Abstract
Background
Conventional MRI cannot be used to identify H3 K27M mutation status. This study aimed to investigate the feasibility of predicting H3 K27M mutation status by applying an automated machine learning (autoML) approach to the MR radiomics features of patients with midline gliomas.
Methods
This single-institution retrospective study included 100 patients with midline gliomas, including 40 patients with H3 K27M mutations and 60 wild-type patients. Radiomics features were extracted from fluid-attenuated inversion recovery images. Prior to autoML analysis, the dataset was randomly stratified into separate 75% training and 25% testing cohorts. The Tree-based Pipeline Optimization Tool (TPOT) was applied to optimize the machine learning pipeline and select important radiomics features. We compared the performance of 10 independent TPOT-generated models based on training and testing cohorts using the area under the curve (AUC) and average precision to obtain the final model. An independent cohort of 22 patients was used to validate the best model.
Results
Ten prediction models were generated by TPOT, and the accuracy obtained with the best pipeline ranged from 0.788 to 0.867 for the training cohort and from 0.60 to 0.84 for the testing cohort. After comparison, the AUC value and average precision of the final model were 0.903 and 0.911 in the testing cohort, respectively. In the validation set, the AUC was 0.85, and the average precision was 0.855 for the best model.
Conclusions
The autoML classifier using radiomics features of conventional MR images provides high discriminatory accuracy in predicting the H3 K27M mutation status of midline glioma.
Keywords: H3 K27M mutation automated machine learning, autoML, midline glioma, Tree-based Pipeline Optimization Tool, TPOT
1. AutoML has excellent diagnostic performance for predicting H3 K27M mutation in patients with midline glioma.
2. The prediction model may help establish a precise diagnosis and guide clinical decisions regarding midline glioma treatment.
Importance of the Study.
Molecular parameters such as H3 K27M mutation status are used in addition to histology to help diagnose tumors. H3 K27M mutation is mainly described in midline structures such as the thalamus, brainstem, and spinal cord. According to previous studies, H3 K27M mutation is an independent predictor of overall survival regardless of age, tumor location, or histopathological grading. However, midline gliomas with H3 K27M mutation display variable radiographic features. The H3 K27M status is difficult to determine via conventional subjective analysis of MR images. Radiomics extraction of quantitative features from radiographic images, such as computed tomography (CT) and magnetic resonance imaging (MRI), has become a non-invasive method to predict the molecular status of tumors. Automated machine learning does not require human intervention and performs better than traditional supervised machine learning approaches, representing a promising tool for the identification of H3 K27M mutation. With this prediction model, we built an optimal model that can be used for preoperative routines in the future.
The 2016 World Health Organization classification of tumors of the central nervous system began to integrate molecular and genetic profiling to assist in diagnoses and evaluate prognoses.1 Thereafter, molecular parameters and histology were used to define tumor entities. Diffuse midline glioma, H3 K27M mutant, is a newly defined group of tumors characterized by a K27M mutation in either H3F3A or HIST1H3B/C.2 In early studies, H3 K27M mutation was detected mainly in diffuse intrinsic pontine gliomas arising in the brainstem of children,3 and it was subsequently detected in adult gliomas of midline structures, such as the thalamus, brainstem, and spinal cord.4,5 Among the midline gliomas, H3 K27M mutation is an independent predictor of overall survival (OS), regardless of age, tumor location, or histopathological grading.6–8 Pediatric patients with high- and low-grade thalamic gliomas carrying H3 K27M mutation have a shorter median survival time than patients carrying wild-type H3 K27M.8 Furthermore, an immunological study postulated that H3 K27M potentially represents a novel target for immunotherapeutic approaches due to its significant immunological effect on gliomas.9
Midline gliomas with H3 K27M mutation display substantial histological variations and radiographic features.10,11 An analysis of fluid-attenuated inversion recovery (FLAIR) and T1-weighted contrast-enhanced images of midline gliomas with H3 K27M mutation in pediatric patients revealed enhanced invasive masses with central necrosis, as well as expansive masses without enhancement or necrosis.10 Similarly, in a study of adult patients with brainstem gliomas, almost all H3 K27M–mutant cases displayed contrast enhancement, but a significant correlation between enhancement and H3 K27M mutation status was not identified.12 Thus, a conventional subjective analysis of MR images by radiologists does not appear to provide adequate diagnostic power to identify H3 K27M mutation status.
With the rapid development of medical image analysis in the past decade, radiomics has become a popular research topic. Using high-throughput computing, innumerable quantitative features have been extracted from radiographic images such as computed tomography (CT) and magnetic resonance imaging (MRI) scans. Radiomics extracts more information from medical images and is able to capture tumor heterogeneity in a non-invasive manner.13 The radiomics model has the potential power to improve predictive accuracy14 and has been used to predict molecular markers in gliomas (ie, isocitrate dehydrogenase [IDH] or TP53 mutations) with high accuracy.15–18 However, the prediction of H3 K27M mutation status using a radiomics analysis has not yet been reported.
During the process of discovering the optimal pattern for data, the selection of the approach and setting of appropriate parameters are variable processes that are researcher dependent. Recent studies have developed several automated machine learning (autoML) methods to address this challenge.19 The Tree-based Pipeline Optimization Tool (TPOT) was one of these methods that was designed to automatically optimize the machine learning pipeline using a genetic algorithm without the need for human intervention.20 The performance of autoML matched or exceeded traditional supervised approaches; thus, it may represent an exciting tool for clinical metabolic profiling.21 In another study of genomics applications, a Python-based TPOT library achieved competitive classification accuracy and showed robustness in classifying the data.22 Thus, TPOT is a valuable tool with which to obtain the optimal performance of a prediction during the construction of the radiomics model.
In the present study, we aimed to develop a radiomics model with TPOT to predict the H3 K27M mutation status of midline gliomas based on the hypothesis that radiomics plus autoML would be very helpful in this domain. Radiomics features were extracted from FLAIR images to generate binary predictions of H3 K27M mutation versus H3 K27M wild-type status. We chose T2-FLAIR images instead of the widely used contrast-enhanced T1-weighted images because midline gliomas displayed variable enhancement (from non-enhancement to intense enhancement and from homogeneous enhancement to heterogeneous enhancement), thereby increasing the difficulty of delineating regions of interests (ROIs).
Materials and Methods
Patient Enrollment
This retrospective study was approved by the local institutional review board, and the requirement to obtain informed consent was waived. Patients were recruited form West China Hospital of Sichuan University between December 2016 and September 2018. All patients had newly diagnosed midline glioma and underwent resection or biopsy of the brain tumor. Immunohistochemical staining was performed to detect the histone H3 K27M–mutant protein. If these analyses were inconclusive, H3 K27M mutation status was determined through pyrosequencing of position K27 of H3F3A and HIST1H3B. Initially, 132 patients with midline glioma were enrolled, including 54 patients with H3 K27M mutation and 78 wild-type patients. Finally, 40 patients with H3 K27M–mutant gliomas (mean age, 23.60 y; range, 4–60 y; male/female = 14/26) and 60 patients with wild-type midline gliomas (mean age, 31.57 y; range, 2–76 y; male/female = 28/32) were included in this study; 28 patients were excluded because of inadequate MR images (13 patients had only CT images and 15 patients did not have T2-FLAIR images) or poor image quality (n = 4). The survival data were collected by the neurosurgery staff, and the follow-up of glioma patients after surgery or biopsy is part of their regular clinical practice. Additionally, we recruited 22 patients who met the inclusion and exclusion criteria as an independent and new cohort to validate the model, including 10 patients with H3 K27M–mutant gliomas (mean age, 33.9 y; range, 4–52 y; male/female = 3/7) and 12 patients with wild-type midline gliomas (mean age, 46.6 y; range, 7–63 y; male/female = 4/8) from West China Hospital of Sichuan University between September 2018 and August 2019. None of the patients enrolled in the study had received any treatment prior to the MR examination.
MR Imaging Acquisition
MR examinations of the brain were performed using 3.0-T clinical scanners (Siemens Healthcare, n = 75 patients; Philips Achieva, n = 22; GE MR 750W, n = 7) or 1.5-T clinical scanners (Toshiba Medical Systems, n = 15; Alltech Medical Systems, n = 3). The imaging protocols included non-enhanced fast T1-weighted spin-echo (T1W), fast T2-weighted spin-echo with fat suppression, and T2-weighted axial FLAIR sequences. The contrast material enhanced T1-weighted fast filled echo sequences (on Philips) or 3D T1W-weighted magnetization-prepared rapid acquisition gradient echo sequences (on Siemens) were performed after the administration of the gadolinium-based contrast agent (0.1 mmol/kg body weight). We focused on whole tumors in FLAIR images because not all tumors display enhancement or edema. The main parameters of the FLAIR sequence were as follows: repetition time/echo time (TR/TE) = 6000/81 ms, flip angle = 150°, slice thickness = 5 mm on the Siemens instrument and TR/TE = 8000/120 ms, flip angle = 90°, slice thickness = 6 mm on the Philips instrument.
Tumor Segmentation and Feature Extraction
The ROIs of whole tumors were manually segmented on FLAIR images by 2 authors (X.R., with 3 years of clinical experience in neuroradiology, and S.M., with 5 years of clinical experience in neuroradiology). The authors were blinded to all clinical data and histopathological information. The outline of the whole tumor was drawn on FLAIR images from which the skull was stripped using the FSL library,23 according to the multimodal brain tumor image segmentation benchmark,24 and segmentation was performed with a widely used software package called ITK-SNAP (http://www.itksnap.org) (see Supplementary Figure 1).25 The final ROI was derived from the overlapping segmentation generated by 2 authors.26 If the overlap rate was less than 80%, the ROI was defined by author Q.Y., who has 20 years of diagnostic experience in neuroradiology in an academic full-service hospital. ROIs with a volume equal to or greater than 125 mm3 were applied in this study to understand the radiomics features,26 and ROIs less than 125 mm3 were excluded.
Radiomics features were defined and extracted with the open-source platform called PyRadiomics, which enables radiomics data to be extracted from MR images and processed.27 Loading images were preprocessed and processed using the proposed default setting. Extracted from this platform were 18 first-order features, 13 shape features, 22 gray-level co-occurrence matrix (GLCM) features, 16 gray level run length matrix (GLRLM) features, and 16 gray level size zone matrix (GLSZM) features.
TPOT Overview and Analysis
Radiomics features extracted from FLAIR images were used to generate autoML models. The creation of autoML pipelines and performance of various steps, including feature selection, model selection, and parameter optimization, were achieved using TPOT (https://github.com/rhiever/tpot).22 Genetic programming implemented in the Python package DEAP was used in the TPOT to automatically generate the tree-based pipelines and maximize the final classification accuracy of the pipeline.28 The classification accuracy was evaluated using a fitness function that selects stronger features over weaker features. Prior to the TPOT analysis, data were randomly stratified into a separate 75% training cohort (75 patients) and 25% testing cohort (25 patients). For every iteration (a generation) during the optimization process, the pipeline with the worst performance among all pipelines evaluated was removed and the TPOT proceeded to the next iteration. In this study, we used the following settings: number of generations 100, population size 100, and 10-fold cross-validation on the training set. The importance of each feature was measured and ranked. Finally, TPOT recommended and printed the best-performing pipelines.
Model Comparison and Validation
We replicated the TPOT pipelines 10 times and generated 10 separate models to obtain the preferred model. Model performance was evaluated by determining the accuracy and generating receiver operating characteristic (ROC) curves. Then, we compared the 10 TPOT models based on the training and testing cohorts using the TPOT process and printed the 10 most important features of the best model. Feature importance was measured and ranked according to importance scores. Metrics such as sensitivity, specificity, accuracy, area under the curve (AUC), and average precision provided estimates of the performance of the TPOT-generated model for various features. We compared the accuracy metrics of the TPOT-generated models to determine the best model. The comparison helped to reveal the best-performing model and the most discriminative features in the dataset. Features clearly prioritized H3 K27M mutation and wild-type status as top radiomics features. An overview of the autoML workflow is shown in Fig. 1. The independent set was used to validate the best model selected after a comparison of the models.
Fig. 1.
The workflow of the automated machine learning pipeline.
Statistical Analysis
A statistical analysis of basic clinical information was performed using the open source R package v3.6. A two-sided chi-square test was performed to determine significant differences in sex between 2 groups. Differences in age distribution were evaluated using the Mann–Whitney U-test or Student’s t-test. The survival analysis was conducted using the univariate Kaplan–Meier method. P < 0.05 was considered statistically significant.
Results
Patient Characteristics
One hundred patients with midline gliomas were recruited in the training and testing cohorts, including 40 patients with H3 K27M mutation and 60 wild-type patients. The characteristics of the cohort participating in this study are shown in Table 1. Significant differences in sex were not observed between the H3 K27M–mutant group and the wild-type group. However, the mean age of the mutant group was less than that of the wild-type group. Additionally, adult patients in the mutant group were significantly younger than patients in the wild-type group (32.71 ± 12.24 y vs 41.88 ± 15.78 y, P = 0.02). Adult patients also accounted for a larger proportion than pediatric patients in both groups (mutant group, 60% vs 40%; wild-type group, 68.33% vs 31.67%). In the validation cohort, patients in the mutant group were also younger than those in the wild-type group (10 patients vs 12 patients, 33.9 ± 16.7 y vs 44.6 ± 16.6 y, P = 0.04). There was no significant difference in sex between the 2 groups (P = 0.62).
Table 1.
The demographics of patients with midline gliomas
| H3 K27M Mutant Group | H3 K27M Wild-type Group | P-value (total mutant vs total wild-type) | |||||
|---|---|---|---|---|---|---|---|
| Demographics | Total (n = 40) | Pediatric/Adolescent (age <18 y, n = 16) | Adult (age ≥18 y, n = 24) | Total (n = 60) | Pediatric/Adolescent (age <18 y, n = 19) | Adult (age ≥18 y, n = 41) | |
| Age, y | 0.006* | ||||||
| Mean | 23.6 | 9.94 | 32.71 | 31.57 | 9.32 | 41.88# | |
| Range | 4–60 | 2–17 | 18–60 | 2–76 | 2–17 | 18–76 | |
| Sex | 0.07 | ||||||
| Male | 26 | 8 | 18 | 28 | 7 | 21 | |
| Female | 14 | 8 | 6 | 32 | 12 | 20 | |
*P < 0.01; #P < 0.05 (P = 0.02), comparison of age between 2 groups of adult patients.
Feature Selection and TPOT Models
The average cross-validation score (100 times in each model) for the training set of each model was calculated, and the pipeline with the best test accuracy for the test cohort was printed. In total, 10 independent TPOT classification models were developed to guide diagnostic feature selection in predicting the H3 K27M mutation status. Overall, 10 models performed well for both the training and testing sets and displayed high accuracy (AC). For each model, the following performance was observed for the training and testing set: model 1 (AC = 0.81488, 0.760), model 2 (AC = 0.82643, 0.760), model 3 (AC = 0.78905, 0.800), model 4 (AC = 0.78905, 0.840), model 5 (AC = 0.80238, 0.760), model 6 (AC = 0.78821, 0.760), model 7 (AC = 0.80155, 0.840), model 8 (AC = 0.78905, 0.800), model 9 (AC = 0.810667, 0.750), model 10 (AC = 0.86643, 0.600), and the average of the 10 TPOT-generated models (AC = 0.810667, 0.756). Fixed training and testing sets were applied to the TPOT process to reduce the randomization of patients and controls.
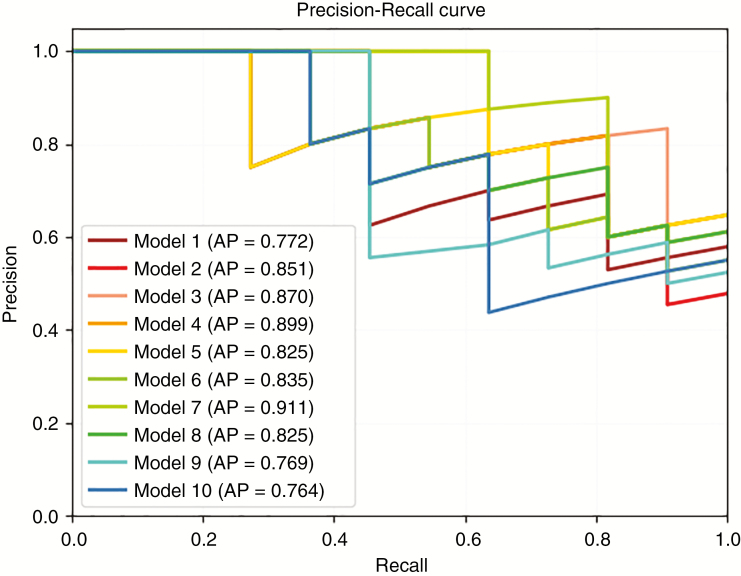
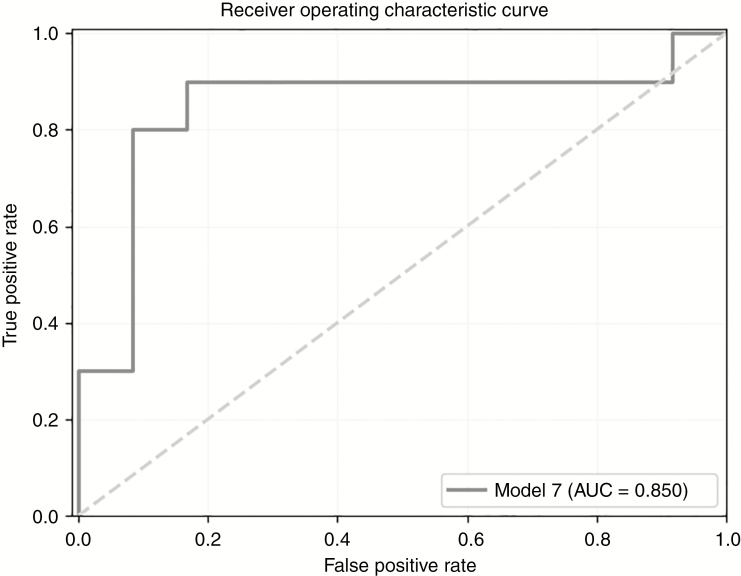
Model Comparison and Final Model
According to the model comparison, model 7 exhibited the best performance, with the highest average precision (0.911) and AUC (0.903) among the 10 models (Table 2 and Figs. 2 and 3; the results for the training set are presented in Supplementary Table 1). The following parameters were used in model 7: model [7] = Gradient Boosting Classifier (learning_rate = 0.1, max_depth = 2, max_features = 0.05, min_samples_leaf = 7, min_samples_split = 14, n_estimators = 100, and subsample = 0.55). The parameters of the other 9 models are described in detail in Supplementary Table 2. The most important features are shown in Supplementary Fig. 2, including the original GLSZM gray level variance, original first-order 10-percentiles, original shape maximum 2D diameter slice, original shape surface volume ratio, original shape volume, original first-order skewness, original first-order minimum, original GLCM inverse difference moment normalized (ldmn), original GLSZM zone variance, and original GLDM dependence non-uniformity normalized.
Table 2.
Comparison results of 10 TPOT models
| Model Index | Sensitivity | Specificity | Accuracy | Kappa Score | Hamming Loss | AUC | Average Precision |
|---|---|---|---|---|---|---|---|
| 1 | 0.545455 | 0.785714 | 0.68 | 0.337748 | 0.32 | 0.792208 | 0.772134 |
| 2 | 0.545455 | 0.928571 | 0.76 | 0.493243 | 0.24 | 0.837662 | 0.850578 |
| 3 | 0.727273 | 0.857143 | 0.8 | 0.590164 | 0.2 | 0.896104 | 0.869712 |
| 4 | 0.727273 | 0.857143 | 0.8 | 0.590164 | 0.2 | 0.896104 | 0.899113 |
| 5 | 0.727273 | 0.857143 | 0.8 | 0.590164 | 0.2 | 0.850649 | 0.82549 |
| 6 | 0.636364 | 0.857143 | 0.76 | 0.503311 | 0.24 | 0.831169 | 0.83455 |
| 7# | 0.636364 | 0.928571 | 0.8 | 0.58194 | 0.2 | 0.902597 | 0.911364 |
| 8 | 0.636364 | 0.857143 | 0.76 | 0.503311 | 0.24 | 0.844156 | 0.824954 |
| 9 | 0.727273 | 0.571429 | 0.64 | 0.290221 | 0.36 | 0.753247 | 0.768781 |
| 10 | 0.545455 | 0.857143 | 0.72 | 0.414716 | 0.28 | 0.733766 | 0.764365 |
#Model 7 was selected as the final TPOT model in this study.
Fig. 2.
Receiver operating characteristic curves for 10 models based on the testing set.
Fig. 3.
Average precision of 10 models for the testing set.
In the validation cohort, the values of validation of the best model (model 7) were as follows: sensitivity = 0.8, specificity = 0.917, accuracy = 0.864, kappa score = 0.722, Hamming loss = 0.136, AUC = 0.85 (Fig. 4) and average precision = 0.855 (see Supplementary Fig. 3).
Fig. 4.
Receiver operating characteristic curves for the best model based on the validation set.
Survival Analysis
The survival status, either dead or alive, was available for 55 patients. Unfortunately, 33 patients were lost to follow-up after surgery, and we lost contact with 12 patients after one or two follow-up sessions. Therefore, 67 patients were included in the survival analysis. For these patients, the median OS of the wild-type group was 33 months, compared with 14 months for the mutant group (P = 0.093). In the subgroup analysis, the median OS of patients in the mutant group who were younger than 18 years old was only 4 months (P = 0.005, median OS of the wild-type group was NA; see Supplementary Fig. 4). However, the survival analysis did not reveal a significant difference in adult patients (P = 0.36, the median OS was NA in the mutant group and 33 mo in the wild-type group).
Discussion
In this study, 40 patients who had a glioma carrying H3 K27M mutation and 60 wild-type patients were recruited. The autoML process based on radiomics was a potential tool for predicting the H3 K27M mutation status of midline gliomas. In the final model, we obtained an AUC of 0.903 and an average precision of 0.911. According to the importance scores of radiomics features, 10 features associated with shape, first-order, and texture features were selected as the most important radiomics features for the prediction of H3 K27M mutation status. Good performance was observed for the best model in the validation set, with an AUC of 0.85 and an average precision of 0.855.
The TPOT method was an exciting tool to optimize traditional machine learning methods and showed great promise for discriminating H3 K27M mutation status in our study. In a previous study, the TPOT pipeline was compared with a basic random tree (random forest, widely used in machine learning studies) with equal numbers of decision trees as the TPOT to validate the classification performance of TPOT. The TPOT not only automatically optimized the model parameters but also discovered useful features to improve the classification accuracy.20 When trained on the midline gliomas with H3 K27M mutation information, our 10 models achieved a prediction accuracy of 0.788–0.867 in the training cohort and an accuracy of 0.60–0.84 in the testing cohort. In the final model selected after model comparison, the AUC of our classifier achieved a value of 0.903. The performance of this autoML classifier was similar to that of a previous machine learning study using a random forest model (highest AUC = 0.82)26 or least absolute shrinkage and selection operator (AUC = 0.763).29 Our study provides compelling evidence for the use of TPOT to construct a prediction model that is able to identify H3 K27M mutation status and to advance further clinical research.
During model construction, autoML was always sensitive to the sample size; thus, a small sample size (n < 50) could lead to overfitting. We included 100 samples and 99 radiomics features in our study to avoid the problem of high variance. Regarding the structure of the dataset, imbalanced datasets or datasets with a large proportion of missing values can result in a biased analysis for machine learning.21 Here, we included generally balanced observations from 2 groups (60 vs 40) and thus reduced the effect of the imbalance. Furthermore, all radiomics features in this study were fully populated, thereby reducing noise in the dataset. The final model in our study required the minimum number of parameters and obtained the best performance in the model comparison.
The 10 most important radiomics features identified through TPOT selection are shown in Supplementary Fig. 2. Each radiomics feature required 10 patients in the model based on a binary classifier to ensure a sufficient power of the classification model.14 In our study, 100 patients and only 10 important features, including 3 shape, 3 first-order, 2 GLSZM, 1 GLCM, and 1 GLDM, were included. Tumor shape features were independent of the gray-level intensity distribution in the ROI. According to the model, tumor shape features played an important role in predicting H3 K27M mutation, among which the maximum 2D diameter of the slice (largest pairwise Euclidean distance between tumor surface mesh vertices in the row-column) of H3 K27M–mutant tumors was smaller than that of wild-type tumors (41.31 vs 59.35, P = 0.007). These results are consistent with those of a previous study11 that used a random forest model to predict the presence of H3 K27M mutation in spinal cord diffuse midline gliomas and found that the maximum length of the tumor was the most important radiological feature in the model, as the mean longest size in the mutant group was smaller than that in the wild-type group (5.3 cm vs 5.9 cm). Another study also suggested that shape information was helpful for improving model performance.24 The relationship between the tumor shape and mutation has not been analyzed extensively and requires further exploration.
The first-order features, which describe the distribution of voxel intensities in the ROI, were also important in the prediction model. These features were also applied to the prediction model for gliomas and lung cancers and yielded good performance.29,30 Texture features, including GLCM, GLSZM, and GLDM, are another group of widely used radiomics features. In a previous study, deep learning radiomics based on texture, shape, and wavelet features achieved good performance, with an AUC of 0.92 for the detection of the IDH1 mutation in gliomas.31 Our study revealed the value of the radiomics features extracted from FLAIR images in detecting H3 K27M mutation status. Consistent with the results from our study, texture features extracted from routine FLAIR images adequately separate IDH mutant low-grade gliomas from IDH wild-type low-grade gliomas.32
In the survival analysis, we did not observe a significant difference in OS between the 2 groups. In the subgroup analysis based on age, differences in OS were observed between the 2 groups in patients aged younger than 18 years but not in adult patients. In a previous study of adult patients with thalamic gliomas, significant differences in OS were not observed between the mutant and wild-type groups.33 Nevertheless, the small sample size subjected to the OS analysis in our study may have affected the results.
Our study has some general and study-specific limitations. First, we recruited only 122 patients in this study because H3 K27M mutation has been analyzable since 2016 in our hospital. Although radiomics can be performed with as few as 100 patients,14 the inclusion of more patients should provide more power and is a better choice in the future. Second, other potential valuable tumor features for radiomics analyses, such as contrast enhancement and edema, were not included in this study, and only the whole tumor (which typically includes regions of contrast enhancement, non–contrast enhancement, and necrosis) delineated on FLAIR images was analyzed. Since some patients did not display contrast enhancement or edema, the combination of these parameters with other features would reduce the number of patients. Third, we did not include clinical data or functional MR imaging data, such as MR spectroscopy and diffusion-weighted imaging, which may add more value to the prediction model. Fourth, many patients were lost to follow-up in the survival analysis, so the quality of follow-up should be improved.
In summary, autoML is able to predict H3 K27M mutation status in patients with midline gliomas with high accuracy based on the radiomics features and the TPOT method, which is able to automatically optimize the machine learning pipeline. This prediction model may aid in providing more precise diagnoses and guide treatment decisions. Further efforts are required to explore the full value of radiomics-based diagnoses using more tumor features, functional-MRI data, and clinical data to build an optimal model for routine clinical use.
Funding
This work was supported by the Sichuan Provincial Foundation of Science and Technology (2019YFS0428, 2013SZ0047, and 2017SZ0006), the Foundation of the National Research Center of Geriatrics, West China Hospital, Sichuan University (Z2018A07), the National Natural Science Foundation of China (81371528, 81621003), Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT, Grant No. IRT16R52) of China, and the Functional and Molecular Imaging Key Laboratory of Sichuan Province (FMIKLSP, Grant No.2019JDS0044).
Conflict of interest statement. The authors declare no potential conflicts of interest.
Authorship statement. Conception and design: X. Su, Q. Yue, H. Sun, N. Chen, X. Yang, J. Su, Q. Tan, Q. Gong, W. Wang, S. Zhang. Development of methodology (acquired and managed patients, provided facilities, etc.): X. Su, Q. Yue, N. Chen, Q. Gong, H. Sun
Acquisition of data: X. Su, H. Sun, Y. Liu. Analysis and interpretation of data (eg, statistical analysis, biostatistics, computational analysis): X. Su, Q. Yue, H. Sun. Writing, review, and/or revision of the manuscript: X. Su, H. Sun, Q. Gong, Q. Yue. Administrative, technical, or material support (ie, reporting or organizing data, constructing databases): X. Su, Q. Yue, H. Sun, Q. Gong, N. Chen, X. Yang, J. Su, Q. Tan, W. Wang, S. Zhang.
Supplementary Material
References
- 1. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 2. Louis DN, Giannini C, Capper D, et al. cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol. 2018;135(4):639–642. [DOI] [PubMed] [Google Scholar]
- 3. Wu G, Broniscer A, McEachron TA, et al. ; St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shows J, Marshall C, Perry A, Kleinschmidt-DeMasters BK. Genetics of glioblastomas in rare anatomical locations: spinal cord and optic nerve. Brain Pathol. 2016;26(1):120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Solomon DA, Wood MD, Tihan T, et al. Diffuse midline gliomas with histone H3-K27M mutation: a series of 47 cases assessing the spectrum of morphologic variation and associated genetic alterations. Brain Pathol. 2016;26(5):569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karremann M, Gielen GH, Hoffmann M, et al. Diffuse high-grade gliomas with H3 K27M mutations carry a dismal prognosis independent of tumor location. Neuro Oncol. 2018;20(1):123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kleinschmidt-DeMasters BK, Mulcahy Levy JM. H3 K27M-mutant gliomas in adults vs. children share similar histological features and adverse prognosis. Clin Neuropathol. 2018;37 (2018)(2):53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ryall S, Krishnatry R, Arnoldo A, et al. Targeted detection of genetic alterations reveal the prognostic impact of H3K27M and MAPK pathway aberrations in paediatric thalamic glioma. Acta Neuropathol Commun. 2016;4(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ochs K, Ott M, Bunse T, et al. K27M-mutant histone-3 as a novel target for glioma immunotherapy. Oncoimmunology. 2017;6(7):e1328340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aboian MS, Solomon DA, Felton E, et al. Imaging characteristics of pediatric diffuse midline gliomas with histone H3 K27M mutation. AJNR Am J Neuroradiol. 2017;38(4):795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jung JS, Choi YS, Ahn SS, Yi S, Kim SH, Lee SK. Differentiation between spinal cord diffuse midline glioma with histone H3 K27M mutation and wild type: comparative magnetic resonance imaging. Neuroradiology. 2019;61(3):313–322. [DOI] [PubMed] [Google Scholar]
- 12. Daoud EV, Rajaram V, Cai C, et al. Adult brainstem gliomas with H3K27M mutation: radiology, pathology, and prognosis. J Neuropathol Exp Neurol. 2018;77(4):302–311. [DOI] [PubMed] [Google Scholar]
- 13. Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shofty B, Artzi M, Ben Bashat D, et al. MRI radiomics analysis of molecular alterations in low-grade gliomas. Int J Comput Assist Radiol Surg. 2018;13(4):563–571. [DOI] [PubMed] [Google Scholar]
- 16. Yu J, Shi Z, Lian Y, et al. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol. 2017;27(8):3509–3522. [DOI] [PubMed] [Google Scholar]
- 17. Altazi BA, Zhang GG, Fernandez DC, et al. Reproducibility of F18-FDG PET radiomic features for different cervical tumor segmentation methods, gray-level discretization, and reconstruction algorithms. J Appl Clin Med Phys. 2017;18(6):32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang X, Tian Q, Wang L, et al. Radiomics strategy for molecular subtype stratification of lower-grade glioma: detecting IDH and TP53 mutations based on multimodal MRI. J Magn Reson Imaging. 2018;48(4):916–926. [DOI] [PubMed] [Google Scholar]
- 19. Hutter F, Lücke J, Schmidtthieme L. Beyond manual tuning of hyperparameters. Künstl Intell. 2015;29(4):329–337. [Google Scholar]
- 20. Olson RS, Moore JH. TPOT: a tree-based pipeline optimization tool for automating machine learning. In: Automated Machine Learning. Cham: Springer; 2019:151–160. [Google Scholar]
- 21. Orlenko A, Moore JH, Orzechowski P, et al. Considerations for automated machine learning in clinical metabolic profiling: altered homocysteine plasma concentration associated with metformin exposure. Pac Symp Biocomput. 2018;23:460–471. [PMC free article] [PubMed] [Google Scholar]
- 22. Olson RS, Urbanowicz RJ, Andrews PC, Lavender NA, Moore JH. Automating biomedical data science through tree-based pipeline optimization. In: European Conference on the Applications of Evolutionary Computation. Cham: Springer; 2016:123–137. [Google Scholar]
- 23. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62(2):782–790. [DOI] [PubMed] [Google Scholar]
- 24. Menze BH, Jakab A, Bauer S, et al. The multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging. 2015;34(10):1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. [DOI] [PubMed] [Google Scholar]
- 26. Kniep HC, Madesta F, Schneider T, et al. Radiomics of brain MRI: utility in prediction of metastatic tumor type. Radiology. 2019;290(2):479–487. [DOI] [PubMed] [Google Scholar]
- 27. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fortin FA, Rainville FMD, Gardner MA, et al. DEAP: evolutionary algorithms made easy. J Mach Learn Res. 2012;13(Jul):2171–2175. [Google Scholar]
- 29. Li Y, Qian Z, Xu K, et al. MRI features predict p53 status in lower-grade gliomas via a machine-learning approach. Neuroimage Clin. 2018;17:306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moran A, Daly ME, Yip SSF, Yamamoto T. Radiomics-based assessment of radiation-induced lung injury after stereotactic body radiotherapy. Clin Lung Cancer. 2017;18(6):e425–e431. [DOI] [PubMed] [Google Scholar]
- 31. Moiseev A, Snopova L, Kuznetsov S, et al. Pixel classification method in optical coherence tomography for tumor segmentation and its complementary usage with OCT microangiography. J Biophotonics. 2018;11(4):e201700072. [DOI] [PubMed] [Google Scholar]
- 32. Jakola AS, Zhang YH, Skjulsvik AJ, et al. Quantitative texture analysis in the prediction of IDH status in low-grade gliomas. Clin Neurol Neurosurg. 2018;164:114–120. [DOI] [PubMed] [Google Scholar]
- 33. Aihara K, Mukasa A, Gotoh K, et al. H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro Oncol. 2014;16(1):140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.