Digital real-time PCR assay rapidly measures single telomere lengths and uncovers telomere maintenance mechanisms in cancer.
Abstract
Telomere length is a promising biomarker for age-associated diseases and cancer, but there are still substantial challenges to routine telomere analysis in clinics because of the lack of a simple and rapid yet scalable method for measurement. We developed the single telomere absolute-length rapid (STAR) assay, a novel high-throughput digital real-time PCR approach for rapidly measuring the absolute lengths and quantities of individual telomere molecules. We show that this technique provides the accuracy and sensitivity to uncover associations between telomere length distribution and telomere maintenance mechanisms in cancer cell lines and primary tumors. The results indicate that the STAR assay is a powerful tool to enable the use of telomere length distribution as a biomarker in disease and population-wide studies.
INTRODUCTION
Telomeres are regions of repetitive nucleotide sequences at the end of each chromosome that protect the ends from degradation and are important structures that regulate chromosome integrity (1). Human telomeres consist of short tandem repeats (5′-TTAGGG-3′) that range from 8 to 15 kb at birth (2), and they shrink by 50 to 200 nucleotides after each replication cycle. After a certain number of cell division, telomeres reach a critical length and trigger an irreversible cell cycle arrest known as cellular senescence (1). Accumulating evidence has shown that patients with abnormal telomere lengths are susceptible to age-associated degenerative diseases (3), diabetes mellitus (4), cardiovascular disease (5), and different prognosis in various cancers (6). These examples support the use of telomere length as a clinically relevant indicator in the management of patients.
Nonetheless, there are still major technical impediments in performing telomere length measurement in clinical settings. Existing telomere length measurement methods are technically demanding and are based on relative measurements taken using arbitrary normalization methods that are poorly standardized across laboratories. Terminal restriction fragment (TRF) analysis involving a Southern blot procedure is considered the gold standard for telomere analysis (7). In this method, telomere fragments are separated by gel electrophoresis, transferred to a nylon membrane, and hybridized to oligonucleotide probes for visualizing telomere fragments by chemiluminescence or radioactivity. Because of the use of a large amount of starting genomic DNA requirement (at least 1 μg) and the laborious process (8), it is unsuitable for clinical applications. On the other hand, real-time quantitative polymerase chain reaction (qPCR)–based telomere length measurement can be performed rapidly using small amounts of sample (~20 ng) (9, 10). However, this method only provides a relative measurement of total telomere length to a single copy gene ratio and does not provide information about the telomere length distribution. Because of the common occurrence of aneuploidy in cancer, qPCR is also inaccurate for quantifying telomere lengths in cancer studies (11).
On the other hand, methods that can quantify the lengths of single telomeres can provide valuable information about telomere length distribution. Metaphase quantitative fluorescence in situ hybridization (Q-FISH) can detect telomere length from the individual chromosome but can only be performed on actively dividing cells that are arrested on metaphase (12). High-throughput Q-FISH (13) can quantify telomere lengths from individual interphase cells, but a distinct disadvantage of interphase Q-FISH is that it does not measure telomere signals of individual chromosome ends but instead aggregations of few telomeres known as “telomere spots” (14). Multiple studies have shown that telomeres tend to cluster spatially, and this had led to a reduction in overall telomere count that could result in the wrong estimation of telomere length distribution (15). Besides, external calibration using cell lines with known telomere length is needed to convert fluorescence signal to telomere length, and extensive controls are needed to correct for experimental variabilities. A recent method, peptide nucleic acid (PNA) hybridization and analysis of single telomere (PHAST) (16), determines telomere length by measuring the fluorescence signal of telomeric DNA hybridized to fluorescent PNA probes as it passes through a microfluidic channel. PHAST requires very specialized equipment and is not suitable for measuring telomere lengths longer than 15 kb. Last, single telomere length analysis (STELA) (17) and its variations [U-STELA (18) and TeSLA (19)] make use of adapter ligation and PCR amplification followed by Southern blot to accurately measure short telomeres. However, these techniques cannot quantify long telomeres (>20 kb) and are too labor intensive (because of the use of Southern blot) for routine clinical and large population studies.
Recent studies on telomere biology have revealed that beyond the classical average telomere length measurements, measuring the telomere length distribution has profound implications in the manifestation and prognosis of cancer and aging-related diseases. The critically short telomeres, rather than average telomere lengths, are causative of age-related pathologies (20). Meanwhile, the longest telomeres are a signature of adult stem cell compartments (21). Reduced heterogeneity of telomere lengths has been related to the clonal growth of cancer cells (22), while extrachromosomal telomere repeats (ECTRs) (23) are commonly found in cancer cells that elongate telomeres via a homologous recombination-based mechanism. These new findings highlight the need to develop a rapid, high-throughput method for telomere length distribution measurement. Here, we developed a method called single telomere absolute-length rapid (STAR) assay. This method can measure the absolute telomere length of individual telomeres in <3 hours for up to 48 samples from low amounts (<1 ng) of extracted DNA. Moreover, the STAR assay has a wide dynamic range of measurement, as it is capable of measuring telomere lengths from 0.2 to 320 kb. We applied the assay to human cancer cell lines and human clinical samples and were able to detect the telomere maintenance mechanism of each sample accurately based on telomere length distributions.
RESULTS
The principle of STAR assay
A schematic illustration of the STAR assay is shown in Fig. 1A. STAR assay is a qPCR-based method to measure the absolute telomere length of single telomere molecules. Stochastic encapsulation of single molecules can be achieved by various methods, with digital PCR platforms providing the highest throughput while minimizing reagent consumption. Sample DNA was first digested with restriction enzymes that do not cut within the telomeric sequence, then added in the telomere-specific primers and qPCR mix, and partitioned all the reagents into individually isolated nanoliter compartments in the digital PCR chip. With limited dilution, most of the compartments will contain either one or zero telomere fragments.
Fig. 1. STAR assay workflow.
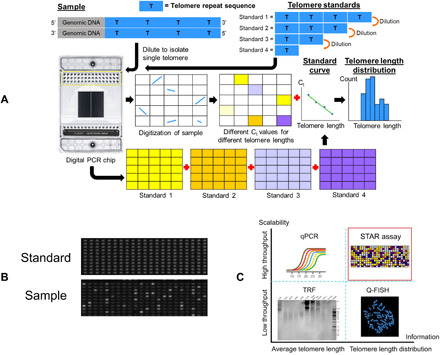
(A) Schematic overview of the STAR assay workflow. Different copies of synthetic telomere repeat sequences are used to generate a standard curve that allows the assay to measure absolute telomere length. By diluting the samples sufficiently, single telomere molecules can be isolated in individual compartments. Different Ct values in each sample will correspond to different telomere lengths. (B) Actual fluorescence (EvaGreen) images of a telomere standard and cell line sample at cycle 27. Telomere standard has uniform fluorescence for all the compartments, while the cell line sample has varying fluorescence intensities among the compartments with some compartments with no fluorescence. (C) Comparison of STAR assay with other common telomere length measurement methods. STAR assay is capable of measuring telomere length distributions with high throughput.
The working principle of the STAR assay is based on the premise that the number of repeat sequences in an individual telomere molecule can be reliably measured via qPCR in nanoliter compartments. Unlike conventional end-point digital PCR assays, real-time monitoring of fluorescence signal is performed in the STAR assay to capture the reaction kinetics in each nanoliter compartment. In compartments that contain telomere molecules, PCR amplification leads to an exponential increase in PCR product that is proportional to the initial length (i.e., copy number) of the telomere repeat fragment. This increase in PCR product is tracked by the changes in fluorescence intensity of a double-stranded DNA binding dye. We used the microfluidic-based Fluidigm Digital PCR system, as it is capable of performing real-time reaction monitoring. The heterogeneous telomere lengths of the sample would be represented by different amplification kinetics of telomere DNA in each compartment captured over time (Fig. 1B and movie S1). The entire process of the digital qPCR assay requires less than 3 hours to complete.
Several key changes from the conventional telomere qPCR assay conditions are needed for robust single-molecule telomere qPCR in nanoliter chambers. The microfluidic digital PCR chip is made out of polydimethylsiloxane (PDMS), a material known to cause nonspecific adsorption of compounds to its surfaces due to its porosity and hydrophobicity (24). Nonspecific adsorption of sample and PCR reagents onto PDMS initially caused nonuniform amplification of the target molecule across the chip. This problem was resolved by replacing SYBR Green I with another double-stranded DNA binding dye EvaGreen and adding Tween 20, a nonionic surfactant, to the qPCR mix. The potassium chloride component in the originally published telomere qPCR mix was also found to be incompatible with the use of EvaGreen and had to be replaced by additional tris-HCl (25). Nonspecific PCR amplification product could be very detrimental to the detection limit of single telomere molecules. We experimentally determined that with a high annealing temperature of 62°C and a low primer concentration of 0.3 μM, minimal nonspecific PCR amplification was observed in the first 27 cycles of PCR.
The ability to measure absolute, rather than relative, telomere length is highly attractive to enable unbiased comparison across laboratories. Absolute telomere length measurement requires well-characterized standards, which include DNA ladders for Southern blot–based telomere length measurement or plasmids containing telomere sequence for Q-FISH–based methods. A current technical bottleneck is generating very long telomeric sequence standards (>20 kb) (16) for absolute measurement of long telomeres, particularly as telomeres exceeding 100 kb are found in certain cancers. Unlike other methods, STAR assay amplifies telomere repeat sequences as short amplicons; therefore, it is possible to achieve a very wide dynamic range by combining multiple short, synthetic telomeric sequences to serve as long telomere standards. We performed digital qPCR using different amounts of 90–base pair synthetic telomere repeat sequences (5 to 320 kb of equivalent lengths) to generate the standard curve of cycle threshold (Ct) value against the total length of synthetic telomere repeats in Fig. 2A. All the compartments with the same amount of telomere repeat standards had uniform amplification kinetics (movie S2), demonstrating excellent reproducibility (coefficient of variation < 4%) in Ct values. The standard curve shows a very good fit to a linear model [coefficient of determination (R2) = 0.995], and we calculated an excellent PCR amplification efficiency (E > 92%) based on this model. These data confirm that telomere qPCR in the nanoliter chamber accurately reflects actual telomere length. With the optimized qPCR conditions, there is negligible PCR amplification in the no template controls up to a Ct value of 27, corresponding to ~0.2 kb of telomere length in the standard curve. Therefore, we determined that the STAR assay has a wide dynamic range (0.2 to 320 kb) for absolute telomere length measurement and would be well suited for a variety of different applications. As a novel method, STAR assay combines the best aspects of current telomere measurement methods (Fig. 1C): (i) the throughput and ease of use of qPCR, (ii) the absolute length measurement of Southern blot methods, and (iii) individual chromosome telomere length measurement of metaphase Q-FISH. We expect this simple yet comprehensive assay to be widely adopted as a standard measurement for telomeres. A detailed comparison of STAR assay with existing telomere length measurement methods can be found in Table 1.
Fig. 2. STAR assay allows single molecule telomere length measurement.
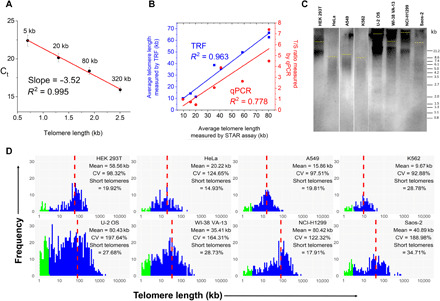
(A) Standard curve generated by plotting Ct values versus telomere length (log scale) for four telomere standards (5 to 320 kb). Data are presented as means ± SD (n = 770 compartments). (B) Correlation of average telomere length of eight cancer cell lines measured by STAR assay with the average telomere lengths measured by TRF (blue) and qPCR (red). (C) TRF image for the eight human cancer cell lines. The yellow line indicates the average telomere length for each cell line. (D) Histograms showing the telomere length distributions for eight human cancer cell lines with the average telomere length (mean), coefficient of variation (CV), and percentage of short telomeres (<3 kb) indicated in each. The proportion of short telomeres is highlighted in green. The average telomere length is marked by the red dotted line.
Table 1. Comparison of STAR assay with other telomere length measurement methods.
TL, telomere length.
| Method |
Amount of starting material |
Processing time | Throughput | Dynamic range | TL distribution | Absolute length |
| TRF (7) | >1 μg | 2 days | 32 samples | 0.8–20 kb | Yes (semiquantitative) |
Yes |
| qPCR (10) | ~20 ng | <2 hours | 96 samples | Measures relative length |
No | No |
| Metaphase Q-FISH (12) |
15 to 20 metaphase cells |
2 days | 10 samples per week |
0.15–80 kb | Yes | Yes |
| High-throughput interphase Q-FISH (13) |
10,000 cells | 2 days | 96 samples | 0.2–80 kb | Yes | Yes |
| STELA, U-STELA, TeSLA (17–19) |
100 to 250 pg | 2 days | 32 samples | 0.4–20 kb | Yes, for short telomeres |
Yes |
| STAR | <1 ng | <3 hours | 48 samples | 0.2–320 kb | Yes | Yes |
STAR assay reveals different telomere length distribution in cancer cell lines
To validate the STAR assay, we chose a panel of human cancer cell lines and determined their telomere lengths by STAR assay. Simultaneously, TRF and telomere qPCR were performed on the same samples for comparison (Fig. 2, B and C). Using STAR assay in conjunction with a standard curve, we obtained the histogram of absolute telomere lengths in each of the cell lines (Fig. 2D), and it is evident that each sample is characterized by a very distinct telomere length distribution profile. To assess the reliability of the telomere lengths measured by STAR assay, we compared the average telomere lengths measured and showed good correlation with TRF (R2 = 0.96), indicating that STAR assay is comparable to the gold standard TRF for average telomere length determination. A more moderate correlation was obtained for the average telomere length measurements between STAR assay and qPCR (R2 = 0.78), but since conventional telomere qPCR relies on normalization upon single-copy gene, this can result in significant deviations due to aneuploidy, which occur in many cancers. Together, consistent results obtained with a large panel of human cell lines validate the STAR assay as a robust tool for absolute telomere length distribution measurements.
The impact of telomere maintenance mechanism on telomere length distribution
Telomere maintenance is a crucial step for cancer cells to bypass senescence (26). Telomerase, the enzyme complex responsible for elongating telomeres through the addition of telomere repeats to chromosome ends, is activated in approximately 90% of all human tumors, but a sizeable fraction of cancer cells use the alternative lengthening of telomeres (ALT) mechanism, which is telomerase independent for telomere maintenance, and patients with ALT positive tumors generally have a poor prognosis (27). With the development of telomerase- and ALT-specific therapies, it would be beneficial to determine whether cancer in any individual is ALT positive or ALT negative to allow stratification of treatment to specific patient groups where it would be most beneficial. Current methods to diagnose ALT include performing TRF to detect long and heterogeneous telomeres or performing combined promyelocytic leukemia immunofluorescence and telomere FISH to detect ALT-associated PML (promyelocytic leukemia) bodies (APBs) (28); both of which are labor intensive and technically challenging. Thus, we investigated whether telomere length profiles obtained through the STAR assay could be used to detect the presence of ALT in samples. Of the eight cell lines used for this STAR assay, four of them are ALT negative [human embryonic kidney (HEK) 293T, HeLa, A549, and K562], while the other four of them are reported to be ALT positive (U-2 OS, WI38-VA13, NCI-H1299, and Saos-2). We found that while most ALT positive cells had longer average telomere lengths and higher heterogeneity than their ALT negative counterparts, there were exceptions. Notably, the ALT negative HEK 293T cells had long telomeres, while the telomere lengths of ALT positive NCI-H1299 cells were less heterogeneous. Therefore, relying on any single telomere parameter alone to determine ALT status lacks specificity.
The subset of very short telomeres (<3 kb) has been reported to be a critical determinant of cell senescence (29). Therefore, we compared the percentage of short telomeres measured by STAR among the different cell lines. Despite the generally longer average telomere lengths of ALT positive cells, three of these cell lines are among the top four samples that have the highest percentage of short telomeres. The abundance of short telomeres in these samples is evident from the bimodal distribution of telomere lengths measured with the STAR assay. The prevalence of short telomeres in ALT cells could be the result of unequal recombination, leading to rapid shortening of telomeres on a subset of chromosomes, which have previously been observed as dim spots or undetected telomeres in Q-FISH (30). Identification of high proportions of short telomeres could be considered as another characteristic determinant in ALT.
Absolute quantification of ECTR
ECTRs are DNA molecules containing telomeric repeat sequences that are not covalently linked to chromosomes. These molecules are thought to be generated by active telomere-trimming events and homologous recombination-mediated telomere maintenance and have been observed in both linear and circular forms in ALT positive cancer cells (23, 31, 32). The C-circle assay, which uses rolling circle amplification coupled with radioactive or qPCR detection, is rapidly becoming an orthogonal method for ALT detection (33). However, the C-circle assay can only detect C-circles and is unable to detect other forms of ECTRs (e.g., linear, t-circles, and G-circles). In a previous study, linear ECTRs were found to be comparable in amount to circular ECTRs and are primarily sequestered in APBs (34). To date, there is no method to determine the copy number of ECTRs, which could indicate the severity of ALT and provide prognostic value when considering drugs targeting the ALT pathway. Since the presence of ECTR is expected to increase the number of physical copies of telomere molecules relative to total DNA mass (Fig. 3A), we investigated whether the presence of ECTR could be detected using the STAR assay. Using the same DNA input amount, the ALT negative samples had relatively constant and low numbers of compartments containing telomere molecules. On the other hand, the ALT positive samples had significantly more compartments containing telomere molecules in the STAR assay. The detection of ECTR using the STAR assay corroborated with C-circle assay results (Fig. 3B) in line with our expectation that the presence of ECTR in ALT positive cells will increase the number of telomere molecules detected in STAR assay. These results validate the existing knowledge that C-circles are a significant and representative component of ECTRs in ALT positive samples (35). ECTR measurement by STAR assay is therefore very useful as an additional dimension for ALT detection. The ability to quantitate both copy numbers and lengths of chromosomal and ECTR makes the STAR assay a very promising tool for tracking the extent of ALT in applications such as screening for drugs targeting the ALT mechanism.
Fig. 3. STAR assay measures ECTR in ALT-positive samples.
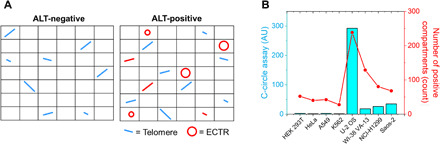
(A) Schematic illustrating how the presence of ECTR (red) in an ALT positive sample will increase the absolute number of physical copies of telomeres being measured by digital PCR. (B) Line graph showing the number of compartments containing telomere molecules, as measured using the STAR assay, while the bar chart shows the C-circle assay when performed for the same samples. Both assays show similar trends. AU, arbitrary units.
Together, STAR assay enables measurement of four key features of ALT: (i) long telomeres, (ii) heterogeneous lengths, (iii) abundance of short telomeres, and (iv) presence of ECTR. When considered individually, these features may be absent in certain samples, but when considered together, they can form a powerful set of parameters to determine ALT status. We calculated the z score of each feature measurements and defined an “ALT score,” which is the sum of all four feature z scores from each sample. While each of the feature z score alone did not determine the ALT status with complete specificity, we showed that the combined ALT score obtained from the STAR assay in the tested cell lines could serve as a highly specific criterion for ALT status determination (Fig. 4).
Fig. 4. STAR assay allows determination of ALT status in cancer.
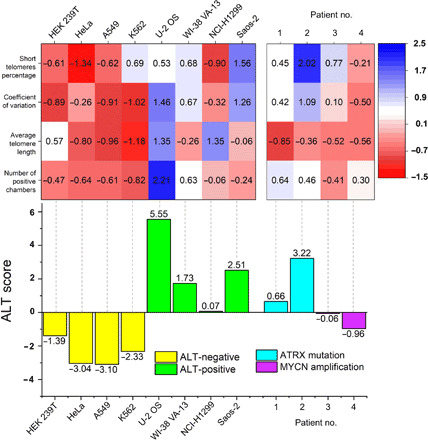
Heatmap of the z scores of the four ALT features for eight cell lines and four patient samples. ALT score is the sum of z scores of all the four ALT features. The ALT score is highly specific for the ALT status in cell lines and patient samples.
STAR assay determines telomere maintenance mechanism in primary tumor
A recent extensive genetic profile of neuroblastoma tumors strongly suggested that telomere maintenance mechanisms are important prognostic indicators for high-risk neuroblastoma (36). Telomerase activity can be detected by measuring human telomerase reverse transcriptase mRNA levels (37) or directly measuring enzyme activity with telomeric repeat amplification protocol (TRAP) (38). Recently, the droplet digital TRAP (ddTRAP) (39) method has been reported to further enhance the quantitation of telomerase activity. On the other hand, ALT determination in the clinic relies on the detection of APB (28) and C-circle assay (40), which are not completely definitive for ALT activity and may underestimate the prevalence of ALT in tumors. In this respect, the STAR assay, which integrates the measurement of multiple ALT characteristics, may be a more sensitive method for determining ALT status. We thus investigated whether the ALT status of clinical pediatric neuroblastoma tumor samples can be determined by STAR assay. We profiled four samples, two of which were known to harbor MYCN amplification and the remaining two had ATRX mutations. Upon measuring the ALT score of each of the clinical samples, we determined that both samples with ATRX mutations were ALT positive, while both samples with MYCN amplification are ALT negative (Fig. 4). This result is in agreement with other studies that showed that recurrent ATRX mutations are found in a subset of ALT positive neuroblastomas (41), whereas MYCN-amplified tumors induce telomerase activity for telomere maintenance (42). These results suggest that STAR assay is a promising platform for rapidly determining telomere maintenance mechanisms in oncology clinics.
DISCUSSION
There is increase utility for telomere length as a biomarker for diseases. Among existing methods, the rapid workflow for telomere qPCR is best suited for clinical implementation, but so far, results have been mixed (43). A major issue with qPCR is that it measures average telomere length, whereas the distribution of telomeres including the subset of short telomeres is found to be more relevant for disease risk stratification. Furthermore, the use of single-copy reference gene (T/S ratio) to obtain relative telomere length leads to substantial biases in the common cases of aneuploidy (11) and the presence of ECTR in cancers that activate ALT pathway (40). Other methods are available for telomere length distribution measurement, but implementation is challenging because of technical difficulties of performing assay and lack of common reference standards (13, 19).
Recently, digital PCR has been gaining a lot of interest for absolute quantification of DNA molecules and has been used for many applications in precision medicine (44). Innovative approaches have also been proposed such as ddTRAP (39) that uses end-point droplet digital PCR to measure telomerase activity. A combination of digital PCR and high-resolution melting analysis has also enabled the measurement of intermolecular heterogeneity of DNA methylation (45). However, so far, end-point digital PCR precludes measurements of structural variations in nucleic acids such as telomere lengths that are keys to chromosomal integrity and important biomarkers for many diseases.
Our work is the first demonstration of a digital real-time PCR assay and its utility for performing single-molecule measurement of telomere length. The combination of the specificity of PCR, single-molecule partition, and absolute length measurement allows a virtual TRF to be performed with the ease and simplicity of a qPCR assay. STAR assay is easy to perform, high throughput (up to 48 samples per chip in current setting), rapid (<3 hours), requires a low amount of sample (<1 ng), and thus represents a crucial improvement toward applying telomere length distribution analysis in large-scale clinical studies. Although the accuracy of the STAR assay was demonstrated in a low number of samples, the high correlation of this assay with the gold standard TRF proves the potential of this assay. Besides telomeres, other tandem repeats are abundant in the genome, and their functional roles are increasingly being recognized. For example, the minisatellite length polymorphisms at the insulin gene VNTR is strongly associated with juvenile obesity and metabolic syndrome (46). Expansions of short tandem repeats (e.g., GGGGCC repeats within C9orf72 gene in amyotrophic lateral sclerosis and CGG repeats within FMR1 gene in fragile X syndrome) are genetic variants that have been implicated in several neuropsychiatric and other disorders, but their assessment remains challenging with Southern blot–based method (47). We envision the principles of STAR assay to be extended for many of these applications for high-throughput screening to replace current cumbersome gel electrophoresis procedures.
The low-sample input requirement and single-molecule sensitivity of the STAR assay suggests that STAR assay can be adapted and scaled to measure individual telomeres in single cells. qPCR-based measurement of relative telomere length (T/S ratio) has been previously reported, and it was demonstrated that individual cells from cancer cell lines showed extensive heterogeneity in relative telomere lengths (48). However, the heterogeneity of individual telomeres within cells and between cells cannot be measured by this method. A single-cell STAR assay would provide the resolution to probe individual telomere lengths in single cells to better understand cellular heterogeneity in telomere length regulation. Although the STAR assay can only process 48 samples per run with the use of Fluidigm’s digital PCR chip, there are other recently developed microfluidic digital PCR systems that have higher throughput (e.g., Qiagen’s QIAcuity system) that the STAR assay can potentially be applied to increase throughput. With a new system such as QIAcuity that can process eight 96-well plates with each well having 8500 chambers, clinical studies involving telomere length from hundreds of patients (49) can be easily processed within a day.
Because of its wide dynamic range, the STAR assay can accurately measure the entire distribution of telomere lengths to provide a comprehensive analysis of telomeres. In addition to providing the same high-quality average telomere length data as conventional methods, STAR assay integrates both the measurement of telomere heterogeneity and the proportion of short telomeres. Moreover, we showed that absolute counting of telomere molecules enables precise enumeration of ECTR molecules, a hallmark of ALT. We also demonstrated that the combination of telomere features measured in the STAR assay improves specificity when determining telomere maintenance mechanisms in cancer cells. We successfully applied this method to determine ALT status in a small number of pediatric neuroblastoma tumor samples. This provides a basis for extending the STAR assay, in combination with ddTRAP simultaneously implemented on a digital PCR platform, to enable a powerful prognosis tool based on the comprehensive determination of telomere maintenance mechanisms in oncology clinics. The ability to diagnose telomere maintenance mechanisms within clinical time scale will facilitate patient stratification for telomere maintenance pathway-targeting drugs (50, 51). The use of rapid molecular diagnostic tests in the clinical setting had been shown to significantly improve the clinical care for patients by decreasing the time taken to initiate treatment (52). In addition, the STAR assay would also provide a useful screening platform for the development of telomere-based therapeutic strategies. With the wide adoption of digital PCR assays in clinical laboratories and the introduction of commercial high-throughput digital PCR platforms that enable real-time reaction monitoring (53), we envision the STAR assay to be a new paradigm for measuring telomere length as a biomarker.
MATERIALS AND METHODS
Cell culture
HEK 293T [human; American Type Culture Collection (ATCC), CRL-3216], HeLa (human; ATCC, CCL-2), A549 (human; ATCC, CCL-185), K562 (human; ATCC, CCL-243), and WI-38 VA-13 (human; ATCC, CCL-75.1) were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, 11965084) supplemented with 10% fetal bovine serum (Gibco, 10270106) and 1% penicillin-streptomycin. U-2 OS (human; ATCC, HTB-96) and Saos-2 (human; ATCC, HTB-85) were grown in McCoy’s 5A (modified) medium (Gibco, 16600082) supplemented with 10% (v/v) fetal bovine serum and 1% penicillin-streptomycin. NCI-H1299 (human; ATCC, CRL-5803) was grown in RPMI 1640 medium (ATCC modification) (Gibco, A1049101) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin.
Genomic DNA extraction
Genomic DNA of the eight cancer cell lines was extracted using the Gentra Puregene Cell Kit (Qiagen) according to the manufacturer’s instructions and dissolved in EB buffer. Clinical tumor samples were first homogenized using a Dounce homogenizer for approximately 100 times before proceeding to extract the genomic DNA using the same Gentra Puregene Cell Kit. Each DNA sample was quantified using a Qubit fluorometer (ThermoFisher Scientific) for concentration, and the integrity of the DNA was analyzed by resolving 100 ng of DNA on a 1% agarose gel.
STAR assay
For the development of STAR, we used the Fluidigm 48.770 digital array integrated fluidic circuit (100-6151) that contains 48 × 770 compartments (0.85 nl each compartment). One microgram of genomic DNA was digested by each of Hin fI and Rsa I (1 U μl−1) in 1× SuRE/Cut buffer A at 37°C for 2 hours. Digested genomic DNA was diluted to 7.689 pg μl−1 to efficiently partition single telomere molecules in individual compartments in the digital PCR chip. The master mix used for the digital real-time PCR contained 1% dimethyl sulfoxide, 1 mM dithiothreitol, 200 μM deoxynucleotide triphosphate mix, 50 mM tris-HCl (pH 8.0), 2 mM MgCl2, 0.5× Rox, 1× EvaGreen dye, 1% Tween 20, AmpliTaq Gold DNA polymerase (0.06 U μl−1), and 0.3 μM each of Tel 1b and 2b primers (Integrated DNA Technologies). The primer sequences are Tel 1b (5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′) and Tel 2b (5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′).
Synthetic telomere repeat sequence (CCCTAA)15 was used to construct a standard curve for absolute telomere length measurement. Synthetic DNA was diluted to achieve the equivalent length of the telomere (5 to 320 kb) in each 0.85 nl of digital PCR compartment. The starting concentration of the synthetic DNA used was 100 μM, and it was diluted to 35 pM to achieve 320 kb. Serial fourfold dilutions were performed to achieve concentrations for 80, 20, and 5 kb. After loading all the standards and samples into the digital PCR chip, digital PCR was performed according to the manufacturer’s instructions in the Fluidigm Biomark HD system using the following thermal conditions (1 cycle of 95°C for 10 min, 40 cycles of 95°C for 15 s, 62°C for 30 s, and 72°C for 30 s). The same sample may be loaded across multiple inlets on the chip to increase the number of telomere length measurements.
Data analysis
The Fluidigm Digital PCR Analysis Program (version 4.1.2) was used to extract Ct values from the real-time PCR. To enable absolute telomere length quantification, a standard curve of mean Ct value against log10 (telomere length) was generated using a fourfold, four-point dilution series of synthetic telomere repeat sequences on the digital PCR chip. A linear equation fitting of this standard curve enables Ct values obtained from the samples to be converted to absolute telomere length.
In no-template controls, no nonspecific PCR amplification was observed until cycle 27. The upper Ct value cutoff was therefore set to 27, which corresponds to ~0.2 kb lower limit of telomere length detection. The average length, coefficient of variation, and percentage of short (<3 kb) telomeres of each sample are calculated on the basis of telomere length distribution obtained from converted Ct values. The average number of compartments with Ct < 27 for each sample was counted to detect the presence of ECTR.
Each feature measurement was converted into a z score by first subtracting from it the mean and dividing by the SD of the eight cell lines. The ALT score is defined as the sum of the four z scores (average length, coefficient of variation, percentage of short telomeres, and number of positive compartments) of each sample. An ALT score greater than zero is associated with an ALT-positive sample.
TRF analysis
TRF was performed using TeloTAGGG telomere length assay (Roche) according to the manufacturer’s instructions. Briefly, extracted genomic DNA for the different cell lines (1.5 μg each) were digested by two restriction enzymes (Hin fI and Rsa I) and separated by size on a 0.8% agarose gel at 3.3 V cm−1 for 3 hours. The digested DNA was then transferred from the agarose gel to a positively charged nylon membrane and fixed by baking at 120°C for 10 min. Hybridization with the DIG (digoxigenin)–labeled telomere-specific probe was done at 42°C for 3 hours with gentle agitation. After washing and blocking, the membrane was incubated with anti-DIG alkaline phosphatase at room temperature for 30 min. After another round of washing, the membrane was incubated with CDP-Star at room temperature for 5 min and detected for telomere signals via chemiluminescence using ChemiDoc imaging system (Bio-Rad). Average telomere length was measured using TESLAQuant program (19).
Quantitative PCR
The telomeric content for the eight cell lines was measured by qPCR using the same master mix and thermal conditions as the STAR assay in AriaMx real-time PCR system (Agilent). The relative telomeric content (T/S ratio) was calculated by, first, normalizing the telomeric content to that of the 36B4 SCG before normalizing it with the reference DNA. The SCG primer sequence is 36B4 (forward, 5′-GGGCGATGGCGCAGCCAATA-3′; reverse, 5′-AACCGCGGATAGCGCTCCTG-3′).
C-circle assay
Genomic DNA was digested with each of Hin fI and Rsa I (1 U μl−1) restriction enzymes at 37°C for 2 hours. Twenty nanograms of samples diluted in 10 mM tris (pH 7.6) was combined with bovine serum albumin (0.2 mg ml−1); 0.1% Tween; 1 mM each deoxyadenosine triphosphate, deoxyguanosine triphosphate, and deoxythymidine triphosphate; 1× Φ29 Buffer (New England Biolabs); and Φ29 DNA polymerase (0.375 U μl−1) (NEB) and incubated at 30°C for 8 hours before inactivating the polymerase at 65°C for 20 min. To quantify the C-circles, qPCR with the STAR assay master mix was used to measure the increase in telomeric DNA produced by the C-circle assay with normalization using the no Φ29 polymerase control.
ATRX sequencing
ATRX AmpliSeq library primers (Life Technologies) were designed with probes for amplicons against the coding region of the human ATRX gene (NM_000489). DNA was extracted from macrodissected tissue sections using the ReliaPrep FFPE gDNA Miniprep System (Promega) following the recommended protocol. DNA concentration was measured using the QuantiFluor ONE dsDNA System (Promega). Ten nanograms of DNA was used for library preparation using the Ion AmpliSeq Library Kit 2.0 (Life Technologies) and DICER1 AmpliSeq panel in the Ion Chef (Life Technologies). The prepared library was templated using the Hi-Q View Chef Kit (Life Technologies) in the Ion Chef. The prepared library was sequenced using an Ion Proton System next-generation sequencer using the Hi-Q View Sequencing Kit (Life Technologies). The Hi-Q View Sequencing Kit, the Hi-Q View Chef Kit, and the Ion AmpliSeq Library Kit 2.0 were used according to the manufacturer’s protocol. Base calling and mapping to hg19 were performed in Torrent Suite (version 5.6), and the variant calling plugin (version 5.6.0.5) was applied to generate VCF (variant call format) files. Data were analyzed using ThermoFisher Scientific’s Ion Reporter (version 5.6).
Clinical samples
The study was approved by the SingHealth Centralised Institutional Review Board (SHS/2014/2079) for the use of excess tumor tissues obtained from pathology specimens from KK Women’s and Children’s Hospital, Singapore. All subjects were recruited with informed consent from parents; verbal assent was obtained from research participants aged 6 to 12 years and written assent from patients older than 12 years. A total of four pediatric neuroblastoma tumor samples were used for this study (tables S1 and S2). All specimens identified for inclusion in the study were transported from the operating room within their original sterile container on ice and sent to the Department of Pathology and Laboratory Medicine, KK Women’s and Children’s Hospital, Singapore. All specimens were reviewed first by the attending pathologist to not compromise clinical diagnosis, and only excess material not required for diagnosis were snap frozen and used for subsequent analysis.
Supplementary Material
Acknowledgments
We thank T. Wu, Y. H. Luah, and H. Womersley for the advice and helpful discussions. We thank K. T. E. Chang for the help with the histological diagnosis, MYCN amplification, and ATRX sequencing. We thank S. H. Tan and C. H. Kuick for the help in specimen management and sequencing. Funding: This work was supported by funding from NUS Research Scholarship, NUS iHealthtech and NMRC OF-YIRG grant (NMRC/OFYIRG/0055/2017), and the VIVA Foundation for Children with Cancer. Author contributions: Y.L. and L.F.C. conceived and designed the assay. R.V. conducted preliminary experiments. Y.L. acquired and analyzed the data. Y.L. and L.F.C. interpreted the data. M.P.H. and A.H.P.L. provided advice. A.H.P.L. provided clinical samples. Y.L. and L.F.C. wrote the manuscript. All authors discussed the results and reviewed the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/34/eabb7944/DC1
REFERENCES AND NOTES
- 1.Blackburn E. H., Structure and function of telomeres. Nature 350, 569–573 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Okuda K., Bardeguez A., Gardner J. P., Rodriguez P., Ganesh V., Kimura M., Skurnick J., Awad G., Aviv A., Telomere length in the newborn. Pediatr. Res. 52, 377–381 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Panossian L. A., Porter V. R., Valenzuela H. F., Zhu X., Reback E., Masterman D., Cummings J. L., Effros R. B., Telomere shortening in T cells correlates with Alzheimer’s disease status. Neurobiol. Aging 24, 77–84 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Sampson M. J., Winterbone M. S., Hughes J. C., Dozio N., Hughes D. A., Monocyte telomere shortening and oxidative DNA damage in type 2 diabetes. Diabetes Care 29, 283–289 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Epel E. S., Merkin S. S., Cawthon R., Blackburn E. H., Adler N. E., Pletcher M. J., Seeman T. E., The rate of leukocyte telomere shortening predicts mortality from cardiovascular disease in elderly men. Aging 1, 81–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frías C., García-Aranda C., De Juan C., Morán A., Ortega P., Gómez A., Hernando F., López-Asenjo J.-A., Torres A.-J., Benito M., Iniesta P., Telomere shortening is associated with poor prognosis and telomerase activity correlates with DNA repair impairment in non-small cell lung cancer. Lung Cancer 60, 416–425 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Kimura M., Stone R. C., Hunt S. C., Skurnick J., Lu X., Cao X., Harley C. B., Aviv A., Measurement of telomere length by the southern blot analysis of terminal restriction fragment lengths. Nat. Protoc. 5, 1596–1607 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Aubert G., Hills M., Lansdorp P. M., Telomere length measurement—Caveats and a critical assessment of the available technologies and tools. Mutat. Res. 730, 59–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cawthon R. M., Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 37, e21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cawthon R. M., Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahlgren P. N., Bishop K., Dey S., Herbert B.-S., Tanaka H., Development of a new monochrome multiplex qPCR method for relative telomere length measurement in cancer. Neoplasia 20, 425–431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. S. S. Poon, P. M. Lansdorp, in Current Protocols in Cell Biology (John Wiley & Sons Inc., 2001), vol. 12, pp. 18.4.1–18.4.21. [DOI] [PubMed] [Google Scholar]
- 13.Canela A., Vera E., Klatt P., Blasco M. A., High-throughput telomere length quantification by FISH and its application to human population studies. Proc. Natl. Acad. Sci. U.S.A. 104, 5300–5305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vera E., Blasco M. A., Beyond average: Potential for measurement of short telomeres. Aging 4, 379–392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adam N., Degelman E., Briggs S., Wazen R.-M., Colarusso P., Riabowol K., Beattie T., Telomere analysis using 3D fluorescence microscopy suggests mammalian telomere clustering in hTERT-immortalized Hs68 fibroblasts. Commun. Biol. 2, 451 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beh C. W., Zhang Y., Zheng Y.-L., Sun B., Wang T.-H., Fluorescence spectroscopic detection and measurement of single telomere molecules. Nucleic Acids Res. 46, e117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baird D. M., Rowson J., Wynford-Thomas D., Kipling D., Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet. 33, 203–207 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Bendix L., Horn P. B., Jensen U. B., Rubelj I., Kolvraa S., The load of short telomeres, estimated by a new method, Universal STELA, correlates with number of senescent cells. Aging Cell 9, 383–397 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Lai T.-P., Zhang N., Noh J., Mender I., Tedone E., Huang E., Wright W. E., Danuser G., Shay J. W., A method for measuring the distribution of the shortest telomeres in cells and tissues. Nat. Commun. 8, 1356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemann M. T., Strong M. A., Hao L. Y., Greider C. W., The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107, 67–77 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Flores I., Canela A., Vera E., Tejera A., Cotsarelis G., Blasco M. A., The longest telomeres: A general signature of adult stem cell compartments. Genes Dev. 22, 654–667 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Britt-Compton B., Lin T. T., Ahmed G., Weston V., Jones R. E., Fegan C., Oscier D. G., Stankovic T., Pepper C., Baird D. M., Extreme telomere erosion in ATM-mutated and 11q-deleted CLL patients is independent of disease stage. Leukemia 26, 826–830 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Chen Y.-A., Shen Y.-L., Hsia H.-Y., Tiang Y.-P., Sung T.-L., Chen L.-Y., Extrachromosomal telomere repeat DNA is linked to ALT development via cGAS-STING DNA sensing pathway. Nat. Struct. Mol. Biol. 24, 1124–1131 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Li N., Schwartz M., Ionescu-Zanetti C., PDMS compound adsorption in context. J. Biomol. Screen. 14, 194–202 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Mao F., Leung W.-Y., Xin X., Characterization of EvaGreen and the implication of its physicochemical properties for qPCR applications. BMC Biotechnol. 7, 76 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddel R. R., The role of senescence and immortalization in carcinogenesis. Carcinogenesis 21, 477–484 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Cesare A. J., Reddel R. R., Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 11, 319–330 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Venturini L., Erdas R., Costa A., Gronchi A., Pilotti S., Zaffaroni N., Daidone M. G., ALT-associated promyelocytic leukaemia body (APB) detection as a reproducible tool to assess alternative lengthening of telomere stability in liposarcomas. J. Pathol. 214, 410–414 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Allsopp R. C., Harley C. B., Evidence for a critical telomere length in senescent human fibroblasts. Exp. Cell Res. 219, 130–136 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Perrem K., Colgin L. M., Neumann A. A., Yeager T. R., Reddel R. R., Coexistence of alternative lengthening of telomeres and telomerase in hTERT-transfected GM847 cells. Mol. Cell. Biol. 21, 3862–3875 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oganesian L., Karlseder J., Mammalian 5′ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol. Cell 42, 224–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cesare A. J., Griffith J. D., Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell. Biol. 24, 9948–9957 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henson J. D., Lau L. M., Koch S., Martin La Rotta N., Dagg R. A., Reddel R. R., The C-Circle Assay for alternative-lengthening-of-telomeres activity. Methods 114, 74–84 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Fasching C. L., Neumann A. A., Muntoni A., Yeager T. R., Reddel R. R., DNA damage induces alternative lengthening of telomeres (ALT) associated promyelocytic leukemia bodies that preferentially associate with linear telomeric DNA. Cancer Res. 67, 7072–7077 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Henson J. D., Cao Y., Huschtscha L. I., Chang A. C., Au A. Y. M., Pickett H. A., Reddel R. R., DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 27, 1181–1185 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Pugh T. J., Morozova O., Attiyeh E. F., Asgharzadeh S., Wei J. S., Auclair D., Carter S. L., Cibulskis K., Hanna M., Kiezun A., Kim J., Lawrence M. S., Lichenstein L., Kenna A. M., Pedamallu C. S., Ramos A. H., Shefler E., Sivachenko A., Sougnez C., Stewart C., Ally A., Birol I., Chiu R., Corbett R. D., Hirst M., Jackman S. D., Kamoh B., Khodabakshi A. H., Krzywinski M., Lo A., Moore R. A., Mungall K. L., Qian J., Tam A., Thiessen N., Zhao Y., Cole K. A., Diamond M., Diskin S. J., Mosse Y. P., Wood A. C., Ji L., Sposto R., Badgett T., London W. B., Moyer Y., Gastier-Foster J. M., Smith M. A., Auvil J. M. G., Gerhard D. S., Hogarty M. D., Jones S. J. M., Lander E. S., Gabriel S. B., Getz G., Seeger R. C., Khan J., Marra M. A., Meyerson M., Maris J. M., The genetic landscape of high-risk neuroblastoma. Nat. Genet. 45, 279–284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulaner G. A., Hu J. F., Vu T. H., Giudice L. C., Hoffman A. R., Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 58, 4168–4172 (1998). [PubMed] [Google Scholar]
- 38.Piatyszek M. A., Kim N. W., Weinrich S. L., Hiyama K., Hiyama E., Wright W. E., Shay J. W., Detection of telomerase activity in human cells and tumors by a telomeric repeat amplification protocol (TRAP). Methods Cell Sci. 17, 1–15 (1995). [Google Scholar]
- 39.A. T. Ludlow, D. Shelton, W. E. Wright, J. W. Shay, in Methods in Molecular Biology (2018), vol. 1768, pp. 513–529. [DOI] [PMC free article] [PubMed]
- 40.Lau L. M. S., Dagg R. A., Henson J. D., Au A. Y. M., Royds J. A., Reddel R. R., Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res. 41, e34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amorim J., Santos G., Vinagre J., Soares P., The role of ATRX in the alternative lengthening of telomeres (ALT) phenotype. Genes 7, 66 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneiderman J., London W. B., Brodeur G. M., Castleberry R. P., Look A. T., Cohn S. L., Clinical significance of MYCN amplification and ploidy in favorable-stage neuroblastoma: A report from the Children’s Oncology Group. J. Clin. Oncol. 26, 913–918 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Lin J., Smith D. L., Esteves K., Drury S., Telomere length measurement by qPCR – Summary of critical factors and recommendations for assay design. Psychoneuroendocrinology 99, 271–278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quan P.-L., Sauzade M., Brouzes E., dPCR: A technology review. Sensors 18, 1271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Keefe C. M., Pisanic T. R. II, Zec H., Overman M. J., Herman J. G., Wang T.-H., Facile profiling of molecular heterogeneity by microfluidic digital melt. Sci. Adv. 4, eaat6459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santoro N., Cirillo G., Amato A., Luongo C., Raimondo P., D’Aniello A., Perrone L., Miraglia del Giudice E., Insulin gene variable number of tandem repeats (INS VNTR) genotype and metabolic syndrome in childhood obesity. J. Clin. Endocrinol. Metab. 91, 4641–4644 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Giesselmann P., Brändl B., Raimondeau E., Bowen R., Rohrandt C., Tandon R., Kretzmer H., Assum G., Galonska C., Siebert R., Ammerpohl O., Heron A., Schneider S. A., Ladewig J., Koch P., Schuldt B. M., Graham J. E., Meissner A., Müller F.-J., Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat. Biotechnol. 37, 1478–1481 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Wang F., Pan X., Kalmbach K., Seth-Smith M. L., Ye X., Antumes D. M. F., Yin Y., Liu L., Keefe D. L., Weissman S. M., Robust measurement of telomere length in single cells. Proc. Natl. Acad. Sci. U.S.A. 110, E1906–E1912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ley B., Newton C. A., Arnould I., Elicker B. M., Henry T. S., Vittinghoff E., Golden J. A., Jones K. D., Batra K., Torrealba J., Garcia C. K., Wolters P. J., The MUC5B promoter polymorphism and telomere length in patients with chronic hypersensitivity pneumonitis: An observational cohort-control study. Lancet Respir. Med. 5, 639–647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu J., Hwang S. S., Liesa M., Gan B., Sahin E., Jaskelioff M., Ding Z., Ying H., Boutin A. T., Zhang H., Johnson S., Ivanova E., Kost-Alimova M., Protopopov A., Wang Y. A., Shirihai O. S., Chin L., DePinho R. A., Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 148, 651–663 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henson J. D., Reddel R. R., Assaying and investigating Alternative lengthening of telomeres activity in human cells and cancers. FEBS Lett. 584, 3800–3811 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Kipiani M., Mirtskhulava V., Tukvadze N., Magee M., Blumberg H. M., Kempker R. R., Significant clinical impact of a rapid molecular diagnostic test (Genotype MTBDRplus Assay) to detect multidrug-resistant tuberculosis. Clin. Infect. Dis. 59, 1559–1566 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dueck M. E., Lin R., Zayac A., Gallagher S., Chao A. K., Jiang L., Datwani S. S., Hung P., Stieglitz E., Precision cancer monitoring using a novel, fully integrated, microfluidic array partitioning digital PCR platform. Sci. Rep. 9, 19606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/34/eabb7944/DC1


