Abstract
Viruses reshape the organization of the cell interior to achieve different steps of their cellular cycle. Particularly, viral replication and assembly often take place in viral factories where specific viral and cellular proteins as well as nucleic acids concentrate. Viral factories can be either membrane-delimited or devoid of any cellular membranes. In the latter case, they are referred as membrane-less replication compartments. The most emblematic ones are the Negri bodies, which are inclusion bodies that constitute the hallmark of rabies virus infection. Interestingly, Negri bodies and several other viral replication compartments have been shown to arise from a liquid-liquid phase separation process and, thus, constitute a new class of liquid organelles. This is a paradigm shift in the field of virus replication. Here, we review the different aspects of membrane-less virus replication compartments with a focus on the Mononegavirales order and discuss their interactions with the host cell machineries and the cytoskeleton. We particularly examine the interplay between viral factories and the cellular innate immune response, of which several components also form membrane-less condensates in infected cells.
Abbreviation: ADV, adenovirus; EBOV, Ebola virus; EBV, Epstein Barr virus; HCMV, human cytomegalovirus; HPV, human papillomavirus; IB, inclusion body; IDD, intrinsically disordered domain; IFN, interferon; ISG, interferon-stimulated genes; LLPS, liquid-liquid phase separation; MeV, measles virus; MNV, Mononegavirales; MuV, mumps virus; NB, Negri bodies; NiV, Nipah virus; PML, promyelocytic leukemia protein; RABV, rabies virus; RNP, ribonucleoprotein; RSV, respiratory syncytial virus; SG, stress granules; VRC, viral replication center; VSV, vesicular stomatitis virus
Keywords: Viral factory, Viral replication, Liquid organelle, Membrane-less compartment, Innate immunity, Interferon
1. Introduction
During their replication cycle, many viruses induce the formation of specialized intracellular compartments in the host cell. These structures often referred as viral inclusions, viral factories or viroplasms, concentrate viral proteins, nucleic acids and specific cellular factors. In many cases, those viro-induced compartments harbor essential steps of the viral cycle and constitute a platform facilitating viral replication and assembly but also protecting the viral genome from cellular defense mechanisms. Such viral factories have been now identified for a variety of non-related viruses [[1], [2], [3]].
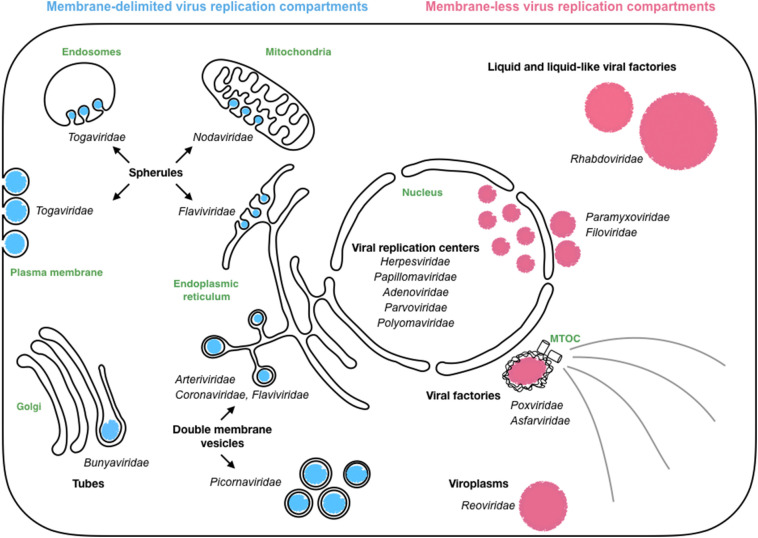
Viral factories are very heterogeneous. They can be associated with membranes from diverse organelles (mainly endoplasmic reticulum -ER-, late endosomes, lysosomes, and mitochondria) that they rearrange. This is the case for positive-strand RNA viruses leading to the formation of double-membrane vesicles (Coronaviridae, Arteriviridae, Picornaviridae and Flaviviridae) or spherules derived from diverse cellular membranes (Togaviridae, Nodaviridae and Flaviviridae) (Fig. 1 ).
Fig. 1.
Diversity of viro-induced compartments.
Those compartments are viral factories that host essential steps of the viral cycle and shield viral components from host defenses. These viral replication compartments can be membrane-delimited or membrane-less compartments.
However, for several negative-strand RNA, double stranded RNA and DNA viruses, viral factories are devoid of membranes. These compartments can be either cytosolic or nuclear and, in several instances, have been demonstrated to have properties like those of liquid organelles (Fig. 1).
Membrane-less liquid organelles contribute to the compartmentalization of the eukaryotic cell interior [[4], [5], [6], [7]]. They are referred as droplet organelles, proteinaceous membrane-less organelles or biomolecular condensates and are involved in a wide range of cell processes. They can be either located in the cytosol (for stress granules - SG [8,9] and P-bodies [5]) or in the nucleus (which is the case for Cajal bodies [10], nucleoli [11,12] and nuclear speckles [13]). They are very dynamic structures, extremely sensitive to their physicochemical environment, which assemble and disassemble much more rapidly than membrane-delimited organelles. Finally, they are highly enriched in some proteins that are much more concentrated in those structures than in the cytosol, as a result of liquid-liquid phase separation (LLPS) [7]. With the rapid identification of cellular membrane-less compartments and proteins that undergo LLPS in vitro, a major challenge in the field is to demonstrate unambiguously that a specific structure is indeed a phase-separated liquid body in the cellular context. Currently, common criteria for defining such a structure are that it is spherical, fuses, reversibly deforms when encountering a physical barrier, and recovers from photobleaching [14].
Here, we will review membrane-less viral compartments with a focus on those that have liquid organelles properties. We will discuss their composition and their interactions with the host cell machineries, particularly those of the innate immune system, of which several components also form membrane-less condensates in infected cells, and of the cytoskeleton.
2. Mononegavirales viral factories
2.1. The Mononegavirales order
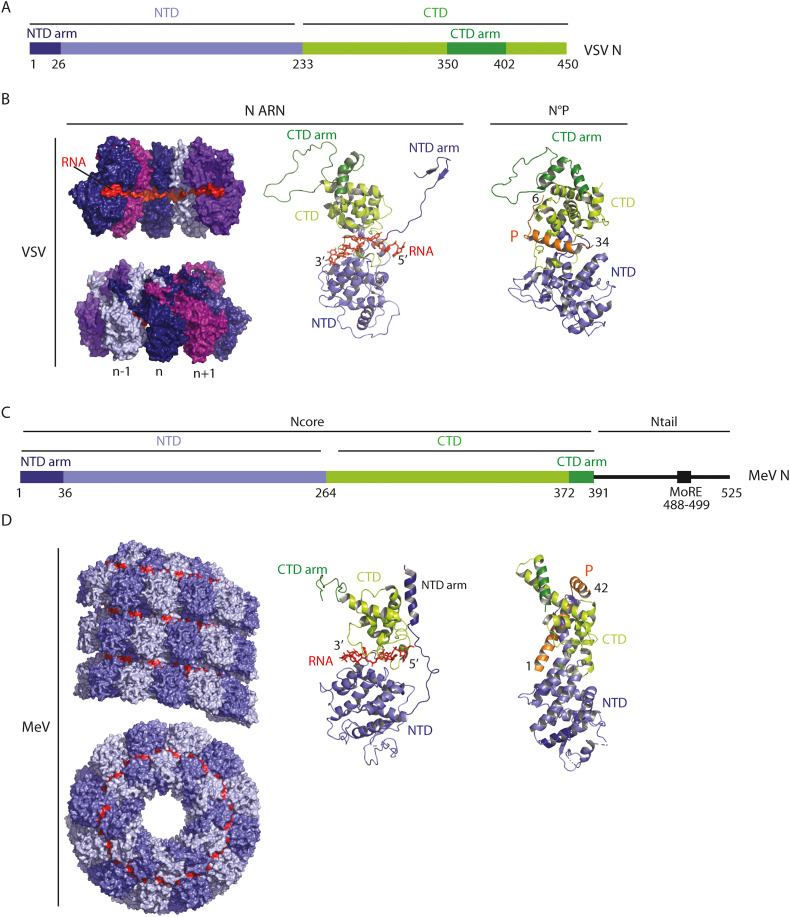
Mononegavirales (MNV) constitute a viral order which contains several viruses causing important human diseases (including rabies virus - RABV, Ebola virus - EBOV, measles virus - MeV, mumps virus - MuV, human respiratory syncytial virus - RSV). Their genome consists of a negative sense, single-stranded RNA molecule ranging from 10 to 15 kb, starting and ending with a non-coding leader and trailer sequence, respectively. The viral RNA is tightly associated with the nucleoprotein (N or NP) to form the helical ribonucleoprotein (RNP) (Fig. 2 ). The RNP recruits the viral RNA-dependent RNA polymerase (RdRp) complex, composed of the protein L (the polymerase which bears polyribonucleotidyltransferase and methyltransferase activities) and its non-enzymatic cofactors (P for rhabdoviruses, paramyxoviruses, and pneumoviruses, VP35 for filoviruses), to form the nucleocapsid. In the nucleocapsid, P acts as a tether between L and the RNP. The nucleocapsid is enwrapped by a lipid bilayer that is derived from a host cell membrane during the budding process. The matrix protein (M, VP40 for filoviruses) is located beneath the viral membrane and bridges the RNP and the lipid bilayer, which contains one or two transmembrane glycoproteins that are involved in viral entry.
Fig. 2.
Structures of rhabdovirus (VSV) and paramyxovirus (MeV) nucleoproteins.
A) Bar diagram showing the domain organization of VSV nucleoprotein.
NTD stands for N-terminal domain CTD and CTD for C-terminal domain.
B) Structure of VSV nucleoprotein.
Left part: Space-filling model of VSV N-RNA complex (X-Ray structure of a 10 N subunit ring (in shades of purple) associated with 90 RNA bases (in red) (2GIC.pdb) [158]. Each VSV N subunit interacts with 9 RNA bases. In this conformation, the RNA molecule is clamped at the interface of the NTD and the CTD. Each N subunit is shown in a different color indicating that the NTD arm from the nth subunit reaches over to the (n − 1)th sub-unit and its CTD arm reaches over to the (n + 1)th sub-unit. This arrangement leads to the interaction of both (n + 1)th NTD and (n − 1)th CTD with each other and the surface of sub-unit nth.
Middle part: Ribbon diagram of a N protomer associated with 9 RNA bases (in orange). Two small subdomains (NTD arm and CTD arm) emerge from NTD (in purple) and CTD (in green) resp.
Right part: Ribbon diagram of VSV N°P RNA free structure (3PMK.pdb) [44]. VSV NΔ21 (lacking the NTD arm) was crystallized in complex with the 60 first residues of VSV P (P60) in orange. P60 folds upon binding to N and avoid RNA binding by filling the RNA-binding groove of N.
C) Bar diagram showing the domain organization of MeV nucleoprotein.
Same abbreviations than in A; MoRE stands for molecular recognition element.
D) Structure of MeV Nucleoprotein.
Left part: Space-filling model of the structure of the MeV Ncore-RNA helical nucleocapsid (side view and top view) obtained by cryoEM (4UFT.pdb) [159]. The top view allows the visualization of the protomers. The RNA molecule is in red.
Middle part: Ribbon diagram of a MeV Ncore promoter associated with 6 RNA bases (in red).
Right part: Ribbon diagram of MeV N°P RNA free complex (5E4V.pdb [45]). Ncore (lacking the 21 first amino acids) was crystallized in complex with the 48 first residues of P (P48). P48 chaperone N°, preventing both binding to RNA and self-assembly.
The cell cycle of MNV is entirely cytoplasmic (with the notable exception of that of the Bornaviridae family members). The negative sense RNP, once released in the cytoplasm, constitutes the template for viral gene expression and replication by the RdRp complex. In this complex, P is bridging L and the template-associated nucleoproteins. Transcription begins at the 3′ end of the genomic RNA and results in the synthesis of a positive, uncapped and short leader RNA and capped and polyadenylated messenger RNAs (mRNAs). Viral mRNAs are then translated by the host cell translation machinery providing a source of N protein necessary to encapsidate the nascent RNA. This results in the switch of the activity of the RdRp complex from transcription to replication to produce RNPs containing full-length antigenomic RNA (positive sense), which in turn serve as templates for the synthesis of genomic RNA (negative sense). During the replication stage, when not bound to the viral genomic or antigenomic RNA, N is kept soluble by binding the amino-terminal region of P thus forming the so-called N0P complex (Fig. 2B, D) [[15], [16], [17]].
2.2. Inclusion bodies formed during MNV infection have liquid properties
Inclusion body (IB) formation is a hallmark of infection by members of the MNV. The most emblematic ones are the Negri bodies (NB) which are formed in the cytoplasm of neurons infected by RABV. They have been discovered by Adelchi Negri in 1903 [18,19] and are easily observed using Seller's staining. These structures have a diameter up to a few micrometers and have been used for decades as a histological proof of RABV infection.
In the case of rhabdoviruses (RABV and vesicular stomatitis virus - VSV) [[20], [21], [22]], filoviruses [23] and RSV [24], these IBs contain all the RNP components (N, L and P) as well as viral RNAs (genomic, antigenomic, and messengers) which are synthesized inside. Therefore, these inclusions are bona fide viral factories. It is also supposed that cytoplasmic IBs formed by paramyxoviruses such as MeV [25], MuV [26] or parainfluenza viruses [27,28] harbor similar viral activities. However, the IBs formed during Nipah virus (NiV, which also belongs to the Paramyxoviridae family but is a member of the Henipavirus genus) infection do not seem to support viral RNA synthesis. They are of two kinds: the first ones are in the periphery of the nucleus and recruit newly synthesized NiV genomes encapsidated by N proteins, which are then further transported to square-shaped crystalline inclusions located beneath the plasma membrane [29].
The viral factories of rhabdoviruses [30,31], paramyxoviruses [25,28,32] and filoviruses [23,33] as well as the peripheral NiV IBs [29] and the nuclear IBs formed by Borna disease virus phosphoprotein [34] are spherical (at least during the initial stages of infection), suggesting that they could be liquid organelles formed by phase separation. This liquid nature was confirmed by live-cell imaging for RABV [30], VSV [31] and MeV [25]. First, it was shown that when two spherical inclusions contact one another, they readily fuse and round up into a single larger spherical one [25,30,31]. They also reversibly deform when encountering a physical barrier and disappear when exposed to an osmotic shock [30]. Finally, fluorescence recovery after photobleaching (FRAP) measurements are also in agreement with the liquid nature of MNV IBs [25,30,31] and reveal that P, although more concentrated in the inclusions, can reversibly exchange with a cytoplasmic pool [25,30,31,34].
2.3. Segmented negative strand RNA viruses also forms IBs
Beyond the MNV order, IBs are also observed during segmented negative strand RNA virus infections. Indeed, it has been shown that cells infected by influenza A virus contain inclusions located in the vicinity of the ER exit site. These inclusions, which have been proposed to be involved in the control of the assembly process, have liquid properties [35]. Similarly, the spherical aspect of the granules, which concentrate the three Bunyavirus genome segments [36], suggests the possibility that they also have liquid properties.
2.4. Minimal systems recapitulating the liquid properties of MNV IBs
Co-expression of N and P proteins of paramyxoviruses and RABV after cell transfection also leads to the formation of spherical inclusions [25,30,37,38]. In the case of VSV, the presence of L protein is also required for such inclusions to be formed [31] whereas in the case of EBOV, NP alone is sufficient for IB generation [39]. For both RABV and MeV, the N-P inclusions formed in this minimal system have the same liquid characteristics as the viral factories [25,30]. In the other cases, the spherical aspect of the inclusions is a strong argument in favor of their liquid nature, but additional experiments are necessary to definitively conclude on this point.
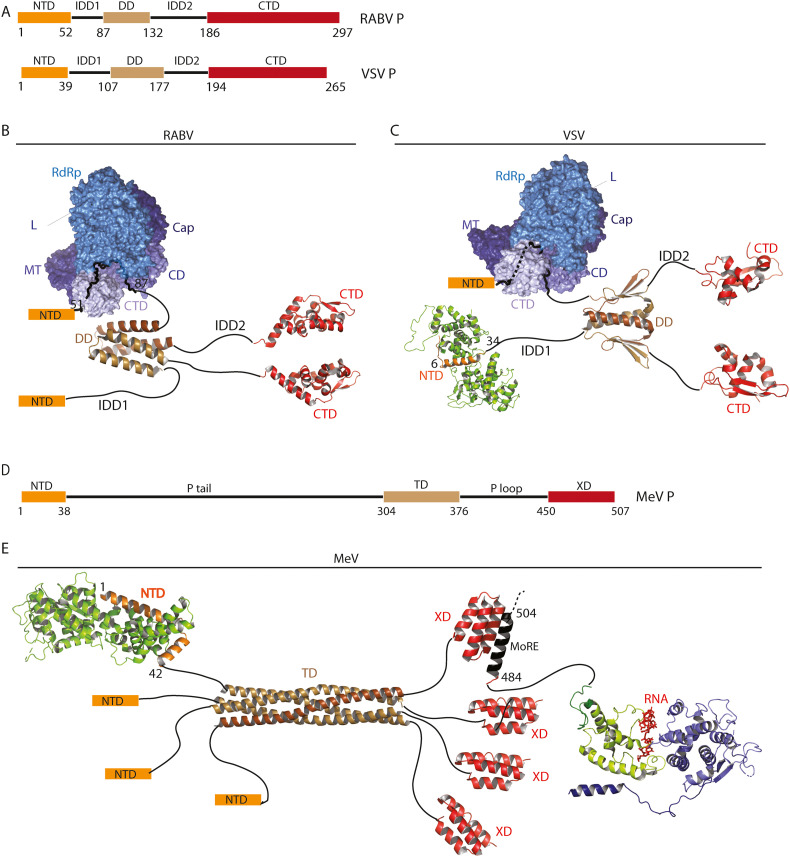
RABV and MeV minimal systems were used to identify P and N domains that are essential for inclusion formation. The phosphoprotein of paramyxoviruses and rhabdoviruses share a common modular organization [[40], [41], [42]] (Fig. 3 ). The N-terminal part of the protein is involved in the formation of the N0P complex [[43], [44], [45]] (Figs. 2B, D, 3C) and, for rhabdoviruses, in the interaction with the L protein [46,47] (Fig. 3B, D). The C-terminal domain (PCTD for RABV and VSV, XD for MeV) binds N associated with RNA [[48], [49], [50]] (Fig. 3). The XD domain of paramyxoviruses can also bind L [51]. Two central intrinsically disordered domains (IDD1 and IDD2 for RABV, Ptail and Ploop for MeV) are flanking an oligomerization domain. Rhabdovirus P is dimeric in solution [52,53] (Fig. 3B, C) whereas paramyxovirus P form tetramers [54] (Fig. 3E). Despite the absence of sequence similarity and the differences in the domain structures (Fig. 3), for both RABV and MeV P, it has been shown that the oligomerization domain, the second intrinsically disordered domain (IDD2/PLoop) and the C-terminal domain (PCTD/XD) are required for IB formation whereas the N-terminal part and the first intrinsically disordered domain are dispensable [25,30].
Fig. 3.
Structures of rhabdoviruses (RABV and VSV) and paramyxovirus (MeV) phosphoproteins.
A) Bar diagram showing the conserved domain organization of RABV and VSV P. NTD stands for N-terminal domain and CTD for C-terminal domain. IDD is for intrinsically disordered domain and DD for dimerization domain.
B) RABV P dimer. Only the X-ray structures of its dimerization domain (3L32.pdb and CTD (1VYI.pdb [50]) are known so far. RABV L interacts with the IDD1 of P. L is composed of an RNA-dependent RNA polymerase (RdRp) domain, capping (CAP) domain, connector domain (CD), methyltransferase (MT) domain, and C-terminal domain (CTD) (6UEB.pdb [47]).
C) VSV P dimer. The X-ray structure of the dimerization domain is indicated (2FQM.pdb [53]). The CTD (2K47.pdb [160]) binds to N-RNA complex. VSV L interacts with the N-terminal part of P (6U1X.pdb [46]). VSV RNA free N sub-units interacts with the P NTD (3PMK.pdb [44]) preventing RNA binding in the N RNA cavity.
D) Bar diagram showing the modular organization of MeV P. Same abbreviations than in A. TD strands for tetramerization domain.
E) MeV P tetramer associates via its oligomerization domain (3ZDO.pdb [161]).The P NTD binds to the N° RNA free sub-units (5E4V.pdb [45]). The XD C-terminal domain of P folds into a small helix bundle that interacts with a MoRE located in the extremity of the Ntail domain of N-RNA nucleocapsid (1T6O.pdb [162]).
More recently, it has been shown that MeV N (produced in association with the 50 first amino-terminal residues of P and thus in the form N0P50) and P expressed in E. coli form liquid-like membrane-less organelles upon mixing in vitro under physiological salt and protein concentrations [55]. As in the cell [25], the oligomerization domain, PLoop and XD are required for LLPS. In this system, it was also shown that the interaction between the disordered C-terminal part of N (NTail residues 400–525) and XD of P is essential for droplet formation. Indeed, a mutation of N (S491L) abrogating the NTail:PXD interaction and known to significantly decrease viral transcription in vivo resulted in suppression of phase separation. Finally, when RNA was added, it concentrated in the N-P droplets and was encapsidated by N protomers to form nucleocapsid-like particles. The rate of encapsidation within droplets was enhanced compared to the dilute phase, which suggests a function of LLPS in MeV replication [55].
Finally, in the case of EBOV, the C-terminal domain of NP (NP-Ct) is necessary for IB formation when NP is expressed alone. However, co-expression of the nucleocapsid component VP35 overcomes deletion of NP-Ct in triggering IB formation. This effect is mediated by an interaction between VP35 and NP implicating a central domain (CD, residues 480–500) of NP enriched in acidic residues and the interferon (IFN) inhibitory domain of VP35 [39].
Globally, MNV IBs share several features with cellular liquid organelles. Indeed, as other liquid organelles, MNV IBs contain RNAs (viral RNAs in infected cells or cellular ones in the minimal systems in which N is associated with cellular RNAs and forms N-RNA rings and short RNP-like structures [43,56]), RNA-binding proteins (N in this case) and proteins containing IDDs (P and sometimes N in this case). For several cellular proteins that phase-separate, cation-pi interactions between arginine and aromatic residues have been shown to be key in the process [57,58]. It is not known whether similar residues mediate the weak interactions that drive LLPS for MNV condensates. A comparison between RABV, MeV and EBOV nucleoprotein and phosphoprotein sequences with a focus on their IDDs do not reveal conserved specificities besides a slight enrichment in positive residues in RABV IDD2 and MeV Ploop (both required to observe LLPS in the minimal systems) (Table 1 ). Furthermore, alignment of the sequences of lyssaviruses phosphoproteins reveal a poor conservation of IDDs compared to the rest of the protein sequence in the viral genus [30].
Table 1.
Some characteristics of nucleoproteins and phosphoproteins of MNV involved in condensates formation.
| Protein (UniProt identifier) | Sequence | Asp/Glu frequencya | Lys/Arg frequencyb | Net charge per residuec | Phe/Tyr/Trp frequencyd |
|---|---|---|---|---|---|
| RABV P (P22363) | Full length 1–297 |
20/29 16.5% |
20/17 12.5% |
−0.040 | 11/6/3 6.7% |
| IDD1 59–90 |
5/6 35.5% |
2/2 12.9% |
−0.218 | 1/1/0 6.5% |
|
| IDD2 132–182 |
1/4 9.8% |
4/5 17.6% |
+0.078 | 2/0/0 3.9% |
|
| MeV P (P03422) | Full length 1–507 |
32/41 14.4% |
31/27 11.4% |
−0.029 | 7/7/2 3.2% |
| P tail 38–304 |
16/24 15% |
8/14 8.3% |
−0.068 | 3/4/1 3.0% |
|
| P loop 376–449 |
5/3 10.8% |
8/5 17.6% |
+0.068 | 1/0/0 1.35% |
|
| EBOV VP35 (Q05127) | Full length 1–340 |
17/20 10.9% |
16/18 10% |
−0.009 | 9/6/3 5.3% |
| IDD1 48–81 |
1/1 5.9% |
2/1 8.8% |
+0,029 | 0/1/0 2.9% |
|
| IDD2 159–213 |
1/9 18.2% |
1/3 7.3% |
−0,109 | 1/2/1 7,3% |
|
| RABV N (P16285) | Full length 1–450 |
22/32 12% |
26/23 10.9% |
−0,011 | 27/21/3 11.3% |
| MeV N (Q89933) | Full length 1–525 |
37/36 13.9% |
17/42 11.2% |
−0.112 | 17/12/5 6.5% |
| Ntail 392–525 |
13/11 17.9% |
3/15 13.43% |
−0,045 | 1/2/0 2.2% |
|
| EBOV NP (P18272) | Full length 1–739 |
59/59 16% |
38/33 9.6% |
−0.063 | 26/21/4 6.9% |
| NTail 413–640 |
41/16 25% |
7/11 7.9% |
−0.171 | 2/5/0 3% |
Both full length and IDD sequences have been analyzed.
RABV N do not contain IDDs. Putative IDDs for EBOV NP and VP35 have been identified using IUPred2A [151].
Number of Asp residues and number of Glu residues. Frequency of negatively charged residues.
Number of Lys residues and number of Arg residues. Frequency of positively charged residues.
Net charge per residue = (number of Arg residues + number of Lys residues − number of Asp residues − number of Glu residues) / total number of residues in the sequence.
Number of Phe residues, number of Tyr residues and number of Trp residues. Frequency of aromatic residues.
3. Beyond MNV: membrane-less replication compartments of DNA and other RNA viruses
3.1. DNA viruses
The first viral factories which were characterized were those of large DNA viruses such as the Poxviridae, the Iridoviridae and the Asfarviridae [[59], [60], [61], [62]]. Those cytoplasmic factories are devoid of membrane and located near the microtubule organizing center. They have several characteristics reminiscent of those of the cellular aggresomes which concentrate misfolded proteins in the cell [63]. They recruit mitochondria in their vicinity, contain molecular chaperones such as heat-shock proteins (HSP) and are surrounded by a vimentin cage [64]. However, no data are supporting the possible liquid nature of those cytoplasmic structures.
Other DNA viruses belonging to Herpesviridae, Adenoviridae, Parvoviridae, Polyomaviridae and Papillomaviridae families induce the formation of membrane-less assemblies inside the nucleus termed viral replication compartments (or centers), hereafter referred as VRCs [65], which concentrate viral proteins and nucleic acids, incoming viral genomes and host proteins. Herpesviruses and adenoviruses VRCs also coalesce as infection progresses and can fuse together in a liquid-like manner [[66], [67], [68], [69]]. However, unlike many other cellular liquid organelles [[70], [71], [72]], HSV-1 VRCs are not disrupted by treatment with 1,6-hexanediol and a model of their formation, not based on LLPS, has been proposed [69]. However, sensitivity to hexanediol is insufficient to unequivocally demonstrate that a structure is formed via LLPS [14] and the identification of the physicochemical principles underlying HSV-1 VRCs remains an open issue. On the other hand, EBNA2 and EBNA-LP proteins of Epstein-Barr virus (EBV) have been shown to form punctate inclusions in the nucleus having properties of liquid organelles. Furthermore, mixing EBNA2 and EBNA-LP in vitro was sufficient to induce liquid phase separation. This phase separation of EBNA2 and EBNA-LP was proposed to be important for their transcription factor activity [73].
The property of DNA viruses to form viro-induced compartments devoid of membranes can be extended to bacteriophages. Indeed, Pseudomonas chlororaphis phage 201φ2-1 assembled a compartment that separates viral DNA from the cytoplasm. However, this compartment in which DNA replication and transcription occur is not formed by LLPS but is rather completely enclosed by an apparently contiguous protein shell [74].
3.2. dsRNA viruses
Double strand RNA (dsRNA) viruses are also known to induce the formation of membrane-less cytosolic electron-dense inclusions. This is particularly documented for the Reoviridae family of viruses which contain 10 (for reoviruses) or 11 (for rotaviruses) segments of genomic dsRNA. Those organelles are referred as viroplasms. They constitute the site of viral genome transcription and replication, as well as the site of packaging of the newly synthesized pregenomic RNA segments into the viral cores [[75], [76], [77], [78]]. Viroplasms appear to be spherical and can fuse together [79,80] which indicates that they have liquid properties. For rotaviruses, viroplasms are nucleated by two essential non-structural proteins, NSP2 and NSP5, and the inner virion capsid protein VP2, with NSP5 being crucial for both the recruitment of viroplasmic proteins and the architectural assembly of the viroplasms. In non-infected cells, co-expression of NSP5 either with NSP2 or VP2 leads to the formation of viroplasm-like structures [81,82]. Interestingly, NSP5 shares several characteristics with the phosphoproteins of MNV as it is phosphorylated, forms oligomers and contains intrinsically disordered segments [83]. Similarly, for reoviruses, the expression of the non-structural protein μNS alone leads to the formation of large inclusions that are similar to the viroplasms [84]. However, unlike NSP5, μNS is not predicted to contain long intrinsically disordered segments.
3.3. Positive strand RNA viruses
Although the viral factories of positive stand RNA viruses are associated with membranes, several of those viruses also form spherical intracytoplasmic inclusions that might have liquid properties. For example, depending on their genus, coronavirus nucleocapsid proteins (N) can form inclusion either in the cytoplasm [85] or in the nucleus in association with the nucleolus [86,87]. In keeping with this idea, it has been shown in vitro that the SARS-CoV-2 N protein contains disordered regions and phase separates with RNA [[88], [89], [90], [91]]. The phase separation is regulated by N phosphorylation [91]. Interestingly, in vitro, N also partitions into liquid phases formed by several human RNA-binding proteins [89], which is reminiscent of coronavirus N ability to associate with the nucleolus. These observations made in acellular systems remain to be consolidated by data obtained in infected cells.
4. Interaction of viral liquid IBs with host-cell machineries
4.1. Viral IB proteomes
LLPS is an efficient process for viral factories enrichment in specific cellular factors. They can be recruited by two different ways. First, their properties can induce their preferential partitioning in the dense phase. Second, they may be recruited by a viral or cellular protein which itself preferentially partitions in the IBs.
For RABV, several cellular proteins are concentrated in NBs. This includes Hsp70 [21,92], the focal adhesion kinase (FAK) [93], chaperonins CCTα and CCTγ [94,95], endothelial nitric oxide synthase [96] and ubiquitinylated proteins [21]. Globally, the cellular proteins identified in MNV IBs are extremely diverse. Another heat shock protein (Hsp72) is found associated with MuV IBs [26]. The WD repeat-containing protein 5 (WDR5), a subunit of histone H3 lysine 4 methyltransferase, as well as the actin-modulating protein cofilin [97,98] concentrate in MeV ones. The phosphorylated mitogen-activated protein kinase p38, a key regulator of cellular inflammatory and stress responses, and the O-linked N-acetylglucosamine (OGN) transferase, an enzyme that catalyzes the posttranslational addition of OGN to protein and is also involved in stress response, are sequestered in RSV IBs [99]. Finally, EBOV NP recruits several proteins into the IBs, which include the nuclear RNA export factor NFX1 to drive viral protein synthesis [100] and the histone-lysine-methyltransferase SMYD3 [101] as well as the CAD protein [102] to facilitate mRNA transcription and replication.
Mass spectrometry analyses of pull-down complexes of tagged NSP2 and NSP5 from human rotavirus incubated with extracts from uninfected rotavirus-permissive MA104 cells revealed the presence of several host heterogeneous nuclear RNPs (hnRNPs) and AU-rich element-binding proteins (ARE-BPs) [103]. Finally, the composition of HSV-1 VRC has been investigated by immunoprecipitation of ICP8 that appears as one of their most important components. This has allowed the identification of more than 50 viral and cellular proteins, mainly involved in DNA replication, DNA repair, chromatin remodelling, transcription, and RNA processing [104].
In conclusion, although a significant number of proteins have been found associated with viral IBs, the precise role of those associations with, or sequestrations in, IBs or VRCs as well as the underlying molecular mechanisms remain to be determined and will certainly constitute a new field of research in the coming years.
4.2. Interactions of viral factories with the cytoskeleton and cellular membranes
The cytoskeleton plays an important role in the morphogenesis and evolution all along the cycle of viral factories. The formation of the cytoplasmic viral factories of large DNA viruses requires an intact microtubule network and induces the redistribution of the intermediate filament protein vimentin that forms a cage around the viral assembly site [64] (Fig. 1).
For MNV, the dependence on the microtubule network may vary depending on the viral family. In the case of RABV, the viral nucleocapsids, once ejected from the NBs, are transported further away along the microtubule network where they can form new viral factories which are detected as NBs of intermediate size. In the presence of Nocodazole (NCZ), a drug that depolymerizes microtubules, RABV NBs of intermediate size are no more detected and a single exceptionally large NB is observed [21,30]. In fact, in the presence of NCZ, RNPs are still ejected from NBs but cannot be transported further away and the newly formed viral factories remain located in the vicinity of, and rapidly fuse with, the initial NB which then becomes much larger [30]. For MeV, the initial small and spherical IBs progressively coalesce and fuse in the nucleus periphery to form bigger inclusions that lose their spherical shape and most probably their liquid properties. In the presence of specific inhibitors of dynein motor function, the vast majority of IBs remains small, spherical, and uniformly distributed throughout the cytoplasm [25]. This suggests a role of the microtubule network in the transport and maturation of the initial IBs. A remarkably similar observation has been made on the Reoviridae family as, here again, the perinuclear condensation of the viral factories is strictly dependent on an intact microtubule network [80].
The role of actin filaments in IB formation and maturation has not been investigated so far but in the presence of the actin depolymerizing agent Cytochalasin D, RABV NBs appear to be more fragmented and smaller [30].
Finally, at the late stages of RABV infection, NBs become wrapped by a double membrane, seemingly derived from rough ER [21]. They lose their spherical shape [21,30] and virions are observed that bud from NBs into the compartment delimited by the associated double membrane [21,105]. The molecular bases of this association between NBs and cell membranes are unknown and whether such an association can be extended to other MNV factories is still an open question. Interestingly, ER contains contact site domains, which tether cytoplasmic liquid organelles such as P-bodies and drive their fission in an active process [106]. It is not excluded that RABV has hijacked a cellular machinery specifically involved in this process or that the same basic physicochemical principles are at work in both cases.
5. Interplay between viral IBs and innate immunity
5.1. MNV proteins counteract innate immunity
The ubiquitous nature of liquid viral factories among negative-strand RNA viruses makes them a signature of infection and one might hypothesize that the cells have evolved a mechanism allowing the sensing of such structures and/or their destabilization by the product of some interferon-stimulated genes (ISG). On the other hand, the sequestration of viral RNAs (and particularly double-stranded RNA exposing 5′triphosphate) inside the viral factories raises the question of their accessibility to cytosolic pathogen recognition receptors such as RIG-I and MDA-5 [107].
The subtle interplay between viral IBs and innate immunity is particularly exemplified by RABV for which P is not only one of the major component of the NBs but also the major viral counteractant of the innate immune response [108]. First, P has a critical role in suppression of IFN production by blocking the phosphorylation of the transcription factor interferon regulatory factor 3 (IRF-3) [109]. Second, the interaction of P with STAT1 leads to the inhibition of IFN signaling by different processes including inhibition of STAT1-DNA binding [110] and STAT1 sequestration away from the nucleus [111].
Similarly, EBOV VP35, which is concentrated in the IBs largely counteracts the innate immunity. It binds double-stranded RNA and inhibits IFN production induced by RIG-I signaling [112,113], prevents PKR activation [114] and impairs the function of IFN regulatory factor-activating kinases IKKε and TBK-1 [115].
In the case of paramyxoviruses, the proteins that inhibit IFN production and counteract IFN response, although expressed from the P gene, are distinct from P [116]. They are either expressed from an alternate reading frame or from an alternate transcript due to the presence of an editing site. C protein of MeV corresponds to the former case and has a completely different amino-acid sequence from P whereas V protein corresponds to the latter case and has the same amino-terminal part as P but a distinct C-terminal domain. Consequently, V is missing the P domain required to associate with IBs and V protein has primarily a diffuse nuclear distribution [117].
Therefore, MNV evolved different strategies to counteract innate immunity. Some viruses may sequester the proteins involved in those pathways inside the viral IBs whereas others keep them away from the viral factory.
5.2. Innate immunity sensors are found in membrane-less condensates
Several cytosolic sensors of the innate immunity are also associated with liquid organelles. The best characterized of those organelles are the SGs which are formed when the cell is under a cytoplasmic stress. They are storage sites containing translationally silenced mRNPs that can be released to resume translation after the stress subsides. SGs can be induced by viral infections [9,118] and are thought to have antiviral activities as they contain RIG-I and MDA-5 [[119], [120], [121]]. Interestingly, during RABV infection, SGs come into close contact with NBs [9] but do not fuse with them [30]. This indicates that NBs and SGs are made of non-miscible liquid phases. However, they exchange some material as the mRNAs (but not the genomes and antigenomes) are transported from NBs, where they are synthesized, into SGs [9]. For VSV, it has been shown that SGs associated proteins such as Poly(RC) Binding Protein 2 (PCBP2), T-cell-restricted intracellular antigen 1 (TIA1), and TIA1-related protein (TIAR) are associated with the viral factories. However, the bona fide SG markers, such as eukaryotic initiation factor 3 (eIF3) or eIF4A were not present in the factory [122]. In accordance with these results, it is worth noting that TIA-1 exerts an antiviral effect on both RABV [9] and VSV [122].
Similarly, mammalian orthoreovirus (MRV) infection also induces the formation of SGs [123]. However, as infection proceeds, normal SG puncta are disrupted, and SG-associated proteins localized to the periphery of viral factories [124]. SGs alteration is due to MRV σNS protein association with Ras-GAP SH3-binding protein 1 (G3BP1) [124], a double-stranded nucleic acid helicase which is a master regulator of SG formation [125]. By comparing MRV replication on wild-type and G3BP1−/− MEFs, it was shown that G3BP1 inhibits viral growth [124]. It has been suggested that G3BP1 relocalization to the viral factory periphery induced by σNS induces SG disruption in order to facilitate MRV replication in the host translational shutoff environment [124]. On the other hand, for rotaviruses, which also belong to the Reoviridae family, it appears that most of the main SG components are present in the viroplasms but that there is a selective exclusion of G3BP1. This sequestration promotes progeny virus production [126]. Taken together, all those examples indicate that there is a general, although remarkably diverse, interplay between SGs and viral factories.
More recently, it has been shown that the cyclic GMP-AMP synthase (cGAS, which leads to the production of the secondary messenger cyclic GMP-AMP), a major sensor of cytosolic DNA from invading viruses which triggers innate immune responses, induced the formation of liquid-like droplets by binding DNA in which cGAS was activated [127]. In a cellular context, the liquid phase separation and the IFN response to intracellular DNA are both dependent on the presence of G3BP1, which binds cGAS. Finally, an RNA-dependent association with PKR promoted the formation of those G3BP1-dependent, membraneless cytoplasmic structures necessary for the DNA-sensing function of cGAS in human cells [128]. Thus, the nucleic acid sensing pathways involved in viral infection detection require the formation of specialized subcellular structures having liquid properties.
5.3. MxA and PML, two ISG products, form membrane-less condensates
Several ISG products also form membrane-less condensates. This is the case of human MxA, a cytoplasmic 70-kDa dynamin-family large GTPase which is induced in cells exposed to type I and III IFNs and has a broad spectrum of antiviral activities [129]. Even in uninfected cells, MxA forms spherical or irregular bodies of variable size, filaments, and sometimes a reticulated network that reversibly disassemble/reassemble within minutes of sequential decrease/increase, respectively, in tonicity of extracellular medium [71]. Furthermore, in VSV-infected Huh7 cells, previously transfected with a plasmid expressing GFP-MxA, N protein is associated with spherical GFP-MxA condensates. Moreover, in the GFP-MxA positive cells, N is much less expressed than GFP-MxA negative cells present in the same cell layer indicating a strong antiviral effect of MxA.
Promyelocytic leukemia protein (PML), which is the organizer of the PML nuclear bodies, is also induced by IFN. PML nuclear bodies also contain two other resident proteins, the ISG product speckled protein of 100 kDa (Sp100) and death-associated dead protein (Daxx), as well as several other proteins transiently recruited in PML bodies in response to different stimuli [130]. PML bodies as other nuclear bodies exhibit liquid-like properties [131,132]. The initiation of VRCs of DNA viruses occurs at sites near PML-nuclear bodies. Human papillomavirus (HPV) infection requires the presence of PML protein suggesting that PML bodies are essential to establish infection [133,134]. However, Sp100 and DAXX, which act as transcriptional repressors, restrict viral gene expression in ADV [135], HPV [136], HSV-1 [137], and HCMV [138] infections. Therefore, it is not surprising that most of the DNA viruses that replicate in the nucleus have evolved strategies to disrupt the PML bodies and to degrade or inhibit their components [[138], [139], [140], [141]]. As a well-characterized example, incoming HSV-1 genomes transiently co-localize with PML nuclear bodies [142], which surround and encapsulate incoming viral genomes before being disrupted by newly synthesized ICP0 viral proteins [137,139,143]. The antiviral action of PML nuclear bodies is also observed on RNA viruses including dengue virus [144], influenza virus [145] and rhabdoviruses [146,147]. Here again, viruses have evolved mechanisms that counteract PML function. As an example, RABV P interacts with PML and retains it in the cytoplasm to alter PML nuclear bodies [148] and consequently is thought to counteract the antiviral effect of isoform PML IV against RABV [146].
6. Conclusion
The discovery that several viro-induced membrane-less compartments have liquid properties and are formed by LLPS is a paradigm shift in the field (Table 2 ). Indeed, these compartments provide a functional microenvironment, of which the physicochemical properties remain to be characterized, for the optimal working of the viral replication machinery. Understanding the molecular basis of the weak interactions that keep the cohesion of those compartments might lead to the development of drugs that destabilize the viral factories or that concentrate inside to target the viral enzymes more efficiently.
Table 2.
Membrane-less viral factories. MeV: measles virus; HPIV3: human parainfluenza virus 3; PIV5: parainfluenza virus 5; NiV: Nipah virus; MuV: mumps virus; SV5: simian virus 5; hRSV: human respiratory syncytial virus; hMPV: human metapneumovirus; VSV: vesicular stomatitis virus; RABV: rabies virus; BoDV: Borna disease virus; EBOV: Ebola virus; IAV: influenza A virus; RVFV: Rift Valley fever virus; ReoV: reovirus; RotaV: rotavirus; EBV: Epstein-Barr virus; HSV-1: herpes virus 1; HCMV: human cytomegalovirus; KSHV: Kaposi's sarcoma herpes virus; VZV: varicella zoster virus; VV: vaccinia virus; ASFV: asfavirus.
| Genome organization | Order | Family | Virus | Liquid properties | Viral proteins found in IBs | Minimal system available (viral proteins required) | References |
|---|---|---|---|---|---|---|---|
| Non-segmented - ssRNA | Mononegavirales | Paramyxoviridae | MeV | Yes | N, P, L | Yes (N, P) | [25,55] |
| HPIV3 | Yes | N, P | Yes (N, P) | [28,152] | |||
| PIV5 | Suspected | N | No | [27] | |||
| NiVa | Suspected | N, P, L | Yes (N, P) | [29] | |||
| NiVa | Crystalline inclusions | N, P, M | Yes (N, P, M) | [29] | |||
| MuV | Suspected | P | No | [26] | |||
| SV5 | Suspected | N, P | No | [32] | |||
| Pneumoviridae | hRSV | Suspected | N, P, L | Yes (N, P, L, M2-1) | [24,153] | ||
| hMPV | Suspected | N, P | Yes (N, P) | [37,154] | |||
| Rhabdoviridae | VSV | Yes | N, P, L | Yes (N, P, L) | [31] | ||
| RABV | Yes | N, P, L | Yes (N, P) | [30] | |||
| Bornaviridae | BoDV | Suspected | P | No | [34] | ||
| Filoviridae | EBOV | Suspected | VP24, VP30, VP35, VP40, NP, L | Yes (NP) | [23,155] | ||
| Segmented - ssRNA | Articulavirales | Orthomyxoviridae | IAV | Yes | RNPs | No | [35] |
| Bunyavirales | Bunyaviridae | RVFV | Suspected | Unknown | No | [36] | |
| Segmented dsRNA | Reovirales | Reoviridae | ReoV | Suspected | σNS, μNS | Yes (μNS) | [78,84] |
| RotaV | Yes | NSP5, NSP2, VP2 | Yes (NSP5, NSP2, VP2) | [78,79,81] | |||
| dsDNA | Herpesvirales | Herpesviridae | EBV | Yes | EBNA2, EBNALP | Yes (EBNA2, EBNA-LP) | [73] |
| EBV | Unknown | BZRF, BNRF1 | No | [156] | |||
| HSV-1 | Unknownb | ICPs 0, 4, 8, 27, UL9, U42 | No | [156] | |||
| HCMV | Unknown | IE1, IE2, UL112–113 | No | [156] | |||
| KSHV | Unknown | LANA, Orfs 6, 9, 59 | No | [156] | |||
| Chitovirales | Poxviridae | VV | Unknown | RNA Pol, VITF-3 | No | [60,62,157] | |
| Asfuvirales | Asfaviridae | ASFV | Unknown | DNA Pol, ligase, helicase | No | [64] |
For NiV, two types of inclusions are observed.
For HSV-1, a model of VRC formation that is not based on liquid-liquid phase separation has been proposed [69].
An exciting field of research is the identification of the proteome of those compartments. Until now, only a few specific cellular factors, which directly interact with viral proteins such as the nucleoproteins and phosphoproteins of MNV, have been shown to concentrate in these structures. The proteome of the viral factories is probably much larger and some proteins might also preferentially partition in the organelle liquid phase due to their physicochemical properties without strongly interacting with any viral protein. As the liquid nature of MNV viral factories precludes their purification, original techniques will be required to map the complete proteome. A promising approach is proximity labeling which has already been used to map the proteome of SGs and processing bodies [125,149,150].
Interactions between viral factories and several cellular components such as the cytoskeleton and the cellular membrane compartments and their role at distinct stages of the cycle also remain to be finely characterized. Here again, understanding those processes may contribute to the development of novel antiviral strategies.
However, it is very likely that the most significant impact of the discovery of the liquid nature of viral factories will be in the area of innate immunity. Indeed, the liquid nature of viral factories as well as the increasing number of innate immunity actors that have been demonstrated to form biomolecular condensates, invite us to revisit the interactions between the viral infection and the cellular defenses. This interplay is certainly much more subtle than currently thought. Therefore, we look forward to an integrated vision of the interactions between viruses and innate immunity that takes into account all the novel data of this booming field.
CRediT authorship contribution statement
All the authors discussed the ideas presented in this review. QN and YG wrote the initial draft. AAA, CLG participated in manuscript editing. AAA, CLG and QN made the figures.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the CNRS, the University of Paris-Saclay, la Fondation Bettencourt Schueller and by the Agence Nationale de la Recherche (ANR-19-CE15-0024-01). QN is a post-doctoral fellow supported by a grant on the ANR contract.
Footnotes
State without borders: Membrane-less organelles and liquid–liquid phase transitions edited by Vladimir N Uversky.
References
- 1.Novoa R.R., Calderita G., Arranz R., Fontana J., Granzow H., Risco C. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell. 2005;97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Netherton C.L., Wileman T. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr Opin Virol. 2011;1:381–387. doi: 10.1016/j.coviro.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Castro I.F., Volonté L., Risco C. Virus factories: biogenesis and structural design. Cell Microbiol. 2013;15:24–34. doi: 10.1111/cmi.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nature Reviews Molecular Cell Biology. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 6.Courchaine E.M., Lu A., Neugebauer K.M. Droplet organelles? The EMBO Journal. 2016;35:1603–1612. doi: 10.15252/embj.201593517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uversky V.N. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Current Opinion in Structural Biology. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 8.Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolic J., Civas A., Lama Z., Lagaudrière-Gesbert C., Blondel D. Rabies virus infection induces the formation of stress granules closely connected to the viral factories. PLOS Pathogens. 2016;12 doi: 10.1371/journal.ppat.1005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Handwerger K.E., Cordero J.A., Gall J.G. Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol Biol Cell. 2005;16:202–211. doi: 10.1091/mbc.E04-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brangwynne C.P., Mitchison T.J., Hyman A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. PNAS. 2011;108:4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marzahn M.R., Marada S., Lee J., Nourse A., Kenrick S., Zhao H., Ben-Nissan G., Kolaitis R., Peters J.L., Pounds S., Errington W.J., Privé G.G., Taylor J.P., Sharon M., Schuck P., Ogden S.K., Mittag T. Higher-order oligomerization promotes localization of SPOP to liquid nuclear speckles. EMBO J. 2016;35:1254–1275. doi: 10.15252/embj.201593169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whelan S.P.J., Barr J.N., Wertz G.W. In: Biology of negative strand RNA viruses: the power of reverse genetics. Kawaoka Y., editor. Springer; Berlin, Heidelberg: 2004. Transcription and replication of nonsegmented negative-strand RNA viruses; pp. 61–119. [DOI] [PubMed] [Google Scholar]
- 16.Albertini A.A.V., Ruigrok R.W.H., Blondel D. In: Advances in Virus Research. Jackson A.C., editor. Academic Press; 2011. Chapter 1 - rabies virus transcription and replication; pp. 1–22. [DOI] [PubMed] [Google Scholar]
- 17.Kolakofsky D. Paramyxovirus RNA synthesis, mRNA editing, and genome hexamer phase: a review. Virology. 2016;498:94–98. doi: 10.1016/j.virol.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 18.Negri A. Contributo allo studio dell’eziologia della rabbia. Boll Soc Med-Chir Pavia. 1903:1459–1460. [Google Scholar]
- 19.Kristensson K., Dasturt D.K., Manghanit D.K., Tsiangt H., Bentivoglio M. Rabies: interactions between neurons and viruses. A review of the history of Negri inclusion bodies. Neuropathol Appl Neurobiol. 1996;22:179–187. [PubMed] [Google Scholar]
- 20.Heinrich B.S., Cureton D.K., Rahmeh A.A., Whelan S.P.J. Protein expression redirects vesicular stomatitis virus RNA synthesis to cytoplasmic inclusions. PLOS Pathogens. 2010;6 doi: 10.1371/journal.ppat.1000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lahaye X., Vidy A., Pomier C., Obiang L., Harper F., Gaudin Y., Blondel D. Functional characterization of Negri bodies (NBs) in rabies virus-infected cells: evidence that NBs are sites of viral transcription and replication. Journal of Virology. 2009;83:7948–7958. doi: 10.1128/JVI.00554-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ménager P., Roux P., Mégret F., Bourgeois J.-P., Le Sourd A.-M., Danckaert A., Lafage M., Préhaud C., Lafon M. Toll-like receptor 3 (TLR3) plays a major role in the formation of rabies virus Negri bodies. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoenen T., Shabman R.S., Groseth A., Herwig A., Weber M., Schudt G., Dolnik O., Basler C.F., Becker S., Feldmann H. Inclusion bodies are a site of ebolavirus replication. Journal of Virology. 2012;86:11779–11788. doi: 10.1128/JVI.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rincheval V., Lelek M., Gault E., Bouillier C., Sitterlin D., Blouquit-Laye S., Galloux M., Zimmer C., Eleouet J.-F., Rameix-Welti M.-A. Functional organization of cytoplasmic inclusion bodies in cells infected by respiratory syncytial virus. Nature Communications. 2017;8:563. doi: 10.1038/s41467-017-00655-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y., Su J.M., Samuel C.E., Ma D. Measles virus forms inclusion bodies with properties of liquid organelles. Journal of Virology. 2019;93 doi: 10.1128/JVI.00948-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh H., Kubota T., Kita S., Nakatsu Y., Aoki N., Mori Y., Maenaka K., Takeda M., Kidokoro M. Heat shock protein 70 regulates degradation of the mumps virus phosphoprotein via the ubiquitin-proteasome pathway. Journal of Virology. 2015;89:3188–3199. doi: 10.1128/JVI.03343-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlos T.S., Young D.F., Schneider M., Simas J.P., Randall R.E. Parainfluenza virus 5 genomes are located in viral cytoplasmic bodies whilst the virus dismantles the interferon-induced antiviral state of cells. J Gen Virol. 2009;90:2147–2156. doi: 10.1099/vir.0.012047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S., Chen L., Zhang G., Yan Q., Yang X., Ding B., Tang Q., Sun S., Hu Z., Chen M. An amino acid of human parainfluenza virus type 3 nucleoprotein is critical for template function and cytoplasmic inclusion body formation. Journal of Virology. 2013;87:12457–12470. doi: 10.1128/JVI.01565-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ringel M., Heiner A., Behner L., Halwe S., Sauerhering L., Becker N., Dietzel E., Sawatsky B., Kolesnikova L., Maisner A. Nipah virus induces two inclusion body populations: identification of novel inclusions at the plasma membrane. PLOS Pathogens. 2019;15 doi: 10.1371/journal.ppat.1007733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikolic J., Le Bars R., Lama Z., Scrima N., Lagaudrière-Gesbert C., Gaudin Y., Blondel D. Negri bodies are viral factories with properties of liquid organelles. Nature Communications. 2017;8 doi: 10.1038/s41467-017-00102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinrich B.S., Maliga Z., Stein D.A., Hyman A.A., Whelan S.P.J. Phase transitions drive the formation of vesicular stomatitis virus replication compartments. MBio. 2018;9 doi: 10.1128/mBio.02290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fearns R., Young D.F., Randall R.E. Evidence that the paramyxovirus simian virus 5 can establish quiescent infections by remaining inactive in cytoplasmic inclusion bodies. Journal of General Virology. 1994;75:3525–3539. doi: 10.1099/0022-1317-75-12-3525. [DOI] [PubMed] [Google Scholar]
- 33.Schudt G., Kolesnikova L., Dolnik O., Sodeik B., Becker S. Live-cell imaging of Marburg virus-infected cells uncovers actin-dependent transport of nucleocapsids over long distances. Proc Natl Acad Sci U S A. 2013;110:14402–14407. doi: 10.1073/pnas.1307681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charlier C.M., Wu Y.-J., Allart S., Malnou C.E., Schwemmle M., Gonzalez-Dunia D. Analysis of Borna disease virus trafficking in live infected cells by using a virus encoding a tetracysteine-tagged P protein. J Virol. 2013;87:12339–12348. doi: 10.1128/JVI.01127-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alenquer M., Vale-Costa S., Etibor T.A., Ferreira F., Sousa A.L., Amorim M.J. Influenza A virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nature Communications. 2019;10:1629. doi: 10.1038/s41467-019-09549-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schreur P.J.W., Kortekaas J. Single-molecule FISH reveals non-selective packaging of Rift Valley fever virus genome segments. PLOS Pathogens. 2016;12 doi: 10.1371/journal.ppat.1005800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derdowski A., Peters T.R., Glover N., Qian R., Utley T.J., Burnett A., Williams J.V., Spearman P., Crowe J.E. Human metapneumovirus nucleoprotein and phosphoprotein interact and provide the minimal requirements for inclusion body formation. J Gen Virol. 2008;89:2698–2708. doi: 10.1099/vir.0.2008/004051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chenik M., Chebli K., Gaudin Y., Blondel D. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. Journal of General Virology. 1994;75:2889–2896. doi: 10.1099/0022-1317-75-11-2889. [DOI] [PubMed] [Google Scholar]
- 39.Miyake T., Farley C.M., Neubauer B.E., Beddow T.P., Hoenen T., Engel D.A. Ebola virus inclusion body formation and RNA synthesis are controlled by a novel domain of NP interacting with VP35. J Virol. 2020;94:e02100-19. doi: 10.1128/JVI.02100-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerard F.C.A., de A. Ribeiro E., Leyrat C., Ivanov I., Blondel D., Longhi S., Ruigrok R.W.H., Jamin M. Modular organization of rabies virus phosphoprotein. Journal of Molecular Biology. 2009;388:978–996. doi: 10.1016/j.jmb.2009.03.061. [DOI] [PubMed] [Google Scholar]
- 41.Habchi J., Longhi S. Structural disorder within paramyxovirus nucleoproteins and phosphoproteins. Mol. BioSyst. 2011;8:69–81. doi: 10.1039/c1mb05204g. [DOI] [PubMed] [Google Scholar]
- 42.Guseva S., Milles S., Jensen M.R., Schoehn G., Ruigrok R.W., Blackledge M. Structure, dynamics and phase separation of measles virus RNA replication machinery. Current Opinion in Virology. 2020;41:59–67. doi: 10.1016/j.coviro.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Mavrakis M., Iseni F., Mazza C., Schoehn G., Ebel C., Gentzel M., Franz T., Ruigrok R.W.H. Isolation and characterisation of the rabies virus N°-P complex produced in insect cells. Virology. 2003;305:406–414. doi: 10.1006/viro.2002.1748. [DOI] [PubMed] [Google Scholar]
- 44.Leyrat C., Yabukarski F., Tarbouriech N., Jr E.A.R., Jensen M.R., Blackledge M., Ruigrok R.W.H., Jamin M. Structure of the vesicular stomatitis virus N0-P complex. PLOS Pathogens. 2011;7 doi: 10.1371/journal.ppat.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guryanov S.G., Liljeroos L., Kasaragod P., Kajander T., Butcher S.J. Crystal structure of the measles virus nucleoprotein core in complex with an N-terminal region of phosphoprotein. J. Virol. 2015;90:2849–2857. doi: 10.1128/JVI.02865-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenni S., Bloyet L.-M., Diaz-Avalos R., Liang B., Whelan S.P.J., Grigorieff N., Harrison S.C. Structure of the vesicular stomatitis virus L protein in complex with its phosphoprotein cofactor. Cell Reports. 2020;30:53–60.e5. doi: 10.1016/j.celrep.2019.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horwitz J.A., Jenni S., Harrison S.C., Whelan S.P.J. Structure of a rabies virus polymerase complex from electron cryo-microscopy. Proc Natl Acad Sci USA. 2020;117:2099–2107. doi: 10.1073/pnas.1918809117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gely S., Lowry D.F., Bernard C., Jensen M.R., Blackledge M., Costanzo S., Bourhis J.-M., Darbon H., Daughdrill G., Longhi S. Solution structure of the C-terminal X domain of the measles virus phosphoprotein and interaction with the intrinsically disordered C-terminal domain of the nucleoprotein. J. Mol. Recognit. 2010;23:435–447. doi: 10.1002/jmr.1010. [DOI] [PubMed] [Google Scholar]
- 49.Green T.J., Luo M. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. PNAS. 2009;106:11713–11718. doi: 10.1073/pnas.0903228106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mavrakis M., McCarthy A.A., Roche S., Blondel D., Ruigrok R.W.H. Structure and function of the C-terminal domain of the polymerase cofactor of rabies virus. J. Mol. Biol. 2004;343:819–831. doi: 10.1016/j.jmb.2004.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abdella R., Aggarwal M., Okura T., Lamb R.A., He Y. Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation. Proc. Natl. Acad. Sci. U.S.A. 2020;117:4931–4941. doi: 10.1073/pnas.1919837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivanov I., Crépin T., Jamin M., Ruigrok R.W.H. Structure of the dimerization domain of the rabies virus phosphoprotein. Journal of Virology. 2010;84:3707–3710. doi: 10.1128/JVI.02557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding H., Green T.J., Lu S., Luo M. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. Journal of Virology. 2006;80:2808–2814. doi: 10.1128/JVI.80.6.2808-2814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarbouriech N., Curran J., Ruigrok R.W.H., Burmeister W.P. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nature Structural Biology. 2000;7:777–781. doi: 10.1038/79013. [DOI] [PubMed] [Google Scholar]
- 55.Guseva S., Milles S., Jensen M.R., Salvi N., Kleman J.-P., Maurin D., Ruigrok R.W.H., Blackledge M. Measles virus nucleo- and phosphoproteins form liquid-like phase-separated compartments that promote nucleocapsid assembly. Sci Adv. 2020;6 doi: 10.1126/sciadv.aaz7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albertini A.A.V., Wernimont A.K., Muziol T., Ravelli R.B.G., Clapier C.R., Schoehn G., Weissenhorn W., Ruigrok R.W.H. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 57.Nott T.J., Craggs T.D., Baldwin A.J. Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nature Chem. 2016;8:569–575. doi: 10.1038/nchem.2519. [DOI] [PubMed] [Google Scholar]
- 58.Qamar S., Wang G., Randle S.J., Ruggeri F.S., Varela J.A., Lin J.Q., Phillips E.C., Miyashita A., Williams D., Ströhl F., Meadows W., Ferry R., Dardov V.J., Tartaglia G.G., Farrer L.A., Schierle G.S.K., Kaminski C.F., Holt C.E., Fraser P.E., Schmitt-Ulms G., et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 2018;173:720–734.e15. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chinchar V.G., Hyatt A., Miyazaki T., Williams T. In: Lesser Known Large DsDNA Viruses. Van Etten J.L., editor. Springer; Berlin, Heidelberg: 2009. Family iridoviridae: poor viral relations no longer; pp. 123–170. [DOI] [PubMed] [Google Scholar]
- 60.Risco C., Rodríguez J.R., López-Iglesias C., Carrascosa J.L., Esteban M., Rodríguez D. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. Journal of Virology. 2002;76:1839–1855. doi: 10.1128/JVI.76.4.1839-1855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rojo G., García-Beato R., Viñuela E., Salas M.L., Salas J. Replication of African swine fever virus DNA in infected cells. Virology. 1999;257:524–536. doi: 10.1006/viro.1999.9704. [DOI] [PubMed] [Google Scholar]
- 62.Schramm B., Locker J.K. Cytoplasmic organization of POXvirus DNA replication. Traffic. 2005;6:839–846. doi: 10.1111/j.1600-0854.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 63.Wileman T. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 2007;61:149–167. doi: 10.1146/annurev.micro.57.030502.090836. [DOI] [PubMed] [Google Scholar]
- 64.Heath C.M., Windsor M., Wileman T. Aggresomes resemble sites specialized for virus assembly. J Cell Biol. 2001;153:449–456. doi: 10.1083/jcb.153.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Charman M., Weitzman M.D. Replication compartments of DNA viruses in the nucleus: location, location, location. Viruses. 2020;12 doi: 10.3390/v12020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomer E., Cohen E.M., Drayman N., Afriat A., Weitzman M.D., Zaritsky A., Kobiler O. Coalescing replication compartments provide the opportunity for recombination between coinfecting herpesviruses. FASEB J. 2019;33:9388–9403. doi: 10.1096/fj.201900032R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang L., Godinez W.J., Kim I.-H., Tektonidis M., de Lanerolle P., Eils R., Rohr K., Knipe D.M. Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proceedings of the National Academy of Sciences. 2011;108:E136–E144. doi: 10.1073/pnas.1103411108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Komatsu T., Quentin-Froignant C., Carlon-Andres I., Lagadec F., Rayne F., Ragues J., Kehlenbach R.H., Zhang W., Ehrhardt A., Bystricky K., Morin R., Lagarde J.-M., Gallardo F., Wodrich H. In vivo labelling of adenovirus DNA identifies chromatin anchoring and biphasic genome replication. Journal of Virology. 2018;92 doi: 10.1128/JVI.00795-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McSwiggen D.T., Hansen A.S., Teves S.S., Marie-Nelly H., Hao Y., Heckert A.B., Umemoto K.K., Dugast-Darzacq C., Tjian R., Darzacq X. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. ELife. 2019;8 doi: 10.7554/eLife.47098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., Taylor J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davis D., Yuan H., Liang F.-X., Yang Y.-M., Westley J., Petzold C., Dancel-Manning K., Deng Y., Sall J., Sehgal P.B. Human antiviral protein MxA forms novel metastable membraneless cytoplasmic condensates exhibiting rapid reversible tonicity-driven phase transitions. J Virol. 2019;93 doi: 10.1128/JVI.01014-19. (/jvi/93/22/JVI.01014-19.atom) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin Y., Mori E., Kato M., Xiang S., Wu L., Kwon I., McKnight S.L. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016;167:789–802.e12. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng Q., Wang L., Qin Z., Wang J., Zheng X., Wei L., Zhang X., Zhang X., Liu C., Li Z., Wu Y., Li G., Yan Q., Ma J. Phase separation of Epstein-Barr virus EBNA2 and its coactivator EBNALP controls gene expression. Journal of Virology. 2020;94 doi: 10.1128/JVI.01771-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaikeeratisak V., Nguyen K., Khanna K., Brilot A.F., Erb M.L., Coker J.K.C., Vavilina A., Newton G.L., Buschauer R., Pogliano K., Villa E., Agard D.A., Pogliano J. Assembly of a nucleus-like structure during viral replication in bacteria. Science. 2017;355:194–197. doi: 10.1126/science.aal2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rhim J.S., Jordan L.E., Mayor H.D. Cytochemical, fluorescent-antibody and electron microscopic studies on the growth of reovirus (ECHO 10) in tissue culture. Virology. 1962;17:342–355. doi: 10.1016/0042-6822(62)90125-3. [DOI] [PubMed] [Google Scholar]
- 76.Silvestri L.S., Taraporewala Z.F., Patton J.T. Rotavirus replication: plus-sense templates for double-stranded RNA synthesis are made in viroplasms. Journal of Virology. 2004;78:7763–7774. doi: 10.1128/JVI.78.14.7763-7774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Patton J.T., Silvestri L.S., Tortorici M.A., Vasquez-Del Carpio R., Taraporewala Z.F. In: Reoviruses: Entry, Assembly and Morphogenesis. Roy P., editor. Springer; Berlin, Heidelberg: 2006. Rotavirus genome replication and morphogenesis: role of the viroplasm; pp. 169–187. [DOI] [PubMed] [Google Scholar]
- 78.Tenorio R., Fernández de Castro I., Knowlton J.J., Zamora P.F., Sutherland D.M., Risco C., Dermody T.S. Function, architecture, and biogenesis of reovirus replication neoorganelles. Viruses. 2019;11 doi: 10.3390/v11030288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eichwald C., Arnoldi F., Laimbacher A.S., Schraner E.M., Fraefel C., Wild P., Burrone O.R., Ackermann M. Rotavirus viroplasm fusion and perinuclear localization are dynamic processes requiring stabilized microtubules. PLOS ONE. 2012;7 doi: 10.1371/journal.pone.0047947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eichwald C., Ackermann M., Nibert M.L. The dynamics of both filamentous and globular mammalian reovirus viral factories rely on the microtubule network. Virology. 2018;518:77–86. doi: 10.1016/j.virol.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 81.Fabbretti E., Afrikanova I., Vascotto F., Burrone O.R. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. Journal of General Virology. 1999;80:333–339. doi: 10.1099/0022-1317-80-2-333. [DOI] [PubMed] [Google Scholar]
- 82.Contin R., Arnoldi F., Campagna M., Burrone O.R. Rotavirus NSP5 orchestrates recruitment of viroplasmic proteins. Journal of General Virology. 2010;91:1782–1793. doi: 10.1099/vir.0.019133-0. [DOI] [PubMed] [Google Scholar]
- 83.Martin D., Ouldali M., Ménétrey J., Poncet D. Structural organisation of the rotavirus nonstructural protein NSP5. Journal of Molecular Biology. 2011;413:209–221. doi: 10.1016/j.jmb.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 84.Broering T.J., Parker J.S.L., Joyce P.L., Kim J., Nibert M.L. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. Journal of Virology. 2002;76:8285–8297. doi: 10.1128/JVI.76.16.8285-8297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zuwała K., Golda A., Kabala W., Burmistrz M., Zdzalik M., Nowak P., Kedracka-Krok S., Zarebski M., Dobrucki J., Florek D., Zeglen S., Wojarski J., Potempa J., Dubin G., Pyrc K. The nucleocapsid protein of human coronavirus NL63. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0117833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hiscox J.A., Wurm T., Wilson L., Britton P., Cavanagh D., Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J Virol. 2001;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wurm T., Chen H., Hodgson T., Britton P., Brooks G., Hiscox J.A. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., Bowman G.R., Hall K.B., Soranno A., Holehouse A.S. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. bioRxiv. 2020 doi: 10.1038/s41467-021-21953-3. 2020.06.17.158121 preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perdikari T.M., Murthy A.C., Ryan V.H., Watters S., Naik M.T., Fawzi N.L. SARS-CoV-2 nucleocapsid protein undergoes liquid-liquid phase separation stimulated by RNA and partitions into phases of human ribonucleoproteins. bioRxiv. 2020 2020.06.09.141101. [Google Scholar]
- 90.Iserman C., Roden C., Boerneke M., Sealfon R., McLaughlin G., Jungreis I., Park C., Boppana A., Fritch E., Hou Y.J., Theesfeld C., Troyanskaya O.G., Baric R.S., Sheahan T.P., Weeks K., Gladfelter A.S. Specific viral RNA drives the SARS CoV-2 nucleocapsid to phase separate. bioRxiv. 2020 2020.06.11.147199. [Google Scholar]
- 91.Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Morgan D.O. Phosphorylation modulates liquid-liquid phase separation of the SARS-CoV-2 N protein. BioRxiv. 2020 2020.06.28.176248. [Google Scholar]
- 92.Lahaye X., Vidy A., Fouquet B., Blondel D. Hsp70 protein positively regulates rabies virus infection. Journal of Virology. 2012;86:4743–4751. doi: 10.1128/JVI.06501-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fouquet B., Nikolic J., Larrous F., Bourhy H., Wirblich C., Lagaudrière-Gesbert C., Blondel D. Focal adhesion kinase is involved in rabies virus infection through its interaction with viral phosphoprotein P. Journal of Virology. 2015;89:1640–1651. doi: 10.1128/JVI.02602-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang J., Wu X., Zan J., Wu Y., Ye C., Ruan X., Zhou J. Cellular chaperonin CCTγ contributes to rabies virus replication during infection. Journal of Virology. 2013;87:7608–7621. doi: 10.1128/JVI.03186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang J., Ye C., Ruan X., Zan J., Xu Y., Liao M., Zhou J. The chaperonin CCTα is required for efficient transcription and replication of rabies virus: role of CCTα in RABV replication. Microbiol Immunol. 2014;58:590–599. doi: 10.1111/1348-0421.12186. [DOI] [PubMed] [Google Scholar]
- 96.Shin T., Weinstock D., Castro M.D., Hamir A.N., Wampler T., Walter M., Kim H.Y., Acland H. Immunohistochemical localization of endothelial and inducible nitric oxide synthase within neurons of cattle with rabies. J. Vet. Med. Sci. 2004;66:539–541. doi: 10.1292/jvms.66.539. [DOI] [PubMed] [Google Scholar]
- 97.Ma D., George C.X., Nomburg J.L., Pfaller C.K., Cattaneo R., Samuel C.E. Upon infection, cellular WD repeat-containing protein 5 (WDR5) localizes to cytoplasmic inclusion bodies and enhances measles virus Replication. J Virol. 2017;92 doi: 10.1128/JVI.01726-17. (/jvi/92/5/e01726-17.atom) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koga R., Sugita Y., Noda T., Yanagi Y., Ohno S. Actin-modulating protein cofilin is involved in the formation of measles virus ribonucleoprotein complex at the perinuclear region. Journal of Virology. 2015;89:10524–10531. doi: 10.1128/JVI.01819-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fricke J., Koo L.Y., Brown C.R., Collins P.L. p38 and OGT sequestration into viral inclusion bodies in cells infected with human respiratory syncytial virus suppresses MK2 activities and stress granule assembly. J. Virol. 2013;87:1333–1347. doi: 10.1128/JVI.02263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wendt L., Brandt J., Bodmer B.S., Reiche S., Schmidt M.L., Traeger S., Hoenen T. The Ebola virus nucleoprotein recruits the nuclear RNA export factor NXF1 into inclusion bodies to facilitate viral protein expression. Cells. 2020;9:187. doi: 10.3390/cells9010187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen J., He Z., Yuan Y., Huang F., Luo B., Zhang J., Pan T., Zhang H., Zhang J. Host factor SMYD3 is recruited by Ebola virus nucleoprotein to facilitate viral mRNA transcription. Emerg Microbes Infect. 2019;8:1347–1360. doi: 10.1080/22221751.2019.1662736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brandt J., Wendt L., Bodmer B.S., Mettenleiter T.C., Hoenen T. The cellular protein CAD is recruited into ebola virus inclusion bodies by the nucleoprotein NP to facilitate genome replication and transcription. Cells. 2020;9:1126. doi: 10.3390/cells9051126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dhillon P., Tandra V.N., Chorghade S.G., Namsa N.D., Sahoo L., Rao C.D. Cytoplasmic relocalization and colocalization with viroplasms of host cell proteins, and their role in rotavirus infection. J Virol. 2018;92 doi: 10.1128/JVI.00612-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Taylor T.J., Knipe D.M. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. Journal of Virology. 2004;78:5856–5866. doi: 10.1128/JVI.78.11.5856-5866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matsumoto S., Schneider L.G., Kawai A., Yonezawa T. Further studies on the replication of rabies and rabies-like viruses in organized cultures of mammalian neural tissues. Journal of Virology. 1974;14:981–996. doi: 10.1128/jvi.14.4.981-996.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee J.E., Cathey P.I., Wu H., Parker R., Voeltz G.K. Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science. 2020;367 doi: 10.1126/science.aay7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gerlier D., Lyles D.S. Interplay between innate immunity and negative-strand RNA viruses: towards a rational model. Microbiol. Mol. Biol. Rev. 2011;75:468–490. doi: 10.1128/MMBR.00007-11. (second page of table of contents) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chelbi-Alix M.K., Vidy A., Bougrini J.E., Blondel D. Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, Stat1, and PML nuclear bodies. Journal of Interferon & Cytokine Research. 2006;26:271–280. doi: 10.1089/jir.2006.26.271. [DOI] [PubMed] [Google Scholar]
- 109.Brzozka K., Finke S., Conzelmann K.-K. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein p interferes with phosphorylation of interferon regulatory factor 3. Journal of Virology. 2005;79:7673–7681. doi: 10.1128/JVI.79.12.7673-7681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vidy A., El Bougrini J., Chelbi-Alix M.K., Blondel D. The nucleocytoplasmic rabies virus p protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. Journal of Virology. 2007;81:4255–4263. doi: 10.1128/JVI.01930-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vidy A., Chelbi-Alix M., Blondel D. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. Journal of Virology. 2005;79:14411–14420. doi: 10.1128/JVI.79.22.14411-14420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cardenas W.B., Loo Y.-M., Gale M., Hartman A.L., Kimberlin C.R., Martinez-Sobrido L., Saphire E.O., Basler C.F. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. Journal of Virology. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ramanan P., Edwards M.R., Shabman R.S., Leung D.W., Endlich-Frazier A.C., Borek D.M., Otwinowski Z., Liu G., Huh J., Basler C.F., Amarasinghe G.K. Structural basis for Marburg virus VP35–mediated immune evasion mechanisms. Proc Natl Acad Sci U S A. 2012;109:20661–20666. doi: 10.1073/pnas.1213559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schümann M., Gantke T., Mühlberger E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. Journal of Virology. 2009;83:8993–8997. doi: 10.1128/JVI.00523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Prins K.C., Cárdenas W.B., Basler C.F. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKɛ and TBK-1. J Virol. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Audsley M.D., Moseley G.W. Paramyxovirus evasion of innate immunity: diverse strategies for common targets. World J Virol. 2013;2:57–70. doi: 10.5501/wjv.v2.i2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Precious B., Young D.F., Bermingham A., Fearns R., Ryan M., Randall R.E. Inducible expression of the P, V, and NP genes of the paramyxovirus simian virus 5 in cell lines and an examination of NP-P and NP-V interactions. Journal of Virology. 1995;69:8001–8010. doi: 10.1128/jvi.69.12.8001-8010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Simpson-Holley M., Kedersha N., Dower K., Rubins K.H., Anderson P., Hensley L.E., Connor J.H. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. Journal of Virology. 2011;85:1581–1593. doi: 10.1128/JVI.02247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yoo J.-S., Takahasi K., Ng C.S., Ouda R., Onomoto K., Yoneyama M., Lai J.C., Lattmann S., Nagamine Y., Matsui T., Iwabuchi K., Kato H., Fujita T. DHX36 enhances RIG-I signaling by facilitating PKR-mediated antiviral stress granule formation. PLOS Pathogens. 2014;10 doi: 10.1371/journal.ppat.1004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Onomoto K., Jogi M., Yoo J.-S., Narita R., Morimoto S., Takemura A., Sambhara S., Kawaguchi A., Osari S., Nagata K., Matsumiya T., Namiki H., Yoneyama M., Fujita T. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLOS ONE. 2012;7 doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Langereis M.A., Feng Q., van Kuppeveld F.J. MDA5 localizes to stress granules, but this localization is not required for the induction of type I interferon. J. Virol. 2013;87:6314–6325. doi: 10.1128/JVI.03213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dinh P.X., Beura L.K., Das P.B., Panda D., Das A., Pattnaik A.K. Induction of stress granule-like structures in vesicular stomatitis virus-infected cells. Journal of Virology. 2013;87:372–383. doi: 10.1128/JVI.02305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Qin Q., Hastings C., Miller C.L. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J. Virol. 2009;83:11090–11101. doi: 10.1128/JVI.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Choudhury P., Bussiere L.D., Miller C.L. Mammalian orthoreovirus factories modulate stress granule protein localization by interaction with G3BP1. J Virol. 2017;91 doi: 10.1128/JVI.01298-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yang P., Mathieu C., Kolaitis R.-M., Zhang P., Messing J., Yurtsever U., Yang Z., Wu J., Li Y., Pan Q., Yu J., Martin E.W., Mittag T., Kim H.J., Taylor J.P. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181:325–345.e28. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dhillon P., Rao C.D. Rotavirus induces formation of remodeled stress granules and P bodies and their sequestration in viroplasms to promote progeny virus production. J Virol. 2018;92 doi: 10.1128/JVI.01363-18. (/jvi/92/24/e01363-18.atom) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Du M., Chen Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hu S., Sun H., Yin L., Li J., Mei S., Xu F., Wu C., Liu X., Zhao F., Zhang D., Huang Y., Ren L., Cen S., Wang J., Liang C., Guo F. PKR-dependent cytosolic cGAS foci are necessary for intracellular DNA sensing. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aav7934. [DOI] [PubMed] [Google Scholar]
- 129.Haller O., Staeheli P., Schwemmle M., Kochs G. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends in Microbiology. 2015;23:154–163. doi: 10.1016/j.tim.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 130.Geoffroy M.-C., Chelbi-Alix M.K. Role of promyelocytic leukemia protein in host antiviral defense. Journal of Interferon & Cytokine Research. 2011;31:145–158. doi: 10.1089/jir.2010.0111. [DOI] [PubMed] [Google Scholar]
- 131.Chen Y.-C.M., Kappel C., Beaudouin J., Eils R., Spector D.L. Live cell dynamics of promyelocytic leukemia nuclear bodies upon entry into and exit from mitosis. MBoC. 2008;19:3147–3162. doi: 10.1091/mbc.E08-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Banani S.F., Rice A.M., Peeples W.B., Lin Y., Jain S., Parker R., Rosen M.K. Compositional control of phase-separated cellular bodies. Cell. 2016;166:651–663. doi: 10.1016/j.cell.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Day P.M., Baker C.C., Lowy D.R., Schiller J.T. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. PNAS. 2004;101:14252–14257. doi: 10.1073/pnas.0404229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Guion L.G., Sapp M. The role of promyelocytic leukemia nuclear bodies during HPV infection. Front Cell Infect Microbiol. 2020;10 doi: 10.3389/fcimb.2020.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schreiner S., Bürck C., Glass M., Groitl P., Wimmer P., Kinkley S., Mund A., Everett R.D., Dobner T. Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res. 2013;41:3532–3550. doi: 10.1093/nar/gkt064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stepp W.H., Meyers J.M., McBride A.A. Sp100 provides intrinsic immunity against human papillomavirus infection. MBio. 2013;4 doi: 10.1128/mBio.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Negorev D.G., Vladimirova O.V., Ivanov A., Rauscher F., Maul G.G. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. Journal of Virology. 2006;80:8019–8029. doi: 10.1128/JVI.02164-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim Y.-E., Lee J.-H., Kim E.T., Shin H.J., Gu S.Y., Seol H.S., Ling P.D., Lee C.H., Ahn J.-H. Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. Journal of Virology. 2011;85:11928–11937. doi: 10.1128/JVI.00758-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Boutell C., Cuchet-Lourenço D., Vanni E., Orr A., Glass M., McFarlane S., Everett R.D. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Berscheminski J., Groitl P., Dobner T., Wimmer P., Schreiner S. The adenoviral oncogene E1A-13S interacts with a specific isoform of the tumor suppressor PML to enhance viral transcription. Journal of Virology. 2013;87:965–977. doi: 10.1128/JVI.02023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Scherer M., Stamminger T. Emerging role of PML nuclear bodies in innate immune signaling. J Virol. 2016;90:5850–5854. doi: 10.1128/JVI.01979-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Alandijany T., Roberts A.P.E., Conn K.L., Loney C., McFarlane S., Orr A., Boutell C. Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Everett R.D., Parada C., Gripon P., Sirma H., Orr A. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. Journal of Virology. 2008;82:2661–2672. doi: 10.1128/JVI.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Giovannoni F., Damonte E.B., García C.C. Cellular promyelocytic leukemia protein is an important dengue virus restriction factor. PLOS ONE. 2015;10 doi: 10.1371/journal.pone.0125690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li W., Wang G., Zhang H., Zhang D., Zeng J., Chen X., Xu Y., Li K. Differential suppressive effect of promyelocytic leukemia protein on the replication of different subtypes/strains of influenza A virus. Biochemical and Biophysical Research Communications. 2009;389:84–89. doi: 10.1016/j.bbrc.2009.08.091. [DOI] [PubMed] [Google Scholar]
- 146.Blondel D., Kheddache S., Lahaye X., Dianoux L., Chelbi-Alix M.K. Resistance to rabies virus infection conferred by the PMLIV isoform. J Virol. 2010;84:10719–10726. doi: 10.1128/JVI.01286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Asmi F.E., Maroui M.A., Dutrieux J., Blondel D., Nisole S., Chelbi-Alix M.K. Implication of PMLIV in both intrinsic and innate immunity. PLOS Pathogens. 2014;10 doi: 10.1371/journal.ppat.1003975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Blondel D., Regad T., Poisson N., Pavie B., Harper F., Pandolfi P.P., de Thé H., Chelbi-Alix M.K. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene. 2002;21:7957–7970. doi: 10.1038/sj.onc.1205931. [DOI] [PubMed] [Google Scholar]
- 149.Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., Luo E.-C., Krach F., Yang D., Sen A., Fulzele A., Wozniak J.M., Gonzalez D.J., Kankel M.W., Gao F.-B., Bennett E.J., Lécuyer E., Yeo G.W. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172:590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Youn J.-Y., Dunham W.H., Hong S.J., Knight J.D.R., Bashkurov M., Chen G.I., Bagci H., Rathod B., MacLeod G., Eng S.W.M., Angers S., Morris Q., Fabian M., Côté J.-F., Gingras A.-C. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Molecular Cell. 2018;69:517–532.e11. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 151.Erdős G., Dosztányi Z. Analyzing protein disorder with IUPred2A. Current Protocols in Bioinformatics. 2020;70 doi: 10.1002/cpbi.99. [DOI] [PubMed] [Google Scholar]
- 152.Zhang S., Jiang Y., Cheng Q., Zhong Y., Qin Y., Chen M. Inclusion body fusion of human parainfluenza virus type 3 regulated by acetylated α-tubulin enhances viral replication. J Virol. 2017;91 doi: 10.1128/JVI.01802-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Richard C.-A., Rincheval V., Lassoued S., Fix J., Cardone C., Esneau C., Nekhai S., Galloux M., Rameix-Welti M.-A., Sizun C., Eléouët J.-F. RSV hijacks cellular protein phosphatase 1 to regulate M2-1 phosphorylation and viral transcription. PLOS Pathogens. 2018;14 doi: 10.1371/journal.ppat.1006920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cifuentes-Muñoz N., Branttie J., Slaughter K.B., Dutch R.E. Human metapneumovirus induces formation of inclusion bodies for efficient genome replication and transcription. Journal of Virology. 2017;91 doi: 10.1128/JVI.01282-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nanbo A., Watanabe S., Halfmann P., Kawaoka Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci Rep. 2013;3:1206. doi: 10.1038/srep01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schmid M., Speiseder T., Dobner T., Gonzalez R.A. DNA virus replication compartments. Journal of Virology. 2014;88:1404–1420. doi: 10.1128/JVI.02046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Katsafanas G.C., Moss B. Linkage of transcription and translation within cytoplasmic poxvirus DNA factories provides a mechanism to coordinate viral and usurp host functions. Cell Host Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Green T.J., Zhang X., Wertz G.W., Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 159.Gutsche I., Desfosses A., Effantin G., Ling W.L., Haupt M., Ruigrok R.W.H., Sachse C., Schoehn G. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science. 2015;348:704–707. doi: 10.1126/science.aaa5137. [DOI] [PubMed] [Google Scholar]
- 160.Ribeiro E.A., Favier A., Gerard F.C.A., Leyrat C., Brutscher B., Blondel D., Ruigrok R.W.H., Blackledge M., Jamin M. Solution structure of the C-terminal nucleoprotein–RNA binding domain of the vesicular stomatitis virus phosphoprotein. Journal of Molecular Biology. 2008;382:525–538. doi: 10.1016/j.jmb.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 161.Communie G., Crépin T., Maurin D., Jensen M.R., Blackledge M., Ruigrok R.W.H. Structure of the tetramerization domain of measles virus phosphoprotein. Journal of Virology. 2013;87:7166–7169. doi: 10.1128/JVI.00487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kingston R.L., Hamel D.J., Gay L.S., Dahlquist F.W., Matthews B.W. Structural basis for the attachment of a paramyxoviral polymerase to its template. PNAS. 2004;101:8301–8306. doi: 10.1073/pnas.0402690101. [DOI] [PMC free article] [PubMed] [Google Scholar]