Summary
The astounding capacity of pluripotent stem cells (PSCs) to differentiate and self-organize has revolutionized the development of 3D cell culture models. The major advantage is its ability to mimic in vivo microenvironments and cellular interactions when compared with the classical 2D cell culture models. Recent innovations in generating embryo-like structures (including blastoids and gastruloids) from PSCs have advanced the experimental accessibility to understand embryogenesis with immense potential to model human development. Taking cues on how embryonic development leads to organogenesis, PSCs can also be directly differentiated to form mini-organs or organoids of a particular lineage. Organoids have opened new avenues to augment our understanding of stem cell and regenerative biology, tissue homeostasis, and disease mechanisms. In this review, we provide insights from developmental biology with a comprehensive resource of signaling pathways that in a coordinated manner form embryo-like structures and organoids. Moreover, the advent of assembloids and multilineage organoids from PSCs opens a new dimension to study paracrine function and multi-tissue interactions in vitro. Although this led to an avalanche of enthusiasm to utilize organoids for organ transplantation studies, we examine the current limitations and provide perspectives to improve reproducibility, scalability, functional complexity, and cell-type characterization. Taken together, these 3D in vitro organ-specific and patient-specific models hold great promise for drug discovery, clinical management, and personalized medicine.
Subject Areas: Bioengineering, Tissue Engineering, Embryology
Graphical Abstract
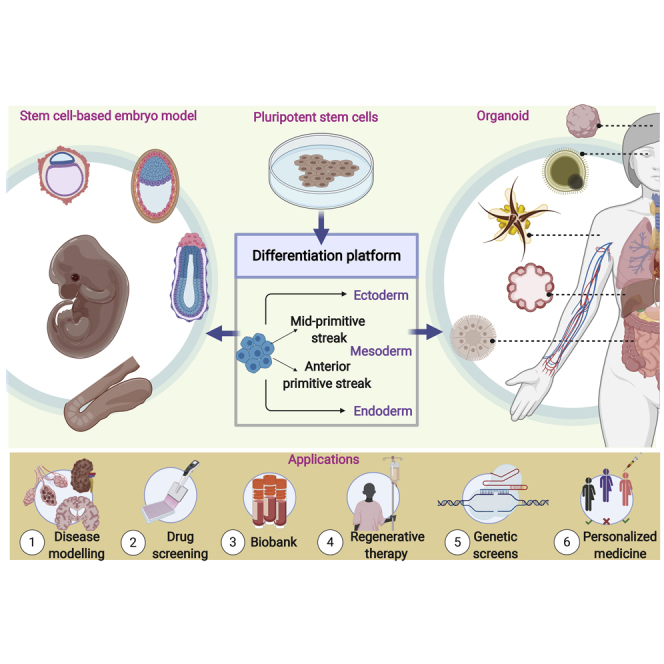
Bioengineering; Tissue Engineering; Embryology
Introduction
Stem cells have a remarkable capacity to not only self-renew and differentiate but also to self-assemble and self-organize into complex and functional tissues and organs. An initially homogeneous population of stem cells can self-organize and undergo an in vivo-like morphogenesis to give rise to three-dimensional (3D) mini-organ-like structures in culture, called organoids (Sasai et al., 2012). They retain both functional and structural complexity of the tissue of origin and enable long-term growth of genetically stable cells. Organoids can be generated from embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) derived from patients that can be used for studying organ development, for modeling human diseases, and for autologous organ replacement. Tissue-specific adult stem cells (ASCs) can also generate organoids, and several protocols have been optimized for the same (Clevers, 2016). Here in this review, we limit our discussion to how pluripotent stem cells (PSCs, including ESCs and iPSCs) form 3D models of the embryo and organs in culture. This process involves recapitulation of implantation, gastrulation, and organogenesis, which altogether guide the key events during embryonic development when PSCs progressively commit to particular lineages. The cross talk between key signaling pathways during these developmental processes is the source of cellular diversity, behaviors, and forms that generate complex organ structure across the animal kingdom (Perrimon et al., 2012). These signaling pathways form the basis for directed differentiation of PSCs to form “mini-organs” (Sasai et al., 2012) or embryo-like structures in vitro (Shahbazi et al., 2019). Although many reviews have addressed the principles of bioengineering to develop 3D cell culture models and protocols to generate organoids from adult tissues, here we present insights into the state-of-the-art knowledge on the self-organization property of PSCs to generate embryo-like structures and organoids, its advantage to model human diseases, and challenges for optimum clinical management.
An Overview of Self-Organization of Stem Cells to Form Embryo-like Structures
Self-organization is a physical property observed in many biological phenomena ranging from collective behavioral traits to embryonic morphogenesis. ESCs derived from pre-implantation embryos are pluripotent and can clonally divide and differentiate into all cell types. ESCs can aggregate together into 3D embryoid bodies (EBs) with the presence of rudimentary cell types. Although EBs can differentiate into eye cups, neural cortical structures, and even cell types from endodermal and mesodermal lineages, they lack proper axial organization, a characteristic of mammalian embryo. Recent studies demonstrate the capability of ESCs to generate self-organizing embryo-like structures that can re-create early embryonic morphogenesis (Shahbazi et al., 2019; Simunovic and Brivanlou, 2017). The mammalian conceptus is composed of the placenta, the fetus, the extra-embryonic tissues to establish feto-maternal interaction, and the “embryo proper,” which forms the main body (Hyun et al., 2020). The formation of extra-embryonic tissues and the generation of embryonic germ layers are the key stages of mammalian embryogenesis, which progress through a series of events from implantation of the conceptus to gastrulation followed by organogenesis (Figure 1A). The relatively small size of the conceptus and lack of protocols to culture along with ethical limitations to acquire human embryos inspired the generation of accessible stem cell-derived embryo models to understand the key stages of embryogenesis. Recent innovations have led to the generation of embryo-like structures that can recapitulate the interaction between embryonic and extra-embryonic tissues (Harrison et al., 2017; Rivron et al., 2018b; Shao et al., 2017a, 2017b; Sozen et al., 2018; Zheng et al., 2019). For example, human ESCs (hESCs) grown in microfluidic chambers can generate epiblast-like structures known as Post-implantation Amniotic Sac Embryoid (PASE), consisting of extra-embryonic tissues like the amniotic ectoderm, embryonic sac, and amniotic cavity, resembling early post-implantation human embryonic landmarks (Shao et al., 2017a, 2017b; Zheng et al., 2019). Mouse stem cells can also self-organize into pre-implantation blastocyst-like structures called blastoids. Blastoids were formed by combining ESCs and extra-embryonic trophoblast stem cells (TSCs) (Rivron et al., 2018b), ESCs/extended pluripotent stem cells (EPSCs) and TSCs (Sozen et al., 2019), or EPSCs alone (Li et al., 2019b). Upon in vitro development, the ESCs (Rivron et al., 2018b; Sozen et al., 2019) and the EPSCs (Li et al., 2019b; Sozen et al., 2019) produced the primitive endoderm cells thus forming a pre-implantation conceptus comprising the three founding cell types. This was confirmed by independent single-cell RNA sequencing (scRNA-seq) analysis and also showed that blastoids formed with EPSCs only, and do not form cells with a proper trophoblast and epiblast transcriptome signature (Posfai et al., 2020). Additional work is needed to capture totipotent stem cells in a dish and form blastoids from only one cell type. Although blastoids are transcriptionally similar to blastocysts and can also trigger the formation of deciduae, they do not support de novo embryonic development beyond a few days (similar to all embryo models that are currently available) (Li et al., 2019b; Rivron et al., 2018b; Sozen et al., 2019). Similarly, self-organizing mouse ESCs (mESCs) in the presence of Wnt agonist and constant agitation can undergo spatial morphogenesis with distinct body axes, germ layer specification, and spatiotemporal gene expression, very much similar to a gastrulating embryo (Beccari et al., 2018; van den Brink et al., 2014; Warmflash et al., 2014). These “gastruloids” can be a complementary system to study early developmental events, and recent scRNA-seq. datasets provide evidence on the emergence of neural tube and somites recapitulating early organogenesis (van den Brink et al., 2020; Veenvliet et al., 2020). Culturing mouse gastruloids with an underlying extracellular matrix led to the formation of structures resembling somites and neural tube (van den Brink et al., 2020; Veenvliet et al., 2020), whereas another report suggested that mixing gastruloids and extra-endodermal cells can lead to the formation of the neural tube (Bérenger-Currias et al., 2020). Recently, gastruloids have been generated from self-organizing hESCs, which can also differentiate to three germ layers (Moris et al., 2020). Somite formation is a rhythmic process that involves molecular oscillation and the expression of certain genes that pattern the embryo, collectively known as the molecular segmentation clock that has been widely characterized in several model systems and PSCs (Diaz-Cuadros et al., 2020; Matsuda et al., 2020; Pourquie, 2003). The presence of an active segmentation clock in gastruloids similar to an in vivo embryo extends the power of this model system to characterize mammalian development (van den Brink et al., 2020; Veenvliet et al., 2020). Nevertheless, the advent of blastoids and gastruloids from mESCs has opened new avenues to understand embryogenesis with wide application in biomedical discoveries especially for understanding developmental malformations. The advantage is its reproducible culture conditions that can be scaled up for any high-throughput studies and its key features strongly resembling an embryo (both in gene expression and axial organization) (Figure 1B, Table 1). The generation of embryo-like structures and organoids is a recent breakthrough of stem cell self-organization with many fundamental questions still unanswered. This will open new avenues to understand germ layer specification in embryos with the possibility to generate organ structures inside embryo models.
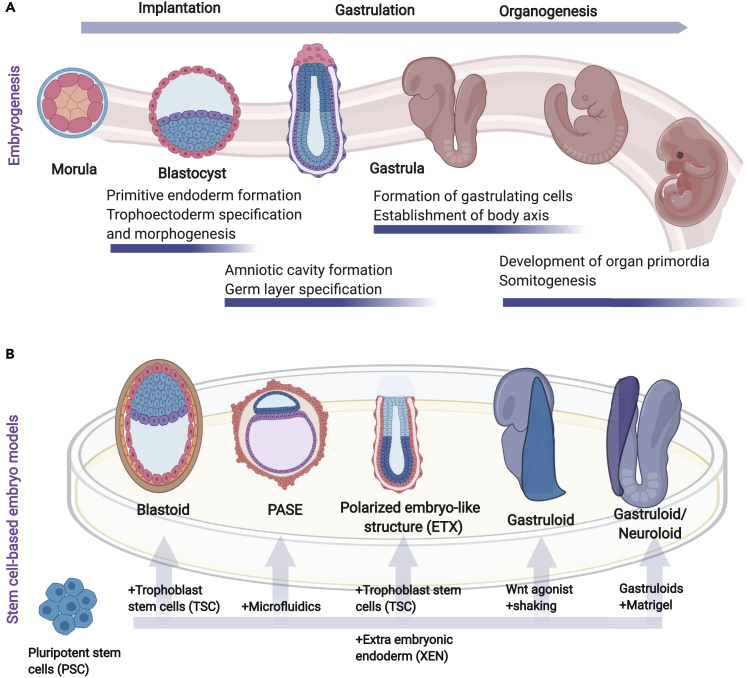
Figure 1.
Stem Cell-Derived Embryo Models Recapitulate Mammalian Embryonic Development
(A) Illustration of the key stages of mammalian embryogenesis, showing the progression of embryonic development of the conceptus inside a reproductive tract in vivo.
(B) Schematic of embryo-like structures generated from pluripotent stem cells in vitro, which can recapitulate the key events of the post-implantation process. These embryo-like structures include the blastoids (modeling the blastocyst stage), Post-implantation Amniotic Sac Embryoid (PASE) and ESC- and TSC-derived (ETS) embryo-like structures (modeling amniotic cavity formation and specification of mesoderm and endodermal layer), and gastruloids (modeling gastrulation and lineage specification with the formation of structures resembling somites and neural tube and early organogenesis). The bottom arrows represent a simplified version of the starting material that is mixed with PSCs to generate the embryo-like structures in a dish (see also Table 1).
Table 1.
A Summary of Different Stem Cell-Based Embryo Models to Study Early Embryonic Development
| Type | PSC Source | Key Features/Cell Types | References |
|---|---|---|---|
| PASE | Human ESC |
|
Shao et al., 2017a, 2017b; Zheng et al., 2019 |
| Blastoid | Mouse ESCs and TSC, or mouse extended pluripotent stem cells alone |
|
Li et al., 2019b; Rivron et al., 2018b; Sozen et al., 2019 |
| ETX model | Mouse ESC + TSC + XEN |
|
Sozen et al., 2018 |
| Gastruloid | Mouse ESC |
|
Beccari et al., 2018; van den Brink et al., 2020; Veenvliet et al., 2020 |
| Human ESC |
|
Moris et al., 2020 |
PASE, Post-implantation Amniotic Sac Embryoid ; XEN, extra endodermal cell; TSC, Trophoblast stem cells.
A collective understanding of how ESCs can be manipulated in vitro to mimic developmental stages of the embryo (Shahbazi and Zernicka-Goetz, 2018) is influential to generate more complex organ type in vitro. For example, the presence of an extracellular matrix, similar to a developing embryo, led to the formation of somites in gastruloids (van den Brink et al., 2020; Veenvliet et al., 2020). The role of the laminin-rich extracellular matrix (also called Matrigel) in the growth and proliferation of intestinal tubules and mammary glands (Li et al., 1987; Montesano et al., 1991) led the foundation to initiate self-organizing structures from ESCs. Seminal discoveries include the generation of bilayered optic cup-like structures from self-organizing mESCs and hESCs (Eiraku et al., 2011; Kuwahara et al., 2015) that can also differentiate in vivo into other retinal cell types after engraftment (Shirai et al., 2016). This triggered the concept that ESCs can be potentially utilized for organogenesis (Murry and Keller, 2008; Sasai et al., 2012).
Mimicking Development to Generate Multi-lineage Organoids
Organoid generation utilizes the spatial and temporal gradients of morphogens that normally orchestrate embryonic development. One of the crucial aspects is the time and precise control of lineage promoting growth factors and expression of tissue-specific transcription factors (TFs). Organoids from tissue-specific ASCs rely on the activation of growth factors necessary for the maintenance of tissue type, whereas the sequential activation of signaling pathways in PSCs lead to the desired organoid type. The precise control of these early developmental processes is still poorly understood and imposes a major challenge for directed differentiation. Here, we present an overview of the concerted efforts of key signaling factors that promote PSC differentiation to self-organize into an organoid of a particular lineage (Figure 2A) and unique features associated with each organoid (Table 2).
Table 2.
Summary of the Organoids Generated from Pluripotent Stem Cells (PSCs) to Study Organ Development
| Type | PSC Source | Key Features/Cell Types | References |
|---|---|---|---|
| Ectodermal Organoids | |||
| Cerebral organoid | Human ESC |
|
Lancaster et al., 2013 |
| Forebrain spheroids (brain assembloid) | Human ESC |
|
Birey et al., 2017 |
| Choroid plexus organoid | Human ESC |
|
Pellegrini et al., 2020 |
| Pituitary organoid | Mouse ESC |
|
Suga et al., 2011 |
| Retinal organoid (optic cup) | Mouse ESC, Human ESC |
|
Eiraku et al., 2011; Völkner et al., 2016 |
| Skin organoid | Mouse ESC |
|
Lee et al., 2018 |
| Human ESC |
|
Lee et al., 2020 | |
| Inner ear organoid | Mouse ESC |
|
Koehler et al., 2013 |
| Human ESC |
|
Koehler et al., 2017 | |
| Mesodermal Organoids | |||
| Cardiac organoid | Human ESC |
|
Hofbauer et al., 2020; Voges et al., 2017 |
| Kidney organoid | Human ESC Human iPSCs |
|
Morizane et al., 2015; Takasato et al., 2015 |
| Endodermal Organoids | |||
| Lung organoid | Human ESC |
|
Dye et al., 2015; Jacob et al., 2017; McCauley et al., 2017 |
| Esophageal organoid | Human ESC |
|
Trisno et al., 2018; Zhang et al., 2018 |
| Liver organoid | Human iPSCs |
|
Takebe et al., 2013 |
| Hepatic-biliary-pancreatic organoid | Human ESC/iPSC |
|
Koike et al., 2019; Wu et al., 2019 |
| Gastric organoid | Mouse ESC Human ESC |
|
McCracken et al., 2014, 2017 |
| Pancreatic organoid | Human ESC |
|
Hohwieler et al., 2017 |
| Intestinal organoid | Human ESC |
|
Spence et al., 2011; Tsai et al., 2017; Watson et al., 2014; Workman et al., 2017 |
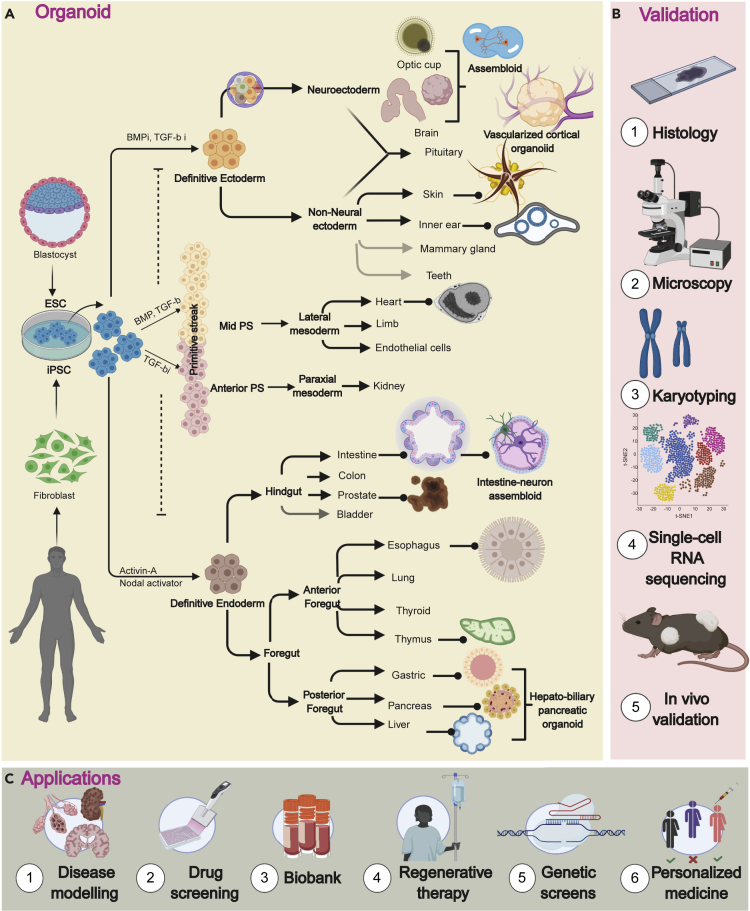
Figure 2.
Directed Differentiation of PSCs to Generate Organoids of Three Different Germ Layers with the Strategies for Functional Validation and Its Biomedical Applications
A simplified roadmap for directed differentiation of PSC using growth factors from different signaling pathways (Wnt, BMP, TGF-β, Notch) to generate 3D cell culture models.
(A) Signaling factors can also promote PSCs to a defined lineage bifurcating from ectoderm and PS. Complex organoid structures (assembloids) can be generated with vascularization or nervous system or coupling multi-endodermal structures. Faded color in the arrow denotes the ability to generate organoids from tissue-specific adult stem cells but not yet from PSCs (see also Table 2 for features of organoids).
(B) An overview of different available tools for functional characterization of 3D culture models.
(C) The potential applications of organoids in biomedical research.
Ectodermal Organoids
During gastrulation, the primitive streak (PS) forms at the midline, where epiblast cells undergo epithelial-to-mesenchymal transition and ingress to form the mesoderm and endoderm. The cells that do not ingress form the ectodermal layer, which patterns to form epidermal cells and most of the central nervous system. Neuro-epithelium is induced in self-organizing PSCs cultured in serum-free media, which evaginates to form neural retina, but a timed notch signaling generates the photoreceptors (Eiraku et al., 2011; Völkner et al., 2016). Inhibiting transforming growth factor (TGF)-β and bone morphogenetic protein (BMP) signaling (blocks mesendodermal commitment) leads to definitive ectoderm formation and neuroectoderm commitment. Furthermore, an additional Wnt activation leads to the formation of cranial neural crest, and a pulse of BMP4 with a block of FGF signaling leads to the formation of non-neural ectodermal fate (Tchieu et al., 2017). Following neuroectodermal fate, PSCs can self-organize into EBs that can be guided to form distinct areas of the brain or can even recapitulate the whole brain containing discrete brain regions with cortical neurons (Lancaster et al., 2013). This has been extended to generate cerebral organoids from human PSCs to recapitulate different brain regions including the hippocampus, midbrain, hypothalamus, cerebellum, and choroid plexus (reviewed in Lancaster and Knoblich, 2014; di Lullo and Kriegstein, 2017; Qian et al., 2019). Choroid plexus organoids derived from PSCs can secrete cerebrospinal fluid and have the ability to form blood-brain-barrier (Pellegrini et al., 2020). The interaction of the hypothalamic neuroectoderm along with non-neural ectoderm also led the path to generate anterior pituitary organoid expressing multiple endocrine lineages (Suga et al., 2011). Non-neural ectoderm also gives rise to inner-ear and the skin appendages. A combination of BMP activation and TGF-β inhibitor can differentiate PSCs into non-neural ectoderm with the formation of more specialized pre-placoidal cells and a distinct population of mechanosensitive hair cells (Koehler et al., 2013, 2017). Skin organoids also utilize a similar differentiation strategy, where individual hair follicles grow with prominent hair shaft, adipocytes, melanocytes, and sebaceous glands (Lee et al., 2018). scRNA-seq analysis of hESC-derived skin organoids also validated the presence of 16 cell clusters that resemble human skin, keratinocytes. Interestingly, grafting these organoids under the skin can generate hair follicles (Lee et al., 2020), a breakthrough in regenerative medicine that can be used to treat skin diseases and rejuvenating burnt skin or scar tissues. Hair follicle development shares a close analogy to the development of other skin appendages like the dental, and the mammary gland (Mikkola, 2007). Although 3D in vitro culture method for culturing teeth (Nakao et al., 2007) and mammary glands (Jamieson et al., 2017) have been developed, generating them from PSCs is still not successful. A deeper investigation of the aforementioned growth factors can be used to generate organoids from teeth, salivary glands, and the mammary gland. Understanding the differentiation hierarchy of mammary, salivary, and dental stem cells (Jussila and Thesleff, 2012; Myllymäki and Mikkola, 2019; Visvader and Stingl, 2014) will be important to generate PSC-derived mammary organoids. Although a mosaic mammary gland is generated by reprogramming dental epithelial stem cells mixed with mammary epithelial cells (MECs) (Jimenez-Rojo et al., 2019) or by mixing mESCs with MECs (Boulanger et al., 2013), generating organoids is not reported. Human iPSCs can be directed to generate mammary organoids; however, they failed to reconstitute in vivo (Qu et al., 2017), suggesting that a fine-tuning of growth factors and timing is critical to recapitulate the mammary microenvironment in organoids.
Intermediate Primitive Streak Specification for Mesodermal and Endodermal Organoids
Organoids of mesodermal and endodermal lineage require an intermediate PS induction in PSCs. PS formation is a major event during gastrulation; anterior PS gives rise to definitive endoderm and paraxial mesoderm (source of bone, cartilage, and fibroblasts), whereas the mid-PS generates the cardiac and limb bud progenitors. Dual SMAD signaling (BMP4 and TGF-β) activation leads to PS formation in ESCs while blocking ectodermal commitment. BMP4 in the presence of FGF2 is the critical regulator that leads to the upregulation of mesoderm-related genes (Brachyury, Cdx2, Tbx3, and Gata4) in hESCs (Gunne-Braden et al., 2020), whereas TGF-β inhibition accelerates mesoderm formation from PS and blocks endoderm specification (Loh et al., 2014). This strategy was used to differentiate ESCs into hematopoietic precursor (Pearson et al., 2008; Zhang et al., 2008) and endothelial progenitors (Goldman et al., 2009) that was instrumental in the formation of cardiac organoids resembling human fetal hearts (Voges et al., 2017). However, differentiated cardiomyocytes do not recapitulate the structural complexity of a mammalian heart (Devalla and Passier, 2018; Mummery et al., 2012) despite showing cardiac electrophysiological response (Mills et al., 2017). This is partly due to the complex architecture of the heart requiring multilineage interactions and paracrine signaling from adjacent tissues. A recent study utilized co-emerging cardiac-like mesoderm and gut-like endoderm from iPSCs that can attain adult heart-type complexity with non-contractile myocytes and blood vessels surrounding it (Silva et al., 2020). Human PSCs can intrinsically self-organize to form cardiac mesoderm without exogenous extracellular matrix proteins and consist of separate myocardial and endothelial layers. The cardiac organoids (cardioids) can also form cavities resembling the heart chambers, which overall opens new avenues to understand cardiogenesis during embryonic development (Hofbauer et al., 2020). Insights from 2D culture suggests that cardioids can be further enhanced to form regionalized cell types by inhibiting Retinoic Acid (RA) and Wnt signaling to generate ventricular cells, or activating them to generate atrial-like cells (Devalla et al., 2015; Zhang et al., 2011). Organoids resembling blood vessel can also be generated that can integrate with the host vasculature (Wimmer et al., 2019), which brings hope to develop organoids of particular lineage coupled with endothelial cells or fused with blood vessel organoids to recapitulate complex organ structure, as recently evidenced in vascularized cortical organoids (Cakir et al., 2019; Shi et al., 2020). Human PSCs can be differentiated into ureteric epithelium or metanephric and mesonephric mesenchyme in vitro that recapitulates embryonic kidney (Taguchi and Nishinakamura, 2017; Taguchi et al., 2014; Takasato et al., 2014, 2015), or even generate kidney organoids from epiblast spheroids (Freedman et al., 2015; Morizane et al., 2015). One of the major feats of kidney-like organoids from PSCs is its incredible ability to vascularize into glomerulus-like structures with interconnecting renal tubules and collecting ducts (Bantounas et al., 2018; van den Berg et al., 2018; Sharmin et al., 2016; Tsujimoto et al., 2020). A reproducible and a scalable 3D organoid culture strategy is developed that supports vascularization in an animal model and therefore brings great promise to regenerative medicine (Koike et al., 2019; Takebe et al., 2015).
Endodermal Organoids
Activating TGF-β signaling at the PS stage leads to the definitive endodermal lineage that gives rise to the internal lining of the lungs, and intestinal tracts and other vital organs like thyroid, thymus, liver, and pancreas. During embryonic development, the endoderm layer gives rise to a primitive gut tube broadly divided into fore, mid, and hindgut, from where other vital organs bud off. This is achieved by using a nodal-related TGF-β signaling molecule, activin-A, which promotes the differentiation of PSCs to the definitive endoderm expressing SOX17 and FOXA2 (D'Amour et al., 2005). Dual inhibition of BMP and TGF-β signaling generates anterior foregut endoderm (AFE), which self-assembles to generate organoids of the esophagus, lung, parathyroid, and thymus. The SOX2+ p63+ progenitors that are responsible for esophageal organoids block the Wnt-BMP signaling required for ventral foregut to generate NKX2-1+ lung progenitors (Trisno et al., 2018; Zhang et al., 2018). However, lung progenitors can also be induced by the addition of FGF10, FGF7, and a low concentration of BMP4 (Huang et al., 2014; Wong et al., 2012) or simultaneous stimulation of WNT and FGF along with high Hedgehog signaling (Dye et al., 2015). Notably, the NKX2-1 progenitors give rise to basal, secretory cells leading to the formation of the airway and alveolar cells, characteristic of the human lung (Huang et al., 2014; Jacob et al., 2017; McCauley et al., 2017), but they mostly resemble a fetal lung, thereby raising the concern of organoid maturity in vitro. NKX2-1+ cells are considered to be a bipotent progenitor population, and Wnt-BMP-FGF signaling differentiates them from being lung versus thyroid (Dame et al., 2017; Kurmann et al., 2015; Serra et al., 2017). The thyroid progenitors can also form organoids and can maintain TH level after xenotransplantation in the mouse model (Kurmann et al., 2015). Thymic organoids are still not successful, but thymic progenitors can be generated by activating BMP4, RA, Wnt3A, and FGF8b, and inhibiting hedgehog signaling can differentiate into T cells in vitro but failed to support prolonged thymogenesis in vivo (Parent et al., 2013; Sun et al., 2013).
The posterior foregut endoderm specifies the formation of the liver, stomach, and pancreas. FGF and BMP4 are pre-hepatic cues that induce the formation of liver progenitors from activin-induced endoderm (Gouon-Evans et al., 2006). Simultaneous inhibition of hedgehog signaling while activating RA results in pancreatic fate (D'Amour et al., 2006). Another strategy utilized the mixing of endothelial and mesenchymal cells that can instruct human iPSC-derived hepatic endoderm to self-organize into liver bud organoids (Takebe et al., 2013, 2017). The liver buds can further differentiate into bile duct cells (activating Notch and TGF-β) or to hepatocytes (blocking Notch and TGF-β) (Ang et al., 2018). A complex architecture can be established by mixing biliary and hepatic cells to generate hepato-biliary organoids that can form bile ducts and hepatic epithelium after transplantation in mice (Wu et al., 2019). Moreover, fusing anterior and posterior gut spheroids from iPSC-derived definitive endoderm can also assemble into multi-endodermal domains to form hepato-biliary-pancreatic organoids (Koike et al., 2019). The generation of the complex organoid system holds tremendous potential as models for studying organogenesis.
While considerable progress has been made in generating multi-organ structures, compartmentalization of organ structures is also evident. For example, gastric organoids can be regionalized into the antral part (Green et al., 2011; McCracken et al., 2014), which upon Wnt and FGF10 activation leads to fundus (McCracken et al., 2017), pancreatic organoids can generate beta cells (β) using keratinocyte growth factor and also resemble the ductal and acinar morphology of human fetal pancreas (Hohwieler et al., 2017). The pancreatic progenitors can form insulin-producing β cells and can maintain glucose levels after transplantation in diabetic mice (Pagliuca et al., 2014). Multiple in vitro differentiation protocols have resulted in pancreatic progenitors, but the critical goal to derive functional β cells is of high biomedical value. However, insulin production is taken as a gold standard to mark β cells; these functional β cells often co-express other hormones leading to the co-occurrence of contaminating lineages (Shahjalal et al., 2018). Although unprecedented progress has been made toward generating β cells in vitro, the existence of unwanted lineages imposes significant concerns that need to be addressed for successful therapeutic application. scRNA-seq on PSC-derived pancreatic organoids compared with the human pancreas will be instrumental to identify the correct lineage to its closest approximation.
A similar level of compartmentalization is evident in intestinal organoids with characteristic villus and crypt-like structures (Spence et al., 2011; Watson et al., 2014), which upon short exposure to FGF4 and Wnt activation recapitulates duodenum and longer exposure generates ileum (Tsai et al., 2017). The colon organoids are generated from the hindgut endoderm in the presence of BMP signaling (Crespo et al., 2017; Múnera et al., 2017), and, embryologically the caudal extension leads to the prostate, bladder, and penile urethra. Not much is known regarding prostate, urethral, and bladder organoid generation from PSCs. Although directed differentiation of hESCs using Wnt10B and FGF expresses prostate-specific transcription factor NKX3.1 and androgen-regulator TMPRSS2 (Calderon-Gierszal and Prins, 2015), this study focused entirely on NKX3.1 expression as a definitive factor for prostate organoid. Several studies on knockout mice have revealed the function of Hoxb1 and Sox9 in prostate commitment irrespective of NKX3.1 status, and detailed characterization of ESC-derived prostate organoids is required with characteristic molecular markers (Toivanen and Shen, 2017).
Overall, the focus has been on generating organoids from a single lineage in vitro that provides insights to organ development, but it fails to recapitulate multi-tissue interactions necessary for organ function. The development of multi-lineage organoids opens new avenues to obtain personalized micro-physiological models for modeling developmental organogenesis in vitro. Few examples include the fusion of neural spheroids or cortical organoids (Bagley et al., 2017; Birey et al., 2017; Xiang et al., 2017, 2019), generating hybrid neuromuscular organoids (Faustino Martins et al., 2020), fusing anterior and foregut endoderm for hepato-biliary and pancreatic organoid structures (Koike et al., 2019), and generating intestinal organoids with a functional enteric nervous system (Workman et al., 2017). Multi-lineage organoids generated from co-emerging cardiac and gut lineage specification from iPSCs can be useful to model heart complexity better than conventional cardiac organoids. These organoids consist of mature cardiomyocytes, stromal epicardial cells, and blood-vessel-like structures along with the primitive gut that resembles the intestine (Silva et al., 2020). The emergence of multilineage organ models will be useful to map paracrine interaction and also understand the role of adjacent organ tissues required for organ maturation. Collectively, these studies present a common set of signaling factors (Wnt, BMP, TGF-β, Hedgehog, Notch) that work in a coordinated and chronological manner to decide the cell fate (Kojima et al., 2014). A thorough identification of the appropriate cocktail of growth factors is crucial to generate correct cell types, and validation using a variety of tools ranging from microscopy to scRNA-seq is necessary (Figure 2B).
Stem Cell-Based Embryo Models and Organoids for Clinical Management
Embryo-like structures and organoids derived from PSCs have diverse scientific applications ranging from modeling mammalian development to understanding disease outcomes (Clevers, 2016; Lancaster and Huch, 2019; Rossi et al., 2018; Sharma et al., 2020). Although in vitro and in vivo models including 2D coculture, microfluidics, tissue engineering, humanized animals, and chimeras are used for decades to model human diseases and drug screening, they have their advantages and associated challenges. Here we present a comprehensive overview of the current applications of PSC-derived 3D culture systems, along with strategies to overcome the challenges to make it accessible for regenerative medicine and cell-based therapeutics (Figure 2C).
Understanding Embryogenesis and Organ Development
iPSCs can be differentiated to form gastruloids and blastoids that can help to understand rare human diseases. “Rare diseases” collectively affect a significant population, which has led to developmental malformations in infants. Despite efforts that have been made to understand the genetic underpinnings of rare diseases, we still have limited clinical progress and lack therapeutic strategies (Anderson and Francis, 2018; Tambuyzer et al., 2020). Given the robustness of iPSC generation from a variety of genetic disorders including Parkinson disease, Down syndrome, and Huntington disease, understanding the defects in early embryogenesis will be helpful from iPSC-derived embryo like structures. Although there is a limitation in the number of patients, the patient-iPSC-derived organoids/gastruloids will be particularly useful to model a genetic disorder, perform drug screening, and understand embryonic development. Mutations in genes associated with the segmentation clock lead to defects in vertebrae. Several other clinically relevant mutations exist that can be useful to study in gastruloids to model developmental anomalies and also perform functional screening. The optimized culture conditions allow in vitro cultures of embryos beyond implantation and provide a broader understanding of pregnancy loss and improve contraception technologies. A major advantage of using stem cell-based embryo models is to utilize them for studying human embryonic development as they represent an ethical alternative to replace the use of embryos. These models should only be used to better understand embryonic development and the origin and treatment of diseases in a dish (Hyun et al., 2020; Rivron et al., 2018a).
Disease Modeling and Functional Screening of Genetic Variants
Organoids have been widely used to model both congenital and acquired diseases including several age-related disorders and infectious diseases (Lancaster and Huch, 2019). Brain organoids from patient-derived iPSCs are utilized to model neurodegenerative disorders and also to recapitulates human brain development (di Lullo and Kriegstein, 2017). To name a few, different types of brain organoids have been used to model microcephaly, Alzheimer disease, and neuro-progenitor abnormality after Zika virus infection (reviewed in Amin and Paşca, 2018). Similarly, gastric organoids are used to model Helicobacter pylori infection (McCracken et al., 2014). A recent study utilized a wide spectrum of organoids generated from human PSCs to understand the viral transmission in different human organs. However, the lack of immune system in organoids may be a limiting factor in these studies to accurately understand the viral immune response (Yang et al., 2020). The generation of thymus epithelial progenitors from PSCs opens another avenue to the ever-growing organoid community. Thymus is one of the specialized organs that plays a major role during infection and other age-related disorders. Thymic organoids capable of T cell differentiation will be particularly applicable for treating several autoimmune diseases (Parent et al., 2013; Sun et al., 2013). On the other hand, a major population globally experiences hearing disorder, and inner ear organoids bring hope to utilize them for personalized medicine. Although transplanting retinal organoids to macular degeneration in primates were successful, the development of ESC-derived inner ear organoids is still in its infancy and needs some fine-tuning before being utilized for regenerative therapies. Furthermore, these organoids can be used for high-throughput drug screening to identify novel targets that promote survival/regeneration of sensory hair cells or show ototoxicity. It also holds great promise to gain insights into genetic mutations affecting inner ear development (Roccio and Edge, 2019; Tang et al., 2019) and model hearing loss defects. Skin organoids are able to regenerate hair follicles under the skin and can be used as bioengineered skin grafts in regenerative surgery Lee et al., 2020. Organoids can be a powerful tool to functionally characterize genetic variants and pancreatic organoids are recently used to study diabetes-causing gene variants (Maxwell et al., 2020), and similar strategies should be implemented to understand other disease-causing mutations.
Around 4.6 million missense variants are reported for disease-causing mutations in the Genome Aggregation Database (GnomAD), of which 99% are rare, and only 2% have a clinical interpretation in ClinVar (Claussnitzer et al., 2020; Starita et al., 2017). Much of our understanding of clinical management comes from ClinVar, but the majority are still considered to be “variants of uncertain significance” (Landrum et al., 2014; Rehm et al., 2015) (Figure 3A). Most of the disease-causing mutations reported in ClinVar are point mutations, and currently, a broad understanding of single-nucleotide variants (SNVs) comes from generating transgenic mice, which is often very laborious and time consuming. To this end, several in vitro functional assays have been developed to screen disease-causing missense variants, but it often fails to recapitulate the disease outcome (Monteiro et al., 2020). Although high-throughput screenings (HTSs) have accelerated the identification of pathogenic and neutral variants of several disease-causing genes (Findlay et al., 2018; Hanna et al., 2020), they fail to comment on its tumorigenic potential in patients. It is not trivial to generate transgenic model organisms to understand the function of individual variants and its disease-causing potential. An alternative approach could be the use of base editors and prime editors (Anzalone et al., 2020) to generate SNVs in ESCs, which can be differentiated to organoids of interest (as described previously) (Figure 3B). This will leverage its potential for in vivo functional studies and reduce the generation of genetically engineered mouse models.
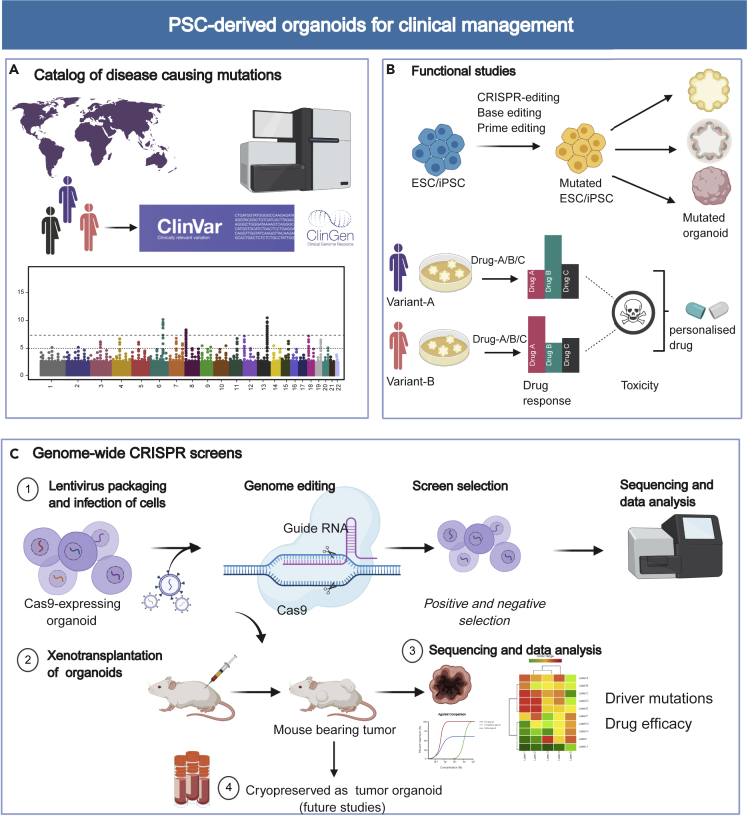
Figure 3.
Applications of PSC-Derived Organoids in Personalized Medicine and Clinical Management
(A) The disease-causing variants are cataloged in several public clinical databases and are used for functional studies.
(B) Different variants can be generated in PSCs using the CRISPR-based genome editing toolbox, which can be further differentiated to the organoid of the desired lineage. The organoids represent each patient/variant, and high-throughput drug screening and cytotoxicity studies can be performed to develop a personalized drug.
(C) Schematic showing the use of organoids in high-throughput genetic screens using CRISPR to identify novel gene targets that get frequently mutated to cause cancer. Furthermore, these organoids can be xenotransplanted to develop an in vivo model for mouse tumorigenesis studies. The resulting tumors can be further cryopreserved as organoids for drug screening and mutagenesis studies.
Drug Screening, Biobanks and the Hope of Using Personalized Medicine
Multiple HTS platforms have been developed using iPSCs, which have advanced our understanding of drug development and toxicity studies (Grskovic et al., 2011). It has just started to be applied in organoids for drug screening (Czerniecki et al., 2018). These screening platforms are very cost effective and reduce any manual error thereby generating uniform organoids more successfully in clinical drug development. During the past decade, tissue engineering and bioprinting technologies have revolutionized our understanding of simulating 3D organ complexity. This allows multiple cell types to be integrated and grown together in 3D culture systems that are compartmentalized by membranes or microchannels to mimic the in vivo environment (Bhatia and Ingber, 2014). Developing an “organ-on-a-chip” concept to generate vascularized multi-lineage organoid along with HTS platforms will play a critical role to improve the scalability and uniformity of organoid generation. As it simulates to its maximum capability, these novel technological insights surely improve our understanding of organ development in vitro, but not all microfluidics can recapitulate the organ complexity. Breakthroughs have been made to integrate multi-organs in a chip (Wu et al., 2020) and coupling these with multilineage organoids will be of high biomedical relevance. Generating organoid biobanks are crucial for personalized medicine, as it brings the ability to perform high-throughput drug screening, epigenomic and transcriptomic analysis, and copy number variations of individual patients at a large scale. While biobanks have been generated for breast cancer, gastric cancer, pediatric kidney cancer, and glioblastoma (Calandrini et al., 2020; Jacob et al., 2020; Sachs et al., 2018; Yan et al., 2018), these can be extended to the development of organoid biobanks with individual disease variants generated in ESCs, or from iPSCs of rare diseases. We hope to see patient-variant organoid biobank in the future that will accelerate personalized drug development.
High-throughput CRISPR-Based Genetic Screens
Recent advances in genome editing, especially with the advent of CRISPR-Cas9 (clustered regularly interspaced sort palindromic repeats-CRISPR-associated protein 9), have revolutionized our understanding of human disease and have been a powerful tool to correct genetic mutations in human organoids (Geurts et al., 2020; Schwank et al., 2013). Although genome editing is a routine technique in 2D cell lines, it requires optimizations to improve its efficiency. Characterizing correctly edited organoids is a major challenge, and some strategies rely on the removal of culture media constituents, antibiotic selection, or fluorescence-activated cell sorting (FACS) after transfection (Artegiani et al., 2020; Driehuis and Clevers, 2017; Fujii et al., 2015; Sasselli et al., 2017). For example, removal of Wnt and R-spondin from culture medium selects survival of APCKO intestinal organoids, and further addition of Nutlin3 allows selection of APC and P53 doubleKO organoids (Drost et al., 2015).
CRISPR and other nucleases have been classically used to understand the function of individual genes by making knockouts or tracking particular cell types using reporter organoids (Artegiani et al., 2020; Matano et al., 2015). Although this “one-gene-at-a-time” approach was crucial to establish the power of genome editing in organoids, it fails to validate a large number of genomic threats underlying a disease. Genome sequencing studies have identified several infrequent mutations that hit together with commonly mutated genes. Some driver mutations that are often considered to be rare can form aggressive cancers and are the guiding force behind genetic heterogeneity in tumors (Nussinov et al., 2019; Vogelstein et al., 2013). To better understand these rare events, genome-wide CRISPR screens are a powerful tool to get an overview of less frequent but actionable genes (Figure 3C). Pooled CRISPR screens are increasingly popular to model these synergistic interactions and identify tumor suppressors and other novel cancer driver genes. The generation of Cas9 knock-in mice was instrumental to extend the in vivo genome editing toolbox that accelerated high-throughput CRISPR screens in organoids (Platt et al., 2014). This led to the initial reports on CRISPR loss-of-function screens in biliary epithelial cell organoids to identify genes necessary for liver regeneration (Planas-Paz et al., 2019) and also for identifying co-occurring mutations in colorectal cancer (Takeda et al., 2019). These studies looked into a specific pool of genes as whole-genome-wide screens in organoids were not successful due to several technical limitations.
Although growth factor selection assay is a powerful method to identify mutated organoids (Drost et al., 2015; Matano et al., 2015), similar niche factor dependence strategies need to be devised for studying other gene perturbations. Moreover, the heterogeneous growth of organoids from transduced single organoid cell suspension introduces variability in guide RNA (gRNA) representation and downstream sequencing analysis. To overcome these shortcomings, the development of barcoded sequencing of individual organoids compared with bulk sequencing has proved to be a good alternative to perform genome-wide knockout screens (Ringel et al., 2020). Furthermore, pooled screens coupled with xenotransplantation of organoids in vivo can be used to perform tumorigenesis studies (Figure 3C). While large-scale mouse mutagenesis screens are a valuable tool for functional genomics, it require large research infrastructure and can be time consuming and laborious (Gondo, 2008). Transplantation of human colonic organoids transduced with a pool of gRNAs led to the identification of recurringly mutated tumor suppressor genes and some novel mediators that were not previously associated with colorectal cancer (Michels et al., 2020). Organoid xenografts in mouse models to study cancer metastasis can be a powerful alternative to mouse mutagenesis screens (O'Rourke et al., 2017; Roper et al., 2017).
Most forward genetic screen studies are performed on ASC-derived organoids, whereas there are no studies reported on CRISPR screens on PSC-derived organoids. Generating missense variants in ESCs or patient iPSC-derived organoids can be utilized in the future to study synergistic gene interactions and also for selective drug response. Several synthetic-lethal relationships have been reported that forms the basis of drug development to treat several cancers; however, multiple studies have also reported on its low efficacy or selective survival (Huang et al., 2020; Nijman, 2011). For example, PARP and ATR inhibitors that have revolutionized cancer treatment have shortcomings and CRISPR activation and knockout screens have identified the genetic basis behind resistance and ways to improve sensitivity. However, 2D cell lines fail to recapitulate the interaction of stem cells and the associated microenvironment, which plays a major role in drug sensitivity. Future studies on genome-wide CRISPR screens will be very useful to understand a particular drug response in patient-derived organoids for personalized medicine (Figure 3C). Apart from disease modeling, forward genetic screens in organoids can be utilized for optimizing niche growth factors and stem cell differentiation strategies and identifying conditions for organoid maturation (Li et al., 2019a).
Challenges and Future Perspectives
Despite paramount success in generating organoids from ESCs and iPSCs, the technologies are still in their infancy for use in the clinic. Genome instability and loss of heterozygosity during extensive rounds of in vitro culture and the introduction of de novo mutations during somatic reprogramming impose serious concerns on utilizing iPSCs. Moreover, the epigenetic memory and DNA methylation status of the parent somatic cells from where iPSCs are generated also affect cell fate (Gore et al., 2011; Hussein et al., 2011; Lister et al., 2011). Collectively these limitations in iPSCs are potential threats to utilize them for robust organoid generation, especially for therapeutics. Recent advances to correct disease-causing mutations using different CRISPR-based genome editing tools in iPSCs followed by organoid generation is a promising avenue for regenerative medicine; however, they need to be tested for CRISPR off-target events that may have other harmful consequences. In vitro differentiation of PSCs into organoids often resembles the embryonic stage and lacks proper maturation, another hurdle to utilize them for functional studies. These organoids can differentiate into mature cell types after orthotopic transplantation, suggesting that cues from the in vivo microenvironment enhance maturity and tissue functionality. Recent advances in organoid protocols have addressed this issue using bioengineering platforms to obtain adult-like tissues (Holloway et al., 2019). The generation of interconnected vasculature has just begun using a mesenchyme-driven self-condensation method. Organ buds are reported from multiple tissue types including the liver, intestine, lung, kidney, heart, and brain (Takebe et al., 2015). The lack of vasculature generates a hypoxic environment leading to increased necrosis or apoptotic cells in organoids. Endothelial cells can be mixed with lineage progenitors to generate vascularization in organoids. Injectable organoids have recently been developed in hydrogels that can also generate perfusive vasculature in mice in vivo (Rossen et al., 2020). Although improved vasculature is currently used, engineering organoids with other surrounding tissue types are necessary. To this end, assembloids have emerged to model the structural interactions between subdomains of an organ that opens the opportunity to couple multi-lineage differentiation. Organ systems are mostly associated by surrounding muscles, nerves, and immune cells that are critical for organ function; engineering organoids containing a neuronal network along with blood vasculature is something to foresee in the future.
Another major challenge is the considerable amount of ambiguity that exists in identifying the correct lineage progenitors, which leads to cell heterogeneity and inter-organoid variability. Spontaneous differentiation due to long-term culture can generate unwanted lineages (Tang et al., 2019) and the high frequency of contaminating progenitors compared with the desired cell types during in vitro differentiation of PSCs can affect organoid reproducibility (Veres et al., 2019; Yao et al., 2017). Therefore “separating the wheat from the chaff” is important to generate organoids, as a minor population of undifferentiated human PSCs has a higher chance to give rise to teratoma (tumor) that often out-competes organ reconstitution in vivo (Fowler et al., 2020). Heterogeneity of both differentiated and undifferentiated PSCs is a common problem, and therefore using specific cell surface markers can efficiently generate the desired lineage. FACS is a powerful method to sort the correct cell type based on surface markers but can often affect the cell population thereby reduces its efficiency (Kelly et al., 2011). A current gold standard to draw an analogy in organ recapitulation is to perform bulk transcriptomic sequencing or correlating protein expression in PSC-derived organoids with respective desired tissue. However, this often outnumbers the exact cell type and fails to detect the presence of a minor population of contaminating lineage. The development of scRNA-seq is instrumental to get a comprehensive view of cell-type composition in an organoid. Although limitations to scRNA-seq exist with overarching challenges of not being able to detect lowly expressed gene clusters (Kharchenko et al., 2014), the high-omics dataset can assess any inter-organoid variability and gives high confidence to utilize them to model a particular organ type. A large-scale transcriptomic profiling study at the single-cell level in brain organoid has identified six distinct clusters to neuroectodermal type and one cluster of cells of mesodermal origin (Quadrato et al., 2017). A stepwise affirmation with scRNA-seq at different phases of differentiation could better mimic organ development in vitro. This led to the generation of a nearly pure cell population of PS that can differentiate to other mesodermal cell types (Loh et al., 2016). Therefore, understanding cell types and state in a particular organoid using diverse technologies ranging from transcriptomics profiling to lineage tracing at single-cell level (McKinley et al., 2020) will be crucial to establish PSC-derived organoids as robust and consistent models for functional studies (Figure 2C).
Not all stem cell lines have similar potency to generate organoids of a particular lineage as observed in the ability to generate retina, inner ear, and skin organoids from ESCs (Lee et al., 2018; Osafune et al., 2008; Völkner et al., 2016). Analysis of 162 differentiation strategies from 61 PSC lines identified Wnt/β catenin signaling leads to the varied differentiation ability, and its activation could alleviate the outcomes (Strano et al., 2020). Future strategies should focus on empirically investigating the organoid-forming efficiency for each ESC line before initiating functional studies. Some common factors that decide the efficiency are self-assembling property of PSCs, effects of Matrigel concentration, precise timings of morphogens, and variable epigenetic status. Matrigel being a biological material may have batch effects, therefore alternative synthetic scaffolds has just begun to be explored (Aisenbrey and Murphy, 2020). Moreover, the cytotoxicity of the chemical inhibitors may also affect cell cycle/growth, giving inconsistencies in cellular differentiation (Laco et al., 2018). Thus, overexpressing tissue-specific TFs can be an alternative to induce differentiation in PSCs as evident in generating hepatocyte-like cells in vitro (Guye et al., 2016), thyroid follicular organoids (Antonica et al., 2012), thymus progenitors (Su et al., 2015), and intestinal organoids from PSCs (Sinagoga et al., 2018). A collaborative effort to fine-tune the timing and concentration of biochemical and biophysical cues will increase the success and reproducibility. The advent of genome editing to rectify a faulty gene with personalized organoids is what we foresee for precision medicine in a personalized era. Although the “bench-to-bedside” transition of organoids may take some time, early pre-clinical studies are very promising with the increasing trend for commercializing organoids for disease modeling and regenerative medicine (Choudhury et al., 2020).
Conclusion
Research on stem cell-based embryo models (blastoids and gastruloids) and organoids will be increasingly popular in the coming years with more opportunities to utilize them for regenerative medicine. These 3D models have already been proved to be an exciting platform for personalized therapies, the above challenges need to be overcome, and efforts have to be made to increase the yield and reproducibility. Future research will look into expanding these platforms to other animal model systems. Some examples include the generation of brain organoids from other non-human primates to study the evolution of the nervous system and brain maturation (Kanton et al., 2019; Pollen et al., 2019), generation of snake venom gland organoids for anti-venom production (Post et al., 2020), and generation of bat intestinal organoids for modeling viral infection (Zhou et al., 2020). Most of our fundamental understanding of signal transduction pathways that we exploit to generate “mini-organs in a dish” comes from studying animal development. Generating self-organizing structures from stem cells in other animal models will provide deep molecular evolutionary insights and a detailed appreciation of developmental networks. The conservation of gene regulatory networks will help to understand the evolution of the structure and function of organs.
Acknowledgments
We sincerely apologize to any authors whose work we were not able to include here due to space restriction. We would like to thank Drs. Kajal Biswas, Manisha Jalan, Suhas Kharat, Satheesh Sengodan, and Terry Sullivan for comments and suggestion. The figures in this review are made using a paid subscription to BioRender.com; schematics of organoids and embryo models are drawn based on the appropriate references explaining them. The Sharan laboratory is funded by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, US National Institutes of Health.
Author Contributions
The authors contributed equally to all aspects of the article. S.S. wrote the first draft of the manuscript.
Contributor Information
Sounak Sahu, Email: sounak.sahu@nih.gov.
Shyam K. Sharan, Email: sharans@mail.nih.gov.
References
- Aisenbrey E.A., Murphy W.L. Synthetic alternatives to matrigel. Nat. Rev. Mater. 2020;5:539–551. doi: 10.1038/s41578-020-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin N.D., Paşca S.P. Building models of brain disorders with three-dimensional organoids. Neuron. 2018;100:389–405. doi: 10.1016/j.neuron.2018.10.007. [DOI] [PubMed] [Google Scholar]
- Anderson R.H., Francis K.R. Modeling rare diseases with induced pluripotent stem cell technology. Mol. Cell. Probes. 2018;40:52–59. doi: 10.1016/j.mcp.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L.T., Tan A.K.Y., Autio M.I., Goh S.H., Choo S.H., Lee K.L., Tan J., Pan B., Lee J.J.H., Lum J.J. A roadmap for human liver differentiation from pluripotent stem cells. Cell Rep. 2018;22:2190–2205. doi: 10.1016/j.celrep.2018.01.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonica F., Kasprzyk D.F., Opitz R., Iacovino M., Liao X.-H., Dumitrescu A.M., Refetoff S., Peremans K., Manto M., Kyba M. Generation of functional thyroid from embryonic stem cells. Nature. 2012;491:66–71. doi: 10.1038/nature11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone A.V., Koblan L.W., Liu D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020;38:824–844. doi: 10.1038/s41587-020-0561-9. [DOI] [PubMed] [Google Scholar]
- Artegiani B., Hendriks D., Beumer J., Kok R., Zheng X., Joore I., Chuva de Sousa Lopes S., van Zon J., Tans S., Clevers H. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat. Cell Biol. 2020;22:321–331. doi: 10.1038/s41556-020-0472-5. [DOI] [PubMed] [Google Scholar]
- Bagley J.A., Reumann D., Bian S., Lévi-Strauss J., Knoblich J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantounas I., Ranjzad P., Tengku F., Silajdžić E., Forster D., Asselin M.-C., Lewis P., Lennon R., Plagge A., Wang Q. Generation of functioning nephrons by implanting human pluripotent stem cell-derived kidney progenitors. Stem Cell Rep. 2018;10:766–779. doi: 10.1016/j.stemcr.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari L., Moris N., Girgin M., Turner D.A., Baillie-Johnson P., Cossy A.C., Lutolf M.P., Duboule D., Arias A.M. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature. 2018;562:272–276. doi: 10.1038/s41586-018-0578-0. [DOI] [PubMed] [Google Scholar]
- Bérenger-Currias N.M.L.P., Mircea M., Adegeest E., van den Berg P.R., Feliksik M., Hochane M., Idema T., Tans S.J., Semrau S. Early neurulation recapitulated in assemblies of embryonic and extraembryonic cells. bioRxiv. 2020 doi: 10.1101/2020.02.13.947655. [DOI] [Google Scholar]
- Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger C.A., Bruno R.D., Mack D.L., Gonzales M., Castro N.P., Salomon D.S., Smith G.H. Embryonic stem cells are redirected to non-tumorigenic epithelial cell fate by interaction with the mammary microenvironment. PLoS One. 2013;8:e62019. doi: 10.1371/journal.pone.0062019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakir B., Xiang Y., Tanaka Y., Kural M.H., Parent M., Kang Y.-J., Chapeton K., Patterson B., Yuan Y., He C.-S. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods. 2019;16:1169–1175. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calandrini C., Schutgens F., Oka R., Margaritis T., Candelli T., Mathijsen L., Ammerlaan C., van Ineveld R.L., Derakhshan S., de Haan S. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat. Commun. 2020;11:1310. doi: 10.1038/s41467-020-15155-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Gierszal E.L., Prins G.S. Directed differentiation of human embryonic stem cells into prostate organoids in vitro and its perturbation by low-dose bisphenol A exposure. PLoS One. 2015;10:e0133238. doi: 10.1371/journal.pone.0133238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury D., Ashok A., Naing M.W. Commercialization of organoids. Trends Mol. Med. 2020;26:245–249. doi: 10.1016/j.molmed.2019.12.002. [DOI] [PubMed] [Google Scholar]
- Claussnitzer M., Cho J.H., Collins R., Cox N.J., Dermitzakis E.T., Hurles M.E., Kathiresan S., Kenny E.E., Lindgren C.M., MacArthur D.G. A brief history of human disease genetics. Nature. 2020;577:179–189. doi: 10.1038/s41586-019-1879-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- Crespo M., Vilar E., Tsai S.Y., Chang K., Amin S., Srinivasan T., Zhang T., Pipalia N.H., Chen H.J., Witherspoon M. Colonic organoids derived from human induced pluripotent stem cells for modeling colorectal cancer and drug testing. Nat. Med. 2017;23:878–884. doi: 10.1038/nm.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerniecki S.M., Cruz N.M., Harder J.L., Menon R., Annis J., Otto E.A., Gulieva R.E., Islas L.V., Kim Y.K., Tran L.M. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell. 2018;22:929–940.e4. doi: 10.1016/j.stem.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- D’Amour K.A., Bang A.G., Eliazer S., Kelly O.G., Agulnick A.D., Smart N.G., Moorman M.A., Kroon E., Carpenter M.K., Baetge E.E. Production of pancreatic hormone–expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006;24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- Dame K., Cincotta S., Lang A.H., Sanghrajka R.M., Zhang L., Choi J., Kwok L., Wilson T., Kańduła M.M., Monti S. Thyroid progenitors are robustly derived from embryonic stem cells through transient, developmental stage-specific overexpression of nkx2-1. Stem Cell Reports. 2017;8:216–225. doi: 10.1016/j.stemcr.2016.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalla H.D., Passier R. Cardiac differentiation of pluripotent stem cells and implications for modeling the heart in health and disease. Sci. Transl. Med. 2018;10:eaah5457. doi: 10.1126/scitranslmed.aah5457. [DOI] [PubMed] [Google Scholar]
- Devalla H.D., Schwach V., Ford J.W., Milnes J.T., El-Haou S., Jackson C., Gkatzis K., Elliott D.A., Chuva de Sousa Lopes S.M., Mummery C.L. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol. Med. 2015;7:394–410. doi: 10.15252/emmm.201404757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Cuadros M., Wagner D.E., Budjan C., Hubaud A., Tarazona O.A., Donelly S., Michaut A., Al Tanoury Z., Yoshioka-Kobayashi K., Niino Y. In vitro characterization of the human segmentation clock. Nature. 2020;580:113–118. doi: 10.1038/s41586-019-1885-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Lullo E., Kriegstein A.R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 2017;18:573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driehuis E., Clevers H. CRISPR/Cas 9 genome editing and its applications in organoids. Am. J. Physiol. Gsatrointest. Liver Physiol. 2017;312:G257–G265. doi: 10.1152/ajpgi.00410.2016. [DOI] [PubMed] [Google Scholar]
- Drost J., Van Jaarsveld R.H., Ponsioen B., Zimberlin C., Van Boxtel R., Buijs A., Sachs N., Overmeer R.M., Offerhaus G.J., Begthel H. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- Dye B.R., Hill D.R., Ferguson M.A., Tsai Y.H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4:1–25. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S., Sekiguchi K., Adachi T., Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- Faustino Martins J.M., Fischer C., Urzi A., Vidal R., Kunz S., Ruffault P.L., Kabuss L., Hube I., Gazzerro E., Birchmeier C. Self-organizing 3D human trunk neuromuscular organoids. Cell Stem Cell. 2020;26:172–186.e6. doi: 10.1016/j.stem.2019.12.007. [DOI] [PubMed] [Google Scholar]
- Findlay G.M., Daza R.M., Martin B., Zhang M.D., Leith A.P., Gasperini M., Janizek J.D., Huang X., Starita L.M., Shendure J. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler J.L., Ang L.T., Loh K.M. A critical look: challenges in differentiating human pluripotent stem cells into desired cell types and organoids. Wiley Interdiscip. Rev. Dev. Biol. 2020;9:1–23. doi: 10.1002/wdev.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman B.S., Brooks C.R., Lam A.Q., Fu H., Morizane R., Agrawal V., Saad A.F., Li M.K., Hughes M.R., Werff R.V. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 2015;6:8715. doi: 10.1038/ncomms9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Matano M., Nanki K., Sato T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc. 2015;10:1474–1485. doi: 10.1038/nprot.2015.088. [DOI] [PubMed] [Google Scholar]
- Geurts M.H., de Poel E., Amatngalim G.D., Oka R., Meijers F.M., Kruisselbrink E., van Mourik P., Berkers G., de Winter-de Groot K.M., Michel S. CRISPR-based adenine editors correct nonsense mutations in a cystic fibrosis organoid biobank. Cell Stem Cell. 2020;26:503–510.e7. doi: 10.1016/j.stem.2020.01.019. [DOI] [PubMed] [Google Scholar]
- Goldman O., Feraud O., Boyer-Di Ponio J., Driancourt C., Clay D., Le Bousse-Kerdiles M.-C., Bennaceur-Griscelli A., Uzan G. A boost of BMP4 accelerates the commitment of human embryonic stem cells to the endothelial lineage. Stem Cells. 2009;27:1750–1759. doi: 10.1002/stem.100. [DOI] [PubMed] [Google Scholar]
- Gondo Y. Trends in large-scale mouse mutagenesis: from genetics to functional genomics. Nat. Rev. Genet. 2008;9:803–810. doi: 10.1038/nrg2431. [DOI] [PubMed] [Google Scholar]
- Gore A., Li Z., Fung H.-L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouon-Evans V., Boussemart L., Gadue P., Nierhoff D., Koehler C.I., Kubo A., Shafritz D.A., Keller G. BMP-4 is required for hepatic specification of mouse embryonic stem cell–derived definitive endoderm. Nat. Biotechnol. 2006;24:1402–1411. doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- Green M.D., Chen A., Nostro M.-C., D’Souza S.L., Schaniel C., Lemischka I.R., Gouon-Evans V., Keller G., Snoeck H.-W. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 2011;29:267–272. doi: 10.1038/nbt.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grskovic M., Javaherian A., Strulovici B., Daley G.Q. Induced pluripotent stem cells — opportunities for disease modelling and drug discovery. Nat. Rev. Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- Gunne-Braden A., Sullivan A., Gharibi B., Sheriff R.S.M., Maity A., Wang Y.-F., Edwards A., Jiang M., Howell M., Goldstone R. GATA3 mediates a fast, irreversible commitment to BMP4-driven differentiation in human embryonic stem cells. Cell Stem Cell. 2020;26:693–706.e9. doi: 10.1016/j.stem.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guye P., Ebrahimkhani M.R., Kipniss N., Velazquez J.J., Schoenfeld E., Kiani S., Griffith L.G., Weiss R. Genetically engineering self-organization of human pluripotent stem cells into a liver bud-like tissue using Gata6. Nat. Commun. 2016;7:10243. doi: 10.1038/ncomms10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna R.E., Hegde M., Fagre C.R., DeWeirdt P.C., Sangree A.K., Szegletes Z., Griffith A., Feeley M.N., Sanson K.R., Baidi Y. Massively parallel assessment of human variants with base editor screens. bioRxiv. 2020 doi: 10.1101/2020.05.17.100818. [DOI] [PubMed] [Google Scholar]
- Harrison S.E., Sozen B., Christodoulou N., Kyprianou C., Zernicka-Goetz M. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science. 2017;356:eaal1810. doi: 10.1126/science.aal1810. [DOI] [PubMed] [Google Scholar]
- Hofbauer P., Jahnel S., Papai N., Giesshammer M., Penc M., Tavernini K., Grdseloff N., Meledeth C., Deyett A., Schmidt C. Cardioids reveal self-organizing principles of human cardiogenesis. bioRxiv. 2020 doi: 10.1101/2020.07.06.189431. [DOI] [PubMed] [Google Scholar]
- Hohwieler M., Illing A., Hermann P.C., Mayer T., Stockmann M., Perkhofer L., Eiseler T., Antony J.S., Müller M., Renz S. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut. 2017;66:473–486. doi: 10.1136/gutjnl-2016-312423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway E.M., Capeling M.M., Spence J.R. Biologically inspired approaches to enhance human organoid complexity. Development. 2019;146:dev166173. doi: 10.1242/dev.166173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A., Garraway L.A., Ashworth A., Weber B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 2020;19:23–38. doi: 10.1038/s41573-019-0046-z. [DOI] [PubMed] [Google Scholar]
- Huang S.X.L., Islam M.N., O’Neill J., Hu Z., Yang Y.-G., Chen Y.-W., Mumau M., Green M.D., Vunjak-Novakovic G., Bhattacharya J. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein S.M., Batada N.N., Vuoristo S., Ching R.W., Autio R., Närvä E., Ng S., Sourour M., Hämäläinen R., Olsson C. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Hyun I., Munsie M., Pera M.F., Rivron N.C., Rossant J. Toward guidelines for research on human embryo models formed from stem cells. Stem Cell Reports. 2020;14:169–174. doi: 10.1016/j.stemcr.2019.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A., Morley M., Hawkins F., McCauley K.B., Jean J.C., Heins H., Na C.-L., Weaver T.E., Vedaie M., Hurley K. Differentiation of human pluripotent stem cells into functional lung alveolar epithelial cells. Cell Stem Cell. 2017;21:472–488.e10. doi: 10.1016/j.stem.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Salinas R.D., Zhang D.Y., Nguyen P.T.T., Schnoll J.G., Wong S.Z.H., Thokala R., Sheikh S., Saxena D., Prokop S. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204.e22. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson P.R., Dekkers J.F., Rios A.C., Fu N.Y., Lindeman G.J., Visvader J.E. Derivation of a robust mouse mammary organoid system for studying tissue dynamics. Development. 2017;144:1065–1071. doi: 10.1242/dev.145045. [DOI] [PubMed] [Google Scholar]
- Jimenez-Rojo L., Pagella P., Harada H., Mitsiadis T.A. Dental epithelial stem cells as a source for mammary gland regeneration and milk producing cells in vivo. Cells. 2019;8:1302. doi: 10.3390/cells8101302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jussila M., Thesleff I. Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb. Perspect. Biol. 2012;4:a008425. doi: 10.1101/cshperspect.a008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanton S., Boyle M.J., He Z., Santel M., Weigert A., Sanchís-Calleja F., Guijarro P., Sidow L., Fleck J.S., Han D. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 2019;574:418–422. doi: 10.1038/s41586-019-1654-9. [DOI] [PubMed] [Google Scholar]
- Kelly O.G., Chan M.Y., Martinson L.A., Kadoya K., Ostertag T.M., Ross K.G., Richardson M., Carpenter M.K., D’Amour K.A., Kroon E. Cell-surface markers for the isolation of pancreatic cell types derived from human embryonic stem cells. Nat. Biotechnol. 2011;29:750–756. doi: 10.1038/nbt.1931. [DOI] [PubMed] [Google Scholar]
- Kharchenko P.V., Silberstein L., Scadden D.T. Bayesian approach to single-cell differential expression analysis. Nat. Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler K.R., Mikosz A.M., Molosh A.I., Patel D., Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500:217–221. doi: 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler K.R., Nie J., Longworth-Mills E., Liu X.-P., Lee J., Holt J.R., Hashino E. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat. Biotechnol. 2017;35:583–589. doi: 10.1038/nbt.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike H., Iwasawa K., Ouchi R., Maezawa M., Giesbrecht K., Saiki N., Ferguson A., Kimura M., Thompson W.L., Wells J.M. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature. 2019;574:112–116. doi: 10.1038/s41586-019-1598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y., Tam O.H., Tam P.P.L. Timing of developmental events in the early mouse embryo. Semin. Cell Dev. Biol. 2014;34:65–75. doi: 10.1016/j.semcdb.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Kurmann A.A., Serra M., Hawkins F., Rankin S.A., Mori M., Astapova I., Ullas S., Lin S., Bilodeau M., Rossant J. Regeneration of thyroid function by transplantation of differentiated pluripotent stem cells. Cell Stem Cell. 2015;17:527–542. doi: 10.1016/j.stem.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara A., Ozone C., Nakano T., Saito K., Eiraku M., Sasai Y. Generation of a ciliary margin-like stem cell niche from self-organizing human retinal tissue. Nat. Commun. 2015;6:6286. doi: 10.1038/ncomms7286. [DOI] [PubMed] [Google Scholar]
- Laco F., Woo T.L., Zhong Q., Szmyd R., Ting S., Khan F.J., Chai C.L.L., Reuveny S., Chen A., Oh S. Unraveling the inconsistencies of cardiac differentiation efficiency induced by the GSK3β inhibitor CHIR99021 in human pluripotent stem cells. Stem Cell Reports. 2018;10:1851–1866. doi: 10.1016/j.stemcr.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Huch M. Disease modelling in human organoids. Dis. Model Mech. 2019;12:dmm039347. doi: 10.1242/dmm.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.-A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Bӧscke R., Tang P.C., Hartman B.H., Heller S., Koehler K.R. Hair follicle development in mouse pluripotent stem cell-derived skin organoids. Cell Rep. 2018;22:242–254. doi: 10.1016/j.celrep.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Rabbani C.C., Gao H., Steinhart M.R., Woodruff B.M., Pflum Z.E., Kim A., Heller S., Liu Y., Shipchandler T.Z. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature. 2020;582:399–404. doi: 10.1038/s41586-020-2352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.L., Aggeler J., Farson D.A., Hatier C., Hassell J., Bissell M.J. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proc. Natl. Acad. Sci. U S A. 1987;84:136–140. doi: 10.1073/pnas.84.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.V., Dixon G., Verma N., Rosen B.P., Gordillo M., Luo R., Xu C., Wang Q., Soh C.L., Yang D. Genome-scale screens identify JNK–JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nat. Genet. 2019;51:999–1010. doi: 10.1038/s41588-019-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zhong C., Yu Y., Liu H., Sakurai M., Yu L., Min Z., Shi L., Wei Y., Takahashi Y. Generation of blastocyst-like structures from mouse embryonic and adult cell cultures. Cell. 2019;179:687–702.e18. doi: 10.1016/j.cell.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Kida Y.S., Hawkins R.D., Nery J.R., Hon G., Antosiewicz-Bourget J., O’Malley R., Castanon R., Klugman S. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.M., Ang L.T., Zhang J., Kumar V., Ang J., Auyeong J.Q., Lee K.L., Choo S.H., Lim C.Y.Y., Nichane M. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14:237–252. doi: 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.M.M., Chen A., Koh P.W.W., Deng T.Z.Z., Sinha R., Tsai J.M.M., Barkal A.A.A., Shen K.Y.Y., Jain R., Morganti R.M.M. Mapping the pairwise choices leading from pluripotency to human bone, heart, and other mesoderm cell types. Cell. 2016;166:451–467. doi: 10.1016/j.cell.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano M., Date S., Shimokawa M., Takano A., Fujii M., Ohta Y., Watanabe T., Kanai T., Sato T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015;21:256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Yamanaka Y., Uemura M., Osawa M., Saito M.K., Nagahashi A., Nishio M., Guo L., Ikegawa S., Sakurai S. Recapitulating the human segmentation clock with pluripotent stem cells. Nature. 2020;580:124–129. doi: 10.1038/s41586-020-2144-9. [DOI] [PubMed] [Google Scholar]
- Maxwell K.G., Augsornworawat P., Velazco-Cruz L., Kim M.H., Asada R., Hogrebe N.J., Morikawa S., Urano F., Millman J.R. Gene-edited human stem cell-derived β cells from a patient with monogenic diabetes reverse preexisting diabetes in mice. Sci. Transl. Med. 2020;12:eaax9106. doi: 10.1126/scitranslmed.aax9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley K.B., Hawkins F., Serra M., Thomas D.C., Jacob A., Kotton D.N. Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of Wnt signaling. Cell Stem Cell. 2017;20:844–857.e6. doi: 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken K.W., Catá E.M., Crawford C.M., Sinagoga K.L., Schumacher M., Rockich B.E., Tsai Y.H., Mayhew C.N., Spence J.R., Zavros Y. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature. 2014;516:400–404. doi: 10.1038/nature13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken K.W., Aihara E., Martin B., Crawford C.M., Broda T., Treguier J., Zhang X., Shannon J.M., Montrose M.H., Wells J.M. Wnt/β-catenin promotes gastric fundus specification in mice and humans. Nature. 2017;541:182–187. doi: 10.1038/nature21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley K.L., Castillo-Azofeifa D., Klein O.D. Tools and concepts for interrogating and defining cellular identity. Cell Stem Cell. 2020;26:632–656. doi: 10.1016/j.stem.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels B.E., Mosa M.H., Streibl B.I., Zhan T., Menche C., Abou-El-Ardat K., Darvishi T., Członka E., Wagner S., Winter J. Pooled in vitro and in vivo CRISPR-cas9 screening identifies tumor suppressors in human colon organoids. Cell Stem Cell. 2020;26:782–792.e7. doi: 10.1016/j.stem.2020.04.003. [DOI] [PubMed] [Google Scholar]
- Mikkola M.L. Genetic basis of skin appendage development. Semin. Cell Dev. Biol. 2007;18:225–236. doi: 10.1016/j.semcdb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Mills R.J., Titmarsh D.M., Koenig X., Parker B.L., Ryall J.G., Quaife-Ryan G.A., Voges H.K., Hodson M.P., Ferguson C., Drowley L. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci.U S A. 2017;114:E8372–E8381. doi: 10.1073/pnas.1707316114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro A.N., Bouwman P., Kousholt A.N., Eccles D.M., Millot G.A., Masson J.-Y., Schmidt M.K., Sharan S.K., Scully R., Wiesmüller L. Variants of uncertain clinical significance in hereditary breast and ovarian cancer genes: best practices in functional analysis for clinical annotation. J. Med. Genet. 2020;57:509–518. doi: 10.1136/jmedgenet-2019-106368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesano R., Schaller G., Orci L. Induction of epithelial tubular morphogenesis in vitro by fibroblast-derived soluble factors. Cell. 1991;66:697–711. doi: 10.1016/0092-8674(91)90115-f. [DOI] [PubMed] [Google Scholar]
- Moris N., Anlas K., van den Brink S.C., Alemany A., Schröder J., Ghimire S., Balayo T., van Oudenaarden A., Martinez Arias A. An in vitro model of early anteroposterior organization during human development. Nature. 2020;582:410–415. doi: 10.1038/s41586-020-2383-9. [DOI] [PubMed] [Google Scholar]
- Morizane R., Lam A.Q., Freedman B.S., Kishi S., Valerius M.T., Bonventre J.V. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 2015;33:1193–1200. doi: 10.1038/nbt.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummery C.L., Zhang J., Ng E.S., Elliott D.A., Elefanty A.G., Kamp T.J. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes. Circ. Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Múnera J.O., Sundaram N., Rankin S.A., Hill D., Watson C., Mahe M., Vallance J.E., Shroyer N.F., Sinagoga K.L., Zarzoso-Lacoste A. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell. 2017;21:51–64.e6. doi: 10.1016/j.stem.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry C.E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Myllymäki S.M., Mikkola M.L. Inductive signals in branching morphogenesis – lessons from mammary and salivary glands. Curr. Opin. Cell Biol. 2019;61:72–78. doi: 10.1016/j.ceb.2019.07.001. [DOI] [PubMed] [Google Scholar]
- Nakao K., Morita R., Saji Y., Ishida K., Tomita Y., Ogawa M., Saitoh M., Tomooka Y., Tsuji T. The development of a bioengineered organ germ method. Nat. Methods. 2007;4:227–230. doi: 10.1038/nmeth1012. [DOI] [PubMed] [Google Scholar]
- Nijman S.M.B. Synthetic lethality: general principles, utility and detection using genetic screens in human cells. FEBS Lett. 2011;585:1–6. doi: 10.1016/j.febslet.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R., Tsai C.J., Jang H. Why are some driver mutations rare? Trends Pharmacol. Sci. 2019;40:919–929. doi: 10.1016/j.tips.2019.10.003. [DOI] [PubMed] [Google Scholar]
- O’Rourke K.P., Loizou E., Livshits G., Schatoff E.M., Baslan T., Manchado E., Simon J., Romesser P.B., Leach B., Han T. Transplantation of engineered organoids enables rapid generation of metastatic mouse models of colorectal cancer. Nat. Biotechnol. 2017;35:577–582. doi: 10.1038/nbt.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K., Caron L., Borowiak M., Martinez R.J., Fitz-Gerald C.S., Sato Y., Cowan C.A., Chien K.R., Melton D.A. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- Pagliuca F.W., Millman J.R., Gürtler M., Segel M., Van Dervort A., Ryu J.H., Peterson Q.P., Greiner D., Melton D.A. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159:428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent A.V., Russ H.A., Khan I.S., LaFlam T.N., Metzger T.C., Anderson M.S., Hebrok M. Generation of functional thymic epithelium from human embryonic stem cells that supports host T cell development. Cell Stem Cell. 2013;13:219–229. doi: 10.1016/j.stem.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson S., Sroczynska P., Lacaud G., Kouskoff V. The stepwise specification of embryonic stem cells to hematopoietic fate is driven by sequential exposure to Bmp4, activin A, bFGF and VEGF. Development. 2008;135:1525–1535. doi: 10.1242/dev.011767. [DOI] [PubMed] [Google Scholar]
- Pellegrini L., Bonfio C., Chadwick J., Begum F., Skehel M., Lancaster M.A. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 2020;5626:1–21. doi: 10.1126/science.aaz5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N., Pitsouli C., Shilo B.Z. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb. Perspect. Biol. 2012;4:a005975. doi: 10.1101/cshperspect.a005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Paz L., Sun T., Pikiolek M., Cochran N.R., Bergling S., Orsini V., Yang Z., Sigoillot F., Jetzer J., Syed M. YAP, but not RSPO-LGR4/5, signaling in biliary epithelial cells promotes a ductular reaction in response to liver injury. Cell Stem Cell. 2019;25:39–53.e10. doi: 10.1016/j.stem.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen A.A., Bhaduri A., Andrews M.G., Nowakowski T.J., Meyerson O.S., Mostajo-Radji M.A., Di Lullo E., Alvarado B., Bedolli M., Dougherty M.L. Establishing cerebral organoids as models of human-specific brain evolution. Cell. 2019;176:743–756.e17. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai E., Schell J.P., Janiszewski A., Rovic I., Murray A., Bradshaw B., Pardon T., El Bakkali M., Talon I., De Geest N. Defining totipotency using criteria of increasing stringency. bioRxiv. 2020 doi: 10.1101/2020.03.02.972893. [DOI] [PubMed] [Google Scholar]
- Post Y., Puschhof J., Beumer J., Kerkkamp H.M., de Bakker M.A.G., Slagboom J., de Barbanson B., Wevers N.R., Spijkers X.M., Olivier T. Snake Venom Gland Organoids. Cell. 2020;180:233–247.e21. doi: 10.1016/j.cell.2019.11.038. [DOI] [PubMed] [Google Scholar]
- Pourquie O. The segmentation clock: converting embryonic time into spatial pattern. Science. 2003;301:328–330. doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- Qian X., Song H., Ming G.L. Brain organoids: advances, applications and challenges. Development. 2019;146:dev166074. doi: 10.1242/dev.166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y., Han B., Gao B., Bose S., Gong Y., Wawrowsky K., Giuliano A.E., Sareen D., Cui X. Differentiation of human induced pluripotent stem cells to mammary-like organoids. Stem Cell Reports. 2017;8:205–215. doi: 10.1016/j.stemcr.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Min Yang S., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L. ClinGen--the clinical genome resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel T., Frey N., Ringnalda F., Janjuha S., Cherkaoui S., Butz S., Srivatsa S., Pirkl M., Russo G., Villiger L. Genome-scale CRISPR screening in human intestinal organoids identifies drivers of TGF-β resistance. Cell Stem Cell. 2020;26:431–440.e8. doi: 10.1016/j.stem.2020.02.007. [DOI] [PubMed] [Google Scholar]
- Rivron N., Pera M., Rossant J., Martinez Arias A., Zernicka-Goetz M., Fu J., van den Brink S., Bredenoord A., Dondorp W., de Wert G. Debate ethics of embryo models from stem cells. Nature. 2018;564:183–185. doi: 10.1038/d41586-018-07663-9. [DOI] [PubMed] [Google Scholar]
- Rivron N.C., Frias-Aldeguer J., Vrij E.J., Boisset J.C., Korving J., Vivié J., Truckenmüller R.K., Van Oudenaarden A., Van Blitterswijk C.A., Geijsen N. Blastocyst-like structures generated solely from stem cells. Nature. 2018;557:106–111. doi: 10.1038/s41586-018-0051-0. [DOI] [PubMed] [Google Scholar]
- Roccio M., Edge A.S.B. Inner ear organoids: new tools to understand neurosensory cell development, degeneration and regeneration. Development. 2019;146:dev177188. doi: 10.1242/dev.177188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper J., Tammela T., Cetinbas N.M., Akkad A., Roghanian A., Rickelt S., Almeqdadi M., Wu K., Oberli M.A., Sánchez-Rivera F. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 2017;35:569–576. doi: 10.1038/nbt.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen N.S., Anandakumaran P.N., zur Nieden R., Lo K., Luo W., Park C., Huyan C., Fu Q., Song Z., Singh-Moon R.P. Injectable therapeutic organoids using sacrificial hydrogels. iScience. 2020;23:101052. doi: 10.1016/j.isci.2020.101052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G., Manfrin A., Lutolf M.P. Progress and potential in organoid research. Nat. Rev. Genet. 2018;19:671–687. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- Sachs N., de Ligt J., Kopper O., Gogola E., Bounova G., Weeber F., Balgobind A.V., Wind K., Gracanin A., Begthel H. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386.e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Sasai Y., Eiraku M., Suga H. In vitro organogenesis in three dimensions: self-organising stem cells. Development. 2012;139:4111–4121. doi: 10.1242/dev.079590. [DOI] [PubMed] [Google Scholar]
- Sasselli V., Blokzijl F., Drost J., Nik-Zainal S., Sasaki N., Clevers H., van Wezel T., Cuppen E., Behjati S., de Ligt J. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017;358:234–238. doi: 10.1126/science.aao3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G., Koo B.-K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Serra M., Alysandratos K.-D., Hawkins F., McCauley K.B., Jacob A., Choi J., Caballero I.S., Vedaie M., Kurmann A.A., Ikonomou L. Pluripotent stem cell differentiation reveals distinct developmental pathways regulating lung- versus thyroid-lineage specification. Development. 2017;144:3879–3893. doi: 10.1242/dev.150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazi M.N., Zernicka-Goetz M. Deconstructing and reconstructing the mouse and human early embryo. Nat. Cell Biol. 2018;20:878–887. doi: 10.1038/s41556-018-0144-x. [DOI] [PubMed] [Google Scholar]
- Shahbazi M.N., Siggia E.D., Zernicka-Goetz M. Self-organization of stem cells into embryos: a window on early mammalian development. Science. 2019;364:948–951. doi: 10.1126/science.aax0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahjalal H.M., Abdal Dayem A., Lim K.M., Jeon T., Cho S.-G. Generation of pancreatic β cells for treatment of diabetes: advances and challenges. Stem Cell Res. Ther. 2018;9:355. doi: 10.1186/s13287-018-1099-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Taniguchi K., Townshend R.F., Miki T., Gumucio D.L., Fu J. A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat. Commun. 2017;8:1–15. doi: 10.1038/s41467-017-00236-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Taniguchi K., Gurdziel K., Townshend R.F., Xue X., Yong K.M.A., Sang J., Spence J.R., Gumucio D.L., Fu J. Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat. Mater. 2017;16:419–427. doi: 10.1038/nmat4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Sances S., Workman M.J., Svendsen C.N. Multi-lineage human iPSC-derived platforms for disease modeling and drug discovery. Cell Stem Cell. 2020;26:309–329. doi: 10.1016/j.stem.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmin S., Taguchi A., Kaku Y., Yoshimura Y., Ohmori T., Sakuma T., Mukoyama M., Yamamoto T., Kurihara H., Nishinakamura R. Human induced pluripotent stem cell-derived podocytes mature into vascularized glomeruli upon experimental transplantation. J. Am. Soc. Nephrol. 2016;27:1778–1791. doi: 10.1681/ASN.2015010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Sun L., Wang M., Liu J., Zhong S., Li R., Li P., Guo L., Fang A., Chen R. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 2020;18:e3000705. doi: 10.1371/journal.pbio.3000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai H., Mandai M., Matsushita K., Kuwahara A., Yonemura S., Nakano T., Assawachananont J., Kimura T., Saito K., Terasaki H. Transplantation of human embryonic stem cell-derived retinal tissue in two primate models of retinal degeneration. Proc. Natl. Acad. Sci. U S A. 2016;113:E81–E90. doi: 10.1073/pnas.1512590113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A.A.C., Matthys O.B., Joy D.A., Kauss M.A., Natarajan V., Lai M.H. Developmental co-emergence of cardiac and gut tissues modeled by human iPSC-derived organoids. bioRxiv. 2020 doi: 10.1101/2020.04.30.071472. [DOI] [PubMed] [Google Scholar]
- Simunovic M., Brivanlou A.H. Embryoids, organoids and gastruloids: new approaches to understanding embryogenesis. Development. 2017;144:976–985. doi: 10.1242/dev.143529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinagoga K.L., McCauley H.A., Múnera J.O., Reynolds N.A., Enriquez J.R., Watson C., Yang H.-C., Helmrath M.A., Wells J.M. Deriving functional human enteroendocrine cells from pluripotent stem cells. Development. 2018;145:dev165795. doi: 10.1242/dev.165795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozen B., Amadei G., Cox A., Wang R., Na E., Czukiewska S., Chappell L., Voet T., Michel G., Jing N. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 2018;20:979–989. doi: 10.1038/s41556-018-0147-7. [DOI] [PubMed] [Google Scholar]
- Sozen B., Cox A.L., De Jonghe J., Bao M., Hollfelder F., Glover D.M., Zernicka-Goetz M. Self-organization of mouse stem cells into an extended potential blastoid. Dev. Cell. 2019;51:698–712.e8. doi: 10.1016/j.devcel.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita L.M., Ahituv N., Dunham M.J., Kitzman J.O., Roth F.P., Seelig G., Shendure J., Fowler D.M. Variant interpretation: functional assays to the rescue. Am. J. Hum. Genet. 2017;101:315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano A., Tuck E., Stubbs V.E., Livesey F.J. Variable outcomes in neural differentiation of human PSCs arise from intrinsic differences in developmental signaling pathways. Cell Rep. 2020;31:107732. doi: 10.1016/j.celrep.2020.107732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M., Hu R., Jin J., Yan Y., Song Y., Sullivan R., Lai L. Efficient in vitro generation of functional thymic epithelial progenitors from human embryonic stem cells. Sci. Rep. 2015;5:9882. doi: 10.1038/srep09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H., Kadoshima T., Minaguchi M., Ohgushi M., Soen M., Nakano T., Takata N., Wataya T., Muguruma K., Miyoshi H. Self-formation of functional adenohypophysis in three-dimensional culture. Nature. 2011;480:57–62. doi: 10.1038/nature10637. [DOI] [PubMed] [Google Scholar]
- Sun X., Xu J., Lu H., Liu W., Miao Z., Sui X., Liu H., Su L., Du W., He Q. Directed differentiation of human embryonic stem cells into thymic epithelial progenitor-like cells reconstitutes the thymic microenvironment in vivo. Cell Stem Cell. 2013;13:230–236. doi: 10.1016/j.stem.2013.06.014. [DOI] [PubMed] [Google Scholar]
- Taguchi A., Nishinakamura R. Higher-order kidney organogenesis from pluripotent stem cells. Cell Stem Cell. 2017;21:730–746.e6. doi: 10.1016/j.stem.2017.10.011. [DOI] [PubMed] [Google Scholar]
- Taguchi A., Kaku Y., Ohmori T., Sharmin S., Ogawa M., Sasaki H., Nishinakamura R. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14:53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Takasato M., Er P.X., Becroft M., Vanslambrouck J.M., Stanley E.G., Elefanty A.G., Little M.H. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat. Cell Biol. 2014;16:118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- Takasato M., Er P.X., Chiu H.S., Maier B., Baillie G.J., Ferguson C., Parton R.G., Wolvetang E.J., Roost M.S., Chuva de Sousa Lopes S.M. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.-R., Ueno Y., Zheng Y.-W., Koike N. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Takebe T., Enomura M., Yoshizawa E., Kimura M., Koike H., Ueno Y., Matsuzaki T., Yamazaki T., Toyohara T., Osafune K. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell. 2015;16:556–565. doi: 10.1016/j.stem.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Kimura M., Yoshizawa E., Ayano S., Koido M., Funayama S., Nakanishi N., Hisai T., Kobayashi T. Massive and reproducible production of liver buds entirely from human pluripotent stem cells. Cell Rep. 2017;21:2661–2670. doi: 10.1016/j.celrep.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Takeda H., Kataoka S., Nakayama M., Ali M.A.E., Oshima H., Yamamoto D., Park J.-W., Takegami Y., An T., Jenkins N.A. CRISPR-Cas9–mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes. Proc. Natl. Acad. Sci. U S A. 2019;116:15635–15644. doi: 10.1073/pnas.1904714116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambuyzer E., Vandendriessche B., Austin C.P., Brooks P.J., Larsson K., Miller Needleman K.I., Valentine J., Davies K., Groft S.C., Preti R. Therapies for rare diseases: therapeutic modalities, progress and challenges ahead. Nat. Rev. Drug Discov. 2020;19:93–111. doi: 10.1038/s41573-019-0049-9. [DOI] [PubMed] [Google Scholar]
- Tang P.-C., Alex A.L., Nie J., Lee J., Roth A.A., Booth K.T., Koehler K.R., Hashino E., Nelson R.F. Defective tmprss3-associated hair cell degeneration in inner ear organoids. Stem Cell Reports. 2019;13:147–162. doi: 10.1016/j.stemcr.2019.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchieu J., Zimmer B., Fattahi F., Amin S., Zeltner N., Chen S., Studer L. A modular platform for differentiation of human PSCs into all major ectodermal lineages. Cell Stem Cell. 2017;21:399–410.e7. doi: 10.1016/j.stem.2017.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivanen R., Shen M.M. Prostate organogenesis: tissue induction, hormonal regulation and cell type specification. Development. 2017;144:1382–1398. doi: 10.1242/dev.148270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trisno S.L., Philo K.E.D., McCracken K.W., Catá E.M., Ruiz-Torres S., Rankin S.A., Han L., Nasr T., Chaturvedi P., Rothenberg M.E. Esophageal organoids from human pluripotent stem cells delineate Sox2 functions during esophageal specification. Cell Stem Cell. 2018;23:501–515.e7. doi: 10.1016/j.stem.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-H., Nattiv R., Dedhia P.H., Nagy M.S., Chin A.M., Thomson M., Klein O.D., Spence J.R. In vitro patterning of pluripotent stem cell-derived intestine recapitulates in vivo human development. Development. 2017;144:1045–1055. doi: 10.1242/dev.138453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto H., Kasahara T., Sueta S.ichi, Araoka T., Sakamoto S., Okada C., Mae S.ichi, Nakajima T., Okamoto N., Taura D. A modular differentiation system maps multiple human kidney lineages from pluripotent stem cells. Cell Rep. 2020;31:107476. doi: 10.1016/j.celrep.2020.03.040. [DOI] [PubMed] [Google Scholar]
- van Den Brink S.C., Alemany A., van Batenburg V., Moris N., Blotenburg M., Vivié J., Baillie-Johnson P., Nichols J., Sonnen K.F., Martinez Arias A. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature. 2020;582:405–409. doi: 10.1038/s41586-020-2024-3. [DOI] [PubMed] [Google Scholar]
- van Den Brink S.C., Baillie-Johnson P., Balayo T., Hadjantonakis A.K., Nowotschin S., Turner D.A., Arias A.M. Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development. 2014;141:4231–4242. doi: 10.1242/dev.113001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C.W., Ritsma L., Avramut M.C., Wiersma L.E., van den Berg B.M., Leuning D.G., Lievers E., Koning M., Vanslambrouck J.M., Koster A.J. Renal subcapsular transplantation of PSC-derived kidney organoids induces neo-vasculogenesis and significant glomerular and tubular maturation in vivo. Stem Cell Reports. 2018;10:751–765. doi: 10.1016/j.stemcr.2018.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenvliet J.V., Bolondi A., Kretzmer H., Haut L., Scholze-Wittler M., Schifferl D., Koch F., Pustet M., Heimann S., Buschow R. Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. bioRxiv. 2020 doi: 10.1101/2020.03.04.974949. [DOI] [PubMed] [Google Scholar]
- Veres A., Faust A.L., Bushnell H.L., Engquist E.N., Kenty J.H.-R., Harb G., Poh Y.-C., Sintov E., Gürtler M., Pagliuca F.W. Charting cellular identity during human in vitro β-cell differentiation. Nature. 2019;569:368–373. doi: 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader J.E., Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28:1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Kinzler K.W. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges H.K., Mills R.J., Elliott D.A., Parton R.G., Porrello E.R., Hudson J.E. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development. 2017;144:1118–1127. doi: 10.1242/dev.143966. [DOI] [PubMed] [Google Scholar]
- Völkner M., Zschätzsch M., Rostovskaya M., Overall R.W., Busskamp V., Anastassiadis K., Karl M.O. Retinal organoids from pluripotent stem cells efficiently recapitulate retinogenesis. Stem Cell Reports. 2016;6:525–538. doi: 10.1016/j.stemcr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmflash A., Sorre B., Etoc F., Siggia E.D., Brivanlou A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 2014;11:847–854. doi: 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C.L., Mahe M.M., Múnera J., Howell J.C., Sundaram N., Poling H.M., Schweitzer J.I., Vallance J.E., Mayhew C.N., Sun Y. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer R.A., Leopoldi A., Aichinger M., Wick N., Hantusch B., Novatchkova M., Taubenschmid J., Hämmerle M., Esk C., Bagley J.A. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565:505–510. doi: 10.1038/s41586-018-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.P., Bear C.E., Chin S., Pasceri P., Thompson T.O., Huan L.-J., Ratjen F., Ellis J., Rossant J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012;30:876–882. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman M.J., Mahe M.M., Trisno S., Poling H.M., Watson C.L., Sundaram N., Chang C.-F., Schiesser J., Aubert P., Stanley E.G. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 2017;23:49–59. doi: 10.1038/nm.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Wu D., Ren Y., Huang Y., Feng B., Zhao N., Zhang T., Chen X., Chen S., Xu A. Generation of hepatobiliary organoids from human induced pluripotent stem cells. J. Hepatol. 2019;70:1145–1158. doi: 10.1016/j.jhep.2018.12.028. [DOI] [PubMed] [Google Scholar]
- Wu Q., Liu J., Wang X., Feng L., Wu J., Zhu X., Wen W., Gong X. Organ-on-a-chip: recent breakthroughs and future prospects. Biomed. Eng. Online. 2020;19:9. doi: 10.1186/s12938-020-0752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Tanaka Y., Patterson B., Kang Y.-J., Govindaiah G., Roselaar N., Cakir B., Kim K.-Y., Lombroso A.P., Hwang S.-M. Fusion of regionally specified hPSC-derived organoids models human brain development and interneuron migration. Cell Stem Cell. 2017;21:383–398.e7. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Tanaka Y., Cakir B., Patterson B., Kim K.-Y., Sun P., Kang Y.-J., Zhong M., Liu X., Patra P. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell. 2019;24:487–497.e7. doi: 10.1016/j.stem.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.H.N., Siu H.C., Law S., Ho S.L., Yue S.S.K., Tsui W.Y., Chan D., Chan A.S., Ma S., Lam K.O. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 2018;23:882–897.e11. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- Yang L., Han Y., Nilsson-Payant B.E., Gupta V., Wang P., Duan X., Tang X., Zhu J., Zhao Z., Jaffré F. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Mich J.K., Ku S., Menon V., Krostag A.-R., Martinez R.A., Furchtgott L., Mulholland H., Bort S., Fuqua M.A. A single-cell roadmap of lineage bifurcation in human ESC models of embryonic brain development. Cell Stem Cell. 2017;20:120–134. doi: 10.1016/j.stem.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Li J., Tan Z., Wang C., Liu T., Chen L., Yong J., Jiang W., Sun X., Du L. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood. 2008;111:1933–1941. doi: 10.1182/blood-2007-02-074120. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Jiang J., Han P., Yuan Q., Zhang J., Zhang X., Xu Y., Cao H., Meng Q., Chen L. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang Y., Jiang M., Huang S.X., Zhang W., Al Alam D., Danopoulos S., Mori M., Chen Y.W., Balasubramanian R. 3D modeling of esophageal development using human PSC-derived basal progenitors reveals a critical role for notch signaling. Cell Stem Cell. 2018;23:516–529.e5. doi: 10.1016/j.stem.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Xue X., Shao Y., Wang S., Esfahani S.N., Li Z., Muncie J.M., Lakins J.N., Weaver V.M., Gumucio D.L. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019;573:421–425. doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]