Abstract
SARS-CoV-2 is the causative agent of COVID-19 and has been declared as pandemic disease by World Health Organization. Lack of targeted therapeutics and vaccines for COVID-2019 have triggered the scientific community to develop new vaccines or drugs against this novel virus. Many synthetic compounds and antimalarial drugs are undergoing clinical trials. The traditional medical practitioners widely use Indian medicinal plant Withania somnifera (Ashwagandha) natural constituents, called withanolides for curing various diseases. The main protease (Mpro) of SARS-CoV-2 plays a vital role in disease propagation by processing the polyproteins which are required for its replication. Hence, it denotes a significant target for drug discovery. In the present study, we evaluate the potential of 40 natural chemical constituents of Ashwagandha to explore a possible inhibitor against main protease of SARS-CoV-2 by adopting the computational approach. The docking study revealed that four constituents of Ashwagandha; Withanoside II (-11.30 Kcal/mol), Withanoside IV (-11.02 Kcal/mol), Withanoside V (-8.96 Kcal/mol) and Sitoindoside IX (-8.37 Kcal/mol) exhibited the highest docking energy among the selected natural constituents. Further, MD simulation study of 100 ns predicts Withanoside V possess strong binding affinity and hydrogen-bonding interactions with the protein active site and indicates its stability in the active site. The binding free energy score also correlates with the highest score of −87.01 ± 5.01 Kcal/mol as compared to other selected compounds. In conclusion, our study suggests that Withanoside V in Ashwagandha may be serve as a potential inhibitor against Mpro of SARS-CoV-2 to combat COVID-19 and may have an antiviral effect on nCoV.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-2019, SARS-CoV-2, Ashwagandha, Withania somnifera, molecular docking, MD simulation
1. Introduction
The novel strain of coronavirus was first identified at the end of December 2019 and emerged during an outbreak in Wuhan, China. Coronavirus disease (COVID-19) is a transmissible infectious disease initiated by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and has become a severe threat to human health (Zhou et al., 2020). On 30 January 2020, World Health Organization WHO declared this outbreak as a Public Health Emergency of international concern and later on 11 February 2020 named it as Coronavirus Diseases-19 (COVID-19). It subsequently declared COVID-19 as a global pandemic on 11 March 2020 (Green, 2020). Till 28 May 2020, WHO reported worldwide 5,593,631 confirmed cases and 353,334 deaths due to SARS-CoV-2 (WHO, 2020).
The term coronavirus was coined from the Latin term corona that means “crown” like shape (Rabi et al., 2020). The suffix 2019 novel coronavirus or “2019-nCoV” coronavirus was given by WHO on 12 January 2020. SARS-CoV-2 transmission is via human-to-human, mainly through direct contact and/or by contact with the infected surface with touching of the face. Additionally, it also spreads through respiratory droplets released by coughs or sneezes (Peng et al., 2020). Common symptoms of coronavirus (SARS-CoV-2) include respiratory symptoms, cough, fever, shortness of breath, and more severe symptoms of this infection can cause pneumonia, severe acute respiratory syndrome, and even death (Qiu et al., 2020). SARS-CoV-2 coronavirus commonly belongs to the beta-CoVs category and has 60–140 nm diameter with a round or elliptical shape (Fehr & Perlman, 2015).
SARS-CoV-2 is a positive-stranded RNA virus wherein part of its genome encodes, 16 non-structural proteins (Nsp1-16), viral RNA dependent RNA polymerase, RNA synthesis materials, and two large non-structural polyproteins. The remaining genome encodes four structural proteins, spike (S), envelope (E), membrane (M), nucleocapsid (N), and the other accessory proteins (Liu et al., 2020; Lu et al., 2020). A maximum number of non-structural proteins are known to play a significant role in the SARS-CoV-2 replication.
The main protease (Mpro) also known as 3-chymotrypsin-like protease (3CLpro) is a key protein required for the proteolytic maturation of the virus (Morse et al., 2020; Stobart et al., 2012). Thus, targeting main protease can provide an effective treatment against SARS-CoV-2 by inhibition of the viral polypeptide cleavage (Zhang et al., 2020). It is one of the best-characterised drug targets among structural and non-structural proteins of coronavirus and currently X-ray structures are reported as both liganded and un-liganded SARS-CoV-2 (Jin et al., 2020; Zhang et al., 2020). Since this protein is absent in humans, it forms an excellent target for drug discovery. Moreover, there is presently no specific medicine or vaccine available for the treatment of COVID-19 infection caused by SARS-CoV-2.
Initial studies have indicated lopinavir, ritonavir, nelfinavir, antimalarial drug chloroquine, hydroxychloroquine and many other antiviral drugs, which have been screened as potential inhibitors against SARS-CoV-2 (Jin et al., 2020; Pant et al., 2020; Wang et al., 2020). Bioactive natural compounds have diverse bioavailability, with relatively less non-toxic property. In the bioactive natural compounds, various phytoconstituents like phenols, steroids, flavonoid molecules are present, that have been reported and screened for their possible therapeutic effects against various viral diseases including COVID-19 (Bhardwaj et al., 2020; Das et al., 2020; A. Kumar et al., 2020; Thuy et al., 2020). Natural compounds such as crocin, digitoxigenin, β-eudesmol, withanone, caffeic acid phenethyl ester and other various bioactive molecules have also been reported to interact with SARS-CoV-2 main protease (Aanouz et al., 2020; Bhardwaj et al., 2020; Gyebi et al., 2020; Islam et al., 2020; V. Kumar et al., 2020).
Thus, in the present pandemic situation, there is an urgent need to investigate bioactive compounds which have the potency to combat this viral infection and provide immunity and strength to our body to combat SARS-CoV-2. Indian medicinal plants have been widely used for various diseases to develop drugs as well as afford strength to our immune system. There are countless benefits of herbal medicine such as naturally available, alleviates hormones and metabolism, anti-inflammatory, and the boosting up of the immune system. Perceiving the significance of immunity boosting actions during the COVID-19 situation, it is very vital to consume supplements in the form of immune nutrients such as vitamin C, B-complex, zinc and copper that will support our body to fight against the SARS-CoV-2. India’s Ministry of AYUSH has also stated, “Ayurveda’s immunity-boosting measures for self-care during COVID 19 crisis” (Ministry of Ayush, 2020). Withania somnifera (Ashwagandha) is one of the most valued Indian ayurvedic medicinal plants that are revered as a biologically active immunomodulator and its ability to balance, energise, rejuvenate, and revitalise (Vetvicka & Vetvickova, 2011). Ashwagandha comprises a rich source of various phytoconstituents such as, Withaferin A, steroidal lactones of the withanolide series, steroidal alkaloids of the withanoside series and some other compounds (Glotter et al., 1973, 1966; Hirayama et al., 1982; Matsuda et al., 2001; Tong et al., 2011). Active constituents of Ashwagandha are known to have a promising anti-influenza activity, against chikungunya, inhibit infectious Bursal Disease virus and are beneficial for the treatment of genital disease caused by Herpes Simplex Virus among African tribes, etc. (Cai et al., 2015; Ganguly et al., 2018; Jain et al., 2018).
In the current study, the aim is to explore the medicinal potential of Withania somnifera against the main protease of SARS-CoV-2 by adopting computational approaches. The data generated is very encouraging and suggests that Withania somnifera has the capability of being effective in the treatment of SARS-CoV-2.
2. Materials and methods
2.1. Compilation of dataset used as ligands
The 40 natural chemical constituents of Withania somnifera (Ashwagandha) were obtained from reported literature and used as ligand dataset for this study (Table 1). The 2D structures of these identified compounds are shown in supplementary table (Table S1). Further, these identified natural compounds were retrieved from PubChem database (https://pubchem.ncbi.nlm.nih.gov/) as .sdf format and imported into the maestro module. Ligprep module was used for the preparation and optimization of these ligands (Schrödinger, 2018) and prepared ligands were used for further studies.
Table 1.
Natural chemical constituents of Withania somnifera (Ashwagandha).
| S.No. | Compound Name | PubChem CID Number | References |
|---|---|---|---|
| 1. | 17alpha-hydroxywithanolide D | 23266161 | (Abraham et al., 1975) |
| 2. | 2,3-Dehydrosomnifericin | 70684083 | (LLanos et al., 2012) |
| 3. | 24,25-dihydrowithanolide D | 23266167 | (Nakano et al., 2013) |
| 4. | 27-Deoxy-14-hydroxywithaferin A | 23266158 | (Glotter et al., 1966) |
| 5. | 27-Deoxywithaferin A | 23266155 | (Hirayama et al., 1982) |
| 6. | 27-Hydroxywithanolide B | 15858981 | (Chaurasiya et al., 2008) |
| 7. | Anaferine | 443143 | (Remya et al., 2016) |
| 8. | Ashwagandhanolide | 16099532 | (Subbaraju et al., 2006) |
| 9. | Beta.-Amyrin | 225689 | (Abou-Douh, 2002) |
| 10. | Coagulin Q | 10100411 | (Zhao et al., 2002) |
| 11. | Scopoletin | 5280460 | (Abou-Douh, 2002) |
| 12. | Sitoindoside IX | 189586 | (Jayaprakasam et al., 2003) |
| 13. | Somnifericin | 101687980 | (Singh & Sharma, 2020) |
| 14. | Somniferine | 14106343 | (Dragar & Bick, 1988) |
| 15. | Withaferin A | 265237 | (Lee et al., 2012; Reddy et al., 2015) |
| 16. | Withanolide A | 11294368 | (Kour et al., 2009; Soman et al., 2013) |
| 17. | Withanolide B | 14236711 | (Turrini et al., 2016) |
| 18. | Withanolide C | 101559583 | (Bessalle & Lavie, 1992) |
| 19. | Withanolide D (AC1L4PUZ) | 118701104 | (Mondal et al., 2012) |
| 20. | Withanolide E | 301751 | (Henrich et al., 2015) |
| 21. | withanolide F | 44562999 | (Ben Bakrim et al., 2018) |
| 22. | Withanolide G | 21679023 | (Glotter et al., 1973) |
| 23. | Withanolide J | 21679022 | (Glotter et al., 1973) |
| 24. | Withanolide L | 179575 | (Ahmad & Dar, 2017) |
| 25. | Withanolide M | 25090669 | (Ahmad & Dar, 2017) |
| 26. | Withanolide N | 23266147 | (Cardenas et al., 2012) |
| 27. | Withanolide O | 23266146 | (Cardenas et al., 2012) |
| 28. | Withanolide P | 21679034 | (Ahlawat et al., 2017) |
| 29. | Withanolide Q | 101281365 | (Kirson et al., 1975) |
| 30. | Withanolide R | 101281364 | (Kirson et al., 1975) |
| 31. | Withanolide S | 11049407 | (White et al., 2016) |
| 32. | Withanone | 21679027 | (Dar et al., 2017; Wadegaonkar & Wadegaonkar, 2013) |
| 33. | Withanoside II | 101168811 | (Matsuda et al., 2001) |
| 34. | Withanoside III | 101168810 | (Matsuda et al., 2001) |
| 35. | Withanoside IV | 71312551 | (Kuboyama et al., 2006) |
| 36. | Withanoside V | 10700345 | (Singh et al., 2016) |
| 37. | WithanosideVIII | 101168805 | (Zhao et al., 2002) |
| 38. | Withanoside X | 101168807 | (Tong et al., 2011) |
| 39. | Withanoside XI | 10952344 | (Zhao et al., 2002) |
| 40. | Withasomnine | 442877 | (Schröter et al., 1966) |
2.2. Protein preparation and grid generation
The glide module of Schrödinger Maestro was used to perform the molecular docking study to analyse the binding mode of identified phyto-compounds of Withania somnifera (Ashwagandha) into the active site of main protease SARS-CoV-2. The crystal structure of SARS-CoV-2 main protease was obtained from the protein data bank (PDB ID:6LU7) (Jin et al., 2020). The crystal structure was further pre-processed, refined, optimized and minimized by the protein preparation wizard module. The missing side chain and loops in the crystal structure were included using the Prime module. Water molecules in the active site more than 5 Å from the hetero atoms including ligands were deleted. Further, the crystal structure was optimized by PROPKA method and minimized by using the OPLS3 force field (Jorgensen et al., 1996). The receptor grid generation module was used for the generation of a grid around the centroid of co-crystallized ligand (N3). Re-docking of the co-crystallized ligand in the active site of main protease of SARS-CoV-2 was performed to validate the docking protocol. The docked complex was superimposed and aligned with the original crystal structure by employing the superimposing tool to calculate the RMSD value. The generated grid was used to perform the molecular docking with the prepared ligands of Ashwagandha obtained from the ligprep module. After docking, the binding affinity of the docked complex was predicted on the basis of the glide score and results analyzed using glide XP visualizer.
2.3. Molecular dynamics simulation
The classical molecular dynamics simulation was carried out for the docked complexes using the Desmond module of Schrodinger suite to affirm the stability and binding interaction of these docked complexes (Bowers et al., 2006). The docked complexes were solvated using TI3P solvent model (specifies a three-site rigid water molecule with charges) in an orthorhombic box of 15 × 15 × 15 Å3 size. For neutralizing the system, appropriate cation (Na+) or anion (Cl-) along with a salt concentration of 0.15 mol/L were added. The prepared system was energy minimized using steepest descent conjugate gradient method, where maximum interaction was set to 2000, and the convergence threshold was set to 1.0 kcal/mol/Å. Prior to a production run, the NPT simulation were performed at 300 K temperature set by Nose-Hoover thermostat and 1.01325 bar pressure to relax the complexes (Kaczor et al., 2015). Finally, a production run of 100 ns was performed and trajectory was recorded at every 20 ps time. After the completion of simulation obtained trajectories were analysed by using various parameters of MD simulation, root mean square deviation (RMSD), protein root mean square fluctuation (RMSF) and protein ligand (PL) contacts with the help of simulation interaction analysis module of Desmond.
2.4. Binding free energy calculation (MM-GBSA)
Molecular mechanics generalized Born surface area (MM-GBSA) calculations are widely performed in computational drug discovery process to estimate the binding free energy prediction of protein-ligand complexes. A total of 100 frames of protein ligand conformations spanned between 60 to 100 ns simulation run were extracted for binding energy calculation. The prime module of Schrodinger was used to perform MM-GBSA calculations (Hou et al., 2011). The binding energy calculation (ΔG bind) occurs as per the below equation
where Gcomplex, Greceptor and Gligand represent the free energies of the complex, receptor and ligand respectively.
3. Results and discussion
The reported literature was utilized to identify 40 phyto-compounds of Ashwagandha, and a database was created of these compounds by downloading their structures from PubChem database. All the identified compounds were docked in the active site of main protease of SARS-CoV-2. Molecular docking yielded four compounds which were selected on the basis of their docking score that was higher than the co-crystallized ligand N3 (-8.12 Kcal/mole). The docking score of the co-crystallized ligand N3 subsequently served as a control in this study. The docking scores of all the remaining compounds are provided in Supplementary material Table S1.
3.1. Molecular docking study
The main protease of SARS-COV-2 is a cysteine protease and is a homodimeric protein where each monomer is designated as a promoter. Each promoter comprises three domains each, domain I, domain II and domain III containing residues 8–101, residues 102–184, and residues 201–303 respectively. Domain II is connected to domain III through along loop from by residues 185 to 200 (Wu et al., 2020). The residues 1 to 7 comprise the N-finger wherein the N-finger of one promoter (Promoter A) sits in the pocket formed in second promoter (Promoter B). The active site dyad amino acid residues, Cys-145 and His-41, are located between the gap formed between domain I and domain II. The binding site is surrounded by hydrophilic as well as hydrophobic amino acid residues with two negatively charged residues, Glu-166 and Asp-187, and one positively charged residue, Arg-188 (Jin et al., 2020; Yang et al., 2003). The hydrophilic and hydrophobic amino acid surrounding the binding site comprises Thr-24, Leu-27, His-41, Met-49, Asn-142, His-163, His-164, Phe-140, Met-145, Gln-189, Pro-168 and Cys-145. The main protease protein is typically conserved amongst the various CoVs, and the absence of the protein in the humans makes it an attractive target for drug design.
The initial docking protocol was validated by cross-docking the inhibitor N3 into the binding site of the protein, main protease of SARS-CoV-2 (PDB ID:6LU7). The docking protocol revealed a root mean square deviation (RMSD) of 1.0 Å between the positions of the ligand N3 observed in the crystal structure and the docked ligand. Subsequently, molecular docking study was performed within the region of main protease protein where the inhibitor bound with the validated grid. The obtained results after the docking study revealed that from all the selected compounds used in the present study, four compounds, Withanoside II (PubChem CID-101168811), Withanoside IV (PubChem CID-71312551) Withanoside V (PubChem CID-10700345) and Sitoindoside IX (PubChem CID-189586) possessed a higher docked score with a range of −8.37 to −11.30 kcal/mole as compared to the standard compound N3 whose docked score was found to −8.12 kcal/mole. The structures of these identified compounds are given in Table 2.
Table 2.
Chemical Structures of selected compounds.
| S.No. | Compound Name | 2D Structure |
|---|---|---|
| 1 | Withanoside II (PubChem CID-101168811) |
 |
| 2 | Withanoside IV (PubChem CID-71312551) |
 |
| 3 | Withanoside V (PubChem CID-10700345) |
 |
| 4 | Sitoindoside IX (PubChem CID -189586) |
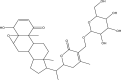 |
3.1.1. Binding mode analysis of withanoside II
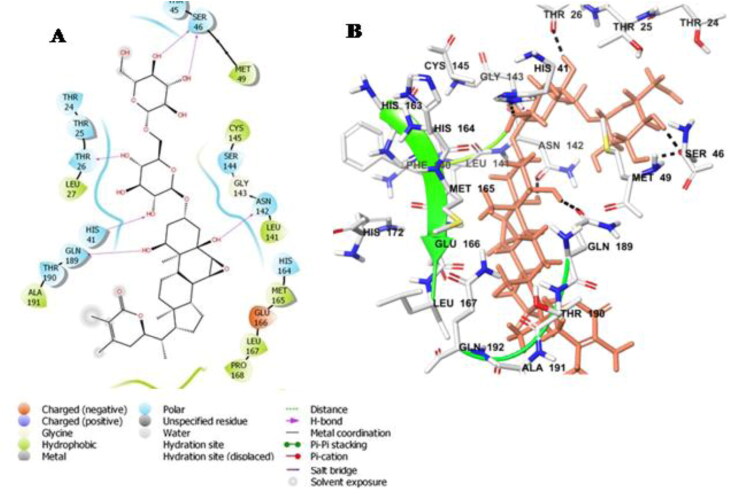
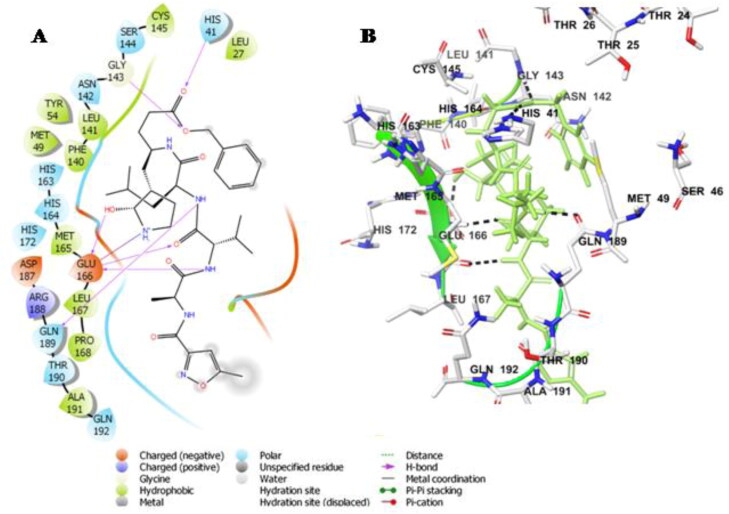
The ligand Withanoside II interacts with the catalytic dyad amino acid residues, His-41 and Cys-145 with a docking score of −11.30 Kcal/mole (Table 3). Withanoside II forms hydrogen bond interaction with His-41 and hydrophobic interaction with Cys-145 amino acid residues. Further, interaction analysis revealed that the compound formed hydrogen bond with Thr-26, Ser-46, Asn-142 and Gln-189 amino acid residues. Withanoside II also interacts through hydrophobic interactions with residues Met-49, Cys-145, Met-165, Pro-168 and charged interaction with His-164, Glu-166 amino acid residues (Figure 1A,B).
Table 3.
Results of molecular docking, interacting residues and binding free energies.
| S.No | Compound Name | Glide G Score (Kcal/mol) | Interacting Residues |
|---|---|---|---|
| 1 | Withanoside II (PubChem CID-101168811) |
−11.30 | Thr-24c, Thr-25c, Thr-26cd, Leu-27b, His-41c,d, Thr-45c, Ser-46cd, Met-49b, Leu-141b, Asn-142c,d, Ser-144c, Cys-145b,His-164c, Met-165b, Glu-166a, Leu-167b, Pro-168b, Gln-189c,d, Thr-190c, Ala-191b, |
| 2 | Withanoside IV (PubChemCID-71312551) |
−11.02 | Thr-25c, Thr-26c,d, Leu-27b, His-41c, Met-49b, Leu-141b,d, Asn-142c, Ser-144c, Cys-145b,His-163c, His-164c, Met-165b, Glu-166a, Leu-167b, Pro-168b, Arg-188a, Gln-189c,d, Thr-190c,d, Ala-191b, Gln-192c,d |
| 3 | Withanoside V (PubChem CID-10700345) |
−8.96 | Thr-24c,d, Thr-25c,d, Thr-26c,d, Leu-27b, His-41c, Cys-44b, Thr-45c, Ser-46c, Met-49b, Leu-50b, Phe-140b, Leu-141b, Asn-142c, Gly-143d, Ser-144c, Cys-145b,His-163c,d, His-164c, Met-165b, Glu-166a,d, Leu-167b, Pro-168b, Arg-188a, Gln-189c, Thr-190c, Ala-191b, Gln-192c |
| 4 | Sitoindoside IX (PubChem CID -189586) |
−8.37 | Thr-24c,d, Thr-25c, Thr-26c,d, Leu-27b, His-41c, Cys-44b, Thr-45c, Ser-46c, Met-49b, Leu-141b, Asn-142c, Gly-143d, Ser-144c, Cys-145b,d, His-163c, His-164c, Met-165b, Glu-166a, Leu-167b, Pro-168b, Arg-188a, Gln-189c, Thr-190c, Ala-191b, Gln-192c |
| 5 | N3 (Control) (PDB ID: 6LU7) |
−8.12 | Leu-27b, His-41c,d, Met-49b, Phe-140b,Leu-141b, Asn-142c, Gly-143d, Ser-144c, Cys-145b, His-163c, His-164c, Met-165b, Glu-166a,d, Leu-167b, Pro-168b, His-172c His-163c Asp-187a, Arg-188a Gln-189c,d, Thr-190c, Ala-191b, Gln-192c |
Superscript: a-Charged Interaction; b-Hydrophobic Interaction; c-Polar Interaction; d-Hydrogen Bond
Figure 1.
Molecular docking simulations analysis on Mpro SARS-CoV-2 complex with Withanoside II (A) Schematic representation of Withanoside II complex with Mpro SARS-CoV-2 (B) Withanoside II (stick, pink) complexed to the protein. The interacting residues of protein are in grey (stick) with the hydrogen bonded interactions being represented by black dashed lines.
3.1.2. Binding mode analysis of withanoside IV
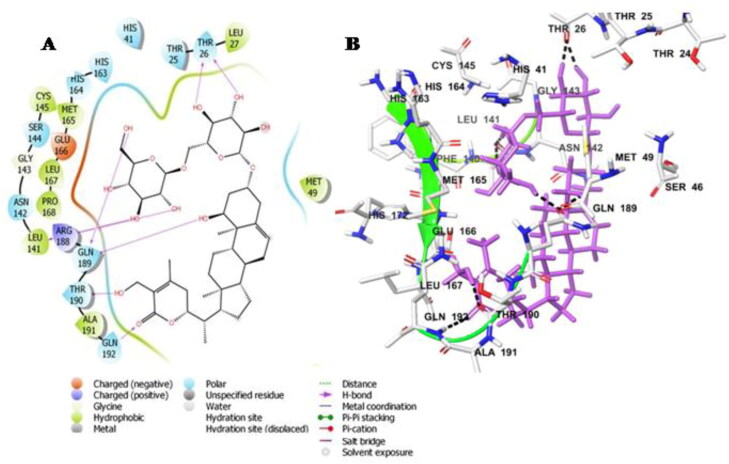
Withanoside IV docked into the protein active site and it formed interactions with the catalytic dyad residues His-41 and Cys-145 with a docking score of −11.02 Kcal/mole (Table 3). It is also involved in hydrogen bonded interaction with the amino acid residues Thr-26, Leu-141, Gln-189, Thr-190 and Gln-192 as well as a number of hydrophobic interactions with Met-49, Pro-168, Leu-167, Met-165, Leu-27. Withanoside IV was further stabilized through polar interaction with amino acid residues His-41, His-163, His-164, Asn-142 and Glu-192, respectively (Figure 2A,B).
Figure 2.
Molecular docking simulations analysis on Mpro SARS-CoV-2 complex with Withanoside IV (A) Schematic representation of Withanoside IV complex with Mpro SARS-CoV-2 (B) Withanoside IV (stick, magenta) complexed to the protein. The interacting residues of protein are in grey (stick) with the hydrogen bonded interactions being represented by black dashed lines.
3.1.3. Binding mode analysis of withanoside V
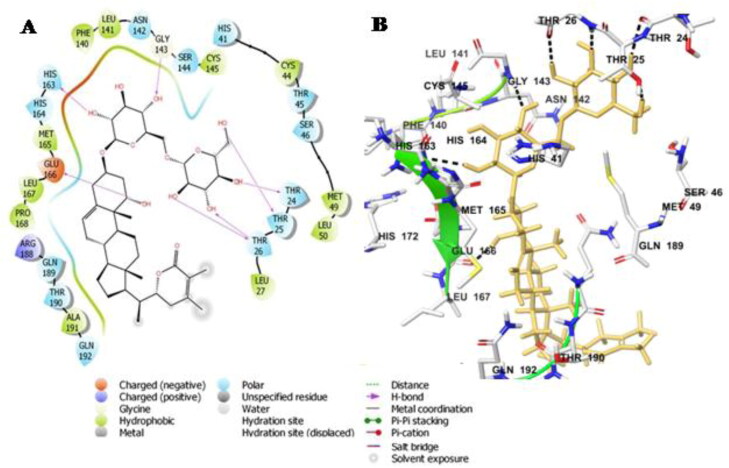
Like Withanoside II and IV, Withanoside V also interacted with the active site dyad amino acid residues His-41 and Cys-145 present between gap of the two domains, domain I and domain II. The docking score of this docked complex was found to be −8.96 Kcal/mole (Table 3). The docked protein-ligand complex was stabilised through hydrogen bonding interactions with residues Thr-24, Thr-25, Thr-26, Gly-143, His-163, Glu-166. A number of hydrophobic interaction were observed between the ligand and protein comprising residue Met-49, Cys-44, Leu-141, Phe-140, Met-165, Leu-167 and Pro-168, respectively (Figure 3A,B).
Figure 3.
Molecular docking simulations analysis on Mpro SARS-CoV-2 complex with Withanoside V(A) Schematic representation of Withanoside V complex with Mpro SARS-CoV-2 (B) Withanoside V (stick, yellow) complexed to the protein. The interacting residues of protein are in grey (stick) with the hydrogen bonded interactions are indicated as black dashed lines.
3.1.4. Binding mode analysis of sitoindoside IX
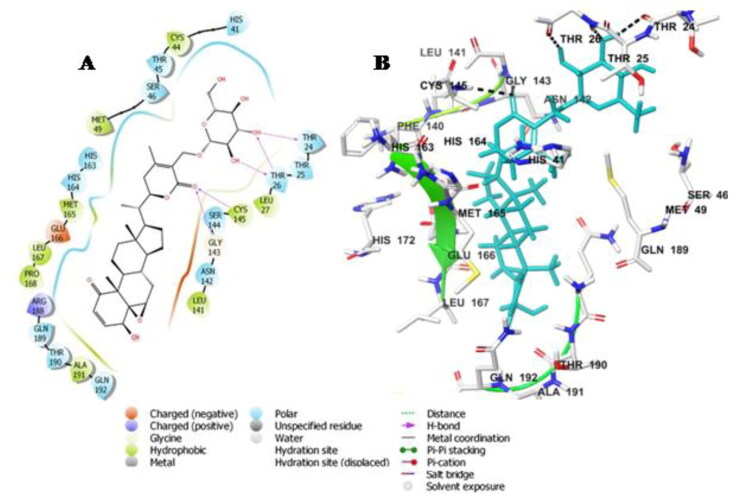
The docking energy of Sitoindoside IX-protease docked complex was −8.37 Kcal/mole (Table 3). Sitoindoside IX disclosed polar interaction with His-41 and hydrogen bonding interaction with Cys-145 amino acid of the catalytic dyad residues. Sitoindoside IX showed hydrogen bond interaction with Thr-24, Thr-26, Gly-143 and Cys-145 amino acid residues. The ligand formed hydrophobic interactions with amino acid residues Leu-27, Met-49, Met-165, Leu-167 and Pro-168 and polar interaction with Thr-24, His-163, His-164, Asn-142, Gln-189, Glu-166 and Arg-166 in the substrate binding region (Figure 4A,B).
Figure 4.
Molecular docking simulations analysis on Mpro SARS-CoV-2complex with Sitoindoside IX (A) Schematic representation of Sitoindoside IX complex with Mpro SARS-CoV-2 (B) Sitoindoside IX (stick, cyan) complexed to the protein. The interacting residues of protein are in grey (stick) with the hydrogen bonded interactions are represented by black dashed lines.
From docking studies, we infer that the residues of the catalytic dyad Cys-145 and His-41 together with other amino acid residues Met-49, Asn-142, His-163, His-164, Met-165, Glu-166, Gln-189, Asp-187, Arg-188 have the highest binding capability for the selected compounds. This is comparable to the binding observed of the co-crystallized ligand N3 with the protein main protease protein (Figure 5A,B).
Figure 5.
Molecular docking simulations analysis on Mpro SARS-CoV-2 complex with N3 compound (A) Schematic representation of N3 complex with Mpro SARS-CoV-2 (B) N3 (stick, green) complexed to the protein. The interacting residues of protein are in grey (stick) with the hydrogen bonded interactions are represented by black dashed lines. The Pi-Pi stacking interaction is not indicated.
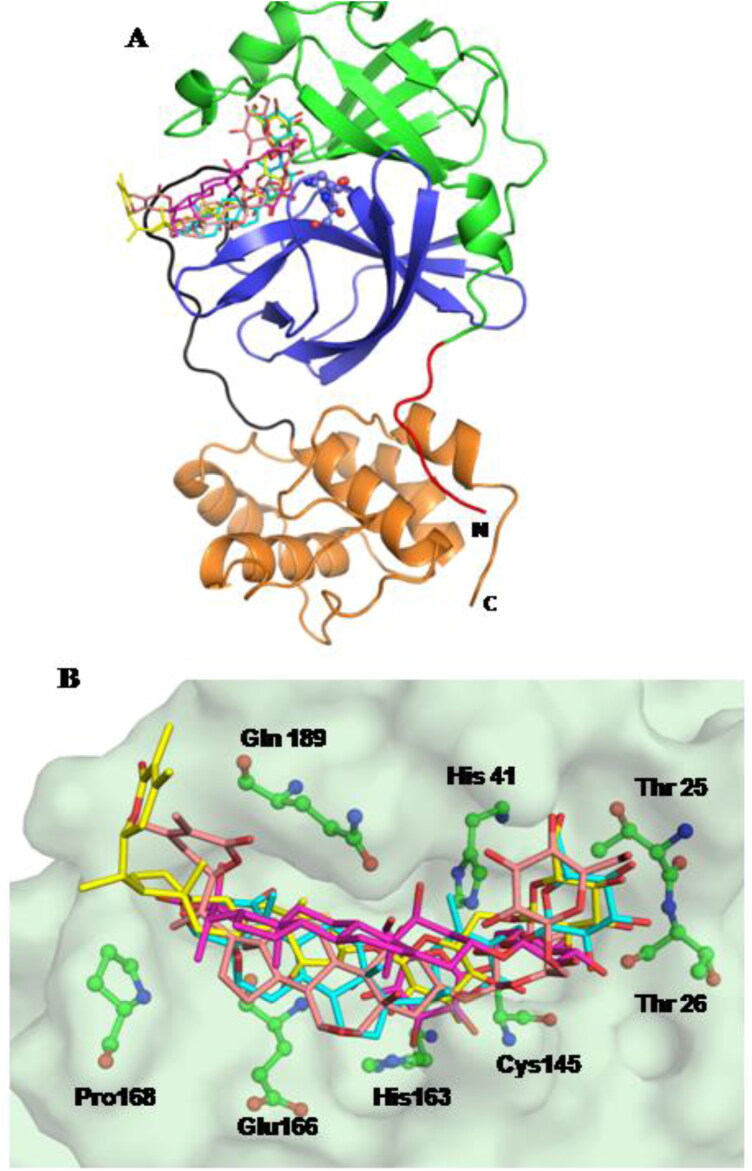
The molecular docking studies pinpoint that these are the most significant residues required for ligand binding situated in the substrate-binding region of the main protease enzyme. The compounds bind with the catalytic dyad residues and occupy the substrate binding region. A superposition of all compounds indicates that they adopt a similar mode of binding except for compound Withanoside IV (Figure 6A,B).
Figure 6.
(A) Three-dimensional structure of MPro protein indicating the various domains: Domain I (green), Domain II (blue) and Domain III (mustard) with loop connecting domain II to domain III (black). All the compounds bind in the substrate binding region; Compounds: Withanoside II (pink); Withanoside IV (magenta); Withanoside V (yellow); Sitoindoside IX (cyan) (B) Surface representation of the protein. The compounds (stick) are superimposed in the binding site with the key interacting residues in green (ball-and-stick).
3.2. Molecular dynamics simulation study
Further, we performed MD simulation study of 100 ns time scale of all these selected docked complex (Withanoside II, Withanoside IV, Withanoside V, Sitoindoside IX) and co-crystallized ligand N3 to check their stability in the active site of main protease SARS-COV-2 protein. The results obtained after the simulation were analysed from the generated equilibrium trajectories by using the various protocol of MD simulation, i.e. root mean square deviation (RMSD), root mean square fluctuations (RMSF) and interaction analysis of hydrogen bond and their stability during the simulation period.
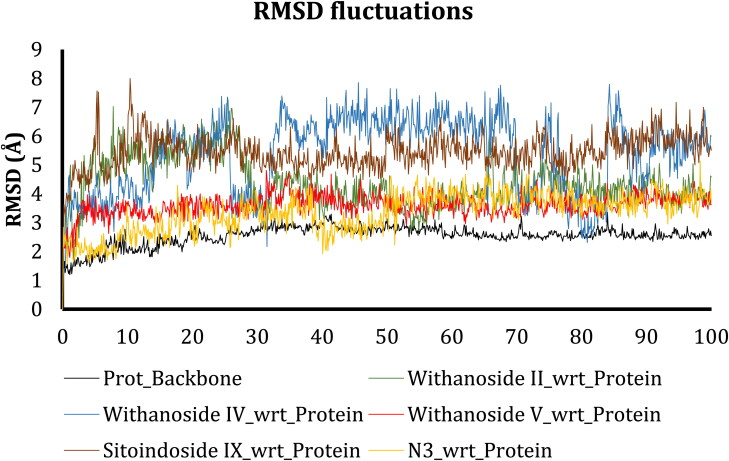
Root mean square deviation measures the stability of the simulation system and calculates its conformational perturbations which occur in the protein backbone during the simulation time (Sargsyan et al., 2017). After the simulation, obtained RMSD results were plotted against the simulation time for the analysis of results (Figure 7).
Figure 7.
RMSD fluctuations of protein backbone (black), Withanoside II (Green), Withanoside IV (Blue), Withanoside V (Red), Sitoindoside IX (Brown), N3 (Yellow) during 100 ns simulation runs on Mpro Protein.
From the RMSD graph, we predict that the protein backbone of main protease was stable during the entire simulation time with a mean value of 2 Å. These lower fluctuations indicate the stable behaviour of protein backbone during the entire simulation. Comparing the docked complex RMSD trajectories, we inferred that Withanoside II, Withanoside IV and Sitoindoside IX manifested fluctuations initially up to 30 ns time scale and after 30 ns the compound Withanoside II did not reveal any significant RMSD fluctuations as compared to the docked complex trajectories of the other compounds. The Withanoside V docked complex trajectory was comparatively more stable with a mean value of 3 Å as compared to the trajectories of other docked complexes. Moreover, the trajectory of the Withanoside V docked complex did not indicate any fluctuating behaviour; this indicates its stable complex formation and least conformational changes during simulation. Thus, from the RMSD trajectory analysis of all docked complexes, we observed that Withanoside V trajectory is steady throughout the simulation and changes in backbone RMSD are within the acceptable range.
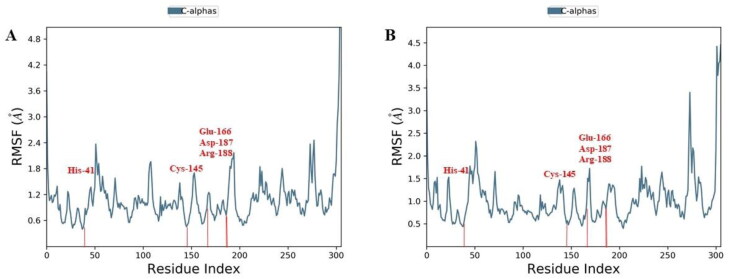
Further, to understand the residue wise fluctuations in the docked complexes, root mean square fluctuations (RMSF) analysis was performed. The RMSF behaviour of all the docked complexes was generated from the stable trajectory behaviour and plotted against the amino acid residue in x-axis vs RMSF(Å) in the y-axis. The RMSF plot for Withanoside V, yielded the least fluctuations at the catalytic active site dyad, His-41 and Cys-145, as well as the substrate-binding pocket residues Glu-166, Asp-187 and Arg-188 with flexibility range of 0.6 to 3.0 Å (Figure 8).
Figure 8.
Comparative RMSF of (A) Withanoside V (B) N3 compound in the protein-ligand complex throughout the simulations period of 100 ns.
The RMSF pattern of Withanoside V is found to be comparable to the co-crystalized ligand N3. The other selected compounds (Withanoside II, Withanoside IV and Sitoindoside IX) demonstrated higher fluctuations behaviour (Supplementary material Figure S1).
The protein-ligand interaction analysis was also assessed for all the docked complexes formed during the simulation time to check the stability of docked complexes. Thus, the hydrogen-bonding pattern of all the identified hits was evaluated and plotted against the simulation time scale along with the percentage interaction of active site residues. The interaction pattern of Withanoside II obtained from MD simulation, divulged hydrogen bonding interaction with His-41 amino acid residue apart from Cys-145 in the catalytic dyad region. The ligand-protein contact analysis revealed that catalytic dyad residue Cys-145 formed a hydrogen bond through a solvated molecule with the main protease protein residues. The histogram representation of interaction fraction and timeline of percentage index is represented in Supplementary material Figure S2. Further, analysis of result at the substrate binding region revealed, interactions with residues Asn-142, Glu-166 and Gln-189. The RMSD trajectory analysis of Withanoside II was also not found to be stable.
The interaction pattern of the selected compound, Withanoside IV obtained from MD simulation indicated hydrogen bonding interactions with the catalytic dyad amino acid residues His-41 and Cys-145 wherein the interaction with Cys-145 residue was maintained during the entire simulation time. The RMSD pattern of this docked complex showed greater fluctuations as compared to all the other docked complexes. Withanoside IV like Withanoside II possessed interactions with the specific substrate-binding region amino acid residues Asn-142, Glu-166 and Gln-189 (Supplementary material Figure S3).
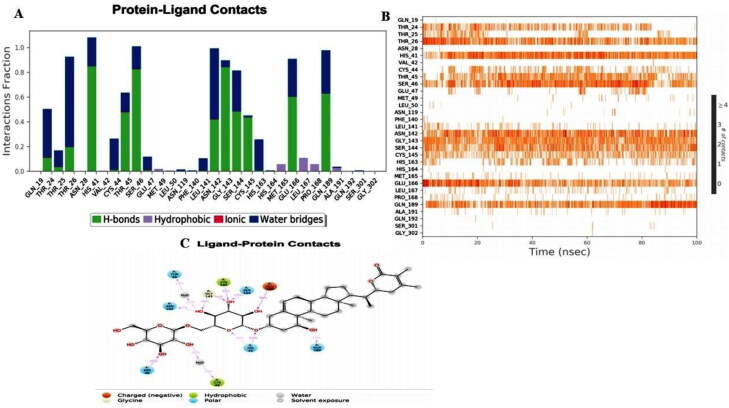
The MD simulation interaction analysis of Withanoside V with main protease protein was also evaluated and plotted as a percentage index of total contacts during the simulation time. The hydrogen bond interactions with catalytic dyad residues His-41 and Cys-145 were seen to be retained during the entire simulation with more than 53% and 40% simulation time. The polar interaction with His-41 and hydrophobic interaction with Cys-145 amino acid residues formed in the docked complex were also maintained throughout the simulation. Moreover, a stable RMSD behaviour of its trajectory was observed during simulation indicating the stability of this protein-ligand complex in the active site. The histogram, in combination with the 2D simulation interaction diagram between Withanoside V with main protease protein is illustrated in Figure 9. The interaction analysis revealed that in addition to the stable interactions present at the catalytic dyad interaction, substrate binding site residues Thr-24, Asn-142, His-163, Glu-166, Gln-189 were also observed to be stable during the entire simulation time.
Figure 9.
Molecular dynamics interaction analysis of Withanoside V docked complex. (A) Histogram showing interaction fractions with active amino acid residues; (B) Timeline representation showing interaction with all the amino acid residues at each time frame; (C) Schematic representation of ligand indicating percentage interactions with active site residues.
The MD simulation of Sitoindoside IX- protein complex revealed that the interaction with His-41 in the catalytic dyad region was not adequately retained during the simulation. The histogram in combination with the 2D simulation interaction diagram indicated that the interactions present at the substrate binding region with residues Asn-142 and Glu-166 were not stable during the entire simulation scale (Supplementary material Figure S4).
The interaction pattern for the co-crystallized ligand N3, obtained from MD simulation showcased hydrogen bonding interactions with catalytic dyad amino acid residues His-41 and Cys-145. However, its interaction with Cys-145 residue was not observed during the entire simulation time. The histogram representation of interaction fraction illustrated interactions with the residues in the specific substrate-binding region, Asn-142, Glu-166 and Gln-189 (Supplementary material Figure S5).
3.3. Binding free energy calculation
The binding free energy calculation was carried out for all the systems (Withanoside II, Withanoside IV, Withanoside V, Sitoindoside IX) along with the co-crystallized ligand N3. The prime MM/GBSA (molecular mechanics energies combined with the generalized Born and surface area continuum solvation) free energy calculation is widely accepted and commonly used to estimate the ligand-binding affinities in the protein systems. To evaluate the binding free energy, 100 conformations that spanned between the 60 to 100 ns time interval of the MD production run were extracted.
The MM-GBSA ΔG bind score obtained after the calculations is provided in Table 4. The calculated binding free energy of these compounds Withanoside II (-62.50 ± 5.25 Kcal/mole), Withanoside IV (-81.29 ± 4.78 Kcal/mole), Withanoside V (-87.01 ± 5.01 Kcal/mole), Sitoindoside IX (-49.90 ± 4.15 Kcal/mole) and for the co-crystallized ligand (-86.60 ± 6.16 Kcal/mole) were then determined. The negative values of binding energy indicate that all the hit compounds interact favourably with the receptor. These results clearly suggest that Withanoside V possessed the maximum negative binding energy (-87.01 ± 5.01 Kcal/mol) as compared to the other selected compounds. The binding energy of Withanoside II, and Sitoindoside IX was not found to be comparable with the co-crystallized ligand N3. Thus, Withanoside V with the least binding energy and better binding affinity against the target could consequently prove to be a potential inhibitor of main protease of SARS-CoV-2.
Table 4.
The computed (MM-GBSA) binding free energies (ΔGbind) of the selected compounds against Mpro enzymes.
| S.No | Compound Name | MM/GBSA (Kcal/mol) |
|---|---|---|
| 1 | Withanoside II (PubChem CID-101168811) |
−62.50 ± 5.25 |
| 2 | Withanoside IV (PubChemCID-71312551) |
−81.29 ± 4.78 |
| 3 | Withanoside V (PubChem CID-10700345) |
−87.01 ± 5.01 |
| 4 | Sitoindoside IX (PubChem CID -189586) |
−49.90 ± 4.15 |
| 5 | N3 (Control) (PDB ID: 6LU7) |
−86.60 ± 6.16 |
The comparison of the interaction patterns of all the selected hit compounds pinpoints Withanoside V, as the compound of choice as it revealed stable interactions with both the residues of catalytic dyad (His-41 and Cys-145) in main protease of SARS-CoV-2. Though Withanoside IV exhibited stable interactions with the catalytic dyad residues, however, its RMSD behaviour was not within the acceptable range as it presented higher fluctuation and its binding energy was also found to be −81.29 ± 4.78 Kcal/mole. In contrast, Withanoside V displayed stable interactions with the substrate binding site amino acid residues, Thr-24, Asn-142, His-163, Glu-166, Gln-189 throughout the MD simulation. The co-crystallized ligand N3 did not retain the interaction with Cys-145 throughout the simulation but its interaction with substrate binding regions was present throughout. The residue Glu-166 in main protease is known to be involved in the formation of its functional dimeric form (Anand et al., 2003). The ability of Withanoside V to bind to the catalytic dyad along with the substrate binding region which houses the Glu-166 residues prevents the dimerization of main protease as the residue Glu-166 of one monomer/promoter is unable to interact with the N-finger residues of other monomer. The MM-GBSA binding energy of Withanoside V was also found to be most negative, −87.01 ± 5.01 Kcal/mole, compared to other selected compounds. Thus, based on these observations, we conclude that Withanoside V present in Withania somnifera (Ashwagandha) can be a potent and efficacious biomolecule which interacts with SARS-CoV-2 and can be further explored as a drug candidate.
4. Conclusion
The molecular docking and dynamics simulation approach was applied for the natural chemical constituents of Withania somnifera (Ashwagandha) to identify their potential against the main protease protein of SARS-CoV-2, a pandemic COVID-19 that is presently wreaking havoc across the world. Our study suggested that out of the forty constituents of Ashwagandha, Withanoside V was the most potent natural inhibitor. Moreover, the RMSD trajectories of Withanoside V exhibited a stable behaviour with comparatively lesser fluctuations as compared to other ligands. The ligand displayed stable interactions with the catalytic dyad residues (His-41 and Cys-145) of main protease as well the substrate binding region which promotes the dimerization of the protein. Further, the MM-GBSA score of Withanoside V presented the maximum negative binding energy (-87.01 ± 5.01 Kcal/mol). Mpro promotes activation for the polyactive site of main protease, which is important for the replication of the SARS-CoV-2. Hence, our study concludes that Withanoside V which is present in the plant Withania somnifera is a potent inhibitor of main protease of SARS-CoV-2. The status of Withania somnifera as an immunity enhancer, anti-diabetic agent and blood pressure and hormonal regulation is well known. The present study suggests that additionally, the Ayurveda herb Ashwagandha could prove to be an alternative to available treatments of COVID-19. Moreover, the backbone structure of Withanoside V could also be further exploited to develop more potent inhibitors of SARS-CoV-2 Mpro. However, further in vitro and in vivo studies are necessary for validation of this compound to combat COVID-19.
Supplementary Material
Disclosure statement
The authors have no conflict of interest regarding the publication of this article.
References
- Aanouz, I., Belhassan, A., El-Khatabi, K., Lakhlifi, T., El-Ldrissi, M., & Bouachrine, M. (2020). Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: Computational investigations. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1758790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou-Douh, A. M. (2002). New withanolides and other constituents from the fruit of Withania somnifera. Archiv Der Pharmazie, 335(6), 267–276. [DOI] [PubMed] [Google Scholar]
- Abraham, A., Kirson, I., Lavie, D., & Glotte, E. (1975). The withanolides of Withania somnifera chemotypes I and II. Phytochemistry, 14(1), 189–194. 10.1016/0031-9422(75)85035-7 [DOI] [Google Scholar]
- Ahlawat, S., Saxena, P., Ali, A., Khan, S., & Abdin, M. Z. (2017). Comparative study of withanolide production and the related transcriptional responses of biosynthetic genes in fungi elicited cell suspension culture of Withania somnifera in shake flask and bioreactor. Plant Physiology and Biochemistry: PPB, 114, 19–28. 10.1016/j.plaphy.2017.02.013 [DOI] [PubMed] [Google Scholar]
- Ahmad, M., & Dar, N. J. (2017). 8 – Withania somnifera: Ethnobotany, pharmacology, and therapeutic functions. In: Bagchi, D. B. T.-S. E. for E. H. F. and A. (Ed.), Academic Press, pp. 137–154. 10.1016/B978-0-12-805413-0.00008-9 [DOI]
- Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R., & Hilgenfeld, R. (2003). Coronavirus main proteinase (3CLPro) structure: Basis for design of anti-SARS drugs. Science, 300(5626), 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Ben Bakrim, W., El Bouzidi, L., Nuzillard, J.-M., Cretton, S., Saraux, N., Monteillier, A., Christen, P., Cuendet, M., & Bekkouche, K. (2018). Bioactive metabolites from the leaves of Withania adpressa. Pharmaceutical Biology, 56(1), 505–510. 10.1080/13880209.2018.1499781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessalle, R., & Lavie, D. (1992). Withanolide C, A chlorinated withanolide from Withania somnifera. Phytochemistry, 31(10), 3648–3651. 10.1016/0031-9422(92)83749-O [DOI] [Google Scholar]
- Bhardwaj, V. K., Singh, R., Sharma, J., Rajendran, V., Purohit, R., & Kumar, S. (2020). Identification of bioactive molecules from Tea plant as SARS-CoV-2 main protease inhibitors. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1766572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, K. J., Chow, D. E., Xu, H., Dror, R. O., Eastwood, M. P., Gregersen, B. A., Klepeis, J. L., Kolossvary, I., Moraes, M. A., Sacerdoti, F. D., Salmon, J. K., Shan, Y., & Shaw, D. E. (2006). Scalable algorithms for molecular dynamics simulations on commodity [Paper presentation]. SC ’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, p. 43. 10.1109/SC.2006.54 [DOI] [Google Scholar]
- Cai, Z., Zhang, G., Tang, B., Liu, Y., Fu, X., & Zhang, X. (2015). Promising anti-influenza properties of active constituent of Withania somnifera ayurvedic herb in targeting neuraminidase of H1N1 influenza: Computational study. Cell Biochemistry and Biophysics, 72(3), 727–739. 10.1007/s12013-015-0524-9 [DOI] [PubMed] [Google Scholar]
- Cardenas, J., Esquivel, B., Gupta, M., Rray, A. B., & Rodriquez-Hahn, L. (2012). Fortschritte der Chemie organischer Naturstoffe Progress in the Chemistry of Organic Natural Products. Springer. [Google Scholar]
- Chaurasiya, N., Das, Uniyal, G. C., Lal, P., Misra, L., Sangwan, N. S., Tuli, R., & Sangwan, R. S. (2008). Analysis of withanolides in root and leaf of Withania somnifera by HPLC with photodiode array and evaporative light scattering detection. Phytochemical Analysis : Pca, 19(2), 148–154. 10.1002/pca.1029 [DOI] [PubMed] [Google Scholar]
- Dar, N. J., Bhat, J. A., Satti, N. K., Sharma, P. R., Hamid, A., & Ahmad, M. (2017). Withanone, an active constituent from Withania somnifera, affords protection against NMDA-induced excitotoxicity in neuron-like cells. Molecular Neurobiology, 54(7), 5061–5073. 10.1007/s12035-016-0044-7 [DOI] [PubMed] [Google Scholar]
- Das, S., Sarmah, S., Lyndem, S., & Singha Roy, A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragar, C., & Bick, I. R. C. (1988). Somniferine, a novel dimeric opium alkaloid. Tetrahedron Letters, 29(25), 3115–3116. 10.1016/0040-4039(88)85100-1 [DOI] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis. Methods in Molecular Biology, 1282, 1–23. 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly, B., Umapathi, V., & Rastogi, S. K. (2018). Nitric oxide induced by Indian ginseng root extract inhibits Infectious Bursal Disease virus in chicken embryo fibroblasts in vitro. Journal of Animal Science and Technology, 60, 2. 10.1186/s40781-017-0156-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotter, E., Kirson, I., Abraham, A., & Lavie, D. (1973). Constituents of Withania somnifera Dun—XIII: The withanolides of chemotype III. Tetrahedron, 29(10), 1353–1364. 10.1016/S0040-4020(01)83156-2 [DOI] [Google Scholar]
- Glotter, E., Waitman, R., & Lavie, D. (1966). Constituents of Withania somnifera Dun. Part VIII. A new steroidal lactone, 27-deoxy-14-hydroxy-withaferin A. Journal of the Chemical Society C: Organic, 19, 1765–1767. 10.1039/j39660001765 [DOI] [Google Scholar]
- Green, M. S. (2020). Did the hesitancy in declaring COVID-19 a pandemic reflect a need to redefine the term? The Lancet, 395(10229), 1034–1035. 10.1016/S0140-6736(20)30630-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi, G. A., Ogunro, O. B., Adegunloye, A. P., Ogunyemi, O. M., & Afolabi, S. O. (2020). Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): An in silico screening of alkaloids and terpenoids from African medicinal plants. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1764868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich, C. J., Brooks, A. D., Erickson, K. L., Thomas, C. L., Bokesch, H. R., Tewary, P., Thompson, C. R., Pompei, R. J., Gustafson, K. R., McMahon, J. B., & Sayers, T. J. (2015). Withanolide E sensitizes renal carcinoma cells to TRAIL-induced apoptosis by increasing cFLIP degradation. Cell Death & Disease, 6, e1666. 10.1038/cddis.2015.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama, M., Gamoh, K., & Ikekawa, N. (1982). Stereoselective synthesis of withafein A and 27-deoxywithaferin A1. Tetrahedron Letters., 23(45), 4725–4728. 10.1016/S0040-4039(00)85697-X [DOI] [Google Scholar]
- Hou, T., Wang, J., Li, Y., & Wang, W. (2011). Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. Journal of Chemical Information and Modeling, 51(1), 69–82. 10.1021/ci100275a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, R., Parves, M. R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., Hossain, M. N., Ali, M. A., & Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, J., Narayanan, V., Chaturvedi, S., Pai, S., & Sunil, S. (2018). In vivo evaluation of Withania somnifera-based Indian traditional formulation (Amukkara Choornam), against Chikungunya virus-induced morbidity and arthralgia. Journal of Evidence-Based Integrative Medicine, 23, 2156587218757661. 10.1177/2156587218757661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaprakasam, B., Zhang, Y., Seeram, N. P., & Nair, M. G. (2003). Growth inhibition of human tumor cell lines by withanolides from Withania somnifera leaves. Life Sciences, 74(1), 125–132. 10.1016/j.lfs.2003.07.007 [DOI] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Jorgensen, W. L., Maxwell, D. S., & Tirado-Rives, J. (1996). Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. Journal of the American Chemical Society, 118(45), 11225–11236. 10.1021/ja9621760 [DOI] [Google Scholar]
- Kaczor, A. A., Targowska-Duda, K. M., Patel, J. Z., Laitinen, T., Parkkari, T., Adams, Y., Nevalainen, T. J., & Poso, A. (2015). Comparative molecular field analysis and molecular dynamics studies of α/β hydrolase domain containing 6 (ABHD6) inhibitors. Journal of Molecular Modeling, 21(10), 250. 10.1007/s00894-015-2789-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirson, I., Cohen, A., & Abraham, A. (1975). Withanolides Q and R, two new 23-hydroxy-steroidal lactones. Journal of the Chemical Society, Perkin Transactions 1, 1(21), 2136–2138. 10.1039/p19750002136 [DOI] [PubMed] [Google Scholar]
- Kour, K., Pandey, A., Suri, K. A., Satti, N. K., Gupta, K. K., & Bani, S. (2009). Restoration of stress-induced altered T cell function and corresponding cytokines patterns by Withanolide A. International Immunopharmacology, 9(10), 1137–1144. 10.1016/j.intimp.2009.05.011 [DOI] [PubMed] [Google Scholar]
- Kuboyama, T., Tohda, C., & Komatsu, K. (2006). Withanoside IV and its active metabolite, sominone, attenuate Abeta(25-35)-induced neurodegeneration. European Journal of Neuroscience, 23(6), 1417–1426. 10.1111/j.1460-9568.2006.04664.x [DOI] [PubMed] [Google Scholar]
- Kumar, A., Choudhir, G., Shukla, S. K., Sharma, M., Tyagi, P., Bhushan, A., & Rathore, M. (2020). Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1772112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V., Dhanjal, J. K., Kaul, S. C., Wadhwa, R., & Sundar, D. (2020). Withanone and caffeic acid phenethyl ester are predicted to interact with main protease (Mpro) of SARS-CoV-2 and inhibit its activity. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1772108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W., Kim, T. H., Ku, S.-K., Min, K., Lee, H.-S., Kwon, T. K., & Bae, J.-S. (2012). Barrier protective effects of withaferin A in HMGB1-induced inflammatory responses in both cellular and animal models. Toxicology and Applied Pharmacology, 262(1), 91–98. 10.1016/j.taap.2012.04.025 [DOI] [PubMed] [Google Scholar]
- Liu, C., Zhou, Q., Li, Y., Garner, L. V., Watkins, S. P., Carter, L. J., Smoot, J., Gregg, A. C., Daniels, A. D., Jervey, S., & Albaiu, D. (2020). Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Central Science, 6(3), 315–331. 10.1021/acscentsci.0c00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LLanos, G. G., Araujo, L. M., Jiménez, I. A., Moujir, L. M., & Bazzocchi, I. L. (2012). Withaferin A-related steroids from Withania aristata exhibit potent antiproliferative activity by inducing apoptosis in human tumor cells. European Journal of Medicinal Chemistry, 54, 499–511. 10.1016/j.ejmech.2012.05.032 [DOI] [PubMed] [Google Scholar]
- Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, H., Murakami, T., Kishi, A., & Yoshikawa, M. (2001). Structures of withanosides I, II, III, IV, V, VI, and VII, new withanolide glycosides, from the roots of Indian Withania somnifera Dunal. and inhibitory activity for tachyphylaxis to clonidine in isolated guinea-pig ileum. Bioorganic & Medicinal Chemistry, 9(6), 1499–1507. 10.1016/S0968-0896(01)00024-4 [DOI] [PubMed] [Google Scholar]
- Ministry of Ayush . (2020)., Ayurveda’s immunity boosting measures for self care during COVID 19 crisis [WWW Document]. https://www.ayush.gov.in/docs/123.pdf.
- Mondal, S., Bhattacharya, K., Mallick, A., Sangwan, R., & Mandal, C. (2012). Bak compensated for Bax in p53-null cells to release cytochrome c for the initiation of mitochondrial signaling during Withanolide D-induced apoptosis. PLoS One, 7(3), e34277. 10.1371/journal.pone.0034277 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Morse, J. S., Lalonde, T., Xu, S., & Liu, W. R. (2020). Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem , 21(5), 730–738. 10.1002/cbic.202000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, D., Ishitsuka, K., Katsuya, H., Kunami, N., Nogami, R., Yoshimura, Y., Matsuda, M., Kamikawa, M., Tsuchihashi, R., Okawa, M., Ikeda, T., Nohara, T., Tamura, K., & Kinjo, J. (2013). Screening of promising chemotherapeutic candidates from plants against human adult T-cell leukemia/lymphoma (II): Apoptosis of antiproliferactive principle (24,25-dihydrowithanolide D) against ATL cell lines and structure-activity relationships with withanolides isolated from solanaceous plants. Journal of Natural Medicines, 67(2), 415–420. 10.1007/s11418-012-0700-9 [DOI] [PubMed] [Google Scholar]
- Pant, S., Singh, M., Ravichandiran, V., Murty, U. S. N., & Srivastava, H. K. (2020). Peptide-like and small-molecule inhibitors against Covid-19. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1757510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, X., Xu, X., Li, Y., Cheng, L., Zhou, X., & Ren, B. (2020). Transmission routes of 2019-nCoV and controls in dental practice. International Journal of Oral Science, 12(1), 9. 10.1038/s41368-020-0075-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, H., Wu, J., Hong, L., Luo, Y., Song, Q., & Chen, D. (2020). Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: An observational cohort study. Lancet Infectious Diseases, 20(6), 689-696. 10.1016/S1473-3099(20)30198-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabi, A. F., Al Zoubi, S. M., Kasasbeh, A. G., Salameh, M. D., Al-Nasser, D. A. (2020). SARS-CoV-2 and coronavirus disease 2019: What we know so far. Pathog. 10.3390/pathogens9030231 [DOI] [PMC free article] [PubMed]
- Reddy, S. V. G., Reddy, K. T., Kumari, V. V., & Basha, S. H. (2015). Molecular docking and dynamic simulation studies evidenced plausible immunotherapeutic anticancer property by Withaferin A targeting indoleamine 2,3-dioxygenase. Journal of Biomolecular Structure and Dynamics, 33(12), 2695–2709. 10.1080/07391102.2015.1004834 [DOI] [PubMed] [Google Scholar]
- Remya, C., Dileep, K. V., Variayr, E. J., & Sadasivan, C. (2016). An in silico guided identification of nAChR agonists from Withania somnifera. Frontiers in Life Science, 9(3), 201–213. 10.1080/21553769.2016.1207569 [DOI] [Google Scholar]
- Sargsyan, K., Grauffel, C., & Lim, C. (2017). How molecular size impacts RMSD applications in molecular dynamics simulations. Journal of Chemical Theory and Computation, 13(4), 1518–1524. 10.1021/acs.jctc.7b00028 [DOI] [PubMed] [Google Scholar]
- Schrödinger . (2018). Release 2018-2: LigPrep. Schrödinger, LLC. [Google Scholar]
- Schröter, H.-B., Neumann, D., Katritzky, A. R., & Swinbourne, F. J. (1966). Withasomnine. A pyrazole alkaloid from Withania somnifera Dun. Tetrahedron, 22(8), 2895–2897. 10.1016/S0040-4020(01)99082-9 [DOI] [Google Scholar]
- Singh, B., & Sharma, R. A. (2020). Secondary metabolites of medicinal plants: Ethnopharmacological properties, biological activity and production strategies. Wiley. [Google Scholar]
- Singh, G., Tiwari, M., Singh, S. P., Singh, S., Trivedi, P. K., & Misra, P. (2016). Silencing of sterol glycosyltransferases modulates the withanolide biosynthesis and leads to compromised basal immunity of Withania somnifera. Scientific Reports, 6, 25562. 10.1038/srep25562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soman, S., Anju, T. R., Jayanarayanan, S., Antony, S., & Paulose, C. S. (2013). Impaired motor learning attributed to altered AMPA receptor function in the cerebellum of rats with temporal lobe epilepsy: Ameliorating effects of Withania somnifera and withanolide A. Epilepsy & Behavior, 27(3), 484–491. 10.1016/j.yebeh.2013.01.007 [DOI] [PubMed] [Google Scholar]
- Stobart, C. C., Lee, A. S., Lu, X., & Denison, M. R. (2012). Temperature-sensitive mutants and revertants in the coronavirus nonstructural protein 5 protease (3CLpro) define residues involved in long-distance communication and regulation of protease activity. Journal of Virology, 86(9), 4801–4810. 10.1128/JVI.06754-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaraju, G. V., Vanisree, M., Rao, C. V., Sivaramakrishna, C., Sridhar, P., Jayaprakasam, B., & Nair, M. G. (2006). Ashwagandhanolide, a bioactive dimeric thiowithanolide isolated from the roots of Withania somnifera. Journal of Natural Products, 69(12), 1790–1792. 10.1021/np060147p [DOI] [PubMed] [Google Scholar]
- Thuy, B. T. P., My, T. T. A., Hai, N. T. T., Hieu, L. T., Hoa, T. T., Thi Phuong Loan, H., Triet, N. T., Anh, T. T., Van, Quy, P. T., Tat, P., Van, Hue, N., Van, Quang, D. T., Trung, N. T., Tung, V. T., Huynh, L. K., & Nhung, N. T. A. (2020). Investigation into SARS-CoV-2 resistance of compounds in garlic essential oil. ACS Omega, 5(14), 8312–8320. 10.1021/acsomega.0c00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, X., Zhang, H., & Timmermann, B. N. (2011). Chlorinated Withanolides from Withania somnifera. Phytochemistry Letters, 4(4), 411–414. 10.1016/j.phytol.2011.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrini, E., Calcabrini, C., Sestili, P., Catanzaro, E., De Gianni, E., Diaz, R. A., Hrelia, P., Tacchini, M., Guerrini, A., Canonico, B., Papa, S., Valdrè, G., & Fimognari, C. (2016). Withania somnifera induces cytotoxic and cytostatic effects on human T leukemia cells. Toxins, 8(5), 147. 10.3390/toxins8050147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetvicka, V., & Vetvickova, J. (2011). Immune enhancing effects of WB365, a novel combination of Ashwagandha (Withania somnifera) and Maitake (Grifola frondosa) extracts. North American Journal of Medical Sciences, 3(7), 320–324. 10.4297/najms.2011.3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadegaonkar, V. P., & Wadegaonkar, P. A. (2013). Withanone as an inhibitor of survivin: A potential drug candidate for cancer therapy. Journal of Biotechnology, 168(2), 229–233. 10.1016/j.jbiotec.2013.08.028 [DOI] [PubMed] [Google Scholar]
- Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., Shi, Z., Hu, Z., Zhong, W., & Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, P. T., Subramanian, C., Motiwala, H. F., & Cohen, M. S. (2016). Natural withanolides in the treatment of chronic diseases. Advances in Experimental Medicine and Biology, 928, 329–373. 10.1007/978-3-319-41334-1_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . (2020). Coronavirus disease (COVID-19) Situation Report – 129 [WWW Document]. Retrieved from https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200528-covid-19-sitrep-129.pdf?sfvrsn=5b154880_2
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766-788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Yang, M., Ding, Y., Liu, Y., Lou, Z., Zhou, Z., Sun, L., Mo, L., Ye, S., Pang, H., Gao, G. F., Anand, K., Bartlam, M., Hilgenfeld, R., & Rao, Z. (2003). The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proceedings of the National Academy of Sciences of the United States of America, 100(23), 13190–13195. 10.1073/pnas.1835675100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368(6489), 409-412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Nakamura, N., Hattori, M., Kuboyama, T., Tohda, C., & Komatsu, K. (2002). Withanolide Derivatives from the Roots of Withania somnifera and Their Neurite Outgrowth Activities. Chemical & Pharmaceutical Bulletin, 50(6), 760–765. 10.1248/cpb.50.760 [DOI] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., … Shi, Z.-L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.