Abstract
Background
The receptor binding domain (RBD) of spike protein S1 domain SARS-CoV-2 plays a key role in the interaction with ACE2, which leads to subsequent S2 domain mediated membrane fusion and incorporation of viral RNA into host cells. In this study we tend to repurpose already approved drugs as inhibitors of the interaction between S1-RBD and the ACE2 receptor.
Methods
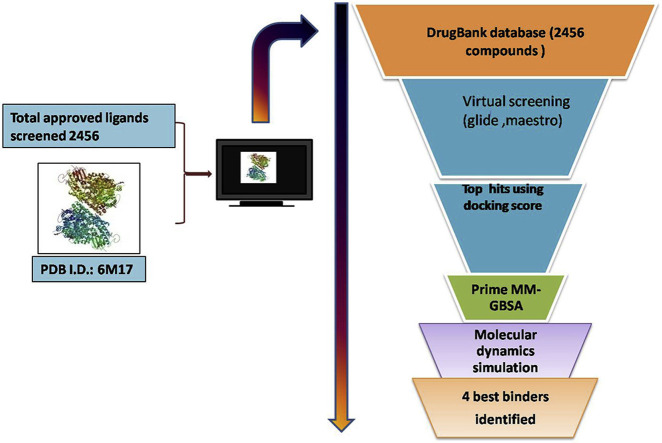
2456 approved drugs were screened against the RBD of S1 protein of SARS-CoV-2 (target PDB ID: 6M17). As the interacting surface between S1-RBD and ACE2 comprises of bigger region, the interacting surface was divided into 3 sites on the basis of interactions (site 1, 2 and 3) and a total of 5 grids were generated (site 1, site 2, site 3, site 1+site 2 and site 2+site 3). A virtual screening was performed using GLIDE implementing HTVS, SP and XP screening. The top hits (on the basis of docking score) were further screened for MM-GBSA. All the top hits were further evaluated in molecular dynamics studies. Performance of the virtual screening protocol was evaluated using enrichment studies.
Result
and discussion: We performed 5 virtual screening against 5 grids generated. A total of 42 compounds were identified after virtual screening. These drugs were further assessed for their interaction dynamics in molecular dynamics simulation. On the basis of molecular dynamics studies, we come up with 10 molecules with favourable interaction profile, which also interacted with physiologically important residues (residues taking part in the interaction between S1-RBD and ACE2. These are antidiabetic (acarbose), vitamins (riboflavin and levomefolic acid), anti-platelet agents (cangrelor), aminoglycoside antibiotics (Kanamycin, amikacin) bronchodilator (fenoterol), immunomodulator (lamivudine), and anti-neoplastic agents (mitoxantrone and vidarabine). However, while considering the relative side chain fluctuations when compared to the S1-RBD: ACE2 complex riboflavin, fenoterol, cangrelor and vidarabine emerged out as molecules with prolonged relative stability.
Conclusion
We identified 4 already approved drugs (riboflavin, fenoterol, cangrelor and vidarabine) as possible agents for repurposing as inhibitors of S1:ACE2 interaction. In-vitro validation of these findings are necessary for identification of a safe and effective inhibitor of S1: ACE2 mediated entry of SARS-CoV-2 into the host cell.
Keywords: 2019 novel corona virus; SARS-CoV-2; Spike protein; 2019-nCoV,receptor binding domain; RBD; S1 domain; ACE2
Abbreviations: SARS, Severe acute respiratory syndrome; CoV, Coronavirus; 2019-nCoV, novel coronavirus; RBD, receptor binding domain; NTD, N terminal domain; CTD, C terminal domains; MD, Molecular-dynamics; S protein, spike protein; COVID-19, Coronavirus disease 2019
Graphical abstract
Highlights
-
•
In this study we tend to repurpose already approved drugs as inhibitors of the interaction between S1-RBD of SARS-CoV-2 spike protein and the ACE2 receptor.
-
•
A virtual screening of approved drugs (n=2456) were performed using GLIDE implementing HTVS, SP and XP screening (target PDB ID: 6M17).
-
•
The top hits (n=42, on the basis of docking score) were further screened for MM-GBSA and molecular dynamics simulation studies. Performance of the virtual screening protocol was evaluated using enrichment studies.
-
•
On the basis of molecular dynamics studies, we come up with 10 molecules (acarbose, riboflavin, levomefolic acid, cangrelor, Kanamycin, amikacin fenoterol, lamivudine, Mitoxantrone and vidarabine) with favorable interaction profile.
-
•
However, while considering the relative side chain fluctuations, riboflavin, fenoterol, cangrelor and vidarabine emerged out as molecules with prolonged relative stability.
1. Introduction
The spike protein of coronavirus plays a major role in attachment to the host and further series of events e.g., membrane fusion and incorporation of the host RNA genome to the host cell [1,2]. The S1 domain of spike protein comprises of one N terminal domain (NTD) and three c terminal domain (CTD 1 to 3) and the RBD resides within the CTD-1 [3]. The interactions between the S1-RBD with the PD (peptidase domain) of ACE2 leads to attachment of the virus to the host [4,5]. which is followed by S2 domain mediated membrane fusion and internalization of viral ribonucleoprotien complex into the host cell [3].
Compared to SARS-CoV, many changes can be noted in the RBD region of SARS-CoV-2 e.g. at the N-terminus, the following variations, Arg426 to Asn439, Tyr484 to Gln498, and Thr487 to Asn501, at equivalent positions were observed between SARS-CoV and SARS-CoV-2 [4]. Similar findings are also reported by Ortega et al., 2020 [6]. On sequence homology comparison between the SARS-CoV and SARS-CoV-2 spike protein, only 76.04% similarity (SARS-CoV sequence taken from UniProtKB - P59594 (SPIKE_CVHSA) and SARS-CoV-2 sequence I.D GeneID: 43740568) was observed. Hence, there is an urgent need for the development of drugs targeting against SARS-CoV-2.
Although, the combination HCQ + Azithromycin showed promise [7] in clinical settings for the treatment of 2019-nCoV in the first clinical study, a subsequent study showed negative results [8]. Here comes the need for rapid development of a safe and effective new drug or repurposing the already existing drugs against 2019-nCoV. For the purpose of our study, we have selected S1: ACE2 interaction because, being the first point of contact between the host and the virus, the Spike protein S1 domain represents a potential target for the development of anti-SARS-CoV-2 drugs which might have the potential to inhibit the entry of SARS-CoV-2 into the host cells [4].
A typical new drug development process takes 10–12 years [9]. However, in the settings of this current pandemic of COVID-19, we need a safe and effective drug at the earliest and thus repurposing seems to be the most effective strategy. Use of approved drugs for the repurposing activity carries the advantage that the preclinical, clinical toxicity information and kinetic data of these agents are already available and thus we can expect a shorter regulatory pathway [10]. In this current study, we have repurposed the approved drugs for finding a novel inhibitor of S1: ACE2 interaction, inspired from the theme of relatable studies from Sarma et al. [64]. We did not target the host protein ACE2, as its a major component of the human rennin angiotensin system (RAS) system and is important for maintaining of blood pressure and other important homeostatic mechanisms [11].
As there are only a few thousands of approved drugs are available, a preliminary in-vitro screening of all these agents for the identification of potential inhibitors may be time-consuming and costly. Here comes the need for computational approaches, which can help us in selection potential agents beforehand and help our selection of molecules for further evaluation in in-vitro, [10]. Although a previous paper has already addressed the similar theme [12], they however, took SARS-CoV spike protein (PDB: 2AJF_E) for their virtual screening studies. In this regard, we used the spike protein of SARS-CoV-2. Additionally, we validated the results of our experiments using molecular dynamics studies. So, this is the first study, which purely targeted SARS-CoV-2 spike protein S1 domain and ACE2 interaction for the identification of inhibitors.
2. Materials and methods
In-silico studies were performed on an ACER Predator Helios 300 laptop running on LINUX Ubuntu OS 18.04.02 LTS using Schrodinger Maestro version 2019–3 for docking and molecular dynamics simulation processes.
2.1. The target PDB structure selection
The target protein considered for this study is the Spike protein S1 specifically the RBD (receptor binding domain of S1 protein) as the key target. In the RCSB database [13], we could find 5 PDB IDs (till date 05-04-2020, which are 6M17, 6VSB, 6VW1, 6LZG and 6MOJ) which has a structure of these Spike protein S1 domain of 2019-nCoV.
Among these different structures, 6M17 was screened as it is a cryo-EM structure with a resolution of 2.9 Å, and had the full length of RBD region (229 amino acid residues) and the interaction details between S1 RBD and ACE2 [4]. The rationale for excluding the 6LZG, 6M0J proteins was the non-availability of details of interface interactions, 6VSB PDB was presenting only a small region of RBD which had no ACE-2 binding region and 6VW1 PDB structure was a chimeric structure. Therefore, PDB structure of PDB I.D. 6M17 was considered for this study [14].
2.2. Validation of target protein
The validity of the target protein structure was evaluated using PROCHECK tool from SAVES v5.0 server. PROCHECK is a useful tool to examine the stereochemical efficiency of target protein structure. By this server we analyzed the geometry of ‘residues by residue’ or overall residues and also determined the Ramachandran plot to check the quality of selected target protein model. This tool was helpful to assure the qulity of protein which were used as a model in our study (PDB: 6M17).
2.3. Preparation of the protein
The selected protein was prepared for further docking purposes. Protein preparation was performed using the “protein preparation wizard” tool of “Maestro” version 10.2 [62]. The missing side chain and loops were filled in pre-processingat OPLS3 force field minimization algorithm where water molecules beyond 5.0 Å to rule out the hindrence [63].
2.4. Retrieval and preparation of ligands
The DrugBank database is a free accessible, comprehensive database of drugs and drug targets, which also contains the approved small molecules, biologics, nutraceuticals and few thousand of experimental drugs. For the purpose of repurposing, we selected the approved small molecule set (2456 drugs, up to date 10-01-2020) from DrugBank database [18]. SDF files of these 2456 approved drugs were obtained from the drug bank database [19] and prepared using ligprep module. This preparation of ligand is a necessary step for further docking and molecular simulations. OPLS 2003 force field was used for the energy minimization process, which is helpful to produce the low-energy isomers and prepare the ligands for docking [20].
2.5. S1- RBD and ACE2 interaction details
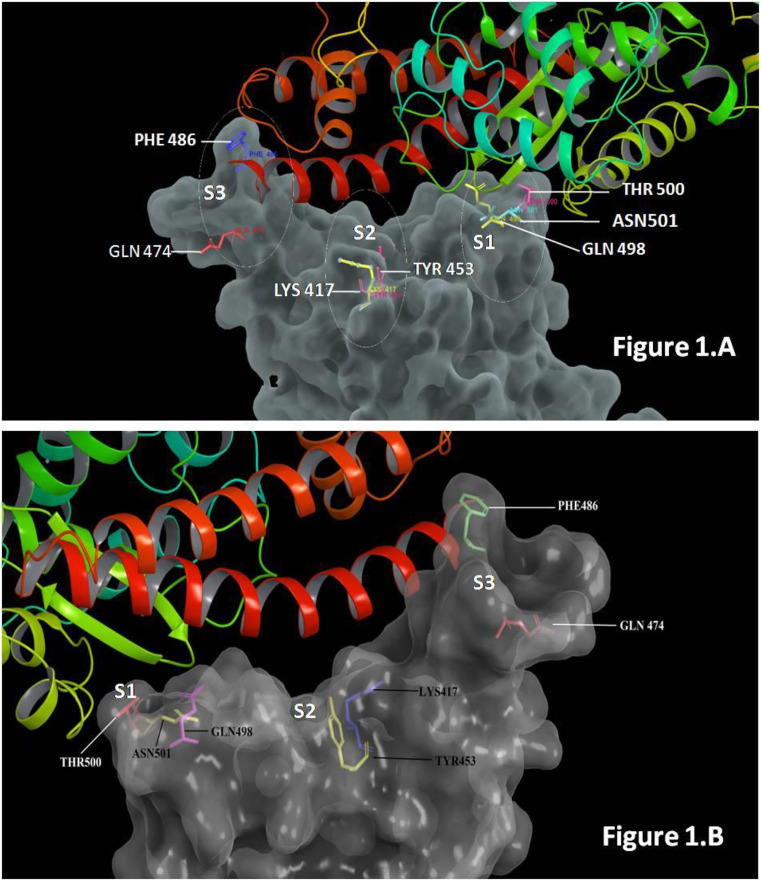
The RBD region of SARS-CoV-2 comprises of a core structure and the RBM (receptor binding motif) [21]. The host recognition by SARS-CoV-2 depends upon the interaction between the claw structured peptidase domain of ACE2 and the RBM of RBD region of S1 of spike protein [22]. In SARS-CoV-2, the S1-RBD extends from amino acid residues 331 to 524 [23]. An extended loop region of the RBD spans the arch-shaped α1 helix of the ACE2-PD like a bridge, moreover, these contacts can be arranged into three distinct clusters to apprehend the interacting domain. At the N terminus of α1 helix, Gln498, Thr500, and Asn501 of the S1-RBD forms a network of hydrogen bonds network with Tyr41, Gln42, Lys353, and Arg357 of ACE2. In the mid region, the “bridge”, Lys417 and Tyr453 of the S1-RBD can be observed interacting to Asp30 and His34 of ACE2, respectively. The C terminus of α1interacts with ACE2 through H-bond (Gln474RBD-----Gln 24ACE2), while the van der Waals forces were seen formed between Phe486 of S1-RBD and Met82 of ACE2 [22]. The details of interaction between S1 and ACE-2 are represented in Fig. 1 and Table 1 .
Fig. 1.
Details of interaction between spike protein (showed in surface diagram) and ACE2 (showed in ribbon diagram). Three points of interactions can be seen between ACE2 and spike protein. Fig. 1A represents interaction between S1 and ACE2 (first view) and figure and 1.B. represents the same in from 180° from the first view.
Table 1.
Roles of important amino acid residues (physiologically important residues) in the S1-RBD: ACE2 interactions.
| Parameter | Important interacting residues |
|---|---|
| N terminus of α1 of ACE2 | The ACE2: α1 (N terminal) residues Tyr 41, Gln 42, Lys 353 and Arg 357 form hydrogen bond with Gln498, Thr500 and Asn501 of S1-RBD region [22]. |
| Middle of the “bridge” region of interacting surface of ACE2 | S1-RBD residues Lys417 and Tyr453 interact with Asp 30 and His 34 of ACE2 [22]. |
| C terminus of α1 of ACE2 | The ACE2: α1 (C terminal) residues Gln 24 and Met 82 interact with Gln 474 and Phe 486 via H-bond and van-der waals forces [22]. |
| Hotspot residues for recognition of host receptor ACE2 | In SARS-CoV, two prime residues 479 and 487 of RBD- CoV are involved in the recognition of ACE2 [24,25]. In the case of SARS-CoV-2, the residues corresponding to N479 is Q493 and T487 is N501. |
| Physiologically important changes in SARS-CoV-2 when compared to SARS-CoV | The residues val 404 (in SARS-CoV) change into Lys417 in SARS-CoV-2, which may result in stronger association with ACE2. This stronger affinity may be attributed to formation of salt bridge between Lys417 of SARS-CoV-2 and ASP30 of ACE2 [22]. |
| The residues Leu 472 change into Phe486 may shown the stronger van der Waals interaction at met82. How-ever, replacement of Arg426 to Asn439 appears to weaken the interaction by losing one important salt bridge with Asp 329 on ACE2 [22]. | |
|
Capping loop residues [6] (Two capping loops in the binding domain, which stabilized the interaction with ACE2 by producing an increased amount of electrostatic interactions) |
The capping loop in case of SARS-CoV-2 comprises of residues N 487, G 485,V 445, F 486, Y 449, E 484, A 475, Q 474 and Y 473 [6]. |
The site of interactions can be divided into three sites: the portion of S-RBD interacting with of N terminus of alpha 1 of ACE2 (in our study, we denoted it as site 1), part of S1-RBD interacting with middle of the bridge region of PD of ACE2 (in our study, we denoted it as site 2) and part of S1-RBD interacting with the c terminus of Alpha 1 of PD of ACE2 (in our study, we denoted it as site 3). Details of site 1, site 2 and site 3 are showed in Fig. 1.
2.6. Generation of grid
It is the very important and essential step for the binding of ligands to the receptor. The three dimensional boundary were generated around actively participating residues on RBD (which are involved in S1-RBD and ACE2 interactions) for ligand binding at desired site by using of “receptor grid generation” module of GLIDE, Schrödinger tool [26].
Site 1: The important interacting residues in the RBD Gln498, Thr500, and Asn501 form a network of hydrogen bonds (H-bonds) with Tyr41, Gln42, Lys353, and Arg357 from ACE2s. Details are showed in Fig. 1.
Site 2: The middle regions of RBD have important residues that are Lys417 and Tyr453 interact with Asp30 and His34 of ACE2, respectively. Details are showed in Fig. 1.
Site 3: The important residues in third region are Gln474, Phe486 form an interaction with GLN24 and Met82 of ACE2 respectively [22]. Details are showed in Fig. 1.
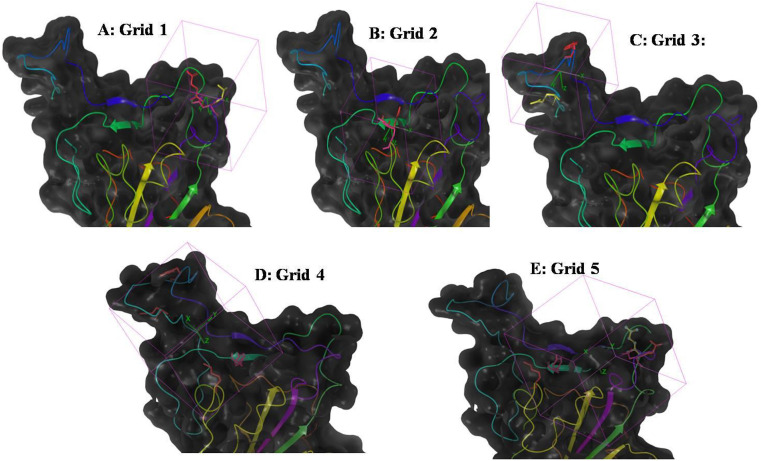
For the purpose of virtual screening, we made 5 grids (grid = site 1, grid 2 = site 2, grid 3 = site 3, grid 4 = Site 1 + site 2 and grid 5 = Site 2 + site 3), considering their importance in formation of S1-RBD: ACE2 interface and were used as distinct target sites in virtual screenings [Fig. 2 (A to E)].
Fig. 2.
Details of grids generated for the purpose of virtual screening.
2.7. Virtual screening
The virtual screening was performed using “Glide” tool of “Maestro”. A total of 2456 approved drugs were screened in a tandem HTVS→ SP→XP progression [16]. For virtual screening, a total of 5 parallel virtual screening was performed against the 5 grids generated.
2.7. MM-GBSA: Binding free energy calculation
The binding free energy between the ligand and receptor target, which were present in the docked structure, can be measured by using Prime MM-GBSA. Maestro, Schrödinger, LLC, New York, NY, USA which computes end-point binding free energy utilizing the receptor -ligand complex pose files [[22], [23], [24]]. The ligand-bound pose-viewer complexes were subjected to MM-GBSA in VSGB solvation within 5.0 Å from flexible residues at OPLS3 force field [29].
2.8. ROC curve plot and enrichment study
The performance of the virtual screening protocol was evaluated by performing enrichment studies using parameters area under the receiver operator characteristics curve (ROC) and Boltzmann enhanced discrimination of receiver operating characteristic (BEDROC). This test differentiates whether the used docking protocol can differentiate between actives and decoys. Y axis represents the sensitivity and X axis represents the 1-specificity. An AUC of 1 represents best performance of the virtual screening protocol and a value of 0 signifies worst performance. As five grids were generated for the purpose of our study, we evaluated the performance of each of the of these protocols by performing enrichment studies using actives and decoy molecules (Schrodinger decoy set, n = 1000) [31].
2.9. Molecular dynamics
The Desmond module was used to carry out MD simulations of the S1 RBD-drug complexes and apo forms. Firstly, water model were developed and then sodium and chloride ions were added to neutralize the ligand protein complex by using “system builder” tool of Desmond, Schrödinger. Liquid simulations optimization was minimized at OPLS3 force field. After creating frames for 100ps interval, the complexes were submitted for 100ns simulations at NPT ensemble at 310 K, where N lied between 34,900–35,000 in the solvation box. Trajectory analysis for the trajectory outputs extracted Cα RMSD, side chain RMSF, ligand contact maps and binding profile [15]. We performed 50 ns molecular dynamics studies of all the ligand-target complexes.
3. Result and discussion
3.1. Validation of target protein
The stereochemical quality of selected protein structure was checked by PROCHECK method. SAVES v5.0 server was used check the model quality of the 3D structure of target protein PDB: 6M17. This server has PROCHECK tool which helps investigate the obtained crystal structure model of protein. Verifying the 3D structure of 6M17 showed that 87.89% of the residues have averaged 3D-1D score≥0.2 (At least 80% of the amino acids have scored≥0.2 in the 3D/1D profile), the qulity of model were assessed by Ramachandran plot which revealed that 83.5% of the residues were in the most favored region and 15.3% were in the allowed region. Overall G factor was −0.05. The maximum deviation was 9.2 and planar groups: 100.0% within limits (Fig. 3 ).
Fig. 3.
Results of protein crystal structure quality check using PROCHECK (PDB: 6M17).
3.2. Virtual screening and MM-GBSA results
A total of five virtual screening was performed against the five grids generated. Ligands used were FDA approved drugs. The top performing ligands on the basis of docking score are shown in Table 2 .
Table 2.
Result of virtual screening (on the basis of Docking score), Glide score and MMGBSA against different grids.
| Grid number | Ligand Name | Docking Score | Glide Score | MMGBSA |
|---|---|---|---|---|
| Grid 1 | Acarbose | −9.948 | −10.054 | −58.104 |
| Lactulose | −8.943 | −8.943 | −42.12 | |
| Riboflavin | −6.779 | −6.779 | −54.187 | |
| Mitoxantrone | −5.988 | −5.988 | −58.173 | |
| Cangrelor | −5.786 | −5.8 | −42.741 | |
| Miglitol | −5.709 | −5.754 | −29.972 | |
| Isoetharine | −5.614 | −5.614 | −31.623 | |
| Epinephrine | −5.586 | −5.598 | −30.399 | |
| Xanthinol | −5.561 | −5.638 | −34.219 | |
| Kanamycin | −5.419 | −5.567 | −34.476 | |
| Fenoterol | −5.102 | −5.116 | −39.063 | |
| Grid 2 | Cangrelor | −6.324 | −6.338 | −43.164 |
| Diosmin | −5.834 | −5.834 | −49.239 | |
| Amphotericin B | −5.746 | −5.784 | −53.656 | |
| Leucovorin | −5.559 | −5.559 | −36.434 | |
| Amikacin | −5.399 | −5.463 | −47.89 | |
| Levomefolic acid | −5.3 | −5.316 | −40.358 | |
| Caspofungin | −5.272 | −5.28 | −57.793 | |
| Fenoterol | −4.399 | −4.413 | −42.208 | |
| Grid 3 | Pantethine | 9.889 | −9.889 | −79.537 |
| Cytarabine | 6.583 | −6.583 | −38.072 | |
| Ferrous ascorbate | −6.467 | −6.467 | −26.162 | |
| Calcium ascorbate | −6.394 | −6.394 | −23.709 | |
| Ascorbic acid | −6.269 | −6.269 | −23.428 | |
| Ribavirin | −5.925 | −5.925 | −32.866 | |
| Norepinephrine | −5.743 | −5.76 | −32.787 | |
| Lamivudine | −5.512 | −5.512 | −22.296 | |
| Levosalbutamol | 5.465 | −5.465 | −30.357 | |
| Epinephrine | −5.361 | −5.373 | −30.305 | |
| Azacitidine | −5.353 | −5.77 | −28.749 | |
| Pirbuterol | −5.211 | −5.574 | −36.495 | |
| Isoprenaline | −5.042 | −5.042 | −36.95 | |
| Grid 4 (Site 1 + Site 2) | Diosmin | −7.488 | −7.488 | −45.818 |
| Mitoxantrone | −6.329 | −6.329 | −59.905 | |
| Amikacin | −6.213 | −6.277 | −41.141 | |
| Cangrelor | −5.582 | −5.596 | −29.969 | |
| Fenoterol | −5.346 | −5.36 | −49.499 | |
| Ribavirin | −5.345 | −5.345 | −28.821 | |
| Gluconolactone | −5.129 | −5.129 | −32.239 | |
| Vidarabine | −5.094 | −5.094 | −36.665 | |
| Grid 5 (site 2 + site 3) | Norepinephrine | −6.375 | −6.393 | −32.356 |
| Miglitol | −6.293 | −6.337 | −23.833 | |
| Nystatin | −6.239 | −6.278 | −47.004 | |
| Tenapanor | −6.203 | −6.258 | −78.259 | |
| Sodium ascorbate | −5.856 | −5.856 | −18.563 |
In our study we have generated 5 grids in target protein and after then virtual screening process were performed. The top XP docking result (on the basis of docking score) were found form each grid. All the ligands which showed good docking score against the respected grids are showed in Table 2.
In case of Grid 1, highest docking score is −9.948 (Acarbose), in grid site 2 have highest docking score is −6.324 of Cangrelor, grid 3 have highest docking score is −9.889 (pentethinin), grid 4 (S1+S2) and grid 5 (S2+S3) were shown highest docking score is −7.488 (diosmin) and −6.375 (norepinephrine) respectively. The results of docking score and glide score of the top performing ligands are showed in Table 2.
In prime MM-GBSA, in grid 1, lowest MM-GBSA score was −58.173 and the highest score was showed −29.972, in the case of grid 2, the lowest MM-GBSA score was −57.793 and highest score was-36.434, In grid 3, lowest MM-GBSA score was −79.537 highest score was −22.296, in the Grid 4 (S1+S2) lowest MM-GBSA score was −59.905 and the highest score was −29.969 and in case of grid 5 (S2+S3), lowest MM-GBSA score was −78.259 and highest score was −18.563. Data showed in Table 2.
3.3. Enrichment studies
Overall, the XP docking protocol showed good AUC-ROC with range being 0.71 to 0.97 (for all the grids). For Grid 1, AUC was 0.94 and BEDROC was 0.813 (at alpha = 160.9), Grid 2, AUC was 0.94 and BDROC was 0.946 (at alpha = 160.9). In case of Grid 3, AUC was 0.97 and BEDROC was 0.842 at (at alpha = 160.9). In case of grid 4, AUC was 0.71 and BEDROC was 0.497 (at alpha = 160.9) and in case of Grid 5, AUC was 0.94 and BEDROC was 0.870 (at alpha = 160.9). (Table 3 ).
Table 3.
Details of enrichment studies.
| Grid No | Site covered | 1% | 2% | 5% | 10% | 20% | Enrichment | Matrix value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Site 1 | Active counts | 7 | 9 | 9 | 9 | 9 | ROC | 0.95 | |
| Area under accumulation | 0.94 | |||||||||
| BEDROC | α = 160.9 | 0.813 | ||||||||
| % of actives | 70.0 | 90.0 | 90.0 | 90.0 | 90.0 | α = 20% | 0.879 | |||
| α = 8% | 0.891 | |||||||||
| 2 | Site 2 | Active counts | 8 | 8 | 8 | 8 | 8 | ROC | 0.94 | |
| Area under accumulation | 0.94 | |||||||||
| BEDROC | α = 160.9 | 0.946 | ||||||||
| % of actives | 88.9 | 88.9 | 88.9 | 88.9 | 88.9 | α = 20% | 0.897 | |||
| α = 8% | 0.892 | |||||||||
| 3 | Site 3 | Active counts | 8 | 16 | 16 | 16 | 16 | ROC | 0.97 | |
| Area under accumulation | 0.96 | |||||||||
| BEDROC | α = 160.9 | 0.842 | ||||||||
| % of actives | 47.1 | 94.1 | 94.1 | 94.1 | 94.1 | α = 20% | 0.914 | |||
| α = 8% | 0.930 | |||||||||
| 4 | Site 1 + Site 2 | Active counts | 5 | 6 | 6 | 6 | 6 | ROC | 0.71 | |
| Area under accumulation | 0.71 | |||||||||
| BEDROC | α = 160.9 | 0.497 | ||||||||
| % of actives | 35.7 | 42.9 | 42.9 | 42.9 | 42.9 | α = 20% | 0.436 | |||
| α = 8% | 0.431 | |||||||||
| 5 | Site 2 + site 3 | Active counts | 7 | 8 | 8 | 8 | 8 | ROC | 0.94 | |
| Area under accumulation | 0.94 | |||||||||
| BEDROC | α = 160.9 | 0.870 | ||||||||
| % of actives | 77.8 | 88.9 | 88.9 | 88.9 | 88.9 | α = 20% | 0.881 | |||
| α = 8% | 0.885 | |||||||||
3.4. Molecular dynamics studies
We performed molecular dynamic simulation experiment at 50ns of all the ligands which showed good docking profile in virtual screening. Data shown in Table 4 .
Table 4.
Result of Molecular dynamics studies.
| Ligand details | Simulation Time (ns) |
RMSD average value | RMSF | Important Interactions (>30%) | H bond residues (30%) | Interaction with physiologically important residues | |
|---|---|---|---|---|---|---|---|
| Grid 1 | Acarbose | 50ns | 3.15 Å | 3.0 Å | Asn501, gly 496, gln493, ser494, tyr505, gly 502, | Gln493, ser494, asn501 | Asn501, gln493 |
| Lactulose | 50ns | 3.0 Å | 5.5 Å | Insignificant interaction | |||
| Riboflavin | 50ns | 3.0 Å | 1.2 Å | Gln498,tyr505,arg403,tyr495,ser494, | Tyr505,gln498,tyr495,ser494 | Gln498 | |
| Mitoxantrone | 50ns | 3.5 Å | 4.8 Å | ARG403,GLN498, ASN501, | Asn501,gln498,gly 496,arg403 | GLN498, ASN501, | |
| Cangrelor | 50ns | 2.1 Å | 3.5 Å | Ser494,gly 496,arg403,lys417 | Arg403,ser494,lys417 | lys417 | |
| Miglitol | 50ns | 2.5 Å | 4.1 Å | Insignificant interactions | |||
| Epinephrine | 50ns | 3.8 Å | 2.8 Å | Asp364 | Asp364 | ||
| Kanamycin | 100ns | 3.45 Å | 3.2 Å | Glu484, | glu484 | Glu484 | |
| Fenoterol | 50ns | 3.2 Å | 0.8 Å | Tyr449,tyr495,tyr505,arg403 | Tyr495,arg403 | Tyr449 | |
| Grid 2 | Cangrelor | 50ns | 2.2 Å | 0.9 Å | Arg403,tyr495,glu406,lys417,arg408,gln409,thr415 | Lys417,gln409,arg408 | Lys417 |
| Diosmin | 50ns | 2.5 Å | 4.0 Å | Insignificant interaction | |||
| Amphotericin B | 50ns | 2.4 Å | Insignificant interaction | ||||
| Leucovorin | 50ns | 2.7 Å | 4.3 Å | Insignificant interaction | |||
| Amikacin | 50ns | 3.1 Å | 2.2 Å | Tyr495,tyr453,arg403lys417,glu406 | Arg403,glu406tyr453 | Lys417,tyr453 | |
| Levomefolic acid | 50ns | 3.0 Å | 1.0 Å | Glu406,ile418, lys417,tyr453 | Glu406,lys417,tyr453,ser494 | Tyr453,lys417 | |
| Caspofungin | 50ns | 2.4 Å | Insignificant interaction | ||||
| Fenoterol | 50ns | 3.3 Å | 3.8 Å | Pro491,tyr489,phe 456 | |||
| Grid 3 | Pantethinone | 50ns | 2.8 Å | 4.8 Å | Ala 372 | Asn481,pro479 | |
| Cytarabine | 50ns | 3.2 Å | 5 Å | Insignificant interaction | |||
| Ferrous ascorbate | 50ns | 2.9 Å | 3.6 Å | Insignificant interaction | |||
| Calcium ascorbate | 50ns | 2.25 Å | 3.3 Å | Insignificant interaction | |||
| Ascorbic acid | 50ns | 2.2 Å | 3.2 Å | Insignificant interaction | |||
| Ribavirin | 50ns | 2.8 Å | 4.0 Å | Insignificant interaction | |||
| Norepinephrine | 50ns | 2.15 Å | 2.5 Å | Insignificant interaction | |||
| Lamivudine | 50ns | 2.95 Å | 3.6 Å | Asn481,val 483,phe486 | Asn481,gly 482,val 483,phe486, | Phe486 | |
| Levosalbutamol | 50ns | 2.3 Å | 3.0 Å | Insignificant interaction | |||
| Epinephrine | 50ns | 2.65 Å | 4.0 Å | Insignificant interaction | |||
| Azacitidine | 50ns | 3 Å | 4.5 Å | Phe486 | Phe486,asn487 | Phe486,asn487 | |
| Pirbuterol | 50ns | 3.1 Å | 7 Å | Glu340 | Glu340 | ||
| Grid 4 | Diosmin | 50ns | 2.15 Å | 3.1 Å | Tyr489 | ||
| Mitoxantrone | 50ns | 2.5 Å | 3.0 Å | Arg403,gln498,asn501 | Asn501,gln498,arg403, | Asn501,gln498 | |
| Amikacin | 50ns | 3.15 Å | 3.8 Å | Tyr495,arg403,gly 496,glu484 | Glu484,ser494,tyr495,gly 496,gln498,asn501 | Asn501,gln498,glu484 | |
| Cangrelor | 50ns | 3.6 Å | 1.2 Å | Arg408,lys417 | Arg403,arg408,lys417, | Lys417 | |
| Fenoterol | 50ns | 3.95 Å | 7 Å | Insignificant interaction | |||
| Ribavirin | 50ns | 2.7 Å | 3.2 Å | Insignificant interaction | |||
| Gluconolactone | 50ns | 2.6 Å | 4.7 Å | Insignificant interaction | |||
| Vidarabine | 50ns | 2.6 Å | 1.0 Å | Gly496,gln498,tyr505 | Gly496,gln498,tyr505 | Gln498 | |
| Grid 5 | Norepinephrine | 50ns | 3.25 Å | 7.0 Å | Lys458,glu471,asp467,ser469 | Lys458,asp467,ser469,glu471 | |
| Miglitol | 50ns | 3.2 Å | 4.1 Å | Insignificant interaction | |||
| Nystatin (anti-fungal) | 50ns | Insignificant interaction | |||||
| Tenapanor (Used in IBS) | 50ns | 2.3 Å | Insignificant interaction | ||||
| Sodium ascorbate | 50ns | 2.9 Å | 7 Å | Leu455 | Leu455, phe 456, gln493 | Gln493 |
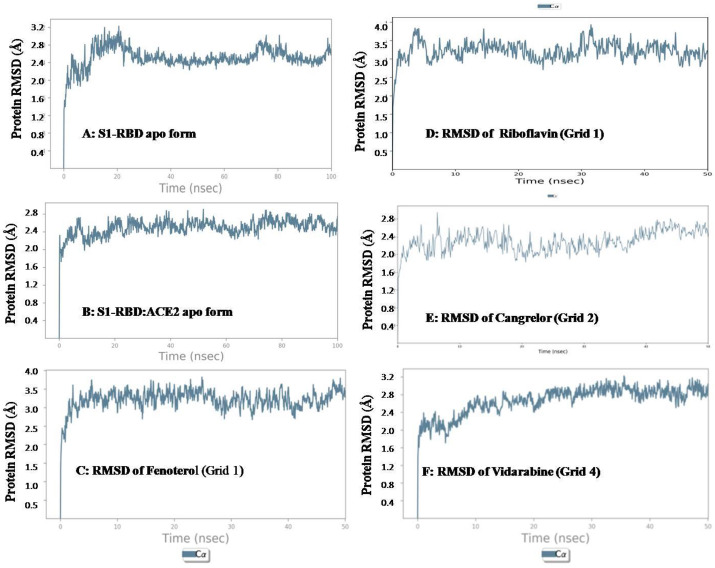
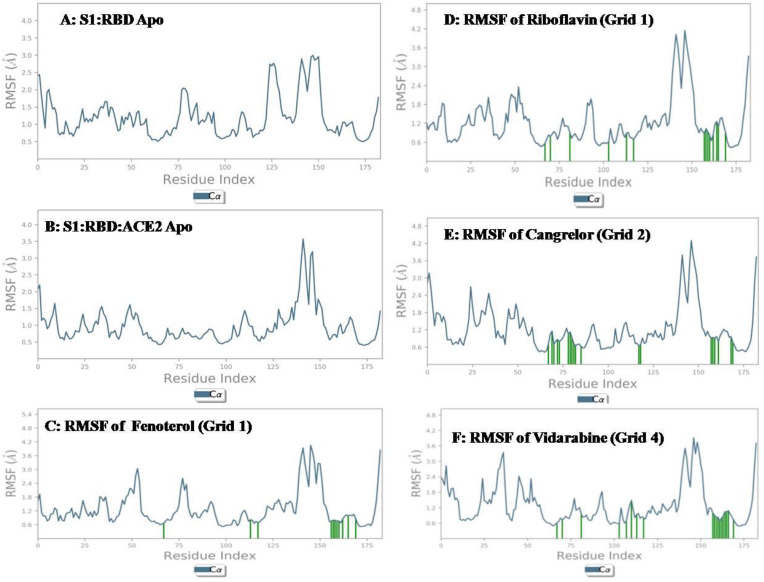
First we performed molecular dynamic simulation study of the apo form of targeted protein to evaluate and analyze the stability of the protein. Here we took two apo forms, first is, S1-RBD: ACE2 complex, and the second is, only RBD region of spike protein (S1-RBD). In our study the RBD region of S1 protein ranged from Cys 336 to Leu 518. We conducted a 100 ns MD simulation study for both the apo forms. The average RMSD value was found to be 2.4 Å for the ACE2: RBD Apo form and 2.7 Å found for the Apo S1– RBD protein. Data shown in Fig. 4, Fig. 5 . When comparing the RMSF value of the S1-RBD apo and the S1-RBD: ACE2 complex apo form, we could find that higher fluctuations were noted in the S1-RBD apo, especially in residue index 0–25, 75–100, 125–150 and 150–175.
Fig. 4.
RMSD of Apo forms and ligand bound forms of selected molecules.
Fig. 5.
RMSF of Apo forms and ligand bound forms of selected molecules.
3.5. Grid 1
Among all the ligands evaluated in molecular dynamics studies, favourable interaction profile was seen in cases of acarbose, riboflavin, cangrelor, epinephrine, Kanamycin and fenoterol. However, among these, only acarbose, riboflavin, cangrelor, Kanamycin and fenoterol showed interaction with physiologically important residues. Data of two best performers in terms of RMSD and RMSF value (riboflavin and fenoterol) are showed in Fig. 4, Fig. 5. The RMSF values of riboflavin and fenoterol were found to be 1.2 Å and 0.8 Å. However, we couldn’t find much difference in fluctuation between the S1-RBD: ACE2 apo form and the ligand bound complex.
3.6. Grid 2
Among all the ligands evaluated in molecular dynamics studies, favourable interaction profile was seen in cases of cangrelor, amikacin, levomefolic acid and fenoterol. However, among these, only cangrelor, amikacin and levomefolic acid showed interaction with physiologically important residues. Among these, in terms of RMSD and RMSF, cangrelor was found to be the best performer in terms of retaining the site. Data showed in Fig. 4, Fig. 5. Compared to the S1-RBD: ACE2 apo form, the cangrelor bound form showed less fluctuation in terms of side chain RMSF values.
3.7. Grid 3
Among all the ligands evaluated in molecular dynamics studies, favourable interaction profile was seen in cases of pantethinone, lamivudine, azacitidine and pirbuterol. However RMSF values were higher in case of pantethinone, azacitidine and pirbuterol. Lamivudine showed interaction with physiologically important residue Phe486, however, the RMSD value was 3.6 and a higher fluctuation (RMSF) was noted when compared to the S1-RBD: ACE2 complex apo form. Data showed in Table 4.
3.8. Grid 4
Among all the ligands evaluated in molecular dynamics studies, favourable interaction profile was seen in cases of diosmin, mitoxantrone, amikacin, cangrelor and vidarabin. However interaction with physiologically important residue was seen cases of mitoxantrone, amikacin, cangrelor and vidarabin. Among these vidarabine performed better in terms of RMSD (2.4 Å) and RMSF (1 Å) result and fluctuations were less in terms of RMSF vale compared to the S1-RBD: ACE2 apo form. Data showed in Table 4 and Fig. 4, Fig. 5.
3.9. Grid 5
Among all the ligands evaluated in molecular dynamics studies, although interaction was seen in cases of nor-epinephrine and sodium ascorbate, however, RMSF value was high for both the cases. Data showed in Table 4.
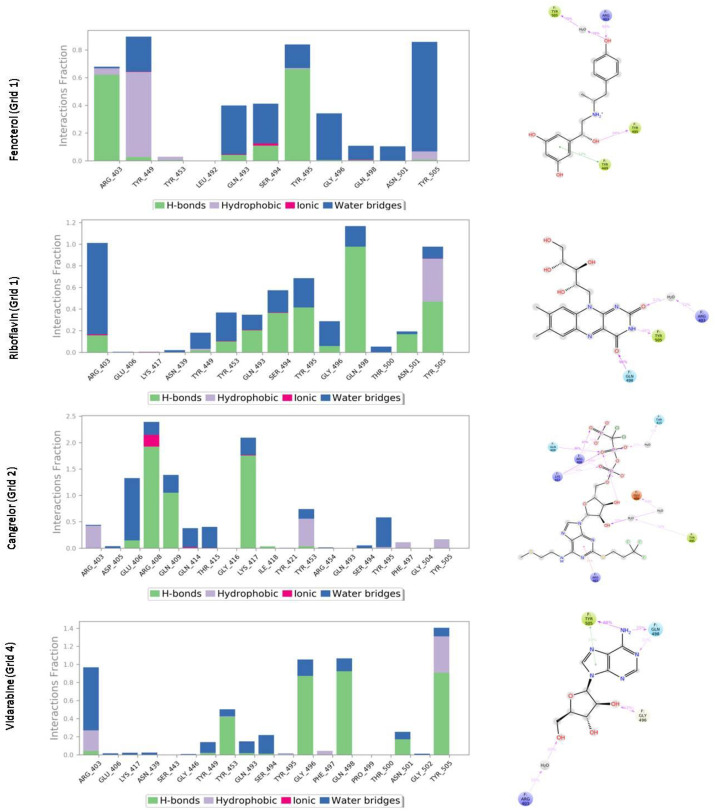
So, while combining the results of all the experiments, we come up with 10 molecules with favourable interaction profile, which also interacted with physiologically important residues. These are antidiabetic (acarbose), vitamins (riboflavin and levomefolic acid), anti-platelet agents (cangrelor), aminoglycoside antibiotics (Kanamycin, amikacin) bronchodilator (fenoterol), immunomodulator (lamivudine), and anti-neoplastic agents (mitoxantrone and vidarabine). However, while considering the relative fluctuation when compared to the S1-RBD: ACE2 complex riboflavin, fenoterol, cangrelor and vidarabine were the best performers in terms of relative stability. Details of interactions of these ligands with the target protein are showed in Fig. 6 .
Fig. 6.
Details of protein Ligand contact profile of selected ligands.
Acarbose: Acarbose is an antidiabetic. Acarbose is effective in delaying or preventing the progression of prediabetes to type 2 diabetes mellitus (T2DM) [32] In our study, acarbose showed interaction with write all residues Asn501, Gly 496, Gln 493, Ser 494, tyrosine 505 and Gly 502. Hydrogen bonding was seen in cases of Gln 493, Ser 494 and Asn501 Among these, residue Asn501 and Gln493 are physiologically important for the S1: ACE2 interaction. The residue Asn 501 of S1 interacts with the N terminus of alpha 1 of ACE2 and plays an important role in host receptor recognition by the virus [22] and both Asn501 and Gln493 are host-spot residues for host recognition by the virus [24,25]. The 501 residue plays a critical part in human infectivity. One study were predicted that a single N501T mutation (corresponding to the S487T mutation in SARS-CoV) may significantly enhance the binding affinity between 2019-nCoV RBD and human ACE2, highlighting the importance of the residue [21]. Data shown in Table 4.
Riboflavin: Riboflavin is also known as vitamin B2. Riboflavin showed interaction with residues Gln 498, Tyr 505, Arg 403, Tyr 495 and Ser 494. Among these, hydrogen bond formation was seen in cases of Tyr 505, Gln 498, Tyr 495 and Ser 494 (Table 4 and Fig. 6). However, among these only Gln 498 plays a strong physiological role in the interaction S1 and ACE2. Interaction between Gln 498 of S1 and N terminus of α1 of ACE2 plays important role in host receptor recognition and further sequelae [22]. Validity of the findings from our study is also highlighted by the fact that in in-vitro level, in human plasma and whole blood, use of riboflavin was associated with reduction of titre of SARS-CoV-2 to below detection level [33]. This highlights the validity of our methodology. In vero cell plaque assay also, use of riboflavin was associated with reduction of the titre of SARS-CoV-2 in both plasma and platelet products to below the limit of detection in tissue culture [34].
Levomefolic acid: Levomefolic acid is a biologically active form of folic acid [35]. In our study, Levomefolic acid showed interactions with residues of Glu406, Ile 418, Lys417, Tyr453 and H bond interaction was seen with Glu406, Lys417, Tyr453 and Ser494. However, among these only Lys 417 and Tyr 453 plays a strong physiological role in the interaction S1 and ACE. In clinical studies, low serum folate levels were associated with a more severe disease [36]. Homocystine is a predictor of radiological progression of COVID-19 [37] Folic acid supplementation also reduces the level of homocystine [[38], [39], [40]]. In in-silico studies, folic acid was also found to bind to active site of coronavirus main proitease [41]. Another multi target based study also indicated that folic acid may be useful in case of COVID-19 [42]. Folic acid also decreases the level of homocystine in clinical studies [43]. Hyper-homocystinemia is associated with more severe disease in case of COVID-19 [44].
Cangrelor: Cangrelor is an adenosine triphosphate analogue that selectively and specifically blocks P2Y12 receptor-mediated platelet activation [45]. In this study, we found that cangrelor showed interaction with Arg403, Tyr495, Glu406, Lys417, Arg408, Gln409, Thr415 residues of RBD region of CoV-2 (Table 4 and Fig. 6). However, among these, H bond interaction was seen with Lys417, Gln409, Arg408 residues. However, only Lys 417 residue has a physiological important role in interaction S1 domain and ACE2. Cangrelor is an antiplatelet agent. Coagulopathy is reported in case of COVID-19 and this is a major cause of mortality and morbidity [46]. Another anti-platelet agent “aspirin” is already under evaluation for its protective action against COVID-19 [47]. In this context, being an anti-platelet agent, the effect of cangrelor will be of interest.
3.10. Kanamycin
Kanamycin, an aminoglycoside class antibiotic is a widely used antibiotic available as intra-muscular, intra-venous and oral formulation [48]. It is a commonly used antibiotic, which is used in many difficult to treat cases also [49]. Regarding antiviral activity, Kanamycin showed antiviral activity in in-vitro against HSV-2 [50], influenza-A virus [51], respiratory syncytial virus [52] and against foot and mouth disease viruses [53]. Important side effects are nephrotoxicty, ototoxicity and vestibular toxicity and toxicity depends upon cumulative life-time exposure [54]. In our study, Kanamycin showed interaction with physiological important residue Glu484 of S1-RBD. This residue plays important role in the interaction between S1 and ACE2.
Amikacin: Amikacin is an antibiotic of aminoglycoside class [55]. In our study amikacin showed interaction with Tyr495, tyr453, arg403, lys417 and glu406 residues of RBD region. H bond was seen with residues Arg403, Glu406 and Tyr453 of S1 RBD. Among all interaction, physiologically important residues showing interaction with amikacin are Lys417, Tyr453. Amikacin is a semi-synthetic aminoglycoside antibiotic that is derived from kanamycin-A [56].
Fenoterol: In our study, fenopterol (inhaled bronchodilator) showed significant interactions with residues Tyr449, tyr495, tyr505 and arg403 of S1 RBD. Among these, H-bond interaction was seen with tyr495 and arg403 residues (Fig. 6). Among all only Tyr449 residue were shown the physiological important residue. Fenoterol is sympatho-mimetic agent. Fenoterol stimulates the beta-2-adrenergic receptors in the lungs, which in turn activates the enzyme adenylyl cyclase which then catalyzes the conversion of ATP to cAMP. The increased cAMP levels relaxes bronchial smooth muscle, relieve bronchial spasms and decline the inflammatory cell mediator release, principally from the mast cells [57].
Lamivudine: Lamivudine have shown intraction with Asn481, val 483, phe486 residues of S1 RBD and H bond intraction was seen with Asn481, gly 482 and phe486 residues. However, among the interacting residues, only phe486 residue was physiologically important in the interaction between S1 RBD and ACE2. Regarding antiviral effect of lamivudine, its a known anti-retroviral agent and also active against hepatitis B virus [58].
Mitoxantrone: It showed interaction with Arg403, Gln498 and Asn501 residue and all residue formed H bond with Mitoxantrone. The physiological important residues among them are gln498, asn501. Mitoxantrone is an anticancer agent and immune suppressor agent used for treating cancer and multiple sclerosis. Interestingly, this drug has also been reported as a potential main protease binder [59]. It also inhibit vaccinia virus replication by blocking virion assembly [60].
Vidarabine: In our study vidarabine have shown H-bond interaction with Gly496, Gln498, and Tyr505 residue of S1-RBD region. Among all interaction, physiological important residues interaction was shown with Gln498 (Fig. 6). One of recent study were found that vidarabine were useful to overcome the SARS-CoV-2 [65].
4. Conclusion
Among all the agents, Fenoterol and riboflavin ligand at Grid 1 (site S1), Cangrelor at grid 2 (site 2) and vidarabine at grid 4 (site 1 + 2) showed good interaction with RBD region of S1. We need in-vitro proof of concept studies of these identified agents to find out a potential inhibitor of S1: ACE2 interaction.
Declaration of competing interest
The authors declare that they have no known competing financialinterestsor personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors thank Dr. Prajwal Nandekar and Mr. Vinod Devaraji of Schrodinger Corporation for their kind help and Mr. Nripendra Bhatta for his logistic help.
References
- 1.Sarma P., Prajapat M., Avti P., Kaur H., Kumar S., Medhi B. Therapeutic options for the treatment of 2019-novel coronavirus: an evidence-based approach. Indian J. Pharmacol. 2020 Jan 1;52(1):1. doi: 10.4103/ijp.IJP_119_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H. Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 2020 Jan 1;52(1):56. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He J., Tao H., Yan Y., Huang S.-Y., Xiao Y. Molecular mechanism of evolution and human infection with the novel coronavirus (2019-nCoV) [Internet] Microbiology. 2020 Feb http://biorxiv.org/lookup/doi/10.1101/2020.02.17.952903 [cited 2020 Mar 27]. Available from: [Google Scholar]
- 4.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020 Mar 27;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prajapat M., Sarma P., Shekhar N., Prakash A., Avti P., Bhattacharyya A. Update on the target structures of SARS-CoV-2: a systematic review. Indian J. Pharmacol. 2020 Mar 1;52(2):142. doi: 10.4103/ijp.IJP_338_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020 Mar 18;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020 Mar 20:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Molina J.M., Delaugerre C., Goff J.L., Mela-Lima B., Ponscarme D., Goldwirt L. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med. Maladies Infect. 2020 doi: 10.1016/j.medmal.2020.03.006. http://www.sciencedirect.com/science/article/pii/S0399077X20300858 [Internet] Mar 30 [cited 2020 Apr 4]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohs R.C., Greig N.H. Drug discovery and development: role of basic biological research. Alzheimers Dement (N Y) 2017 Nov 11;3(4):651–657. doi: 10.1016/j.trci.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discovery. 2020 Mar 16;6(1):1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Junyi, Zheng Huang, Lin Li, Lv Jiagao. Coronavirus disease 2019 (COVID-19) and cardiovascular disease: a viewpoint on the potential influence of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers on onset and severity of severe acute respiratory syndrome coronavirus 2 infection. Journal of the American Heart Association. 2020 Apr 7;9(7) doi: 10.1161/JAHA.120.016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senathilake K., Samarakoon S., Tennekoon K. Virtual screening of inhibitors against spike glycoprotein of 2019 novel corona virus: a drug repurposing approach. 2020. https://www.preprints.org/manuscript/202003.0042/v1 Mar 3 [cited 2020 Apr 4]; Available from:
- 13.Bank R.P.D. Rcsb PDB: homepage. http://www.rcsb.org/ [Internet]. [cited 2020 Apr 4]. Available from:
- 14.Rcsb Pdb 6M17: the 2019-nCoV RBD/ACE2-B0AT1 complex. https://www.rcsb.org/structure/6m17 [Internet]. [cited 2020 Mar 27]. Available from:
- 15.Desmond Schrödinger. https://www.schrodinger.com/desmond [Internet]. [cited 2020 Mar 27]. Available from:
- 16.Molecular docking: a powerful approach for structure-based drug discovery. https://www.ncbi.nlm.nih.gov/pubmed/21534921 PubMed - NCBI [Internet]. [cited 2020 Mar 27]. Available from: [DOI] [PMC free article] [PubMed]
- 18.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018 Jan 4;46(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DrugBank. https://www.drugbank.ca/ [Internet]. [cited 2020 Mar 27]. Available from:
- 20.LigPrep Schrödinger. https://www.schrodinger.com/ligprep [Internet]. [cited 2020 Mar 27]. Available from:
- 21.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020 Mar 17 doi: 10.1128/JVI.00127-20. https://jvi.asm.org/content/94/7/e00127-20 [Internet] [cited 2020 Apr 4];94(7). Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020 Mar 27;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0400-4. http://www.nature.com/articles/s41423-020-0400-4 [Internet] Mar 19 [cited 2020 Apr 4]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou J., Yin J., Fang L., Yang M., Wang T., Wu W. Computational prediction of mutational effects on the SARS-CoV-2 binding by relative free energy calculations. 2020. https://chemrxiv.org/articles/Computational_Prediction_of_Mutational_Effects_on_the_SARS-CoV-2_Binding_by_Relative_Free_Energy_Calculations/11902623 Mar 3 [cited 2020 Apr 5]; Available from: [DOI] [PubMed]
- 25.Lu G., Wang Q., Gao G.F. Bat-to-human: spike features determining “host jump” of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015 Aug;23(8):468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.GlideScore/Docking Score doesn’t correlate with my known activities. What is wrong? | Schrödinger. https://www.schrodinger.com/kb/144 [Internet]. [cited 2020 Mar 27]. Available from:
- 29.Prime S. LLC. New York, NY. 2017.
- 31.Lu S.-H., Wu J.W., Liu H.-L., Zhao J.-H., Liu K.-T., Chuang C.-K. The discovery of potential acetylcholinesterase inhibitors: a combination of pharmacophore modeling, virtual screening, and molecular docking studies. J. Biomed. Sci. 2011 Jan 21;18(1):8. doi: 10.1186/1423-0127-18-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu R., Li Y., Lv Q., Wu T., Tong N. Acarbose monotherapy and type 2 diabetes prevention in eastern and western prediabetes: an ethnicity-specific meta-analysis. Clin. Therapeut. 2015 Aug;37(8):1798–1812. doi: 10.1016/j.clinthera.2015.05.504. [DOI] [PubMed] [Google Scholar]
- 33.Ragan I., Hartson L., Pidcoke H., Bowen R., Goodrich R. Pathogen reduction of SARS-CoV-2 virus in plasma and whole blood using riboflavin and UV light. PloS One. 2020 May 29;15(5) doi: 10.1371/journal.pone.0233947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keil SD, Ragan I, Yonemura S, Hartson L, Dart NK, Bowen R. Inactivation of severe acute respiratory syndrome coronavirus 2 in plasma and platelet products using a riboflavin and ultraviolet light-based photochemical treatment. Vox Sang. [Internet]. [cited 2020 Jul 3];n/a(n/a). Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/vox.12937. [DOI] [PMC free article] [PubMed]
- 35.PubChem Levomefolic acid. https://pubchem.ncbi.nlm.nih.gov/compound/135398561 [Internet]. [cited 2020 Jul 3]. Available from:
- 36.Itelman E., Wasserstrum Y., Segev A., Avaky C., Negru L., Cohen D. Clinical characterization of 162 COVID-19 patients in Israel: preliminary report from a large tertiary center. Isr. Med. Assoc. J. 2020 May;22(5):271–274. [PubMed] [Google Scholar]
- 37.Yang Z., Shi J., He Z., Lü Y., Xu Q., Ye C. Predictors for imaging progression on chest CT from coronavirus disease 2019 (COVID-19) patients. Aging (N Y) 2020 Apr 10;12(7):6037–6048. doi: 10.18632/aging.102999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rydlewicz A., Simpson J.A., Taylor R.J., Bond C.M., Golden M.H.N. The effect of folic acid supplementation on plasma homocysteine in an elderly population. QJM. 2002 Jan 1;95(1):27–35. doi: 10.1093/qjmed/95.1.27. [DOI] [PubMed] [Google Scholar]
- 39.Modaghegh M.H.S., Ravari H., Haghighi M.Z., Rajabnejad A. Effect of folic acid therapy on homocysteine level in patients with atherosclerosis or buerger’s disease and in healthy individuals: a clinical trial. Electron. Physician. 2016 Oct 25;8(10):3138–3143. doi: 10.19082/3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Homocysteine lowering with folic acid and B vitamins in vascular disease. N. Engl. J. Med. 2006 Apr 13;354(15):1567–1577. doi: 10.1056/NEJMoa060900. [DOI] [PubMed] [Google Scholar]
- 41.Serseg T., Benarous K., Yousfi M. Hispidin and lepidine E: two natural compounds and folic acid as potential inhibitors of 2019-novel coronavirus main protease (2019-nCoVMpro), molecular docking and SAR study. Curr. Comput. Aided Drug Des. 2020 Apr 21 doi: 10.2174/1573409916666200422075440. [DOI] [PubMed] [Google Scholar]
- 42.Manikyam H.K., Joshi S.K. Whole genome analysis and targeted drug discovery using computational methods and high throughput screening tools for emerged novel coronavirus (2019-nCoV) J Pharm Drug Res. 2020;3(2):341–361. [PMC free article] [PubMed] [Google Scholar]
- 43.van Oort F.V., Melse-Boonstra A., Brouwer I.A., Clarke R., West C.E., Katan M.B. Folic acid and reduction of plasma homocysteine concentrations in older adults: a dose-response study. Am. J. Clin. Nutr. 2003 May 1;77(5):1318–1323. doi: 10.1093/ajcn/77.5.1318. [DOI] [PubMed] [Google Scholar]
- 44.Ponti G., Maccaferri M., Ruini C., Tomasi A., Ozben T. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020 Jun 5:1–11. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferreiro J.L., Ueno M., Angiolillo D.J. Cangrelor: a review on its mechanism of action and clinical development. Expert Rev. Cardiovasc Ther. 2009 Oct;7(10):1195–1201. doi: 10.1586/erc.09.101. [DOI] [PubMed] [Google Scholar]
- 46.Becker R.C. COVID-19 update: covid-19-associated coagulopathy. J. Thromb. Thrombolysis. 2020 May 15:1–14. doi: 10.1007/s11239-020-02134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xijing Hospital . clinicaltrials.gov; 2020 Apr. Protective Effect of Aspirin on COVID-19 Patients.https://clinicaltrials.gov/ct2/show/NCT04365309 [Internet] [cited 2020 Jul 1]. Report No.: NCT04365309. Available from: [Google Scholar]
- 48.Kanamycin https://www.drugbank.ca/drugs/DB01172 [Internet]. [cited 2020 Apr 5]. Available from:
- 49.Vardanyan R.S., Hruby V.J. Synthesis of Essential Drugs. Elsevier; 2006. Antimycobacterial drugs.https://linkinghub.elsevier.com/retrieve/pii/B9780444521668500340 [Internet] [cited 2020 Apr 5]. pp. 525–34. Available from: [Google Scholar]
- 50.Rahman M.M., Yamamoto N., Morishima T., Maeno K., Nishiyama Y. Inhibition of herpes simplex virus type 2 replication in vitro by 1-N-pentadecanoyl-3’’-N-trifluoroacetyl kanamycin A. Antivir. Res. 1988 Feb;9(1–2):11–22. doi: 10.1016/0166-3542(88)90063-0. [DOI] [PubMed] [Google Scholar]
- 51.Yamada Y., Shimokata K., Yamada Y., Yamamoto N., Goshima F., Nishiyama Y. Inhibition of influenza A virus replication by a kanamycin derivative. Antivir. Res. 1991 Apr;15(3):171–182. doi: 10.1016/0166-3542(91)90064-x. [DOI] [PubMed] [Google Scholar]
- 52.(1) Anti-respiratory syncytial virus activity of kanamycin sulfate in vitro [Internet]. ResearchGate. [cited 2020 Mar 26]. Available from: https://www.researchgate.net/publication/288707279_Anti-respiratory_syncytial_virus_activity_of_kanamycin_sulfate_in_vitro.
- 53.Mohamed A.A. Study of in-vitro antiviral activity of aminoglycosides on foot and mouth disease virus. ARC J. Anim. Vet. Sci. 2019;5(1):24–30. [Google Scholar]
- 54.Clark C.H. Toxicity of aminoglycoside antibiotics. Mod. Vet. Pract. 1977 Jul;58(7):594–598. [PubMed] [Google Scholar]
- 55.Magallón J., Chiem K., Tran T., Ramirez M.S., Jimenez V., Tolmasky M.E. Restoration of susceptibility to amikacin by 8-hydroxyquinoline analogs complexed to zinc. bioRxiv. 2019 Apr 5:598912. doi: 10.1371/journal.pone.0217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amikacin https://www.drugbank.ca/drugs/DB00479 [Internet]. [cited 2020 Jul 3]. Available from:
- 57.PubChem Fenoterol. https://pubchem.ncbi.nlm.nih.gov/compound/3343 [Internet]. [cited 2020 Jul 3]. Available from:
- 58.Sherman K.E. Management of the hepatitis B virus/HIV–coinfected patient. Top Antivir Med. 2016 Nov 28;23(3):111–114. [PMC free article] [PubMed] [Google Scholar]
- 59.Potential inhibitors of SARS-CoV-2 main protease (mpro) identified from the library of FDA approved drugs using molecular docking studies - abstract - europe PMC. https://europepmc.org/article/ppr/ppr148893 [Internet]. [cited 2020 Jul 4]. Available from: [DOI] [PMC free article] [PubMed]
- 60.Deng L., Dai P., Ciro A., Smee D.F., Djaballah H., Shuman S. Identification of novel antipoxviral agents: mitoxantrone inhibits vaccinia virus replication by blocking virion assembly. J. Virol. 2007 Dec 15;81(24):13392–13402. doi: 10.1128/JVI.00770-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maestro S. Llc. New York, NY. 2017.
- 63.Release S. 4: Schrödinger Suite 2017-4 Protein Preparation Wizard. Epik, Schrödinger, LLC, New York, NY. 2017.
- 64.Sarma P, Shekhar N, Prajapat M, Avti P, Kaur H, Kumar S, Singh S, Kumar H, Prakash A, Dhibar DP, Medhi B. In-silico homology assisted identification of inhibitor of RNA binding against 2019-nCoV N-protein (N terminal domain) Journal of Biomolecular Structure and Dynamics. 2020 May;16:1–9. doi: 10.1080/07391102.2020.1753580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H, Saravanan KM, Yang Y, Hossain MT, Li J, Ren X, Pan Y, Wei Y. Deep learning based drug screening for novel coronavirus 2019-nCov. Interdisciplinary Sciences, Computational Life Sciences. 2020 Jun;1:1. doi: 10.1007/s12539-020-00376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]