Short abstract
The National Institutes of Health’s (National Cancer Institute) precision medicine initiative emphasizes the biological and molecular bases for cancer prevention and treatment. Importantly, it addresses the need for consistency in preclinical and clinical research. To overcome the translational gap in cancer treatment and prevention, the cancer research community has been transitioning toward using animal models that more fatefully recapitulate human tumor biology. There is a growing need to develop best practices in translational research, including imaging research, to better inform therapeutic choices and decision-making. Therefore, the National Cancer Institute has recently launched the Co-Clinical Imaging Research Resource Program (CIRP). Its overarching mission is to advance the practice of precision medicine by establishing consensus-based best practices for co-clinical imaging research by developing optimized state-of-the-art translational quantitative imaging methodologies to enable disease detection, risk stratification, and assessment/prediction of response to therapy. In this communication, we discuss our involvement in the CIRP, detailing key considerations including animal model selection, co-clinical study design, need for standardization of co-clinical instruments, and harmonization of preclinical and clinical quantitative imaging pipelines. An underlying emphasis in the program is to develop best practices toward reproducible, repeatable, and precise quantitative imaging biomarkers for use in translational cancer imaging and therapy. We will conclude with our thoughts on informatics needs to enable collaborative and open science research to advance precision medicine.
Keywords: co-clinical trial, preclinical PET, MR, CT, quantitative imaging, informatics, precision medicine, patient-derived tumor xenograft (PDX), genetically engineered mouse model (GEMM), cell transplant model (CTM)
Background
Co-clinical trials are an emerging area of investigation in which a clinical trial is coupled with a preclinical study to inform the corresponding clinical trial (1–7). The preclinical arm of the co-clinical trial generally uses genetically engineered mouse models (GEMMs), cell transplant models (CTMs) of human cancers or patient-derived tumor xenografts (PDXs) to aid in therapeutic efficacy assessment, patient stratification, and optimal treatment strategies designing (8, 9). The emergence of GEMMs, CTMs, and PDXs as co-clinical platforms is largely motivated by the realization that established cell lines do not recapitulate the heterogeneity of human tumors and the diversity of tumor phenotypes (10) and that better oncology models are needed to support high-impact translational cancer research. To that end, the National Cancer Institute’s (NCI) Patient-Derived Models Repository (https://pdmr.cancer.gov), EuroPDX (https://www.europdx.eu), academic institutions, and numerous commercial entities have launched wide-ranging animal model repositories to advance the biological and molecular bases for cancer prevention and treatment toward realization of precision medicine. In light of the prominent role of preclinical imaging in cancer research, the NCI has recently launched the Co-Clinical Imaging Research Resource Program (CIRP) (https://nciphub.org/groups/cirphub).
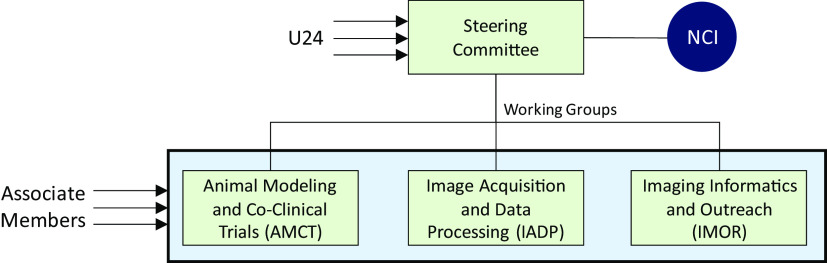
The purpose of this communication is to present our involvement in the CIRP and highlight its objective and scope to the imaging community. CIRP’s mission is to advance the practice of precision medicine by establishing consensus-based best practices for co-clinical imaging and developing optimized state-of-the-art translational quantitative imaging methodologies to enable disease detection, risk stratification, and assessment/prediction of response to therapy. Operationally, CIRP is structured as a steering committee (SC) and three working groups (WGs) focused on practical aspects of co-clinical imaging (Figure 1). Investigators of the NCI-funded U24 award make up the steering committee and the WGs. The WGs include animal models and co-clinical trials (AMCT), image-acquisition and data processing (IADP), and informatics and outreach (IMOR). The animal models and co-clinical trials focuses on topics relevant to co-clinical oncology models and co-clinical trial design where animal models are used in therapeutic screening, patient stratification and to inform the clinical trial. The image-acquisition and data processing focuses on optimization and standardization of image acquisition and data processing pipelines. Finally, the informatics and outreach addresses resource-sharing and informatics needs for preclinical and clinical imaging to support co-clinical studies. Investigators not directly funded by the U24 mechanism may petition to join the CIRP network as associate members. Associate members are then affiliated with one or more of the WGs. In line with the objective of the CIRP, the SC and the WGs tackle key issues in co-clinical trials, translational quantitative imaging, and informatics.
Figure 1.
Organizational structure of Co-Clinical Imaging Resource Program (CIRP). Members of the steering committee (SC) comprise investigators from within U24 awardees. The SC oversees the activities of 3 working groups (WGs) (animal models and co-clinical trials, image-acquisition and data processing [IADP], and informatics and outreach [IMOR]) focused on practical aspects of co-clinical imaging. Individuals not directly funded by the U24 mechanism may petition to join the CIRP network as associate members. Associate members are then affiliated with one or more of the WGs.
In the ensuing sections, we will detail key considerations in designing co-clinical imaging trials in terms of selection of animal models, considerations in designing co-clinical imaging studies, standardization of instruments, and harmonization of preclinical and clinical quantitative imaging pipelines. An underlying emphasis is to develop best practices toward reproducible, repeatable, and precise quantitative imaging biomarkers for use in translational cancer imaging and therapy. We will conclude with informatics needs to enable collaborative and open science research to advance precision medicine.
Animal Models and Co-Clinical Trials
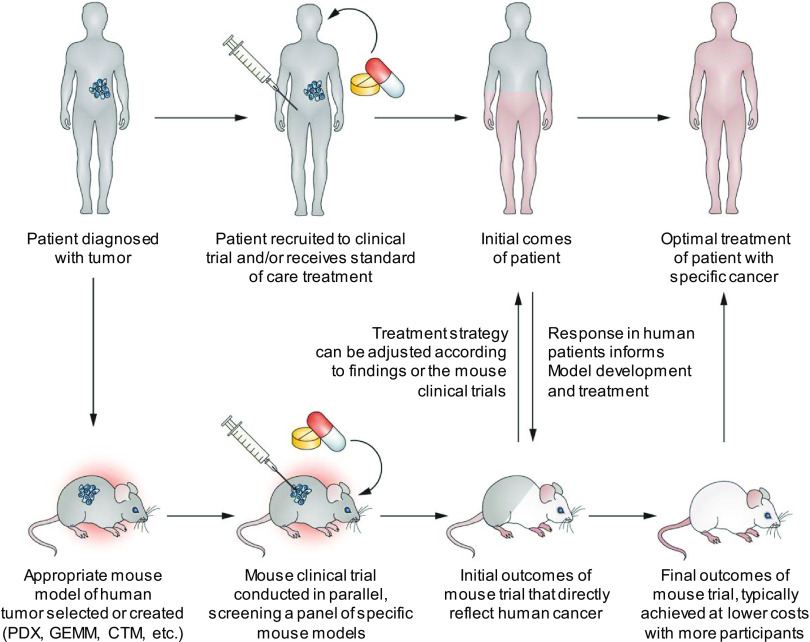
NCI’s precision medicine initiative emphasizes the use of translational oncology models to address the biological and molecular bases of cancer prevention and treatment. As noted above, translational oncology models considered in this context include (but are not limited to) PDXs, GEMMs, and CTMs. The advantages and disadvantages of these models are summarized in Table 1. Rapid disease progression presents a limitation for essentially all mouse models of cancer used in co-clinical trials. However, co-clinical animal models offer the opportunity for streamlined assessment of tumor sensitivity to drugs being tested in the human clinical study, as well as to evaluate mechanisms of treatment resistance and evaluate novel drug combinations (Figure 2). Initial outcomes in the human clinical trial can influence treatment strategies used in the mouse model and indicate whether new models are needed for more faithful recapitulation of the human disease process. Similarly, findings in the mouse trial can inform selection of patients most likely to benefit from an intervention, provide guidance for optimal imaging and biospecimen collection for correlative studies, and lead to adjustments in therapeutic approach. In an ideal co-clinical paradigm, the mouse trials will enable rapid transfer of information from mouse experiments to human trials to provide information to optimize treatment regimens with a focus on ultimately improving clinical care and patient outcomes (9).
Table 1.
Use of Co-Clinical Models in Preclinical Imaging Research
| Advantages | Disadvantages | Considerations in Imaging | |
|---|---|---|---|
| PDX | Ability to accurately reflect patients’ tumors in terms of the histomorphology, gene mutation and expression profiles, and gene copy number alterations Ability to predict therapeutic response in patients |
Variable take rate Immuneocompromised background, although efforts are underway to develop humanized PDX models |
Need to be credentialed/validated to match human tumor Need to document clinical information regarding the tumor of origin Genetic drift with subsequent passages may impact phenotype Experiments should be performed using low passage numbers Relative age of diseased mice is youngerthan corresponding patients |
| GEMMs | Gradual disease development Intact immune system Significant inter- and intra-tumor heterogeneity Recapitulate histopathological features of human tumors |
High total cost Potential long time to tumor latency Single genetic alterations that may not match the genetic heterogeneity of human disease. Variability in penetrance and latency Potentially low mutational load |
Relative age of diseased mice younger than corresponding patients Relatively large group size to address inter-mice heterogeneity |
| CTMs ofcancer | Match of driver mutations present in patients Faithfully maintaining underlying genetic basis of disease present in patients Rapidly producing large numbers of immunocompetent mice for treatment studies using syngeneic bone marrow transplants |
Myeloablative conditioning regimens used to facilitate engraftment of transplanted HSCs in recipient animals Accelerated course of disease relative to patients. |
Relative age of diseased mice younger than corresponding patients Accelerated disease progression makes some manifestations of disease, such as fibrosis, easier to reverse |
Figure 2.
The co-clinical precision medicine design paradigm. In the co-clinical study design, mouse models (patient-derived tumor xenograft [PDX], genetically engineered mouse model [GEMMs], or cell transplant models [CTMs]) are developed to match the patient’s tumor genotype or subtype. In parallel with the clinical trials, these patient-matched co-clinical models are used to assess the sensitivity of tumors to drugs or drug combinations, and thus inform the clinical trial. (Adapted from Clohessy and Pandolfi (9).)
Patient-Derived Tumor Xenografts
To date, PDX models perhaps come closest to addressing the co-clinical, paradigm (11). The key advantages of PDXs include the following:
(1) ability to accurately reflect patients’ tumors in terms of the histomorphology, gene expression profiles, and gene copy number alterations (12–16); and
(2) ability to predict therapeutic response in patients, especially when a clinically relevant drug dosage is used (17–19).
In this model, a sample of viable tumor tissue obtained from surgical resection of the tumor—or a solid (20) or liquid biopsy (21)—is implanted onto immune-compromised mice, either subcutaneously or orthotopically (11, 22–24). While not depicted in Figure 2, established PDXs can be used as a renewable source of tumor cells for generation of patient-derived organoids (PDOs) (25–27). There is also an interesting variation of the in vivo PDX method, mini-PDX, where patient-derived tumor cells are seeded into hollow fiber capsules that are implanted subcutaneously into mice. The mini-PDX-bearing mice are treated with therapeutics of interest for 7 days and tumor cells in tubes are assessed for therapeutic effect (28). The authenticity of PDX models is crucial to the validity of the studies performed with them. Meehan et al. (29) described several key considerations for investigators interested in generating PDX models. Special attention should be given to documenting clinical information regarding the tumor of origin, as this can aid in identification of potential biomarkers of therapeutic response or resistance. Patient information should also be tracked, such as age, sex, diagnosis, race, ethnicity, treatment history and response, as well as virology status (presence of HIV, HBV, HCV, HTLV, EBV, and other viral pathogens). It is also important to document information about the primary tumor, such as whether tissue originated from a primary, metastatic, or recurrent tumor, as well as specific features of histology, stage, grade, and presence of driver mutations and loss or mutation of key tumor suppressor genes.
Once the PDX model has been developed, details of the mouse strain used, engraftment procedure, rate of engraftment, tumor preparation before injection, passage number, and injection site should be recorded. PDX tumors need to be carefully validated for quality assurance (QA) based upon histology, special stains, and short tandem repeat to ensure authenticity of PDX samples. Genetic analysis should be performed to validate any genetic drift. In particular, evaluation of next-generation sequencing data generated from PDX samples requires special considerations. It is important to perform RNA sequence analysis of the primary tumor and to compare that analysis to the RNA sequence analysis of the PDX, as well as to accurately distinguish between sequencing reads that originate from the host versus those arising from the xenograft itself. Failure to correctly identify contaminating host reads can lead to incorrect mutation and expression calls (30). It is also important to note that PDX tumors do undergo some evolution as they are forced to adapt to grow in a mouse host. As a result of these mouse host-induced changes, PDX tumors can diverge from the primary patient’s tumor from which they were derived. This genetic drift is especially evident by distinct copy number alterations in PDXs that accumulate with each PDX passage (31). Thus, it is important to perform experiments with low-passage PDXs to ensure faithful representation of the primary tumor genome.
Because PDX tumors use immune-compromised mice, one main disadvantage is the lack of an immune component to test the contribution of the immune response in therapeutic studies. To address this critical need in studies of immune-oncology, numerous mouse models have been implanted with human CD34+ cells to reconstitute the mouse with a “human immune system”; however, there are weaknesses for each model as detailed elsewhere [NSG-SGM3 (32, 33), NSG-β2µ (34), MISTR (35), and NOG-EXL (36, 37)]. Despite the nuances mentioned above, PDXs hold great value for testing novel therapeutic regimens, as well as for studying mechanisms of therapeutic response and resistance in a variety of hematological and solid tumors (11, 38, 39).
Genetically Engineered Mouse Models of Cancer
Genetically engineered mouse models (GEMMs) typically use targeted delivery or expression of a recombinase to trigger genetic recombination events that lead to spatially and temporally restricted tumorigenesis. The Cre-lox and Flp-frt are the most commonly used systems. Cre is a site-specific recombinase that deletes DNA flanked by loxP sites [ie, “floxed alleles”, FL (40)] and similarly flippase (FLP) recognizes FLP recombinase target sequences (ie, “frted alleles”, FRT) to facilitate targeted mutations (41). Numerous tissue-specific Cre drivers are available to localize mutations in particular tissues, and inducible Cre strains offer an additional layer of temporal control. For example, CreER(T2) recombinase is a tamoxifen-dependent Cre recombinase that can be activated by systemic administration of tamoxifen or localized administration of 4-hydroxytamoxifen (42). Alternatively, viruses can be used to deliver recombinase to a particular site to induce recombination of the mutant alleles. Multiple genes can be altered simultaneously in the Cre-lox system, with oncogene expression triggered by deletion of a floxed stop cassette (loxP-STOP-loxP, “LSL”) preceding an oncogene (eg, LSL-KrasG12D) and tumor suppressor knockout from deletion of floxed exons (43). GEMMs offer several advantages, including autochthonous and gradual disease development in the presence of an intact immune system (44) recapitulating the inter- and intratumor heterogeneity and histopathological features of the human tumor and microenvironment (9, 45). However, disadvantages of GEMMs include their high cost, relatively long time to tumor onset, and use of genetic alterations that may not exactly mimic the heterogeneity of the individual patient’s disease. These models also lack the genetic heterogeneity typified by most human tumors (38). Furthermore, relatively large treatment groups are typically needed for GEMM experiments owing to the degree of variability among tumors. GEMMs have shown utility in co-clinical trials of immunotherapy of pancreatic cancer (46) and non–small cell lung cancer (47). However, most GEMMs have low mutational load, which can pose a challenge for immunotherapy studies that require tumors to express neoantigens to engender an immune response. This challenge has recently been overcome by combining exposure to the carcinogen 3-methylcholanthrene with Cre-mediated p53 knockout in p53fl/fl mice to generate a relatively high mutational load soft tissue sarcoma (48).
Cell Transplant Models of Cancer
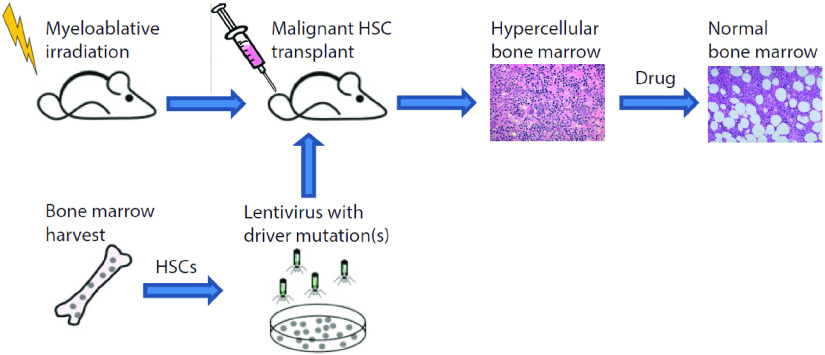
Cell transplant models (CTMs) represent a distinct subset of GEMMs applied primarily to studies of hematologic cancers, such as myeloproliferative neoplasms and leukemia that arise from mutations arising in hematopoietic stem cells (HSCs) and progenitor cells. To generate a CTM model of cancer, investigators isolate bone marrow from a donor animal and transduce enriched HSCs or the total population of bone marrow cells with recombinant retroviruses or lentiviruses expressing critical oncogenes for a target disease (49). These types of viral vectors integrate into the genome of cells, ensuring stable transmission of key oncogenic mutations from HSCs to more differentiated hematopoietic cells (Figure 3). Retroviral and lentiviral vectors for CTMs commonly include a coexpressed fluorescent protein or other reporter molecules to facilitate detection of transduced cells ex vivo and in recipient mice. After conditioning with whole-body irradiation or high-dose chemotherapy to ablate endogenous HSCs, investigators transplant transduced HSCs intravenously into recipient mice which hone to bone marrow niches through signaling pathways analogous to stem cell transplants in humans. The hematopoietic system is subsequently reconstituted with malignant progenitor and differentiated cell lineages over several weeks. Recipient mice progressively develop features of disease that recapitulate key pathologies evident in patients, including increased cellularity in bone marrow (hypercellular marrow), splenomegaly, hepatomegaly, and/or inflammatory constitutional symptoms.
Figure 3.
Generation of CTMs for use in co-clinical trials. Investigators isolate bone marrow from a donor animal and transduce enriched hematopoietic stem cells (HSCs) or the total population of bone marrow cells with recombinant retroviruses or lentiviruses expressing critical oncogenes for a target disease. These types of viral vectors integrate into the genome of cells, ensuring stable expression of key oncogenic mutations in HSCs and transmission of mutations to more differentiated hematopoietic cells.
CTMs typically use syngeneic (genetically identical), immunocompetent murine models for both donors and recipients of HSC transplants, although studies have successfully transduced human HSCs with a driver oncogene and established xenograft models of hematologic cancer in immunocompromised mice. Using syngeneic mice avoids complications secondary to mismatch of donor and recipient, including graft-versus-host and graft rejection. CTMs offer several advantages in the context of co-clinical trials:
(1) generating cohorts of mice that match frequencies of driver mutations present in patients receiving the same treatment;
(2) studying disease progression and response to therapy in mice with a full range of hematopoietic cells (syngeneic all murine models);
(3) faithfully maintaining underlying genetic basis of disease present in patients; and
(4) rapidly producing large numbers of mice for treatment studies, which minimizes delays in assessing effects of therapy in studies with sufficient animals for high statistical power and rigorous validation (eg, histology).
Limitations of the model include potential adverse effects of myeloablative conditioning regimens used to facilitate engraftment of transplanted HSCs in recipient animals and accelerated course of disease relative to patients.
Co-clinical Imaging Study Design, Instruments, and Standardization
Standardization of clinical quantitative imaging (QI) has been realized to a large extent by numerous initiatives, such as the Quantitative Biomarker Alliance and NCI’s Quantitative Imaging Network to implement advanced QI methods in clinical practice (50–53). While these and other initiatives have had a great impact in advancing clinical applications of QI, preclinical imaging remains a critical component in the translational pipeline of validating QI methods and imaging agents for applications in drug discovery, cancer detection, and response to therapy assessment. An inherent challenge in preclinical imaging is lack of standardization in terms of study design and animal logistics, image acquisition, and analysis and instrument quality control/assurance. While generally applicable to other modalities, in the ensuing sections, these considerations will be discussed in relation to co-clinical magnetic resonance imaging (MRI), computed tomography (CT), and positron emission tomography (PET) in the context of co-clinical trials.
Co-clinical Imaging Study Design and Animal Logistics
Small animals are noncompliant subjects and as such the vast majority of small-animal imaging studies use general anesthesia, typically isoflurane in oxygen. As mice are not able to maintain their core body temperature while under general anesthesia, it is necessary to use vital signs’ monitoring in combination with active maintenance of core body temperature for preclinical studies. Many quantitative imaging parameters are temperature-dependent; therefore, it is critical that the animal achieves a stable core body temperature and physiological state before initiation of any quantitative imaging studies (54, 55). Numerous other factors involved in the setup for preclinical imaging have been documented to impact imaging parameters, including anesthesia, animal handling, and diet (duration of fasting), among other factors (54–61). Parameters related to animal husbandry, including housing conditions, acclimation, chow, strain of animal, and physiological stress, may also impact the outcome of imaging studies (56). Imaging studies typically performed during day times, disrupt the animal’s circadian rhythms which modifies disease metabolism in some cases (62, 63). An important consideration in multicenter preclinical trials is institutional variability in housing. A recent analysis of data derived from Mouse Metabolic Phenotyping Centers suggests that the location (and corresponding institution) at which a study was performed contributed to differences in energy expenditure in rodents, even when the same diet was used across institutions (64). Thus, institutional differences in animal housing may also impact preclinical imaging studies. To enhance the reproducibility and the translational impact of preclinical imaging studies, these factors need to be considered and recorded to facilitate interpretation of co-clinical trials.
MRI
MRI provides superb soft tissue contrast that can be manipulated by suitable adjustment of the acquisition parameters. Image contrast can be sensitized to several properties of tissue water including nuclear relaxation rate, water diffusion, blood flow, or perfusion and chemical exchange. These properties can be quantitatively measured by magnetic resonance, resulting in numerous biometric markers of disease state (65). Typically, an MRI examination includes generation of a series of images with different contrast weighting, thereby facilitating multiparametric analysis (66–68) and improved specificity relative to single-parameter imaging techniques. MRI may also be combined with injectable contrast agents to provide additional information. The reader is referred to numerous treaties describing MRI methodologies for additional details (69–71). Although a majority of MRI techniques available clinically may be applied in preclinical studies, there are a number of significant differences between the available tools and methodologies that pose challenges to the development of co-clinical studies (65).
Motion Control in Co-clinical MRI.
In the clinic, physiological motion must be addressed when imaging certain regions of the anatomy. Various techniques, including breath-holding, parallel acquisition (72), and fast acquisition methods such as echo planar imaging (73, 74) and fast imaging with steady-state precession (75), are often used alone or in combination to effectively freeze the motion. Lightly anesthetized mice have physiological motion rates that are an order of magnitude greater than those of humans (heart rates in the range of 400–600 bpm; respiratory rates of 60–90 breaths/min). The methods used in the clinic to address physiological motion are either not available or not fast enough to suppress motion artifacts in small-animal studies. It is therefore necessary to use other means of motion correction in preclinical studies of the abdomen or thorax. Prospective gating (ECG and/or respiration) (76–78), retrospective gating (79, 80), navigator echoes (81–83), and “self-gated” methods (84–86) are commonly used to minimize motion artifacts in preclinical studies. These methods are often used in combination with sampling schemes that have reduced sensitivity to motion including radial (87) and spiral (88, 89) k-space trajectories.
Differences in Preclinical and Clinical MR Instrumentation.
There currently is a trend among preclinical instrument vendors to produce static fields (B0) that mimic those used in the clinic (1.5, 3, and 7 T). However, the B0 used in preclinical systems varies over a broader range (1–21 T) and is typically higher than those used in the clinical setting. The push to higher fields is driven by the fact that the MRI signal scales somewhere between linearly and the square of the B0 (70). The additional signal strength is necessary to offset the limited signal-to-noise ratio owing to the smaller voxel size required for preclinical imaging. However, the most commonly used MRI contrast parameters (relaxation times) are known to be B0-dependent. Both clinical and preclinical protocols must therefore be optimized for the target field strength. This poses many challenges when developing co-clinical trials that use significantly different field strengths. The higher B0 used in preclinical studies is also problematic for methods that are sensitive to magnetic susceptibility effects, as these effects also scale with B0. Gradient echo and echo planar imaging techniques often suffer geometric distortions and/or signal dropout owing to magnetic susceptibility effects. These effects make single-shot techniques extremely difficult at field strengths commonly used for preclinical studies.
The gradient system for any MRI scanner provides a magnetic field gradient that is within a few percent of linear over a prescribed spherical volume at the isocenter of the magnet. Nonlinearities in the gradient system outside of this volume lead to reproducible geometric distortions in images acquired in this region of space. On clinical scanners, the gradient nonlinearities are fully characterized during installation and suitable nonlinear deformations are applied to acquired data in order to correct for these effects, including deformation corrections for apparent diffusion coefficient maps (90). These corrections are generally not available on preclinical scanners. It is therefore critical to assure that the volume of interest is limited to the linear volume of the gradient system when performing volumetric studies on preclinical instrumentation. Alternatively, the nonlinearities could be characterized and corrected as is done in the clinical case.
Calibration and QA.
The American College of Radiology (ACR) provides detailed requirements for accreditation of clinical sites (www.acraccreditation.org/modalities/). This includes detailed protocols for instrumentation calibration, QA tests and preventative maintenance. The ACR also routinely performs site visits to assure that facilities adhere to recommended best practices. No such accreditation exists for preclinical sites. Many of the best practices provided by the ACR could be extrapolated to preclinical sites. However, this is thwarted to some degree by the lack of standard phantoms to be used of QA tests. Though numerous phantoms have been described in the literature (91–93), there is a lack of consensus among preclinical MRI sites as to the optimal phantoms to be used for QA.
CT
CT is a fast and powerful anatomical x-ray-based imaging method in which contrast is based on x-ray attenuation which is dependent on the x-ray energy and the composition of the subject. A clinical CT system includes an x-ray tube and detector assembled in a gantry which rotates around the subject to create hundreds of radiographic projections. The projections are used by a reconstruction algorithm to produce (3D) tomographic data. The most used reconstruction algorithm for cone beam CT is filtered backprojection. The contrast is based on x-ray attenuation which is dependent on the x-ray energy and the composition of the subject. CT is the modality of choice for bone and lung imaging. If used with contrast agents, CT can provide perfusion information and/or cardiac function. One of the major limitations of x-ray CT imaging is exposure to radiation. More information on the basics of CT imaging is provided elsewhere (69–71, 94).
Differences in Preclinical and Clinical CT Instrumentation.
Preclinical CT also known as micro-CT is a volumetric imaging method based upon the same principles and components as clinical CT scanners, but delivering much higher spatial resolution (95). There are 2 possible system design geometries in micro-CT imaging:
(1) rotating gantry (tube and detector), and
(2) rotating specimen.
All of the current commercial systems for in vivo scanning use the rotating gantry geometry, that is, they are scaled versions of the clinical CT scanners. The micro-CT scanners with a rotating specimen geometry are mostly used for ex vivo imaging.
The higher resolution achievable with micro-CT is linked to the use of microfocus x-ray tubes with small focal spots (eg, 10 µm for 8 W power) and x-ray detectors with small voxel size (eg, 20 µm). The smaller voxels of micro-CT (relative to clinical CT) require much higher dose, as any voxel—independent of its size—needs to interact with a certain number of x-ray photons for adequate image quality (96, 97). If the photon noise (measured by its variance) is kept constant and the linear dimension of the voxel is reduced by 2 (ie, a reduction of voxel volume by 8), the dose must be increased by up to 16 times (98, 99). Thus, the acquisition protocols should be customized to minimize the x-ray dose but this comes usually as a penalty to image quality. X-ray radiation exposure can lead to biological damage and long-term health effects (100). The LD50/30 whole-body radiation dose in mice (the dose required to kill 50% of mice within 30 days) depends on many factors, but tends to be between 5 and 8 Gy (101, 102). The typical radiation dose for a single micro-CT scan can vary widely and reported values in the literature range from 0.017 Gy to 0.78 Gy (101). Rodents have the ability to repair damage from low doses of radiation (up to ∼0.3 Gy) over the course of several hours (103), so most low-dose micro-CT scans should have limited biological impact, even when the same animals are longitudinally scanned over the course of a study. But for higher-dose scans, longitudinal micro-CT imaging can potentially lead to a cumulative dose that could affect biological function (particularly immune function and tumor response) and long-term health (100). Therefore, careful consideration must be made to determine the optimal imaging protocol for each individual application to minimize the effects of radiation dose on the experiment.
For small-animal micro-CT imaging, the use of clinical contrast agents is particularly difficult. Mice have much higher renal clearance rates than humans, so injected contrast agents are rapidly excreted. To overcome the rapid clearance of traditional contrast agents, micro-CT can benefit from blood pool contrast agents, exhibiting prolonged blood residence time and stable enhancement from minutes to hours. Blood pool agents are made up of a wide variety of high-molecular-weight compounds or nanoparticles that avoid renal clearance owing to their large size. Their use for micro-CT imaging has been reviewed previously (104).
Motion Control in Co-clinical CT Imaging.
An important aspect of in vivo imaging with micro-CT when imaging the cardiopulmonary system is related to physiological gating (105). Unlike in clinical chest CT, which is performed in a single breath hold, preclinical projection data in micro-CT must be acquired over many breaths, requiring respiratory gating. However, both respiratory and cardiac motion can lead to artifacts and blurry appearance in reconstructed images. To compensate, gating approaches have been developed to synchronize the projection acquisition with physiological motion, ensuring that all projection images are acquired during the same phase of motion. There are 3 types of gating strategies: prospective, retrospective, and image-based gating (106). Prospective gating is being used in scanners that operate under step-and-shoot mode, in which after the gantry rotates to any new angle, the x-ray tube waits for a trigger signal before acquiring the next projection image. The trigger is based on a signal provided by a pneumatic cushion positioned on the animal’s diaphragm or an optical measurement (107). Typically, only 1 projection is acquired at each projection angle (108). However, it is also possible to acquire multiple projections (called frames) at each angle, and then sum the frames into a single projection (ie, multiframe acquisitions) to improve signal-to-noise ratio. Retrospective (109) and image-based methods of gating have also been for micro-CT (110).
Calibration and QA.
To ensure that a CT system performs well, it is important to assess the image quality and dose routinely using phantoms (111) especially as quantitative CT image features are widely being investigated in radiomics for tissue phenotype characterization. In the animal imaging space, 1 example of a comercially available micro-CT phantom consists of 6 separate modular sections (resolution coils, slanted edge, geometric accuracy, CT number evaluation, linearity, and uniformity and noise), each designed to evaluate 1 aspect of image quality (112). This phantom has been used for performance evaluation for various scan protocols with micro-CT system (113, 114). Other custom phantoms to assess specific imaging tasks have been also reported, for example, a phantom to assess the the voxel scaling accuracy and it has been tested for a variety of micro-CT scanners covering a range of image resolutions (115). Some anatomically correct simulated phantoms have been introduced (116) (117). Moreover, a 4D digital mouse phantom (MOBY) exists, providing not only anatomical detail but also realistic motion owing to the cardiac and respiratory cycles (118).
PET
Over the years, numerous reports have highlighted considerations in small-animal PET imaging (54–56). Unfortunately, despite some progress, there are still gaps in standardization of small-animal imaging protocols and QI methods to produce consistent results as highlighted by recent works (119, 120). Importantly, in line with the theme of this communication, there is a need to harmonize preclinical and clinical PET QI pipelines so as to enhance the translational impact of developments in PET imaging. To harmonize clinical and small-animal PET images, instrumentation and software factors affecting spatial resolution and scanner sensitivity should be considered.
Factors Affecting Spatial Resolution and Sensitivity in PET.
PET physics dictates that photon nonacolinearity and positron range negatively contribute to the spatial resolution a system can achieve (121). Photon noncolinearity effect on spatial resolution is proportional to scanner radius and thus will be of importance in human imaging system only. 18F is the most widely used nuclide in PET imaging. The positron from this nuclide has a maximum energy of 0.63 MeV and its range contributes to a loss of 0.5 mm in spatial resolution (121). This value is negligible compared with other factors in human imaging but has a small impact in small-animal PET imaging. From a camera design standpoint, the crystal size ultimately determines the intrinsic spatial resolution, and the detector technology has constantly evolved over the past few decades with progressively smaller crystals, from 6 mm in the 1990’s to ∼3 mm nowadays. Absolute system sensitivity depends of the scanner diameter, axial field of view (FOV), and crystal thickness. Ultimately, the performance of a system is a compromise between sensitivity and resolution, not a simple choice for most applications.
System Design—Clinical vs Preclinical.
The current generation of clinical PET/CT scanners are designed for whole-body imaging and thus have a diameter of ∼80 cm, allowing for a patient port of ∼70 cm, and are composed of a cylindrical configuration of PET detectors. At such radius, high sensitivity is achieved with more crystals, either thicker (2–3 cm) for increased detection efficiency and/or with the use of longer-axis scanner. The current generation of PET scanner will typically use a crystal size of ∼4-mm, having a thickness of 20–30 mm and an axial FOV of ∼25 cm. Recent camera designs such as the Siemens Biograph Vision pushes this limit with the use of 3.2-mm crystals and 26 cm axial FOV. The absolute sensitivity of clinical PET/CT systems ranges from ∼8 to 23 cps/KBq (0.8%–2.3%) with best spatial resolution at ∼4–6 mm of full-width half-max at the central portion of the FOV. Small-animal systems, on the other hand, can achieve better resolution, in part, because of the smaller diameter making the photon nonacolinearity a nonfactor, but mostly from the use of smaller crystal size. State-of-the-art systems use ∼1-mm crystal or achieve ∼1-mm spatial sampling and typically claim to less than ∼1-mm spatial resolution with iterative image reconstruction. Because the camera radius can be kept small, mouse sensitivity of 4%–8% is typically achieved. Those values are reported for typically wide energy acceptance windows of 350–650 keV or even 250–750 keV. This acceptance energy window is much wider than that in the clinical setting. In mice, the scatter fraction is small, at least smaller than in human setting. The positron range is an additional remaining factor not to be neglected in small-animal imaging and it is unlikely that further progress can be made in small-animal PET without consideration for positron range even when imaging with 18F.
Image Reconstruction.
Most systems use statistical-iterative image reconstruction and implement the 3D-OSEM (ordered subset expectation-maximization) algorithm (122), which is based on the maximum likelihood (ML-EM) algorithm (123) with a subdivision of the projection views into subsets for accelerated image reconstruction. Commonly all manufacturers will implement point spread function modeling that has the effect of improving spatial resolution and reducing imaging noise. In the clinical arena, point spread function modeling (124) is now commonly available and time of flight image reconstruction is available for most systems. In the latter, the critical parameter is the coincidence timing resolution that most systems achieve in <400 ps (125–127). The time of flight image reconstruction brings the benefit of improved signal to noise. In light of differences in system design, clinical and preclinical systems offer widely different performance levels in terms of spatial resolution and typically, small-animal PET will have a 2- to 4-fold improved system sensitivity over clinical PET systems. However, the imaging scales of the subjects to be imaged are widely different. In terms of resolution-to-scale, clinical systems have a significant advantage over preclinical systems.
Co-clinical Radiotracer Considerations.
Factors related to radiotracers that could potentially affect the harmony of molecular imaging co-clinical trials are numerous and worthy of a more in-depth discussion than space allows here. However, it is worth noting a number of issues that dovetail with current trends in tracer development for cancer-specific imaging. First, specific tracer retention mechanisms should ideally be identical, spanning model system to human. Whether tracers are aimed at probing classic ligand–receptor interactions or targeting enzymes that might portend intracellular concentration, a first assumption is that similar mechanisms are in place in both the model system and humans. Parallels between the 2 systems, however, are complex. For example, the biochemical rates that might enzymatically concentrate a radiotracer within cells may be quite different between humans and models and result in different trapping rates. Even more critical, the specific targets of radiotracers might not be expressed to the same level, or even at all, in both species. Because targets of radiotracers may be expressed at different levels (concentrations), of particular consideration are mass effects and specific activity considerations (128) which may impact quantitative imaging measurement in co-clinical trial settings. Moreover, immune-oncology provides the latest examples where species-specific selectivity renders certain radiotracers active in one system but agnostic in others. Described elsewhere in this publication are humanized mouse model systems that aim to address the conundrum of species selectivity. Other factors that must be considered but are not necessarily insurmountable include species-specific physiology and metabolism. However, sophisticated imaging and parallel biochemical analyses can help in normalizing differences across species and scale. Imaging protocols can also be developed that minimize the effect of diet on results, which could be of particular importance with respect to cancer metabolism.
Phantoms for Calibration and QA
Clinical phantoms.
The ACR (American College of Radiology) has developed a widely accepted phantom used for quality control and site qualification for clinical trials in PET/CT (129). The phantom consists of specially designed top flange to the widely adopted Jaszczak phantom (Peter Esser flange). The contains 4 fillable cylinders, 25 mm in length, 8, 12, 16, and 25 mm in diameter (hot lesions), in addition to 2 additional fillable 25-mm-diameter cylinders (air and nonradioactive water), and a solid polytetrafluoroethylene (PTFE) cylinder (mimicking bones) are included. A cold-rod section is inserted at the other end of the phantom to provide a means to estimate the scanner spatial resolution through visual inspection. The phantom is typically prepared in a protocol trying to emulate a clinical FDG PET imaging scenario to define the activity levels in the hot lesions and background area. A ratio of 4:1 of activity concentration in the hot lesions relative to background is typically chosen. The phantom is then imaged as per the site clinical imaging protocol in use for FDG oncology PET/CT patients. At this set activity ratio and typical scan time, and standard image reconstruction algorithm and parameter set for clinical applications, the smallest hot lesion would be typically barely visible. Maximum values in the hot cylinders provide a measurement of count recovery as a function of object size, but these values are used for only site qualification and not for scanner performance. The uniform water section of the phantom allows the measurement of absolute scanner quantification accuracy, in plane uniformity and across plane uniformity (at least for a few centimeters of its axial section). This phantom was designed to allow easy preparation and to allow the measurement of a number of parameters useful to compare imaging performance of a scanner (and the chosen reconstruction) for the clinical task of clinical oncological FDG-PET imaging in the setting of clinical trials.
Other phantoms have been reported to investigate the dependence of PET image bias on CT-based attenuation correction (130). In the clinical setting, the NEMA IEC phantom is used routinely (131). The NEMA IEC phantom is composed of a hollow chamber, is water-fillable, and contains 6 fillable spheres (10–37 mm) in its midsection. Cylindrical insert (50-mm-diameter) filled with a mixture of Styrofoam beads and water is inserted in the phantom to represent lung material. This phantom is typically used for acceptance testing according to NEMA NU-2 standard and EANM/EARL accreditation (132). Typical filling procedure consists of a 10:1 ratio between the sphere and background at total activity commensurate to standard 18F-FDG oncologic FDG PET/CT applications. This phantom allows measuring size-dependent (contrast)-recovery curve, background variability, absolute scanner calibration, and scattering correction accuracy. More recently the SNMMI-CTN PET phantom was developed to validate scanners at sites that wish to participate in oncology clinical trials (133). The CTN oncology clinical simulator phantom is an anthropomorphic chest phantom with lung fields and 6 spherical objects with inner diameters ranging from 7 to 20 mm reproducibly secured at specific locations within the phantom. This phantom allows the measurement of contrast recovery curves, and their reproducibility, for realistic lesion and operational clinical image reconstruction settings.
Preclinical phantoms.
For small-animal scanners, the NEMA NU-4 2008 proposes a mouse size image quality phantom that has been used to compare preclinical PET imaging systems (134). The phantom consists of a Lucite cylinder with, at one end, a 5 fillable rod pattern with diameter 1–5 mm for count recovery measurement, and at the other end, 2 small 8-mm-diameter lung inserts to evaluate scatter correction efficiency. A fillable water section in the middle allows the measurement of in-plane uniformity. The design was chosen to allow for a robust and easy-to-fill phantom and was initially designed for the purpose of scanner comparison. The fillable hot rod simplifies construction and avoids the problem of spheres with a cold wall. The hot rod-like lesions were chosen to be of diameter commensurate to organ sizes in mice; however, these are cylindrical in shape, not spherical. The recovery values are to be expected to be larger in rod-like objects relative to sphere-like objects. Importantly, the rod-like lesion sizes are of appropriate size to challenge most small-animal PET imaging systems and thus one can evaluate the quantitative performance for imaging small objects with this phantom for the purpose of comparing animal scanners. Preclinical phantoms play a critical role in harmonizing preclinical instruments across multiple sites (120).
Precision and Accuracy In Quantitative Imaging
There are a variety of considerations that must be made to acquire preclinical imaging data that best serve a study. Although the endpoints and acquisition goals will vary by study, all imaging data sets should be analyzed with a functional understanding of sources of variability and uncertainty in the data. The National Institutes of Standards and Technology cites 3 underlying sources of uncertainty in the clinical study and implementation of quantitative imaging biomarkers (135). The first is variability caused by the devices used to capture images, or instrumentation variance. The second is variability in image interpretation by clinicians/technicians, or reader variance. The third is variability owing to intrinsic properties of the biology, or biological variance. These uncertainties exist in both preclinical and clinical imaging domains, highlighting the need to define the appropriate methods by which data are measured, interpreted, and validated (136). Unfortunately, the deficit in standardized metrics for preclinical imaging and analysis is even greater than that faced by clinical imaging scientists and technicians. Although certain challenges are specific to modality, a general foundation for defining and assessing the utility of imaging biomarkers is assessing their reproducibility and repeatability.
Numerous methods are used to assess reproducibility of image metrics including Lin’s concordance correlation coefficient (137) and Bland–Altman analysis (BA) (138). The Lin’s concordance correlation coefficient, is the product of the Pearson correlation coefficient and the bias correction factor and accounts for both precision and accuracy. The method outlined by Watson and Petrie (139) is typically used to calculate these metrics. The procedure used to calculate the statistical parameters for the BA plots are summarized by Galbraith (140) and Raunig (141). To assess reproducibility between image metrics derived from consecutive days, it is important to test that the “day 1” vs “day 2” absolute differences are independent of the means using Kendall tau test for correlation (140). Let Δ denote the within-mouse difference between the measurements, and N denote the number of paired measurements. The standard deviation for the mean difference is calculated using the following equation:
and the within-subject standard deviation (wSD), using the following equation:
The 95% confidence limits in the BA plots are the limits of agreement defined as the mean difference ± the repeatability coefficient (RC).
These limits are independent of the sample size so that the results from an individual test–retest experiment is expected to fall within these boundaries 95% of the time. Guidelines for the implementation of these techniques in evaluating QI biomarkers (142) and for improved precision in multicenter trials has been reported recently (143). Importantly, there have been numerous applications of these techniques in biomedical imaging in both preclinical (55, 144–148) and clinical (149–153) settings.
Correlative Biology in Validation of QI Biomarkers
The value proposition of medical imaging is that it can interrogate human biology in vivo, noninvasively, spatially, or longitudinally, and thus provides diagnostic, predictive, and therapeutic insights to manage patient outcome. In addition to validating the precision and accuracy of QI imaging metrics (as outlined in Precision and Accuracy of Quantitative Imaging section), ideally QI metrics for a given biomarker need to be validated against the underlying biology. A “biomarker” is defined as a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or responses to a therapeutic intervention (154). Thus, an imaging biomarker is an objective QI metric derived from an in vivo image for a given biomarker. Traditionally, clinical QI biomarkers have been validated by statistical analyses against measures of outcome. More often, tissue biopsies are available and can be used to correlate in vivo QI metrics to measures pathology or OMICS (among others) measures. In contrast to biopsies, in vivo images can provide information about the spatial heterogeneity of the whole tumor, albeit at lower resolution. With the advent of radiomics (155, 156), there is an underlying effort to validate image features against pathological or genomic features of tumor heterogeneity derived from biopsies (157–165). However, there are numerous complicating nuances in correlating a QI metric derived from a clinical image to features derived from a biopsy; these nuances include mismatches in scale, discrepancies in coregistration, and importantly single point (needle) biopsies may not reflect the pathobiology of the whole tumor. The use of co-clinical models thus enables correlation and validation QI metrics against the underlying heterogeneous biology of co-clinical human tumor models when human specimen is scarce.
Informatics Needs to Support Co-clinical Research
The preclinical imaging workflow is somewhat complex, routinely using multiple instruments to characterize the physiology and biology of a given tumor in vivo. To validate in vivo imaging measurements, in vitro or ex vivo multiscale assays such as pathology, -OMICS (genomics, proteomics, metabolomics, etc.), immunohistochemistry, multiplexed immunofluorescence are used to correlate quantitative image–derived measurements to ex vivo measures. When multiplied by the number of subjects (animals) per group in a given experiment, multitudes of interventions (eg, drugs), descriptive data (weight, diet, tumor volume, blood, metabolic panel data, etc.) and the number of time-points in a longitudinal imaging protocol, the resulting data sets are vast and prohibitive to track and manage long term. As a consequence, nontractable data result in poor reproducibility and present obstacles for open science collaboration and data mining.
To that end, informatics solutions are needed to support co-clinical imaging research including collection of metadata to qualify co-clinical imaging studies. For example, a recent guideline on the use of PDX in preclinical research (29), lists ∼45 fields (metadata) to capture to qualify research results. Importantly, there is a need for harmonization and integration of preclinical cancer imaging data, imaging acquisition protocols, and annotation. Legacy preclinical imaging databases are not equipped to support big data science and collection of metadata/annotations to support NCI’s precision medicine initiative. Although some institutions have developed databases to house preclinical imaging data, many such legacy databases are not compatible with the complexity and growing demands in preclinical cancer imaging which include big data needs and collection of metadata/annotation to support NCI’s precision medicine initiative. Importantly, the increasing prevalence of quantitative acquisition and analysis approaches depend on sophisticated computational methods that generate additional derived data. Given these “big data” challenges, informatics tools are needed that have the capacity to organize data structures, enforce QA practices, generate audit trails and provenance records, provide detailed reports and data tracking tools, and ultimately facilitate data analysis.
Lack of reproducibility in preclinical cancer research, including imaging, has been highlighted by numerous publications (119, 166). Other than promoting open science, data sharing has been suggested as one solution to address reproducibility. Similarly, sharing of quantitative imaging pipelines is expected to enhance reproducibility, as it will allow for testing of multiple analytic pipelines using a common data set for comparison and validation. The NCI has chosen to establish an open environment in which the oncology community can collaborate to tackle the sundry issues that pertain to reproducibility of animal model research as required for precision medicine. Prominent among those issues is transparency of details that document imaging experiments and their application to translational research. Thus, informatics tools and platforms are needed to enhance reproducibility in preclinical imaging, enable data mining with collection of metadata and annotations tools, and promote open science.
Summary
Advances in clinical QI have been realized to a large extent by numerous initiatives such as the Quantitative Imaging Network and the Quantitative Imaging Biomarker Alliance to standardize and implement advanced QI methods in clinical practice. Although these and other initiatives have had a significant impact in advancing clinical applications of QI, preclinical imaging plays a critical role in developing in vivo translational imaging strategies to interrogate disease mechanisms, detect disease, and asses/predict response to therapy. The use of co-clinical animal models of cancer ushers-in new paradigms involving co-clinical trials where biological and molecular mechanisms of disease as well as therapeutic strategies can be investigated in relevant human cancer models in parallel with clinical trials to support translational imaging investigations. In this context, the NCI’s precision medicine initiative emphasizes the biological and molecular bases for cancer prevention and treatment, as well as consistency/harmonization in preclinical and clinical research, including QI. The CIRP, therefore, was organized to devise best practices for co-clinical imaging and to develop optimized state-of-the-art translational quantitative imaging methodologies to enable disease detection, risk stratification, and assessment/prediction of response to therapy. It is expected that the quest for best practices will neither result in reduced creativity nor hamper progress in preclinical imaging science, as some may conjecture. Rather such creativity should be viewed as investment towards progress in translational imaging and its role in guiding precision medicine into the next decade.
Acknowledgments
The research agenda outlined in this communication is supported by the Washington University Co-Clinical Imaging Research Resource, NCI Grant # U24CA209837 and Siteman Cancer Center Support Grant P30CA091842; the Duke Preclinical Research Resources for Quantitative Imaging Biomarkers, NCI Grant # U24CA220245; Integrating OMICS and Quantitative Imaging Data in Co-Clinical Trials to Predict Treatment Response in Triple Negative Breast Cancer, NCI Grant # U24CA226110 and CPRIT RR160005 (T.E.Y. is a CPRIT Scholar of Cancer Research); University of Michigan Quantitative Imaging Research, NCI Grant # U24CA237683; Vanderbilt University-PET imaging Resource to Enhance Delivery of Individualized Cancer Therapeutics, NCI Grant # U24CA220325; and PENN Quantitative MRI Resource for Pancreatic Cancer, NCI Grant # U24CA231858.
Footnotes
- CIRP
- Co-Clinical Imaging Research Resource Program
- GEMM
- genetically engineered mouse models
- CTM
- cell transplant models
- PDX
- patient-derived tumor xenograft
- FLP, flippase; QA
- quality assurance
- SC
- steering committee
- WG
- working group
- HSCs
- hematopoietic stem cells
- QI
- quantitative imaging
- NCI
- National Cancer Institute
- MRI
- magnetic resonance imaging
- CT
- computed tomography
- PET
- positron emission tomography
- ACR
- American College of Radiology
- FOV
- field of view
- BA
- Bland–Altman analysis
References
- 1.Chen Z, Akbay E, Mikse O, Tupper T, Cheng K, Wang Y, Tan X, Altabef A, Woo S-A, Chen L, Reibel JB, Janne PA, Sharpless NE, Engelman JA, Shapiro GI, Kung AL, Wong K-K. Co-clinical trials demonstrate superiority of crizotinib to chemotherapy in ALK-rearranged non-small cell lung cancer and predict strategies to overcome resistance. Clin Cancer Res. 2014;20:1204–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim HR, Kang HN, Shim HS, Kim EY, Kim J, Kim DJ, Lee JG, Lee CY, Hong MH, Kim SM, Kim H, Pyo KH, Yun MR, Park HJ, Han JY, Youn HA, Ahn MJ, Paik S, Kim TM, Cho BC. Co-clinical trials demonstrate predictive biomarkers for dovitinib, an FGFR inhibitor, in lung squamous cell carcinoma. Ann Oncol. 2017;28:1250–1259. [DOI] [PubMed] [Google Scholar]
- 3.Kwong LN, Boland GM, Frederick DT, Helms TL, Akid AT, Miller JP, Jiang S, Cooper ZA, Song X, Seth S, Kamara J, Protopopov A, Mills GB, Flaherty KT, Wargo JA, Chin L. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J Clin Invest. 2015;125:1459–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lunardi A, Ala U, Epping MT, Salmena L, Clohessy JG, Webster KA, Wang G, Mazzucchelli R, Bianconi M, Stack EC, Lis R, Patnaik A, Cantley LC, Bubley G, Cordon-Cardo C, Gerald WL, Montironi R, Signoretti S, Loda M, Nardella C, Pandolfi PP. A co-clinical approach identifies mechanisms and potential therapies for androgen deprivation resistance in prostate cancer. Nat Genet. 2013;45:747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishino M, Sacher AG, Gandhi L, Chen Z, Akbay E, Fedorov A, Westin CF, Hatabu H, Johnson BE, Hammerman P, Wong KK. Co-clinical quantitative tumor volume imaging in ALK-rearranged NSCLC treated with crizotinib. Eur J Radiol. 2017;88:15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owonikoko TK, Zhang G, Kim HS, Stinson RM, Bechara R, Zhang C, Chen Z, Saba NF, Pakkala S, Pillai R, Deng X, Sun S-Y, Rossi MR, Sica GL, Ramalingam SS, Khuri FR. Patient-derived xenografts faithfully replicated clinical outcome in a phase II co-clinical trial of arsenic trioxide in relapsed small cell lung cancer. J Transl Med. 2016;14:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sia D, Moeini A, Labgaa I, Villanueva A. The future of patient-derived tumor xenografts in cancer treatment. Pharmacogenomics. 2015;16:1671–1683. [DOI] [PubMed] [Google Scholar]
- 8.Cho SY, Kang W, Han JY, Min S, Kang J, Lee A, Kwon JY, Lee C, Park H. An integrative approach to precision cancer medicine using patient-derived xenografts. Mol Cells. 2016;39:77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clohessy JG, Pandolfi PP. Mouse hospital and co-clinical trial project—from bench to bedside. Nat Rev Clin Oncol. 2015;12:491–498. [DOI] [PubMed] [Google Scholar]
- 10.Sulaiman A, Wang L. Bridging the divide: preclinical research discrepancies between triple-negative breast cancer cell lines and patient tumors. Oncotarget. 2017;8:113269–113281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, Zhang C, Schnell C, Yang G, Zhang Y, Balbin OA, Barbe S, Cai H, Casey F, Chatterjee S, Chiang DY, Chuai S, Cogan SM, Collins SD, Dammassa E, Ebel N, Embry M, Green J, Kauffmann A, Kowal C, Leary RJ, Lehar J, Liang Y, Loo A, Lorenzana E, Robert McDonald E, McLaughlin ME, Merkin J, Meyer R, Naylor TL, Patawaran M, Reddy A, Röelli C, Ruddy DA, Salangsang F, Santacroce F, Singh AP, Tang Y, Tinetto W, Tobler S, Velazquez R, Venkatesan K, Von Arx F, Wang HQ, Wang Z, Wiesmann M, Wyss D, Xu F, Bitter H, Atadja P, Lees E, Hofmann F, Li E, Keen N, Cozens R, Jensen MR, Pryer NK, Williams JA, Sellers WR. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21:1318–1325. [DOI] [PubMed] [Google Scholar]
- 12.DeRose YS, Wang G, Lin Y-C, Bernard PS, Buys SS, Ebbert MTW, Factor R, Matsen C, Milash BA, Nelson E, Neumayer L, Randall RL, Stijleman IJ, Welm BE, Welm AL. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao X, Liu Z, Yu L, Zhang Y, Baxter P, Voicu H, Gurusiddappa S, Luan J, Su JM, Leung HC. E, Li XN. Global gene expression profiling confirms the molecular fidelity of primary tumor-based orthotopic xenograft mouse models of medulloblastoma. Neuro Oncol. 2012;14:574–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morton CL, Houghton PJ. Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc. 2007;2:247–250. [DOI] [PubMed] [Google Scholar]
- 15.Reyal F, Guyader C, Decraene C, Lucchesi C, Auger N, Assayag F, De Plater L, Gentien D, Poupon M-F, Cottu P, De Cremoux P, Gestraud P, Vincent-Salomon A, Fontaine J-J, Roman-Roman S, Delattre O, Decaudin D, Marangoni E. Molecular profiling of patient-derived breast cancer xenografts. Breast Cancer Res. 2012;14:R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krepler C, Xiao M, Spoesser K, Brafford PA, Shannan B, Beqiri M, Xu W, Garman B, Nathanson KL, Xu X, Karakousis G, Mills GB, Lu Y, Caponigro G, Boehm M, Peters M, Schuchter L, Herlyn M. Personalized pre-clinical trials in BRAF inhibitor resistant patient derived xenograft models identify second line combination therapies. Clin Cancer Res. 2016;22:1592–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerbel RS. Human tumor xenografts as predictive preclinical models for anticancer drug activity in humans: better than commonly perceived-but they can be improved. Cancer Biol Ther. 2003;2:S134–9. [PubMed] [Google Scholar]
- 18.Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S, Kalyandrug S, Christian M, Arbuck S, Hollingshead M, Sausville EA. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer. 2001;84:1424–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scholz CC, Berger DP, Winterhalter BR, Henss H, Fiebig HH. Correlation of drug response in patients and in the clonogenic assay with solid human tumour xenografts. Eur J Cancer. 1990;26:901–905. [DOI] [PubMed] [Google Scholar]
- 20.Allaway RJ, Fischer DA, de Abreu FB, Gardner TB, Gordon SR, Barth RJ, Colacchio TA, Wood M, Kacsoh BZ, Bouley SJ, Cui J, Hamilton J, Choi JA, Lange JT, Peterson JD, Padmanabhan V, Tomlinson CR, Tsongalis GJ, Suriawinata AA, Greene CS, Sanchez Y, Smith KD. Genomic characterization of patient-derived xenograft models established from fine needle aspirate biopsies of a primary pancreatic ductal adenocarcinoma and from patient-matched metastatic sites. Oncotarget. 2016;7:17087–17102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girotti MR, Gremel G, Lee R, Galvani E, Rothwell D, Viros A, Mandal AK, Lim KHJ, Saturno G, Furney SJ, Baenke F, Pedersen M, Rogan J, Swan J, Smith M, Fusi A, Oudit D, Dhomen N, Brady G, Lorigan P, Dive C, Marais R. Application of sequencing, liquid biopsies, and patient-derived xenografts for personalized medicine in melanoma. Cancer Discov. 2016;6:286–299. [DOI] [PubMed] [Google Scholar]
- 22.Huang S, Lira P, Wang K, Zhang C, Jackson-Fisher A, Ching K. Molecular profiling of AML patient derived xenograft models with deep sequencing using a 109 AML associated gene panel and a 409 gene comprehensive cancer panel. Cancer Res. 2015:75. [Google Scholar]
- 23.Qi L, Kogiso M, Du Y, Lindsay H, Zhang H, Zhao S, Braun FK, Shu Q, Yu L, Zhao X, Liu Z, Huang Y, Perlaky L, Su J, Baxter P, Adesina A, Parsons DW, Chintagumpala M, Li XN. A comprehensive panel of patient-derived orthotopic xenograft mouse models of malignant pediatric brain tumors. Neuro Oncol. 2016;18:139-. [Google Scholar]
- 24.Krepler C, Sproesser K, Brafford P, Beqiri M, Garman B, Xiao M, Shannan B, Watters A, Perego M, Zhang G, Vultur A, Yin X, Liu Q, Anastopoulos IN, Wubbenhorst B, Wilson MA, Xu W, Karakousis G, Feldman M, Xu X, Amaravadi R, Gangadhar TC, Elder DE, Haydu LE, Wargo JA, Davies MA, Lu Y, Mills GB, Frederick DT, Barzily-Rokni M, Flaherty KT, Hoon DS, Guarino M, Bennett JJ, Ryan RW, Petrelli NJ, Shields CL, Terai M, Sato T, Aplin AE, Roesch A, Darr D, Angus S, Kumar R, Halilovic E, Caponigro G, Jeay S, Wuerthner J, Walter A, Ocker M, Boxer MB, Schuchter L, Nathanson KL, Herlyn M. A comprehensive patient-derived xenograft collection representing the heterogeneity of melanoma. Cell Rep. 2017;21:1953–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romero-Calvo I, Weber CR, Ray M, Brown M, Kirby K, Nandi RK, Long TM, Sparrow SM, Ugolkov A, Qiang W, Zhang Y, Brunetti T, Kindler H, Segal JP, Rzhetsky A, Mazar AP, Buschmann MM, Weichselbaum R, Roggin K, White KP. Human organoids share structural and genetic features with primary pancreatic adenocarcinoma tumors. Mol Cancer Res. 2019;17:70–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okazawa Y, Mizukoshi K, Koyama Y, Okubo S, Komiyama H, Kojima Y, Goto M, Habu S, Hino O, Sakamoto K, Orimo A. High-sensitivity detection of micrometastases generated by GFP lentivirus-transduced organoids cultured from a patient-derived colon tumor. J Vis Exp. 2018;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beshiri ML, Tice CM, Tran C, Nguyen HM, Sowalsky AG, Agarwal S, Jansson KH, Yang Q, McGowen KM, Yin J, Alilin AN, Karzai FH, Dahut WL, Corey E, Kelly K. A PDX/Organoid Biobank of advanced prostate cancers captures genomic and phenotypic heterogeneity for disease modeling and therapeutic screening. Clin Cancer Res. 2018;24:4332–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang FF, Wang WJ, Long Y, Liu H, Cheng JJ, Guo L, Li R, Meng C, Yu S, Zhao Q, Lu S, Wang L, Wang H, Wen D. Characterization of drug responses of mini patient-derived xenografts in mice for predicting cancer patient clinical therapeutic response. Cancer Commun (Lond). 2018;38:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meehan TF, Conte N, Goldstein T, Inghirami G, Murakami MA, Brabetz S, Gu Z, Wiser JA, Dunn P, Begley DA, Krupke DM, Bertotti A, Bruna A, Brush MH, Byrne AT, Caldas C, Christie AL, Clark DA, Dowst H, Dry JR, Doroshow JH, Duchamp O, Evrard YA, Ferretti S, Frese KK, Goodwin NC, Greenawalt D, Haendel MA, Hermans E, Houghton PJ, Jonkers J, Kemper K, Khor TO, Lewis MT, Lloyd KCK, Mason J, Medico E, Neuhauser SB, Olson JM, Peeper DS, Rueda OM, Seong JK, Trusolino L, Vinolo E, Wechsler-Reya RJ, Weinstock DM, Welm A, Weroha SJ, Amant F, Pfister SM, Kool M, Parkinson H, Butte AJ, Bult CJ. PDX-MI: minimal information for patient-derived tumor xenograft models. Cancer Res. 2017;77:e62-e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khandelwal G, Girotti MR, Smowton C, Taylor S, Wirth C, Dynowski M, Frese KK, Brady G, Dive C, Marais R, Miller C. Next-generation sequencing analysis and algorithms for PDX and CDX models. Mol Cancer Res. 2017;15:1012–1016. [DOI] [PubMed] [Google Scholar]
- 31.Ben-David U, Ha G, Tseng Y-Y, Greenwald NF, Oh C, Shih J, McFarland JM, Wong B, Boehm JS, Beroukhim R, Golub TR. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49:1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rau RE. CMML/JMML PDXs: as easy as 1, 2, NSG-SGM3. Blood. 2017;130:385–386. [DOI] [PubMed] [Google Scholar]
- 33.Yoshimi A, Balasis ME, Vedder A, Feldman K, Ma Y, Zhang H, Lee SC-W, Letson C, Niyongere S, Lu SX, Ball M, Taylor J, Zhang Q, Zhao Y, Youssef S, Chung YR, Zhang XJ, Durham BH, Yang W, List AF, Loh ML, Klimek V, Berger MF, Stieglitz E, Padron E, Abdel-Wahab O. Robust patient-derived xenografts of MDS/MPN overlap syndromes capture the unique characteristics of CMML and JMML. Blood. 2017;130:397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brehm MA, Kenney LL, Wiles MV, Low BE, Tisch RM, Burzenski L, Mueller C, Greiner DL, Shultz LD. Lack of acute xenogeneic graft- versus-host disease, but retention of T-cell function following engraftment of human peripheral blood mononuclear cells in NSG mice deficient in MHC class I and II expression. FASEB J. 2019;33:3137–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, Saito Y, Marches F, Halene S, Palucka AK, Manz MG, Flavell RA. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32:364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver JL, Zadrozny LM, Gabrielson K, Semple KM, Shea KI, Howard KE. BLT-immune humanized mice as a model for nivolumab induced immune-mediated adverse events: comparison of the NOG and NOG-EXL strains. Toxicol Sci. 2019:169:194.–. [DOI] [PubMed] [Google Scholar]
- 37.Ito R, Takahashi T, Katano I, Kawai K, Kamisako T, Ogura T, Ida-Tanaka M, Suemizu H, Nunomura S, Ra C, Mori A, Aiso S, Ito M. Establishment of a human allergy model using human IL-3/GM-CSF-transgenic NOG mice. J Immunol. 2013;191:2890–2899. [DOI] [PubMed] [Google Scholar]
- 38.Richmond A, Su Y. Mouse xenograft models vs GEM models for human cancer therapeutics. Dis Model Mech. 2008;1:78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, He X, Liu S, Hoog J, Lu C, Ding L, Griffith OL, Miller C, Larson D, Fulton RS, Harrison M, Mooney T, McMichael JF, Luo J, Tao Y, Goncalves R, Schlosberg C, Hiken JF, Saied L, Sanchez C, Giuntoli T, Bumb C, Cooper C, Kitchens RT, Lin A, Phommaly C, Davies SR, Zhang J, Kavuri MS, McEachern D, Dong YY, Ma C, Pluard T, Naughton M, Bose R, Suresh R, McDowell R, Michel L, Aft R, Gillanders W, DeSchryver K, Wilson RK, Wang S, Mills GB, Gonzalez-Angulo A, Edwards JR, Maher C, Perou CM, Mardis ER, Ellis MJ. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4:1116–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kirsch DG. Using genetically engineered mice for radiation research. Radiat Res. 2011;176:275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee CL, Moding EJ, Huang X, Li Y, Woodlief LZ, Rodrigues RC, Ma Y, Kirsch DG. Generation of primary tumors with Flp recombinase in FRT-flanked p53 mice. Dis Mod Mech. 2012;5:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feil S, Valtcheva N, Feil R. Inducible Cre mice. Methods Mol Biol. 2009;530:343–363. [DOI] [PubMed] [Google Scholar]
- 43.Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, Nielsen GP, Quade BJ, Chaber CJ, Schultz CP, Takeuchi O, Bronson RT, Crowley D, Korsmeyer SJ, Yoon SS, Hornicek FJ, Weissleder R, Jacks T. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–997. [DOI] [PubMed] [Google Scholar]
- 44.Day CP, Merlino G, Van Dyke T. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell. 2015;163:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kersten K, Visser KE, Miltenburg MH, Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med. 2017;9:137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beatty GL, Chiorean EG, Fishman MP, Saboury B, Teitelbaum UR, Sun W, Huhn RD, Song W, Li D, Sharp LL, Torigian DA, O'Dwyer PJ, Vonderheide RH. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331:1612–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T, Liu Y, Tupper T, Ouyang J, Li J, Gao P, Woo MS, Xu C, Yanagita M, Altabef A, Wang S, Lee C, Nakada Y, Peña CG, Sun Y, Franchetti Y, Yao C, Saur A, Cameron MD, Nishino M, Hayes DN, Wilkerson MD, Roberts PJ, Lee CB, Bardeesy N, Butaney M, Chirieac LR, Costa DB, Jackman D, Sharpless NE, Castrillon DH, Demetri GD, Jänne PA, Pandolfi PP, Cantley LC, Kung AL, Engelman JA, Wong K-K. A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature. 2012;483:613–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee C-L, Mowery YM, Daniel AR, Zhang D, Sibley AB, Delaney JR, Wisdom AJ, Qin X, Wang X, Caraballo I, Gresham J, Luo L, Van Mater D, Owzar K, Kirsch DG. Mutational landscape in genetically engineered, carcinogen-induced, and radiation-induced mouse sarcoma. JCI Insight. 2019;4. pii: 128698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen TK, Morse SJ, Fleischman AG. Transduction-transplantation mouse model of myeloproliferative neoplasm. J Vis Exp. 2016;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mountz JM, Yankeelov TE, Rubin DL, Buatti JM, Erikson BJ, Fennessy FM, Gillies RJ, Huang W, Jacobs MA, Kinahan PE, Laymon CM, Linden HM, Mankoff DA, Schwartz LH, Shim H, Wahl RL. Letter to cancer center directors: progress in quantitative imaging as a means to predict and/or measure tumor response in cancer therapy trials. J Clin Oncol. 2014;32:2115–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yankeelov TE. The Quantitative Imaging Network: a decade of achievement. Tomography. 2019;5:A8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clarke LP, Nordstrom RJ, Zhang H, Tandon P, Zhang Y, Redmond G, Farahani K, Kelloff G, Henderson L, Shankar L, Deye J, Capala J, Jacobs P. The Quantitative Imaging Network: NCI’s historical perspective and planned goals. Transl Oncol. 2014;7:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nordstrom RJ. The Quantitative Imaging Network in precision medicine. Tomography. 2016;2:239–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tseng JR, Dandekar M, Subbarayan M, Cheng Z, Park JM, Louie S. Reproducibility of 3′-deoxy-3′-(18)F-fluorothymidine microPET studies in tumor xenografts in mice. J Nucl Med. 2005;46:1851–1857. [PMC free article] [PubMed] [Google Scholar]
- 55.Dandekar M, Tseng JR, Gambhir SS. Reproducibility of 18F-FDG microPET studies in mouse tumor xenografts. J Nucl Med. 2007;48:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fueger BJ, Czernin J, Hildebrandt I, Tran C, Halpern BS, Stout D, Phelps ME, Weber WA. Impact of animal handling on the results of 18F-FDG PET studies in mice. J Nucl Med. 2006;47:999–1006. [PubMed] [Google Scholar]
- 57.Hartley CJ, Reddy AK, Madala S, Michael LH, Entman ML, Taffet GE. Effects of isoflurane on coronary blood flow velocity in young, old and ApoE(-−/−) mice measured by Doppler ultrasound. Ultrasound Med Biol. 2007;33:512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hildebrandt IJ, Su H, Weber WA. Anesthesia and other considerations for in vivo imaging of small animals. ILAR J. 2008;49:17–26. [DOI] [PubMed] [Google Scholar]
- 59.Kober F, Iltis I, Cozzone PJ, Bernard M. Cine-MRI assessment of cardiac function in mice anesthetized with ketamine/xylazine and isoflurane. MAGMA. 2004;17:157–161. [DOI] [PubMed] [Google Scholar]
- 60.Kober F, Iltis I, Cozzone PJ, Bernard M. Myocardial blood flow mapping in mice using high-resolution spin labeling magnetic resonance imaging: influence of ketamine/xylazine and isoflurane anesthesia. Magn Reson Med. 2005;53:601–606. [DOI] [PubMed] [Google Scholar]
- 61.Zhang H, Qiao H, Frank R, Eucker S, Lu M, Huang B, et al. Endothelial progenitor cells mediated improvements in post-infarct left ventricular myocardial blood flow estimated by spin label CMR. Circulation. 2010;122:A20415. [Google Scholar]
- 62.Kress GJ, Liao F, Dimitry J, Cedeno MR, FitzGerald GA, Holtzman DM, Musiek ES. Regulation of amyloid-β dynamics and pathology by the circadian clock. J Exp Med. 2018;215:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang G, Chen L, Grant GR, Paschos G, Song W-L, Musiek ES, Lee V, McLoughlin SC, Grosser T, Cotsarelis G, FitzGerald GA. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med. 2016;8:324ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Corrigan JK, Ramachandran D, He Y, Palmer C, Jurczak Mj, Li B, et al. A big-data approach to understanding metabolic rate and response to obesity in laboratory mice. bioRxiv. 2019:839076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hormuth DA, Sorace AG, Virostko J, Abramson RG, Bhujwalla ZM, Enriquez‐Navas P, Gillies R, Hazle JD, Mason RP, Quarles CC, Weis JA, Whisenant JG, Xu J, Yankeelov TE. Translating preclinical MRI methods to clinical oncology. J Magn Reson Imaging. 2019;50:1377–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Y, Reynolds HM, Parameswaran B, Wraith D, Finnegan ME, Williams S, Haworth A. Multiparametric MRI and radiomics in prostate cancer: a review. Australas Phys Eng Sci Med. 2019;42:3–25. [DOI] [PubMed] [Google Scholar]
- 67.Luna A, Pahwa S, Bonini C, Alcala-Mata L, Wright KL, Gulani V. Multiparametric MR Imaging in abdominal malignancies. Magn Reson Imaging Clin N Am. 2016;24:157–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gurses B, Boge M, Altinmakas E, Balik E. Multiparametric MRI in rectal cancer. Diagn Interv Radiol. 2019;25:175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wehrli F, Shaw D, Kneeland J. Biomedical Magnetic Resonance Imaging: Principles, Methodology, and Applications. New York, NY: VCH Publishers; 1988. [Google Scholar]
- 70.Brown RW, Venkatesan R, Thompson MR, Haacke EM, Norman Cheng YC. Magnetic Resonance Imaging: Physical Principles and Sequence Design. NY, New York: Wiley; 1999. [Google Scholar]
- 71.Liang ZP. Principles of Magnetic Resonance Imaging: A Signal Processing Perspective. IEEE Press; 2000. [Google Scholar]
- 72.Deshmane A, Gulani V, Griswold MA, Seiberlich N. Parallel MR imaging. J Magn Reson Imaging. 2012;36:55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu O, Koroshetz WJ, Østergaard L, Buonanno FS, Copen WA, Gonzalez RG, Rordorf G, Rosen BR, Schwamm LH, Weisskoff RM, Sorensen AG. Predicting tissue outcome in acute human cerebral ischemia using combined diffusion- and perfusion-weighted MR imaging. Stroke. 2001;32:933–942. [DOI] [PubMed] [Google Scholar]
- 74.Porter DA, Heidemann RM. High resolution diffusion-weighted imaging using readout-segmented echo-planar imaging, parallel imaging and a two-dimensional navigator-based reacquisition. Magn Reson Med. 2009;62:468–475. [DOI] [PubMed] [Google Scholar]
- 75.Chavhan GB, Babyn PS, Jankharia BG, Cheng H-L, Shroff MM. Steady-state MR imaging sequences: physics, classification, and clinical applications. Radiographics. 2008;28:1147–1160. [DOI] [PubMed] [Google Scholar]
- 76.Wiesmann F, Ruff J, Hiller K-H, Rommel E, Haase A, Neubauer S. Developmental changes of cardiac function and mass assessed with MRI in neonatal, juvenile, and adult mice. Am J Physiol Heart Circ Physiol. 2000;278:H652–657. [DOI] [PubMed] [Google Scholar]
- 77.Zhou R, Pickup S, Glickson JD, Scott C, Ferrari VA. Assessment of global and regional myocardial function in the mouse using cine- and tagged MRI. Magn Reson Med. 2003;49:760–764. [DOI] [PubMed] [Google Scholar]
- 78.Zhou R, Pickup S, Yankeelov TE, Springer CS Jr., Glickson JD. Simultaneous measurement of arterial input function and tumor pharmacokinetics in mice by dynamic contrast enhanced imaging: effects of transcytolemmal water exchange. Magn Reson Med. 2004;52:248–257. [DOI] [PubMed] [Google Scholar]
- 79.Bishop J, Feintuch A, Bock NA, Nieman B, Dazai J, Davidson L, Henkelman RM. Retrospective gating for mouse cardiac MRI. Magn Reson Med. 2006;55:472–477. [DOI] [PubMed] [Google Scholar]
- 80.Heijman E, de Graaf W, Niessen P, Nauerth A, van Eys G, de Graaf L, Nicolay K, Strijkers GJ. Comparison between prospective and retrospective triggering for mouse cardiac MRI. NMR Biomed. 2007;20:439–447. [DOI] [PubMed] [Google Scholar]
- 81.Sachs TS, Meyer CH, Hu BS, Kohli J, Nishimura DG, Macovski A. Real-time motion detection in spiral MRI using navigators. Magn Reson Med. 1994;32:639–645. [DOI] [PubMed] [Google Scholar]
- 82.Kim WS, Mun CW, Kim DJ, Cho ZH. Extraction of cardiac and respiratory motion cycles by use of projection data and its applications to NMR imaging. Magn Reson Med. 1990;13:25–37. [DOI] [PubMed] [Google Scholar]
- 83.Anderson AW, Gore JC. Analysis and correction of motion artifacts in diffusion weighted imaging. Magn Reson Med. 1994;32:379–387. [DOI] [PubMed] [Google Scholar]
- 84.Pipe JG. Motion correction with PROPELLER MRI: application to head motion and free-breathing cardiac imaging. Magn Reson Med. 1999;42:963–969. [DOI] [PubMed] [Google Scholar]
- 85.Larson AC, White RD, Laub G, McVeigh ER, Li D, Simonetti OP. Self-gated cardiac cine MRI. Magn Reson Med. 2004;51:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brau AC, Brittain JH. Generalized self-navigated motion detection technique: preliminary investigation in abdominal imaging. Magn Reson Med. 2006;55:263–270. [DOI] [PubMed] [Google Scholar]
- 87.Subashi E, Qi Y, Johnson GA. Dynamic contrast-enhanced MR microscopy identifies regions of therapeutic response in a preclinical model of colorectal adenocarcinoma. Med Phys. 2015;42:2482–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Castets CR, Ribot EJ, Lefrançois W, Trotier AJ, Thiaudière E, Franconi J-M, Miraux S. Fast and robust 3D T1 mapping using spiral encoding and steady RF excitation at 7 T: application to cardiac manganese enhanced MRI (MEMRI) in mice. NMR Biomed. 2015;28:881–889. [DOI] [PubMed] [Google Scholar]
- 89.Janiczek RL, Blackman BR, Roy RJ, Meyer CH, Acton ST, Epstein FH. Three-dimensional phase contrast angiography of the mouse aortic arch using spiral MRI. Magn Reson Med. 2011;66:1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Malyarenko DI, Wilmes LJ, Arlinghaus LR, Jacobs MA, Huang W, Helmer KG, Taouli B, Yankeelov TE, Newitt D, Chenevert TL. QIN DAWG validation of gradient nonlinearity bias correction workflow for quantitative diffusion-weighted imaging in multicenter trials. Tomography. 2016;2:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O’Callaghan C. Respiratory gated PET/CT in-house phantom design—a mission statement. Eur J Nucl Med Mol Imaging. 2011;38:S447. [Google Scholar]
- 92.Hubbard PL, Zhou FL, Eichhorn SJ, Parker G. Biomimetic phantom for the validation of diffusion magnetic resonance imaging. Magn Reson Med. 2015;73:299–305. [DOI] [PubMed] [Google Scholar]
- 93.Yoshimaru E, Totenhagen J, Alexander GE, Trouard TP. Design, manufacture, and analysis of customized phantoms for enhanced quality control in small animal MRI systems. Magn Reson Med. 2014;71:880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buzug TM. Computed tomography In: Kramme R, Hoffmann KP, Pozos RS (eds), Springer Handbook of Medical Technology. Berlin, Heidelberg: Springer; 2011:311–342. [Google Scholar]
- 95.Badea C, Drangova M, Holdsworth D, Johnson G. In vivo small-animal imaging using micro-CT and digital subtraction angiography. Phys Med Biol. 2008;53:R319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Faulkner K, Moores BM. Noise and contrast detection in computed tomography images. Phys Med Biol. 1984;29:329–339. [DOI] [PubMed] [Google Scholar]
- 97.Brooks RA, Di Chiro G. Statistical limitations in x-ray reconstructive tomography. Med Phys. 1976;3:237–240. [DOI] [PubMed] [Google Scholar]
- 98.Ford NL, Thornton MM, Holdsworth DW. Fundamental image quality limits for microcomputed tomography in small animals. Med Phys. 2003;30:2869–2877. [DOI] [PubMed] [Google Scholar]
- 99.Kalender W. Computed Tomography: Fundamentals, System Technology, Image Quality, Applications. Munich, Germany: Publicis MCD Verlag; 2000. [Google Scholar]
- 100.Boone JM, Velazquez O, Cherry SR. Small-animal X-ray dose from micro-CT. Mol Imaging. 2004;3:149–158. [DOI] [PubMed] [Google Scholar]
- 101.Carlson SK, Classic KL, Bender CE, Russell SJ. Small animal absorbed radiation dose from serial micro-computed tomography imaging. Mol Imaging Biol. 2007;9:78–82. [DOI] [PubMed] [Google Scholar]
- 102.Ritman EL. Micro-computed tomography-current status and developments. Annu Rev Biomed Eng. 2004;6:185–208. [DOI] [PubMed] [Google Scholar]
- 103.Parkins CS, Fowler JF, Maughan RL, Roper MJ. Repair in mouse lung for up to 20 fractions of X rays or neutrons. Br J Radiol. 1985;58:225–241. [DOI] [PubMed] [Google Scholar]
- 104.Ashton JR, West JL, Badea CT. In vivo small animal micro-CT using nanoparticle contrast agents. Front Pharmacol. 2015;6:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Blocker SJ, Holbrook MD, Mowery YM, Sullivan DC, Badea CT. The impact of respiratory gating on improving volume measurement of murine lung tumors in micro-CT imaging. PLoS One. 2020;15:e0225019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clark DP, Badea CT. Micro-CT of rodents: state-of-the-art and future perspectives. Phys Med. 2014;30:619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Burk LM, Lee YZ, Wait JM, Lu J, Zhou OZ. Non-contact respiration monitoring for in-vivo murine micro computed tomography: characterization and imaging applications. Phys Med Biol. 2012;57:5749–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Badea CT, Hedlund LW, Johnson GA. Micro-CT with respiratory and cardiac gating. Med Phys. 2004;31:3324–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Song J, Liu QH, Johnson GA, Badea CT. Sparseness prior based iterative image reconstruction for retrospectively gated cardiac micro-CT. Med Phys. 2007;34:4476–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Johnston SM, Perez BA, Kirsch DG, Badea CT. Phase-selective image reconstruction of the lungs in small animals using Micro-CT. Proc SPIE Int Soc Opt Eng. 2010;7622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu L, Liang Y, Schwartz LH, Zhao B. Reliability of radiomic features across multiple abdominal CT image acquisition settings: a pilot study using ACR CT phantom. Tomography. 2019;5:226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Du LY, Umoh J, Nikolov HN, Pollmann SI, Lee TY, Holdsworth DW. A quality assurance phantom for the performance evaluation of volumetric micro-CT systems. Phys Med Biol. 2007;52:7087–7108. [DOI] [PubMed] [Google Scholar]
- 113.Bretin F, Warnock G, Luxen A, Plenevaux A, Seret A, Bahri MA. Performance evaluation and x-ray dose quantification for various scanning protocols of the GE eXplore 120 Micro-CT. IEEE Trans Nucl Sci. 2013;60:3235–3241. [Google Scholar]
- 114.Blocker SJ, Mowery YM, Holbrook MD, Qi Y, Kirsch DG, Johnson GA, Badea CT. Bridging the translational gap: implementation of multimodal small animal imaging strategies for tumor burden assessment in a co-clinical trial. PLoS One. 2019;14:e0207555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Waring CS, Bax JS, Samarabandu A, Holdsworth DW, Fenster A, Lacefield JC. Traceable micro-CT scaling accuracy phantom for applications requiring exact measurement of distances or volumes. Med Phys. 2012;39:6022–6027. [DOI] [PubMed] [Google Scholar]
- 116.Zhang X, Tian J, Feng J, Zhu S, Yan G. An anatomical mouse model for multimodal molecular imaging. Conf Proc IEEE Eng Med Biol Soc. 2009;2009:5817–5820 [DOI] [PubMed]
- 117.Wang H, Stout DB, Chatziioannou AF. Estimation of mouse organ locations through registration of a statistical mouse atlas with micro-CT images. IEEE Trans Med Imaging. 2012;31:88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Segars WP, Tsui BMW, Frey EC, Johnson GA, Berr SS. Development of a 4-D digital mouse phantom for molecular imaging research. Mol Imaging Biol. 2004;6:149–159. [DOI] [PubMed] [Google Scholar]
- 119.McDougald W, Vanhove C, Lehnert A, Lewellen B, Wright J, Mingarelli M, Corral CA, Schneider JE, Plein S, Newby DE, Welch A, Miyaoka R, Vandenberghe S, Tavares AAS. Standardization of preclinical PET/CT imaging to improve quantitative accuracy, precision and reproducibility: a multi-center study. J Nucl Med. 2020;61:461.–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mannheim JG, Mamach M, Reder S, Traxl A, Mucha N, Disselhorst JA, Mittelhäuser M, Kuntner C, Thackeray JT, Ziegler S, Wanek T, Bankstahl JP, Pichler BJ. Reproducibility and comparability of preclinical PET imaging data: a multicenter small-animal PET study. J Nucl Med. 2019;60:1483–1491. [DOI] [PubMed] [Google Scholar]
- 121.Moses WW. Fundamental limits of spatial resolution in PET. Nucl Instrum Methods Phys Res A. 2011;648:S236–S40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hudson HM, Larkin RS. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans Med Imaging. 1994;13:601–609. [DOI] [PubMed] [Google Scholar]
- 123.Lange K, Carson R. EM reconstruction algorithms for emission and transmission tomography. J Comput Assist Tomogr. 1984;8:306–316. [PubMed] [Google Scholar]
- 124.Panin VY, Kehren F, Michel C, Casey M. Fully 3-D PET reconstruction with system matrix derived from point source measurements. IEEE Trans Med Imaging. 2006;25:907–921. [DOI] [PubMed] [Google Scholar]
- 125.Pan T, Einstein SA, Kappadath SC, Grogg KS, Lois Gomez C, Alessio AM, Hunter WC, El Fakhri G, Kinahan PE, Mawlawi OR. Performance evaluation of the 5-Ring GE Discovery MI PET/CT system using the national electrical manufacturers association NU 2-2012 Standard. Med Phys. 2019;46:3025–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Sluis J, de Jong J, Schaar J, Noordzij W, van Snick P, Dierckx R, Borra R, Willemsen A, Boellaard R. Performance characteristics of the digital biograph vision PET/CT system. J Nucl Med. 2019;60:1031–1036. [DOI] [PubMed] [Google Scholar]
- 127.Zhang J, Maniawski P, Knopp MV. Performance evaluation of the next generation solid-state digital photon counting PET/CT system. EJNMMI Res. 2018;8:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kung MP, Kung HF. Mass effect of injected dose in small rodent imaging by SPECT and PET. Nucl Med Biol. 2005;32:673–678. [DOI] [PubMed] [Google Scholar]
- 129.MacFarlane CR, American College of Radiologists . ACR accreditation of nuclear medicine and PET imaging departments. J Nucl Med Technol. 2006;34:18–24. [PubMed] [Google Scholar]
- 130.Byrd DW, Sunderland JJ, Lee TC, Kinahan PE. Bias in PET images of solid phantoms due to CT-based attenuation correction. Tomography. 2019;5:154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Daube-Witherspoon ME, Karp JS, Casey ME, DiFilippo FP, Hines H, Muehllehner G, Simcic V, Stearns CW, Adam L-E, Kohlmyer S, Sossi V. PET performance measurements using the NEMA NU 2-2001 standard. J Nucl Med. 2002;43:1398–1409. [PubMed] [Google Scholar]
- 132.Boellaard R, Delgado-Bolton R, Oyen WJG, Giammarile F, Tatsch K, Eschner W, Verzijlbergen FJ, Barrington SF, Pike LC, Weber WA, Stroobants S, Delbeke D, Donohoe KJ, Holbrook S, Graham MM, Testanera G, Hoekstra OS, Zijlstra J, Visser E, Hoekstra CJ, Pruim J, Willemsen A, Arends B, Kotzerke J, Bockisch A, Beyer T, Chiti A, Krause BJ. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol Imaging. 2015;42:328–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sunderland JJ, Christian PE. Quantitative PET/CT scanner performance characterization based upon the society of nuclear medicine and molecular imaging clinical trials network oncology clinical simulator phantom. J Nucl Med. 2015;56:145–152. [DOI] [PubMed] [Google Scholar]
- 134.Goertzen AL, Bao Q, Bergeron M, Blankemeyer E, Blinder S, Canadas M, Chatziioannou AF, Dinelle K, Elhami E, Jans H-S, Lage E, Lecomte R, Sossi V, Surti S, Tai Y-C, Vaquero JJ, Vicente E, Williams DA, Laforest R. NEMA NU 4-2008 comparison of preclinical PET imaging systems. J Nucl Med. 2012;53:1300–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Clarke LP, Sriram RD, Schilling LB. Imaging as a Biomarker: standards for change measurements in therapy workshop summary. Acad Radiol. 2008;15:501–530. [DOI] [PubMed] [Google Scholar]
- 136.Prescott JW. Quantitative imaging biomarkers: the application of advanced image processing and analysis to clinical and preclinical decision making. J Digit Imaging. 2013;26:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. [PubMed] [Google Scholar]
- 138.Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8:135–160. [DOI] [PubMed] [Google Scholar]
- 139.Watson PF, Petrie A. Method agreement analysis: a review of correct methodology. Theriogenology. 2010;73:1167–1179. [DOI] [PubMed] [Google Scholar]
- 140.Galbraith SM, Lodge MA, Taylor NJ, Rustin GJS, Bentzen S, Stirling JJ, Padhani AR. Reproducibility of dynamic contrast-enhanced MRI in human muscle and tumours: comparison of quantitative and semi-quantitative analysis. NMR Biomed. 2002;15:132–142. [DOI] [PubMed] [Google Scholar]
- 141.Raunig DL, McShane LM, Pennello G, Gatsonis C, Carson PL, Voyvodic JT, Wahl RL, Kurland BF, Schwarz AJ, Gönen M, Zahlmann G, Kondratovich MV, O’Donnell K, Petrick N, Cole PE, Garra B, Sullivan DC. Quantitative imaging biomarkers: a review of statistical methods for technical performance assessment. Stat Methods Med Res. 2015;24:27–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Obuchowski NA, Bullen J. Quantitative imaging biomarkers: effect of sample size and bias on confidence interval coverage. Stat Methods Med Res. 2018;27:3139–3150. [DOI] [PubMed] [Google Scholar]
- 143.Shukla‐Dave A, Obuchowski NA, Chenevert TL, Jambawalikar S, Schwartz LH, Malyarenko D, Huang W, Noworolski SM, Young RJ, Shiroishi MS, Kim H, Coolens C, Laue H, Chung C, Rosen M, Boss M, Jackson EF. Quantitative imaging biomarkers alliance (QIBA) recommendations for improved precision of DWI and DCE-MRI derived biomarkers in multicenter oncology trials. J Magn Reson Imaging. 2019;49:e101–e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Barnes SL, Whisenant JG, Loveless ME, Ayers GD, Yankeelov TE. Assessing the reproducibility of dynamic contrast enhanced magnetic resonance imaging in a murine model of breast cancer. Magn Reson Med. 2013;69:1721–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ge X, Quirk JD, Engelbach JA, Bretthorst GL, Li S, Shoghi KI, Garbow JR, Ackerman JJH. Test-retest performance of a 1-hour multiparametric MR image acquisition pipeline with orthotopic triple-negative breast cancer patient-derived tumor xenografts. Tomography. 2019;5:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Savaikar MA, Whitehead T, Roy S, Strong L, Fettig N, Prmeau T, Luo J, Li S, Wahl RL, Shoghi KI. SUV25 and µPERCIST: precision imaging of response to therapy in co-clinical FDG-PET imaging of triple negative breast cancer (TNBC) patient-derived tumor xenografts (PDX). J Nucl Med. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Whisenant JG, Ayers GD, Loveless ME, Barnes SL, Colvin DC, Yankeelov TE. Assessing reproducibility of diffusion-weighted magnetic resonance imaging studies in a murine model of HER2+ breast cancer. Magn Reson Imaging. 2014;32:245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Whisenant JG, Peterson TE, Fluckiger JU, Tantawy MN, Ayers GD, Yankeelov TE. Reproducibility of static and dynamic (18)F-FDG, (18)F-FLT, and (18)F-FMISO MicroPET studies in a murine model of HER2+ breast cancer. Mol Imaging Biol. 2013;15:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bane O, Hectors SJ, Wagner M, Arlinghaus LL, Aryal MP, Cao Y, Chenevert TL, Fennessy F, Huang W, Hylton NM, Kalpathy-Cramer J, Keenan KE, Malyarenko DI, Mulkern RV, Newitt DC, Russek SE, Stupic KF, Tudorica A, Wilmes LJ, Yankeelov TE, Yen YF, Boss MA, Taouli B. Accuracy, repeatability, and interplatform reproducibility of T1 quantification methods used for DCE-MRI: results from a multicenter phantom study. Magn Reson Med. 2018;79:2564–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fraum TJ, Fowler KJ, Crandall JP, Laforest RA, Salter A, An H, Jacobs MA, Grigsby PW, Dehdashti F, Wahl RL. Measurement repeatability of (18)F-FDG PET/CT versus (18)F-FDG PET/MRI in solid tumors of the pelvis. J Nucl Med. 2019;60:1080–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lodge MA. Repeatability of SUV in Oncologic (18)F-FDG PET. J Nucl Med. 2017;58:523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Newitt DC, Zhang Z, Gibbs JE, Partridge SC, Chenevert TL, Rosen MA, Bolan PJ, Marques HS, Aliu S, Li W, Cimino L, Joe BN, Umphrey H, Ojeda‐Fournier H, Dogan B, Oh K, Abe H, Drukteinis J, Esserman LJ, Hylton NM. Test-retest repeatability and reproducibility of ADC measures by breast DWI: results from the ACRIN 6698 trial. J Magn Reson Imaging. 2019;49:1617–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sorace AG, Wu C, Barnes Sl, Jarrett Am, Avery S, Patt D, Goodgame B, Luci JJ, Kang H, Abramson RG, Yankeelov TE, Virostko J. Repeatability, reproducibility, and accuracy of quantitative mri of the breast in the community radiology setting. J Magn Reson Imaging. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Abramson RG, Burton KR, Yu J-PJ, Scalzetti EM, Yankeelov TE, Rosenkrantz AB, Mendiratta-Lala M, Bartholmai BJ, Ganeshan D, Lenchik L, Subramaniam RM. Methods and challenges in quantitative imaging biomarker development. Acad Radiol. 2015;22:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJWL, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Grossmann P, Gutman DA, Dunn WD Jr., Holder CA, Aerts HJ. Imaging-genomics reveals driving pathways of MRI derived volumetric tumor phenotype features in glioblastoma. BMC Cancer. 2016;16:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kotrotsou A, Zinn PO, Colen RR Radiomics in brain tumors: an emerging technique for characterization of tumor environment. Magn Reson Imaging Clin N Am. 2016;24:719–729. [DOI] [PubMed] [Google Scholar]
- 160.Sala E, Mema E, Himoto Y, Veeraraghavan H, Brenton JD, Snyder A, Weigelt B, Vargas HA. Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol. 2017;72:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zinn PO, Singh SK, Kotrotsou A, Hassan I, Thomas G, Luedi MM, Elakkad A, Elshafeey N, Idris T, Mosley J, Gumin J, Fuller GN, de Groot JF, Baladandayuthapani V, Sulman EP, Kumar AJ, Sawaya R, Lang FF, Piwnica-Worms D, Colen RR. A co-clinical radiogenomic validation study - Conserved magnetic resonance radiomic appearance of Periostin expressing Glioblastoma in patients and xenograft models. Clin Cancer Res. 2018;24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Permuth JB, Choi J, Balarunathan Y, Kim J, Chen DT, Chen L, Orcutt S, Doepker MP, Gage K, Zhang G, Latifi K, Hoffe S, Jiang K, Coppola D, Centeno BA, Magliocco A, Li Q, Trevino J, Merchant N, Gillies R, Malafa M, Collaborative O. B O T F P. Combining radiomic features with a miRNA classifier may improve prediction of malignant pathology for pancreatic intraductal papillary mucinous neoplasms. Oncotarget. 2016;7:85785–85797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Katsila T, Matsoukas MT, Patrinos GP, Kardamakis D. Pharmacometabolomics Informs Quantitative Radiomics for Glioblastoma Diagnostic Innovation. OMICS. 2017;21:429–439. [DOI] [PubMed] [Google Scholar]
- 164.Kickingereder P, Neuberger U, Bonekamp D, Piechotta PL, Gotz M, Wick A, Sill M, Kratz A, Shinohara RT, Jones DTW, Radbruch A, Muschelli J, Unterberg A, Debus J, Schlemmer HP, Herold-Mende C, Pfister S, von Deimling A, Wick W, Capper D, Maier-Hein KH, Bendszus M. Radiomic subtyping improves disease stratification beyond key molecular, clinical and standard imaging characteristics in patients with glioblastoma. Neuro Oncol. 2018;20:848.–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lee G, Lee HY, Park H, Schiebler ML, van Beek EJR, Ohno Y, Seo JB, Leung A. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: state of the art. Eur J Radiol. 2017;86:297–307. [DOI] [PubMed] [Google Scholar]
- 166.Samsa G, Samsa L. A guide to reproducibility in preclinical research. Acad Med. 2019;94:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]