Abstract
We propose here that one of the potential mechanisms for the relapse of the COVID-19 infection could be a cellular transport pathway associated with the release of the SARS-CoV-2-loaded exosomes and other extracellular vesicles. It is possible that this “Trojan horse” strategy represents possible explanation for the re-appearance of the viral RNA in the recovered COVID-19 patients 7–14 day post discharge, suggesting that viral material was hidden within such exosomes or extracellular vesicles during this “silence” time period and then started to re-spread again.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, COVID-19, reinfection, exosome, extracellular vesicle
Introduction
Are we ready to the fact that we must adapt ourselves to live with COVID-19 and, perhaps, for a long time? Now, we know that not all people exposed to SARS-CoV-2 are infected, not all infected patients show symptoms, and not all showing symptoms develop severe respiratory illness. Although the world-wide daily case fatality rate (CFR) of the ongoing COVID-19 pandemic is declining, there are many countries where CFR is still increasing, and the rate of new infections is still high. In fact, within May month only, the number of new COVID-19 cases almost doubled (increased from 3.3 to 6.3 million), whereas June witnessed the highest ever number of new COVID-19 cases recorded in one day (194,191 on June 26, 2020). COVID-19 is characterized by a basic reproductive number (R0, which is the number of people infected by each sick person) ranging from 3.8 to 8.9 (Sanche et al., 2020). All these are rather disturbing news, especially for countries, which are coming out or a planning to come out of the COVID-19-related quarantine.
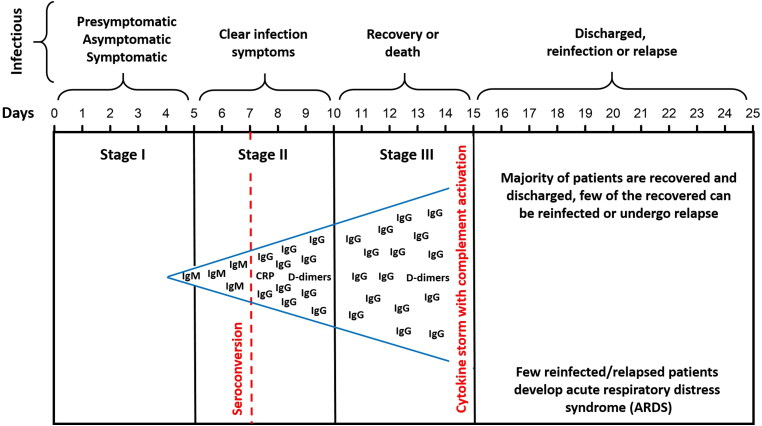
For about 80% of the SARS-CoV-2 infected patients, the disease is mild, being mostly restricted to the upper and conducting airways. Although on average, ∼15% of the confirmed cases progress to the severe phase, for patients over 65, the chance to progress into the severe phase is noticeably higher (Shi et al., 2020). SARS-CoV-2 infection can be roughly divided into three stages (see Figure 1): stage I, an asymptomatic incubation period with or without detectable virus; stage II, non-severe symptomatic period with the presence of virus; stage III, severe respiratory symptomatic stage with high viral load. In more than 50% of patients, the seroconversion take place by day 7, and for the remaining patients – by day 13–14 (Korber et al., 2020; Mason, 2020; Shi et al., 2020; Wölfel et al., 2020). One should keep in mind that since SARS-CoV2 coronavirus is a new very aggressive causative agent, and novel data are released on an hourly basis, there are multiple classifications for the disease progression. However, the classification outlined here is the most clinically sound, being assembled based on the clinical data for more than 1000 patients (Korber et al., 2020; Mason, 2020; Shi et al., 2020; Wölfel et al., 2020).
Figure 1.
Schematic representation of stages of the COVID-19 infection and infectivity of corresponding patients. Here, seroconversion corresponds to the transition from the initial (primary infection) phase of the infection, where immunoglobulin M (IgM) antibodies are produced to the phase, where IgM levels drop (and become undetectable) and the immunoglobulin G (IgG) levels rise and remain detectable. C-reactive protein (CRP) is an acute inflammatory protein that increases up to 1,000-fold at sites of infection or inflammation. D-dimer is a degradation product of the cross-linked fibrin resulting from plasmin cleavage. In the blood of most healthy individuals, D-dimer is present in negligible amounts, whereas the elevated blood levels of D-dimer are the reflection of the intravascular coagulation and venous thromboembolism (VTE), which can present as either deep vein thrombosis (DVT) or pulmonary embolism (PE). Elevated D-dimer levels in COVID-19 patients are associated with the severity of COVID-19 infection and correlate with higher mortality.
The acute respiratory distress syndrome (ARDS) is the main cause of the COVID-19-related mortality, which actually represents a common immunopathological event for SARS-CoV-2, SARS-CoV, and MERS-CoV infections. One of the main ARDS mechanisms is the cytokine storm, the deadly uncontrolled systemic inflammatory response resulting from the release of large amounts of pro-inflammatory cytokines (such as IFN-α, IFN-γ, IL-1b, IL-6, IL-12, IL-18, IL-33, TNF-α, TGFβ, etc.) and chemokines (CCL2, CCL3, CCL5, CXCL8, CXCL9, CXCL10, etc.) by immune effector cells (Li, Geng, et al., 2020). Furthermore, the patients with the severe COVID-19 showed widespread complement activation, characterized by the C3a generation and C3-fragment deposition (Risitano et al., 2020).
Prolonged SARS‐CoV‐2 RNA shedding, COVID-19 reinfection and reactivation
Currently, the number of people recovered from COVID-19 exceeds the half of the total cases of SARS-CoV-2 infection. Despite this positive dynamics, the reinfection and/or infection reactivation in the recovered patients represent a potential hurdle, as recovered/discharged patients usually mix with their community representing a hidden source for new infections. Some of the recovered/discharged patients showed a positive viral RNA burden for as long as 10 to 27 days after the discharge (see, e.g. Korber et al., 2020; Ye et al., 2020), and although the median viral shedding duration was 20 days, in some cases, it was observed for 37 days (Zhou et al., 2020). It was demonstrated recently that the situation can be even worse, since in some COVID-19 patients, the duration of the SARS‐CoV‐2 RNA shedding could be much longer than 30 days. In fact, it was reported that in some such patients, the prolonged SARS-CoV-2 RNA shedding occurred with a median duration of 53 days and a maximum of 83 days (Li, Wang, et al., 2020). In addition to this prolonged carriage of SARS-CoV-2, some patients who had recovered from COVID-19 demonstrated recurrence of SARS-CoV-2 (Xiao et al., 2020; Ye et al., 2020; Yuan et al., 2020). In fact, in one study, as much as 9.1% of the discharged COVID-19 patients were shown to be presented with the SARS-CoV-2 reactivation (Ye et al., 2020). Another study revealed that 14.5% of discharged COVID-19 patients with negative (polymerase chain reaction) PCR, had a later positive reverse transcription PCR (RT-PCR) test for SARS-CoV-2 (Yuan et al., 2020), while still another study indicated that the number of such recurring patients can be as high as 21.4% (Xiao et al., 2020).
Although the PCR-based methods cannot distinguish between the infectious virus and the non-infectious nucleic acid of the same virus, the positive PCR of recovered/discharged patients or even the non-survivors represents an important way of the infection diagnosis. A very interesting case was recently reported, where a 78-year-old woman that was ready to be discharged after the three consecutive PCR tests on her nasopharyngeal swab samples indicated that she was SARS-CoV-2 negative, her conditions were significantly improved, and CT examination showed absorption of pulmonary exudation, but suddenly she fell into the cardiac arrest and died (Yao et al., 2020). This case raised a fundamental question, from what she has died: Was it from the virus or some other cause? To answer these questions, digital PCR was performed on tissue sections from the lung, liver, heart, intestine, and skin (Yao et al., 2020). This analysis unexpectedly found positive PCR of SARS-CoV-2 RNA only in the lung, but not in other tissues. Consistently, electron microscopic analysis clearly revealed the presence of the coronavirus particles in the lung biopsy (Yao et al., 2020). Based on these important observations the authors recommended that the PCR detection of the SARS-CoV-2 nucleic acid should be conducted on broncho-alveolar lavage fluid. They also suggested the extension of quarantine time and the timely follow-up medical examination on discharged patients, especially aged ones (Yao et al., 2020).
The clinical data collected for these reactivated patients were similar to the data of patients with primary infection. Importantly, the originally asymptomatic patients were among the reactivated patients, suggesting the noticeable reactivation potential of asymptomatic or minimally symptomatic patients (Korber et al., 2020; Ye et al., 2020). The viral loads in presymptomatic and asymptomatic cases were similar to those of symptomatic cases, indicating that the transmission could occur during the incubation period (Arteaga-Livias et al., 2020; Gatto et al., 2020).
To deal with the potential reactivation and/or reinfection, the China National Health Commission (National Health Commission) has issued four criteria for discharge of those seem recovered: (a) no fever for at least 3 days; (b) significant improvement in respiratory symptoms; (c) substantial improvement of the radiological abnormalities on chest computed tomography (CT) or X-ray; and (d) two consecutive negative RT-PCR results of SARS-CoV-19 nucleic acid (at least with a 24-hour interval). This is also in line with the WHO’s guidelines on clinical management, according to which a COVID-19 patient can be discharged from the hospital after two consecutive negative PCR results at least 24 h apart in a clinically recovered patient (https://www.ecdc.europa.eu/en/publications-data/covid-19-guidance-discharge-and-ending-isolation). However, despite meeting all these criteria, some patients still show positive results of their RT-PCR tests sometime after the discharge, which may be caused by the incomplete viral clearance, incorrect sampling, false-negative, and/or false-positive RT-PCR results, as well as by reinfection/reactivation. On average, these recurring patients were hospitalized for 15.36 ± 3.81 days during their primary infection, and the average period between the previous discharge and the positive test for SARS-CoV-19 was ranged from 4 to 17 days (Ye et al., 2020; Yuan et al., 2020).
The presence of a relatively high potential for the discharged COVID-19 patient to develop reinfection/reactivation also indicates that a virus-eliminating immune response to SARS-CoV-2 may be difficult to achieve, at least in some patients. Reinfection is unlikely under the strict quarantine measures, which require additional 14-day isolation and medical observation after discharge. It is necessary to isolate patients with recurrent positive RT-PCR results and continue treatment until they meet the discharge criteria again. In a recent study, 25 patients with reoccurrence of RT-PCR results were reported, and the median time from their last negative result to turning positive was 6 days. This is the reason why strict quarantine and follow-up of COVID-19 patients is of upmost importance. Patients with high-risk factors for COVID-19 should be monitored carefully and subjected to a full diagnostic procedure, with comprehensive treatment (Dong, Cao, et al., 2020; Lan et al., 2020; Shi et al., 2020; Xing et al., 2020). There is no available methodology, which would help physicians to differentiate or distinguish between the recovered patients who have protective immunity and the recovered patients that do not have it. Obviously, patients who developed more robust immune responses with formation of memory CD8+ T-cells and helper CD4+ T-cells would be the most equipped if exposed to the SARS-CoV-2 again. There are three main mechanisms for reinfection, where the immune response can be (i) ineffective, (ii) strain specific, or (iii) short-lived. Furthermore, the SARS-CoV-2 was shown to develop “escape mutants” (Chaturvedi et al., 2020).
Although there is a possibility that some recurrence cases actually might be attributed to persistent infection, where PCR result were falsely negative at discharge (Hoang et al., 2020), there is still the rising concern that patients who recovered from the COVID-19 may be at risk of reinfection or reactivation. This opportunity raised several critical questions (Kang et al., 2020), such as what are the reasons for the reappearance of SARS-CoV-2 RNA in some patients recovered from COVID-19 and what are the mechanisms of such reinfection, reactivation, and relapse? Are these reinfected/reactivated/relapsed COVID-19 patients infectious? How should such recovered patients which were retested positive for SARS-CoV-2 be managed? Are there neutralized antibodies or protective reaction induced by the SARS-CoV-2 infection? Will the SARS-CoV-2 vaccines work on such reinfected, relapsed, or reactivated COVID-19 patients?
To in vivo test the possibility of reinfection, the acquired immunity to SARS-CoV-2 in rhesus macaques was investigated (Bao et al., 2020). Four rhesus monkeys were infected with SARS-CoV-2, and two were reinfected after confirmed recovery. After primary infection, viral replication was detected in the nose, pharynx, lungs and gut, with histopathological evidence of lung damage. Sera collected from recovered monkeys before reinfection exhibited neutralizing activity against SARS-CoV-2. Upon reinfection, viral replication was not detected in nasopharyngeal or anal swabs, and reinfected monkeys did not show any signs of COVID-19 disease recurrence. This suggests that in some cases, immunity acquired following primary infection with SARS-CoV-2 may protect from the reinfection (Bao et al., 2020; Ota, 2020), and this immunity may be responsible for the protection of the recovered patients from transiting to the severe complications by controlling the virus replication (Vardhana & Wolchok, 2020).
The numerical experiments using the Susceptible-Exposed-Infectious-Removed-Undetectable-Susceptible (SEIRUS) model were conducted to evaluate the probability of the reinfection in the recovered individuals (Victor Okhuese, 2020). This study came to the conclusion that in the absence of a curative vaccination, the fraction of the infected population will continue to increase worldwide, whereas the recovery rate will continue to increase slowly but steadily over a long period of time, and the death rate will be determined by the ratio of the infection rate to the recovery rate (Victor Okhuese, 2020). The model also suggested that the reinfection rate within the recovered population will decline to zero over time as the virus is cleared clinically from the system of the recovered population (Victor Okhuese, 2020). However, contrary to this conclusion, there is a clear evidence that at the current stage of pandemics with the continuing raise of the COVID-19 infection cases the possibility for reinfection/relapse/reactivation of SARS-CoV-2 is rather high.
COVID-19 can be transmitted via person-to-person contact from three hidden sources, which are presymptomatic and asymptomatic patients, or people with COVID-19 who was not diagnosed, likely because they did not feel very sick. These patients can spread the virus and may represent a population that can be easily neglected in epidemic prevention (Arons et al., 2020; Lai et al., 2020; Shi et al., 2020; Wei et al., 2020). The fourth source of infection is non-hidden, which is a symptomatic COVID-19 patient. The reinfection of a recovered person with the same SARS-CoV-2 strain can originate from all the aforementioned sources, or it may be caused by a new viral strain, and in this case the reinfected persons may show complex symptoms.
Many factors are combined to define the extremely aggressive nature of SARS-CoV-2, which is highly transmissible and is characterized by high mortality rate. Although we have a long history of co-existence with coronaviruses that has started as early as the 1930s (Weiss, 2020), there is no population immunity against SARS-CoV-2 as of yet, which makes us more susceptible to this infection. One also should remember that this virus is not only efficiently transmitted horizontally, but also vertically, being passed by the infected mothers to their newborns (Dong, Tian, et al., 2020; Ferrazzi et al., 2020). And, of course, there is a chance for reinfection.
Exosomes and extracellular vesicles as potential mediators of viral infection, reinfection, and reactivation
In relation to the reinfected patients (i.e. the discharged recovered patients who became SARS-CoV-2 RNA positive again), an important question is where (or what cite) the hidden SARS-CoV-2 RNA is coming from? Many viruses are known to entry the extracellular double-membrane vesicle (EDMV) or exosome avenue during synthesis and intra-host spreading (Badierah et al., 2020). There is no literature covering this scenario for SARS-CoV-2. However, there is an in vitro study on SARS-CoV cultured in the AT2 cells, which reported that the viral particles can be seen within the double membrane vesicles (Mason, 2020; Qian et al., 2013). Coronavirus RNA is synthesized in virus-induced double membrane vesicles in the cytoplasm of the infected cells. The virions bud intracellularly from the membranes of the ER–Golgi-intermediate compartment, are transported through the cytoplasm in secretory vesicles, and are released from cells by an exocytic process (Qian et al., 2013). The fact that SARS-CoV-2 can be present within the vacuoles or double membrane vesicles (DMVs) within the host cells was proven by the careful post-mortem histopathological analysis of the renal samples of patients with COVID-19 by light microscopy, electron microscopic examination, and immunostaining (Farkash et al., 2020; Su et al., 2020). In fact, these analyses revealed the presence of the clusters of coronavirus-like particles with distinctive spikes in the renal tubular epithelium (Su et al., 2020). These intracellular viruses were organized into arrays, indicating that they were manufactured and assembled intracellularly. Furthermore, DMVs with possible viral assembly were found near the rough endoplasmic reticulum (RER), suggesting that the mechanism of the SARS-CoV-2 viral assembly is analogous to that of SARS-CoV (Farkash et al., 2020). In these studies, individual viruses ranged in size from 65 to 91 nm (mean 76 nm). Mature-appearing viruses were predominantly located within the cytoplasm, focally organized into small arrays. Viral particles with crown-like morphology were composed of cores with intermediate electron density surrounded by an envelope studded with abundant crown-like, electron-dense spikes. Adjacent vacuoles contained abundant ovoid double-membrane vesicles associated with rough endoplasmic reticulum, representing possible viral assembly (Farkash et al., 2020). These findings also suggested that there is a possibility for the use of the exosomal cellular transport as a mode of systemic SARS-CoV-2 viral dissemination and serve as a potential means for the COVID-19 reactivation.
A primary site of SARS-CoV-2 infection appears to be the lung, which may be a source for viral spread to other tissues such as the kidney and intestine, where virus has been found in stool (Wang, Xu, et al., 2020) and urine (Ling et al., 2020). Although the viremia is established during the course of the disease, although viral RNA in blood is only infrequently observed (Wang, Xu, et al., 2020). However, the virus has a size of 80–100 nm indicating that viremic SARS-CoV-2 must first infect blood vessels prior to local tissue infections (Monteil et al., 2020). The infected alveolar units tend to be peripheral and subpleural. SARS-CoV propagates within type II cells, where the large number of viral particles is released, and the cells undergo apoptosis and die (Mason, 2020). The pathologic changes accompanying of SARS-CoV and SARS-CoV-2 infection is diffuse alveolar damage (DAD) with fibrin-rich hyaline membranes (as the first target organ) and the free viral particles or vacuolated or double membrane vesicles containing the viral particles released from the infected to the adjacent new cells/tissues and expanded to circulate systemically and disseminate to reaching distant tissues (Monkemuller et al., 2020). In this way, the virus can be disseminated into many organs, including the vasculature system (Monteil et al., 2020; Varga et al., 2020). It could attack the endothelium causing endotheliitis, which may explain impaired microcirculatory function across different organs and the frequently observed prothrombotic state with in-situ clot formation (Puelles et al., 2020; Su et al., 2020; Varga et al., 2020; Ye et al., 2007, 2020).
Lipid membranes define the boundaries of many cellular organelles, such as mitochondria, the endoplasmic reticulum (ER), nucleus, and the Golgi apparatus. They are also indispensable for the biochemical functions performed by these highly specialized microcompartments, which require both structural support and physical separation from the cytosol. Along the same lines, all positive-strand RNA (+RNA) viruses characterized to date induce the formation of dedicated membrane structures to support the cytoplasmic replication of their genomes (Knoops et al., 2008, 2012). Member of the coronaviruses family induce typical double-membrane vesicles (DMVs), with average diameter of the 300 nm, and convoluted membranes (CM) (Knoops et al., 2008, 2012). Usually, DMV clusters are derived from the endoplasmic reticulum (ER) or ER-Golgi intermediate compartment and contain newly synthesized viral genome (as electron dense inclusions), viral RNA, and viral replicase proteins, as well as the viral particles. This diversity of DMVs may explain the disbranching of the reinfected patients, with some of them being PCR positive without any COVID-19 symptoms and the other having acute to mild symptoms.
Cells infected with coronavirus (such as SARS‐CoV (Knoops et al., 2008), MERS-CoV (de Wilde et al., 2013), or the mouse hepatitis virus (MHV) (Ulasli et al., 2010)) display early membrane rearrangements in the form of ER‐derived convoluted membranes interconnected with DMVs (about 250 nm in diameter) (Blanchard & Roingeard, 2015; Knoops et al., 2012). Furthermore, fusion of the outer membranes of some CoV‐induced DMVs can lead to the formation of larger vesicle packets (1–5 μm in size) (Knoops et al., 2008). The ability to hijack cytoplasmic cellular membrane compartments in order to establish a “replication/transcription complex” capable of amplifying and further expressing their genome is a characteristic feature of all studied positive-strand RNA viruses that often utilizes for this purpose viral non-structural proteins capable of coordinating accessory functions (Ahlquist, 2006; Mackenzie, 2005; Moriel-Carretero, 2020).
The SARS-CoV replicative machinery consists of up to sixteen non-structural proteins (Nsp1 to Nsp16), with the Nsp3, Nsp4, and Nsp6 being engaged in the induction of DMVs (Angelini et al., 2013). Mutations in the Nsp4 have been linked to the formation of aberrant DMVs with apparent defects in membrane pairing. This may explain the appearance of the single-membrane vesicles and double-membrane vesicles possessing several single-membrane vesicles enclosed within an outer membrane and containing SARS-CoV in Vero E6 9 (Goldsmith et al., 2004). On the other hand, mutations in Nsp6 from several coronaviruses cause formation of single-membrane vesicles (Al-Mulla et al., 2014). These observations indicate that the membrane rearrangement (redirect and rearrange host cell membranes for use as part of the viral genome replication and transcription machinery) represents a tactic used by all known positive-sense single-stranded RNA viruses.
Electron microscopy was used to identify murine coronaviruses with mutations in Nsp3 and Nsp14 that replicated normally, while producing only half the normal amount of DMVs under the low-temperature growth conditions (Al-Mulla et al., 2014). Viruses with mutations in Nsp5 and Nsp16 produced small DMVs but also replicated normally. Quantitative RT-PCR confirmed that the most strongly affected of these, the Nsp3 mutant, produced more viral RNA than the wild-type virus (Al-Mulla et al., 2014). In the same line, it was demonstrated (Oudshoorn et al., 2017) that for both MERS-CoV and SARS-CoV, co-expression of Nsp3 and Nsp4 is required and sufficient to induce DMVs, and co-expression of MERS-CoV Nsp3 and Nsp4 either as individual proteins or as a self-cleaving Nsp3-4 precursor resulted in the appearance of very similar DMVs, and in both setups, proliferation of zippered ER was observed that appeared to wrap into nascent DMVs (Oudshoorn et al., 2017). The detailed new knowledge about the roles of Nsp7-10 in virus replication was recently established (Gildenhuys, 2020). It seems that the order of the Nsp7-10 cleavage is important for controlling various viral processes and seems to have relevance in terms of the protein–protein complexes formation (Gildenhuys, 2020). These important details can be used for the design of the effective interventions (Gildenhuys, 2020; Krichel et al., 2020).
DMV formation, regulation, and reshaping as well as the remodelling of the intracellular membranous machinery became a rich area for drug discovery, specifically against coronaviruses. Some of the compounds that can be used for targeting the DMVs as a means to combat viral infection were recently reviewed (Shahmohamadnejad et al., 2020). The authors first considered several compounds, such as K22, oxysterol-binding protein (OSBP), OSW-1, T-00127-HEV-2, and TTP-8307, that can be used to fight against many positive sense RNA viruses by working on their Nsp proteins, and then recommended to use OSBP against SARS-CoV-2 (Shahmohamadnejad et al., 2020). Recently it was pointed out that the single-membrane vesicles that appear at the early stages of viral infection and some of which seem to be embedded in the ER clearly resemble nascent lipid droplets (LDs), which are the membrane-enclosed organelles that are formed by the progressive nucleation of non-polar lipids, such as triacylglycerols and steryl esters, within the hydrophobic core of the ER bilayer (Moriel-Carretero, 2020). In fact, among the specific morphological features of the DMVs induced during the SARS-CoV infection is the fact that although their inner layer constitutes a closed circle, the outer lipid layer is continuous and wraps around other DMVs, making the whole structure strikingly similar to entangled LD trapped within the ER as a consequence of a local excess in phosphatidic acid (Moriel-Carretero, 2020).
Although the extracellular vesicles (EVs) represent a family of natural carriers in the human body and play a number of critical roles in cell-to-cell communications, they were recently suggested to be used as unique drug carriers to deliver protease inhibitors to treat COVID-19 (Kumar et al., 2020). On the other hand, there is evidence that viruses can use EV endocytic routes to enter uninfected cells and hijack the EV secretory pathway to exit infected cells, thereby illustrating that EVs and viruses share common cell entry and biogenesis mechanisms (Badierah et al., 2020; Urbanelli et al., 2019). Numerous publications dedicated to the analysis of the roles of EV in viral infection in general and in infectivity of HIV, HCV, and SARS specifically have been recently reviewed (Badierah et al., 2020; Giannessi et al., 2020). Although the role of EVs in HCV and HIV infectivity had been extensively document, the ability of SARS to hijack the EV endocytic and secretory pathways represents a novel development in the field analysing the involvement of the EVs in viral infections (Giannessi et al., 2020). Overall, since the composition and biological activity of exosomes and other EVs change during a variety of bacterial, fungal, and viral infections (Schorey et al., 2015) and since these exosomes/EVs may incorporate viral proteins and/or fragments of viral RNAs to carry this viral materials from infected cells to target ones they are expected to play important roles in viral infections (Raab-Traub & Dittmer, 2017; Thery et al., 2009; Urbanelli et al., 2019).
There are several pieces of evidence supporting these important conclusions. For example, recently, it was reported that the exosomes originating from the transplanted lungs may contribute to the immune pathogenesis of the lung allograft failure from chronic lung allograft dysfunction (CLAD) after lung transplant (LTx) (Gunasekaran et al., 2017). In fact, the lungs transplanted from the donor contained numerous pathogenic virus antigen and RNAs originated from rhinovirus, coronavirus (types HKU1, NL63, 229E, OC43), and respiratory syncytial virus (RSV), whereas exosomes isolated from these patients contained specific viral antigens, lung-associated self-antigens (SAgs), as well as mismatched donor human leukocyte antigen (HLA) and SAgs (such as collagen-V (Col-V) and K-alpha-1 tubulin (Kα1T)) (Gunasekaran et al., 2020). This study revealed circulating exosomes containing viral antigens were transiently present in 3 of 5 lung transplant recipients (LTxRs), whereas 2 of 5 LTxRs with respiratory viral infections (RVI) had persistently circulating exosomes with lung SAgs and viral antigens (Gunasekaran et al., 2020). Recently, the evidence was presented that the exosomes can transfer angiotensin converting enzyme 2 (ACE2) to the recipient cells (Wang, Chen, et al., 2020), thereby making them susceptible to virus docking and suggesting a supportive function of the exosomes for the SARS-CoV-2 internalization and infection (Hassanpour et al., 2020).
Furthermore, several cases considered below indicated the presence of virus-containing DMVs in the post-mortem, biopsy, autopsy, and endoscopy samples ultrastructure of which was analysed by electron microscopy. EM histological analysis (see Table 1) clearly showed that the viral particles consistent with coronavirus (SARS-CoV) were detected in all 5 small intestine tissue samples obtained on autopsy, as well as in the terminal ileal and colonic biopsy specimens obtained on colonoscopy, which were isolated, in vitro cultured, and propagated then confirmed by RT-PCR. Viral particles were confined to the epithelial cells, primarily in the apical surface enterocytes and rarely in the glandular epithelial cells. Intracellularly, viral particles were contained within dilated cytoplasmic vesicles consistent with dilated endoplasmic reticulum. The vesicles containing the viral particles were often seen toward the apical cytoplasm. Clusters of coronaviruses were also detected on the surface microvilli, which may suggest virus leaving from the luminal surface of enterocytes. There was no evidence of villous atrophy despite viral adhesion and colonization (Leung et al., 2003). Analogously, Table 1 shows that free viral particles, viral particles and/or viral particle within the single- or double-membrane vesicles, multiple viral particles with variable virions diameter within a single DMV were consistently fund in samples derived from the patients infected with SARS-CoV, MERS-CoV, and SARS-CoV2 (Alsaad et al., 2018; Goldsmith & Miller, 2009; Kissling et al., 2020; Larsen et al., 2020; Martines et al., 2020; Menter et al., 2020; Qinfen et al., 2004; Shieh et al., 2005; Su et al., 2020; Ye et al., 2007; Zhu et al., 2020).
Table 1.
Electron microscopy characterized ultrastructure features of the SARS-CoV, SARS-CoV-2, and MERS-CoV viral particles.
| Reference | Sample type | Sample source | Viral particle size | Virus features |
|---|---|---|---|---|
| Leung et al. (2003); Monkemuller et al. (2020) | Post-mortem colon biopsy | Human colon | 60–90nm | Crown-like structure, vesicle containing viruses, viral particles on the microvilli surface of enterocytes, viral particle appeared detached from the cells. |
| Goldsmith et al. (2004); Ksiazek et al. (2003); Rota et al. (2003) | Cell-line, BAL |
Vero E6 Human bronchial alveolar lavage (BAL) |
78 nm (average) | Virus-containing vesicles (double-membrane vesicle), vesicles composed of multiple single membrane vesicles containing SARS-CoV of Vero E6, Virions covered the exterior surface of the BAL cells, Diffuse, extracellular virions in BAL sample |
| Shieh et al. (2005) | Lung tissue autopsy | Human lung | 51 nm (average) | Pneumocytes cytoplasm containing viral particles, extracellular SARS-CoV virions is embedded in fibrin within alveolar space, SARS-CoV antigen detected in alveolar macrophage phagosome, free viral particles presented in alveolar space |
| Bulfamante et al. (2020) | Autopsy within three hours post-mortem | Human . olfactory nerve, . Gyrus, . brainstem |
98–160 nm | Spherical particle with crown-like shape and inner dense core and electron-dense periphery, double nuclear envelope, severe damage in the olfactory nerve, autophagy phenomena appeared in the cytoplasm |
| Varga et al. (2020) | Post-mortem autopsy | Human transplanted kidney | 150 nm | Viral inclusion bodies in peipenilubular space and viral particles in endothelial cells, aggregates of viral particles with dense circular surface and lucid centre, capillaries containing viral particles |
| Yao et al. (2020) | Post-mortem was ready-for discharge | Human pulmonary biopsy | 70–100 nm | SARS-CoV2 particles in bronchiolar epithelial cells marked by cilia and type II alveolar epithelial cells featured with lamellar body, featured with lamellar body, IHC staining presented the SARS-CoV-2 nucleocapsid. |
| Ng et al. (2016) | Post-mortem, body was kept refrigerated at 4°C Autopsy 10 days after death. | Human Lung tissue I.M. samples were excised from a paraffin block |
50–50 nm | Fragmented pneumocyte infected with MERS-CoV, Spherical clusters or individualized of viral particles within membrane-bound vesicles in pneumocytes, |
| Menter et al. (2020) | Autopsy of <12 hours post-mortem | Kidney and lung tissue | 70–110 nm | Virus-like particles within vesicles and not in the cytoplasm, Activated podocytes and endothelial cells, podocytes cytoplasm contained multiple vesicles, some with attached ribosomes and double membranes virus-like particles with electron dense granules detected within these vesicles, Sporadically, these particles present in endothelial cells and proximal tubular epithelial cells, the is vesicle close to the luminal border with virus-like particles, multiple cytoplasmic vesicles one of which contains virus-like particles. |
| Su et al. (2020) | Post-mortem autopsy | Kidney | 65–136 nm | clusters of coronavirus-like particles with distinctive spikes in the tubular epithelium and podocytes and to less in distal tubules, double membrane with surface projections, nucleocapsid apposing to the viral envelope, and the interior electron-lucent of the particles, viral particle occasional vacuolation and detachment of podocytes from the glomerular basement membrane |
| Carsana et al. (2020) | Post-mortem autopsy | Lung | 82 nm | The virions were mainly localised along plasmalemmal membranes and within cytoplasmic vacuoles, viral particles detected in type 1 and type 2 pneumocytes, and alveolar macrophages cells in nine out of ten samples |
| Kissling et al. (2020) | Kidney biopsy on day 8 | Kidney | 50–110 nm | Cytoplasmic vacuoles containing numerous spherical particles in the podocytes, the viral particles surrounded by spikes measuring 9–10nm, the particles have the typical appearance (“solar corona”) of viral inclusion bodies. |
Similarly, the electron microscopy analysis of the Vero E6 cells infected with SARS-CoV revealed the early formation and accumulation of typical DMVs, which probably carried the viral replication complex (Snijder et al., 2006). Furthermore, visualization of virus particles that were secreted from the infected cells displayed a strikingly well-preserved features, including clearly visible spikes (Snijder et al., 2006). However, these authors argued against the previously proposed involvement of the autophagic pathway as the source for the vesicles (Snijder et al., 2006).
It was previously discovered that the SARS-CoV was able to attach to Vero E6 cultured cells, enter host cells, and uncoat the nucleocapsids, all within a 30-min period (Ng et al., 2003a). Many informative EM images were taken during the entry events. At 5 min after infection, several virus particles lined the Vero cell plasma membrane. After entry (10 and 15 min), spherical core particles moved into the cytoplasm within the large vacuoles (DMVs). Then, they found that even at such early stages of infection (20 min), the noticeable virus-induced changes in the infected cells were evident (where the induction of myelin-like membrane whorls was obvious within the same vacuoles as the core particles) (Ng et al., 2003a). However, by 25–30 min post-infection, the spherical core particles appeared to be disassociating and, in their place, doughnut-shaped electron-dense structures were observed, which could represent the viral genomes embedded within the helical nucleocapsids (Ng et al., 2003a). They were no longer in large vacuoles but packaged into smaller vacuoles in the cytoplasm, and occasionally were observed in small groups (Ng et al., 2003a). Currently, these important virus-induced changes are rather well-known (Al-Mulla et al., 2014; Angelini et al., 2013; Knoops et al., 2012; Oudshoorn et al., 2017).
In a subsequent comprehensive analysis, the entire replication cycle of the SARS-CoV (strain 2003VA2774 isolated from a patient) in Vero E6 cells was followed from 1 to 30 h post-infection (p.i.) (Ng et al., 2003b). This study showed that the proliferation of the Golgi complexes and related vesicles was the most obvious ultrastructural change observed within the first hour of infection, and swelling of some of the trans-Golgi sacs was observed, which contained virus nucleocapsids at different stages of maturation. By 5 h p.i., ∼ 5% of the cell population contained extracellular virus particles, and this number such cells rapidly increased to ∼ 30% within the next hour. In addition to be found in swollen Golgi sacs the virus precursors were also observed in the large vacuoles and were in close association with membrane whorls that could represent the replication complexes, since they appeared rather early in the replication cycle (Ng et al., 2003b). As aforementioned, the formation of such membranous whorls represents a part of viral tactic to redirect and rearrange host cell membranes for use as part of the viral genome replication and transcription machinery (Al-Mulla et al., 2014; Angelini et al., 2013; Knoops et al., 2012; Oudshoorn et al., 2017). At the later infection stages (on the 12 to 21 h p.i.), the cytoplasm of the infected cells was shown to contain numerous large, smooth membrane-embedded vacuoles possessing a mixture of mature viruses and spherical cores (Ng et al., 2003b). Some of these virus-containing vacuoles were in a close proximity to the cell periphery, prepared for the exocytosis-based export of the mature progeny virus particles. Finally, at the last infection stage (by 24 to 30 h p.i.), the cell surface commonly contained crystalline arrays of the extracellular virus particles (Ng et al., 2003b).
These studies showed that the entry events for SARS-CoV are really fast. But what one can say about the duration of these entry events for SARS-CoV-2 (COVID-19), whose spike protein has 10–20 fold higher affinity for ACE2 more than that of SARS-CoV (Wrapp et al., 2020)? If one would use the factor of 15 (which is the mean of the aforementioned 10–20-fold higher affinity for receptor) then divided the time required for SARS-CoV entry (30 min) by this value, the one would see that the time required for SARS-CoV-2 to entry the host cell would be just 2 min. Of course, these back-of-the-envelope calculations need an experimental validation. However, such estimations are important, since they show why the COVID-19 infection is more transmissible and aggressive than the infection with other human CoVs.
One could also speculate that the SARS-CoV-2 infection through nasopharyngeal route, where the virus is rapidly attached to specific cellular receptors, swiftly enters the host cell, and vigorously and aggressively replicate within the epithelial tissue cells within 20–30 min. Within next 12–24 h, the mature viral particle are released from the infected cells and diffuse to the nearby cells or invade the lower respiratory tract and in some persons also into digestive system. Is such scenario could be mimicked by the Vero E6 infection with SARS-CoV?
Generally speaking, multiple aspects of SARS-CoV-2 shows strong similarity to those of the SARS-CoV, and this is especialyl applicable to the remarkable resemblance of their ultrastructural histopathologies. SARS-CoV infection of cultured cells has shown features similar to those of previously characterized coronaviruses. SARS-specific characteristics include large granular cytoplasmic areas, presence of the nucleocapsid inclusions and typical DMVs.
In some SARS autopsies, EM examination revealed the presence of cytoplasmic viral particles in pneumocytes of lung tissues, as well as within the cells of nervous system and intestinal tissues (Table 1). The majority of these viral particles were within membrane-bound vesicles. Viral particles have also been observed in macrophages in lung tissue. Furthermore, the presence of inclusion bodies within the infected cells has been reported in several studies, with the viral origin of such inclusion bodies being confirmed by immunogold labelling (Gu & Korteweg, 2007; Zeng et al., 2020).
Data considered in this article indicate that similar to many other viruses, SARS-CoV-2 utilizes exosomal and extracellular vesicle cellular transport avenues for reproduction and intra-host spreading as a mode of systemic virus dissemination (see Figure 2). Is this “Trojan horse” strategy of the release of the SARS-CoV-2-loaded exosomes or EDMVs represent a reasonable explanation for the appearance of the viral RNA in the recovered COVID-19 patients 7–14 day post discharge? What one can find in such hidden exosomes/EDMVs, if they do exists, and where do they hide during the “silence” time period; i.e. after the recovered patients discharge from the hospitals and before the viral RNA starts to reappear again, reflecting the COVID-19 reinfection/reactivation but now under the conditions of the primed acquired immune response? These are important questions, which are waiting for their answers.
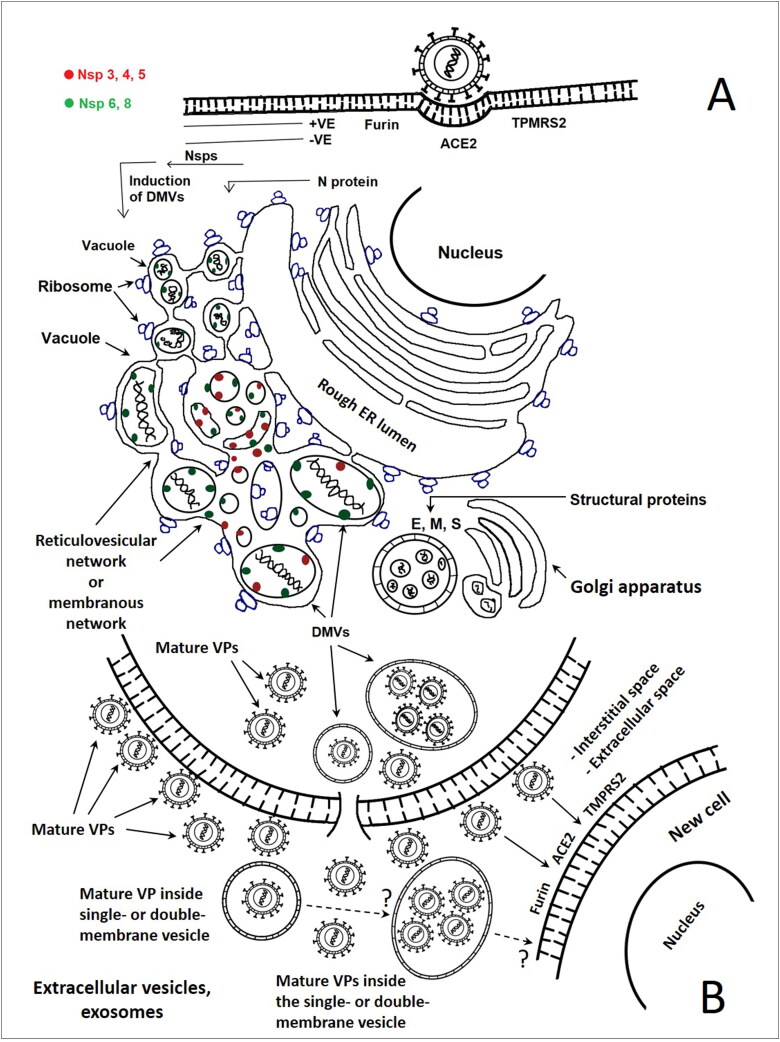
Figure 2.
A. Putative life cycle of the SARS-CoV and SARS-CoV-2 in the human host cell (in vivo) and/or in Vero E6 cell (in vitro). Virus-induced double membrane vesicles in the cytoplasm of infected cells represent platforms for coronaviruses replication, assembling, trafficking, extrusion, and shedding the mature viral particles (free and/or inside vesicles). Cell infected with the virus demonstrated the formation of a reticulovesicular network of modified membranes, which included single/multiple double-membrane vesicles, representing the site where the virus replicate. All are and contiguous with the rough endoplasmic reticulum. The viral + RNA is released into the cytoplasm and primarily translated into viral polyproteins encoding the Nsps, which stimulate/induce the DMVs to proceed and complete the virus life cycle in association with the Golgi stacks to produce the virus particles in the vesicles, which eventually fuse with the plasma membrane. The DMV may contain the mature or immature viral particle, or the non-assembled viral apparatus. The Nsp 3–8 are present on the CM, while some of Nsp8 can be detected inside the DMVs. The histological and ultrastructural analysis of the appearance of the samples from the SARS-CoV-2 infected patients demonstrated the presence of mature viral particles as well as the immature viral particles or non-assembled viral apparatus inside DMVs. The illustration depends on the data from (Alsaad et al., 2018; Angelini et al., 2013; Bulfamante et al., 2020; Goldsmith & Miller, 2009; Knoops et al., 2008; Menter et al., 2020; Oudshoorn et al., 2017; Perlman & Netland, 2009; Qinfen et al., 2004; Shieh et al., 2005; Sims et al., 2008; Su et al., 2020). B. The mature and immature viral particles spread/disseminated into new neighbouring cells as documented in the text for SARS-CoV-2, while the extracellular vesicles (exosomes) introduce the SARS-CoV-2 virus particles into the cells still needs to be documented.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Ahlquist, P. (2006). Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nature Reviews. Microbiology, 4(5), 371–382. 10.1038/nrmicro1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mulla, H. M. N., Turrell, L., Smith, N. M., Payne, L., Baliji, S., Züst, R., Thiel, V., Baker, S. C., Siddell, S. G., & Neuman, B. W. (2014). Competitive fitness in coronaviruses is not correlated with size or number of double-membrane vesicles under reduced-temperature growth conditions. mBio, 5(2), e01107–e01113. 10.1128/mBio.01107-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaad, K. O., Hajeer, A. H., Al Balwi, M., Al Moaiqel, M., Al Oudah, N., Al Ajlan, A., AlJohani, S., Alsolamy, S., Gmati, G. E., Balkhy, H., Al-Jahdali, H. H., Baharoon, S. A., & Arabi, Y. M. (2018). Histopathology of Middle East respiratory syndrome coronovirus (MERS-CoV) infection - clinicopathological and ultrastructural study. Histopathology, 72(3), 516–524. 10.1111/his.13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini, M. M., Akhlaghpour, M., Neuman, B. W., & Buchmeier, M. J. (2013). Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio, 4(4), e00524–e0013. 10.1128/mBio.00524-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arons, M. M., Hatfield, K. M., Reddy, S. C., Kimball, A., James, A., Jacobs, J. R., Taylor, J., Spicer, K., Bardossy, A. C., Oakley, L. P., Tanwar, S., Dyal, J. W., Harney, J., Chisty, Z., Bell, J. M., Methner, M., Paul, P., Carlson, C. M., McLaughlin, H. P., … Jernigan, J. A. (2020). Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. New England Journal of Medicine, 382(22), 2081–2090. 10.1056/NEJMoa2008457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga-Livias, K., Pecho-Silva, S., Panduro-Correa, V., & Rodriguez-Morales, A. J. (2020). The dilemmas of the classification of SARS-CoV-2 infection without clinical manifestations: Asymptomatic or presymptomatic. Journal of the Formosan Medical Association, 119(7), 1237–1238. 10.1016/j.jfma.2020.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badierah, R. A., Uversky, V. N., & Redwan, E. M. (2020). Dancing with Trojan horses: An interplay between the extracellular vesicles and viruses. Journal of Biomolecular Structure and Dynamics, 1–27. 10.1080/07391102.2020.1756409 [DOI] [PubMed] [Google Scholar]
- Bao, L., Deng, W., Gao, H., Xiao, C., Liu, J., Xue, J., Lv, Q., Liu, J., Yu, P., Xu, Y., Qi, F., Qu, Y., Li, F., Xiang, Z., Yu, H., Gong, S., Liu, M., Wang, G., Wang, S.,… Qin, C. (2020). Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. bioRxiv. 10.1101/2020.03.13.990226 [DOI]
- Blanchard, E., & Roingeard, P. (2015). Virus-induced double-membrane vesicles. Cellular Microbiology, 17(1), 45–50. 10.1111/cmi.12372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulfamante, G., Chiumello, D., Canevini, M. P., Priori, A., Mazzanti, M., Centanni, S., & Felisati, G. (2020). First ultrastructural autoptic findings of SARS-Cov-2 in olfactory pathways and brainstem. Minerva Anestesiol, 86(6), 678–679. 10.23736/S0375-9393.20.14772-2 [DOI] [PubMed] [Google Scholar]
- Carsana, L., Sonzogni, A., Nasr, A., Rossi, R. S., Pellegrinelli, A., Zerbi, P., Rech, R.., Colombo, R., Antinori, S., Corbellino, M., Galli, M., Catena, E., Tosoni, A., Gianatti, A., & Nebuloni, M. (2020). Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: A two-centre descriptive study. Lancet Infectious Diseases. 10.1016/S1473-3099(20)30434-5 [DOI] [PMC free article] [PubMed]
- Chaturvedi, R., Naidu, R., Sheth, S., & Chakravarthy, K. (2020). Efficacy of serology testing in predicting reinfection in patients with SARS-CoV-2. Disaster Medicine and Public Health Preparedness, 1–7. 10.1017/dmp.2020.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde, A. H., Raj, V. S., Oudshoorn, D., Bestebroer, T. M., van Nieuwkoop, S., Limpens, R. W. A. L., Posthuma, C. C., van der Meer, Y., Bárcena, M., Haagmans, B. L., Snijder, E. J., & van den Hoogen, B. G. (2013). MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-α treatment. The Journal of General Virology, 94(Pt 8), 1749–1760. 10.1099/vir.0.052910-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, L., Tian, J., He, S., Zhu, C., Wang, J., Liu, C., & Yang, J. (2020). Possible vertical transmission of SARS-CoV-2 from an infected mother to her newborn. JAMA, 323(18), 1846–1848. 10.1001/jama.2020.4621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, X., Cao, Y. Y., Lu, X. X., Zhang, J. J., Du, H., Yan, Y. Q., Akdis, C. A., Gao, Y. D. (2020). Eleven faces of coronavirus disease 2019. Allergy. 10.1111/all.14289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash, E. A., Wilson, A. M., & Jentzen, J. M. (2020). Ultrastructural evidence for direct renal infection with SARS-CoV-2. Journal of the American Society of Nephrology. 10.1681/ASN.2020040432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrazzi, E., Frigerio, L., Savasi, V., Vergani, P., Prefumo, F., Barresi, S., Bianchi, S., Ciriello, E., Facchinetti, F., Gervasi, M.T., Iurlaro, E., Kustermann, A., Mangili, G., Mosca, F., Patane, L., Spazzini, D., Spinillo, A., Trojano, G., Vignali, M., … Cetin, I. (2020). Vaginal delivery in SARS-CoV-2 infected pregnant women in Northern Italy: A retrospective analysis. BJOG. 10.1111/1471-0528.16278 [DOI] [PMC free article] [PubMed]
- Gatto, M., Bertuzzo, E., Mari, L., Miccoli, S., Carraro, L., Casagrandi, R., & Rinaldo, A. (2020). Spread and dynamics of the COVID-19 epidemic in Italy: Effects of emergency containment measures. Proceedings of the National Academy of Sciences of the United States of America, 117(19), 10484–10491. 10.1073/pnas.2004978117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannessi, F., Aiello, A., Franchi, F., Percario, Z. A., & Affabris, E. (2020). The role of extracellular vesicles as allies of HIV, HCV and SARS viruses. Viruses, 12(5), 571. 10.3390/v12050571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildenhuys, S. (2020). Expanding our understanding of the role polyprotein conformation plays in the coronavirus life cycle. The Biochemical Journal, 477(8), 1479–1482. 10.1042/BCJ20200223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith, C. S., & Miller, S. E. (2009). Modern uses of electron microscopy for detection of viruses. Clinical Microbiology Reviews, 22(4), 552–563. 10.1128/CMR.00027-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith, C. S., Tatti, K. M., Ksiazek, T. G., Rollin, P. E., Comer, J. A., Lee, W. W., Rota, P. A., Bankamp, B., Bellini, W. J., & Zaki, S. R. (2004). Ultrastructural characterization of SARS coronavirus. Emerging Infectious Diseases, 10(2), 320–326. 10.3201/eid1002.030913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, J., & Korteweg, C. (2007). Pathology and pathogenesis of severe acute respiratory syndrome. The American Journal of Pathology, 170(4), 1136–1147. 10.2353/ajpath.2007.061088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran, M., Bansal, S., Ravichandran, R., Sharma, M., Perincheri, S., Rodriguez, F., Hachem, R., Fisher, C. E., Limaye, A. P., Omar, A., Smith, M. A., Bremner, R. M., & Mohanakumar, T. (2020). Respiratory viral infection in lung transplantation induces exosomes that trigger chronic rejection. The Journal of Heart and Lung Transplantation, 39(4), 379–388. 10.1016/j.healun.2019.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekaran, M., Xu, Z., Nayak, D. K., Sharma, M., Hachem, R., Walia, R., Bremner, R. M., Smith, M. A., & Mohanakumar, T. (2017). Donor-derived exosomes with lung self-antigens in human lung allograft rejection. American Journal of Transplantation: Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons, 17(2), 474–484. 10.1111/ajt.13915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanpour, M., Rezaie, J., Nouri, M., & Panahi, Y. (2020). The role of extracellular vesicles in COVID-19 virus infection. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 85, 104422. 10.1016/j.meegid.2020.104422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, V. T., Dao, T. L., & Gautret, P. (2020). Recurrence of positive SARS-CoV-2 in patients recovered from COVID-19. Journal of Medical Virology. 10.1002/jmv.26056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H., Wang, Y., Tong, Z., & Liu, X. (2020). Retest positive for SARS-CoV-2 RNA of “recovered” patients with COVID-19: Persistence, sampling issues, or re-infection? Journal of Medical Virology. 10.1002/jmv.26114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissling, S., Rotman, S., Gerber, C., Halfon, M., Lamoth, F., Comte, D., Lhopitallier, L., Sadallah, S., & Fakhouri, F. (2020). Collapsing glomerulopathy in a COVID-19 patient. Kidney International, 98(1), 228–231. 10.1016/j.kint.2020.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops, K., Barcena, M., Limpens, R. W., Koster, A. J., Mommaas, A. M., & Snijder, E. J. (2012). Ultrastructural characterization of arterivirus replication structures: Reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. Journal of Virology, 86(5), 2474–2487. 10.1128/JVI.06677-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops, K., Kikkert, M., Worm, S. H. E. v d., Zevenhoven-Dobbe, J. C., van der Meer, Y., Koster, A. J., Mommaas, A. M., & Snijder, E. J. (2008). SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biology, 6(9), e226. 10.1371/journal.pbio.0060226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber, B. Fischer, W. M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., Foley, B., Giorgi, E. E., Bhattacharya, T., Parker, M. D., Partridge, D. G., Evans, C. M., Freeman, T. M., de Silva, T. I., LaBranche, C. C., & Montefiori, D. C. (2020). Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 10.1101/2020.04.29.069054 [DOI]
- Krichel, B., Falke, S., Hilgenfeld, R., Redecke, L., & Uetrecht, C. (2020). Processing of the SARS-CoV pp1a/ab nsp7-10 region. The Biochemical Journal, 477(5), 1009–1019. 10.1042/BCJ20200029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek, T. G., Erdman, D., Goldsmith, C. S., Zaki, S. R., Peret, T., Emery, S., Tong, S., Urbani, C., Comer, J. A., Lim, W., Rollin, P. E., Dowell, S. F., Ling, A.-E., Humphrey, C. D., Shieh, W.-J., Guarner, J., Paddock, C. D., Rota, P., Fields, B., … Anderson, L. J. (2003). A novel coronavirus associated with severe acute respiratory syndrome. New England Journal of Medicine, 348(20), 1953–1966. 10.1056/NEJMoa030781 [DOI] [PubMed] [Google Scholar]
- Kumar, S., Zhi, K., Mukherji, A., & Gerth, K. (2020). Repurposing antiviral protease inhibitors using extracellular vesicles for potential therapy of COVID-19. Viruses, 12(5), 486. 10.3390/v12050486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C. C., Liu, Y. H., Wang, C. Y., Wang, Y. H., Hsueh, S. C., Yen, M. Y., Ko, W. C., & Hsueh, P. R. (2020). Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. Journal of Microbiology, Immunology and Infection, 53(3), 404–412. 10.1016/j.jmii.2020.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, L., Xu, D., Ye, G., Xia, C., Wang, S., Li, Y., & Xu, H. (2020). Positive RT-PCR test results in patients recovered from COVID-19. JAMA, 323(15), 1502. 10.1001/jama.2020.2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, C. P., Bourne, T. D., Wilson, J. D., Saqqa, O., & Sharshir, M. A. (2020). Collapsing glomerulopathy in a patient with coronavirus disease 2019 (COVID-19). Kidney International Reports, 5(6), 935–939. 10.1016/j.ekir.2020.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, W. K., To, K.-F., Chan, P. K. S., Chan, H. L. Y., Wu, A. K. L., Lee, N., Yuen, K. Y., & Sung, J. J. Y. (2003). Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology, 125(4), 1011–1017. 10.1016/s0016-5085(03)01215-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N., Wang, X., & Lv, T. (2020). Prolonged SARS-CoV-2 RNA shedding: Not a rare phenomenon. Journal of Medical Virology. 10.1002/jmv.25952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Geng, M., Peng, Y., Meng, L., & Lu, S. (2020). Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis, 10(2), 102–108. 10.1016/j.jpha.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling, Y., Xu, S. B., Lin, Y. X., Tian, D., Zhu, Z. Q., Dai, F. H., Wu, F., Song, Z. G., Huang, W., Chen, J., Hu, B. J., Wang, S., Mao, E. Q., Zhu, L., Zhang, W. H., & Lu, H. Z. (2020). Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chinese Medical Journal (Engl), 133(9), 1039–1043. 10.1097/CM9.0000000000000774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, J. (2005). Wrapping things up about virus RNA replication. Traffic (Copenhagen, Denmark), 6(11), 967–977. 10.1111/j.1600-0854.2005.00339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martines, R. B., Ritter, J. M., Matkovic, E., Gary, J., Bollweg, B. C., Bullock, H., Goldsmith, C. S., Silva-Flannery, L., Seixas, J. N., Reagan-Steiner, S., Uyeki, T., Denison, A., Bhatnagar, J., Shieh, W. J., Zaki, S. R., & COVID-Pathology Working Group. (2020). Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerging Infectious Diseases, 26(9). 10.3201/eid2609.202095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, R. J. (2020). Pathogenesis of COVID-19 from a cell biology perspective. European Respiratory Journal, 55(4), 2000607. 10.1183/13993003.00607-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menter, T., Haslbauer, J. D., Nienhold, R., Savic, S., Hopfer, H., Deigendesch, N., Frank, S., Turek, D., Willi, N., Pargger, H., Bassetti, S., Leuppi, J. D., Cathomas, G., Tolnay, M., Mertz, K. D., & Tzankov, A. ( 2020). Post-mortem examination of COVID19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings of lungs and other organs suggesting vascular dysfunction. Histopathology. 10.1111/his.14134 [DOI] [PMC free article] [PubMed]
- Monkemuller, K., Fry, L., & Rickes, S. (2020). COVID-19, coronavirus, SARS-CoV-2 and the small bowel. Revista Espanola de Enfermedades Digestivas: Organo Oficial de la Sociedad Espanola de Patologia Digestiva, 112(5), 383–388. 10.17235/reed.2020.7137/2020 [DOI] [PubMed] [Google Scholar]
- Monteil, V., Kwon, H., Prado, P., Hagelkrüys, A., Wimmer, R. A., Stahl, M., Leopoldi, A., Garreta, E., Hurtado Del Pozo, C., Prosper, F., Romero, J. P., Wirnsberger, G., Zhang, H., Slutsky, A. S., Conder, R., Montserrat, N., Mirazimi, A., & Penninger, J. M. (2020). Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell, 181(4), 905–913. 10.1016/j.cell.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriel-Carretero, M. (2020). The hypothetical role of phosphatidic acid in subverting ER membranes during SARS-CoV infection. Traffic. 10.1111/tra.12738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, D. L., Al Hosani, F., Keating, M. K., Gerber, S. I., Jones, T. L., Metcalfe, M. G., Tong, S., Tao, Y., Alami, N. N., Haynes, L. M., Mutei, M. A., Abdel-Wareth, L., Uyeki, T. M., Swerdlow, D. L., Barakat, M., & Zaki, S. R. (2016). Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of middle east respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. The American Journal of Pathology, 186(3), 652–658. 10.1016/j.ajpath.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, M. L., Tan, S. H., See, E. E., Ooi, E. E., & Ling, A. E. (2003. a). Early events of SARS coronavirus infection in vero cells. Journal of Medical Virology, 71(3), 323–331. 10.1002/jmv.10499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, M. L., Tan, S. H., See, E. E., Ooi, E. E., & Ling, A. E. (2003. b). Proliferative growth of SARS coronavirus in Vero E6 cells. The Journal of General Virology, 84(Pt 12), 3291–3303. 10.1099/vir.0.19505-0 [DOI] [PubMed] [Google Scholar]
- Ota, M. (2020). Will we see protection or reinfection in COVID-19? Nature Reviews. Immunology, 20(6), 351–351. 10.1038/s41577-020-0316-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudshoorn, D., Rijs, K., Limpens, R. W. A. L., Groen, K., Koster, A. J., Snijder, E. J., Kikkert, M., & Bárcena, M. (2017). Expression and cleavage of Middle East respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio, 8(6), 1–17. 10.1128/mBio.01658-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman, S., & Netland, J. (2009). Coronaviruses post-SARS: Update on replication and pathogenesis. Nature Reviews. Microbiology, 7(6), 439–450. 10.1038/nrmicro2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles, V. G., Lutgehetmann, M., Lindenmeyer, M. T., Sperhake, J. P., Wong, M. N., Allweiss, L., Chilla, S., Heinemann, A., Wanner, N., Liu, S., Braun, F., Lu, S., Pfefferle, S., Schroder, A. S., Edler, C., Gross, O., Glatzel, M., Wichmann, D., Wiech, T., ... Huber, T. B. (2020). Multiorgan and renal tropism of SARS-CoV-2. New England Journal of Medicine. 10.1056/NEJMc2011400 [DOI] [PMC free article] [PubMed]
- Qian, Z., Travanty, E. A., Oko, L., Edeen, K., Berglund, A., Wang, J., Ito, Y., Holmes, K. V., & Mason, R. J. (2013). Innate immune response of human alveolar type II cells infected with severe acute respiratory syndrome-coronavirus. American Journal of Respiratory Cell and Molecular Biology, 48(6), 742–748. 10.1165/rcmb.2012-0339OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qinfen, Z., Jinming, C., Xiaojun, H., Huanying, Z., Jicheng, H., Ling, F., Kunpeng, L., & Jingqiang, Z. (2004). The life cycle of SARS coronavirus in Vero E6 cells. Journal of Medical Virology, 73(3), 332–337. 10.1002/jmv.20095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Traub, N., & Dittmer, D. P. (2017). Viral effects on the content and function of extracellular vesicles. Nature Reviews. Microbiology, 15(9), 559–572. 10.1038/nrmicro.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risitano, A. M., Mastellos, D. C., Huber-Lang, M., Yancopoulou, D., Garlanda, C., Ciceri, F., & Lambris, J. D. (2020). Complement as a target in COVID-19? Nature Reviews. Immunology, 20(6), 343–344. 10.1038/s41577-020-0320-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota, P. A., Oberste, M. S., Monroe, S. S., Nix, W. A., Campagnoli, R., Icenogle, J. P., Peñaranda, S., Bankamp, B., Maher, K., Chen, M.-H., Tong, S., Tamin, A., Lowe, L., Frace, M., DeRisi, J. L., Chen, Q., Wang, D., Erdman, D. D., Peret, T. C. T., … Bellini, W. J. (2003). Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science (New York, N.Y.), 300(5624), 1394–1399. 10.1126/science.1085952 [DOI] [PubMed] [Google Scholar]
- Sanche, S., Lin, Y. T., Xu, C., Romero-Severson, E., Hengartner, N., & Ke, R. (2020). High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerging Infectious Diseases, 7, 26. 10.3201/eid2607.200282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorey, J. S., Cheng, Y., Singh, P. P., & Smith, V. L. (2015). Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Reports, 16(1), 24–43. 10.15252/embr.201439363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmohamadnejad, S., Nabavi, S. F., Habtemariam, S., Sarkar, K., Sil, P. C., Dowran, R., & Nabavi, S. M. (2020). May we target double membrane vesicles and oxysterol-binding protein to combat SARS-CoV-2 infection? Cell Biology International. 10.1002/cbin.11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., Wang, Y., Shao, C., Huang, J., Gan, J., Huang, X., Bucci, E., Piacentini, M., Ippolito, G., & Melino, G. (2020). COVID-19 infection: The perspectives on immune responses. Cell Death and Differentiation, 27(5), 1451–1454. 10.1038/s41418-020-0530-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh, W.-J., Hsiao, C.-H., Paddock, C. D., Guarner, J., Goldsmith, C. S., Tatti, K., Packard, M., Mueller, L., Wu, M.-Z., Rollin, P., Su, I.-J., & Zaki, S. R. (2005). Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Human Pathology, 36(3), 303–309. 10.1016/j.humpath.2004.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims, A. C., Burkett, S. E., Yount, B., & Pickles, R. J. (2008). SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Research, 133(1), 33–44. 10.1016/j.virusres.2007.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E. J., van der Meer, Y., Zevenhoven-Dobbe, J., Onderwater, J. J., van der Meulen, J., Koerten, H. K., & Mommaas, A. M. (2006). Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. Journal of Virology, 80(12), 5927–5940. 10.1128/JVI.02501-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, H., Yang, M., Wan, C., Yi, L. X., Tang, F., Zhu, H. Y., Yi, F., Yang, H. C., Fogo, A. B., Nie, X., & Zhang, C. (2020). Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney International, 98(1), 219–227. 10.1016/j.kint.2020.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery, C., Ostrowski, M., & Segura, E. (2009). Membrane vesicles as conveyors of immune responses. Nature Reviews. Immunology, 9(8), 581–593. 10.1038/nri2567 [DOI] [PubMed] [Google Scholar]
- Ulasli, M., Verheije, M. H., de Haan, C. A., & Reggiori, F. (2010). Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cellular Microbiology, 12(6), 844–861. 10.1111/j.1462-5822.2010.01437.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanelli, L., Buratta, S., Tancini, B., Sagini, K., Delo, F., Porcellati, S., & Emiliani, C. (2019). The role of extracellular vesicles in viral infection and transmission. Vaccines (Vaccines), 7(3), 102. 10.3390/vaccines7030102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhana, S. A., & Wolchok, J. D. (2020). The many faces of the anti-COVID immune response. Journal of Experimental Medicine, 217(6), e20200678. 10.1084/jem.20200678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga, Z., Flammer, A. J., Steiger, P., Haberecker, M., Andermatt, R., Zinkernagel, A. S., Mehra, M. R., Schuepbach, R. A., Ruschitzka, F., & Moch, H. (2020). Endothelial cell infection and endotheliitis in COVID-19. Lancet (London, England), 395(10234), 1417–1418. 10.1016/S0140-6736(20)30937-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor Okhuese, A. (2020). Estimation of the probability of reinfection with COVID-19 by the susceptible-exposed-infectious-removed-undetectable-susceptible model. JMIR Public Health and Surveillance, 6(2), e19097. 10.2196/19097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Chen, S., & Bihl, J. (2020). Exosome-mediated transfer of ACE2 (Angiotensin-converting enzyme 2) from endothelial progenitor cells promotes survival and function of endothelial cell. Oxidative Medicine and Cellular Longevity, 2020, 4213541. 10.1155/2020/4213541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Xu, Y., Gao, R., Lu, R., Han, K., Wu, G., & Tan, W. (2020). Detection of SARS-CoV-2 in different types of clinical specimens. JAMA, 323(18), 1843–1844. 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, W. E., Li, Z., Chiew, C. J., Yong, S. E., Toh, M. P., & Lee, V. J. (2020). Presymptomatic transmission of SARS-CoV-2 - Singapore, January 23–March 16, 2020. MMWR Morbidity and Mortality Weekly Report, 69(14), 411–415. 10.15585/mmwr.mm6914e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, S. R. (2020). Forty years with coronaviruses. Journal of Experimental Medicine, 217(5), e20200537. 10.1084/jem.20200537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel, R., Corman, V. M., Guggemos, W., Seilmaier, M., Zange, S., Müller, M. A., Niemeyer, D., Jones, T. C., Vollmar, P., Rothe, C., Hoelscher, M., Bleicker, T., Brünink, S., Schneider, J., Ehmann, R., Zwirglmaier, K., Drosten, C., & Wendtner, C. (2020). Virological assessment of hospitalized patients with COVID-2019. Nature, 581(7809), 465–469. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.), 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, A. T., Tong, Y. X., & Zhang, S. (2020). False-negative of RT-PCR and prolonged nucleic acid conversion in COVID-19: Rather than recurrence. Journal of Medical Virology. 10.1002/jmv.25855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y., Mo, P., Xiao, Y., Zhao, O., Zhang, Y., & Wang, F. (2020). Post-discharge surveillance and positive virus detection in two medical staff recovered from coronavirus disease 2019 (COVID-19), China, January to February 2020. Eurosurveillance, 25(10), 2000191. 10.2807/1560-7917.ES.2020.25.10.2000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X.-H., He, Z.-C., Li, T.-Y., Zhang, H.-R., Wang, Y., Mou, H., Guo, Q., Yu, S.-C., Ding, Y., Liu, X., Ping, Y.-F., & Bian, X.-W. (2020). Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Research, 30(6), 541–543. 10.1038/s41422-020-0318-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, G., Pan, Z., Pan, Y., Deng, Q., Chen, L., Li, J., Li, Y., & Wang, X. (2020). Clinical characteristics of severe acute respiratory syndrome coronavirus 2 reactivation. Journal of Infection, 80(5), e14–e17. 10.1016/j.jinf.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J., Zhang, B., Xu, J., Chang, Q., McNutt, M. A., Korteweg, C., Gong, E., & Gu, J. (2007). Molecular pathology in the lungs of severe acute respiratory syndrome patients. The American Journal of Pathology, 170(2), 538–545. 10.2353/ajpath.2007.060469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J., Kou, S., Liang, Y., Zeng, J., Pan, Y., & Liu, L. (2020). PCR assays turned positive in 25 discharged COVID-19 patients. Clinical Infectious Diseases. 10.1093/cid/ciaa398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z., Xu, L., Xie, X. Y., Yan, H. L., Xie, B. J., Xu, W. Z., Liu, X. A., Kang, G. J., Jiang, W. L., & Yuan, J. P. (2020). Pulmonary pathology of early phase COVID-19 pneumonia in a patient with a benign lung lesion. Histopathology. 10.1111/his.14138 [DOI] [PMC free article] [PubMed]
- Zhou, F., Yu, T., Du, R., Fan, G., Liu, Y., Liu, Z., Xiang, J., Wang, Y., Song, B., Gu, X., Guan, L., Wei, Y., Li, H., Wu, X., Xu, J., Tu, S., Zhang, Y., Chen, H., & Cao, B. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. The Lancet, 395(10229), 1054–1062. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]