Abstract
Herein, the DrugBank database which contains 10,036 approved and investigational drugs was explored deeply for potential drugs that target SARS-CoV-2 main protease (Mpro). Filtration process of the database was conducted using three levels of accuracy for molecular docking calculations. The top 35 drugs with docking scores > −11.0 kcal/mol were then subjected to 10 ns molecular dynamics (MD) simulations followed by molecular mechanics–generalized Born surface area (MM-GBSA) binding energy calculations. The results showed that DB02388 and Cobicistat (DB09065) exhibited potential binding affinities towards Mpro over 100 ns MD simulations, with binding energy values of −49.67 and −46.60 kcal/mol, respectively. Binding energy and structural analyses demonstrated the higher stability of DB02388 over Cobicistat. The potency of DB02388 and Cobicistat is attributed to their abilities to form several hydrogen bonds with the essential amino acids inside the active site of Mpro. Compared to DB02388 and Cobicistat, Darunavir showed a much lower binding affinity of −34.83 kcal/mol. The present study highlights the potentiality of DB02388 and Cobicistat as anti-COVID-19 drugs for clinical trials.
Communicated by Ramaswamy H. Sarma.
Keywords: COVID-19, SARS-CoV-2 main protease, drug repurposing, in-silico drug discovery, molecular dynamics
Graphical Abstract
Introduction
The case of a recent outbreak of coronavirus infectious disease (COVID-19) has been identified and attributed to the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2, namely 2019nCOV), which originated from the meat market in Wuhan, China (Cao et al., 2020). The World Health Organization (WHO) has announced COVID-19 as a potential global health threat because of high mortality rate, high basic reproduction number (R0), and lack of clinically approved drugs and vaccines for this disease. Due to the increasing number of deaths all over the world caused by COVID-19, there is an urgent demand for potent anti-COVID-19 drugs. Traditional de novo drug discovery is a time-consuming and expensive process. Drug repurposing is an alternative time-efficient and cost-effective strategy, in which new uses for approved or investigational drugs are identified (Elmezayen et al., 2020; Khan et al., 2020; Khan et al., 2020; Pushpakom et al., 2019). Experimental and computational drug repurposing have attracted interests from both the pharmaceutical industry and the research community over the last decades (Khan et al., 2020; Pushpakom et al., 2019). Several successful attempts of drug repurposing have been reported for treatment of various diseases (Karaman & Sippl, 2019; Yella et al., 2018).
All recent studies showed that the usage of therapeutic agents as anti-viral (Lai et al., 2020), anti-malarial (Gao et al., 2020), and herbal treatments (Luo et al., 2020) to treat COVID-19 infection is unclear. The re-investigation of available drugs is urgently needed for COVID-19 treatment and saving of lives all over the world.
Coronavirus polyproteins include two main proteases, main protease (3C-like protease, Mpro) and a papain-like protease (PLpro) (Hilgenfeld, 2014). Both coronaviral proteases play a vital role in the replication and transcription of the virus and, in turn, are considered as charming anti-viral targets for the discovery of novel drugs (Xue et al., 2008; Zhao et al., 2013). The first crystal structure of SARS-CoV-2 Mpro with a peptidomimetic inhibitor (N3) has been recently reported (Jin et al., 2020), opening a new horizon for the utilization of structure-based drug discovery approaches towards the inhibition of SARS-CoV-2 Mpro.
The aim of this research project has therefore been to evaluate the reuse of approved and clinical trial drugs as anti-COVID-19 drugs by combining molecular docking and molecular dynamics simulation methods. 10,036 drug candidates in the DrugBank database were first prepared and their binding affinities were filtered based on their docking scores against SARS-CoV-2 Mpro. Three levels of docking calculations were utilized towards verification of docking scores accuracy. The top 35 drugs with highest docking scores with Mpro were then subjected to molecular dynamics simulation of 10 ns. Molecular mechanics–generalized Born surface area (MM-GBSA) approach was employed to estimate drug-Mpro binding energies (ΔGbinding) over the simulated time. The stability of the most potent drugs inside the active site of Mpro was further investigated over 100 ns molecular dynamics simulations. The study presented sheds light on DB02388 and Cobicistat which should be clinically investigated as anti-COVID-19 drugs.
Computational methodology
Database preparation
Prior to molecular docking calculations, the DrugBank database containing 10,036 approved and investigational drugs was prepared (Wishart et al., 2006, 2018). The database was retrieved in SDF format and the 3D structures of drug molecules were then generated using Omega2 software (Hawkins et al., 2010; OMEGA, 2013). 3D chemical structures of the top potent eight DrugBank drugs showing high MM-GBSA binding energies with SARS-CoV-2 Mpro are illustrated in Figure 1. The geometrical structures of all drugs were minimized with MMFF94S force field using SZYBKI software (SZYBKI, 2016) and the partial atomic charges were assigned using Gasteiger method (Gasteiger & Marsili, 1980). Only neutral drug molecules containing C, H, N, O, S, P, F, Cl, Br and I were considered in the current study. The prepared files of DrugBank database may be accessed via www.compchem.net/ccdb.
Figure 1.
3D chemical structures of the top potent eight DrugBank drugs showing high MM-GBSA binding energies with SARS-CoV-2 main protease (Mpro).
Mpro preparation
The crystal structure of SARS-CoV-2 main protease (Mpro) complexed with peptidomimetic inhibitor (N3) (PDB code: 6LU7 (Jin et al., 2020)) was taken as a template for all molecular docking and molecular dynamics calculations. Water molecules and ions were stripped out. The protonation state of Mpro was investigated using H++ server, and all missing hydrogen atoms were added (Gordon et al., 2005). For molecular docking calculations, pdbqt file for Mpro was prepared according to AutoDock protocol (Forli et al., 2016).
Molecular docking
All molecular docking calculations were carried out using Autodock4.2 software (Morris et al., 2009). Three different levels of docking accuracy were conducted, namely: standard, moderate and expensive. In the three calculations, all docking parameters were kept to default values, except the number of genetic algorithm (GA) run and the maximum number of energy evaluation (eval). The latter two variables were set to 50, 100 and 250 and 5,000,000, 10,000,000 and 25,000,000 for standard, moderate and expensive molecular docking calculations, respectively. A schematic representation of the utilized calculations is depicted in Figure 2. The docking grid was set to 60Å x 60Å x 60Å with a grid spacing value of 0.375 Å, and the grid center was placed at the center of the Mpro active site.
Figure 2.
Schematic representation of the utilized techniques.
Molecular dynamics simulations
Molecular dynamics (MD) simulations were carried out on top potent drugs in complex with Mpro using AMBER16 software (Case et al., 2016). General AMBER force field (GAFF) (Wang et al., 2004) and AMBER force field 14SB (Maier et al., 2015) were used to describe the studied drugs and Mpro, respectively. The atomic partial charges of the studied drugs were calculated using the restrained electrostatic potential (RESP) approach (Bayly et al., 1993) at the HF/6-31G* level with the help of Gaussian09 software (Frisch et al., 2009). The docked drug-Mpro complexes were solvated in a cubic water box with 15 Å distances between the edges of the box and any atom of drug or drug-Mpro complexes. The solvated drug-Mpro complexes were then minimized for 5000 step and subsequently gently annealed from 0 K to 300 K throughout 50 ps. The systems were equilibrated for 1 ns and production stages were conducted over simulation times of 10, 50 and 100 ns. Periodic boundary conditions and the NPT ensemble were adopted in all MD simulations, including both the equilibration and production stages. Long-range electrostatic interactions under periodic conditions with a direct space cut-off 12 Å was treated with Particle Mesh Ewald (PME) method (Darden et al., 1993). Langevin dynamics with the collision frequency gamma_ln set to 1.0 was used to keep the temperature constant at 298 K. Berendsen barostat with a relaxation time of 2 ps was employed to control the pressure (Berendsen et al., 1984). A time step of 2 fs and the SHAKE option to constrain all bonds involving hydrogen atoms were used. Coordinates and energy values were collected every 10 ps over the production stage for binding energy calculations and post-dynamics analyses. All MD simulations were performed with the GPU version of pmemd (pmemd.cuda) in AMBER16 on CompChem GPU/CPU cluster (hpc.compchem.net). Visual Molecular Dynamics (VMD) (Humphrey et al., 1996) and LigPlot+ (Laskowski & Swindells, 2011) software were used to visualize drug-Mpro complexes in 3D and 2D, respectively.
Binding energy calculations
Molecular mechanics–generalized Born surface area (MM-GBSA) (Massova & Kollman, 2000) approach implemented in AMBER16 software was used to evaluate the binding energy of the studied drugs with Mpro. The MM-GBSA binding free energies were estimated as follows:
where the energy term (G) is estimated as:
with and as the van der Waals, electrostatic, General Born solvation, and surface area energies, respectively. For the drugs, entropy contributions were neglected.
Results and discussion
Molecular docking
Molecular docking is utilized as a substantial tool in the drug discovery process (Ferreira et al., 2015; Guedes et al., 2014; Meng et al., 2011). Towards drug repurposing for discovering potent Mpro inhibitor, the DrugBank database was deeply screened (Wishart et al., 2006, 2018). The database which contains 10,036 approved and investigational drugs was first retrieved and prepared as described in Computational Methodology. Due to a large number of investigated drugs, three levels of docking calculations were performed to reduce computing time.
First, standard molecular docking calculations with GA = 50 and eval = 2,500,000 were conducted for 10,036 DrugBank molecules against Mpro and their docking scores were evaluated. Drugs were ranked based on the calculated docking scores, and the top 1,000 potent ones were subjected to further calculations. The top 1,000 potent drugs identified from standard docking calculations were redocked against Mpro using GA and eval values of 100 and 10,000,000, respectively. Based on their binding scores, the drugs were ranked and the top 250 drugs were selected. From the latter calculations, the top potent 250 drugs were finally re-docked using expensive docking parameters of GA = 250 and eval = 25,000,000. The predicted expensive docking scores for the top 250 potent drugs with Mpro are listed in Table S1.
Table 1.
Standard (Std.), moderate (Mod.) and expensive (Exp.) docking scores (in kcal/mol) for the selected eight potent drugs and their binding features with SARS-CoV-2 main protease (Mpro).a
| No. | Drug Name/Code | Docking Score
(kcal/mol) |
Binding Featuresb (Hydrogen bond length in Å) | ||
|---|---|---|---|---|---|
| Std. | Mod. | Exp. | |||
| 1 | DB02388 | −10.21 | −10.36 | −11.74 | GLU166 (2.10 Å), TYR54 (2.19 Å), ASP187 (2.11 Å) |
| 2 | Pavinetant (DB11692) | −11.66 | −11.74 | −11.74 | GLU166 (1.95 Å, 2.09 Å), HIS164 (1.99 Å) |
| 3 | Cobicistat (DB09065) | −10.55 | −10.68 | −11.59 | GLU166 (1.94 Å), THR26 (3.01 Å), GLY143 (2.22 Å), CYS145 (2.79 Å), GLN189 (3.06 Å) |
| 4 | DB07618 | −10.97 | −11.27 | −11.31 | GLU166 (1.78 Å, 2.11 Å, 2.12 Å), ARG188 (2.09 Å) |
| 5 | Pyronaridine (DB12975) | −11.00 | −11.12 | −11.14 | GLU166 (2.65 Å, 2.81 Å), GLN189 (2.16 Å) |
| 6 | BMS-986158 (DB15435) | −10.72 | −11.03 | −11.11 | GLU166 (2.84 Å), GLY143 (2.01 Å), HIS163 (2.36 Å), THR190 (1.96 Å), GLN192 (2.92 Å) |
| 7 | MK-6186 (DB12999) | −10.69 | −10.83 | −11.09 | TYR54 (2.37 Å), SER144 (2.28 Å), HIS163 (1.91 Å) |
| 8 | CUDC-907 (DB11891) | −10.51 | −10.99 | −11.04 | GLU166 (2.03 Å, 2.26 Å, 2.41 Å), GLN192 (2.06 Å) |
The potent drugs were selected based on later MM-GBSA calculations.
Determined based on the docked drug-Mpro complex using expensive docking parameters.
As seen from data listed in Table S1, 35 drugs showed docking scores > −11.0 kcal/mol. Accordingly, these 35 drugs were subjected to further investigation using MD techniques. The binding affinities and features for these 35 drugs are summarized in Table S2. The binding modes of eight of the most promising drugs, selected based on later MM-GBSA calculations, inside the active site of Mpro are depicted in Figure 3, and their binding features are listed in Table 1.
Table 2.
Average MM-GBSA binding energies (in kcal/mol) and drug-GLU166 hydrogen bond length (Å) for the top eight potent drugs.a
| No. | Drug Name/Code | 10 ns
Molecular Dynamics |
50 ns
Molecular Dynamics |
||
|---|---|---|---|---|---|
| Binding Energy (kcal/mol) | Hydrogen Bond Length (Å) | Binding Energy (kcal/mol) | Hydrogen Bond Length (Å) | ||
| 1 | Cobicistat (DB09065) |
−59.84 | 2.17 | −53.35 | 3.01 |
| 2 | DB02388 | −50.72 | 2.49 | −51.07 | 2.31 |
| 3 | DB07618 | −42.78 | 2.77 | −45.25 | 2.55 |
| 4 | Pyronaridine (DB12975) |
−44.91 | 2.75 | −44.31 | 2.72 |
| 5 | BMS-986158 (DB15435) |
−40.81 | 2.43 | −43.25 | 2.24 |
| 6 | MK-6186 (DB12999) |
−42.18 | 2.33b | −41.12 | 2.85 |
| 7 | Pavinetant (DB11692) |
−42.53 | 2.86 | −40.81 | 3.06 |
| 8 | CUDC-907 (DB11891) | −40.30 | 2.41 | −40.37 | 2.49 |
Drugs ranked based on their binding energies over 50 ns MD simulation.
Measured with HIS163.
Figure 3.
Cartoon backbone representation of predicted binding modes of the selected eight potent drugs with SARS-CoV-2 main protease (Mpro).
According to data given in Table S2 and Figure 3, most of the drugs share the same binding modes, forming an essential hydrogen bond with GLU166 and other hydrogen bonds with various amino acid residues inside the active site. For instance, the high docking score of −11.74 kcal/mol for DB02388 with Mpro is attributed to its ability to form three short hydrogen bonds with GLU166, TYR54 and ASP187 with bond lengths of 2.10, 2.19 and 2.11 Å, respectively.
Molecular dynamics simulations
Although molecular docking is a fast and widely-used technique, reliability of its predicted drug-target binding affinity is still under debate (Pagadala et al., 2017; Pantsar & Poso, 2018). To achieve reliable predictions for binding affinities, drug-receptor intermolecular interactions and conformational flexibilities of drug-receptor complexes, solvent effects and dynamics must be considered. This can be accomplished using all-atom molecular dynamics (MD) simulations combined with binding energy calculations over reasonable simulation time. Considering computational costs and time, molecular mechanics combined with Generalized Born and surface area continuum solvation (MM-GBSA) methods is a popular approach for the estimation of drug-target binding energies.
Therefore, the 35 identified drugs retrieved from expensive molecular docking calculations were further investigated using MD techniques. The docked drug-Mpro complexes were prepared and subjected to 10 ns molecular dynamics simulations and the corresponding binding energies were evaluated using MM-GBSA approach. The estimated average MM-GBSA binding energies over the 10 ns MD simulations are listed in Table S3.
Table 3.
Decomposition of MM-GBSA binding energies for Cobicistat and DB02388 in complex with SARS-CoV-2 main protease (Mpro) through 100 ns MD simulations.
| Drug Name/Code | Calculated
MM-GBSA binding energy (kcal/mol) |
||||||
|---|---|---|---|---|---|---|---|
| ΔEVDW | ΔEele | ΔEGB | ΔESUR | ΔGgas | ΔGsolv | ΔGbinding | |
| DB02388 | −63.78 | −10.93 | 31.58 | −6.54 | −74.70 | 25.03 | −49.67 |
| Cobicistat (DB09065) |
−61.88 | −12.85 | 35.63 | −7.50 | −74.73 | 28.13 | −46.60 |
According to the calculated MM-GBSA binding energies (ΔGbinding), only eight out of the investigated 35 drugs showed considerable binding energies (ΔGbinding > −40.0 kcal/mol); while 9 and 18 drugs were observed with relatively weak binding energies in ranges of −40.0≥ ΔGbinding > −30.0 and −30.0≥ ΔGbinding > −23.0 kcal/mol, respectively. Interestingly, Cobicistat (DB09065) and DB02388 exhibited promising binding affinities towards Mpro with binding energies (ΔGbinding) of −59.84 and −50.72 kcal/mol, respectively. Measuring the essential hydrogen bond length formed between the drug and the key amino acid GLU166 would reflect the drug-Mpro binding affinity. Therefore, the average hydrogen bond length for the eight drugs (with ΔGbinding > −40.0 kcal/mol) were calculated and listed in Table 2. According to the average hydrogen bond length, the eight drugs form a strong hydrogen bond with GLU166 with bond length ranging from 2.17 to 2.86 Å, except for MK-6186 which forms hydrogen bond with HIS163 with a length of 2.33 Å.
In an attempt to achieve reliable results and spot-light the most potent drug, the 8 drugs with ΔGbinding > −40.0 kcal/mol were further investigated over 50 ns molecular dynamics simulations, and their corresponding MM-GBSA binding energies were consequently estimated. The MM-GBSA binding energies obtained from the 50 ns MD simulations and the average hydrogen bond lengths with GLU166 are listed in Table 2.
According to the results presented in Table 2, there was no significant difference between MM-GBSA binding energies estimated over 50 ns and those over 10 ns, except in the case of Cobicistat. For the latter drug, the ΔGbinding decreased from −59.84 to −53.34 kcal/mol over 10 ns and 50 ns, respectively. Inspecting the hydrogen bond length between Cobicistat and GLU166 revealed an increase in the bond length (from 2.17 Å to 3.01 Å for 10 ns and 50 ns MD simulations, respectively). This finding raises intriguing questions regarding the stability of Cobicistat inside the active site of Mpro. Interestingly, DB02388 maintained its binding affinity with Mpro over the 50 ns MD simulations, with an average hydrogen bond length of 2.31 Å with GLU166.
DB02888 vs Cobicistat
Molecular docking and molecular dynamics results demonstrated promising binding affinity of DB02388 and Cobicistat (DB09065) for Mpro. DB02388, the imidazole-pyrimidine compound, is an investigational drug developed for the treatment of neurological disorders via inhibition mitogen-activated protein kinase 10 (MAPK10, known as c-Jun N-terminal kinase 3(JNK3))(Scapin et al., 2003; Wishart et al., 2006, 2018). Cobicistat, marketed under the name Tybost (formerly GS-9350), acts as a pharmacokinetic enhancer by inhibiting cytochrome P450 3A isoforms (CYP3A) for treating the infection with human immunodeficiency virus (HIV) (Deeks, 2014; Wishart et al., 2006, 2018). It is worth mentioning that Cobicistat, which was identified and highlighted in the current manuscript, has undergone in two clinical trials (ChiCTR2000029544 trial started on the third of February 2020 and ChiCTR2000029548 trial started on the fourth of February 2020)(Harrison, 2020). The trials’ results are not yet available to us.
Therefore, a comparison between the affinity and stability of DB02388 and Cobicistat was performed over 100 ns MD simulations. Snapshots for DB02388- and Cobicistat-Mpro complexes were collected every 10 ps over the 100 ns MD simulations, giving 10,000 snapshots in total. Based on the extracted snapshots, several energetic and structural analyses were performed, including binding energy per frame, hydrogen bond length, center-of-mass (CoM) distance and root-mean-square-deviation (RMSD).
Correlation between the binding-energy and time would give a deeper insight into the stability of drug-Mpro interactions. Therefore, MM-GBSA binding energy was calculated per frame for each of DB02388- and Cobicistat-Mpro complexes and plotted vs time in Figure 4.
Figure 4.
Variations in the MM-GBSA binding energies for Cobicistat (in black) and DB02388 (in red) with SARS-CoV-2 main protease (Mpro) through 100 ns MD simulation.
According to data in Figure 4, DB02388 showed narrow-fluctuated binding energies with an average value of −49.67 kcal/mol over the 100 ns MD simulations. However, higher fluctuations in the binding energy for Cobicistat with Mpro were noticed, ranging from −22.12 to −72.23 kcal/mol with an average value of −46.60 kcal/mol. These findings indicated that DB02388 bounds more tightly with Mpro complex than Cobicistat. The decomposition of MM-GBSA binding energy was also performed to reveal the nature of dominant interactions in DB02388- and Cobicistat-Mpro complexes (Table 3). Energy decomposition results showed that Evdw is the dominant force in the drug-Mpro binding affinity, −63.78 and −61.88 kcal/mol for DB02388-Mpro and Cobicistat-Mpro complexes, respectively. Besides, Eele interaction of DB02388 and Cobicistat with Mpro is favorable (−10.93 and −12.85 kcal/mol, respectively).
The hydrogen bond lengths between the DB02388 and Cobicistat and H-N of the key amino acid GLU166 residue were measured through 100 ns MD simulations and depicted in Figure 5(a). As shown in Figure 5(a), DB02388 displayed higher stability inside the active site of Mpro with an average hydrogen bond length of 2.32 Å. However, Cobicistat showed stable hydrogen bond through the first 33 ns with an average length of 1.95 Å. After 33 ns MD simulations, there was a dramatic increase in the hydrogen bond length, giving an average bond length of 5.35 Å throughout the rest 67 ns MD time. This reflects the downturn of Cobicistat-Mpro binding energy from −59.84 kcal/mol over the first 10 ns to −46.60 kcal/mol over 100 ns MD simulation.
Figure 5.
(a) Hydrogen bond lengths and (b) center-of-mass (CoM) distances between Cobicistat (in black) and DB02388 (in red) and the key residue amino acid GLU166 of SARS-CoV-2 main protease (Mpro) through 100 ns MD simulations.
The stability of DB02388- and Coblistat-Mpro complexes was evaluated through measuring the center-of-mass (CoM) distance between the drug and that of GLU166 amino acid residue over the simulated 100 ns (Figure 5(b)). According to Figure 5(b), the average CoM distance between DB02388 and GLU166 was nearly constant around 6.3 Å throughout the 100 ns MD simulations, while the corresponding distance for Cobicistat was much higher indicating instability of the latter complex.
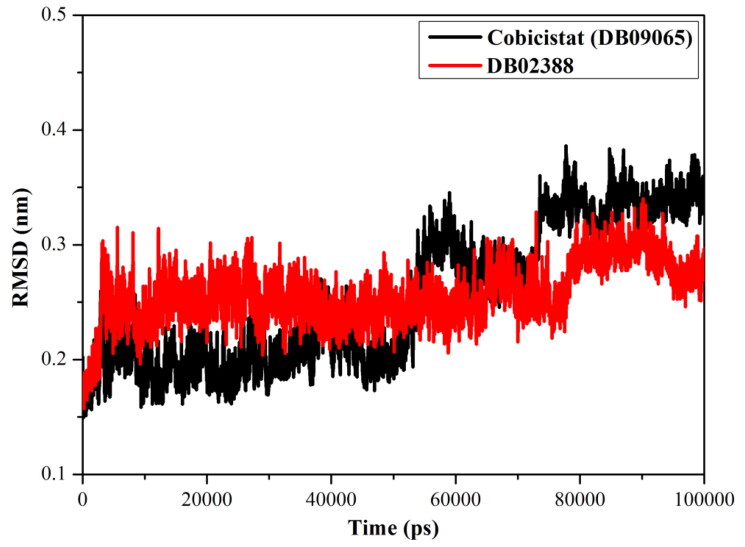
Structural changes in the Mpro were analyzed using root-mean-square deviation (RMSD) for the backbone atoms of the complexes relative to the starting structures. The RMSD for DB02388 and Cobicistat-Mpro over 100 ns MD simulations were calculated and plotted in Figure 6(a). As shown in Figure 6(a), the backbone of DB02388-Mpro complex was well stabilized after the first 5 ns period, giving RMSD with less than 0.05 nm over the rest 95 ns period. This confirms that DB02388 is tightly bonded in the active site of the Mpro and does not affect the overall topology of the protein. On the other hand, Cobicistat-Mpro complex showed considerable stability in the first 45 ns period, followed by a dramatic instability for the rest 55 ns, with RMSD 0.05 and 0.17 nm, respectively. The RMSD results are appropriate with lower stability of Cobicistat-Mpro compared with DB02388-Mpro complex.
Figure 6.
Root-mean-square-deviation (RMSD) of the backbone atoms from the initial structure for Cobicistat (in black) and DB02388 (in red) with the SARS-CoV-2 main protease (Mpro) through 100 ns MD simulations.
In summary, the energetic and structural analyses showed the high stability of DB02388-Mpro complex over Cobicistat-Mpro complex.
DB02888 vs Darunavir
Darunavir (DB01264) is a HIV protease inhibitor and has been recently subjected to clinical trial as COVID-19 drug (Harrison, 2020). Therefore, expensive molecular docking and MD simulations followed by MM-GBSA binding energy calculations were conducted for Darunavir with Mpro. The docking score and MM-GBSA binding energy over 100 ns MD simulations were estimated and compared to that of DB02388 (Table 4).
Table 4.
Expensive molecular docking scores and MM-GBSA binding energies for DB02388 and Darunavir in complex with SARS-CoV-2 main protease (Mpro).
| Drug Name/Code | Docking Score (kcal/mol) | Calculated
MM-GBSA binding energy (kcal/mol) |
Binding Features (Hydrogen bond length in Å) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| ΔEVDW | ΔEele | ΔEGB | ΔESUR | ΔGgas | ΔGSolv | ΔGbinding | |||
| DB02388 | −11.74 | −63.78 | −10.93 | 31.58 | −6.54 | −74.70 | 25.03 | −49.67 | GLU166 (2.10 Å), TYR54 (2.19 Å), ASP187 (2.11 Å) |
| Darunavir (DB01264) | −8.19 | −47.37 | −15.12 | 33.82 | −6.15 | −62.50 | 27.67 | −34.83 | GLU166 (1.94 Å, 2.88 Å), LEU167 (1.96 Å) |
Interestingly, the predicted binding mode of Darunavir with Mpro showed the ability of Darunavir to form three short hydrogen bonds with GLU166 and LEU167, compared to DB02388 which forms three hydrogen bonds (Figure 7). Despite its larger number of formed hydrogen bonds with active site’s amino acids, Darunavir-Mpro docking score was lower than of DB02388 (−8.19 and −11.74 kcal/mol, respectively). MM-GBSA binding energy decomposition of Darunavir-Mpro complex revealed that van der Waals (ΔEVDW) interaction between Darunavir and Mpro was only −47.37 kcal/mol, compared to −63.78 kcal/mol for DB02388, giving total binding energy (ΔGbinding) of −34.83 and −49.67 kcal/mol, respectively.
Figure 7.
2D LigPlot + representation of interactions of (a) DB02388 and (b) Darunavir with important amino acid residues of SARS-CoV-2 main protease (Mpro). Hydrogen bonds (in Å) and hydrophobic contacts are illustrated by green dashed lines and red arcs, respectively.
Conclusion
SARS-CoV-2 main protease (Mpro) is one of the most charming targets for the discovery of potent anti-COVID-19 drugs. In the presented study, in-silico drug repurposing technique was utilized to identify potent Mpro inhibitors. Filtration of DrugBank database was performed using molecular docking and molecular dynamics (MD) followed by molecular mechanics–generalized Born surface area (MM-GBSA) binding energy calculations. According to molecular docking calculations, 35 drugs showed docking scores higher than −11.0 kcal/mol with Mpro. Investigation of binding features revealed the ability of the identified drugs to form essential hydrogen bond with the key amino acid GLU166 besides other hydrogen bonds with active site’s amino acids. MM-GBSA binding energy calculations through MD simulations over 50 ns revealed that DB02388 and Cobicistat exhibited promising binding affinities of −49.67 and −46.60 kcal/mol, respectively, with Mpro. Energetic and structural analyses throughout 100 ns MD revealed the promising binding affinity and stability of DB02388 with Mpro over Cobicistat. Therefore, tenable conclusions from the study are i) DB02388 forms stable hydrogen bond with GLU166 with an average bond length of 2.32 Å, ii) center-of-mass distance between DB02388 and GLU166 was nearly constant over the 100 ns MD simulations, iii) narrow-fluctuated root-mean-square-deviation (RMSD) below 0.4 nm was observed over the 100 ns MD. The current results establish that DB02388 and Cobicistat hold promise as inhibitors against SARS-CoV-2 Mpro and are ready for in vitro inhibition against COVID-19.
Supplementary Material
Funding Statement
The computational work was completed with resources supported by the Science and Technology Development Fund, STDF, Egypt, Grants No. 5480 & 7972.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Bayly, C. I., Cieplak, P., Cornell, W., & Kollman, P. A. (1993). A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. The Journal of Physical Chemistry, 97(40), 10269–10280. 10.1021/j100142a004 [DOI] [Google Scholar]
- Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A., & Haak, J. R. (1984). Molecular-dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G., Ruan, L., Song, B., Cai, Y., Wei, M., Li, X., Xia, J., Chen, N., Xiang, J., Yu, T., Bai, T., Xie, X., Zhang, L., Li, C., … Wang, C. (2020). A trial of Lopinavir-Ritonavir in adults hospitalized with severe Covid-19. The New England Journal of Medicine, 382(19), 1787–1799. 10.1056/NEJMoa2001282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case, D. A., Betz, R. M., Cerutti, D. S., Cheatham, T. E., Darden, T. A., Duke, R. E., Giese, T. J., Gohlke, H., Goetz, A. W., Homeyer, N., Izadi, S., Janowski, P., Kaus, J., Kovalenko, A., Lee, T. S., LeGrand, S., Li, P., Lin, C., Luchko, T., ... Kollman, P. A. (2016). AMBER 2016. University of California. [Google Scholar]
- Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald: AnN⋅log(N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98(12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Deeks, E. D. (2014). Cobicistat: A review of its use as a pharmacokinetic enhancer of Atazanavir and Darunavir in patients with HIV-1 infection. Drugs, 74(2), 195–206. 10.1007/s40265-013-0160-x [DOI] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., Şahin, A. T., & Yelekçi, K. (2020). Drug repurposing for coronavirus (COVID-19): In silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 1–13. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, L. G., Dos Santos, R. N., Oliva, G., & Andricopulo, A. D. (2015). Molecular docking and structure-based drug design strategies. Molecules (Basel, Switzerland), 20(7), 13384–13421. 10.3390/molecules200713384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forli, S., Huey, R., Pique, M. E., Sanner, M. F., Goodsell, D. S., & Olson, A. J. (2016). Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nature Protocols, 11(5), 905–919. 10.1038/nprot.2016.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G. A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H. P., Izmaylov, A. F., Bloino, J., Zheng, G., Sonnenberg, J. L., Hada, M., Ehara, M., ... Fox, D. J. (2009). Gaussian 09 (Version Revision E01). Gaussian Inc. [Google Scholar]
- Gao, J., Tian, Z., & Yang, X. (2020). Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Bioscience Trends, 14(1), 72–73. 10.5582/bst.2020.01047 [DOI] [PubMed] [Google Scholar]
- Gasteiger, J., & Marsili, M. (1980). Iterative partial equalization of orbital electronegativity - a rapid access to atomic charges. Tetrahedron, 36(22), 3219–3228. 10.1016/0040-4020(80)80168-2 [DOI] [Google Scholar]
- Gordon, J. C., Myers, J. B., Folta, T., Shoja, V., Heath, L. S., & Onufriev, A. (2005). H++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Research, 33(Web Server issue), W368–W371. 10.1093/nar/gki464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedes, I. A., de Magalhães, C. S., & Dardenne, L. E. (2014). Receptor-ligand molecular docking. Biophysical Reviews, 6(1), 75–87. 10.1007/s12551-013-0130-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, C. (2020). Coronavirus puts drug repurposing on the fast track. Nature Biotechnology, 38(4), 379–381. 10.1038/d41587-020-00003-1 [DOI] [PubMed] [Google Scholar]
- Hawkins, P. C. D., Skillman, A. G., Warren, G. L., Ellingson, B. A., & Stahl, M. T. (2010). Conformer generation with OMEGA: Algorithm and validation using high quality structures from the protein databank and Cambridge structural database. Journal of Chemical Information and Modeling, 50(4), 572–584. 10.1021/ci100031x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgenfeld, R. (2014). From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. The FEBS Journal, 281(18), 4085–4096. 10.1111/febs.12936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., Jiang, R., Yang, X., You, T., Liu, X., … Yang, H. (2020). Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature, 582(7811), 289–293. 10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- Karaman, B., & Sippl, W. (2019). Computational drug repurposing: Current trends. Current Medicinal Chemistry, 26(28), 5389–5409. 10.2174/0929867325666180530100332 [DOI] [PubMed] [Google Scholar]
- Khan, R. J., Jha, R. K., Amera, G. M., Jain, M., Singh, E., Pathak, A., Singh, R. P., Muthukumaran, J., & Singh, A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, S. A., Zia, K., Ashraf, S., Uddin, R., & Ul-Haq, Z. (2020). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1751298 [DOI] [PubMed] [Google Scholar]
- Lai, C.-C., Shih, T.-P., Ko, W.-C., Tang, H.-J., & Hsueh, P.-R. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. International Journal of Antimicrobial Agents, 55(3), 105924–105932. 10.1016/j.ijantimicag.2020.105924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R. A., & Swindells, M. B. (2011). LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. Journal of Chemical Information and Modeling, 51(10), 2778–2786. 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- Luo, H., Tang, Q.-L., Shang, Y.-X., Liang, S.-B., Yang, M., Robinson, N., & Liu, J.-P. (2020). Can Chinese medicine be used for prevention of Corona Virus disease 2019 (COVID-19)? A review of historical classics, research evidence and current prevention programs. Chinese Journal of Integrative Medicine, 26(4), 243–250. 10.1007/s11655-020-3192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, J. A., Martinez, C., Kasavajhala, K., Wickstrom, L., Hauser, K. E., & Simmerling, C. (2015). ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. Journal of Chemical Theory and Computation, 11(8), 3696–3713. 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massova, I., & Kollman, P. A. (2000). Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspectives in Drug Discovery and Design, 18(1), 113–135. 10.1023/A:1008763014207 [DOI] [Google Scholar]
- Meng, X.-Y., Zhang, H.-X., Mezei, M., & Cui, M. (2011). Molecular docking: A powerful approach for structure-based drug discovery. Current Computer-Aided Drug Design, 7(2), 146–157. 10.2174/157340911795677602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OMEGA . (2013). OMEGA (Version 2.5.1.4). OpenEye Scientific Software. [Google Scholar]
- Pagadala, N. S., Syed, K., & Tuszynski, J. (2017). Software for molecular docking: A review. Biophysical Reviews, 9(2), 91–102. 10.1007/s12551-016-0247-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantsar, T., & Poso, A. (2018). Binding affinity via docking: Fact and fiction. Molecules, 23(8), 1899–1909. 10.3390/molecules23081899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakom, S., Iorio, F., Eyers, P. A., Escott, K. J., Hopper, S., Wells, A., Doig, A., Guilliams, T., Latimer, J., McNamee, C., Norris, A., Sanseau, P., Cavalla, D., & Pirmohamed, M. (2019). Drug repurposing: Progress, challenges and recommendations. Nature Reviews. Drug Discovery, 18(1), 41–58. 10.1038/nrd.2018.168 [DOI] [PubMed] [Google Scholar]
- Scapin, G., Patel, S. B., Lisnock, J., Becker, J. W., & LoGrasso, P. V. (2003). The structure of JNK3 in complex with small molecule inhibitors: Structural basis for potency and selectivity. Chemistry & Biology, 10(8), 705–712. 10.1016/S1074-5521(03)00159-5 [DOI] [PubMed] [Google Scholar]
- SZYBKI . (2016). SZYBKI (Version 1.9.0.3). OpenEye Scientific Software. [Google Scholar]
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., & Case, D. A. (2004). Development and testing of a general amber force field. Journal of Computational Chemistry, 25(9), 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., Sajed, T., Johnson, D., Li, C., Sayeeda, Z., Assempour, N., Iynkkaran, I., Liu, Y., Maciejewski, A., Gale, N., Wilson, A., Chin, L., Cummings, R., Le, D., … Wilson, M. (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Research, 46(D1), D1074–D1082. 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart, D. S., Knox, C., Guo, A. C., Shrivastava, S., Hassanali, M., Stothard, P., Chang, Z., & Woolsey, J. (2006). DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Research, 34(Database issue), D668–D672. 10.1093/nar/gkj067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, X., Yu, H., Yang, H., Xue, F., Wu, Z., Shen, W., Li, J., Zhou, Z., Ding, Y., Zhao, Q., Zhang, X. C., Liao, M., Bartlam, M., & Rao, Z. (2008). Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design. Journal of Virology, 82(5), 2515–2527. 10.1128/JVI.02114-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yella, J., Yaddanapudi, S., Wang, Y., & Jegga, A. (2018). Changing trends in computational drug repositioning. Pharmaceuticals (Pharmaceuticals), 11(2), 57–78. 10.3390/ph11020057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Q., Weber, E., & Yang, H. (2013). Recent developments on coronavirus main protease/3C like protease inhibitors. Recent Patents on anti-Infective Drug Discovery, 8(2), 150–156. 10.2174/1574891x113089990017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.