Abstract
Process analytical technology (PAT) for the manufacture of monoclonal antibodies (mAbs) is defined by an integrated set of advanced and automated methods that analyze the compositions and biophysical properties of cell culture fluids, cell-free product streams, and biotherapeutic molecules that are ultimately formulated into concentrated products. In-line or near-line probes and systems are remarkably well developed, although challenges remain in the determination of the absence of viral loads, detecting microbial or mycoplasma contamination, and applying data-driven deep learning to process monitoring and soft sensors. In this review, we address the current status of PAT for both batch and continuous processing steps and discuss its potential impact on facilitating the continuous manufacture of biotherapeutics.
Keywords: continuous manufacturing, monoclonal antibodies, process analytical technology, sensors, spectroscopy, spectrometry
Monoclonal Antibodies
mAbs have evolved from being scientific tools derived from murine hybridomas in 1975 to biotherapeutic molecules based on humanized antibodies (see Glossary). The first mAb for therapeutic use in humans was approved in 1986 and the first bispecific mAb (bsAb; catumaxomab) was approved in 2009 [1]. Humanized antibodies include IgG1s, 2s, and 4s grafted onto the Fc and Fv regions, which comprise human sequences. Currently, there are ~570 antibody therapeutics at various clinical phases, with 62 in late-stage trials [2]. Global mAb sales have grown from US $18.5 billion in 2010 [1] to US $98 billion in 2017 with 57 mAbs and 11 biosimilars in clinical use as of the end of 2017’ [3]. Of these, 93% are produced in USA and Europe and half are based on fully human genetic sequences [3].
Over time, several classes of mAb have evolved [1]. Early products (Erbitux, Remicade, and rituximab) were obtained by grafting antigen-specific variable domains of mouse antibodies onto constant domains of a human antibody. Humanized mAbs (e.g., Avastin, Mylotarg, and Herceptin), based on a murine hypervariable region grafted onto a human antibody framework, resulted in decreased immunogenic properties and reduced formation of antidrug antibodies. Ultimately, human mAbs emerged from research that utilized phage display technology and transgenic mouse strains that express human variable domains (i.e., Humira, Simponi, and Yervoy).
Approximately 20 bsAbs for non-oncology indications have entered various stages of testing since 2000 [4,5]. Candidates have the potential to attack Pseudomonas aeruginosa, treat type 2 diabetes mellitus, or provide postexposure protection against Ebola viruses. Approximately 500 fully human antibodies have been identified in the blood of a survivor of coronavirus disease 2019 (COVID-19), and are being assessed for effectiveness against COVID-19 with the goal of rapidly developing a therapeutic antibodyi. Both Biogen and GlaxoSmithKline have separately partnered with Vir to produce mAbs found capable of binding to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)ii.
Administration of biologics at concentrations of 150 mg/ml or higher in formulated matrices suitable for injection [6] opens the possibility of subcutaneous home self-administration as the number of biotherapeutic mAbs increases. Dosages range from 50 to 1000 mg per patient per treatment. The patient’s need for the therapeutic may last from months to years, and some mAbs or biosimilars will apply to a large population of patients. These combined requirements will necessitate total global production of metric ton quantities annually [4,7]. PAT will be an important component of achieving enhanced productivity.
PAT is a framework for ensuring the quality of a pharmaceutical product by monitoring process streams and unit operations, thereby providing a real-time understanding of the manufacturing process. Determining the sources of variability in a process, how the variability is managed by the process, and whether the product quality may be predicted from process parameters is central to PAT. This knowledge is used to decide which material and process attributes need to be measured and controlled during manufacturing. Implementation of PAT tools (e.g., multivariate analytics, process analyzers, process controllers, and continuous improvement tools) for these critical attributes helps to ensure product quality. The FDA introduced the PAT framework in their Guidance to the Industry document in 2004iii , iv.
Since its introduction, the PAT framework has been implemented for the development of small-molecule active pharmaceutical ingredients (APIs) to improve the understanding of process chemistry, as highlighted by the International Consortium for Innovation and Quality in Pharmaceutical Development (IQ Consortium) [8]. Implementation of PAT for the development and manufacture of mAbs is now gaining momentum with pilot-scale demonstrations of multiattribute monitoring and potential for process control [9]. The need for reliable scalable manufacturing of mAbs (and other biologics) continues to increase (e.g., antibody-based therapies for the COVID-19 pandemic). In this review, we highlight the current status of mAb manufacturing, associated challenges, and how PAT and data analytics can help overcome these challenges to develop a new therapeutic product.
Manufacture of mAbs: Batch and Continuous Processing
Chinese hamster ovary (CHO) cells, introduced over 30 years ago and widely used in batch processes, have become a major cell line for the commercial production of mAbs in submerged culture using bioreactors with volumes of up to 15 000 l. CHO cells produce 60% of all mAbs produced, with myeloma cells producing the remainder [3].
Improvements in cell lines used in batch culture and development of end-to-end continuous manufacturing of mAbs (Figure 1A,E) has been proposed as a way to increase productivity, decrease equipment footprint, and control cost [10., 11., 12., 13., 14.]. PAT is key to tracking the health, productivity, and titer of large-scale CHO cell cultures over a period of months in a manufacturing environment and is welldeveloped for monitoring the recovery and purification of the mAb product regardless of whether batch or continuous production is used [15].
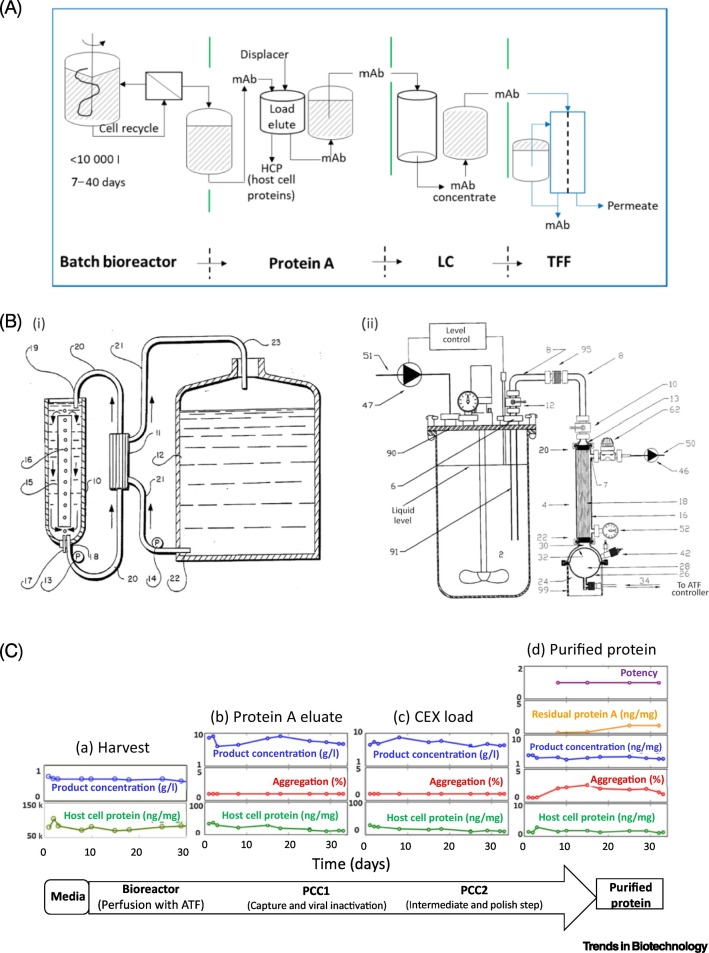
Figure 1.
Unit Operations for the Manufacture of mAbs.
(A) Schematic of batch process sequence for monoclonal antibody (mAb) manufacture; (B) perfusion culture system: (i) reactor on left side of drawing; (ii) membrane-based separation system on right side of drawing; (for details of numbers, see [16]); (C) downstream processing sequence for recovery and purification of mAb; (D) product capacities of mAb processes; (E) comparison with a continuous process; (F) tangential flow filtration (TFF) in a continuous process configuration. For both chromatographic separations (E,F), TFF batch processes operate continuously by cycling discrete unit operations between service and regeneration steps. Reproduced, with permission, from [16] (A), [16] (B), [17] (C), and [7] (D). Abbreviations: CEX, cation exchange chromatography; DF, diafiltration; LC, liquid chromatography; UF, ultrafiltration.
Once an expression system is selected, production is carried out in a sequence of cell culture, recovery, and purification (Figure 1A). Cell culture entails that batch processes that are carried out in specially designed glass or stainless-steel bioreactors or, alternately, in single-use systems (see Box 1 for the evolution of single-use bioreactors). Perfusion culture, first patented in 1989 [16] (Figure 1B), is semicontinuous, with media being replenished at the same rate that the bioreactor fluid is separated from cells. The cells return to the bioreactor (Figure 1B) and the mAb product is subsequently harvested, recovered, and purified (Figure 1C), with detailsof the overall sequence given in Figure 1D [17]. As summarized in Box 1, Kelly correctly anticipated increases in production capacity and bioreactor volumes (Figure 1D [7]).
Box 1. Single-use bioreactors.
Single-use bioreactors are functionally equivalent to stainless steel reactors except the wetted surfaces are made of disposable materials that contact media, cell culture fluids, and process streams. These materials are constructed of multilayer films comprising a structural layer, a barrier layer, and a fluid contact layer. The polymeric films have the requisite inertness and sealing properties [64]. Single-use bioreactors consist of disposable filter capsules and presterilized containers resembling large plastic bags with volumes of up to 2000 l. Introduced to the industry ~35 years ago, single-use technology has evolved to include fittings, presterilized, prepackaged sensors, and plastic tubing connections comprising‘welded’ junctions, where tubing is spliced together and then bonded during set-up of a run. Sterility is achieved during the manufacture of single-use components by beta or gammairradiation.
Alt-text: Box 1
The potential impact of the combined application of biology with continuous manufacturing technology is particularly relevant for treating infectious diseases, such as COVID-19. As succinctly stated by Walker [18]: ‘An antibody drug could be developed far more quickly’ (than a small molecule drug)… ‘because the cure for the new coronavirus likely already exists in the blood of survivors.’ Once antibodies are identified and an expression system developed, their combination with continuous manufacturing of mAbs is arguably a key step in scaling availability to accelerate release of the final product for broad clinical use. In the meantime, blood plasma of survivors could serve a limited role for treating patients with COVID-19. Economic impacts could be large, given the US$2 trillion cost of COVID-19 to the US economy as of 10 April 2020.
Process Analytical Technology
PAT is a set of analytical techniques for which probes are in contact with the process flow and provide frequent and fully automated measurements in realtime for process control with minimal operator intervention [15]. These tools are required for maintaining the product quality and for helping to understand and identify critical process attributes. The FDA guidance document intends to encourage innovation in the development, manufacturing, and quality assurance of pharmaceuticalsiii , iv. Key elements of PAT fall into several basic categories, as illustrated for granulocyte colony-stimulating factor (GCSF) by Hebbi and colleagues [19]. PAT applies to: process sensors and process analyzers; process control tools (hardware and software); and, more broadly, utilizes multivariate design; data acquisition and analysis; and continuous improvement and knowledge management tools.
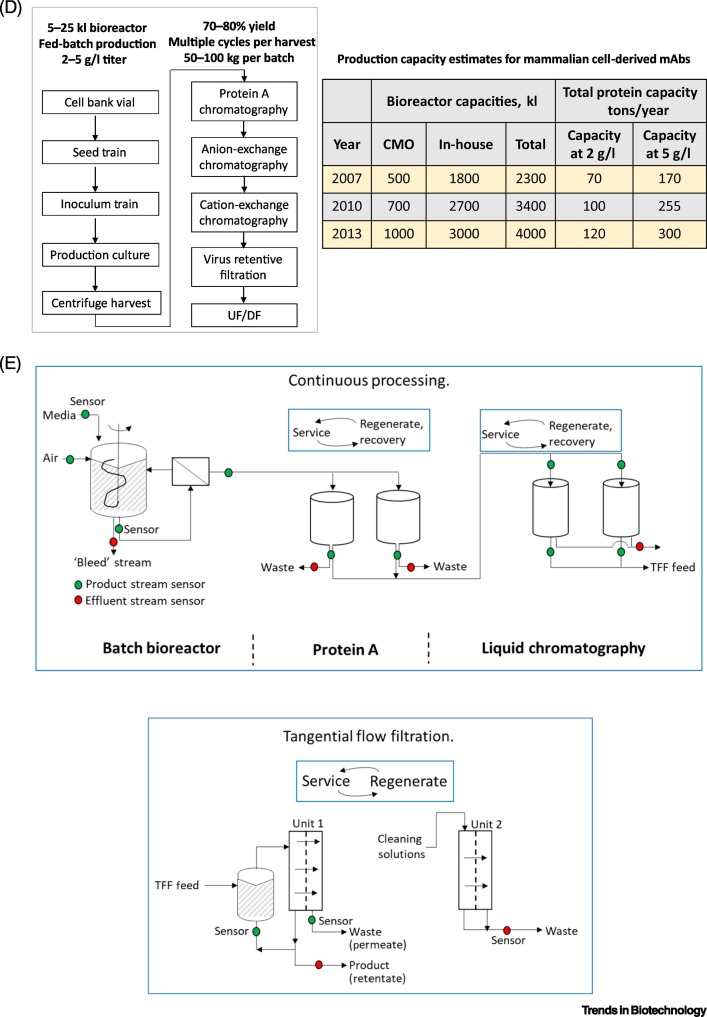
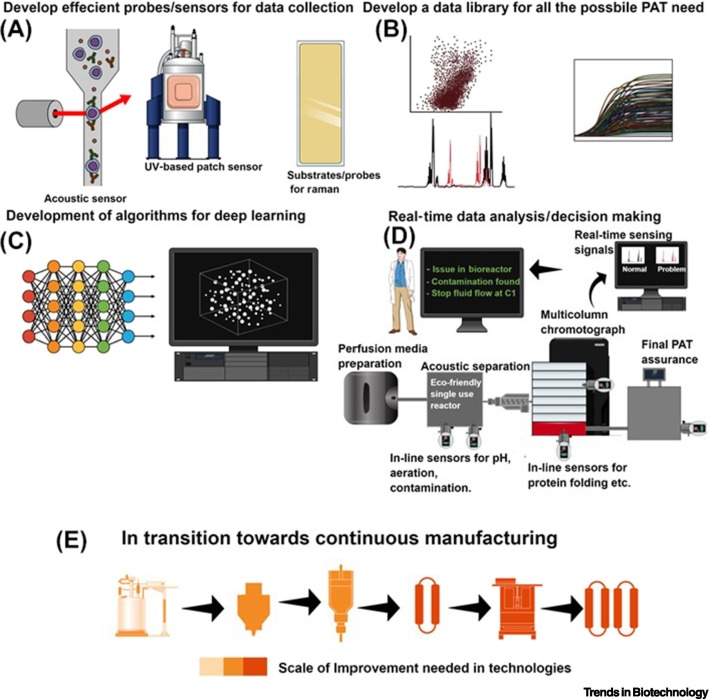
The upstream and downstream components of a mAb manufacturing sequence, together with identification of PAT most likely to be associated with the specific unit operations,are depicted in Figure 2 . Figure 2 summarizes key areas for continued advances in PAT, including efficient probes and sensors for data collection, comprehensive data libraries, algorithms for deep learning, and real-time data analysis for making real-time decisions about process operations. Combined with improvements in cell lines and unit operations throughout the entire sequence of manufacturing unit operations, PAT has the potential to have a catalytic role in accelerating the transition from batch to continuous manufacture of mAbs.
Figure 2.
Steps for Implementing Process Analytical Technology (PAT) for the Continuous Manufacture of Monoclonal Antibodies (mAbs).
(A) Sensor development; (B) data library based on model or historical process data; (C) development of deep learning algorithms; (D) real-time data analysis/decision-making for continuous processes; (E) scale of improvement needed in the transition of upstream and downstream unit operations in the transition from batch to continuous manufacturing. Figure created using Mind the Graph (www.mindthegraph.com).
Upstream Processing
Upstream processes for batch and single-use bioreactors have well-developed cell culture media and clonal cell lines [20,21]. Culture conditions that maintain appropriate cell density, stability, and integrity are well defined and readily monitored [22,23]. Viable cell count, cell metabolites, changes in media components, and mAb accumulation may be measured near-line by liquid chromatography-mass spectrometry (LC-MS), MS-MS, Coulter counters, osmometers, and rheometers within a timeframe of 30 min to several hours. In addition, chemometrics may be applied for chemical analyses combined with data analytics to help guide the analysis of the manufacturing processes.
Perfusion Culture
Perfusion processes operate for up to 40 days and retain cells in the bioreactor while product is removed and media is replenished, resulting in high cell densities (108/ml) within a small footprint. A fed-batch reactor run for 10–21 days results in 10× lower cell counts(i.e., 107 cells/ml). A perfusion reactor may be operated for a longer period of time since waste products, proteins, and spent media are continually removed in several ways, including a membrane separation system (Figure 1Bii). Run time in a fed-batch reactor is limited by the bioreactor volume available for the addition of nutrients after the run has started, as well as the concentration of nutrients needed for optimal cell growth.Batch and perfusion reactors have evolved to encompass single-use components, thereby helping to enable contract manufacture where products change from one run to the next.
Cell culture media required to reach high cell densities (such as Ham’s F10 or Dulbecco’s Modified Eagle Media) are free of animal blood serum supplements and are chemically defined [24]. One example is an off-the-shelf chemically defined medium for CHO cells (EX-CELL® Advanced™ HD Perfusion Medium Cell Vento CHO 100, and others)vi. A comparison of initial amino acid compositions by Reinhart and colleagues [19] for commercially available media summarized the changes in amino acid compositions that occur during fed-batch culture. Their comprehensive discussion of culture media indicated components that could be monitored by PAT, and also captured the complexity of cell culture fluids, resulting in the introduction of background that may interfere with online analysis of target molecules and that must be accounted for if PAT is to be utilized.
Clonal Selection (Productivity, Stability, Quality)
The whole-genome sequence for CHO cells, completed in 2011 [25], enabled more facile applications of gene-editing tools, including clustered regularly interspaced short palindromic repeats (CRISPR) coupled to the enzyme Cas9 [21]. Improved cell lines resulted from accurate annotations for the CHO genome, including introns, exons, transcription start sites, promoter regions, and noncoding RNAs [26]. Transgene integration improved productivity by achieving a high gene transcript for the target protein (e.g., mAb) [27].
Systems Biology and Process Sensor Requirements
PAT sensors (Table 1 ) monitor media conditions (pH and dissolved oxygen) and measure cellular processes, media supplementation, and extrinsic factors that influence protein expression [28]. In addition to genome editing during cell line research and development, process control of the quantity and quality of expressed proteins achieved by directing the metabolism of the cellsin a scalable system depends on measurements from in-line sensors or near-line analytical methods [20]. Since cells are suspended in a medium into which air is introduced, sensors for three phases are needed: air, liquid, and solid (Table 1) [29]. Temperature, shear force, conductivity, pH, pO2, pCO2, leachables, bioburden, secondary metabolites, and cell morphology, as well as cell viability, density, stability, and productivity [30], protein aggregation and secondary structure [21], osmolality, and ammonia are measurable, and measured, by in-line sensors or near-line analysis of culture fluids. However, determination of viral load [31] or mycoplasmamay require several days and, thus, is currently less amenable to online methods. We focus on in-line sensors in Table 1 because defining the state of a new process during development requires the use of as many attributes as possible to explain the sources of variability. When moving from research to production, the number of parameters to be monitored in-line may be reduced if a lower number of distinct parameters are shown to be sufficient to describe the state of the process. While some components in single-use reactors may be used only once and standard sensors for temperature, pH, and pO2 are now also disposable, many sensors are reused after cleaning and recalibration.
Table 1.
| Type of measurement | Sensor technique | Segment of installation | Mode of measurement |
|---|---|---|---|
| Physical parameters | |||
| Temperature | Thermometer | USP and DSP | In-line |
| Pressure | Thermocouple | USP and DSP | In-line |
| Redox potential | Membrane pressure sensors, redox (Pt) electrode | USP and DSP | In-line |
| Chemical parameters | |||
| pH | pH electrodes | USP and DSP | In-line |
| Dissolved gas | Amperometry electrodes, CO2 electrodes | Mostly USP | In-line |
| Gas phase | Paramagnetic CO2 sensors | Mostly USP | In-line |
| Dissolved compounds | Spectroscopic sensors (glucose, lactate) | USP and DSP | In-line (mostly in research) |
| Biological parameters | |||
| Biomass |
Spectroscopic sensors | USP and DSP | |
| Impedance sensors | Mostly USP | ||
| Cell morphology | Fluorescence sensors | USP | In-line (mostly in research) |
| Impedance sensors | USP | ||
| Viability | In situ microscopy | USP | |
| DNA/RNA/content | Spectroscopic sensors | USP and/or DSP | |
Downstream Processing
Downstream processes (Figure 1C,D)comprise cell harvesting, viral clearance, chromatography, buffer exchange, and ultrafiltration (UF) to obtain the final product. Unlike upstream processing with time constants of hours to days, residence times of product in downstream processes are measured in hours or less (not including hold tanks). Rapid analytical measurements are feasible [e.g., continuous monitoring of chromatography effluent by UV and refractive index detectors] [32,33]. Given the capital costs and operational expenses required to produce, recover, and purify the biologic molecule, maximal yields from recovery and purification processes are critical to process economics. The most likely approaches that will assist continuous monitoring will be based on optical methods (e.g., UV, conductivity, refractive index, and fluorescence) since these are minimally invasive, give rapid response, and probe known protein properties. In-line or near-line measurements of product purity are particularly important for the effective pooling of products [23,34]. However, absence of virus must be demonstrated, since detection of virus downstream results in the entire batch being discarded.
Product Recovery
Cell recovery (i.e., harvest) may occur simultaneously with cell culture in a perfusion reactor (Figure 1B) or sequentially after a batch culture is completed, using centrifugation or microfiltration to separate cells from media. Media that contain the protein product are further processed by chromatography and membrane-based concentration (Figure 1D,F).
Chromatography
Column-based protein-A affinity chromatography is used to capture the mAb products and remove host cell proteins (HCP) that flow through the columns (second step in Figure 1C) [35]. The protein A column comprises a ligand (native, recombinant, or alkalistabilized) immobilized to polymeric or agarose media, with particle sizes ranging from 45 to 125 μm. The load of mAb, buffer wash, and desorption steps are carried out in a co-current flow [32]. Binding capacities under flow conditions (i.e., dynamic capacity) are equivalent to 20–80 mg human IgG/mlresin at residence times of <10 min [12,35]. Protein A resins are costly (>US$10 000/l) and are estimated to account for >50% of separation costs; thus, these adsorbents are cleaned and reused for up to 200 cycles. Protein A is typically followed by ion exchange chromatography to remove components introduced with mobile phases, and to remove other macromolecules, including nucleic acids (Figure 1C,D) [35].
Viral Clearance
Removal of viruses from mAb product streams is essential to ensure the safety of the product [36]. The general requirement is that there are two orthogonal steps for viral clearance. These usually comprise inactivation (by low pH, detergent, or solvent treatment) and removal (by filtration). Four to five steps are evaluated for viral reduction, including protein A and other column chromatography steps where the mAb is bound and other components flow through (see Box 2 for difficulties in viral detection).
Box 2. Rapid Viral Detection: A Difficult Challenge.
Current methods for detecting viruses are based on PCR assays, animal infection susceptibility, immune responses, or cell culture infectivity assays [36,69]. Methods, such as reverse transcription loop-mediated isothermal amplification [70] and next-generation sequencing, hold the potential to decrease the amount of time required for detecting viruses while maintaining the same limits of detection as traditional methods (one viral copy per cell). Spectroscopic technologies (e.g., Raman spectroscopy) could be used in detecting viruses, and these are under development [71]. Thus, if a robust method for the online detection of virus were to be developed, it would be an important advance in the industry. This is a difficult challenge since detection would need to be at very low levels, detection of DNA does not speak to infectivity, and a measure of whether a virus is replication competent would be needed. However, the necessary methodologies and technologies still need to be developed.
Alt-text: Box 2
Tangential Flow Filtration
Injectable biologics are formulated with added excipients to stabilize the protein and control viscosity. UF/diafiltration (DF) by tangential flow filtration (TFF) is used to remove salts or other solubles introduced during the purification process and toreplace them with formulation matrix components to increase the concentration of the biologic. Achieving high concentrations requires the capability to handle diminishing volume with multiple units if continuous manufacture were to be carried out (Figure 1F) [12]. TFF through a 30-kDa membrane removes salt and water while retaining the mAb product (molecular weight of 140–160 kDa) [37] to achieve high protein concentrations and is inherently a batch process. TFF uses recirculation to obtain high protein titers. Typically, the last steps before storage entail viral filtration and often sterile filtration. Scale-up is achieved through modular addition of the membrane area [33]. PAT for at-line product analysis is based on an emerging technology for monitoring the concentration of proteins using a nanofluidic platform that enables size-based analysis of biologics to quantify protein concentration in about an hour [38].
Downstream Processing and Sensor Requirements
The specification of sensors for downstream processing depends on both the unit operations (i.e., chromatography vs. TFF) and the target protein. In chromatography, in-line sensors are designed to detect proteins and nucleic acids (UV absorbance and fluorescence), lipids and carbohydrates (refractive index), and salts (conductivity), and also to control flowrates and pressure (flow and pressure sensors, respectively) and mobile phase compositions [32]. Off-line analysis typically includes MS to identify structures and, in the case of proteins, molecular weights or detection of aggregates. Other analyses target detection of the presence of contaminants, including bacteria and mycoplasma [39,40]. Near-line methods may also include tests for pyrogens (derived from bacteria). While wellestablished, improvements in sensitivity, signal:noise ratios, and robustness of these methods are needed for PAT to be used in continuous monitoring of downstream processes. One possibility to achieve such improvement is the use of spectroscopic sensors [41]. A critical review of recent trends [41] and a future perspective of optical spectroscopy as found in PAT in biopharmaceutical downstream processing for simultaneous measurements of multiple attributes is described in the section ‘Spectroscopy and Sensors for PAT’.
Data Analytics
Data analytics promises future advances in our ability to design mAbs, their expression systems, and purification. The ability to know in near real time the cell culture conditions that are favorable for the regulation of specific genes that influence antibody product quality may have a significant impact on process control through soft sensors. Laboratory-based scale-down models for process development may enable direct testing of impacts of gene regulation to inform manufacturing processes [42]. However, massive data analysis is required to correlate and understand the interactions between cell culture environment and gene regulation leading to protein expression and protein processing.
An example is given by the CHO genome,which has 24 383 predicted genes. Only 11 099 of these have been confirmed as transcripts [25,43]. Of those 11 099 transcripts, only 6164 proteins have been confirmed [44]. To identify this relatively small number of proteins, 682 097 mass spectra were mapped to 93 548 unique peptides that were then overlaid onto the 6164 unique proteins.The data analysis challenges in handling this information were daunting. The authors state ‘One of the biggest challenges in proteomics is the data analysis, which can be a limiting step for protein identification’ [44]. It would appear that data analytics capabilities are beginning to catch up.
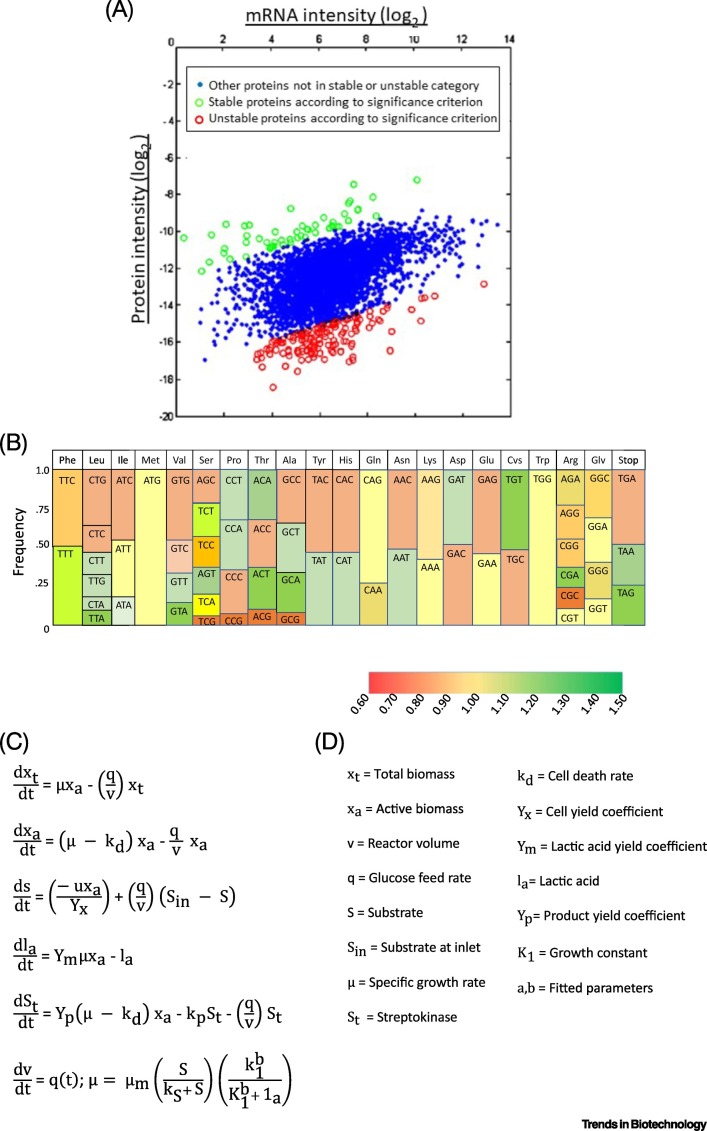
Following on from protein expression and post-translational modification control, the transcriptomic and proteomic data can be used to find highly transcribed regions of the genome. Codon usage for specific cell lines suitable for targeted gene insertion with higher frequency of transcription and more efficient translation through tRNAs may also occur (Figure 3A,B) that compare stable and unstable proteins and codon frequencies in CHO cells relative to human cells [44]. Data analytics and machine learning also have the potential for the design of better proteins and affinity ligands, thus impacting downstream processing and purification of mAb products.
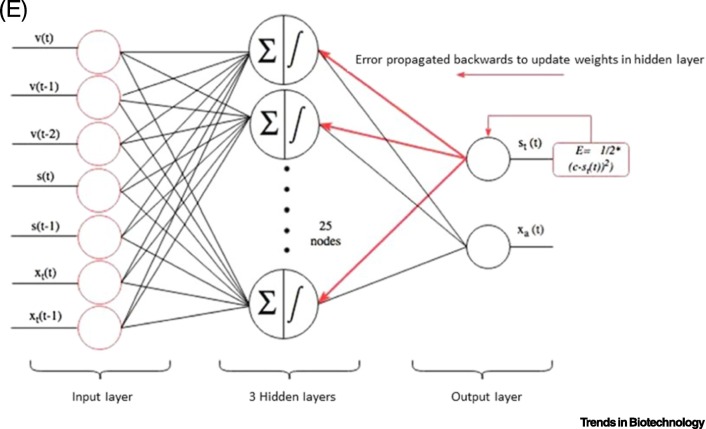
Figure 3.
Representation of Transcriptomic and Proteomic Data, Phenomenological Model and Data Analytic Methods.
(A) Protein versus RNA intensities for Chinese hamster ovary (CHO) cells; (B) codon frequency for CHO cells. It is important to optimize the codons of human proteins to increase their expression; (C) equations for a phenomena-logical model of microbial production of streptokinase based on the original concepts of specific growth rates, as first proposed by Monod in 1941 [47]; the variables represented in this formulation of ordinary differential equations are based on active (xa) and total (xt) microbial biomass, as listed in (D); (E) depiction of error back propagation in a deep learning model developed to estimate important parameters in the fermentation of streptokinase. The semisupervised deep neural network-based soft sensor has a superior overall performance to that of support vector regression specifically for cases with a larger unlabeled dataset. Reproduced, with permission, from [51] (A), [44] (B), and [46] (D,E).
The design of a putative optimized channel rhodopsin was published recently in which four protein attributes were optimized [45]. This work could provide a roadmap to creating a Protein A ligand for capture chromatography that is basestable, proteaseresistant, highaffinity, and highly pHtuned.When there are four protein attributes requiring optimization, creating variants is laboratory intensive and screening variants is data intensive.In the work by Bedbrooket al., a theoretical library of 2 × 310 chimeric variants (~120 000) was simulated using the properties from 76 actual chimeric variants.The results of the machine-optimized channel rhodopsin were validated with invitro and invivo tests.
Finally, the possibilities for cataloging and understanding the behavior of proteins in downstream purification are significant. While the behaviors of target proteins are usually understood in a localized sense, the fate of the large number of impurities found in antibody purification is not known in detail. Data derived from proteomics and transcriptomics can be used to identify the contaminants and predict their fate. The identification of phenomena such as nonspecific binding in affinity purification steps, which is difficult to predict from first principles, may not be as random as supposed when data are collected and analyzed for a range of conditions from a range of processes, particularly those for the production of mAbs.
Soft Sensors
Soft sensors for monitoring or controlling the production of mAbs have potential since some process indicators, particularly in cellculture, such as specific growth rates (i.e., cellular biomass) and active protein product are difficult to measure online. Soft sensors correlate readily measured variables with variables obtained from a ‘first principles’ model based on knowledge of the host cell and expression systems, or from measurements collected during previous or current runs [46]. In the case of streptokinase, a phenomenological model that calculated production of streptokinase by combining initial cell inoculum, specific growth rates, substrate depletion, lactate accumulation, and measured yield coefficients, was used to generate training sets in lieu of experiments (Figure 3C). The unstructured (or phenomenological) model was based on assumptions and specific growth rate equations originally proposed by Monod (PhD Thesis, Universite de Paris, 1941) for Escherichia coli [47].
Data-driven models represent a second approach for the development of soft sensors and utilize accumulated historic data from operation of the process absent in a mechanistic or phenomenological model. This is referred to as a black box. The data are fitted to obtain a polynomial expression for product generation as a function of known inputs (i.e., input layer; Figure 3C), (cell mass, substrate concentration, and initial product i.e., streptokinase). Given sufficient numbers of data sets obtained under a variety of conditions, fit of the data with automatically generated error correction factors is used to calculate key outputs such as the number of viable cells and level of product based on minimal measured input variables, such as inoculum and substrate.The black box accounts for the effects of other factors, such as agitation, individual key media components, mixing rate, and changes in metabolite concentrations, even though the mechanistic relation of these parameters to cell metabolism is unknown.
The availability of sophisticated computational algorithms combined with computational resources able to collect and manage large data sets makes such an approach practical. This is particularly useful for the production of mAbs through mammalian cell culture, where mechanistic (or phenomenological) models have yet to be fully developed, and the cellular metabolism leading to the final product is complicated. As hardware for sensor technology is developed, software for relating sensor-based measurements to process performance will become even more valuable for control of bioprocesses as they evolve from batch to continuous manufacturing platforms. Huge process databases, catalogued and rendered into useful formats will help make soft sensors of practical value to process scientists. Data analytics coupled to soft sensors will be an important development in the application of PAT to continuous processes for the manufacture of biotherapeutics.
Invasive versus Non-invasive Sensors
Non-invasive in-line sensors are required to provide real-time measurements without affecting the production process. Optical methods hold the greatest potential for PAT. Spectroscopy and sensors combined with appropriate analytical tools are particularly important for the continuous manufacturing of biologics. Applicable spectroscopic methods include UV-visible (UV-vis), fluorescence, infrared [near-infrared (NIR) and mid-infrared (MIR)], Raman spectroscopy, and Rayleigh [41], and include detection of viruses (Box 2).
Conventional sensors used in stainless-steel reactors are invasive; they need to be sterilized, calibrated, and validated before use [41]. Single-use bioreactors utilize established sensors that are packaged in disposable housings. Optically transparent windows allow for the transmission of optical signals from culture media to external detectors through in situ probes or LED-based flat sensors [48] that measure several cm2 in area. These sensors operate in UV to MIR wavelengths and enable fluorescence and Raman spectroscopies [28].The most recent developments in application of various spectroscopic sensors and dynamic light scattering (DLS) and static light scattering (SLS) [9,28,49., 50., 51.] are summarized in Table 2 .
Table 2.
Potential Spectroscopic PAT Sensors
| Optical technique | Wavelength range | Attributes for PAT sensing | Companies implementing in-line sensors in their process | Refsa |
|---|---|---|---|---|
| UV-vis spectroscopy | 240–340 nm | Cell density, aromatic amino acids | – | [48] |
| Circular dichroism | 190–260 nm | Proteins secondary structure, aggregation. | – | [49] |
| FT-NIR spectroscopy | 750–2500 nm | Chemical/biological compositions | Brucker, USA | [48] |
| Raman spectroscopy | 100–3500 cm–1 | antibody concentration | Pfizer, USA; GlaxoSmithKline, USA | [50] |
| Fluorescence spectroscopy | Excitation: 240–300 nm; emission: 260–450 nm | Cell metabolism, fluorescent compounds in media. | Sartorius Stedim Biotech GmbH, Germany | [28] |
| DLS | Visible light | Macromolecule diffusion | Colloid Metrix GmbH, Germany; Sanofi-Aventis Deutschland GmbH, Germany | [9] |
| SLS | Visible light | Size of macromolecules in solution | Sanofi-Aventis | [9] |
| Multiangle light scattering | Visible light | Protein aggregation | Merck & Co., Inc., USA; Wyatt Technology Corporation, USA | [51] |
The transition of manufacturing of mAbs from batch to continuous processes (Figure 2) will require changes in the unit operations, particularly downstream of product recovery [8]. The PAT associated with upstream cell culture is by now welldefined (Figure 2A) and utilizes commercially available sensor and measurement technology, operational experience in the research laboratory and in industry, as well as acceptance by regulators (Box 3 provides recent examples of continuous processes).
Box 3. Continuous Manufacturing.
Both batch-processing and perfusion cultures are characterized by operational times of between 1 week and 1 month for batch processing and longer for continuous culture, with product titers of 5 g/ after 2 weeks for some types of CHO cell [65]. Continuous manufacturing for pharmaceuticals has potential benefits of improving product quality and process control [14], as exemplified bysmall-molecule (drug) manufacture. Current approvals for continuous processes for small molecules include Orkambi and Symdeko, developed by Vertex, in 2015 for treating cystic fibrosis, and Prezista, developed by Jannsen, in 2016 for treating HIV [11,66]v. FDA approval of these processes included definition and regulation of PAT (see Box 4 in the main text). Process and product parameters for protein biologics are more complex than for small-molecule drugs, and, hence, PAT has special significance in the manufacture of these therapeutics [67,68].
Continuous bioprocessing markets for biotherapeutics are expected to grow ~10% annually between now (US$6.7 billion) and 2025 (US$12.1 billion), when fully integrated continuous biomanufacturing becomes routine, according to Mayix. Challenges that must be addressed include finding equipment suitable for good manufacturing practice (GMP) environments, maintaining sterility, and determining acceptability of different media harvests before these are pooled for downstream processing. Examples of continuous processes are for the manufacture of omalizumab (by BiosanaPharma, The Netherlands, for a Phase I clinical trial), and Sanofi’s recently opened, research-scale continuous manufacturing, facility (awarded ‘2020 Facility of the Year Award’ in the Facility of the Future category by the International Society for Pharmaceutical Engineeringviii). This facility is designed and operated for ‘process automation and on-line analytics’ that ‘enable automated control of steady-state conditions, and the generation of massive process data sets’… ‘with over 770 million datapoints sampled per day across the process and facility (attributed to Dean Morris by Mike Mayix). PAT will be a key factor in the emergence of this sector of biopharmaceutical manufacturing.
Alt-text: Box 3
Selecting the Appropriate Sensors for PAT
Selection of appropriate sensors or methods for PAT follows a rational sequence. First, the analyte and its relationship to the process are determined. Then, the most suitable analytical technology is defined. This decision is based on attributes of range, in-line or on-line integration possibilities, accuracy, specificity, speed, and sensitivity. Once defined, the analytical methods are developed, sensors with proper calibration deployed, and data acquisition software for multivariate, univariate, or deep learning strategies is selected to minimize the PAT decision time.
A recent example of this approach was demonstrated by Rolinger and colleagues [9], where the goal was to monitor process parameters during TFF (also known as cross-flow filtration). To measure the protein concentration, buffer exchange, apparent molecular weight, and hydrodynamic radius, Rolinger and colleagues used an in-line variable pathlength UV-vis spectrometer and an on-line measurement loop with a light-scattering photometer and a liquid density sensor. This set-up was then used to monitor TFF processes for lysozyme, glucose oxidase, and a mAb while demonstrating in-line measurement of protein concentration in the range of 3–120 g. The authors compared the results from in-line/on-line and off-line methods to exemplify excellent correlation (with coefficients exceeding 0.92). In the cases where discrepancy was observed between on-line and off-line methods, on-line methods were potentially more reliable because the wait time between sample collection and off-line measurement led to aggregation of proteins. In this example, data analytics was mainly based on first principles and measurements from one sensor (liquid density sensor) were used for analyzing data from another (light-scattering photometer) to improve the accuracy of estimating the targeted process parameter(i.e., hydrodynamic radius).
Spectroscopy and Sensors for PAT
The complexity of mAb production requires monitoring of several critical quality attributes (CQAs). On-line analytical chromatography enables monitoring of the production process of biologics. Sampling and separation may require up to an hour, while spectroscopic and spectrometric methods may be faster (seconds to minutes) [52]. However, these methods analyze multiple components, and provide a fingerprint rather than single component peaks. Hence, multivariate analysis (or chemometrics) is necessary to extract information from spectroscopic measurements. Multivariate projection methods, principal component analysis (PCA), and partial least square regression (PLSR) are currently being investigated for spectroscopy in PAT. For example, turbidity measurement combined with Raman spectroscopy and macroscopic kinetic modeling is able to estimate glucose concentrations [15]. As the amount of data available by spectroscopic methods increases, machine learning and neural networks will be increasingly important in transforming measurements to information.
UV-Vis spectroscopy measures the absorption from the aromatic amino acids (phenylalanine, tyrosine, and tryptophan) in proteins in the wavelength range of 240–340 nm. While 280 nm quantifies total protein, multiwavelength UV measurements help discriminate between different proteins in a mixture [53]. By comparison, at the other end of the spectrum, Fourier transforms infrared spectroscopy(FTIR) characterizes proteins by the vibration of amide bonds in the polypeptide backbone, which are dependent on the secondary structure of the protein. FTIR allows measurement of changes in protein structure. However, FTIR has a strong absorption peak for water, which is removed either computationally or by using attenuated total reflection (ATR)-FTIR [8].
Raman spectroscopy has a weak signal from water and a high molecular specificity because the signal arises from a change in polarizability during molecular motion. Combined with a capability to directly interrogate the sample, Raman may be used in-line, although interference from fluorescence must be considered, which could be overcome by using longer wavelengths or time-gated instrumentation [54]. The first demonstration of in-line measurement, in 2011, showed the measurement of multiple parameters to be feasible. These included glutamine, glutamate, glucose, lactate, ammonium, viable cell density, and total cell density [55]. Raman spectroscopy for the production of mAbs has also been demonstrated [56]. Raman spectra for complex mixtures must be deconvoluted to enable their interpretation. Analyzing these spectra requires multivariate analysis for extracting decision-making parameters.
The use of single-use, disposable technologies for the manufacturing of mAbs is expanding. Thus, sensors also need to be versatile and compatible with single-use bioreactors [57]. Soft-sensors, discussed earlier, combine the input of readily measured process attributes with algorithms to estimate the state of the process [46,47,57., 58., 59.].
Statistical Approaches for PAT
Statistical approaches based on multivariate data analysis (MVDA), such as PCA combined with PLSR, will evolve to be an important part of bioprocess data analytics [60,61]. PCA is generally used in process engineering to study critical process parameters (CPP) and to develop a hypothesis based on the pattern. While PLSR is a technique to study the relationship between product and process, it is also used as a calibration system for various spectroscopic tools [62]. The main limitation of PCA and PLSR is that they are linear methods and, thus, inefficient at predicting complex nonlinear relationships between the quality attributes [60]. Mammalian cell cultures represent complex systems where the MVDA is limited; to overcome this, data-driven and hybrid models using deep learning strategies have been proposed [46]. Analysis of PAT for downstream processing shows opportunities for in-line methods that could be used to monitor chromatography column effluents and inform systems operations with respect to changing buffer compositions or gradients, thereby maximizing recovery of purified product at each stage. This capability will be of increasing importance as batch processes that define downstream processing are operated in a continuous manner.
Concluding Remarks
The overall industrial application of PAT in the production of mAbs is of great industrial interest and a subject of FDA research (Box 4 )vii. The field is rich with possibilities but lean on applicable technologies beyond those able to measure basic process properties and parameters.To discern the intricate structural and chemical properties of an antibody as it is produced, it is clear that PAT systems will have to integrate measurements with models and machine learning based on multivariate analysis or other validatable algorithmsto predict and control these product attributes. In addition to real-time understanding of product-related substances and impurities, there is a need for real-time detection of exogenous product contamination. Significant improvement in the confirmation of absence of viruses and real-time detection of bacteria or mycoplasma contamination will greatly improve patient safety, and data analytics to support these improvements are still evolving in a manner that will expand the range of PAT operations within the production of biotherapeutics (see Outstanding Questions). Speed, accuracy, and cost effectiveness will be key to the adoption of PAT.
Box 4. PAT, Regulatory Framework, and the IQ Consortium.
The role of PAT in the continuous production of mAbs is both as a monitoring tool and as a tool for addressing FDA guidance. The opportunity comes in defining product characteristics within a regulatory framework at different points of the manufacturing process, where different criteria would apply (i.e., cell robustness and metabolites early in the process; product activity and absence of immunogenic substances, viral particles, nucleic acids, and media components late in the process). The challenge is to assemble the resulting data sets and perform analytics to define conditions that fit within operational envelopes that are set by regulatory processes. Since PAT is positioned to provide results within the residence times defined by unit operations of cell culture, protein recovery, and product purification, it is possible to envision transition of mAb manufacture to a continuous basis, where rates of inputs and outputs are controlled to be constant over periods of time of weeks to months. This has been achieved for some types of small-molecule drug (i.e., chemically synthesized entities) and for which PAT was identified as an important technology on the pathway to continuous manufactureiii[59].
The role of PAT can help to reduce personnel hazards related to sampling during process validation. PAT also contributes to understanding the measurements during various parameters in the manufacturing of mAbs without disturbing the flow. These criteria are explained in reports of the IQ Consortium that address the current state of PAT for API development in branded pharmaceutical companies. A recent report used an API process workflow (process steps from raw material identification through to finished API) to provide representative examples, including why and how the pharmaceutical industry uses PAT tools in API development [8].
Alt-text: Box 4
Given the huge amounts of data that are readily obtained (Box 3), soft sensors and general user interfaces that display information that is gathered during process operation will catalyze an expanded application of PAT to the continuous manufacture of biologics. Requirements of scale, fidelity, and hands-off operations will necessitate PAT systems that are capable of being run remotely. This capability is envisioned as offering potential to achieve a new class of protein product that has the necessary safety, high consistency, structure, and bioactivity associated with an approved biotherapeutic, while being manufactured continuously.
Outstanding Questions.
How quickly will deep learning algorithms for PAT applications develop and start to have a major role in the process control of continuous mAb production?
What are the detection limits of in-line sensors for in situ measurements in the presence of background signals from biological and biochemical components in culture media?
How much specificity can be achieved by a combination of advanced spectroscopic techniques and data analytics when analyzing a complex process stream, such as perfusion bioreactor effluent?
Can soft sensors help overcome the limits (response time, signal:noise ratio, and dynamic range) imposed by the hardware of current analytical methods for in situ measurements?
What is the maximum productivity (rate, titer, and yield) of mAbs when PAT, advanced cell lines obtained with gene editing tools, deep learning, and soft sensors are combined to monitor and control a continuous biologic manufacturing process?
Alt-text: Outstanding Questions
Acknowledgments
This work was funded by grants from the College of Engineering and the College of Agriculture at Purdue University and the Purdue University Office of Agricultural Research and Graduate Education through its 150th Anniversary Elevating the Visibility of Agricultural Research. Other support is from (Hatch Act 10677, 10646) and ‘Continuous Manufacturing of Biologics,’ funded by Purdue College of Engineering Faculty Conversations (EFC).M.L. expresses appreciation to Ruben Carbonell and Gary Gilleskie for providing an excellent experience, and for facilitating his participation in courses in the BTEC Center at NC State University in 2019, during his change in duty station, and to Carla Carie for her preparation of figures and working through the numerous revisions of this manuscript.
Glossary
- Active pharmaceutical ingredient (API)
found in biologics; denotes only the biologically active chemical that is found along with the other chemicals that make the drug work.
- Biosimilars
biologic that is highly similar to another already approved product.
- Bispecific antibodies (bsAbs)
antibodies that bind to tumor cell surface antigens as well as to immune cells to recruit T cells or natural killer cells; have two different variable regions targeting two different antigens.
- Channel rhodopsin
light-gated trans-membrane protein ion channels found in photosynthetic algae; serve as sensory photoreceptors and control movement in response to light, such as in blue-green algae.
- Coronavirus disease 2019 (COVID-19)
infectious pandemic disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
- Critical process parameter (CPP)
process variable that has a direct relationship with a product quality attribute. These attributes are monitored to maintain product quality and consistency.
- Critical quality attributes (CQAs)
attributes that comprise biological, chemical, and physical characteristics that should be measured continuously to ensure the desired product quality; used to ensure the safety, efficacy, and stability of pharmaceutical products.
- Dynamic light scattering (DLS)
technique used to measure the particle size in nm range in a colloidal solution based on the Brownian motion of dispersed particles. The distribution of diffusion coefficients can be used to determine hydrodynamic diameter and, thus, monitor the folding, aggregation, and conformation changes of proteins.
- Fourier transform infrared spectroscopy (FTIR)
measures the absorption of IR radiation by sample as a function of wavelength. Changes in a spectrum are used for monitoring product quality.
- Host cell proteins (HCP)
proteins from the host cell, generated or present as a function of cellular metabolism, and which are expressed in addition to product molecule.
- Humanized antibodies
monoclonal antibodies, the protein sequences of which are modified to replicate sequences of antibodies produced in humans.
- International Consortium for Innovation and Quality in Pharmaceutical Development (IQ Consortium)
represents a pharmaceutical and biotechnology consortium for advancing innovation and quality in the biopharmaceutical industry.
- Multivariate data analysis (MVDA)
statistical technique to analyze more than one variable or type of variable for the study of process characterization, monitoring, and fault diagnosis.
- Near-infrared spectroscopy (NIR)
optical imaging methods that give ‘fingerprints’ of food, pharmaceutical, and biotechnology products at wavelengths of 780–2500 nm.
- Principal component analysis (PCA)
statistical approach to reduce multidimensional data to a few orthogonal components that are weighted linear combinations of the original data (as determined by the variance explained by each variable). It is an unsupervised learning method most commonly used to find patterns of clustering.
- Process analytical technology (PAT)
framework defined by the US Food and Drug Administration (FDA) for building and implementing innovative methods of pharmaceutical development, manufacturing, and quality assurance.
- Raman spectroscopy
technique that uses a laser to illuminate a sample that scatters electromagnetic radiation, causing a shift in energy, therefore enabling material properties to be determined.
- Single-use systems
disposed after use in the manufacture of biopharma products rather than being cleaned, sterilized, and reused. The term applies to single-use bioreactors (SUB), connecting tubing, valves, sensors, feed and reagent containers, and downstream process components.
- Soft sensors
describes the methodology for predicting ‘difficult-to-measure variables by correlating them with easily available sensors’ for application to bioprocess control [46].
- Static light scattering (SLS)
physiochemical technique that measures the intensity and angle of scattered light (i.e., Rayleigh scattering) and correlates it to the molecular weight or size of proteins in aqueous solution.
- Tangential flow filtration (TFF)
system that facilitates continuous cross-flow over membranes for microfiltration (removes particulates) or ultrafiltration (retains protein or other molecules >30 kDa in molecular weight) in a manner that minimizes fouling.
Resources
iwww.prnewswire.com/news-releases/abcellera-and-lilly-to-co-develop-antibody-therapies-for-the-treatment-of-covid-19-301022689.htmliiwww.statnews.com/2020/04/06/glaxosmithkline-and-vir-aim-to-take-on-covid-19-with-antibodies-and-crispr/iiiwww.fda.gov/media/71012/downloadivwww.fda.gov/media/121314/downloadvwww.biopharmadive.com/news/pharmas-slow-embrace-of-continuous-manufacturing/532811/viwww.prnewswire.com/news-releases/milliporesigma-launches-industrys-first-off-the-shelf-cell-culture-media-for-perfusion-processes-300457240.htmlviihttps://doi.org/10.1016/J.IJPHARM.2016.10.038viiiIspe.org/news/2020-ispe-facility-year-awards-winners-announcedixwww.genengnews.com/insights/biomanufacturers-cope-with-challenges-of-going-continuous/References
- 1.Buss N.A., et al. Monoclonal antibody therapeutics: history and future. Curr. Opin. Pharmacol. 2012;12:615–622. doi: 10.1016/j.coph.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Kaplon H., Reichert J.M. Antibodies to watch in 2019. MAbs. 2019;11:219–238. doi: 10.1080/19420862.2018.1556465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grilo A.L., Mantalaris A. The increasingly human and profitable monoclonal antibody market. Trends Biotechnol. 2019;37:9–15. doi: 10.1016/j.tibtech.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Udpa N., Million R.P. Monoclonal antibody biosimilars. Nat. Rev. Drug Discov. 2016;15:13–14. doi: 10.1038/nrd.2015.12. [DOI] [PubMed] [Google Scholar]
- 5.Mullard A. Bispecific antibody pipeline moves beyond oncology. Nat. Rev. Drug Discov. 2017;16:667–668. doi: 10.1038/nrd.2017.187. [DOI] [PubMed] [Google Scholar]
- 6.Whitaker N., et al. A formulation development approach to identify and select stable ultra-high-concentration monoclonal antibody formulations with reduced viscosities. J. Pharm. Sci. 2017;106:3230–3241. doi: 10.1016/j.xphs.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Kelly B. Industrialization of mAb production technology: the bioprocessing industry at a crossroads. mAbs. 2009;1:443–452. doi: 10.4161/mabs.1.5.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chanda A., et al. Industry perspectives on process analytical technology and development. Org. Process R&D. 2015;19:63–83. [Google Scholar]
- 9.Rolinger L., et al. Multi-variate PAT for UF / DF of proteins – monitoring concentration, particle sizes, and buffer exchange. Anal. Bioanal. Chem. 2020;412:2123–2136. doi: 10.1007/s00216-019-02318-8. [DOI] [PubMed] [Google Scholar]
- 10.Alper H.S., Wittmann C. Systems metabolic engineering approaches for rewiring cells. Biotechnol. J. 2019;14:1–2. doi: 10.1002/biot.201900312. [DOI] [PubMed] [Google Scholar]
- 11.Fisher A.C., et al. The current scientific and regulatory landscape in advancing integrated continuous biopharmaceutical manufacturing. Trends Biotechnol. 2018;37:253–267. doi: 10.1016/j.tibtech.2018.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Somasundaram B., et al. Progression of continuous downstream processing of monoclonal antibodies: current trends and challenges. Biotechnol. Bioeng. 2018;115:2893–2907. doi: 10.1002/bit.26812. [DOI] [PubMed] [Google Scholar]
- 13.Pollock J., et al. Integrated continuous bioprocessing: economic, operational, and environmental feasibility for clinical and commercial antibody manufacture. Biotechnol. Prog. 2017;33:854–866. doi: 10.1002/btpr.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Academies of Sciences, Engineering, and Medicine, et al. National Academies Press; 2019. Continuous Manufacturing for the Modernization of Pharmaceutical Production: Proceedings of a Workshop. [PubMed] [Google Scholar]
- 15.Kornecki M., Strube J. Process analytical technology for advanced process control in biologics manufacturing with the aid of macroscopic kinetic modeling. Bioengineering. 2018;5:25. doi: 10.3390/bioengineering5010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrossian, A. and DeGiovanni, A. Porton International. Perfusion airlift reactor. US4806484
- 17.Godawat R., et al. End-to-end integrated fully continuous production of recombinant monoclonal antibodies. J. Biotechnol. 2015;213:13–19. doi: 10.1016/j.jbiotec.2015.06.393. [DOI] [PubMed] [Google Scholar]
- 18.Walker J. Lack of blood samples stalls virus-drug work. Wall Street J. 2020 18 March. [Google Scholar]
- 19.Hebbi V., et al. Process analytical technology implementation for protein refolding: GCSF as a case study. Biotechnol. Bioeng. 2019;116:1039–1052. doi: 10.1002/bit.26900. [DOI] [PubMed] [Google Scholar]
- 20.Jayapal K.P., et al. Recombinant protein therapeutics from CHO Cells - 20 years and counting. Chem. Eng. Prog. 2007;103:40–47. [Google Scholar]
- 21.Grav L.M., et al. Application of CRISPR/Cas9 genome editing to improve recombinant protein production in CHO cells. Methods Mol. Biol. 2017;1603:101–118. doi: 10.1007/978-1-4939-6972-2_7. [DOI] [PubMed] [Google Scholar]
- 22.Zydney A.L. Continuous downstream processing for high value biological products: a Review. Biotechnol. Bioeng. 2016;113:465–475. doi: 10.1002/bit.25695. [DOI] [PubMed] [Google Scholar]
- 23.Rudge S.R., Ladisch M.R. Industrial challenges of recombinant proteins. Adv. Biochem. Eng. Biotechnol. 2020;171:1–22. doi: 10.1007/10_2019_120. [DOI] [PubMed] [Google Scholar]
- 24.Gronemeyer P., et al. Trends in upstream and downstream process development for antibody manufacturing. Bioengineering (Basel) 2014;1:188–212. doi: 10.3390/bioengineering1040188. [DOI] [PubMed] [Google Scholar]
- 25.Xu X., et al. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 2011;29:735–741. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo C.-C., et al. The emerging role of systems biology for engineering protein production in CHO cells. Curr. Opin. Biotechnol. 2018;51:64–69. doi: 10.1016/j.copbio.2017.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C., et al. Upstream process intensification and continuous manufacturing. Curr. Opin. Chem. Eng. 2018;22:191–198. [Google Scholar]
- 28.Claβen J., et al. Spectroscopic sensors for in-line bioprocess monitoring in research and pharmaceutical industrial application. Anal. Bioanal. Chem. 2017;409:651–666. doi: 10.1007/s00216-016-0068-x. [DOI] [PubMed] [Google Scholar]
- 29.Bluma A., et al. Process analytical sensors and image-based techniques for single-use bioreactors. Eng. Life Sci. 2011;11:550–553. [Google Scholar]
- 30.Li F., et al. Cell culture processes for monoclonal antibody production. MAbs. 2010;2:466–477. doi: 10.4161/mabs.2.5.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillespie C., et al. Continuous In-line virus inactivation for next generation bioprocessing. Biotechnol. J. 2019;14 doi: 10.1002/biot.201700718. [DOI] [PubMed] [Google Scholar]
- 32.Ladisch M.R. Wiley; 2001. Bioseparations Engineering: Principles, Practice and Economics. [Google Scholar]
- 33.Harrison R.G., et al. 2nd edn. Oxford University Press; 2015. Bioseparations Science and Engineering. [Google Scholar]
- 34.Mendhe R., et al. Comparison of PAT based approaches for making real-time pooling decisions for process chromatography – use of feed forward control: PAT based approaches for real-time pooling. J. Chem. Technol. Biotechnol. 2015;90:341–348. [Google Scholar]
- 35.Pathak M., et al. Re-use of protein A resin: fouling and economics. BioPharm Int. 2015;28:28–33. [Google Scholar]
- 36.Johnson S.A., et al. Adapting viral safety assurance strategies to continuous processing of biological products. Biotechnol. Bioeng. 2017;114:21–32. doi: 10.1002/bit.26060. [DOI] [PubMed] [Google Scholar]
- 37.Binabaji E., et al. Ultrafiltration of highly concentrated antibody solutions: experiments and modeling for effects of module and buffers conditions. Biotechnol. Prog. 2016;32:692–701. doi: 10.1002/btpr.2252. [DOI] [PubMed] [Google Scholar]
- 38.Ko S.H., et al. Nanofluidic device for continuous multiparameter quality assurance of biologics. Nat. Nanotech. 2017;12:804–812. doi: 10.1038/nnano.2017.74. [DOI] [PubMed] [Google Scholar]
- 39.Zhi Y., et al. Validation of a PCR method for the detection of mycoplasma according to European Pharmacopaeia section 2.6.7. Biologicals. 2010;38:232–237. doi: 10.1016/j.biologicals.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Angart P., et al. Considerations for risk and control of mycoplasma in bioprocessing. COChE. 2018;22:161–166. [Google Scholar]
- 41.Rolinger, et al. A critical review of recent trends, and a future perspective of optical spectroscopy as PAT in biopharmaceutical downstream processing. Anal. Bioanal. Chem. 2020;412:2047–2064. doi: 10.1007/s00216-020-02407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertrand V., et al. Proteomic analysis of microscale bioreactors as a scale-down model for a mAb producing CHO industrial fed-batch platform. J. Biotechnol. 2018;279 doi: 10.1016/j.jbiotec.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 43.Becker J., et al. Unraveling the Chinese hamster ovary cell line transcriptome by next-generation sequencing. J. Biotechnol. 2011;156:227–235. doi: 10.1016/j.jbiotec.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 44.Bayein-Hizal D., et al. Proteomic analysis of Chinese Hamster Ovary Cells. J. Proteome Res. 2012;11:5265–5276. doi: 10.1021/pr300476w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bedbrook C.N., et al. Machine learning-guided channelrhodopsin engineering enables minimally invasive optogenetics. Nat. Methods. 2019;16:1176–1184. doi: 10.1038/s41592-019-0583-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gopakumar V., et al. A deep learning based data driven soft sensor for bioprocesses. Biochem. Eng. J. 2018;136:28–39. [Google Scholar]
- 47.Mosier N., Ladisch M.R., editors. Modern Biotechnology: Connecting Innovations in Microbiology and Biochemistry to Engineering Fundamentals. John Wiley and Sons; 2009. pp. 73–96. 112–133. [Google Scholar]
- 48.Biechele P., et al. Sensor systems for bioprocess monitoring. Eng. Life Sci. 2015;15:469–488. [Google Scholar]
- 49.Joshi V., et al. Circular dichroism spectroscopy as a tool for monitoring aggregation in monoclonal antibody therapeutics. Anal. Chem. 2014;66:11606–11613. doi: 10.1021/ac503140j. [DOI] [PubMed] [Google Scholar]
- 50.Yilmaz D., et al. Application of Raman spectroscopy in monoclonal antibody producing continuous system for downstream process. Biotechnol. Prog. 2020;36 doi: 10.1002/btpr.2947. [DOI] [PubMed] [Google Scholar]
- 51.Patel B.A., et al. Multi-angle light scattering as a process analytical technology measuring real-time molecular weight for downstream process control. mAbs. 2018;10:945–950. doi: 10.1080/19420862.2018.1505178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rüdt M., et al. Advances in downstream processing of biologics – spectroscopy: an emerging process analytical technology. J. Chromatogr. A. 2017;1490:2–9. doi: 10.1016/j.chroma.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 53.Hanson S.K., et al. Selective high throughput protein quantification based on UV absorption spectra. Biotechnol. Bioeng. 2013;110:448–460. doi: 10.1002/bit.24712. [DOI] [PubMed] [Google Scholar]
- 54.Buckley K., Ryder A.G. Applications of Raman spectroscopy in biopharmaceutical manufacturing: a short review. Appl. Spectosc. 2017;71:1085–1116. doi: 10.1177/0003702817703270. [DOI] [PubMed] [Google Scholar]
- 55.Abu-Absi N.R., et al. Real time monitoring of multiple parameters in mammalian cell culture bioreactors using an in-line Raman spectroscopy probe. Biotechnol. Bioeng. 2018;108:1215–1221. doi: 10.1002/bit.23023. [DOI] [PubMed] [Google Scholar]
- 56.Santos R.M., et al. Monitoring mAb cultivations with in-situ Raman spectroscopy: the influence of spectral selectivity on calibration models and industrial use as reliable PAT tool. Biotechnol. Prog. 2018;34:659–670. doi: 10.1002/btpr.2635. [DOI] [PubMed] [Google Scholar]
- 57.Holzberg T.R., et al. Sensors for biomanufacturing process development: facilitating the shift from batch to continuous manufacturing. Curr. Opin. Chem. Eng. 2018;22:115–127. [Google Scholar]
- 58.Funatsu K. In: Applied Chemoinformatics. Engel T., Gasteiger J., editors. John Wiley & Sons; 2018. Process control and soft sensors; pp. 571–584. [Google Scholar]
- 59.Guerra A., et al. Toward biotherapeutic product real-time quality monitoring. Crit. Rev. Biotechnol. 2019;39:289–305. doi: 10.1080/07388551.2018.1524362. [DOI] [PubMed] [Google Scholar]
- 60.Narayanan H., et al. Bioprocessing in the digital age: the role of process models. Biotechnol. J. 2019;15 doi: 10.1002/biot.201900172. [DOI] [PubMed] [Google Scholar]
- 61.Mandenius C.-F., Gustavsson R. Mini-review: Soft sensors as means for PAT in the manufacture of bio-therapeutics. J. Chem. Technol. Biotechnol. 2014;90:215–227. [Google Scholar]
- 62.Sokolov M., et al. Enhanced process understanding and multivariate prediction of the relationship between cell culture process and monoclonal antibody quality. Biotech. Progress. 2017;33:1368–1380. doi: 10.1002/btpr.2502. [DOI] [PubMed] [Google Scholar]
- 63.Rathore A.S., et al. Fermentanomics: relating quality attributes of a monoclonal antibody to cell culture process variables and raw materials using multivariate data analysis. Biotechnol. Prog. 2015;31:1586–1599. doi: 10.1002/btpr.2155. [DOI] [PubMed] [Google Scholar]
- 64.Barbarous M., Setie A. Properties of materials used in single-use flexible containers: requirements and analysis. Biopharm Int. 2006;2006 (Suppl.) [Google Scholar]
- 65.Reinhart D., et al. Benchmarking of commercially available CHO cell culture media for antibody production. Microbiol. Biotechnol. 2015;99:4645–4657. doi: 10.1007/s00253-015-6514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vargas J.M., et al. Process analytical technology in continuous manufacturing of a commercial pharmaceutical product. Int. J. Pharm. 2018;538:167–178. doi: 10.1016/j.ijpharm.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Engisch W., Muzzio F. Using residence time distributions (RTDs) to address the traceability of raw materials in continuous pharmaceutical manufacturing. J. Pharm. Innov. 2016;11:64–81. doi: 10.1007/s12247-015-9238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mercier S.M., et al. Multivariate PAT solutions for biopharmaceutical cultivation: current progress and limitations. Trends Biotechnol. 2014;32:329–336. doi: 10.1016/j.tibtech.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 69.Chiang M.-J., et al. Validation and optimization of viral clearance in a downstream continuous chromatography setting. Biotechnol. Bioeng. 2019;116:2292–2302. doi: 10.1002/bit.27023. [DOI] [PubMed] [Google Scholar]
- 70.Mattila J., et al. Retrospective evaluation of low-pH viral inactivation and viral filtration data from a multiple company collaboration. PDA J. Pharm. Sci. Technol. 2016;70:293–299. doi: 10.5731/pdajpst.2016.006478. [DOI] [PubMed] [Google Scholar]
- 71.Yeh T., et al. A rapid and label-free platform for virus capture and identification from clinical samples. PNAS. 2020;117:895–901. doi: 10.1073/pnas.1910113117. [DOI] [PMC free article] [PubMed] [Google Scholar]