Abstract
In this study, we developed a multi-scale Convolutional neural network based Automated hippocampal subfield Segmentation Toolbox (CAST) for automated segmentation of hippocampal subfields. Although training CAST required approximately three days on a single workstation with a high-quality GPU card, CAST can segment a new subject in less than 1 min even with GPU acceleration disabled, thus this method is more time efficient than current automated methods and manual segmentation. This toolbox is highly flexible with either a single modality or multiple modalities and can be easily set up to be trained with a researcher’s unique data. A 3D multi-scale deep convolutional neural network is the key algorithm used in the toolbox. The main merit of multi-scale images is the capability to capture more global structural information from down-sampled images without dramatically increasing memory and computational burden. The original images capture more local information to refine the boundary between subfields. Residual learning is applied to alleviate the vanishing gradient problem and improve the performance with a deeper network. We applied CAST with the same settings on two datasets, one 7T dataset (the UMC dataset) with only the T2 image and one 3T dataset (the MNI dataset) with both T1 and T2 images available. The segmentation accuracy of both CAST and the state-of-the-art automated method ASHS, in terms of the dice similarity coefficient (DSC), were comparable. CAST significantly improved the reliability of segmenting small subfields, such as CA2, CA3, and the entorhinal cortex (ERC), in terms of the intraclass correlation coefficient (ICC). Both ASHS and manual segmentation process some subfields (e.g. CA2 and ERC) with high DSC values but low ICC values, consequently increasing the difficulty of judging segmentation quality. CAST produces very consistent DSC and ICC values, with a maximal discrepancy of 0.01 (DSC-ICC) across all subfields. The pre-trained model, source code, and settings for the CAST toolbox are publicly available.
Keywords: Hippocampal subfields, Automated segmentation, Convolutional neural network, Residual learning
1. Introduction
The segmentation of hippocampal subfields in MRI has gained great interest in the last decade. These anatomic subregions were found in recent studies to contribute distinct functional roles, for example, CA1 in memory integration and inference (Schlichting et al., 2014), CA3 in memory recall (Chadwick et al., 2014), dentate gyrus (DG) and CA3 in pattern separation (Baker et al., 2016; Berron et al., 2016; Leutgeb et al., 2007). The highly specialized function suggests that these subregions are potentially affected differently by normal aging, Alzheimer’s disease, schizophrenia, epilepsy, major depressive disorder, and posttraumatic stress disorder. However, manual delineation of hippocampal subregions is extremely labor-intensive and time-consuming, consequently limiting studies to a small sample size. The inter-rater and intra-rater reliability also can influence the statistical power of a study. Wisse et al. (2016) observed that CA2 has an intra-rater reliability with a mean dice coefficient as low as 0.66, and the entorhinal cortex (ERC) has inter-rater reliability with an intraclass correlation coefficient as low as 0.27. In addition, the segmentation protocols vary between research centers, thereby presenting undue challenges to comparing results reported in published studies.
The limitations mentioned above could be overcome with the use of automated algorithms. Two major (semi-)automated methods were proposed for hippocampal subfield segmentation from distinct perspectives, parametric and non-parametric. The parametric method proposed in Iglesias (2015) (publicly available in FreeSurfer 6.0 https://surfer.nmr.mgh.harvard.edu/fswiki/HippocampalSubfields) first combines the manual labels from the in vivo and ex vivo data into a single computational atlas and then uses a generative, parametric method to model the spatial distribution of hippocampal subregions and surrounding brain structures from the labeled training data. Because segmentation is connected to the observed image data through a generative process of image formation without assumptions about MRI acquisition, this method was claimed to be adaptive to any MRI pulse sequence and resolution. The non-parametric method developed by Yushkevich et al. (2015b) (publicly available at https://www.nitrc.org/projects/ashs) applies strict nonlinear whole-brain and hippocampus-regional registration, multi-atlas joint label fusion, and voxel-wise learning-based error correction to propagate manual labels from a set of atlases to an unlabeled image. This non-parametric method takes advantage of the prior information about the distribution of image intensities derived from training data for automated labelling, which is ignored in FreeSurfer 6.0.
Both methods are applicable for multiple modalities, for example, T1 and T2 weighted images together for segmentation. However, each method has its own limitations. The atlas in FreeSurfer 6.0 was built using in vivo and ex vivo data from elderly subjects. Thus its reliability in studies of younger populations is required to be tested. Further improving the segmentation performance by training this method with a set of imaging data is impossible, because the MRI contrast is discarded (Mueller et al., 2018). ASHS has a few atlas sets, and more effort was dedicated to expanding the population from elderly healthy subjects and mild cognitive impairment patients to young adults (Berron et al., 2017). ASHS achieved reasonable results with data acquired from multiple magnets, e.g. ADNI data (http://adni.loni.usc.edu). However, optimal segmentation results could require a customized population specific atlas from the same magnet (Mueller et al., 2018). In addition, these two methods are computationally expensive and thus infeasible for applications requiring instantaneous segmentation.
In this study, we present a multi-scale Convolutional neural network based Automated hippocampal subfield Segmentation Toolbox (CAST) for fast and automated segmentation of hippocampal subfields. Deep learning has emerged as a promising technique for neuroimaging analysis in the last decade. Multiple studies have demonstrated the capability of deep learning in segmenting the whole hippocampus in both healthy subjects and disease populations (Goubran et al., 2020; Liu et al., 2020; Nogovitsyn et al., 2019; Novosad et al., 2020; Thyreau et al., 2018). Zhu et al. (2019) successfully applied dilated dense U-net for segmenting the hippocampal subfields of infants and adults. Nevertheless, a robust and fast hippocampus subfield segmentation pipeline remains unavailable. CAST is designed to provide a publicly available, computationally efficient, and flexible toolbox for segmenting hippocampal subfields with the capability for multiple imaging modalities. When CAST is applied to segment a new subject with an optimized model, this toolbox requires only a raw image as input and outputs the segmented image in less than 1 min. Its computational efficiency is applicable for large sample size and instantaneous segmentation. CAST can be easily trained on a single workstation with a high-quality GPU card to generate a customized segmentation model for a unique dataset.
A 3D multi-scale deep convolutional neural network (CNN) is the key algorithm used in the CAST toolbox. This network is based on the Tensorflow package (https://www.tensorflow.org) and the DeepMedic project (Kamnitsas et al., 2017). Residual learning is applied to alleviate the degradation problem and improve the performance with a deeper network depth. In contrast to the ASHS technique, the CAST toolbox does not rely on a strict nonlinear transformation to register individual subjects to multiple atlases during both training and segmentation pipelines. Instead, CAST tolerates the variability between subjects, thereby eliminating the time cost for nonlinear registration. This property becomes more valuable when the deformation is severe and the segmentation could fail at the nonlinear coregistration step. While training the network in CAST remains computationally expensive, the absence of any nonlinear coregistration in CAST has substantially improved its time efficiency compared to ASHS and Freesurfer when segmenting a new subject. In fact, since the imaging data in each dataset used in this study were acquired with the same acquisition protocol and roughly in the same orientation, no linear or nonlinear transformations are applied in either the training or segmentation process. An affine transformation is sufficient when a new subject with different acquisition orientation is used in the segmentation process. When training the multi-scale CNN, the original image is down-sampled by two different sampling factors. Then, the original and two down-sampled images are fed to their corresponding 8-layer convolutional pathways (the same architecture but distinct parameters). The main merit of multi-scale data is its ability to capture more global structural information from down-sampled data, because of its larger field of view, without dramatically increasing the memory and computational burden. The down-sampled images achieve coarse labelling and reduce the risk of mislabeling non-hippocampus voxels as subregions, and the original images can capture more local information to refine the boundary between subregions. The outputs from these three convolutional blocks are concatenated as the input to three more layers, with the final layer as classification layer. We have provided the pre-trained model for multiple data at github (https://github.com/pipiyang). The source code and settings for the CAST toolbox are also publicly available to researchers who would prefer to train the model with their own unique dataset.
2. Material and methods
2.1. Data acquisition and preprocessing
We validated the CAST segmentation method on two publicly available datasets, one 7T dataset with T2 structural images as the single input imaging modality and one 3T dataset with both T1 and T2 structural images available to demonstrate the multi-modality capability. The first dataset (Package UMC Utrecht 7T in https://www.nitrc.org/projects/ashs/) contained 26 subjects (59 ± 9 years; male/female 12/14), named as UMC dataset (Wisse et al., 2016) and were collected on a 7T Philips MRI scanner (Philips Healthcare, Best, the Netherlands) with a 32-channel head coil (Nova Medical, Wilmington, MA). 3D T2-weighted TSE images were acquired with 0.7 × 0.7 × 0.7 mm3 isotropic voxel size, TR = 3158 ms, TE = 301 ms, flip angle 120°, TSE factor = 182, and matrix size 356 × 357 × 272. The T2 images were interpolated to a spatial resolution of 0.35 × 0.35 × 0.35 mm3 isotropic voxel size by zero-filling during reconstruction. While cropped T1 images are also available in this dataset, because the cropped T1 image is in the template space but the T2 and manual segmentation map is in the subject’s space, T1 is not used in CAST segmentation for this dataset to avoid potential registration error. The tse_native_chunk images (available in the ASHS data repository), which are cropped T2-weighted images including the entire hippocampus in the native space, were used in our analysis. These images were demeaned and normalized to eliminate imaging intensity differences before being input to the neural network. Note that the CAST method does not enforce all subjects in the same space. Thus, no nonlinear coregistration is involved in the automated segmentation process. Affine transformation is also not required because the imaging orientation is roughly in the same direction, and resolution is the same for all subjects. The cornu ammonis (CA) fields CA1, CA2, CA3, DG, subiculum (SUB), ERC (even though ERC is usually not considered part of the hippocampus), cyst and tail were manually segmented by the corresponding investigators. CA4 is also included in the DG label.
The second dataset (https://www.nitrc.org/projects/mni-hisub25/) contained 25 healthy adult subjects (21–53 years, mean ± std age = 31.2 ± 7.5 years; male/female = 12/13), named as MNI dataset (Kulaga-Yoskovitz et al., 2015) and were collected on a 3T Siemens Tim Trio MRI scanner (Siemens, Erlangen, Germany) with a 32-channel head coil. Submillimeter T1 and T2 images were acquired for all subjects. The 3D MPRAGE T1 image was acquired with spatial resolution of 0.6 × 0.6 × 0.6 mm3 (isotropic voxel size), TR = 3000 ms, TE = 4.32 ms, TI = 1500 ms, flip angle = 7°, matrix size = 336 × 384, FOV 201 mm × 229 mm, 240 axial slices with 0.6 mm slice thickness. The T2 image was acquired with a 2D turbo spin-echo sequence with acquisition parameters TR = 10, 810 ms, TE = 81 ms, flip angle = 119°, matrix size 512 × 512, FOV = 203 mm × 203 mm, 60 coronal slices angled perpendicular to the hip pocampal long axis with slice thickness = 2 mm, resulting in 0.4 × 0.4 × 2.0 mm3 voxel size. The T1 and T2 images were linearly registered to MNI-ICBM152 template and resampled to a resolution of 0.4 × 0.4 × 0.4 mm3, which are the images available in the dataset. Then the T1 and T2 images in the template space were cropped to include the left and right hippocampus separately. These images were demeaned and normalized to eliminate imaging intensity differences before being input to the neural network. Because these images are in template space, no nonlinear or linear registration is applied in our analysis. The manual segmentation protocol for this dataset separated hippocampus into three labels, namely SUB, a combination of CA1, CA2, and CA3 (CA1–3), and a combination of CA4 and DG (CA4+DG).
The delineation of the subregions varies with different automated segmentation methods and the manual segmentation protocols from different centers. Mueller et al. (2018) and the Supplementary Fig.S1 have demonstrated that both have different subregion names and distinct boundaries between subregions were observed across their manual segmentation and automated segmentation methods, including Freesurfer 6.0, and ASHS, due to their varying segmentation protocols. Using the manual segmentation maps from the UMC and MNI datasets described above as ground truth is inappropriate for evaluating Freesurfer’s performance. Therefore, Freesurfer 6.0 is not used for comparison in our study, and each dataset is analyzed separately. Given the potential structural difference between the left and right hippocampus, the left and right hippocampus are trained and segmented separately.
2.2. CNN: A multi-scale residual neural network
A 3D three-scale convolutional neural network is the key deep learning algorithm used in this toolbox for hippocampal subfield segmentation. Residual learning is implemented in the algorithm to alleviate the degradation problem and improve the performance with a deeper network depth.
2.2.1. Deep residual learning
For classification, deep neural networks traditionally stack multiple layers of neurons in sequential order, leading to a more complex feature representation encoded at a deeper layer, with the classifier as the last layer (Goodfellow et al., 2016; LeCun et al., 2015). While stacking more layers may provide more comprehensive features, simply adding more layers does not always improve classification accuracy. A deeper neural network has gradients that vanish more rapidly than its shallower counterpart, and thus becomes more difficult to train and even stops the neural network from further training. Deep residual learning using residual connections was recently shown to be a simple but powerful approach to overcoming the vanishing gradient problem in a deeper neural network and thus facilitate optimizing the network (He et al., 2016). and Jégou et al. (2017) successfully trained a very deep residual network with over 100 layers with increasing accuracy. The residual connection allows the output from one specific layer to skip multiple layers and directly add to the output from another layer without adding extra parameters or increasing the computational burden. Fig. 1 shows one convolutional layer with residual connections in the CAST neural network framework. The upper plot in Fig. 1 indicates that the input to the L-1st convolutional layer is directly added to the output of the L-th convolutional layer and forms the final output of the convolutional layer with a residual unit. Mathematically, the output from the L-th layer can be expressed as
| (1) |
where and xL = xL–2 + xl denote the output from the L-th convolutional layer before and after residual connection, and xL–2 is the input to the L-1st layer. WL is a set of weight parameters in the L-th convolutional layer. For simplicity, we marked the output from the Lth layer with residual connection with a ⊕ symbol, as shown in the bottom plot in Fig. 1. This simplified notation was used to describe the architecture of CAST.
Fig. 1.
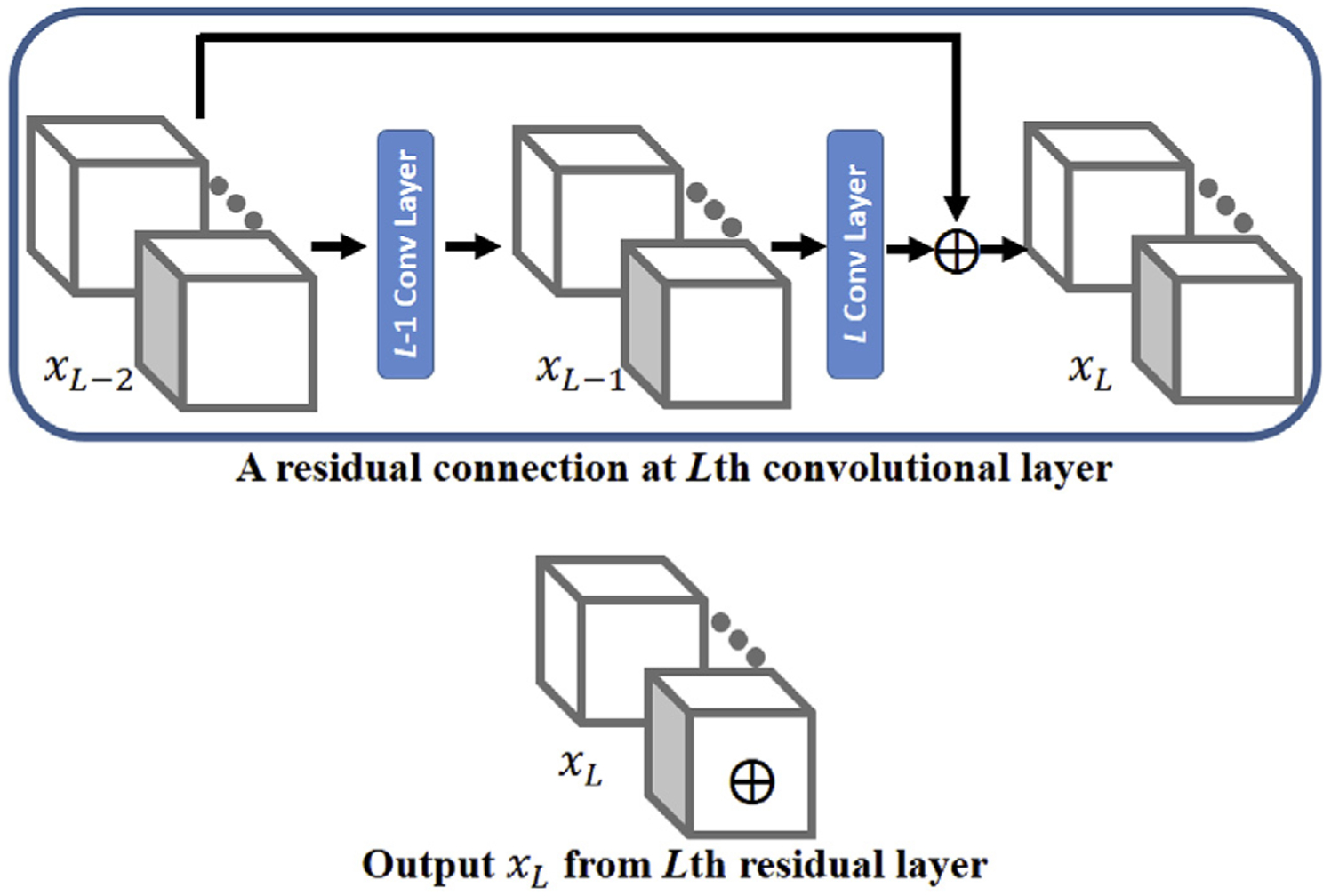
An example plot of residual connection in the convolutional neural network. The top panel shows that the input to the L-1st convolutional layer (xL–2) is connected (added) to the output of the Lth layer to form the output xL. The bottom panel indicates the output after a residual connection is marked with a symbol ⊕ for simplicity in the architecture of the deep neural network.
2.2.2. 3D CNN for semantic segmentation
Semantic segmentation refers to the process of classifying each voxel in a 2D image or a 3D volume to a certain label. Convolutional neural networks (CNNs) predict voxel-wise labels in semantic segmentation by taking the information from a voxel and its neighboring voxels into account. The element involved in carrying out the convolution operation in a convolutional layer is called a filter. Numerous CNNs have been developed with 2D filters because most of these studies examine natural images. With volumetric MRI data, the CNN should strengthen the volumetric feature representation learning. Thus, 3D filters are implemented in the network. Observing that a small filter has the advantage of computational efficiency and is capable to encode the features in the data (Simonyan and Zisserman, 2014), we have employed small convolutional filters (filter size as 33 or 13) in the CAST network. During the convolution operation, the filter is applied on a proportion of the image over which the filter is hovering and then the filter shifts to another location simultaneously across all channels. This process repeats until the entire volume is traversed. The shifting step along each dimension is defined as a stride, and a stride length of 1 along x, y, and z direction is used in our analysis. The number of filters in the convolutional layers determines the number of output channels. Therefore, the weight parameters for a convolutional layer are of the dimension input channels × filter size × output channels. With the input data of 313 resolution and 40 channels in Fig. 1, the weight parameters WL for the convolutional layer is 40 × 33 × 50 if this layer is specified with 50 filters. When a filter with a size of 13 is used in a convolutional layer, this layer is equivalent to a fully connected layer, where each voxel’s output channels are all connected to its own input channels, without considering any neighboring voxels.
2.2.3. Architecture of the CNN
The CNN has the original resolution image (blue) and two down-sampled images with factor of 3 and 5 (green and purple, respectively) as input. The architecture of the network with both T1 and T2 as input imaging modalities is shown in Fig. 2. The green and purple boxes indicate the size of the down-sampled images in the original resolution, and the cropped input images have a dimension of 373 × 2, 233 × 2 and 213 × 2. The factor of 2 corresponds to the two input imaging channels in the network, as in MNI dataset. The factor becomes 1 for the single-modality UMC dataset. Image cropping is a commonly used technique in deep learning to reduce the GPU memory cost, and the imaging dimensions are specified to maximally use available memory. These three cropped images are fed to three separate pathways with the same network architecture but independent parameters. Each pathway consists of eight convolutional layers with a filter size of 33 and filters are numbered 30, 30, 40, 40, 40, 40, 50 and 50 in this order. The network is designed with unary filter stride and no padding. The 4th, 6th, and 8th layers are the convolutional layers with residual connection, as explained in Fig. 1. Because the outputs from these three 8-layer pathways have different sizes, the outputs from the two down-sampled pathways are up-sampled to match the dimension of the output from the original pathway and concatenated with a dimension of 150 × 213 for the following concatenated convolutional block. The concatenated convolutional block consists of three convolutional layers, and residual connection is enabled for the second layer. The first layer has a filter size of 33, and the second and the final layer has filter size 13, which significantly reduces the memory and computational requirements compared to filter size 33. The final layer is the classification layer, and each channel outputs the probabilistic maps for a certain subfield. A 50% dropout rate is used to alleviate over-fitting, and pReLU (He et al., 2015) is the activation function used in the network. The red box indicates the final output image size of 213 for this image patch, and the corresponding manual labelling is used to calculate multi-class cross-entropy, which is the loss function minimized to optimize the network parameters. While using zero padding in the intermediate layers can make the network segment all voxels within the original resolution image (blue box) instead of only the centering voxels (red box), zero padding may lead to less accurate labelling for the voxels between the blue and red box because less valid information is used for segmentation. In addition, when there is no zero padding, the input image size can be slightly larger to contain more information because of the memory limit. Therefore, no zero padding is used in the network. Zero padding is directly added to the input image patch, but not to the network, to enable estimating the voxels on the boundary of the cropped images. Note that applying the network in Fig. 2 with multiple imaging modalities (for example, the MNI dataset), instead of single imaging modality (for example, the UMC dataset), has negligible influence on memory cost. Only the input data are required to update from a single channel to multiple channels, the rest of the network remains the same, and the size of the input data is much smaller than the output of intermediate layers. Theoretically, the input to the network can extend beyond two modalities.
Fig. 2.
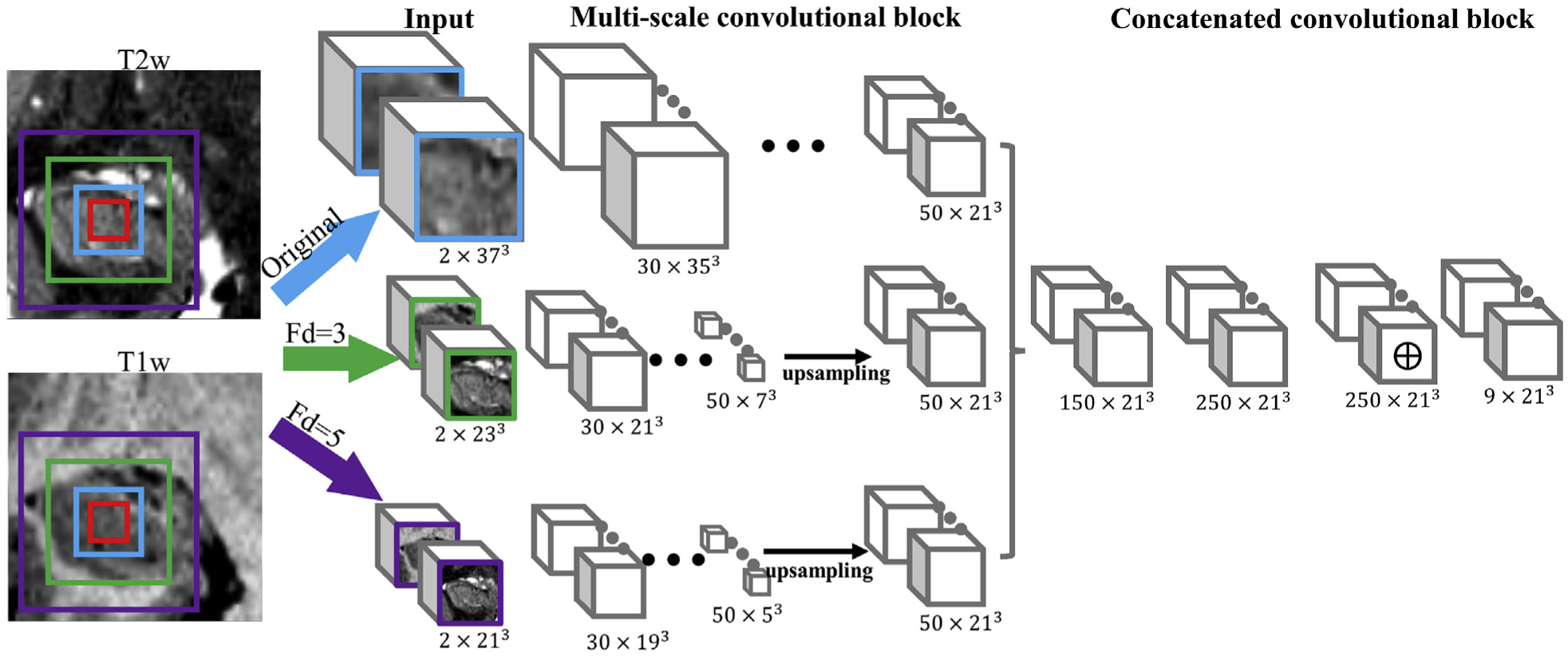
Architecture of the multi-scale 3D convolutional neural network with two imaging modalities as input. The original and two down-sampled image patches are fed to three pathways. Each pathway consists of 8 sequential 3D convolutional layers but independent parameters, where the 4th, 6th, and 8th layers have residual connections as described in Fig. 1. The output from these down-sampled pathways are up-sampled to match the size of the original pathway. Then, these three outputs are concatenated for the following three layers in the concatenated convolutional block. The red box indicates the final output image size.
This multi-scale CNN consists of approximately 2 million parameters and was developed based on the TensorFlow package (https://www.tensorflow.org) and the DeepMedic project (https://github.com/deepmedic/deepmedic) (Kamnitsas et al., 2017). TensorFlow is an open source platform for machine learning, particularly deep learning. DeepMedic provides a general framework for a multi-scale convolutional neural network.
2.3. CAST training and segmentation pipeline
For the UMC dataset, a single imaging channel using T2 weighted data and the manual segmented hippocampal subfields treated as ground truth was used to train the network. For the MNI dataset, instead, two imaging channels containing T1 and T2 weighted data were used together to train the network. The CAST training and segmentation pipeline with a single T2 imaging modality as example was shown in Fig. 3. For both training and segmentation, common processing steps, including affine transformation, image cropping, intensity normalization and down-sampling were carried out. Affine transformation (e.g. ANTS 3 −m MI[…,…,1,32] −i 0) using the publicly available software ANTS (Avants et al., 2008) is applied to acquire data from different subjects in approximately the same orientation using a certain subject as reference. Other software packages, such as FSL (https://fsl.fmrib.ox.ac.uk/fsl/fslwiki) and SPM (https://www.fil.ion.ucl.ac.uk/spm/), also can be used for the linear transformation. We observed that CNN can handle variability between subjects and did not use nonlinear co-registration in the entire pipeline, regardless of training or segmenting on a new subject. Affine transformation is not required if data are acquired with the same acquisition orientation. To reduce computational cost, the whole-brain images are cropped to regional images with the entire hippocampus included. The cropped images are intensity-normalized before fed to the network to eliminate the inter-subject variation of images. CAST automatically partitions the input cropped images to small imaging patches (purple, green and blue boxes in Fig. 2) and iterates through the entire cropped image for training and segmentation, thus the input images for the training and segmentation process are not required to have the same size and shape for each subject in the dataset (e.g. the UMC dataset has different sizes for each subject). Partitioning input images to small-size image patches is a technique commonly used in deep learning to reduce GPU memory overload. In our implementation, the original resolution image patch has a size of 373, and two down-sampled images have sizes of 233 and 213 after being down-sampled by a factor of 3 and 5, respectively. The CNN is trained with these image patches as input and optimized by minimizing the cross-entropy between the output of soft probabilistic maps from the network and manual segmentation. Training a model on a Tesla K40c GPU card requires approximately three days.
Fig. 3.
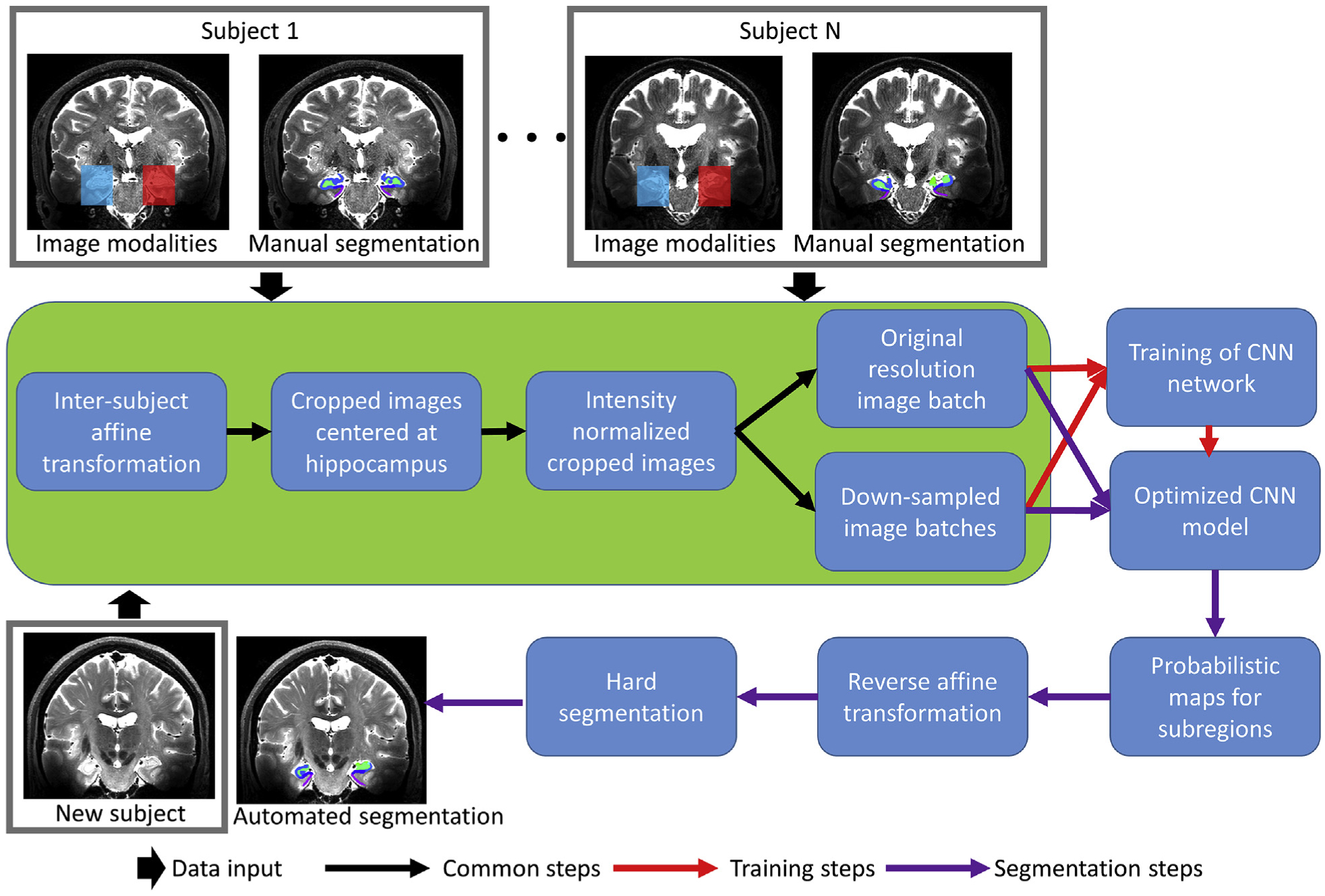
CAST training and segmentation pipeline with a single T2 imaging modality as input.
The input to the segmentation pipeline is the raw imaging data (the same input imaging modalities as used in the training pipeline) from a new subject. When the CAST is applied on a new subject for automated segmentation, after passing through the common processing steps, the trained network is applied and outputs the probabilistic maps for each subregion. Then, a reverse affine transformation is applied to have the probabilistic map projected to the subject space. Finally, a hard segmentation map is generated by assigning each voxel to the subfield with the highest probability. The segmentation pipeline can be applied with or without GPU support. CAST segments a new subject in about 10 s with GPU enabled and less than 1 min when GPU is unavailable.
2.4. Statistical analysis
A five-fold cross validation technique was used to evaluate the performance of CAST. The subjects are divided into five folds, each fold approximately (in case the number of subjects are not integrally divisible by five) has the same number of subjects. The subjects within four folds were used to train the network and the subjects within the remaining one fold were automatically segmented. Among the training data, one subject was randomly selected for validation to monitor if severe overfitting occurred in the training process. The process was repeated until all subjects were automatically segmented once. The dice similarity coefficient (DSC) was applied to evaluate the accuracy of CAST. The DSC was calculated for each subfield separately, and a generalized DSC score (Crum et al., 2006) was computed with all subfields considered jointly. The reliability of automated segmentation was measured with an intraclass correlation coefficient (ICC), which measures the absolute agreement under a two-way random effects from a single measurement (Koo and Li, 2016). The Jaccard similarity coefficient was also used to assess the performance of automated segmentation. Note that these metrics (e.g. ICC and DSC) cannot be compared across different datasets, even for the same subregion, because the criteria for delineating subregions vary in the manual segmentation protocols.
Unlike traditional machine learning, which usually considers each subject as a sample, CAST is trained with each voxel as a sample with a “ground-truth” labelling from manual segmentation. Each voxel has its own unique neighboring voxel environment, and overfitting to the voxels seen in the training data is unlikely to perform well with the large number of samples (voxels) in segmenting a new subject. In traditional machine learning applications, leave-one-out cross validation analysis was observed to have a high risk of overfitting to the priors and achieve unrealistic results because only a single sample was tested in each trained model. We have tested CAST with leave-one-out and five-fold cross validation on left hippocampus of the UMC dataset with the observation that five-fold cross validation had average generalized DSC score 0.005 lower than the score of the leave-one-out technique. While the ICC and DSC scores for ASHS segmentation reported in Wisse et al. (2016) and compared in this study was computed with leave-one-out technique, five-fold cross validation was used and reported for CAST segmentation to reduce the repetitions of training the model, leading to decreased computational time.
3. Results
3.1. Automated segmentation for UMC data
By running CAST with a five-fold cross validation technique, the DSC and ICC coefficient for the 26 subjects in UMC data are shown in Fig. 4. The ICC and DSC of ASHS-automated versus manual rater (named as ASHS) with a leave-one-out technique, of 2 independent raters (named as inter-rater) and of a single rater (named as intra-rater) were directly extracted from Table 2 in Wisse et al. (2016). Note that the values for manual segmentation are based on a proportion of subjects instead of the entire data. For UMC dataset, ASHS is trained by Wisse et al. (2016) with both T1 and T2 weighted data, and CAST is trained with only T2 weighted data. The intra-rater metrics represent the upper limit of automated segmentation, because the automated method is trained with manual segmentation. As expected, the intra-rater metrics have higher DSC and ICC scores than CAST, ASHS, and inter-rater metrics of manual segmentation except the ICC of CA2 in the right hippocampus. Overall, automated methods CAST and ASHS achieve comparable performance in terms of DSC for all subregions. However, the ICC value for each method differs. For both CAST and ASHS, the mean generalized DSC across all subfields is 0.80 ± 0.03 in the left hippocampus, and 0.78 ± 0.03 and 0.79 ± 0.03 respectively for the right hippocampus. Along with calculating the DSC for subfields, we also calculated the DSC for background (label = 0, non-hippocampus region). The DSC of background in CAST is above 99.4% for all subjects, thus indicating that few background voxels were misclassified as hippocampal subfields. The remaining maximum 0.6% discrepancy could be explained by more or less regions being segmented than by manual segmentation.
Fig. 4.
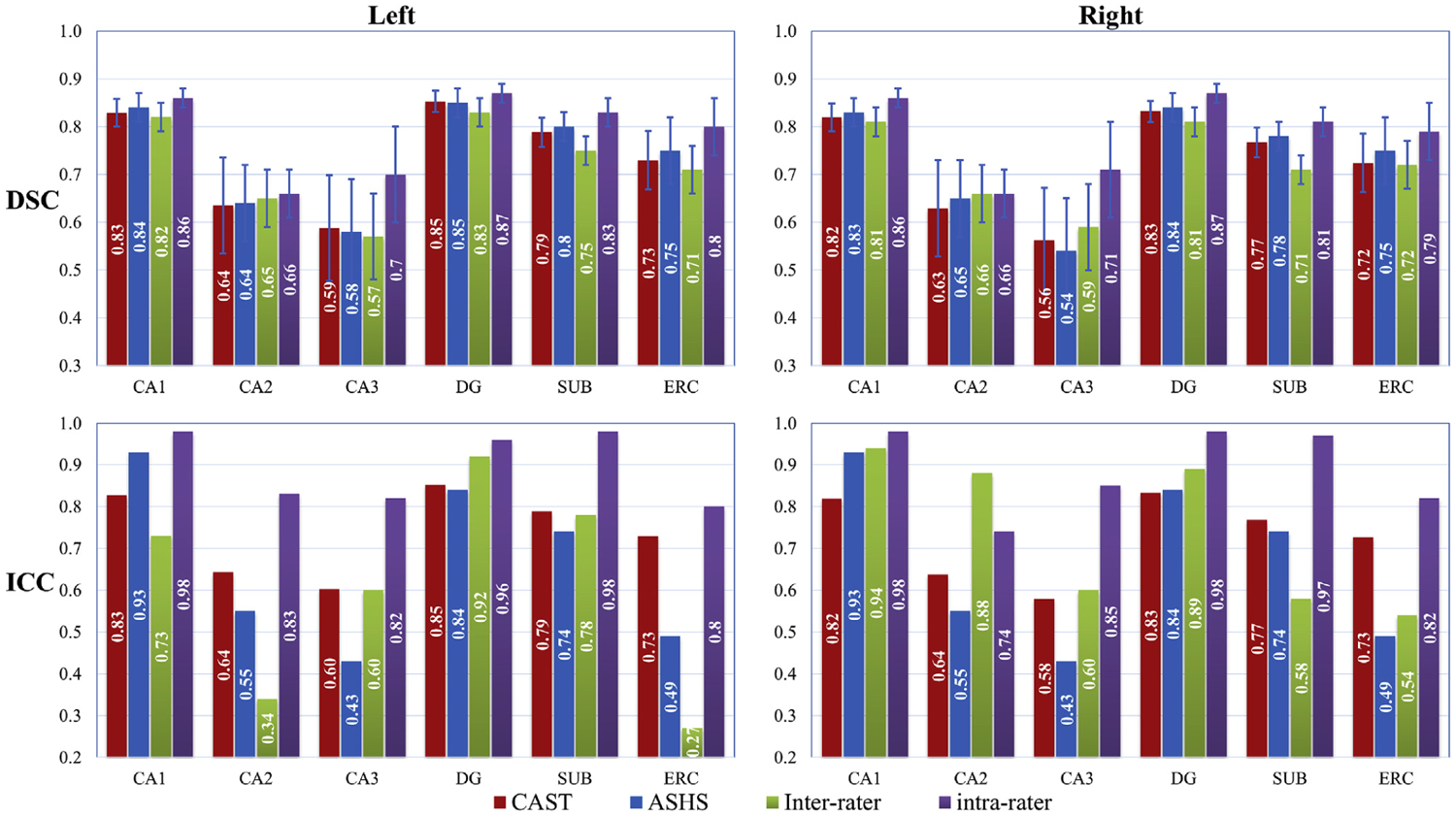
Dice coefficients (DSC) and intraclass correlation coefficient (ICC) for all subfields from UMC dataset. The DSC and ICC values for ASHS-automated versus manual rater (named as ASHS) with a leave-one-out technique, of 2 independent raters (named as inter-rater) and of a single rater (named as intra-rater) are directly taken from Wisse et al. (2016). The intra-rater and inter-rater metrics are based on a part of subjects instead of the entire data. Five-fold cross validation, instead of leaving one out as in ASHS, is used in CAST to reduce the repetitions of training the model. Both T1 and T2 weighted images are used in ASHS, but only T2 weighted is used in CAST. Compared to ASHS segmentation, CAST substantially improved the ICC coefficients for CA2, CA3, SUB, and ERC in the left hippocampus relatively by 17%, 40%, 7%, and 49%, respectively, and for CA3 and ERC in the right hippocampus by 29% and 42%, respectively, based on the measure (ICC(CAST)-ICC(ASHS))/ICC(ASHS) × 100%. However, CAST had a worse ICC coefficient for CA1 compared to ASHS by 11% and 16% for the left and the right hippocampus. The DSC and ICC values are highly consistent for CAST segmentation with a maximal discrepancy of 0.01 for DSC - ICC.
Table 2.
A summary of the comparison between Freesurfer 6.0, ASHS and CAST automated segmentation methods.
| Automated segmentation methods | Time cost to segment a new subject | Feasible to train a new model? | Hardware requirement to train a new model | Required coregistration | Support multiple modalities | Adaptive to different MRI pulse sequence |
|---|---|---|---|---|---|---|
| Freesurfer 6.0 | A few hours up to one day with -itkthreads 8 setting | Not feasible | Not applicable | Affine and nonlinear | Yes | The same model is used because MRI contrast is discarded |
| ASHS | Approximately 30 min on a 8-core machine | Feasible | Single workstation or computer clusters | Affine and nonlinear | Yes | Performs best when trained on images acquired with the same sequence |
| CAST | Less than 1 min | Feasible | Single workstation with a descent GPU card (e.g. Tesla K40c; GPU is not required for segmentation) | Affine | Yes | Performs best when trained on images acquired with the same sequence |
Compared to ASHS segmentation, CAST substantially improved the ICC coefficients for CA2, CA3, SUB, and ERC in left hippocampus relatively by 17%, 40%, 7%, and 49%, respectively and for CA3 and ERC in right hippocampus by 29% and 42%, respectively, based on the measure (ICC(CAST)-ICC(ASHS))/ICC(ASHS) × 100%. However, compared to CAST, ASHS appears less sensitive to the variation of CA1 in the atlas, which allows ASHS to tolerate the disagreement between subjects and make subregion CA1 have much higher ICC score than CAST segmentation. CAST had a worse ICC coefficient for CA1 compared to ASHS by 11% (ICC of CA1: CAST 0.83, ASHS 0.93) for left hippocampus and 16% (ICC of CA1: CAST 0.82, ASHS 0.97) for right hippocampus. By comparing the difference between ICC and DSC scores (DSC - ICC) in Fig. 4, the inter-rater metrics of manual segmentation shows discrepant DSC and ICC values for CA2 (left: DSC 0.65, ICC 0.34; right: DSC 0.66 ICC 0.88), DG (left: DSC 0.83, ICC 0.92; right: DSC 0.81, ICC 0.89) and ERC (left: DSC 0.71, ICC 0.27; right: DSC 0.72, ICC 0.54). ASHS shows discrepant DSC and ICC values for CA2 (left: DSC 0.64, ICC 0.55), CA3 (left: DSC 0.58, ICC 0.43; right: DSC 0.54, ICC 0.45) and ERC (left: DSC 0.75, ICC 0.49; right DSC 0.75, ICC 0.51). In contrast, the DSC and ICC values are highly consistent for CAST segmentation with a maximal discrepancy of 0.01. The example plot of the CAST-automated and manual segmentation from the subject with the best, median, and worst generalized DSC score as 0.85, 0.79 and 0.74 respectively across left and right hippocampus are shown in Fig. 5. The 3D rendering plot of CAST and manual segmentations with median generalized DSC score is shown in Fig. 6. The automated segmentation overall is very similar to the manual segmentation but with small localized differences observed at the boundary among subfields or between subfields and background.
Fig. 5.
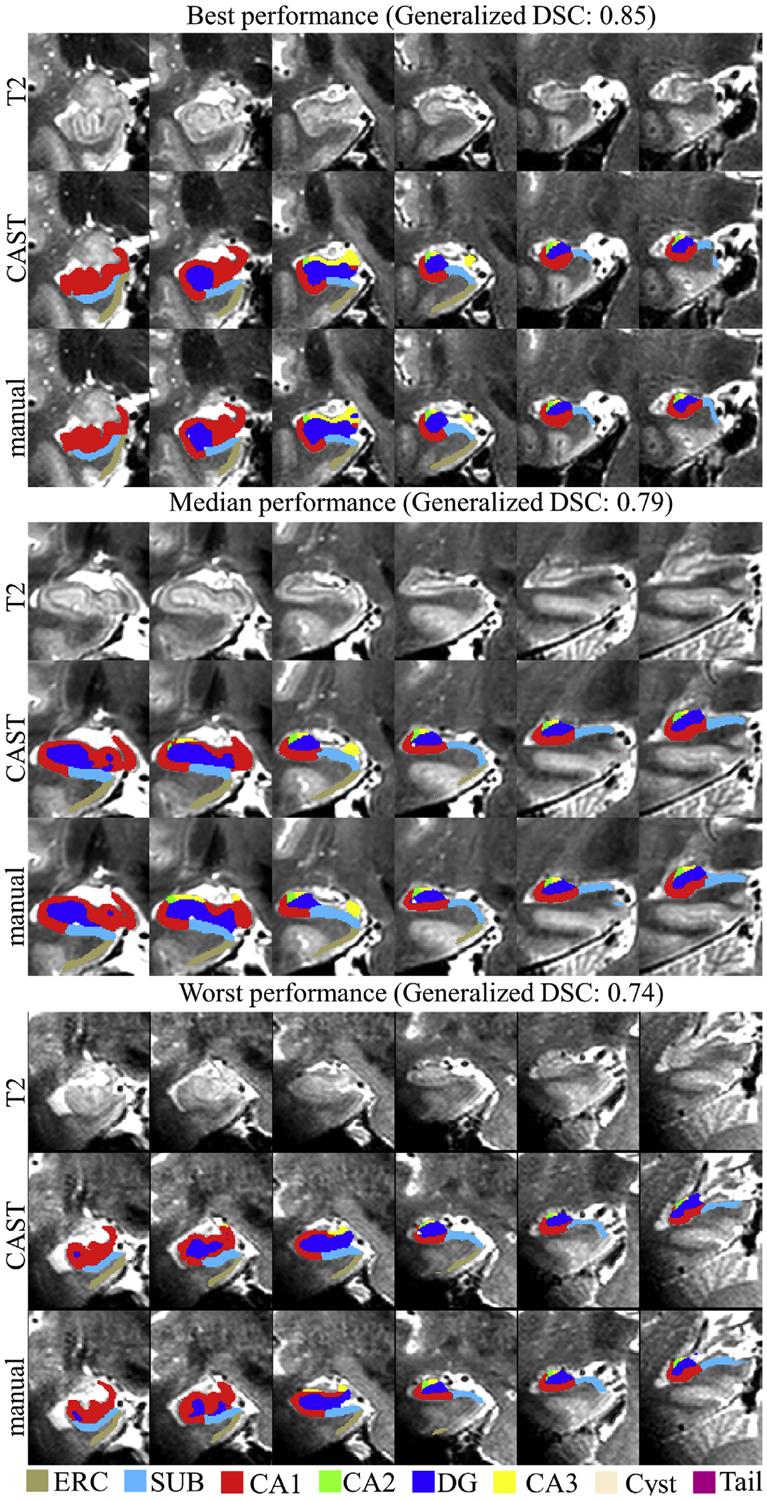
Example plot of T2 weighted image, manual and CAST segmentations from the subject having best (right hippocampus), median (right hippocampus) and worst (right hippocampus) generalized DSC coefficient across left and right hippocampus in UMC dataset. Top panel: generalized DSC 0.85, right hippocampus; middle panel: generalized DSC 0.79, right hippocampus; lower panel: generalized DSC 0.74, right hippocampus.
Fig. 6.
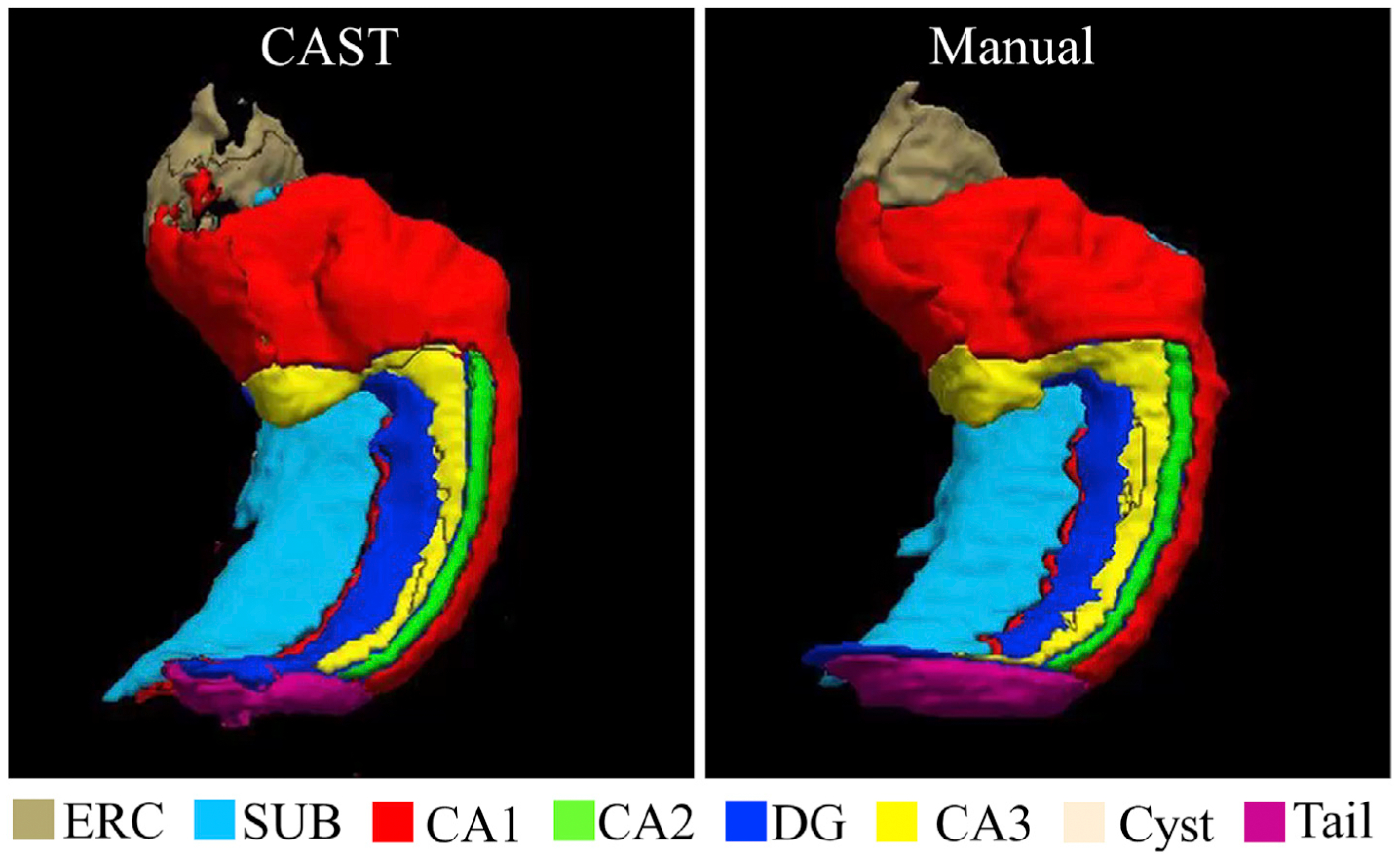
3D rendering of manual and CAST segmentation with median generalized DSC score of the UMC dataset. Note that the manual segmentation is blind to CAST when using CAST to segment the corresponding subject. The video of the 3D rendering is attached as Supplementary information.
To further investigate the different ICC values for the CA1 subfield, we have visually inspected the manual segmentations and observed that the boundary between CA1 and SUB was defined differently across subjects based on the geometric rule in the protocol. The manual segmentations from two subjects are shown in Fig. 7, and the different boundaries between CA1 and SUB are pointed out by white arrows. The challenge for CAST to learn some criteria used in manual segmentation is also observed with UMag dataset (see Supplementary), where distinct segmentation rules are applied when a certain hard threshold is met, e.g. the depths of the collateral sulcus in the range >10 mm, 7–10 mm, 4–7 mm or <4 mm for Area 35. The difficulty to strictly follow some criteria in manual segmentation could explain why manually enforcing some criteria on CAST segmented maps (CAST-post in Supplementary) improves the segmentation performance slightly.
Fig. 7.
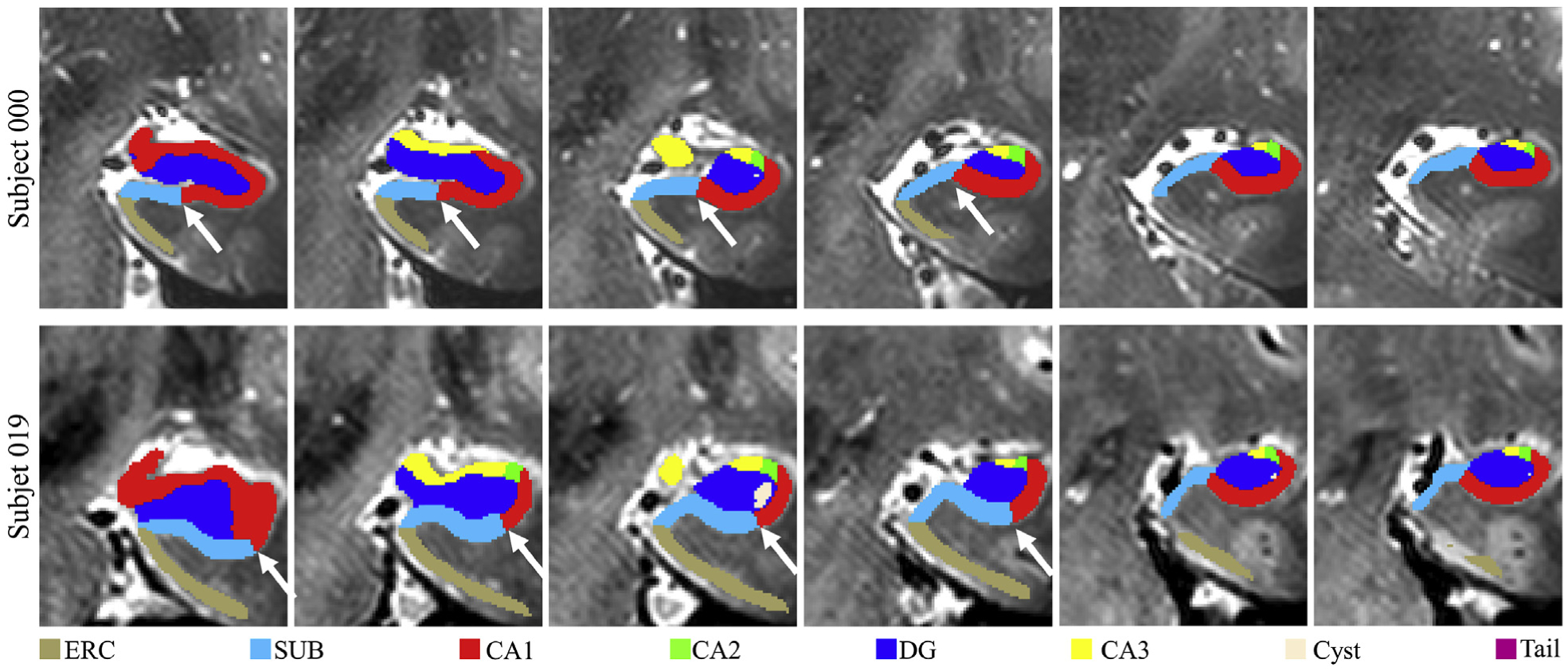
Example plots of the boundary between CA1 and SUB varying across subjects in UMC dataset. The white arrows point to the distinct boundary between CA1 and SUB in two subjects.
For UMC dataset, the mean Jaccard similarity coefficient over all subjects for CA1, CA2, CA3, DG, SUB, and ERC are 0.71,0.47, 0.42, 0.74, 0.65 and 0.58 respectively for the left hippocampus and 0.70, 0.47, 0.40 0.71, 0.62, and 0.57 respectively for the right hippocampus.
The DSC and ICC scores for hippocampus cyst and tail were not reported in Wisse et al. (2016). CAST had the scores of these two subregions varying considerably between subjects. We concatenated the manual and CAST segmentation maps of all subjects and evaluated CAST segmentation of cyst with DSC score as 0.55/0.50 and of tail with DSC score as 0.67/0.59 for left/right hippocampus. Consistent with all other subregions, the difference between DSC and ICC scores is below 0.01 for cyst and tail.
3.2. Automated segmentation for the MNI dataset
Five-fold cross validation technique was also applied to the MNI dataset, and automated segmentation maps for 25 subjects were obtained. The DSC, ICC, and Jaccard coefficient for CAST segmentation are shown in Table 1. The DSC and ICC scores of a single rater and two independent raters were directly extracted from Kulaga-Yoskovitz et al. (2015) and listed in Table 1 with gray background. The left and right hippocampus were analyzed together in Kulaga-Yoskovitz et al. (2015), thus a single score for each subject was obtained for each subfield. Consistent with the performance in the UMC dataset, the DSC and ICC scores achieved by CAST overall were lower than the corresponding intra-rater scores of manual segmentation. However, the ICC scores for CA1–3 in bilateral hippocampus were comparable between CAST and intra-rater manual segmentation (CAST: 0.912/0.914 for left and right hippocampus; manual segmentation: 0.91/0.91 for inter-rater and intra-rater scores). Notably, automated segmentation of SUB had ICC score 0.16 higher than the inter-rater ICC score (CAST: 0.887/0.886; inter-rater: 0.73), and automated segmentation of CA4+DG had ICC score 0.02 less than the inter-rater ICC score (CAST: 0.880/0.875; inter-rater: 0.90). CAST had DSC score slightly (approximately 0.01) higher than the inter-rater DSC score for all the other subfields, except CA4+DG in right hippocampus (CAST: 0.875 for CA4+DG in right hippocampus; inter-rater: 0.876 for CA4+DG in bilateral hippocampus). Compared to the surface patch-based segmentation method (Caldairou et al., 2016), CAST achieved substantially improved DSC score. CAST had the mean (standard deviation) Jaccard coefficient of 0.848(0.019)/0.850(0.022) for CA1–3, 0.802(0.029)/0.800(0.032) for SUB and 0.787(0.032)/0.779(0.039) for CA4+DG in left/right hippocampus. For all the subfield-specific metrics except inter-rater and intra-rater ICC, CA1–3 always had the highest score, and CA4+DG had the lowest score regardless of whether automated or manual segmentation is carried out. While CAST and intra-rater ICC scores are very close, CAST has the ICC scores of SUB and CA4+DG considerably lower than the corresponding intra-rater ICC, which could be because of the difficulty of delineating the boundary between SUB and CA4+DG, especially considering the slightly lower DSC scores for these two labels. The left and right hippocampus had the same generalized DSC score 0.906 with results for the left hippocampus being slightly more stable (less standard deviation). As with the UMC dataset, the discrepancy between DSC and ICC was below 0.01 for all subregions. The plot of best, median, and the worst automated segmentation based on generalized DSC score is shown in Fig. 8.
Table 1.
Evaluation of CAST automated segmentation in the MNI dataset.
| Left/Right hippocampus | |||
|---|---|---|---|
| CAST DSC | 0.917(0.011)/0.919(0.013) | 0.890(0.017)/0.889(0.020) | 0.881(0.021)/0.875(0.025) |
| Inter-rater DSCa | 0.903 (0.036) | 0.871 (0.053) | 0.876 (0.048) |
| Intra-rater DSCa | 0.929 (0.010) | 0.905 (0.016) | 0.900 (0.019) |
| CAST ICC | 0.912/0.914 | 0.887/0.886 | 0.880/0.875 |
| Inter-rater ICCa | 0.91 | 0.73 | 0.90 |
| Intra-rater ICCa | 0.91 | 0.94 | 0.96 |
| CAST Jaccard coefficient | 0.848(0.019)/0.850(0.022) | 0.802(0.029)/0.800(0.032) | 0.787(0.032)/0.779(0.039) |
| CAST | 0.906(0.011)/0.906(0.014) | ||
| Generalized | |||
| DSC | |||
The DSC and ICC scores of a single rater (intra-rater) and two independent raters (inter-rater) were directly extracted from Kulaga-Yoskovitz et al. (2015) and marked with gray background. All the rest metrics are calculated from CAST segmentation.
Fig. 8.
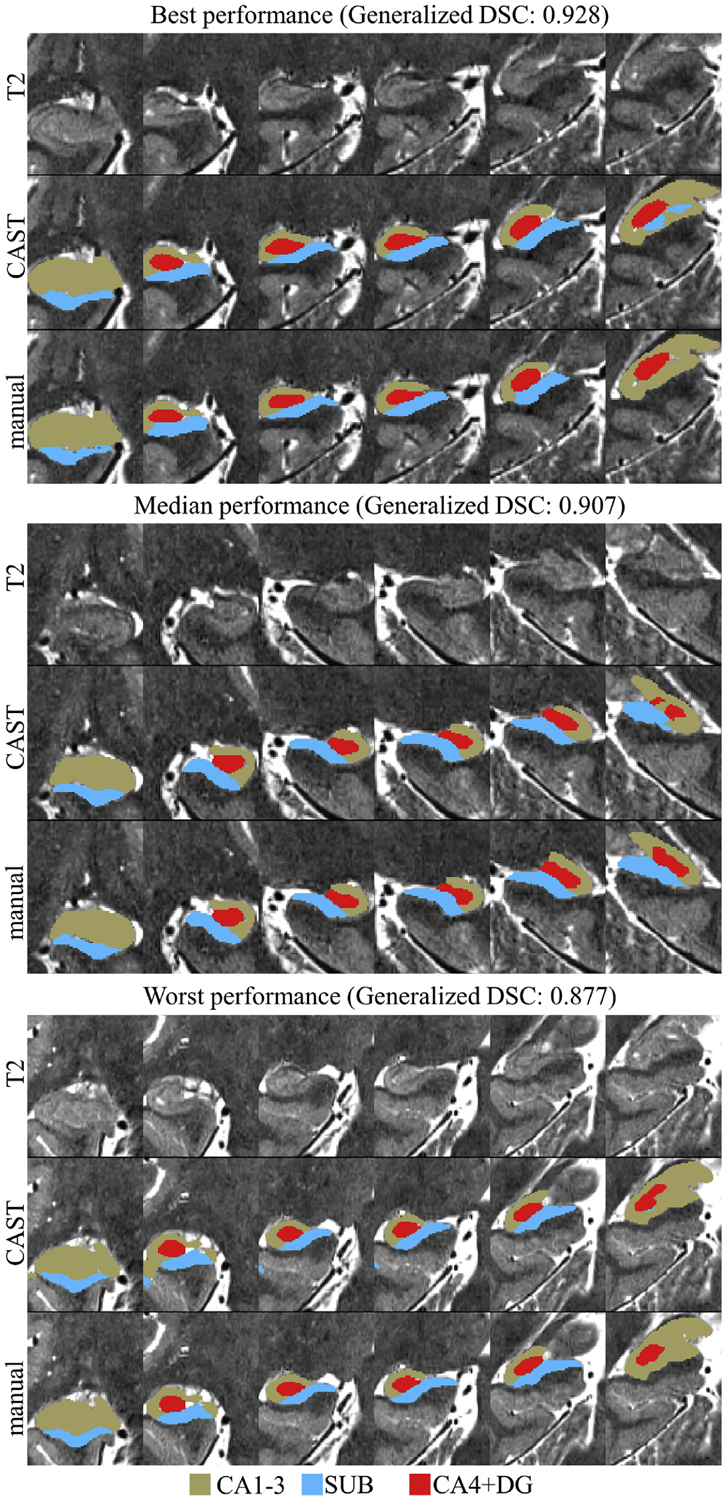
Plot of the automated segmentation from the five-fold cross validation method with the best, median and the worst generalized dice similarity coefficient (DSC) scores. Upper panel: generalized DSC 0.928, right hippocampus; middle panel: generalized DSC 0.907, left hippocampus; lower panel: generalized DSC 0.877, right hippocampus.
4. Discussion
In this study, a Convolutional neural network based Automated hippocampal subfield Segmentation Toolbox (CAST) is developed. We have demonstrated that CAST is feasible for automated segmentation on one 7T dataset and one 3T MRI dataset without revising the network architecture and parameters settings, suggesting that CAST is generalizable and could potentially be used to train other data without modification. The CAST segmentation achieves DSC scores comparable with the inter-rater score of manual segmentation and even has lower disagreement than 2 independent raters following the same segmentation protocol for some subfields, particularly CA2 and ERC in the UMC dataset and SUB in the MNI dataset. No nonlinear registration occurs in the entire process, and, once a model is trained, CAST can segment one subject in less than 1 min. Therefore, CAST is useful for studies with large sample sizes and applications requiring fast processing time. In contrast, ASHS and Freesurfer 6.0 are computationally intensive and more time-consuming for segmenting a single subject. As with the automated segmentation methods in ASHS and Freesurfer 6.0, CAST is also applicable for multiple imaging modalities. Theoretically, more than two modalities can be fed to CAST for training and segmentation without substantially increasing memory and computational cost. Furthermore, the parameters in the deep neural network are independent of the input image size. Therefore, CAST does not restrict input images to the same size in both training and segmentation pipelines, as long as these images have the same resolutions. ASHS and manual segmentation are observed to have distinct DSC and ICC values for certain subfields (e.g. CA3 and ERC), the discrepancy makes judging the quality of segmentation difficult. In contrast, CAST has very consistent DSC and ICC values, with a maximal discrepancy of 0.01 for all subregions in both the UMC and MNI datasets. A summary of the comparison between these three automated segmentation methods is shown in Table 2.
To evaluate the necessity of each down-sampled pathway in the multi-scale CNN, we tested the network with only one down-sampled pathway. Regions were mislabeled as hippocampal subfields when the pathway with input down-sampled by a factor of 5 (the third pathway) or 3 (the second pathway) was removed. We also tested the CNN with different architectures, and no statistically significant improvement was observed when further increasing the number of layers or pathways in the network.
Instead of down-sampling input images and using multiple pathways to extract more global information from far-distance pixels/voxels, using a dilated convolutional filter (Yu and Koltun, 2015) and an intermediate down-sampling layer are common techniques used in deep learning to extract multi-scale features in the application of natural images. Both a dilated convolutional filter and an intermediate down-sampling layer require a larger input image size to acquire more global information with the cost of an exponentially increased memory requirement. In contrast, down-sampling input images allows extracting global information from coarse images with the memory cost linearly increasing with the number of pathways in the network. In this study, we have attempted to segment hippocampal subfields using other deep neural networks with a dilated convolutional filter or an intermediate down-sampling layer enabled. VoxResNet (Chen et al., 2018) is a 25-layer voxelwise residual network for 3D medical image segmentation, which down-samples images three times at intermediate layers and concatenates outputs from four intermediate layers as a final feature set to segment brain as white matter, gray matter, or ventricle. VoxResNet achieved the best performance in the MICCAI MRBrainS challenge. However, VoxResNet had a high false positive rate when applied for hippocampal segmentation and misclassified non-hippocampus voxels as hippocampal subfields. Similar phenomena were observed when we extended the 2D tiramisu network (Jégou et al., 2017), a very deep, fully convolutional neural network, to 3D with a dilated convolutional layer enabled for hippocampal subfield segmentation. For both networks, maximal input image size within the limit of memory is used for both the training and segmentation processes.
Although a high field MRI scanner is used, the hippocampus has a much lower contrast between subfields than the contrast in natural images or the contrast between gray matter, white matter, and ventricles. In addition, the structure of the hippocampal subfield is highly anisotropic and delicate, which makes the segmentation more challenging. Furthermore, some criteria in manual segmentation of hippocampal subfields are based on information from a third-party region relatively far away from the hippocampus itself, e.g. appearance of colliculi (Berron et al., 2017). While using another region as a landmark is common for segmenting hippocampal subfields, this third-party information is barely considered for either natural image processing (e.g. identifying humans, cars, traffic lights and so on) or other brain segmentation applications (e.g. segmenting brain images into gray matter, white matter and cerebrospinal fluid) in deep learning segmentation. These three reasons, along with memory limit on the input image size and the unconventional design of multi-scale CNN, could explain why CAST has superior performance in hippocampal subfield segmentation over the two other deep learning algorithms. Conditional random field (CRF) is a commonly used post-processing step in natural imaging processing for refining segmented images with neighboring information. We implemented a 3D fully-connected CRF with pairwise Gaussian potential (Krähenbühl and Koltun, 2011) to further process the probabilistic maps for subfields (see Fig. 3) before the hard segmentation. Surprisingly, no statistically significant improvement was observed based on either the DSC or the ICC values, possibly because CAST has captured enough neighboring information, and the effect of CRF becomes less prominent.
One drawback in CAST is that CAST appears more sensitive to the anatomical variability of CA1 between subjects and achieves lower ICC scores for the UMC dataset. The border between CA1 and SUB shifts to the most medial point of the most anterior DG as soon as DG becomes visible in the segmentation protocol (Wisse et al., 2012). Since the location of the DG differs between subjects, the border between CA1 and SUB differs as well. The superior performance of ASHS in segmenting CA1 may be because ASHS registers a new subject to all atlases separately and then uses a joint label fusion method to determine the final segmentation (Wisse et al., 2016), which allows ASHS to tolerate the variations between subjects in the atlases. Instead, CAST achieves a single model by learning the common rules from all atlases, which may compromise the variation between subjects and lead to lower ICC values. In addition, the hard threshold based on the anatomical variability, e.g. the depths of the collateral sulcus used in UMag dataset (see Supplementary), is easy to measure and improves the reliability in manual segmentation, but learning such a hard threshold is challenging in machine learning or deep learning algorithm, which is the reason why enforcing some criteria used in manual segmentation on CAST segmented map (namely CAST-post in Supplementary) improves the performance slightly. More manually segmented subjects to train the network potentially can be beneficial for further improving the performance of CAST segmentation. However, that CAST does not strictly follow criteria in manual segmentation may be because it is adaptive to anatomical variation. In UMag dataset (see Supplementary), the manual segmentation of CA2 and CA3 is fixed to start 4 slices (4.4 mm) before the end of the hippocampal head, CAST segments the anterior border of CA3 in the head 3 to 5 slices (3.3–5.5 mm) away from the start of the body, which is consistent with the finding from Ding and Van Hoesen (2015). Certainly, there is no evidence to prove if the variation in the anterior border of CA3 as segmented by CAST matches the underlying anatomy since no histological annotations are available for these subjects.
CAST segmentation in no way underemphasizes the importance of manual segmentation. Instead, we cannot overemphasize the critical nature of manual segmentation. While CAST does not know the exact criteria in the manual segmentation protocols, CAST encodes the manual labelling information in the network by minimizing the discrepancy between estimated and manual labelling and then generalizes the labelling to a new subject. The performance of automated segmentation, in fact, heavily relies on the reliability of manual segmentation. As with UMC dataset, low DSC scores for CA2 and CA3 in measuring intra-rater reliability directly leads to low DSC scores for both CAST and ASHS segmentation. Because of the varying segmentation protocols across different institutes, CAST-segmented subfields cannot be directly compared with the result from Freesurfer 6.0 or a different atlas set in the ASHS repository (e.g. the results from UMC and UMag datasets are not comparable). Harmonizing different protocols for hippocampal and parahippocampal subfields (Wisse et al., 2017; Yushkevich et al., 2015a) will facilitate comparison between studies and is the key to eliminating the discrepancy between automated segmentation toolboxes. Another limitation of both CAST segmentation and ASHS is that automated segmentation may not perform well when a pre-trained model is directly applied to images with distinct acquisition parameters or from different scanners, especially having different magnetic strength. A set of subjects with imaging data acquired from different scanners (e.g. 7T and 3T scanners) with different acquisition parameters (e.g. flip angles and TE) but segmented by following the same manual segmentation protocol would be particularly valuable to create a more generalizable model for both CAST and ASHS segmentation. However, CAST easily can be trained on a single workstation with a decent GPU card (e.g. Tesla K40c) when a set of manual segmentations for that dataset is available, likely without revising the network architecture. A transfer learning technique, which initializes a new model with the parameters from the optimized model of other data, could further shorten training time. Instead of using only T1 and T2 structural images for segmenting hippocampal subfields, Wu et al. (2018) demonstrated the possibility of combining resting-state functional magnetic resonance imaging together with structural images for improved segmentation performance. We will explore the possibility of embedding functional and structural information into CAST architecture in our future research.
5. Conclusion
In this study, we present a fast automated hippocampal subfield segmentation method based on a multi-scale deep convolutional neural network, which can segment a new subject in less than 1 min. Compared to current state-of-art methods, this method achieves comparable accuracy in terms of dice coefficient and is more reliable in terms of the intraclass correlation coefficient for most subfields. CAST segmentation is a flexible and adaptive method, and we successfully applied this method to datasets from two different MRI scanner vendors with different acquisition parameters without revising any settings and network architecture. Both the scripts and trained model are publicly available at https://github.com/pipiyang/CAST.
Supplementary Material
Acknowledgment
We would like to thank the three anonymous reviewers for their insightful comments and suggestions. This research study was supported by the NIH (grant 1R01EB014284 and COBRE grant 5P20GM109025), a private grant from Peter and Angela Dal Pezzo, a private grant from Lynn and William Weidner, and a private grant from Stacie and Chuck Matthewson. The atlases and data for this study were generously shared by corresponding investigators, and publicly available on NeuroImaging Tools & Resources Collaboratory (NITRC) website (UMC and UMag datasets: https://www.nitrc.org/projects/ashs; MNI dataset: https://www.nitrc.org/projects/mni-hisub25/).
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neuroimage.2020.116947.
References
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal 12, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S, Vieweg P, Gao F, Gilboa A, Wolbers T, Black SE, Rosenbaum RS, 2016. The human dentate gyrus plays a necessary role in discriminating new memories. Curr. Biol 26, 2629–2634. [DOI] [PubMed] [Google Scholar]
- Berron D, Schütze H, Maass A, Cardenas-Blanco A, Kuijf HJ, Kumaran D, Düzel E, 2016. Strong evidence for pattern separation in human dentate gyrus. J. Neurosci 36, 7569–7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berron D, Vieweg P, Hochkeppler A, Pluta J, Ding S-L, Maass A, Luther A, Xie L, Das S, Wolk D, 2017. A protocol for manual segmentation of medial temporal lobe subregions in 7 Tesla MRI. Neuroimage: Clin 15, 466–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldairou B, Bernhardt BC, Kulaga-Yoskovitz J, Kim H, Bernasconi N, Bernasconi A, 2016. A surface patch-based segmentation method for hippocampal subfields In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 379–387. [Google Scholar]
- Chadwick MJ, Bonnici HM, Maguire EA, 2014. CA3 size predicts the precision of memory recall. Proc. Natl. Acad. Sci. Unit. States Am 111, 10720–10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Dou Q, Yu L, Qin J, Heng P-A, 2018. VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage 170, 446–455. [DOI] [PubMed] [Google Scholar]
- Crum WR, Camara O, Hill DL, 2006. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans. Med. Imag 25, 1451–1461. [DOI] [PubMed] [Google Scholar]
- Ding S, Van Hoesen GW, 2015. Organization and detailed parcellation of human hippocampal head and body regions based on a combined analysis of cyto-and chemoarchitecture. J. Comp. Neurol 523, 2233–2253. [DOI] [PubMed] [Google Scholar]
- Goodfellow I, Bengio Y, Courville A, 2016. Deep Learning. MIT press. [Google Scholar]
- Goubran M, Ntiri EE, Akhavein H, Holmes M, Nestor S, Ramirez J, Adamo S, Ozzoude M, Scott C, Gao F, 2020. Hippocampal segmentation for brains with extensive atrophy using three-dimensional convolutional neural networks. Hum. Brain Mapp 41, 291–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2015. Delving deep into rectifiers: surpassing human-level performance on imagenet classification. In: Proceedings of the IEEE international conference on computer vision, pp. 1026–1034. [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2016. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 770–778. [Google Scholar]
- Iglesias JE, 2015. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. NeuroImage 115, 117–137. 10.1016/j.neuroimage.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jégou S, Drozdzal M, Vazquez D, Romero A, Bengio Y, 2017. The one hundred layers tiramisu: fully convolutional densenets for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, pp. 11–19. [Google Scholar]
- Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B, 2017. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal 36, 61–78. [DOI] [PubMed] [Google Scholar]
- Koo TK, Li MY, 2016. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropract. Med 15, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krähenbühl P, Koltun V, 2011. Efficient inference in fully connected crfs with Gaussian edge potentials. Adv. Neural Inf. Process. Syst 109–117. [Google Scholar]
- Kulaga-Yoskovitz J, Bernhardt BC, Hong S-J, Mansi T, Liang KE, Van der Kouwe AJ, Smallwood J, Bernasconi A, Bernasconi N, 2015. Multi-contrast submillimetric 3 Tesla hippocampal subfield segmentation protocol and dataset. Sci. Data 2, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G, 2015. Deep learning. Nature 521, 436. [DOI] [PubMed] [Google Scholar]
- Leutgeb JK, Leutgeb S, Moser M-B, Moser EI, 2007. Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science 315, 961–966. [DOI] [PubMed] [Google Scholar]
- Liu M, Li F, Yan H, Wang K, Ma Y, Shen L, Xu M, Initiative A.s.D.N., 2020. A multi-model deep convolutional neural network for automatic hippocampus segmentation and classification in Alzheimer’s disease. Neuroimage 208, 116459. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Yushkevich PA, Das S, Wang L, Van Leemput K, Iglesias JE, Alpert K, Mezher A, Ng P, Paz K, 2018. Systematic comparison of different techniques to measure hippocampal subfield volumes in ADNI2. Neuroimage: Clin. 17, 1006–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogovitsyn N, Souza R, Muller M, Srajer A, Hassel S, Arnott SR, Davis AD, Hall GB, Harris JK, Zamyadi M, 2019. Testing a deep convolutional neural network for automated hippocampus segmentation in a longitudinal sample of healthy participants. Neuroimage 197, 589–597. [DOI] [PubMed] [Google Scholar]
- Novosad P, Fonov V, Collins DL, Initiative A.s.D.N., 2020. Accurate and robust segmentation of neuroanatomy in T1-weighted MRI by combining spatial priors with deep convolutional neural networks. Hum. Brain Mapp 41, 309–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlichting ML, Zeithamova D, Preston AR, 2014. CA1 subfield contributions to memory integration and inference. Hippocampus 24, 1248–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K, Zisserman A, 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition arXiv preprint arXiv:1409.1556.
- Thyreau B, Sato K, Fukuda H, Taki Y, 2018. Segmentation of the hippocampus by transferring algorithmic knowledge for large cohort processing. Med. Image Anal 43, 214–228. [DOI] [PubMed] [Google Scholar]
- Wisse L, Gerritsen L, Zwanenburg JJ, Kuijf HJ, Luijten PR, Biessels GJ, Geerlings MI, 2012. Subfields of the hippocampal formation at 7 T MRI: in vivo volumetric assessment. Neuroimage 61, 1043–1049. [DOI] [PubMed] [Google Scholar]
- Wisse LE, Daugherty AM, Olsen RK, Berron D, Carr VA, Stark CE, Amaral RS, Amunts K, Augustinack JC, Bender AR, 2017. A harmonized segmentation protocol for hippocampal and parahippocampal subregions: why do we need one and what are the key goals? Hippocampus 27, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse LE, Kuijf HJ, Honingh AM, Wang H, Pluta JB, Das SR, Wolk DA, Zwanenburg JJ, Yushkevich PA, Geerlings MI, 2016. Automated hippocampal subfield segmentation at 7T MRI. Am. J. Neuroradiol 37, 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Gao Y, Shi F, Ma G, Jewells V, Shen D, 2018. Segmenting hippocampal subfields from 3T MRI with multi-modality images. Med. Image Anal 43, 10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Koltun V, 2015. Multi-scale Context Aggregation by Dilated Convolutions arXiv preprint arXiv:1511.07122.
- Yushkevich PA, Amaral RS, Augustinack JC, Bender AR, Bernstein JD, Boccardi M, Bocchetta M, Burggren AC, Carr VA, Chakravarty MM, 2015a. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: towards a harmonized segmentation protocol. Neuroimage 111, 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Pluta JB, Wang H, Xie L, Ding SL, Gertje EC, Mancuso L, Kliot D, Das SR, Wolk DA, 2015b. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum. Brain Mapp 36, 258–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shi F, Wang L, Hung S-C, Chen M-H, Wang S, Lin W, Shen D, 2019. Dilated dense U-Net for infant hippocampus subfield segmentation. Front. Neuroinf 13, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


