Abstract
Ongoing COVID-19 outbreak has raised a drastic challenge to global public health security. Most of the patients with COVID-19 suffer from mild flu-like illnesses such as cold and fever; however, few percentages of the patients progress from severe illness to death, mostly in an immunocompromised individual. The causative agent of COVID-19 is an RNA virus known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Despite these debilitating conditions, no medication to stop the disease progression or vaccination is available till now. Therefore, we aimed to formulate a multi-epitope vaccine against SARS-CoV-2 by utilizing an immunoinformatics approach. For this purpose, we used the SARS-CoV-2 spike glycoprotein to determine the immunodominant T- and B-cell epitopes. After rigorous assessment, we designed a vaccine construct using four potential epitopes from each of the three epitope classes such as cytotoxic T-lymphocytes, helper T-lymphocyte, and linear B-lymphocyte epitopes. The designed vaccine was antigenic, immunogenic, and non-allergenic with suitable physicochemical properties and has higher solubility. More importantly, the predicted vaccine structure was similar to the native protein. Further investigations indicated a strong and stable binding interaction between the vaccine and the toll-like receptor (TLR4). Strong binding stability and structural compactness were also evident in molecular dynamics simulation. Furthermore, the computer-generated immune simulation showed that the vaccine could trigger real-life-like immune responses upon administration into humans. Finally, codon optimization based on Escherichia coli K12 resulted in optimal GC content and higher CAI value followed by incorporating it into the cloning vector pET28+(a). Overall, these results suggest that the designed peptide vaccine can serve as an excellent prophylactic candidate against SARS-CoV-2.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2, multi-epitope vaccine, COVID-19, immunoinformatics, immune simulation, dynamics simulation
1. Introduction
Coronavirus disease 2019 (COVID-19) is an acute highly infectious disease. Patients with COVID-19 mostly feel like flu-like symptoms including cold and fever; a few percentages of the patients suffer from the respiratory tract infection leading to severe atypical pneumonia that eventually ends up in a case of fatality (Boopathi et al., 2020; Joshi et al., 2020; Vankadari & Wilce, 2020). Moreover, patients admitted to the intensive care unit were likely to report cardiovascular, respiratory disease, cerebrovascular, abdominal pain, endocrine, anorexia and digestive diseases (Chan et al., 2020; Wang et al., 2020). Acute cardiac injury and acute respiratory distress syndrome are commonly observed in severe cases and is strongly associated with the mortality rate (Abdelli et al., 2020; Wang et al., 2020; Wu et al., 2020). The infected patients can transmit the virus through coughs, sneezes, exhales and many other ways, hence, playing an essential role in human to human transmission (Chan et al., 2020). Infection with the virus is sometimes asymptotic, which also plays a vital role in the transmission process (Shen et al., 2020).
The COVID-19 outbreak has already been taken place all over the world with a total of 7,732,952 confirmed cases and 428,248 death cases (13 June 2020, 03:33 GMT) over 213 countries and territories around the world (www.worldometers.info). As of 13 June 2020, the outbreak in Bangladesh includes 81,523 confirmed and 1095 death cases, and the number of cases is increasing drastically day by day. The causative agent of the COVID-19 outbreak is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which is a positive-sense single-stranded RNA virus that belongs to the family of Coronaviridae (Boopathi et al., 2020; Umesh et al., 2020), and genus beta-coronavirus (Huang et al., 2020; Li & De Clercq, 2020). This virus was first identified in patients with a cluster of pneumonia in the province Wuhan of China on 29 December 2019 (Das et al., 2020; Kumar et al., 2020; Zhu et al., 2020). The World Health Organization (WHO) country office in China announced the official declaration of this virus on 31 December 2019 (Calisher et al., 2020; Heymann & Shindo, 2020). The new coronavirus was named ‘SARS-CoV-2’ by the International Committee on Taxonomy of Viruses (ICTV), and the disease caused by the pathogen was announced as COVID-19 by the WHO on 11 February 2020 (Gupta et al., 2020; Muralidharan et al., 2020; Wu et al., 2020). Epidemiological investigations of the Wuhan zoonotic virus revealed 89.1% nucleotide similarity between SARS-CoV-2 and previously originated group of SARS-like coronavirus (Khan et al., 2020; Vankadari & Wilce, 2020; Wu et al., 2020).
During the SARS-CoV-2 infection, the counts of CD4+ and CD8+ T-cells are increased in the peripheral blood, and cytotoxic granules are upturned with high concentration (Xu et al., 2020). The over-activation of T-cells causes injury to the immune system of the infected patients resulting in the characteristic feature of ‘Lymphopenia’ increasing the disease severity. In contrast, less effective T-cell responses may allow the progression of viral pathology and thus increased mortality in SARS-CoV-2 infected patients (Ahammad et al., 2019; Xu et al., 2020). The CD4+ and CD8+ responses provide a long-lasting protection against COVID-19 (Enayatkhani et al., 2020). Moreover, antibody-mediated immune response along with cellular immunity plays a critical role to induce protectivity against these infections (Enayatkhani et al., 2020). In recent studies, it has been illustrated that the nucleotide structure of this virus particle has a similarity with SARS-like coronavirus. Their genome was encoded with 16 different non-structural proteins and four main structural proteins, including spike (S), envelope (E), nucleocapsid (N), and membrane (M) proteins (Hasan et al., 2020; Sarma et al., 2020; Wahedi et al., 2020; Wu et al., 2020). The S-protein formed the viral outer layer; the N-protein helps in the viral replication, genome construction and host cellular response; the M-protein determined the envelope shape and the E-protein functions in production and maturation of the SARS-CoV-2 (Astuti & Ysrafil, 2020). The S-protein of the virus contains two subunit, S1 and S2; the S1 subunit of the S-protein recognizes the host T-cells while S2 subunit mediates fusion between the viral and host T-cells (Astuti & Ysrafil, 2020) and characterizes as a highly antigenic and surface exposure (Shang et al., 2020; Wrapp et al., 2020). The CD8+ and CD4+ T-cells recognize viral epitopes presented by the major histocompatibility complex class I (MHC I) and class II (MHC II), respectively (Abdellrazeq et al., 2020; Borthwick et al., 2020). The heterogeneity in T-cells responses to SARS-CoV-2 may, in part, be related to the capacity to recognize the viral antigens in the context of MHC I and MHC II proteins (Astuti & Ysrafil, 2020). It has been found that T-cell epitopes of SARS-CoV-2 spike protein elicit a T-cells immune response in patients who recover from the disease, and most of these immunogenic epitopes were localized to the S protein of the virus (Astuti & Ysrafil, 2020). The S-protein has strong interactions and binding affinity to the human angiotensin-converting enzyme 2 (ACE2) receptor and facilitates viral entry into the target cell (Sinha et al., 2020; Xu et al., 2020). The S-protein of the SARS-CoV-2 is the major host interacting protein, which causes cell adhesion and virulence to the human host (Vankadari & Wilce, 2020; Wu et al., 2020). The virus S-protein entry is mediated by ACE2, and results in an inflammatory cascade initiation by the innate immune system of the host (Astuti & Ysrafil, 2020). So, targeting S-protein can provide an immunogenic response in the human host, and has been chosen for designing a multi-epitopes vaccine candidate against the SARS-CoV-2. The structural pattern of SARS-CoV-2 protein can be recognized by the transmembrane toll-like receptor 4 (TLR4) which induces inflammatory cytokines or chemokines reaction. The TLR4 protein plays a vital role in the host pathogenesis. Moreover, the involvement of the receptors has also been reported in various immune protective responses by the host. Immune responses are a crucial step to the pathophysiology of the SARS-CoV-2 virus-related disease, and initiation of immune response targeting TLR4 can trigger the anti-viral host defense mechanisms necessary for the elimination of the COVID-19 related infection (Astuti & Ysrafil, 2020).
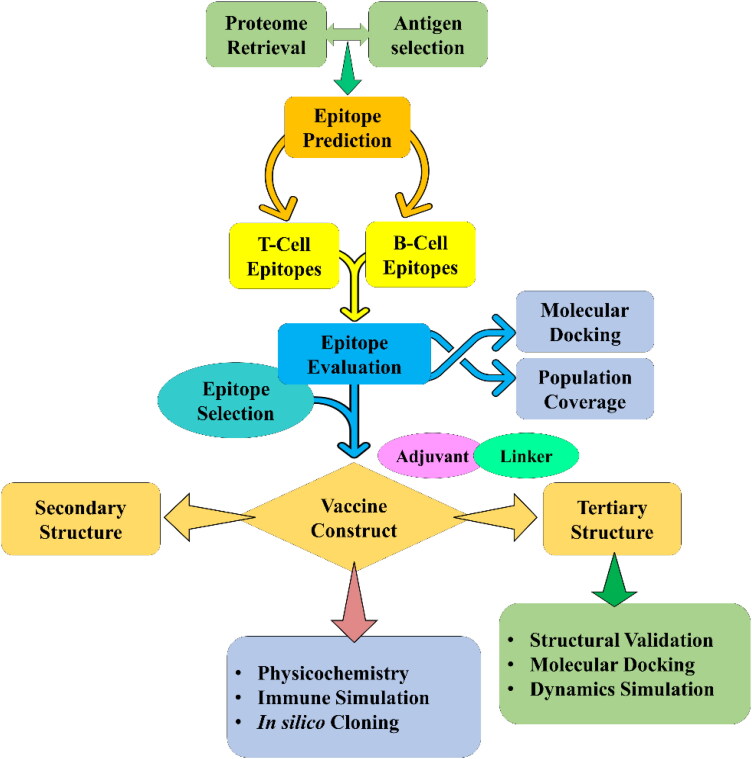
Vaccine is an immune-modulatory preparation that triggers a specific immune response against a foreign particle within the host body. A vaccine is now the primary demand to save millions of people from the COVID-19 pandemic. The current world situation is releasing the necessity of an implausible and effectiveness of different anti-viral drugs or vaccine candidates against the SARS-CoV-2. However, no effective drug or vaccine candidates have been developed that can fight against the SARS-CoV-2 (Elfiky, 2020). Therefore, a multi-epitope vaccine consisting of potential T- and B-cell epitopes can be an ideal approach for the prevention of COVID-19 (Astuti & Ysrafil, 2020). The vaccine can produce both cellular and humoral immune responses against specific pathogens without producing any immune complications. Besides, it is very easy to control, cause the effectiveness of the vaccine to be regulated by choosing the specific and desired allelic interactions, which provide robust and diverse immune response over a large group of people (Elfiky, 2020). In multi-epitope vaccine, the biohazard risk is lower as compared to other types of immunizations. In this research, a multi-epitope vaccine has been constructed using the immunoinformatics approach. Epitopes used for the vaccine construction were non-toxic, non-allergenic, highly immunogenic and antigenic. A sufficient number of linkers were used to combine those selected epitopes resulting in busting the immunogenic activity of SARS-CoV-2 vaccine (Gaafar et al., 2019; Li et al., 2014). A flow chart representing the overall procedure from the antigen selection to vaccine construction and evaluation is illustrated in Figure 1.
Figure 1.
Schematic representation of the overall workflow applied in the current study.
2. Materials and methods
2.1. Proteome retrieval and antigen selection
For antigen selection, we collected available SARS-CoV-2 proteomes from the ViPR (https://www.viprbrc.org/) database (Pickett et al., 2012). The outer membrane of the SARS-CoV-2 is formed by the spike glycoproteins. With the help of these glycoproteins, they adhere to the human host and enter into the host immune system (Peng et al., 2020). Due to the direct involvement of glycoproteins in pathogenesis, we considered the spike glycoprotein of the SARS-CoV-2 for multi-epitope vaccine design. Initially, we isolated all the spike glycoprotein, and the selected protein sequences of the virus were downloaded in FASTA format. The protective antigens of the surface glycoprotein were checked by VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/) server (Doytchinova & Flower, 2007) and ANTIGENpro (http://scratch.proteomics.ics.uci.edu/) server with a threshold value 0.4 was set for both of them (Magnan et al., 2010). Finally, we selected the spike glycoprotein with the highest antigenic score for further investigations.
2.2. Prediction and assessment of cytotoxic T-lymphocyte epitopes
Cytotoxic T-lymphocytes (CTLs) represent one of several types of cells of the immune system that have the capacity to kill other infectious cells directly (Xu et al., 2020). They go right away inside the virus-cell and play an important role in the host defense mechanism. For the prediction of CTLs epitope, the sequence of the selected protein was submitted into the NetCTL v1.2 server available at http://www.cbs.dtu.dk/services/NetCTL/ (Larsen et al., 2007). The predicted epitopes were further assessed through the VaxiJen v2.0 (Doytchinova & Flower, 2007), MHC class I immunogenicity (http://tools.iedb.org/immunogenicity/) (Calis et al., 2013), ToxinPred (http://crdd.osdd.net/raghava/toxinpred/) (Gupta et al., 2013), and AllerTop v2.0 (https://ddg-pharmfac.net/AllerTOP/) (Dimitrov et al., 2013) servers. The default parameters of those servers were used for all the predictions.
2.3. Prediction and evaluation of helper T-lymphocyte epitopes
Helper T-cells (HTLs) are an integral part of adaptive immunity that recognizes foreign antigens and activates B and cytotoxic T-cells resulting in destruction of the infectious pathogen (Xu et al., 2020). To determine the HTL epitopes, we used the IEDB’s MHC class II binding allele prediction tool, available at http://tools.iedb.org/mhcii/. The HTL epitopes were selected based on a percentile rank of 5% using the CONSENSUS method (Wang et al., 2010). The predicted epitopes were further evaluated based on their antigenicity and cytokine, i.e. interferon-γ (IFNγ), interleukin-4 (IL4) and interleukin-10 (IL10) inducing abilities. The antigenicity was anticipated with the VaxiJen v2.0 server while IFNγ, IL4, and IL10 features were predicted using IFNepitope (http://crdd.osdd.net/raghava/ifnepitope/) (Dhanda et al., 2013), IL4pred (http://crdd.osdd.net/raghava/il4pred/) (Dhanda et al., 2013) and IL10pred (http://crdd.osdd.net/raghava/IL-10pred/) (Nagpal et al., 2017) servers, respectively, with default parameters.
2.4. Prediction and assessment of linear B-lymphocyte epitopes
B-cell epitopes are essential to induce humoral or antibody-mediated immunity. B-cells consist of groups of amino acids that interact with the secreted antibodies and activate the immune system to destroy the pathogens (Nain et al., 2019). Therefore, we predicted the linear B-lymphocyte (LBL) epitopes using the iBCE-EL server, available at http://www.thegleelab.org/iBCE-EL/ with default parameters (Manavalan et al., 2018). The predicted LBL epitopes were also evaluated using the VaxiJen v2.0, ToxinPred, and AllerTop v2.0 servers.
2.5. Estimation of population coverage
In computational vaccine design, the population coverage directly indicates the worldwide effectiveness of the vaccine by evaluating the prevalence of HLA (Human Leukocyte Antigen) alleles related to the epitope of interest. Therefore, the population coverage was calculated using the T-cell epitopes with their respective HLA binding alleles. To accomplish this, selected epitopes along their allelic information was submitted to the IEDB population coverage tool (Bui et al., 2006).
2.6. Peptide modeling and molecular docking
For modeling, the selected epitopes of CTL and HTL were submitted into PEP-FOLD v3.0 (https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/) server. The sOPEP sorting scheme with 200 simulations was selected for the operation (Latysheva & Babu, 2016). By analyzing the epitope-wise HLA binding alleles, allele HLA-B*15:01 and HLA-C*06:02 were considered for selected CTL epitopes, while DRB1*01:01 and DRB1*15:01 were selected for HTL epitopes. The crystal structures of the HLA alleles were retrieved from the Protein Data Bank (PDB) (https://www.rcsb.org/) (Berman et al., 2000) followed by processing with BIOVIA Discovery Studio 2017. For molecular docking, a grid-box around the active site of each HLA allele was defined by the AutoDock tool. Finally, molecular docking was performed between the epitopes and respective HLA alleles using the AutoDock Vina script (Trott & Olson, 2010). The respective co-crystal ligands were used as the positive control to compare the epitope binding efficiency. The docked complex was visualized in BIOVIA Discovery Studio 2017.
2.7. Formulating of multi-epitope vaccine
The vaccine construct was designed by using the selected CTL, HTL, and LBL epitopes as well as a suitable adjuvant that was linked by the appropriate linkers (Dorosti et al., 2019; Nain et al., 2019). Here, we used TLR4 agonist as the adjuvant since TLR4 was recognized by viral glycoproteins, and the adjuvant is required for optimal translation and maximal rate of synthesis of the target vaccine candidate (Olejnik et al., 2018; Pandey et al., 2018). Therefore, 50S ribosomal protein L7/L12 (NCBI ID: P9WHE3) was considered as the adjuvant to improve the immunogenicity of the vaccine candidate. The adjuvant was linked to the vaccine front with a bi-functional linker EAAAK that has the ability of several lengths of helix-forming peptides to separate two weakly interacting β domains. In contrast, the selected CTL was linked with the help of Ala-Ala-Tyr (AAY) linkers, the HTL was linked with Gly-Pro-Gly-Pro-Gly (GPGPG) linkers and the LBL was linked with Lys-Lys (KK) linker (Dorosti et al., 2019; Nain et al., 2019). The AAY linker is a type of cleavage site of proteasomes that was used to influence protein stability, reduce less immunogenicity and enhance epitope presentation (Abdellrazeq et al., 2020; Borthwick et al., 2020). The GPGPG, known as the glycine–proline linker, prevents the formation of ‘junctional epitopes’ and facilitates the immune processing, where the bi-lysine KK linker helps to preserve their independent immunogenic activities of the vaccine construct.
2.8. Physicochemical and immunological evaluation
The physiochemistry indicates the basic properties of a protein. The physicochemical features of the vaccine were anticipated using the ProtParam server available at https://web.expasy.org/protparam/ to understand the fundamental nature of the vaccine (Gasteiger et al., 2005). We also evaluated the immunological properties through VaxiJen v2.0 (Doytchinova & Flower, 2007), MHC-I immunogenicity (Calis et al., 2013), AllerTop (Dimitrov et al., 2013), and SOLpro (Magnan et al., 2009) servers.
2.9. Secondary structure prediction
The two-dimensional (2D) structural features such as alpha-helix, beta-turn, and random coils of the construct were identified by SOPMA (Self-Optimized Prediction Method with Alignment) server at https://npsa-prabi.ibcp.fr/NPSA/npsa_seccons.html (Geourjon & Deléage, 1995) and PSIPRED v4.0 (PSI-blast based secondary structure prediction) server at http://bioinf.cs.ucl.ac.uk/psipred/ (Buchan et al., 2013) with default parameters. SOPMA has more than 80% prediction accuracy (Geourjon & Deléage, 1995). The 2D structural features were retrieved and evaluated to understand the composition quality of the vaccine.
2.10. Homology modeling, 3D structure refinement and validation
The constructed vaccine was submitted into I-TASSER (Iterative Threading Assembly Refinement) online web portal (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) for three-dimensional (3D) structure prediction (Roy et al., 2010). The I-TASSER web produces the structure of the protein and its functions most accurately using a state-of-the-art algorithm in the form of a 3D structure (Roy et al., 2010). This web server can predict and determine the C-score, TM-score value, RMSD and top five models of the given protein sequence. The produced 3D structure was downloaded into the PDB format, which was chosen based on the C-score value. The server contains a C-score ranging from –5 to 2, where a higher value indicates a protein model with high confidence. The identified 3D structure was submitted into the GalaxyRefine (http://galaxy.seoklab.org/refine) online web-based server for the refinement of the vaccine structure. This webserver was run by the CASP10 refine technique (Nugent et al., 2014). The GalaxyRefine website provides the RMSD, energy score and overall quality score. The refined structure was downloaded, and the selected structure was identified depending on the energy score of the lowest and highest RMSD value. The refined and identified structure was visualized using the PyMOL v2.3.4 software (DeLano, 2002). The resulted 3D structure was evaluated depending on the Ramachandran plot score (vaccine structure validity) and Z-score value that determine the standard deviations from the mean value (Z-score within the known native protein range indicating the good quality of the prepared model). The Ramachandran plot was analyzed by the Rampage server (http://mordred.bioc.cam.ac.uk/∼rapper/rampage.php), which runs considering allowed and disallowed regions of amino acid (Lovell et al., 2003; Ramachandran et al., 1963); and Z-score plot was analyzed by the ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php) tool (Wiederstein & Sippl, 2007).
2.11. Molecular docking studies
Molecular docking studies can reveal the binding interactions between modeled protein and receptor molecules. For this purpose, we submitted the refined vaccine model as ligand and TLR4 protein as immunological receptor into the ClusPro v2.0 server, available at https://cluspro.bu.edu/, for molecular docking (Kozakov et al., 2017). The TLR4 receptor (PDB ID: 4G8A) was selected and downloaded from the PDB server (Sussman et al., 1998). Initially, the receptor was prepared by separating the attached ligand from the protein, followed by the removal of waters and other chemicals. All these processes were performed in PyMOL v2.3.4 software (DeLano, 2002). Binding interactions and residues involved in the interacting plane were analyzed with Discovery Studio 2017.
2.12. Molecular dynamics simulation
For molecular dynamics (MD) simulation, we used both software and server-based tools to evaluate the dynamics and stability of the vaccine–receptor complex critically. The stability of the docked complex was evaluated by a highly intuitive and accurate molecular dynamic simulation tool YASARA, where parameters for macromolecules to facilitate simulations were generated using the AMBER14 force field (Nain et al., 2020). We evaluated the stability, fluctuation and compactness of the vaccine–receptor complex in terms of root mean square deviation (RMSD), root mean square fluctuation (RMSF), and radius of gyration (Rg) values, respectively. Also, the complex was submitted to the iMODS server, available at http://imods.chaconlab.org/ (López-Blanco et al., 2014). Based on the normal mode analysis (NMA), this server provides eigenvalues, deformability, B-factors, and elastic network model to clarify the aggregate protein movement in the inside directions.
2.13. Immune response simulation
To evaluate the possible immune response of the vaccine, the whole construct was submitted into C-IMMSIM v10.1 server available at http://www.cbs.dtu.dk/services/C-ImmSim-10.1/, and the generated responses were retrieved for detailed observation (Rapin et al., 2010). In this case, we considered a minimum interval period of 30 days between two doses, as described earlier (Castiglione et al., 2012). In silico administration of three injections were given with time steps of 1, 84, and 168, respectively, where one-time step is equal to 8 h in real life. The maximum value for simulation steps was set to 300, while the rest of the stimulation parameters were kept default.
2.14. Codon adaptation and in silico cloning
For the expression of a foreign gene in a host organism, codon optimization is necessary according to the specific host organism (Grote et al., 2005). Therefore, the construct was submitted into the JCat server (http:/jcat.de/) for the codon adaptation. Herein, we considered widely used E. coli K12 as the host, and the whole operation is carried out by avoiding the following three criteria: (1) restriction enzymes cleavage sites, (2) binding sites of the prokaryotic ribosome and (3) rho-independent termination of transcription. The adapted sequence was evaluated based on the codon adaptation index (CAI) value and guanine–cytosine (GC) content (Grote et al., 2005). Finally, the adapted nucleotide sequence was used for in silico cloning into the pET28a (+) expression vector. The whole in silico cloning operation was executed in SnapGene v4.2 software (Goldberg et al., 2018).
3. Results
3.1. Highest antigenic protein selection
We found 250 S-proteins from all the retrieved SARS-CoV-2 proteomes. Based on the antigenicity, we selected a spike protein with an antigenic score of 0.4646 (VaxiJen) and 0.717 (ANTIGENpro), which was the highest among all tested proteins. The length of the selected S-protein was 1273 amino acids long while the GenBank accession was QIC53213. The primary sequence of the selected protein was used for further analysis.
3.2. Potential CTL epitopes
A total of 270 CTL epitopes, each with a length of 9 amino acids, were predicted from the selected spike protein. Assessment revealed that twenty-nine CTL epitopes were antigenic, immunogenic, non-toxic and non-allergenic (Supplementary Table S1). Due to the high number of potential epitopes, we selected the top four CTL epitopes for the final vaccine construction based on the antigenicity score (Table 1).
Table 1.
The selected CTL epitopes for the final vaccine construction.
| Epitope | C-score | Antigenicity | Immunogenicity | Toxicity | Allergenicity |
|---|---|---|---|---|---|
| RQIAPGQTG | 0.8698 | 1.7890 | Positive | Negative | Negative |
| VVFLHVTYV | 1.0304 | 1.5122 | Positive | Negative | Negative |
| IAIVMVTIM | 1.1592 | 1.1339 | Positive | Negative | Negative |
| VRFPNITNL | 1.0215 | 1.1141 | Positive | Negative | Negative |
C-score is the combined score provided by the NetCTL server.
3.3. Potential HTL epitopes
A total of 478 HTL epitopes, each with a length of 15 amino acids, were identified initially using the IEDB server. Among them, only 16 HTL epitopes were able to induce the evaluated three types of cytokines, such as IFNγ, IL4, and IL10 (Supplementary Table S2). Likewise, we considered the top four HTL epitopes for incorporating into the final vaccine construct based on the antigenic score (Table 2).
Table 2.
The selected HTL epitopes for the final vaccine construction.
| Epitope | Antigenicity | IFNγ | IL4 | IL10 | Toxicity | Allergenicity |
|---|---|---|---|---|---|---|
| QYIKWPWYIWLGFIA | 1.1541 | Positive | Inducer | Inducer | Negative | Negative |
| LLLQYGSFCTQLNRA | 0.9471 | Positive | Inducer | Inducer | Negative | Negative |
| VVLSFELLHAPATVC | 0.8618 | Positive | Inducer | Inducer | Negative | Negative |
| NDPFLGVYYHKNNKS | 0.8199 | Positive | Inducer | Inducer | Negative | Negative |
IFNγ stands for interferon-gamma while IL-4 and IL-10 indicate the interleukin-4 and interleukin-10, respectively.
3.4. Potential LBL epitopes
Preliminary analysis revealed a total of 61 LBL epitopes, each with a length of 12 amino acids. Later with further evaluation, 14 epitopes were found as antigenic, non-toxic and non-allergenic (Supplementary Table S3). Out of 14 LBL epitopes, we selected the top four LBL epitopes for vaccine design purposes based on the antigenicity score (Table 3).
Table 3.
The selected LBL epitopes for the final vaccine construction.
| Epitope | Probability | Antigenicity | Allergenicity | Toxicity |
|---|---|---|---|---|
| IAPGQTGKIADY | 0.8088 | 1.6626 | Negative | Negative |
| YQTSNFRVQPTE | 0.8172 | 1.1569 | Negative | Negative |
| TRTQLPPAYTNS | 0.8391 | 0.9332 | Negative | Negative |
| EPLVDLPIGINI | 0.8227 | 0.7968 | Negative | Negative |
3.5. Worldwide population coverage
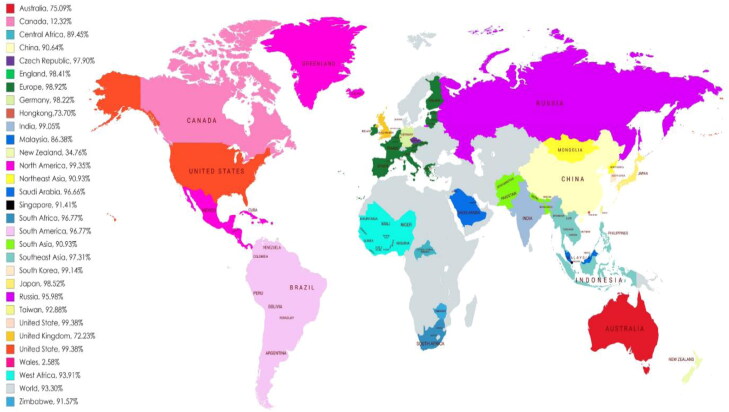
The selected CTL and HTL epitopes, along with their binding alleles, were used to evaluate the population coverage, as shown in Figure 2. Both CTL and HTL epitopes provided a high percentage (93.30%) of population coverage across the world. The selected epitopes showed interactions with a high number of HLA alleles from different countries such as the United States (99.38%), North America (99.35%), South Korea (99.14%), South Asia (99.10%) and India (99.05%). This result suggests that the vaccine designed with these epitopes could be effective on most of the population in the world (Figure 2 and Supplementary Table S4).
Figure 2.
Worldwide population coverage map predicted based on the selected T-cell epitopes.
Table 4.
Binding affinities and interaction between selected epitopes and HLA alleles.
| T-cell epitope | HLA allele | Epitope affinity (kcal/mol) | Control affinity (kcal/mol) | Number of hydrogen bonds (CHB) | Residues involved in CHB networks (n) |
|---|---|---|---|---|---|
| IAIVMVTIM | HLA-B*15:01 | –7.1 | –9.2 | 11 (9) | Tyr9, Rg62, Gln65, Asn70, Ser77, Trp147, Thr7, Ile8, Met9, Ile1, Ala2, Ile3, Tyr74 (13) |
| RQIAPGQTG | HLA-B*15:01 | –7.9 | –9.2 | 9 (6) | Arg62, Ser77, Asn80, Trp147, Gln2, Gly9, Gly6, Arg1, Gln2, Ala4 (10) |
| VRFPNITNL | HLA-C*06:02 | –8.3 | –8.2 | 13 (12) | Arg69, Gln70, Asn77, Lys80, Tyr84, Lys146, Trp147, Trp156, Thr7, Leu9, Asn5, Asn8, Ile6, Thr143 (15) |
| VVFLHVTYV | HLA-C*06:02 | –9.0 | –8.2 | 12 (8) | Lys80, Tyr84, Lys146, Val2, Thr7, Tyr8, Val9, Lys66, Asn77, Thr143 (10) |
| NDPFLGVYYHKNNKS | DRB1*01:01 | –6.9 | –7.7 | 15(11) | Gln9, Asn62, Trp61, Gln64, Arg71, His81, Asn82, Asn1, Tyr9, Lys14, Tyr8, Pro3, Asp2, Ser53, Glu28 (15) |
| VVLSFELLHAPATVC | DRB1*01:01 | –6.6 | –7.7 | 11(8) | Glu55, Asn69, Gln70, Arg71, Thr77, Asn82, Ala12, Thr13, Val14, Val1, Glu6, Ser4 (12) |
| LLLQYGSFCTQLNRA | DRB1*15:01 | –5.8 | –7.4 | 11(9) | Gln9, Ser53, Asn62, Trp308, His328, Asn329, Leu3, Asn13, Arg14, Gln11, Arg14, Phe8, Gln11, Leu1 (14) |
| QYIKWPWYIWLGFIA | DRB1*15:01 | –6.9 | –7.4 | 9(6) | Ser53, Glu55, Asn62, His328, Trp7, Ala15, Phe13, Tyr8, Ile14, Ile3 (10) |
CHB denotes the classical hydrogen bonds while ‘n’ is the number of interacting residues.
3.6. Docking studies of epitope and alleles
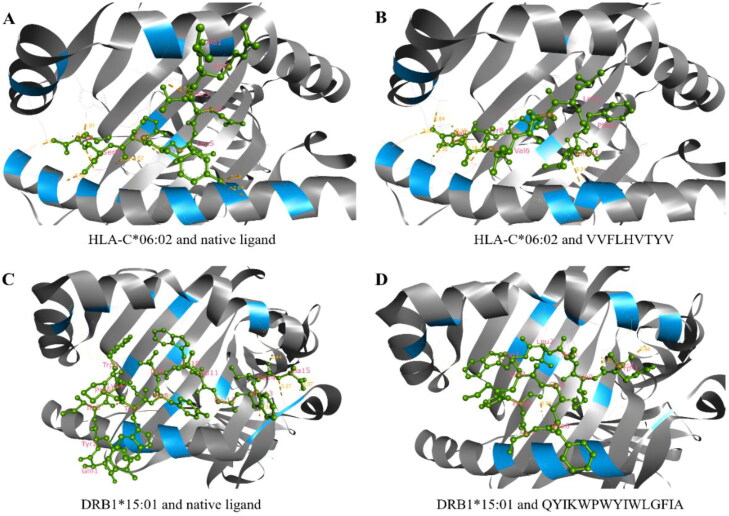
We used the docking method to validate the efficacy of selected epitopes in binding their respective HLA alleles. The epitopes, along with their respective docking allele, binding affinities, interactions and residues involved in the hydrogen bonds, are described in Table 4. The binding affinities of CTL epitopes were between –7.1 and –9.0 kcal/mol, while for HTL epitopes, it was between –5.8 and –6.9 kcal/mol. The binding affinities were either very close or even higher than that of the positive control (Table 4). In addition to the tabulated details, we presented the best interacting CTL (VVFLHVTYV) and HTL (QYIKWPWYIWLGFIA) epitopes in Figure 3. Herein, the best CTL epitope produced a total of 12 hydrogen bonds, in which 8 were classical interactions involved with the active site residue Lys80, Tyr84, Lys146, Val2, Thr7, Tyr8, Val9, Lys66, Asn77, and Thr143. On the other hand, the best HTL epitope showed nine hydrogen bonds, including six classical interactions while it interacted with Ser53, Glu55, Asn62, His328, Trp7, Ala15, Phe13, Tyr8, Ile14, and Ile3 residues.
Figure 3.
Molecular docking between epitopes and their respective binding alleles. Herein, we provided the docking interactions of the best HTL and CTL epitopes as the representative of the all selected epitopes, where (A) docking between the HLA-C*06:02 alleles and its native ligand as a positive control, (B) docking between the HLA-C*06:02 alleles and CTL epitope VVFLHVTYV, (C) docking between the DRB1*15:01 alleles and its native ligand as a positive control and (D) docking between the DRB1*15:01 alleles and HTL epitope QYIKWPWYIWLGFIA. Both epitopes scored higher binding affinity toward the allele than that of their respective native ligands.
3.7. Vaccine construct and basic properties
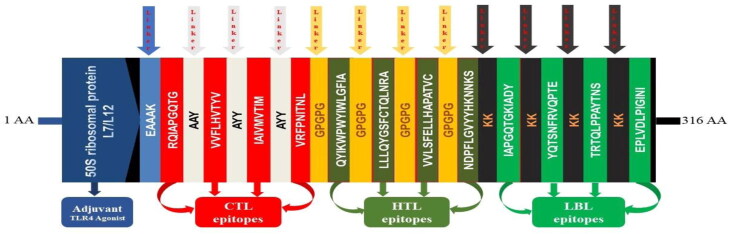
The vaccine construct was formulated using the previously selected 12 epitopes belonging to three different classes (4 CTL, 4HTL, and 4 LBL). The epitopes were added together with AAY, GPGPG and KK linkers, respectively, as shown in Figure 4. An adjuvant was added ahead of the construct to improve the immunogenicity. The TLR4 agonist 50S ribosomal protein L7/L12 was linked to the first CTL epitope as an adjuvant by using EAAAK linker. The final vaccine construct was 316 amino acids long (Figure 4).
Figure 4.
Graphical map of the formulated multi-epitope vaccine construct. The vaccine constructs included (left to right) an adjuvant, CTL, HTL and LBL epitopes are shown in the dark blue, red, olive green and green rectangular boxes. Herein, the adjuvant and the first CTL epitope were linked by EAAAK linker (blue), CTL epitopes were added together by AYY linkers (off-white), HTL epitopes by GPGPG linkers (orange), and LBL epitopes by KK linkers (black).
3.8. Physicochemical properties and immunological evaluation
The physicochemical properties of the vaccine construct were assessed as shown in Table 5. The molecular weight of the construct was found to be 33,614.95 Da. At the same time, other properties such as theoretical isoelectric point (pI) was 8.28, chemical formula was C1539H2432N390O439S6, instability index was 24.33, the aliphatic index was 93.64, and grand average of hydropathicity was 0.035. In addition, physicochemical features and the immunological potency of the construct were evaluated. For instance, the antigenicity of the construct was 0.6166, while immunogenicity was 1.58298. Furthermore, the vaccine was non-allergenic and soluble, with a score of 0.871753 out of 1 (Table 5).
Table 5.
Antigenic, allergenic and physicochemical characteristics of the construct.
| Characteristics | Finding | Remark |
|---|---|---|
| Number of amino acids | 316 | Suitable |
| Molecular weight | 33,614.95 Da | Average |
| Theoretical pI | 8.28 | Slightly basic |
| Chemical formula | C1539H2432N390O439S6 | – |
| Extinction coefficient (at 280 nm in H2O) | 34505 | – |
| Estimated half-life (mammalian reticulocytes, in vitro) | 30 h | – |
| Estimated half-life (yeast-cells, in vivo) | >20 h | – |
| Estimated half-life (E. coli, in vivo) | >10 h | – |
| Instability index of vaccine | 24.33 | Stable |
| Aliphatic index of vaccine | 93.64 | Thermostable |
| Grand average of hydropathicity (GRAVY) | 0.035 | Hydrophobic |
| Antigenicity | 0.6166 | Antigenic |
| Immunogenicity | 1.58298 | Immunogenic |
| Allergenicity | No | Non-allergen |
| Solubility | 0.871753 | Soluble |
The secondary structural features include α-helix, β-strand and random coils that were evaluated using two different servers. The SOPMA server predicted 39.56% α-helix, 23.42% β-strand and 37.03% random coils in the construct (Table 6, Supplementary Figure S1). On the other hand, the PSIPRED server anticipated the features as 42.41% α-helix, 10.44% β-strand, and 47.15% random coils (Table 6, Supplementary Figure S1).
Table 6.
The secondary structural features of the vaccine construct.
| SOMPA
server |
PSIPRED
server |
|||
|---|---|---|---|---|
| Features | Amino acid | Percentage | Amino acid | Percentage |
| Alpha helix | 125 | 39.56% | 134 | 42.41% |
| Beta strand | 74 | 23.42% | 33 | 10.44% |
| Random coil | 117 | 37.03% | 149 | 47.15% |
3.9. Tertiary structure, refinement and validation
In homology modeling, the I-TASSER server used 1DD4 (PDB ID) as the best template to generate the top five models. Among the five models, we considered the model with the lowest C-score (–4.82) as recommended by the server (Supplementary Figure S4). A structural representation of the designed vaccine is provided in Figure 5. After refinement, the vaccine (model 3) showed 84.7% residues in the favorable region in the Ramachandran plot, with GDT-HA score 0.9146, RMSD value 0.526, MolProbity 2.597, Clash score 27.9 and Poor rotamers score 0.8 (Supplementary Table S5).
Figure 5.
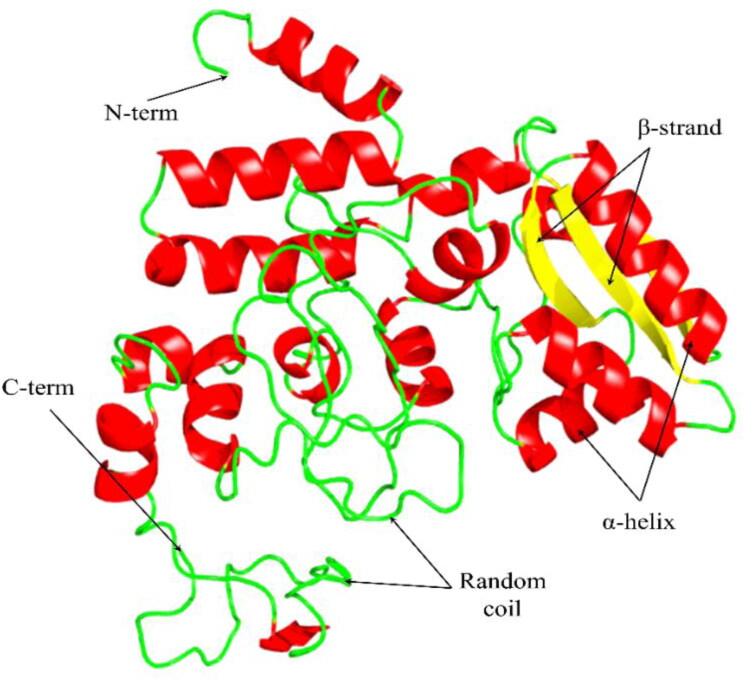
The tertiary structure of the designed vaccine construct. The three domains of the construct have indicated by red (α-helix), yellow (β-strand), and green (random coil) colors.
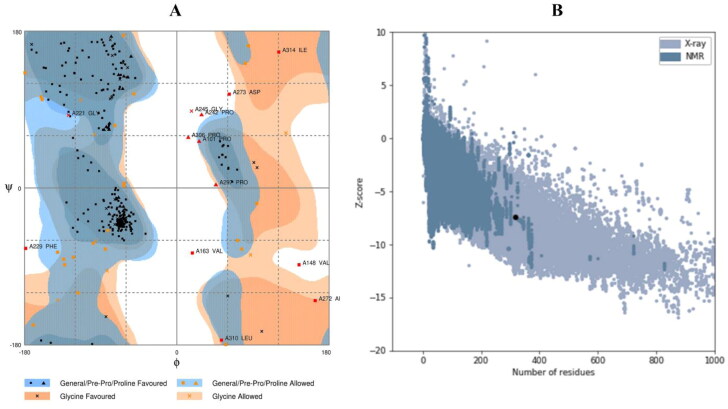
The refined 3D vaccine model was further validated with the RAMPAGE and ProSA-web servers. Before refinement, the Ramachandran plot of the vaccine showed 61.1% residues in the favorable region and 27.4% in allowed regions, while 11.5% residues in disallowed regions (Supplementary Figure S3). The Ramachandran plot of the refined vaccine model showed 86.3% residues in the favorable region and 9.6% in allowed regions, while 4.1% residues in disallowed regions (Figure 6(A)). Likewise, the crude model showed a Z-score value of –7.17, while the refined model provided a value of –7.4 (Figure 6(B), Supplementary Figure S4).
Figure 6.
Validation of the tertiary structure of the vaccine. (A) The Ramachandran plot statistics represent the most favorable, accepted, and disallowed regions with a percentage of 86.3, 9.6, and 4.1%, respectively, and (B) The ProSA-web representing the Z-score of –7.4 for the refined vaccine model.
3.10. Molecular docking studies
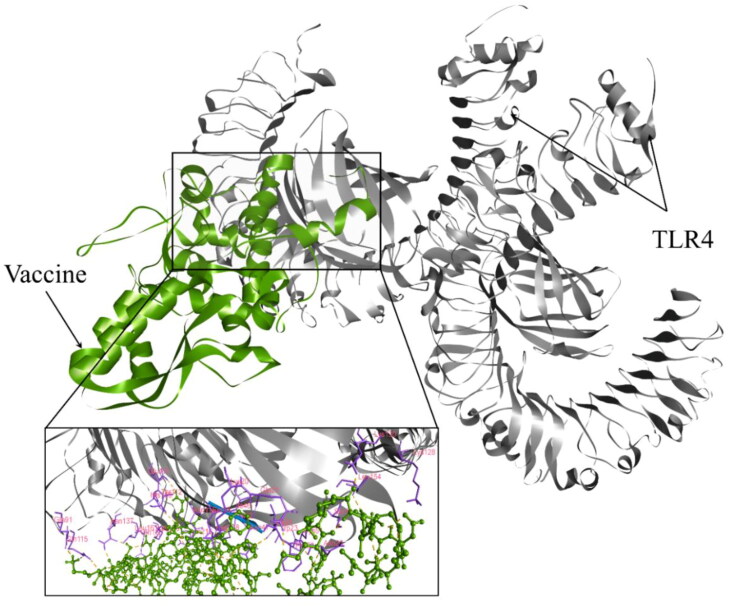
The docking between the vaccine (ligand) and TLR4 (receptor) was performed to anticipate their binding affinity and interactions. In doing so, the ClusPro v2.0 server provided 30 docked complexes with different poses. Among them, we selected the complex with the least energy score and binding pose with functional interactions (Supplementary Table S6). Thus, model 1 fulfilled the inclined criteria. Hence, it was picked as the best vaccine–TLR4 complex, which had an energy score of –964.6 (Figure 7). The selected complex was analyzed for binding interactions and involved in active site residues. The number of hydrogen and hydrophobic bonds present in the interaction plane was 35 and 9, respectively. Among the hydrogen bonds, 28 were classical hydrogen bonds (CHB). The interacting residues in the CHB from the vaccine were Ala36, Arg283, Asn281, Gln267, Gln285, Glu30, Glu33, Lys3, Met1, Pro184, Pro265, Thr268, Thr279, Thr31, Thr6, Leu186, Lys275 and Ser280. Moreover, associated TLR4 active site residues were Gln115, Asn137, Gln163, Lys186, Lys20, Gln21, Ser45, Asn47, Cys51, Asn114, Lys130, Gln91, His159, Asn26, Glu111, Leu154, Thr112 and Trp23 (Figure 7). Other hydrogen bond interactions were as follows: four were electrostatic salt bridges, two were carbon–hydrogen bonds and a single Pi-donor hydrogen bond.
Figure 7.
Molecular docking between the vaccine and the TLR4 receptor. The interacting residues from the vaccine were Ala36, Arg283, Asn281, Gln267, Gln285, Glu30, Glu33, Lys3, Met1, Pro184, Pro265, Thr268, Thr279, Thr31, Thr6, Leu186, Lys275, and Ser280 while associated TLR4 active site residues were Gln115, Asn137, Gln163, Lys186, Lys20, Gln21, Ser45, Asn47, Cys51, Asn114, Lys130, Gln91, His159, Asn26, Glu111, Leu154, Thr112, and Trp23.
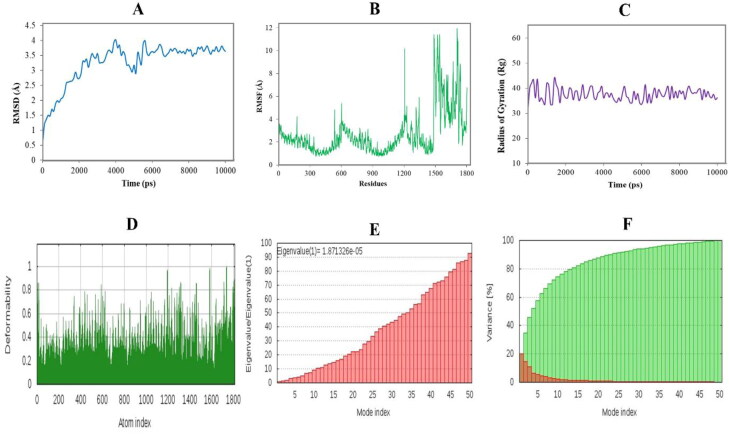
3.11. Molecular dynamics simulation
We executed software-based MD simulation where trajectories from 10 ns long simulation showed structural stabilization around 6.2 ns and light fluctuation afterward (Figure 8(A)). The calculated average RMSD value was 3.25 Å, while the average RMSF score was 2.65 Å. The fluctuation was higher in the vaccine part from AA1500 to AA1800 (Figure 8(B)). Furthermore, the average simulation energy was –7,229,534.41 kJ/mol. The average Rg score was 37.62 that fluctuates between 30.90 and 44.07 (Figure 8(C)). The relatively higher pick at 1500 to 1800 amino acid residues was due to the flexible regions of the docked complex, which was the vaccine part. The MD simulation was also carried out in the iMODS server, where NMA assessment was applied to the internal coordinates of the complex. The deformability builds up the independent distortion of each residue portrayed by the method of chain hinges (Figure 8(D)). The eigenvalue determined for the complex was found to be 1.871 e–05 (Figure 8(E)). The variance of each typical mode was gradually decreased (Figure 7(F)). All these results suggest stable binding interactions with compact conformation and minor fluctuations in the vaccine–TLR4 complex.
Figure 8.
Molecular dynamics simulation of the vaccine–TLR4 complex. Herein, different MD simulation plots show (A) root mean square deviation, (B) root mean square fluctuation, (C) radius of gyration, (D) deformability index, (E) eigenvalue, and (F) NMA variance.
3.12. Immune response simulation
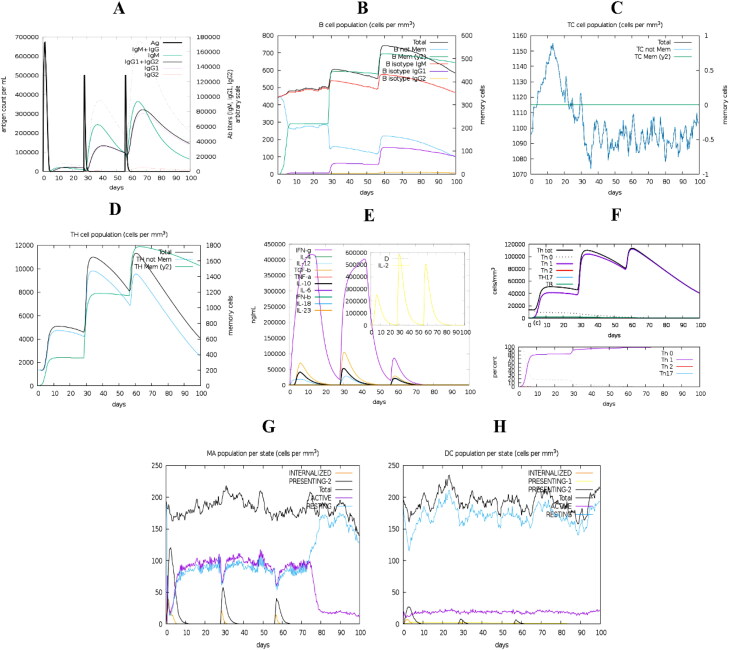
The simulated immune response showed similar to actual immunological phenomena provoked by specific pathogens as shown in Figure 9. For instance, secondary and tertiary immune responses were higher than the primary immune response (Figure 9(A)). Secondary and tertiary responses showed higher levels of antibodies (i.e. IgG1 + IgG2, IgM, and IgG + IgM), which coincided with an antigen extenuation indicating the development of memory cells, thus, intensified antigen clearance upon successive exposures (Figure 9(A)). Additionally, a prolonged period of viability in B-cells, cytotoxic T-cells and helper T-cells were noticed, indicating the class switching between immune cells and IgM memory formation (Figure 9(B–D)). The elevated levels of IFNγ, IL-4 and IL-10 were additionally apparent (Figure 9(E)). The percentage (%) and amount (cells/mm3) of Th0 type immune reaction were lower than the Th1 type reaction (Figure 9(F)). During the presentation, expanded macrophage movement was illustrated, while dendritic cell movement was predictable (Figure 9(G,H)).
Figure 9.
Immune response triggered by the designed vaccine. The graph shows (A) primary, secondary and tertiary immune responses, (B) B-cell population, (C) cytotoxic T-cell population, (D) helper T-cell population, (E) induction of cytokines and interleukins, (F) Th1 mediated immune response, (G) macrophage population per state, and (H) dendritic cell population per state.
3.13. Codon adaptation and in silico cloning
We optimized the codons present in the vaccine construct according to the E. coli K12 in the JCat server to increase their translation efficiency. The peptide vaccine construct (316 AA residues) produced 948 lengths of nucleotide sequences. Moreover, the adapted nucleotide sequence has GC content and CAI value of 59.04% and 1.0, respectively. To insert the adapted sequence into the pET28a (+) vector, we selected XhoI and BamHI restriction sites as the start and end cut points, respectively. Thus, the optimized vaccine construct was cloned into the pET28a (+) cloning vector with the SnapGene software (Figure 10). The final size of the cloning vector was 6281 nucleotide base pairs (bp).
Figure 10.
The in silico cloning of the designed vaccine into the pET-28a (+) vector. Herein, purple color represents the vector DNA, while the red color indicates the adapted DNA sequence of the designed vaccine.
4. Discussion
The present demonic appearance of COVID-19 generates a life-threatening situation to the global public health (Vankadari & Wilce, 2020), which influences us to design this multi-epitope vaccine applying immunoinformatics approach. The glycoprotein-based vaccine demonstrated an extraordinary significance ordained by immunoinformatics and revealed our attempt trustworthy. A vaccine is a safe and effective way to protect against infectious diseases (Li et al., 2014). It should have the ability to provide acquired immunity against contagious diseases (Bol et al., 2016). In this study, we designed an epitope-based vaccine that could provide a strong immune response against SARS-CoV-2, thereby, preventing the COVID-19 pandemic. A vaccine can prevent future outbreaks (Melief et al., 2015). However, in the absence of an effective vaccine, control and prevention of COVID-19 infection and transmission are very difficult. Besides, effective vaccination is yet to be developed in controlling the current situation. Thus, a new strategy of vaccine development is a prime need that will contribute to finding a solution to solve this present life-threatening public health issue.
Since S-protein of SARS-CoV-2 plays a major role in immune invasion as well as the human to human transmission, our purpose was to design an epitope vaccine by targeting the S-protein. The location of the antigenic region of the surface of S-protein was evaluated so that this protein can be recognized by cellular and humoral immune systems. First, all potential CTL, HTL and LBL epitopes were identified and evaluated. Then, the vaccine was designed with the top four antigenic CTL, HTL and LBL epitopes with their desired linkers. They were incorporated in vaccine construction as a part of the essential element that enhances the stability, folding and expression patterns of our vaccine candidate (Shamriz et al., 2016). The adjuvant was attached to the CTL epitope by EAAAK linker, which helps to induce high levels of both cellular and immunogenic humoral responses for particular antigens, and amplify the vaccine’s stability and longevity (Bonam et al., 2017; Lee & Nguyen, 2015). Finally, the vaccine construction was found to accumulate 316 amino acid residues long. Solubility, a type of physicochemical property of a vaccine candidate, is counted as a vital characteristic of any recombinant vaccine (Khatoon et al., 2017). Hence, the solubility of the vaccine construct was predicted by using a solubility assessing tool to determine the quality of being solvability of the construct inside the host E. coli, and the vaccine construct was found to be solvable inside the host E. coli. The nature of the vaccine determined by theoretical PI value was found to be acidic. Instability index suggested by server tools indicate that the protein would remain stable after synthesis. In contrast, the GRAVY value and aliphatic index portrayed the vaccine to be the hydrophilic and thermostable, respectively. A favorable physicochemical property predicted for the vaccine and all the scores on different parameters relies on a high possibility to confer this vaccine as a valid candidate against SARS-CoV-2.
In our approach, we found the best population coverage all-over the world (93.30%) for the combined results that make this vaccine construct a good candidate and significant weapon. The most cases of infection and mortality were found in the United States and the United Kingdom, and we find the parentage of population coverage for those countries in a significant level (United States (99.38%), Europe (98.92%)). After the 3D structure prediction (based on c-score), the identified models were refined and selected the best model (based on the lowest energy score). In the validation test of 3D structure, we found a good number of Z-score (–7.4) and the superior features of most favored, accepted and disallowed regions for the Ramachandran plot. Molecular docking between the peptide vaccine and virus glycoprotein binding favorable receptor of TLR4 with lowest energy score of −964.6 confirmed the possibility of infection inhibitory activity of the vaccine and suggested a possible tight interaction between the modeled vaccine as a ligand and the TLR4 receptor surface. Molecular dynamics simulation is a potential technique for accessing the physical ground of the protein structure and function of biological macromolecules. Protein dynamic simulations can provide certain information regarding individual atomic movement as a function of time. For the dynamics evaluation of the vaccine candidate, 10 ns dynamic simulations have been performed, and results have been analyzed based on the RMSD and RMSF score. RMSD value is used to compare different atomic conformations of a given molecular system. In this study, the RMSD value was used to determine the significant flexibility and departure of vaccine candidates from the receptor structure, where the RMSF of the complex structure was determined to measure the displacement of our particular vaccine candidate’s atom relative to the receptor structure. The calculated average RMSD and RMSF value was 3.25 Å and 2.65 Å, respectively. The fluctuation was found higher in the vaccine part, but it smoothly became stable after 600 ps suggesting possible stability of the modeled vaccine and the receptor. Finally, we performed an immune simulation to observe the optimal behavior and cell density parameters for successful target clearance and find the best immunological response against the pathogen. The vaccine doses were upgraded immunological reaction causing memory B-cell (having a half-life of several months) and T-cell. Sustained generation of IFN-γ and IL-2 were seen after immunization because of expanded aide T-cell initiation. In this manner, the vaccine effectively simulated a humoral immunological response to increasing immunoglobulin creation. The MD simulation was performed to evaluate the stability of the vaccine candidate with the receptor, where codon optimization was performed to stabilize the construct vaccine within the host for optimum multi-epitope vaccine production. Finally, the codon was optimized, and desired vaccine candidate in silico cloning was performed successfully into the pET28a (+) cloning vector of E. coli K12 expression host.
5. Conclusion
In this study, a series of computational approaches led to the discovery of potential T- and B-cell epitopes in S-protein of SARS-CoV-2 that eventually embroidered into a multi-epitope vaccine. The newly designed vaccine has desired immunodominant properties with high population coverage. Importantly, it was able to bind with the immune receptor TLR4 strongly as well as to elicit robust immune response upon SARS-CoV-2 infection. Based on our findings, we believe that the vaccine candidate can be an important starting point for developing a potent vaccine against the etiological agent of COVID-19 outbreak. Moreover, the potential epitopes identified in this study can be used in future studies as well. However, further experimental assessments are required to confirm our formulated vaccine as an effective prophylactic against SARS-CoV-2.
Supplementary Material
Acknowledgments
We extend our gratitude towards the Deanship of Scientific Research (DSR) at King Abdulaziz University and Biological Solution Centre (BioSol Centre) for providing technical support. Special thanks go to Monokesh Kumer Sen, School of Medicine, Western Sydney University, Australia for his contribution in revising the whole manuscript.
Glossary
Abbreviations
- 2D
two dimensional
- 3D
three dimensional
- AAY
Ala-Ala-Tyr
- ACE2
angiotensin converting enzyme 2
- ARDS
acute respiratory disease syndrome
- COVID-19
coronavirus disease 2019
- Combined epitope
CTL and HTL epitopes
- CTL
cytotoxic T-lymphocyte
- E. coli
Escherichia coli
- GPGPG
Gly-Pro-Gly-Pro-Gly
- HTL
helper T-lymphocyte
- I-TASSER
iterative threading assembly refinement
- JCAT
Java codon adaptation tool
- KK
Lys-Lys
- LBL
linear B-lymphocyte
- MD
molecular dynamic
- MHC-I
major histocompatibility complex-I
- MHC-II
major histocompatibility complex-II
- Rg
radius of gyration
- RMSD
root-mean-square deviation
- RMSF
root mean square fluctuation
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SOPMA
self-optimized prediction method with alignment
- TLR4
toll-like receptor 4
- WHO
World Health Organization
Disclosure statement
The authors declare no conflict of interest.
Author contributions
AS, FA, and MSR designed the project; AS, ZN, RA, and MH performed the experiments; AS, FA, ZN, and RA evaluated and interpreted the data; AS, FA, RRI, and ZN prepared the draft manuscript; FA, ZN, AS, and MSR finalized the manuscript. All authors approved the final version of the manuscript.
References
- Abdelli, I., Hassani, F., Bekkel Brikci, S., & Ghalem, S. (2020). In silico study the inhibition of Angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from western Algeria. Journal of Biomolecular Structure and Dynamics, 1–17. 10.1080/07391102.2020.1763199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdellrazeq, G. S., Fry, L. M., Elnaggar, M. M., Bannantine, J. P., Schneider, D. A., Chamberlin, W. M., Mahmoud, A. H. A., Park, K.-T., Hulubei, V., & Davis, W. C. (2020). Simultaneous cognate epitope recognition by bovine CD4 and CD8 T cells is essential for primary expansion of antigen-specific cytotoxic T-cells following ex vivo stimulation with a candidate Mycobacterium avium subsp. paratuberculosis peptide vaccine. Vaccine, 38(8), 2016–2025. 10.1016/j.vaccine.2019.12.052 [DOI] [PubMed] [Google Scholar]
- Ahammad, F., Rashid, T. A., Rogayah, T., Mohamed, M., Tanbin, S., Fuad, A., & Adyani, F. (2019). Contemporary strategies and current trends in designing antiviral drugs against dengue fever via targeting host-based approaches. Microorganisms, 7(9), 296. 10.3390/microorganisms7090296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astuti, I. & Ysrafil, (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): An overview of viral structure and host response. Diabetes & Metabolic Syndrome, 14(4), 407–412. 10.1016/j.dsx.2020.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The Protein Data Bank. Nucleic Acids Research, 28(1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bol, K. F., Aarntzen, E. H. J. G., Pots, J. M., Olde Nordkamp, M. A. M., van de Rakt, M. W. M. M., Scharenborg, N. M., de Boer, A. J., van Oorschot, T. G. M., Croockewit, S. A. J., Blokx, W. A. M., Oyen, W. J. G., Boerman, O. C., Mus, R. D. M., van Rossum, M. M., van der Graaf, C. A. A., Punt, C. J. A., Adema, G. J., Figdor, C. G., de Vries, I. J. M., & Schreibelt, G. (2016). Prophylactic vaccines are potent activators of monocyte-derived dendritic cells and drive effective anti-tumor responses in melanoma patients at the cost of toxicity. Cancer Immunology, Immunotherapy, 65(3), 327–339. 10.1007/s00262-016-1796-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonam, S. R., Partidos, C. D., Halmuthur, S. K. M., & Muller, S. (2017). An overview of novel adjuvants designed for improving vaccine efficacy. Trends in Pharmacological Sciences, 38(9), 771–793. 10.1016/j.tips.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Boopathi, S., Poma, A. B., & Kolandaivel, P. (2020). Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1758788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthwick, N., Silva-Arrieta, S., Llano, A., Takiguchi, M., Brander, C., & Hanke, T. (2020). Novel nested peptide epitopes recognized by CD4+ T cells induced by HIV-1 conserved-region vaccines. Vaccines, 8(1), 28. 10.3390/vaccines8010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan, D. W. A., Minneci, F., Nugent, T. C. O., Bryson, K., & Jones, D. T. (2013). Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Research, 41(Web Server issue), W349–W357. 10.1093/nar/gkt381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui, H. H., Sidney, J., Dinh, K., Southwood, S., Newman, M. J., & Sette, A. (2006). Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics, 7, 153. 10.1186/1471-2105-7-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calis, J. J. A., Maybeno, M., Greenbaum, J. A., Weiskopf, D., De Silva, A. D., Sette, A., Keşmir, C., & Peters, B. (2013). Properties of MHC class I presented peptides that enhance immunogenicity . PLoS Computational Biology, 9(10), e1003266. 10.1371/journal.pcbi.1003266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher, C., Carroll, D., Colwell, R., Corley, R. B., Daszak, P., Drosten, C., Enjuanes, L., Farrar, J., Field, H., Golding, J., Gorbalenya, A., Haagmans, B., Hughes, J. M., Karesh, W. B., Keusch, G. T., Lam, S. K., Lubroth, J., Mackenzie, J. S., Madoff, L., … Turner, M. (2020). Statement in support of the scientists, public health professionals, and medical professionals of China combatting COVID-19. The Lancet, 395(10226), e42–e43. 10.1016/S0140-6736(20)30418-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglione, F., Mantile, F., De Berardinis, P., & Prisco, A. (2012). How the interval between prime and boost injection affects the immune response in a computational model of the immune system. Computational and Mathematical Methods in Medicine, 2012, 842329. 10.1155/2012/842329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J. F.-W., Yuan, S., Kok, K.-H., To, K. K.-W., Chu, H., Yang, J., Xing, F., Liu, J., Yip, C. C.-Y., Poon, R. W.-S., Tsoi, H.-W., Lo, S. K.-F., Chan, K.-H., Poon, V. K.-M., Chan, W.-M., Ip, J. D., Cai, J.-P., Cheng, V. C.-C., Chen, H., … Yuen, K.-Y. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. The Lancet, 395(10223), 514–523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S., Sarmah, S., Lyndem, S., & Singha Roy, A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1763201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W. (2002). Pymol: An open-source molecular graphics tool. CCP4 Newsletter on Protein Crystallography, 40(1), 82–92. [Google Scholar]
- Dhanda, S. K., Gupta, S., Vir, P., & Raghava, G. P. (2013). Prediction of IL4 inducing peptides . Clinical Developmental Immunology, 2013, 263952. 10.1155/2013/263952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda, S. K., Vir, P., & Raghava, G. P. (2013). Designing of interferon-gamma inducing MHC class-II binders. Biology Direct, 8, 30. 10.1186/1745-6150-8-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov, I., Flower, D. R., & Doytchinova, I. (2013). AllerTOP – A server for in silico prediction of allergens. BMC Bioinformatics, 14(Suppl 6), S4. 10.1186/1471-2105-14-S6-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorosti, H., Eslami, M., Negahdaripour, M., Ghoshoon, M. B., Gholami, A., Heidari, R., Dehshahri, A., Erfani, N., Nezafat, N., & Ghasemi, Y. (2019). Vaccinomics approach for developing multi-epitope peptide pneumococcal vaccine . Journal of Biomolecular Structure and Dynamics, 37(13), 3524–3535. 10.1080/07391102.2018.1519460 [DOI] [PubMed] [Google Scholar]
- Doytchinova, I. A., & Flower, D. R. (2007). VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics, 8(1), 4. 10.1186/1471-2105-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1761882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A., & Azzam, E. B. (2020). Novel guanosine derivatives against MERS CoV polymerase: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1758789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enayatkhani, M., Hasaniazad, M., Faezi, S., Guklani, H., Davoodian, P., Ahmadi, N., Einakian, M. A., Karmostaji, A., & Ahmadi, K. (2020). Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: An in silico study. Journal of Biomolecular Structure and Dynamics, 1–16. 10.1080/07391102.2020.1756411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaafar, B. B. M., Ali, S. A., Abd-Elrahman, K. A., & Almofti, Y. A. (2019). Immunoinformatics approach for multiepitope vaccine prediction from H, M, F, and N proteins of Peste des Petits ruminants virus. Journal of Immunology Research, 2019, 6124030. 10.1155/2019/6124030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., & Bairoch, A. (2005). Protein identification and analysis tools on the ExPASy server. In John M. Walker (Ed.), The proteomics protocols handbook(pp. 571–607). Springer. [Google Scholar]
- Geourjon, C., & Deléage, G. (1995). SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Computer Applications in the Biosciences, 11(6), 681–684. 10.1093/bioinformatics/11.6.681 [DOI] [PubMed] [Google Scholar]
- Goldberg, M. F., Roeske, E. K., Ward, L. N., Pengo, T., Dileepan, T., Kotov, D. I., & Jenkins, M. K. (2018). Salmonella persist in activated macrophages in T cell-sparse granulomas but are contained by surrounding CXCR3 ligand-positioned Th1 cells. Immunity, 49(6), 1090–1102.e1097. 10.1016/j.immuni.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote, A., Hiller, K., Scheer, M., Münch, R., Nörtemann, B., Hempel, D. C., & Jahn, D. (2005). JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Research, 33(Web Server issue), W526–W531. 10.1093/nar/gki376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, M. K., Vemula, S., Donde, R., Gouda, G., Behera, L., & Vadde, R. (2020). In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1751300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Kapoor, P., Chaudhary, K., Gautam, A., Kumar, R., Raghava, G. P., & Open Source Drug Discovery Consortium . (2013). In silico approach for predicting toxicity of peptides and proteins. PLoS One, 8(9), e73957. 10.1371/journal.pone.0073957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, A., Paray, B. A., Hussain, A., Qadir, F. A., Attar, F., Aziz, F. M., Sharifi, M., Derakhshankhah, H., Rasti, B., Mehrabi, M., Shahpasand, K., Saboury, A. A., & Falahati, M. (2020). A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann, D. L., & Shindo, N. (2020). COVID-19: What is next for public health? The Lancet, 395(10224), 542–545. 10.1016/S0140-6736(20)30374-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., Zhang, L., Fan, G., Xu, J., Gu, X., Cheng, Z., Yu, T., Xia, J., Wei, Y., Wu, W., Xie, X., Yin, W., Li, H., Liu, M., … Cao, B. (2020). Clinical features of patients infected with 2019 novel coronavirus in Wuhan. The Lancet, 395(10223), 497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, R. S., Jagdale, S. S., Bansode, S. B., Shankar, S. S., Tellis, M. B., Pandya, V. K., Chugh, A., Giri, A. P., & Kulkarni, M. J. (2020). Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. Journal of Biomolecular Structure and Dynamics, 1-16. 10.1080/07391102.2020.1760137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, R. J., Jha, R. K., Amera, G. M., Jain, M., Singh, E., Pathak, A., Singh, R. P., Muthukumaran, J., & Singh, A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 1-14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatoon, N., Pandey, R. K., & Prajapati, V. K. (2017). Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Scientific Reports, 7(1), 8285. 10.1038/s41598-017-08842-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov, D., Hall, D. R., Xia, B., Porter, K. A., Padhorny, D., Yueh, C., Beglov, D., & Vajda, S. (2017). The ClusPro web server for protein-protein docking. Nature Protocols, 12(2), 255–278. 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D., Kumari, K., Jayaraj, A., Kumar, V., Kumar, R. V., Dass, S. K., Chandra, R., & Singh, P. (2020). Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. Journal of Biomolecular Structure and Dynamics, 1–14. 10.1080/07391102.2020.1752310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, M. V., Lundegaard, C., Lamberth, K., Buus, S., Lund, O., & Nielsen, M. (2007). Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics, 8, 424. 10.1186/1471-2105-8-424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latysheva, N. S., & Babu, M. M. (2016). Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Research, 44(10), 4487–4503. 10.1093/nar/gkw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., & Nguyen, M. T. (2015). Recent advances of vaccine adjuvants for infectious diseases. Immune Network, 15(2), 51–57. 10.4110/in.2015.15.2.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., & De Clercq, E. (2020). Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nature Reviews Drug Discovery, 19(3), 149–150. 10.1038/d41573-020-00016-0 [DOI] [PubMed] [Google Scholar]
- Li, W., Joshi, M. D., Singhania, S., Ramsey, K. H., & Murthy, A. K. (2014). Peptide vaccine: Progress and challenges. Vaccines, 2(3), 515–536. 10.3390/vaccines2030515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Blanco, J. R., Aliaga, J. I., Quintana-Ortí, E. S., & Chacón, P. (2014). iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Research, 42(Web Server issue), W271–W276. 10.1093/nar/gku339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell, S. C., Davis, I. W., Arendall, W. B., de Bakker, P. I. W., Word, J. M., Prisant, M. G., Richardson, J. S., & Richardson, D. C. (2003). Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins: Structure, Function, and Bioinformatics, 50(3), 437–450. 10.1002/prot.10286 [DOI] [PubMed] [Google Scholar]
- Magnan, C. N., Randall, A., & Baldi, P. (2009). SOLpro: Accurate sequence-based prediction of protein solubility. Bioinformatics, 25(17), 2200–2207. 10.1093/bioinformatics/btp386 [DOI] [PubMed] [Google Scholar]
- Magnan, C. N., Zeller, M., Kayala, M. A., Vigil, A., Randall, A., Felgner, P. L., & Baldi, P. (2010). High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics (Oxford, England)), 26(23), 2936–2943. 10.1093/bioinformatics/btq551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavalan, B., Govindaraj, R. G., Shin, T. H., Kim, M. O., & Lee, G. (2018). iBCE-EL: A new ensemble learning framework for improved linear B-cell epitope prediction. Frontiers in Immunology, 9, 1695. 10.3389/fimmu.2018.01695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melief, C. J., van Hall, T., Arens, R., Ossendorp, F., & van der Burg, S. H. (2015). Therapeutic cancer vaccines . Journal of Clinical Investigation, 125(9), 3401–3412. 10.1172/JCI80009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan, N., Sakthivel, R., Velmurugan, D., & Gromiha, M. M. (2020). Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–6. 10.1080/07391102.2020.1752802 [DOI] [PubMed] [Google Scholar]
- Nagpal, G., Usmani, S. S., Dhanda, S. K., Kaur, H., Singh, S., Sharma, M., & Raghava, G. P. (2017). Computer-aided designing of immunosuppressive peptides based on IL-10 inducing potential. Scientific Reports, 7, 42851. 10.1038/srep42851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nain, Z., Abdulla, F., Rahman, M. M., Karim, M. M., Khan, M. S. A., Sayed, S. B., Mahmud, S., Raihan Rahman, S. M., Moinuddin Sheam, Md., Haque, Z., & Adhikari, U. K. (2020). Proteome-wide screening for designing a multi-epitope vaccine against emerging pathogen Elizabethkingia anophelis using immunoinformatic approaches. Journal of Biomolecular Structure and Dynamics, 1–18. 10.1080/07391102.2019.1692072 [DOI] [PubMed] [Google Scholar]
- Nain, Z., Karim, M. M., Sen, M. K., & Adhikari, U. K. (2020). Structural basis and designing of peptide vaccine using PE-PGRS family protein of Mycobacterium ulcerans – An integrated vaccinomics approach. Molecular Immunology, 120, 146–163. 10.1016/j.molimm.2020.02.009 [DOI] [PubMed] [Google Scholar]
- Nugent, T., Cozzetto, D., & Jones, D. T. (2014). Evaluation of predictions in the CASP10 model refinement category. Proteins, 82(Suppl 2), 98–111. 10.1002/prot.24377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejnik, J., Hume, A. J., & Mühlberger, E. (2018). Toll-like receptor 4 in acute viral infection: Too much of a good thing. PLoS Pathogens, 14(12), e1007390. 10.1371/journal.ppat.1007390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, R. K., Bhatt, T. K., & Prajapati, V. K. (2018). Novel immunoinformatics approaches to design multi-epitope subunit vaccine for malaria by investigating anopheles salivary protein. Scientific Reports, 8(1), 1125. 10.1038/s41598-018-19456-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, X., Xu, X., Li, Y., Cheng, L., Zhou, X., & Ren, B. (2020). Transmission routes of 2019-nCoV and controls in dental practice. International Journal of Oral Science, 12(1), 9. 10.1038/s41368-020-0075-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett, B. E., Sadat, E. L., Zhang, Y., Noronha, J. M., Squires, R. B., Hunt, V., Liu, M., Kumar, S., Zaremba, S., Gu, Z., Zhou, L., Larson, C. N., Dietrich, J., Klem, E. B., & Scheuermann, R. H. (2012). ViPR: An open bioinformatics database and analysis resource for virology research. Nucleic Acids Research, 40(Database issue), D593–D598. 10.1093/nar/gkr859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran, G. N., Ramakrishnan, C., & Sasisekharan, V. (1963). Stereochemistry of polypeptide chain configurations. Journal of Molecular Biology, 7, 95–99. 10.1016/S0022-2836(63)80023-6 [DOI] [PubMed] [Google Scholar]
- Rapin, N., Lund, O., Bernaschi, M., & Castiglione, F. (2010). Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLoS One, 5(4), e9862. 10.1371/journal.pone.0009862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, A., Kucukural, A., & Zhang, Y. (2010). I-TASSER: A unified platform for automated protein structure and function prediction. Nature Protocols, 5(4), 725–738. https://doi.org/10.10./nprot.2010.5 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma, P., Sekhar, N., Prajapat, M., Avti, P., Kaur, H., Kumar, S., Singh, S., Kumar, H., Prakash, A., Dhibar, D. P., & Medhi, B. (2020). In-silico homology assisted identification of inhibitor of RNA binding against2019-nCoV N-protein (N terminal domain). Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1753580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamriz, S., Ofoghi, H., & Moazami, N. (2016). Effect of linker length and residues on the structure and stability of a fusion protein with malaria vaccine application. Computers in Biology and Medicine, 76, 24–29. 10.1016/j.compbiomed.2016.06.015 [DOI] [PubMed] [Google Scholar]
- Shang, W., Yang, Y., Rao, Y., & Rao, X. (2020). The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vaccines, 5(1), 1–3. 10.1038/s41541-020-0170-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, K., Yang, Y., Wang, T., Zhao, D., Jiang, Y., Jin, R., Zheng, Y., Xu, B., Xie, Z., Lin, L., Shang, Y., Lu, X., Shu, S., Bai, Y., Deng, J., Lu, M., Ye, L., Wang, X., Wang, Y., Gao, L., & China National Clinical Research Center for Respiratory Diseases . (2020). Diagnosis, treatment, and prevention of 2019 novel coronavirus infection in children: Experts’ consensus statement. World Journal of Pediatrics, 16(3), 223–231. 10.1007/s12519-020-00343-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, S. K., Shakya, A., Prasad, S. K., Singh, S., Gurav, N. S., Prasad, R. S., & Gurav, S. S. (2020). An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. Journal of Biomolecular Structure and Dynamics, 1–12. 10.1080/07391102.2020.1762741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman, J. L., Lin, D., Jiang, J., Manning, N. O., Prilusky, J., Ritter, O., & Abola, E. E. (1998). Protein Data Bank (PDB): Database of three-dimensional structural information of biological macromolecules. Acta Crystallographica. Section D, Biological Crystallography, 54(6), 1078–1084. 10.1107/s0907444998009378 [DOI] [PubMed] [Google Scholar]
- Trott, O., & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesh, K. D., Selvaraj, C., Singh, S. K., & Dubey, V. K. (2020). Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. Journal of Biomolecular Structure and Dynamics, 1–9. 10.1080/07391102.2020.1763202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vankadari, N., & Wilce, J. A. (2020). Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerging Microbes & Infections, 9(1), 601–604. 10.1080/22221751.2020.1739565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahedi, H. M., Ahmad, S., & Abbasi, S. W. (2020). Stilbene-based natural compounds as promising drug candidates against COVID-19. Journal of Biomolecular Structure and Dynamics, 1–10. 10.1080/07391102.2020.1762743 [DOI] [PubMed] [Google Scholar]
- Wang, C., Horby, P. W., Hayden, F. G., & Gao, G. (2020). A novel coronavirus outbreak of global health concern. The Lancet, 395(10223), 470–473. 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Hu, B., Hu, C., Zhu, F., Liu, X., Zhang, J., Wang, B., Xiang, H., Cheng, Z., Xiong, Y., Zhao, Y., Li, Y., Wang, X., & Peng, Z. (2020). Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA, 323(11), 1061–1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Sidney, J., Kim, Y., Sette, A., Lund, O., Nielsen, M., & Peters, B. (2010). Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics, 11(1), 568. 10.1186/1471-2105-11-568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein, M., & Sippl, M. (2007). ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research, 35(Web Server issue), W407–W410. 10.1093/nar/gkm290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., Graham, B. S., & McLellan, J. S. (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Song, Z.-G., Hu, Y., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E. C., & Zhang, Y.-Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X., Chen, P., Wang, J., Feng, J., Zhou, H., Li, X., Zhong, W., & Hao, P. (2020). Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences, 63(3), 457–460. 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z., Shi, L., Wang, Y., Zhang, J., Huang, L., Zhang, C., Liu, S., Zhao, P., Liu, H., Zhu, L., Tai, Y., Bai, C., Gao, T., Song, J., Xia, P., Dong, J., Zhao, J., & Wang, F.-S. (2020). Pathological findings of COVID-19 associated with acute respiratory distress syndrome. The Lancet. Respiratory Medicine, 8(4), 420–422. 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., Tan, W., & China Novel Coronavirus Investigating and Research Team . (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.