure of Mpro from COVID-19 virus and discovery of its inhibitors. BioRxiv. 10.1101/2020.02.26.964882 [DOI] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., & Duan, Y. (2020. b). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 1–5. [DOI] [PubMed] [Google Scholar]
- Kadam, R. U., & Roy, N. (2007). Recent trends in drug-likeness prediction: A comprehensive review of in silico methods. Indian Journal of Pharmaceutical Sciences, 69(5), 609. 10.4103/0250-474X.38464 [DOI] [Google Scholar]
- Kahn, J. S., & McIntosh, K. (2005). History and recent advances in coronavirus discovery. The Pediatric Infectious Disease Journal, 24(11), 223–227. 10.1097/01.inf.0000188166.17324.60 [DOI] [PubMed] [Google Scholar]
- Khan, M. T., Ali, A., Wang, Q., Irfan, M., Khan, A., Zeb, M. T., Zhang, Y. J., Chinnasamy, S., & Wei, D.-Q. (2020). Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2. A molecular dynamic study. Journal of Biomolecular Structure and Dynamics, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., & Lynn, A. (2014). g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations. Journal of Chemical Information and Modeling, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Lee, Y. S., Lee, S., Lee, H. S., Kim, B.-K., Ohuchi, K., & Shin, K. H. (2005). Inhibitory effects of isorhamnetin-3-O-$β$-D-glucoside from Salicornia herbacea on rat lens aldose reductase and sorbitol accumulation in streptozotocin-induced diabetic rat tissues. Biological & Pharmaceutical Bulletin, 28(5), 916–918. 10.1248/bpb.28.916 [DOI] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder, R., Parida, P., Paul, S., & Basak, P. (2019). In vitro and in silico study of Aloe vera leaf extract against human breast cancer. Natural Product Research, 1–4. 10.1080/14786419.2018.1534848 [DOI] [PubMed] [Google Scholar]
- Mittal, L., Kumari, A., Srivastava, M., Singh, M., & Asthana, S. (2020). Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. Journal of Biomolecular Structure and Dynamics, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muley, B. P., Khadabadi, S. S., & Banarase, N. B. (2009). Phytochemical constituents and pharmacological activities of Calendula officinalis Linn (Asteraceae): A review. Tropical Journal of Pharmaceutical Research, 8(5). 10.4314/tjpr.v8i5.48090 [DOI] [Google Scholar]
- Nejadi Babadaei, M. M., Hasan, A., Haj Bloukh, S., Edis, Z., Sharifi, M., Kachooei, E., & Falahati, M. (2020). The expression level of angiotensin-converting enzyme 2 determine the severity of COVID-19: Lung and heart tissue as targets. Journal of Biomolecular Structure and Dynamics, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma, P., Shekhar, N., Prajapat, M., Avti, P., Kaur, H., Kumar, S., Singh, S., Kumar, H., Prakash, A., Dhibar, D. P., & Medhi, B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. John, S. E., Tomar, S., Stauffer, S. R., & Mesecar, A. D. (2015). Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4 - The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS). Bioorganic and Medicinal Chemistry, 23(17), 6036–6048. 10.1016/j.bmc.2015.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temerk, Y. M., Ibrahim, H. S. M., & Schuhmann, W. (2006). Cathodic adsorptive stripping voltammetric determination of the antitumor drug rutin in pharmaceuticals, human urine, and blood serum. Microchimica Acta, 153(1–2), 7–13. 10.1007/s00604-005-0451-3 [DOI] [Google Scholar]
- Tyrrell, D. A., & Bynoe, M. L. (1966). Cultivation of viruses from a high proportion of patients with colds. Lancet (London, England), 1(7428), 76–77. 10.1016/s0140-6736(66)92364-6 [DOI] [PubMed] [Google Scholar]
- van Aalten, D. M., Bywater, R., Findlay, J. B., Hendlich, M., Hooft, R. W., & Vriend, G. (1996). PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. Journal of Computer-Aided Molecular Design, 10(3), 255–262. 10.1007/BF00355047 [DOI] [PubMed] [Google Scholar]
- Verma, S., Grover, S., Tyagi, C., Goyal, S., Jamal, S., Singh, A., & Grover, A. (2016). Hydrophobic interactions are a key to MDM2 inhibition by polyphenols as revealed by molecular dynamics simulations and MM/PBSA free energy calculations. PloS One, 11(2), e0149014. 10.1371/journal.pone.0149014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C., & Zhang, Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, X., Yu, H., Yang, H., Xue, F., Wu, Z., Shen, W., Li, J., Zhou, Z., Ding, Y., Zhao, Q., Zhang, X. C., Liao, M., Bartlam, M., & Rao, Z. (2008). Structures of two coronavirus main proteases: Implications for substrate binding and antiviral drug design. Journal of Virology, 82(5), 2515–2527. 10.1128/JVI.02114-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.