Abstract
Background and Aims
The distribution of cytotypes and its potential correlation with environmental variables represent a cornerstone to understanding the origin and maintenance of polyploid lineages. Although many studies have addressed this question in single species at a regional scale, only a few have attempted to decipher this enigma in groups of closely related species at a broad intercontinental geographical scale. Here, we consider approx. 20 species of a diploid–polyploid complex (Veronica subsect. Pentasepalae) of recent and rapid diversification represented in Europe and North Africa to study the frequency and distribution of cytotypes and their relationship to environmental variables.
Methods
A total of 680 individuals (207 populations) were sampled. Ploidy levels were determined using flow cytometry. Ecological differentiation among cytotypes was tested using climatic and environmental variables related to temperature, precipitation, vegetation and biogeographical region, among others, and by performing univariate and multivariate (constrained principal coordinates analysis) analyses.
Key Results
Four ploidy levels (2x, 4x, 6x and 8x) were found and genome downsizing was observed to occur within the group. Plants of different ploidy level are ecologically differentiated, with hexaploids and octoploids occurring in wetter and colder habitats with a higher seasonality than diploids. A south to north distribution pattern was found, with diploids occupying southern refugial areas and octoploids being more frequent in northern regions of Europe above the permafrost boundary.
Conclusions
The distribution of cytotypes can be explained by ecological differentiation, the geographical position of refuge areas during the Quaternary climatic oscillations as well as by ice and permafrost retreat patterns. The Balkan Peninsula constitutes the most important contact zone between cytotypes. This work provides the first comprehensive ploidy screening within V. subsect. Pentasepalae at a broad scale and indicates that polyploidy and genome downsizing might have contributed to the colonization of new habitats in a recently diverged polyploid complex.
Keywords: Cytotype distribution patterns, ecological differentiation, genome downsizing, historical biogeography, minority cytotype exclusion, ploidy level, polyploidy, Quaternary glaciations, Veronica subsection Pentasepalae
INTRODUCTION
Polyploidy or whole-genome duplication (WGD) plays a major role in the evolution of angiosperms and other eukaryotes (Stebbins, 1950; Otto and Whitton, 2000; Wendel, 2000; Soltis et al., 2015). Molecular studies have demonstrated that repeated rounds of polyploidization have modelled the evolutionary history of all flowering plants since their origin (Jiao et al., 2011; Wendel, 2015). While some authors consider polyploids as ‘evolutionary dead-ends’ (Mayrose et al., 2011; Arrigo and Barker, 2012), other studies associate polyploidy with high diversification rates (Fawcett et al., 2009; Soltis et al., 2014; Meudt et al., 2015; Kellogg, 2016; Levin and Soltis, 2018; Han et al., 2019). Genome downsizing, i.e. the reduction of DNA per cell without numerical loss of chromosomes (Leitch and Bennet, 2004; Ramsey and Ramsey, 2014), is likewise ubiquitous in angiosperms and has also been correlated with an increased diversification rate in polyploid complexes (Kraaijeveld, 2010; Meudt et al., 2015). Indeed, recent work has corroborated the contribution of genome downsizing to the origin of physiological novelties that lead to the diversification of flowering plants and their expansion to new ecological habitats (Pandit et al., 2014; Simonin and Roddy, 2018). Furthermore, changes in genome size may affect different aspects of polyploids regarding ecology, physiology, reproduction (e.g. meiosis) or genetic patterns (Bennett and Smith, 1972; Cavalier-Smith, 1978; Gregory, 2001; Leitch and Bennett, 2004; Comai, 2005; Pandit et al., 2014; Simonin and Roddy, 2018).
The frequency and geographical distribution of cytotypes are important for exploring the origins of polyploids and their maintenance in nature (Duchoslav et al., 2010). Differences in cytotype distribution patterns have been explained by different but often complementary processes. The adaptive evolutionary scenario assumes that the successful persistence of polyploids is determined by ecological factors (Rodriguez, 1996; Husband and Schemske, 1998; Ramsey, 2011; Fowler and Levin, 2016). Under this hypothesis, novel phenotypic, physiological and genetic combinations associated with polyploidy – mainly allopolyploidy – seem to be responsible for different ecological tolerances (Paun et al., 2011; Ramsey, 2011) or broader geographic ranges of polyploids, as compared with diploid congeners (Hijmans et al., 2007; Meimberg et al., 2009). Thus, genome duplication may allow polyploids to expand to new habitats previously unavailable to their diploid counterparts (Stebbins, 1984, 1985; Thompson and Lumaret, 1992; Maceira et al., 1993; te Beest et al., 2012). Numerous studies that have recently assessed this hypothesis have found ecological differences among ploidy levels (see Ramsey and Ramsey, 2014; Mairal et al., 2018). However, other studies failed to detect habitat differentiation among cytotypes, suggesting that niche differentiation patterns are not universal (Glennon et al., 2014; Visser and Molofsky, 2015; Marchant et al., 2016; Castro et al., 2018, 2019). The lack of ecological niche differentiation among cytotypes might occur due to several reasons. First, ecological divergence may arise with time and, thus, the effects of WGD on ecological differentiation can be obscured when studies are focused on a single species, in which new cytotypes may have not had enough time to diverge (e.g. Schranz et al., 2012; Godsoe et al., 2013). Secondly, differences between cytotypes could occur at a microenvironmental scale, thus being difficult to detect by the large-scale climate data, particularly when the ranges of the different cytotypes overlap (Glennon et al., 2014; Kirchheimer et al., 2016). Thirdly, broader geographical scale studies have suggested that species’ ranges are also influenced by their phylogenetic history (Levin, 2000; Losos, 2008; Burns and Strauss, 2011) due to the inheritance of ecological preferences (Martin and Husband, 2009). According to this idea, phylogenetically related taxa would be prone to occupy more similar environments due to the so-called phylogenetic niche conservatism (Harvey and Pagel, 1991; Lord et al., 1995).
Nonetheless, certain distribution patterns (e.g. higher number of polyploids inhabiting higher latitudes) can also be generated by other processes, such as biogeographic history (Avise, 2000; Duchoslav et al., 2010; Paule et al., 2017). For example, the climatic oscillations that occurred during the Quaternary have induced range shifts of plant species that found refuge at lower latitudes or altitudes (Hewitt, 1999; Thompson, 2005). The existence of new and unexploited areas after ice retreat probably represented an opportunity for polyploids to expand and establish outside the range of their diploid counterparts (Stebbins, 1950, 1984; Brochmann et al., 2004; Marchant et al., 2016). Thus, the distribution of cytotypes may also be explained by the geographical position of the refuge areas, as well as by ice and permafrost retreat patterns (van Dijk et al., 1992; Mandáková and Münzbergová, 2006; Duchoslav et al., 2010). Importantly for the analysis of distribution patterns in European plants, permafrost reached even the lowland regions at mid-latitudes within Europe during the last permafrost maximum (LPM; Fig. 1), and active mountain glaciers were present in the central and southern mountain ranges of Europe (Fig. 1). Apart from migration patterns out of refuge areas, Quaternary climatic oscillations might have triggered secondary contacts among otherwise geographically isolated taxa (Stebbins, 1984, 1985; Brochmann et al., 2004; Kadereit, 2015). Accordingly, the biogeographic history of species may have been responsible for the origin of allopolyploid taxa and/or mixed ploidy populations in secondary contact areas.
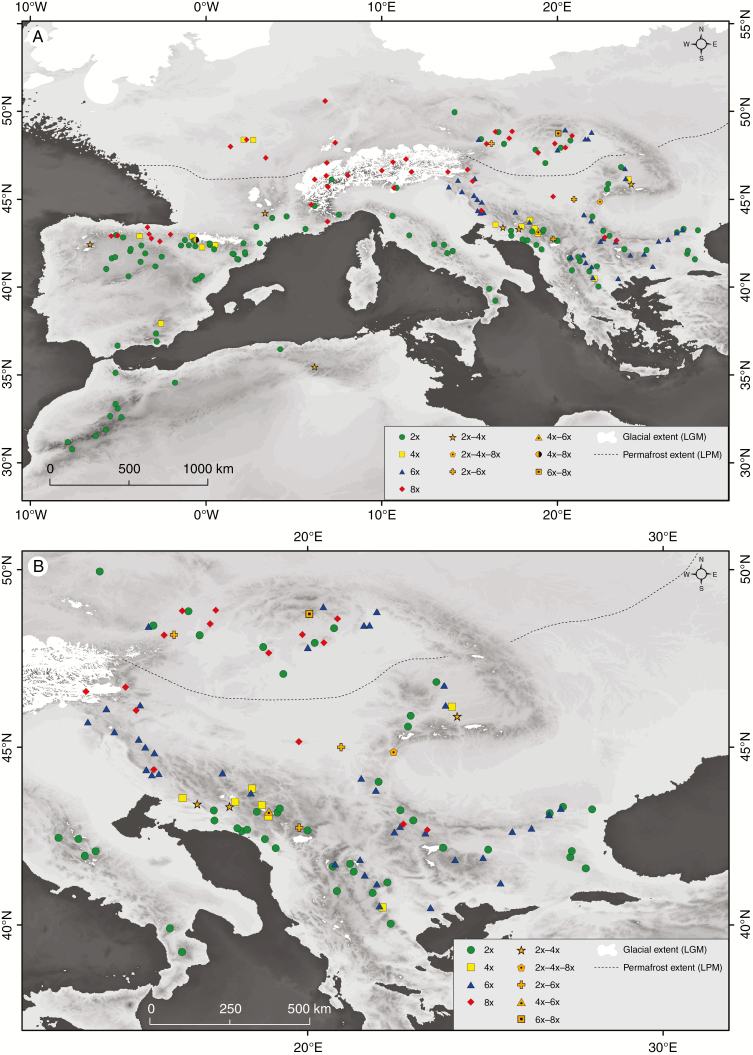
Fig. 1.
Map showing the geographical distribution of cytotypes of Veronica subsect. Pentasepalae during the last glacial maximum (LGM). (A) General view of the study area. The line indicates the limit of the continuous permafrost during the last permafrost maximum (LPM) according to Vanderberghe et al. (2014). (B) Detail of the Balkan Peninsula.
The distribution of cytotypes can also be explained by other locally operating processes, such as the frequency-dependent mating success (Ramsey and Schemske, 1998). According to the minority cytotype exclusion (MCE) model, the reproductive disadvantage of the minority cytotype would lead to its extinction from the population (Levin, 1975). However, several mechanisms may counteract MCE, allowing neopolyploids to establish. On the one hand, neopolyploids may persist outside the range of the parental cytotype by eco-geographical differentiation, which will result in different cytologically uniform regions (Levin, 1975). On the other hand, mixed-ploidy populations have been reported for many plant species (e.g. Husband and Schemske, 2000; Duchoslav et al., 2010; Bardy et al., 2010, 2011; Krejčíková et al., 2013; McAllister et al., 2015; Etterson et al., 2016; Čertner et al., 2017), suggesting the existence of mechanisms that enable the persistence of a new cytotype within the population of origin. Genome duplication may promote the occurrence of reproductive barriers among cytotypes within the same populations (Ramsey, 2011), favouring assortative mating. These include changes in selfing (Fausto et al., 2001; Comai, 2005), shifts in flowering time (Fowler and Levin, 1984), pollinator preferences (Thompson and Merg, 2008) and/or ecological differences at a microscale (Ramsey, 2011), which alone, or in concert, can contribute to the coexistence of cytotypes at the local scale. Cytotype coexistence can also be favoured by asexual reproduction (Paule et al., 2011), clonal propagation (Baldwin and Husband, 2013), a better competitive ability of neopolyploids (Keeler and Davis, 1999) and/or a higher resistance to herbivory (Segraves and Anneberg, 2016). Finally, mixed-ploidy populations originated by autopolyploid events (in the so-called primary contact zones) can also be maintained by the recurrent production of unreduced gametes (Rodriguez, 1996), a process which is in turn influenced by environmental stress (McHale, 1983; Hao et al., 2013).
In this study, we focus on the diploid–polyploid complex Veronica subsection Pentasepalae. This monophyletic group is composed of approx. 20 phylogenetically closely related species with ploidy levels ranging from 2x to 8x, and more rarely 10x (Peev, 1972; Rojas-Andrés et al., 2015). It is a recently diverged polyploid complex [mean crown age of 2.87 Mya (1.37–4.53 Mya); Meudt et al., 2015] mainly distributed over Europe and North Africa, which allows a study at a large geographical scale, enabling us to have the appropriate spatial and environmental data resolution for the model system. All these attributes make V. subsect. Pentasepalae an ideal system to evaluate whether independent polyploidization events within a single monophyletic lineage have led to ecological differentiation. For this, an extensive screening on ploidy levels by flow cytometry was performed for the whole subsection without considering taxonomy (i.e. irrespective of the assignment of individuals to particular taxa under this rank) and statistical analyses were used to address the following questions. (1) What are the frequencies and geographic distribution of cytotypes within V. subsect. Pentasepalae? (2) Do the polyploids show genome downsizing? (3) Are there environmental variables associated with the occurrence of different ploidy levels? (4) Which processes can explain the currently observed cytotype distribution patterns?
MATERIALS AND METHODS
Study group and sampling strategy
Veronica subsection Pentasepalae is a group of perennial herbs with a very branched rhizome indicating that, besides sexual reproduction, asexual vegetative propagation is common in the group. Little is known about the mating system of these plants, whose fruits and seeds lack dispersal organs or evident adaptations to anemochory or zoochory. Both xenogamy and self-pollination have been demonstrated to occur within the group (Scheerer, 1937; Scalone et al., 2013, 2014). Regarding its evolutionary history, much progress has been made in the last years, yet there are still many open questions. Although some species of V. subsect. Pentasepalae have had time to diverge as independent lineages, reproductive isolation among others seems to be weak, as evidenced by hybridization and/or introgression patterns shown by some morphologically intermediate populations of uncertain taxonomic identity (Rojas-Andrés et al., 2015; Rojas-Andrés and Martínez-Ortega, 2016; Padilla-García et al., 2018). While it has been possible to identify the auto- or allopolyploid genesis of some populations, the origin of many polyploids is still not well understood or needs further confirmation (Padilla-García et al., 2018). Genome designation of some polyploids would require extensive genome sequencing to find nucleotide polymorphisms that correlate with ancestral genomes and/or the use of advanced molecular cytogenetic methods such as genomic or fluorescent in situ hybridization. Thus, based on the rapid evolution under comparable environmental and historical conditions, we considered these taxa to be closely related, yet independent and equivalent enough for the purpose of our study. In addition, the existence of some narrow endemic species, for which a very low number of populations is known, hampers a robust statistical analysis at the species level. Given these limitations, our study is designed as an initial ploidy screening to explore cytotype distribution patterns within the subsection at an intercontinental geographical scale and their relationship with environmental conditions.
With this aim, a total of 680 individuals from 207 populations (Supplementary Data Table S1) of V. subsect. Pentasepalae were collected throughout Europe and North Africa (Fig. 1). Given that the area covered by our study is >2 million km2, we tried to find a trade-off between the number of populations analysed and individuals per population. Therefore, we sampled >200 populations and, in general, three individuals per population, which provides an exceptional overview, usually unavailable for most polyploid complexes. Leaf material was collected in the field and immediately stored in silica gel for flow cytometric analyses. Geographical co-ordinates for each sampling location were registered using GPS. Vouchers were deposited at the herbaria MGC, SALA and MA (acronyms following Thiers, 2017).
Genome size and ploidy level estimations
Ploidy levels were estimated by flow cytometry and related to the chromosome counts available from previous studies (Martínez-Ortega et al., 2004; Albach et al., 2008; Delgado et al., 2018). Direct chromosome counts were previously available for 29 sampled populations, so the corresponding data were incorporated in this study. For 18 of these 29 populations, individuals from the same origin were included in our flow cytometric measurements to ensure correspondence between chromosome number and genome size estimations. Genome size was estimated using silica gel-dried leaves for a total of 668 individuals from 196 populations (Supplementary Data Table S1). In total, 207 populations were included in the data analyses (196 plus 11 with chromosome counts only).
Leaf tissue from each individual was chopped together with fresh leaf material from an internal standard using a sharp razor blade in a Petri dish containing 1.1 mL of Woody Plant Buffer (WPB; Loureiro et al., 2007). Depending on the sample C-value and standard availability, Solanum pseudocapsicum (2C = 2.589 pg; Temsch et al., 2010), Zea mays L. ‘CE-777’ (2C = 5.43 pg; Lysak and Dolezel, 1998), Pisum sativum ‘Ctirad’ (2C = 9.09 pg; Dolezel et al., 1998) and Pisum sativum ‘Kleine Rheinländerin’ (2C = 8.84 pg; Greilhuber and Ebert, 1994) were used as internal standards. A 1 mL aliquot of the nuclear suspension was filtered through a 48 μm nylon gauze, mixed with RNase (Sigma) to a final concentration of 0.15 mg mL–1 and digested at 37 °C for 30 min. A total of 450 μL of the nuclear suspension was then mixed with 2 mL of the propidium iodide (PI) staining solution (60 μg mL–1 PI in doubled-distilled water), incubated for at least 10 min and analysed. For each individual, one run of 5000 counts was made on a CyFlow SL (Partec GmbH, Münster, Germany) equipped with a solid-state laser featuring blue excitation at 488 nm.
For each sample, an estimate of genome size was obtained using the following formula: Genome size estimate (pg) = (Veronica G1 peak mean/Reference standard G1 peak mean) × Reference standard genome size. Samples were classified as diploid, tetraploid, hexaploid or octoploid according to the estimates of genome size and previous chromosome counts (Martínez-Ortega et al., 2004; Albach et al., 2008; Delgado et al., 2018). Sixty-two per cent of the measurements presented a sample coefficient of variation (CV) of G1 peaks below 5 %. For the remaining samples, CV values were between 5 and 9 %, even after sample repetition, most probably because the material was stored in silica gel for several months or years before being analysed and/or because of the presence of cytosolic compounds. As it was possible to assign ploidy levels unambiguously for all samples, these values were accepted for the estimation of ploidy levels. However, only measurements with CVs <5 % were considered to evaluate genome size values among different ploidy levels (Supplementary Data Table S2).
Environmental variables
Environmental parameters for 207 populations were extracted using ArcGIS 10.1 (ESRI, Redlands, CA, USA) from different sources (Supplementary Data Table S3). Precipitation and temperature were obtained from WorldClim (http://www.worldclim.org/current) (Hijmans et al., 2005) and additional environmental parameters related to vegetation, slope, solar radiation, human footprint, soil type, land cover and biogeographical region were also included in the analyses. Spatial resolution and sources for all environmental parameters are given in Supplementary Data Table S3. Geographical data were directly obtained from GPS. We could not add any information regarding potential vegetation for the populations of North Africa given that no map of potential vegetation was found for this area. In total, 33 layers were considered: 19 related to precipitation and temperature, five to vegetation index, one to tree cover, one to slope, two to solar radiation, one to human footprint, one to soil type, one to potential natural vegetation, one to land type and one to biogeographical region (Supplementary Data Table S3).
Statistical analyses
All statistical analyses were performed using R 3.2.2 (R Development Core Team, 2015). Ecological differentiation among ploidy levels was first tested using univariate analyses. All variables were analysed to test their significance. Contingency tables were used for qualitative variables, whereas for quantitative variables analysis of variance (ANOVA) and the Kruskal–Wallis test were used (for parametric and non-parametric data, respectively). Significant variables in the overall test were subsequently analysed using Dunn’s test to check for significant differences among ploidy levels for each environmental parameter. The Bonferroni correction of α = 0.0003 for multiple tests was applied to avoid type I errors (Shaffer, 1995). Although it is a very conservative method, it limits the number of significant models to more robust associations (Narum, 2006).
Environmental variables were subsequently subjected to multivariate analysis. Given that some variables might be highly correlated, analysis of correlation and reduction of collinearity are recommended to avoid type II errors and to find which variables are driving the system (Zuur et al., 2010). Therefore, first, a Pearson product correlation was used to identify quantitative variables correlated at a level of |r| ≥ 0.8. From each pair of correlated variables, one was excluded from subsequent analyses. However, looking at correlations only among pairs of predictors is limiting because a linear dependence among three or more variables may exist. Thus, the variance inflation factor (VIF) on the environmental variables was applied, using 6.0 as a threshold value for acceptable collinearity (Chatterjee and Hadi, 2006). The VIF analyses were done with the R package ‘HH’ v. 1.4 (Heiberger, 2015), which was iteratively applied. Regarding association among qualitative variables, a Cramer’s V test was performed with the R package ‘DescTools’ v. 0.99.22 (Signorell et al., 2017). None of the variables considered showed a high level of dependence (≥0.7).
After correlation analyses, the 33 variables initially considered were reduced to 17 variables representing climatic, vegetation, topographic, solar irradiance, anthropic, edaphic and biogeographical variability (Supplementary Data Table S3). Due to different types of descriptors (qualitative and quantitative), the Gower general coefficient of dissimilarity (Gower, 1985) was used to obtain a distance matrix with the R package ‘FD’ v. 1.0-12 (Laliberté et al., 2014). The distance matrix was subjected to principal coordinates analysis (PCoA). The results obtained were subsequently subjected to constrained PCoA [distance-based redundancy analysis (db-RDA); Legendre and Anderson, 1999], where the PCoA scores of the localities containing the environmental information were linked to the ploidy levels (constraints). Calculations were performed with the R package ‘vegan’ v. 2.2-1 (Oksanen et al., 2013). Given that the overall test was significant, pairwise tests between ploidy levels with reduced matrices containing only populations of the ploidy levels being tested were conducted to clarify which of the cytotypes are different from each other regarding environmental conditions. The significance level of the db-RDA was assessed by an ANOVA-like permutation test by which the original environmental matrix was randomly permuted 999 times and the results were compared with the real model (original environmental matrix). The Bonferroni correction was applied for multiple tests (α = approx. 0.001). To identify the environmental variables that differ most between pairs of cytotypes from those significant models, we correlated each environmental variable value with the values of population scores in the first axis of the models: each one containing a pair of cytotypes. Pearson correlations were used for this purpose, and good correlation was considered when |r| > 0.5.
RESULTS
Genome size and cytotype diversity in Veronica subsect. Pentasepalae
Ploidy level estimates were obtained for 668 individuals from 207 localities. Genome size values obtained for individuals of V. subsect. Pentasepalae were arranged in four groups, corresponding to four different ploidy levels: 2x, 4x, 6x and 8x (Supplementary Data Fig. S1). Nuclear DNA contents (1C-values) ranged between 0.70 and 1.03 pg for diploids, 1.13 and 1.58 pg for tetraploids (with approx. 1.72 pg for a tetraploid population from north Spain), 1.71 and 2.06 pg for hexaploids, and 2.24 and 2.88 pg for octoploids (considering only measurements with CVs <5 %). The genome size of tetraploids was not double that of diploids, nor were hexaploids or octoploids three or four times, respectively, the size of diploids (Supplementary Data Table S1; Fig. S1). The monoploid genome size (1Cx-value; Greilhuber et al., 2005) of tetraploids was 17.86 % lower than that of diploids, and 25.00 and 26.20 % lower in the case of hexaploids and octoploids, respectively (Supplementary Data Table S2). Most of the populations included a single cytotype (194 populations; 93.72 %), while there were differences in ploidy level among individuals collected at 13 (6.28 %) populations (Table 1). The latter are hereafter referred as mixed-ploidy populations. Among single ploidy populations (93.72 % of the total), diploid was the most frequent cytotype (49.76 %), followed by octoploids (18.84 %) and hexaploids (18.36 %), and finally by tetraploids (6.76%).
Table 1.
Cytotype diversity found within sampled populations of Veronica subsect. Pentasepalae
| Number of ploidy levels per population | Number of populations/frequency |
|---|---|
| 1 | 194/93.72 % |
| 2 | 12/5.79 % |
| 3 | 1/0.48 % |
| Total | 207 |
| Populations containing one ploidy level | |
| 2x | 103/49.76 % |
| 4x | 14/6.76 % |
| 6x | 38/18.36 % |
| 8x | 39/18.84 % |
| Populations containing different ploidy levels | |
| 2x + 4x | 6/2.90 % |
| 2x + 6x | 3/1.45 % |
| 4x + 6x | 1/0.48 % |
| 4x + 8x | 1/0.48 % |
| 6x + 8x | 1/0.48 % |
| 2x + 4x + 8x | 1/0.48 % |
Percentages are always calculated relative to the total number of sampled populations.
Distribution of cytotypes
Populations of V. subsect. Pentasepalae showed a clear geographic structuring of cytotypes (Fig. 1). Diploids were preferentially found at low latitudes, except for a few populations extending to central Europe. Tetraploids represented the rarest cytotype and were found in specific locations in the Balkan Peninsula, Spain, France and Algeria. Hexaploids were distributed towards the east and south-east of Europe, and octoploids were mainly found at higher latitudes in central Europe, with only some populations in the north of Spain and the Balkan Peninsula. The Balkan Peninsula exhibited the highest diversity in ploidy levels, and it is the only area where all cytotypes were found.
Despite low intra-population sampling, some mixed-ploidy populations were found (Table 1; Fig. 1). Seven of them were found in the Balkans and were composed of 2x + 4x (three), 2x + 6x (two), 4x + 6x (one) and 2x + 4x + 8x (one) cytotypes. Furthermore, six mixed-ploidy populations were found scattered in the remaining area, in north-eastern Algeria (2x + 4x), north-western Spain (2x + 4x), Austria (2x + 6x), Slovakia (6x + 8x), southern France (2x + 4x) and northern Spain (4x + 8x). No uneven ploidy levels were detected.
Relationships between environmental variables and ploidy level distribution
Univariate analysis
From the 33 environmental variables analysed, 14 variables related to temperature, precipitation, vegetation index, potential natural vegetation and biogeographical region were found to be significantly associated with the distribution of ploidy levels (Supplementary Data Table S4; Figs S2, S3 and S4). Diploids and tetraploids occur in areas characterized by higher values of isothermality (Bio 3) in comparison with hexaploids (Supplementary Data Fig. S2A), whereas hexaploids occur in areas characterized by higher temperature seasonality (Bio 4) compared with the areas where diploids, tetraploids and octoploids are present (Supplementary Data Fig. S2B). Diploids occur in areas where the maximum temperature of the warmest month (Bio 5) is higher than in those places occupied by octoploids (Supplementary Data Fig. S2C), and where the minimum temperature of the coldest month (Bio 6), the mean temperature of the driest quarter (Bio 9) and the mean temperature of the coldest quarter (Bio 11) are higher than in those locations where hexa- and octoploids are found (Supplementary Data Fig. S2D−F). Locations occupied by tetra-, hexa- and octoploids are characterized by higher values of precipitation of the driest month (Bio 14) in comparison with those where diploids are present (Supplementary Data Fig. S2G). Hexa- and octoploids are found in locations where the precipitation of the warmest quarter (Bio 18), as well as the average and range values of NDVI (normalized difference vegetation index) are higher than in those areas occupied by diploids (Supplementary Data Fig. S2H, I, L). Likewise, values of the standard deviation of NDVI are higher in areas occupied by hexa- and octoploids than in those occupied by di- and tetraploids (Supplementary Data Fig. S2J). Regarding NDVI annual maximum, significant differences were found between areas occupied by diploids and hexaploids (Supplementary Data Fig. S2K).
Finally, the distribution of cytotypes was found to be significantly affected by the potential vegetation type and biogeographic region (Supplementary Data Fig. S3). Diploids occur in areas with different potential types of vegetation, but they tend to be more frequent in thermophilous mixed deciduous broadleaved forests, with 35 % of populations occurring in this potential vegetation type, followed by potential Mediterranean sclerophyllous forests and scrub (17 %) and potential mesophytic broadleaved deciduous and mixed broadleaved/conifer forests (17 %). Tetraploids are preferentially found in areas with potential thermophilous mixed deciduous broadleaved forests (43 %) and potential mesophytic broadleaved deciduous and mixed broadleaved/conifer forests (35 %). Hexaploids and octoploids are mainly found in potential mesophytic broadleaved deciduous and mixed broadleaved/conifer forests (65 and 50 %, respectively) and to a lesser extent in potential thermophilous mixed deciduous broadleaved forests (28 and 26 %, respectively). Regarding biogeographical regions, diploids occur preferentially in the Mediterranean region, whereas octoploids are mainly found in alpine regions. Hexaploids are preferentially distributed in the continental region and tetraploids are evenly distributed in the Mediterranean and alpine regions.
Multivariate analysis
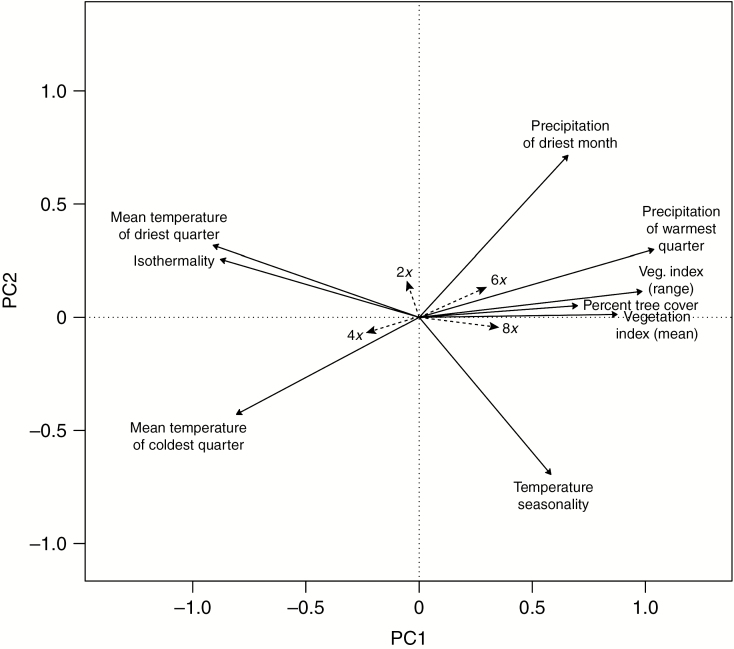
The overall and paired tests showed that plants of different ploidy levels are ecologically differentiated, with the largest niche differences between di- and hexaploids (Tables 2 and 3; Fig. 2). Significant differences were also found between di- and octoploids, and between tetra- and hexaploids. In general, diploids occur predominantly in areas characterized by higher isothermality (Bio 3), and higher values of mean temperature of the driest and coldest quarter (Bio 9 and Bio 11, respectively), as compared with those areas where hexaploids and octoploids grow. Tetraploids tend to occupy locations with similar climatic characteristics to that of diploids [i.e. high isothermality (Bio 3) and high values of mean temperature during the driest quarter (Bio 9)]. No significant differences were found between the areas occupied by hexaploids and octoploids, defined by high levels of precipitation both during the warmest quarter (Bio 18) and during the driest month (Bio 14), as well as vegetation index (NDVI) values. Areas occupied by hexaploids were also characterized by higher temperature seasonality (Bio 4) and higher tree coverage (Tree Cover) than those of diploids and tetraploids.
Table 2.
Summary of the multivariate analyses using constrained PCoA (db-RDA) applied to environmental variables recorded in populations of Veronica subsect. Pentasepalae
| Model | Trace (first first axis) | F | P |
|---|---|---|---|
| Overall model | 1.212 | 5.93 | <0.001 |
| 2x vs. 4x | 0.161 | 1.819 | 0.047 |
| 2x vs. 6x | 0.969 | 11.638 | <0.001 |
| 2x vs. 8x | 0.759 | 9.441 | <0.001 |
| 4x vs. 6x | 0.384 | 4.596 | <0.001 |
| 4x vs. 8x | 0.195 | 2.346 | 0.016 |
| 6x vs. 8x | 0.148 | 1.851 | 0.046 |
Significance level was assessed by an ANOVA-like permutation test by which the environmental matrix was randomly permuted 999 times.
P-values in bold are significant after Bonferroni correction (P < 0.001).
Table 3.
Survey of environmental variables which are best correlated (|r| > 0.5) with occurrence of ploidy levels in constrained PCoA (db-RDA) applied to environmental variables recorded in populations of Veronica subsect. Pentasepalae
| 2x (–) vs. 6x (+) | 2x (–) vs. 8x (+) | 4x (–) vs. 6x (+) | ||||||
|---|---|---|---|---|---|---|---|---|
| Code | Variable | r | Code | Variable | r | Code | Variable | r |
| Bio 18 | Precipitation of warmest quarter | 0.78 | Bio 18 | Precipitation of warmest quarter | 0.86 | Bio 4 | Temperature seasonality | 0.67 |
| NDVI ran | Vegetation index (range) | 0.77 | NDVI ran | Vegetation index (range) | 0.75 | NDVI ran | Vegetation index (range) | 0.67 |
| NDVI av | Vegetation index (average) | 0.63 | NDVI av | Vegetation index (average) | 0.67 | NDVI av | Vegetation index (average) | 0.57 |
| Bio 4 | Temperature seasonality | 0.58 | Bio 14 | Precipitation of driest month | 0.61 | Tree cover | Tree cover | 0.55 |
| Tree cover | Tree cover | 0.55 | Bio 3 | Isothermality | –0.61 | Bio 9 | Mean temperature of driest quarter | –0.57 |
| Bio 14 | Precipitation of driest month | 0.55 | Bio 11 | Mean temperature of coldest quarter | –0.67 | Bio 3 | Isothermality | –0.60 |
| Bio 9 | Mean temperature of driest quarter | –0.64 | Bio 9 | Mean temperature of driest quarter | –0.71 | |||
| Bio 11 | Mean temperature of coldest quarter | –0.65 | ||||||
| Bio 3 | Isothermality | –0.70 |
Within each analysis, variables show the positive or negative correlation with the first axis. The sign of correlation coefficients corresponds to the position of the respective ploidy level along the first axis within each analysis.
Fig. 2.
Constrained PCoA (db-RDA) showing the first and the second axes. Vectors of the environmental variables that were found to be correlated with the distribution of ploidy levels at |r| > 0.5 were used as supplementary data to help in the interpretation of the ordination.
DISCUSSION
Ploidy levels and estimates of genome size in V. subsection Pentasepalae
This study represents the first comprehensive ploidy screening for V. subsect. Pentasepalae covering its whole geographic distribution in Europe and North Africa. The results indicate that diploids are the most common cytotype in the group, whereas the tetraploid cytotype is the least frequent. Although previously reported by chromosome counts (Peev, 1972; Strid, 1986), the decaploid level was not detected, which indicates that this cytotype is not frequent in nature.
Genome size values range between 0.70 and 2.88 pg (1C-value), fitting in the middle range of genome sizes known for Veronica (Meudt et al., 2015). As found for other groups of the genus, comparison of monoploid genome size values (1Cx-values) obtained for V. subsect. Pentasepalae show that the polyploids have experienced genome downsizing with respect to the diploids (Supplementary Data Fig. S1). It is also worth noting that hexaploids and octoploids show stronger genome downsizing and are more widespread than tetraploids, which are the least frequent cytotype within the group. Thus, polyploidy and genome downsizing might be associated with the successful colonization of new habitats in V. subsect. Pentasepalae. Although Meudt et al. (2015) found no significant genome downsizing within V. subsect. Pentasepalae, these results are not in contradiction to those obtained here. These authors considered DNA loss for a sub-set of our sampling across the whole genus and with respect to the ancestral genome size of the genus Veronica, while here genome downsizing is detected for the polyploid taxa within the subsection (Supplementary Data Fig. S1).
The genome size of one population from north-western Spain (Supplementary Data Table S1; population 129) was slightly higher than most of the genome size values of the tetraploid group (Supplementary Data Table S1; Fig. S1). We are, nevertheless, confident that this population is tetraploid. Previous analyses showed an autopolyploid origin of these tetraploids (Martínez-Ortega et al., 2004) from diploids that show a relatively higher genome size (0.96 pg) compared with other diploids from the subsection (0.85 pg). Thus, higher values for those tetraploids are expected. The tetraploid level assigned to these individuals is also based on chromosome counts for other tetraploid individuals from the same area (Martínez-Ortega et al., 2004).
Although the absence of odd ploidies and aneuploidy in V. subsect. Pentasepalae could be due to the low intra-population sampling, our results are consistent with the lack of aneuploids or intermediate ploidies in >200 chromosome counts made to date for V. subsect. Pentasepalae (Krpač, 2000; Martínez-Ortega et al., 2004; Albach et al., 2008; chromosome numbers from the literature are available at: https://www.researchgate.net/publication/258769259_cariologia2013;Delgado et al., 2018). Experimental crosses carried out by Scheerer (1949) demonstrated that normal seeds (in number and morphology) were only produced from crosses between plants of the same ploidy level. Thus, as previously suggested for other plant species (Duchoslav et al., 2010), it is possible that intermediate ploidies or aneuploids have never been detected within V. subsect. Pentasepalae due to their non-viability. For example, gametic barriers are usually strong in inter-cytotype crosses and may include pollen–pistil interactions (e.g. Husband et al., 2002) and/or or genomic imprinting (e.g. unbalanced ratio of maternal to paternal genomes in endosperm tissue; Grossniklaus et al., 2001). The absence of triploids and other odd ploidies should be further confirmed by analysing a higher number of individuals per population and the reproductive interactions between ploidy levels to evaluate whether strong reproductive (pre- or post-zygotic incompatibilities) or ecological barriers (caused by habitat differentiation) exist among cytotypes as is the case for other species (Lumaret et al., 1987; Lumaret and Barrientos, 1990; Bretagnolle and Thompson, 1996; Thompson, 2005; Kolář et al., 2009; Husband et al., 2016).
Patterns of cytotype ecogeographic distribution
Cytotypes of V. subsect. Pentasepalae show a general pattern of geographic distribution at a broad scale, with diploids preferentially distributed in southern Europe and North Africa, and octoploids in central and northern areas of Europe (Fig. 1A). Additionally, hexaploids are geographically restricted to eastern and south-eastern Europe, while tetraploids are restricted to specific areas without clear biogeographic distribution patterns. The complex geographic distribution of cytotypes within V. subsect. Pentasepalae can be explained by different mechanisms: ecological differentiation of plants with different ploidy level, biogeographic history associated with the Quaternary glaciations and other local-level processes. These processes, that are not mutually exclusive and might have acted in combination, are discussed in detail below.
Ecological differentiation among ploidy levels
Univariate and multivariate analyses demonstrate a significant association between the distribution of cytotypes and climatic variables related to temperature and precipitation, as well as vegetation variables (Fig. 2; Tables 2 and 3; Supplementary Data Table S4, Figs S2, S3A and S4A). Consistently, the values of these variables are congruent with the association of ploidy levels with specific biogeographical regions (Supplementary Data Figs S3B and S4B).
Diploids are associated with higher values of isothermality and temperature, whereas hexa- and octoploids are positively correlated with higher values of precipitation and vegetation indexes (Table 3). Niche divergence in hexaploids is also determined by tree coverage and high temperature seasonality (Fig. 2; Table 3). Accordingly, diploids are mainly found in the Mediterranean region and hexaploids in the continental region of southern and south-eastern Europe (Supplementary Data Fig. S3B). Octoploids occur mainly in the alpine region, in areas characterized by mild and humid summers with high vegetation coverage (Fig. 2; Supplementary Data Fig. S3B; Table 3). The occurrence of polyploids in colder and wetter environments than those of their diploid progenitors has been demonstrated in other plant groups, such as Primula sect. Aleuritia or Tolmiea, among others (Martin and Husband, 2009; Theodoridis et al., 2013; Visger et al., 2016; Muñoz-Pajares et al., 2018). These results contrast with the idea that polyploids are more tolerant to water stress (Levin, 2002; Li et al., 2009; Manzaneda et al., 2012; Paule et al., 2017) and show higher fitness than diploids in drought conditions (Ramsey, 2011). Due to larger cell sizes, polyploids usually have larger but fewer stomata, which reduce transpiration rates and water loss, enabling polyploids to inhabit dry environments (Li et al., 2009; Liu et al., 2011). However, this is not always the case, as found in Tolmiea, where stomata size and number did not differ between cytotypes (Visger et al., 2016). Within V. subsect. Pentasepalae, previous studies have shown that stomata were larger in tetraploid than in diploid plants (Brandt, 1961), but this study was restricted to a couple of taxa, and more exhaustive analyses should be performed including all the species from the subsection. Wider xylem diameters have also been found to improve water use efficiency in some polyploids (Pockman and Sperry, 1997; Maherali et al., 2009), but no information about this character is available in Veronica yet. Physiological and anatomical features related to drought stress need to be investigated in V. subsect. Pentasepalae, preferably coupled with reciprocal transplant studies to demonstrate adaptation of diploids to drier regions.
With respect to tetraploids, our analyses demonstrated that they are ecologically differentiated only with respect to hexaploids. Tetraploids occur mainly in two biogeographical regions: the Mediterranean (e.g. western coast of the Balkan Peninsula) and the alpine region (e.g. Pyrenees and Dinaric Alps) (Supplementary Data Figs S3B and S4B), which are the regions where diploids and octoploids, respectively, are mainly found. This overlap in the biogeographical region might explain the lack of differences in the ecological preferences of diploids vs. tetraploids, and tetraploids vs. octoploids. Despite the common assumption that WGD confers competitive advantages to polyploids (Maceira et al., 1993; te Beest et al., 2012; Thompson et al., 2014), this is not always the case. For instance, tetraploids of Ranunculus kuepferi Greuter & Burdet are outcompeted by diploids in warmer conditions (Kirchheimer et al., 2016). Within V. subsect. Pentasepalae, the ecological requirements of high polyploids are different when compared with low ploidy cytotypes in the subsection. Moreover, tetraploids are found in a much smaller number of potential vegetation types than diploids and octoploids. It might be possible that tetraploids exhibit small advantages over diploids, and they are outcompeted by hexa- or octoploids. Further experiments comparing fitness rates of the different ploidy levels will be necessary to fully demonstrate whether the correlation found among environmental parameters and the distribution patterns of cytotypes observed is due to adaptive advantages of polyploids.
Our results demonstrate that polyploidy has been able to counteract phylogenetic niche conservatism in a recently diverged diploid–polyploid complex composed of numerous closely related species. Similar observations have been found in other polyploid species groups within particular genera of angiosperms (Brochmann and Elven, 1992; Theodoridis et al., 2013; Paule et al., 2017). Interestingly, subgenus Pentasepalae has supposedly evolved in xeric habitats from the Middle East, thus suggesting that the ancestors of V. subsect. Pentasepalae were adapted to xeric conditions (Albach et al., 2004). Polyploidization, alone or together with hybridization, could have enhanced environmental adaptations to cold and moist environments, which suggests that the ecological differentiation of cytotypes observed in the group might be explained by the adaptive evolutionary scenario.
Biogeographical history
Species divergence at the diploid level within V. subsect. Pentasepalae has been estimated to have started by the end of the Pliocene–early Pleistocene (Meudt et al., 2015), with polyploids arising later, although no exact date for the origin of the polyploids is available yet. Thus, Quaternary glaciations have probably had an impact on species diversification as well as on their current distribution. In fact, the observed distribution of cytotypes in V. subsect. Pentasepalae partially conforms to patterns of glacial recolonization, with diploids or lower ploidy levels mainly occupying southern refugial areas and octoploids being more frequent in northern areas (Fig. 1). This south to north distribution pattern has been frequently shown in other plant polyploid complexes in Europe (e.g. van Dijk and Bakx-Schotman, 1997; Luttikhuizen et al., 2007; Balao et al., 2009; Bardy et al., 2010; Theodoridis et al., 2013; Kolář et al., 2016), while the opposite pattern seems to be quite uncommon (but see Durand, 1963). It has been recently proposed that at the time of the LGM, the southern extent of continuous permafrost (i.e. occurring everywhere in a given region) reached approximately the latitudes 44–47°N in Europe (Vandenberghe et al., 2014). Within V. subsect. Pentasepalae, octoploids are the main cytotype that is, nowadays, distributed in areas predicted to have been under permafrost during the LPM towards the end of the last ice age (Fig. 1). Thus, the permafrost extent, which is obviously related to Quaternary climatic oscillations, is probably also crucial to understand the geographical patterns observed nowadays for groups of plants distributed in Europe. The results obtained here suggest that octoploids of V. subsect. Pentasepalae might have been successful at establishing new populations in lowland regions that are assumed to have experienced continuous permafrost conditions (i.e. a mean annual air temperature of –8 °C or lower) during the LPM, and in mountain areas that became available after glacial retreat (Fig. 1). The fact that octoploids of V. subsect. Pentasepalae seem to be better colonizers in these environments could be related to the effects of WGD (i.e. genetic and physiological changes). This pattern has been found in other plant species such as Achillea borealis Bong. (Ramsey, 2011), among others (Brochmann et al., 2004; Wefferling et al., 2017). While many studies have demonstrated an influence of the ice retreat on the distribution patterns of polyploids (see references in Ramsey and Ramsey, 2014; Rice et al., 2019), more emphasis should be put on permafrost coverage as a driver that shaped the biogeographic patterns of polyploid groups in temperate regions of Europe.
Additionally, Quaternary climatic oscillations may have promoted recurrent events of polyploidization or repeated secondary contacts among otherwise geographically isolated taxa of V. subsect. Pentasepalae giving rise to homoploid and/or allopolyploid hybrids. The recurrent formation of polyploids has been demonstrated in multiple plant groups (Soltis and Soltis, 1999; McAllister and Miller, 2016), including species of the genus Veronica (Albach, 2007; Bardy et al., 2010). Within V. subsect. Pentasepalae, recent studies have indicated that octoploid plants from western Europe have probably resulted from multiple polyploidization events involving a tetraploid from western Europe (Padilla-García et al., 2018). A higher genetic variability derived from gene flow among new octoploid populations might have contributed to the successful establishment of these octoploid entities in new environments.
Differential rate of cytotype formation
Other processes affecting the mode and frequency of polyploid formation certainly influenced the cytotype distribution patterns of V. subsect. Pentasepalae. Thus, a crucial factor in the emergence of new cytotypes of V. subsect. Pentasepalae is the production rate of unreduced gametes. Previous studies support the link between environmental stress (i.e. extreme temperature, water stress, nutritional stress or herbivory) and the production of unreduced gametes (Brochmann and Elven, 1992; Ramsey and Schemske, 1998; Ramsey, 2007, Parisod et al., 2010). The presence of octoploids mainly in areas covered by permafrost during the LGM may be linked to increased rates of production of unreduced gametes under climatic fluctuation scenarios (Ramsey and Schemske, 1998; McAllister et al., 2015) which, in turn, would have favoured the recurrent formation of octoploids in the subsection (Padilla-García et al., 2018). It should be pointed out that a higher production of unreduced gametes under environmental stress might be common in all cytotypes. Consequently, it is worth noting, once again, that octoploids might have some ecological advantage over other cytotypes that allowed them to colonize central and northern Europe.
Mixed-ploidy populations
Despite the small intra-population sample size, we still observed cytotype variation in 6.27 % of populations (Table 1). The possibility that our results have underestimated the rate of cytotype admixture cannot be neglected since only three individuals per population were sampled for most of the populations studied. However, subsequent analyses carried out in the western Balkans using an average of approx. 20 individuals per population found similar within-population homogeneous ploidy levels (N. López-González et al., University of Salamanca, Spain, unpubl. res.). According to the MCE model, the establishment of a new cytotype within a population is frequency dependent (Levin, 1975). Thus, the coexistence of different cytotypes within a population depends on, among other factors, the development of reproductive barriers, different microhabitat requirements, recurrent polyploidization events (through unreduced gamete production) or differential competitiveness among cytotypes counteracting the MCE (Levin, 1975; Husband and Sabara, 2004; Thompson, 2005; Collins et al., 2011; Husband et al., 2016; Čertner et al., 2017). Vegetative propagation is an important mode of expansion for plants of V. subsect. Pentasepalae. All species have a very branched rhizome that produces numerous adventitious roots and new vegetative shoots (Watzl, 1910). In addition, the production of unreduced gametes in sub-optimal ecological conditions might be an additional factor favouring the local coexistence of ploidy levels. It would be very interesting to explore to what extent and under which circumstances these factors are able to counteract MCE in V. subsect. Pentasepalae.
Five of the 13 mixed-ploidy populations found in our study are located in the western Balkans. The Balkan Peninsula, at the crossroads of several major floras, represents one of the most important hotspots of biodiversity in Europe (Griffiths et al., 2004). According to morphology, two of these mixed-ploidy populations (Supplementary Data Table S1; populations 5 and 114) consist of putative hybrids or taxonomically unresolved entities that may have resulted from secondary contacts among diverging lineages. They are found in the southern Dinaric Alps, an area of special topographic complexity that acted as a Quaternary microrefugium for plant species (e.g. Surina et al., 2011; Kutnjak et al., 2014). Thus, these mixed-ploidy populations are assumed to be the result of repeated admixture of lineages during the Quaternary climatic oscillations up until today, as seen for other morphologically intermediate populations of V. subsect. Pentasepalae from the western Balkans (Padilla-García et al., 2018). In contrast, spontaneous polyploidization (i.e. primary contact) might be responsible for the other two mixed-ploidy populations (Supplementary Data Table S1; populations 22 and 24), which have been demonstrated to have autopolyploid origins (Padilla-García et al., 2018). Altogether, these findings point to the existence of primary and secondary contact zones mainly in the Balkan Peninsula for V. subsect. Pentasepalae.
CONCLUSIONS
This work provides the first comprehensive ploidy screening within V. subsect. Pentasepalae at a broad scale and represents a solid baseline for future studies on polyploid evolution in this subsection. Four ploidy levels (2x, 4x, 6x and 8x) at different frequencies have been found in the group. Polyploids exhibit genome downsizing, suggesting that this process might also play a role in the evolution of the group and its expansion to new or alternative ecological niches. The geographic distribution of cytotypes can be explained by ecological differentiation mainly between diploids and octoploids, and diploids and hexaploids, suggesting that high polyploids might cope better in colder and wetter regions. Our findings also indicate that phylogenetic niche conservatism has been overcome in a diploid–polyploid complex composed of closely related species. The extent of permafrost during the Quaternary glaciations, as well as a high production rate of unreduced gametes associated with present or historical stressful climatic conditions might also have influenced the frequency and distribution patterns of cytotypes in V. subsect. Pentasepalae. In particular, the distribution of octoploids could be related to the LPM extension. Cytotype coexistence is mainly detected in the western Balkans, indicating that it is an important contact zone among cytotypes. Future work should focus on these contact areas at a finer scale including higher intra-population sampling, reciprocal transplants, common garden experiments and synthetic polyploids, together with genomic analyses, to confirm if ecological differentiation of cytotypes is effectively caused by polyploidy.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: observed and expected genome size values in Veronica subsect. Pentasepalae. Figure S2: box plots of the 12 quantitative environmental variables that are significantly associated with the distribution of cytotypes. Figure S3: relative frequencies of cytotypes in relation to the three most common vegetation types and the three most common biogeographical regions. Figure S4: distribution of cytotypes in relation to vegetation type, and biogeographical regions. Table S1: details of individuals of Veronica subsect. Pentasepalae included in this study. Table S2: mean and standard deviation of genome sizes for each ploidy. Table S3: environmental variables used in univariate and multivariate analyses. Table S4: associations between ploidy levels and environmental variables in populations of Veronica subsect. Pentasepalae found by the univariate analyses.
ACKNOWLEDGEMENTS
We thank A. Abad de Blas, S. Andrés, S. Barrios, A. Juan, V. Lucía, J. Peñas, D. Pinto, E. Rico, T. Romero, M. Santos and all other colleagues who helped to collect material for this study. We are particularly grateful to X. Giráldez who joined us during every field campaign. We thank S. Kempen and D. Pinto for help with flow cytometry. We thank Martin Lysak, Trude Schwarzacher and three anonymous reviewers for their constructive comments and suggestions, which greatly improved this work. M.M.O. and B.R.A. designed the research. M.M.O., B.R.A., N.L.G., N.P.G. and L.D. collected samples. B.R.A. and N.L.G. performed ploidy level estimations. M.P. performed statistical analyses. M.P., B.R.A., N.P.G., N.L.G. and L.D. prepared tables and figures. M.M.O. co-ordinated and supervised all these tasks, except for ploidy level estimations that were supervised by S.C., J.L. and D.C.A. N.P.G., B.R.A., M.P. and M.M.O. wrote the manuscript and all authors revised and actively improved the paper.
FUNDING
This work was supported by the Spanish Ministerio de Economía y Competitividad [projects CGL2009-07555 and CGL2012-32574]; the Spanish Ministerio de Ciencia e Innovación [PhD grants AP2008-03434 to B.R.A. and AP2010-2968 to N.L.G.]; and the University of Salamanca [PhD grant to N.P.G. co-financed by Banco Santander].
LITERATURE CITED
- Albach DC. 2007. Amplified fragment length polymorphisms and sequence data in the phylogenetic analysis of polyploids: multiple origins of Veronica cymbalaria (Plantaginaceae). New Phytologist 176: 481–498. [DOI] [PubMed] [Google Scholar]
- Albach DC, Martínez-Ortega MM, Chase MW. 2004. Veronica: parallel morphological evolution and phylogeography in the Mediterranean. Plant Systematics and Evolution 246: 177–194. [Google Scholar]
- Albach DC, Martinez-Ortega MM, Delgado L, Weiss-Schneeweiss H, Ozgokce F, Fischer MA. 2008. Chromosome numbers in Veroniceae (Plantaginaceae): review and several new counts. Annals of the Missouri Botanical Garden 95: 543–566. [Google Scholar]
- Arrigo N, Barker MS. 2012. Rarely successful polyploids and their legacy in plant genomes. Current Opinion in Plant Biology 15: 140–146. [DOI] [PubMed] [Google Scholar]
- Avise JC. 2000. Phylogeography: the history and formation of species. Cambridge, MA: Harvard University Press. [Google Scholar]
- Balao F, Casimiro-Soriguer R, Talavera M, Herrera J, Talavera S. 2009. Distribution and diversity of cytotypes in Dianthus broteri as evidenced by genome size variations. Annals of Botany 104: 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin SJ, Husband BC. 2013. The association between polyploidy and clonal reproduction in diploid and tetraploid Chamerion angustifolium. Molecular Ecology 22: 1806–1819. [DOI] [PubMed] [Google Scholar]
- Bardy KE, Albach DC, Schneeweiss GM, Fischer MA, Schönswetter P. 2010. Disentangling phylogeography, polyploid evolution and taxonomy of a woodland herb (Veronica chamaedrys group, Plantaginaceae s.l.) in southeastern Europe. Molecular Phylogenetics and Evolution 57: 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy KE, Schönswetter P, Schneeweiss GM, Fischer MA, Albach DC. 2011. Extensive gene flow blurs species boundaries among Veronica barrelieri, V. orchidea and V. spicata (Plantaginaceae) in southeastern Europe. Taxon 60: 108–121. [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, et al. 2012. The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany 109: 19–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M, Smith J. 1972. The effects of polyploidy on meiotic duration and pollen development in cereal anthers. Proceedings of the Royal Society B: Biological Sciences 181: 81–107. [Google Scholar]
- Brandt JP. 1961. Cytotaxomomie et cytogéographie de Veronica prostrata L. Bulletin de la Société Neuchâteloise des Sciences Naturelles 84: 35–80. [Google Scholar]
- Bretagnolle F, Thompson JD. 1996. An experimental study of ecological differences in winter growth between sympatric diploid and autotetraploid Dactylis glomerata. Journal of Ecology 84: 343–351. [Google Scholar]
- Brochmann C, Elven R. 1992. Ecological and genetic consequences of polyploidy in arctic Draba (Brassicaceae). Evolutionary Trends in Plants 6: 111–124. [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, et al. 2004. Polyploidy in arctic plants. Biological Journal of the Linnean Society 82: 521–536. [Google Scholar]
- Burns JH, Strauss SY. 2011. More closely related species are more ecologically similar in an experimental test. Proceedings of the National Academy of Sciences, USA 108: 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro M, Castro S, Figueiredo A, Husband B, Loureiro J. 2018. Complex cytogeographical patterns reveal a dynamic tetraploid–octoploid contact zone. AoB Plants 10: ply012. doi: 10.1093/aobpla/ply012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro M, Loureiro J, Serrano M, Husband B, Catarina S, Castro S. 2019. Mosaic distribution of cytotypes in a mixed-ploidy plant species, Jasione montana: nested environmental niches but low geographical overlap. Botanical Journal of the Linnean Society 190: 51–66. [Google Scholar]
- Cavalier-Smith T. 1978. Nuclear volume control by nucleoskeletal DNA, selection for cell volume and cell growth rate, and the solution of the DNA C-value paradox. Journal of Cell Science 34: 247–278. [DOI] [PubMed] [Google Scholar]
- Čertner M, Fenclova E, Kúr P, et al. 2017. Evolutionary dynamics of mixed-ploidy populations in an annual herb: dispersal, local persistence and recurrent origins of polyploids. Annals of Botany 120: 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Hadi AS. 2006. Regression analysis by example, 4th edn.Hoboken, NJ: John Wiley and Sons, Inc. [Google Scholar]
- Collins AR, Naderi R, Mueller-Schaerer H. 2011. Competition between cytotypes changes across a longitudinal gradient in Centaurea stoebe (Asteraceae). American Journal of Botany 98: 1935–1942. [DOI] [PubMed] [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nature Reviews. Genetics 6: 836–846. [DOI] [PubMed] [Google Scholar]
- van Dijk P, Bakx-Schotman T. 1997. Chloroplast DNA phylogeography and cytotype geography in autopolyploid Plantago media. Molecular Ecology 6: 345–352. [Google Scholar]
- van Dijk P, Hartog M, Delden WV. 1992. Single cytotype areas in autopolyploid Plantago media L. Biological Journal of the Linnean Society 46: 315–331. [Google Scholar]
- Delgado L, Rojas-Andrés BM, López-González N, Padilla-García N, Martínez-Ortega MM. 2018. Veronica angustifolia (Vahl) Bernh.; Veronica austriaca subsp. jacquinii (Baumg.) Watzl; Veronica dalmatica N.Pad.Gar., Rojas-Andrés, López-González & M.M.Mart.Ort.; Veronica kindlii Adam.; Veronica orsiniana Ten.; Veronica prostrata L.; Veronica rosea Desf.; Veronica sennenii (Pau) M.M.Mart.Ort. & E.Rico; Veronica tenuifolia subsp. javalambrensis (Pau) Molero & J.Pujadas; Veronica tenuifolia Asso subsp. tenuifolia; Veronica teucrium L.; Veronica thracica Velen. In: Marhold K, ed. IAPT/IOPB chromosome data 28. Taxon, 67: 1235–1236, E2–E7. [Google Scholar]
- Dolezel J, Greilhuber J, Lucretti S, et al. 1998. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Annals of Botany 82: 17–26. [Google Scholar]
- Duchoslav M, Safárová L, Krahulec F. 2010. Complex distribution patterns, ecology and coexistence of ploidy levels of Allium oleraceum (Alliaceae) in the Czech Republic. Annals of Botany 105: 719–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B. 1963. Le complexe Mercurialis annua L. s.l.: une étude biosystématique. Annales des Sciences Naturelles, Botanique, Paris: 12: 579–736. [Google Scholar]
- Etterson JR, Toczydlowski RH, Winkler KJ, Kirschbaum JA, McAulay TS. 2016. Solidago altissima differs with respect to ploidy frequency and clinal variation across the prairie–forest biome border in Minnesota. American Journal of Botany 103: 22–32. [DOI] [PubMed] [Google Scholar]
- Fausto JA, Eckhart VM, Geber MA. 2001. Reproductive assurance and the evolutionary ecology of self-pollination in Clarkia xantiana (Onagraceae). American Journal of Botany 88: 1794–1800. [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. 2009. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proceedings of the National Academy of Sciences, USA 106: 5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler NL, Levin DA. 1984. Ecological constraints on the establishment of a novel polyploid in competition with its diploid progenitor. The American Naturalist 124: 703–711. [Google Scholar]
- Fowler NL, Levin DA. 2016. Critical factors in the establishment of allopolyploids. American Journal of Botany 103: 1–16. [DOI] [PubMed] [Google Scholar]
- Glennon KL, Ritchie ME, Segraves KA. 2014. Evidence for shared broad-scale climatic niches of diploid and polyploid plants. Ecology Letters 17: 574–582. [DOI] [PubMed] [Google Scholar]
- Godsoe W, Larson MA, Glennon KL, Segraves KA. 2013. Polyploidization in Heuchera cylindrica (Saxifragaceae) did not result in a shift in climatic requirements. American Journal of Botany 100: 496–508. [DOI] [PubMed] [Google Scholar]
- Gower JC. 1985. Properties of Euclidean and non-Euclidean distance matrices. Linear Algebra and its Applications 67: 81–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biological Reviews 76: 65–101. [DOI] [PubMed] [Google Scholar]
- Greilhuber J, Ebert I. 1994. Genome size variation in Pisum sativum. Genome 37: 646–655. [DOI] [PubMed] [Google Scholar]
- Greilhuber J, Dolezel J, Lysák MA, Bennett MD. 2005. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Annals of Botany 95: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths HI, Krystufek B, Reed JM. 2004. Balkan biodiversity. Pattern and process in the European hotspot. Dordrecht, The Netherlands: Kluwer Academic Publishers. [Google Scholar]
- Grossniklaus U, Spillane C, Page DR, Köhler C. 2001. Genomic imprinting and seed development: endosperm formation with and without sex. Current Opinion in Plant Biology 4: 21–27. [DOI] [PubMed] [Google Scholar]
- Han T-S, Zheng Q-J, Onstein RE, et al. 2019. Polyploidy promotes species diversification of Allium through ecological shifts. New Phytologist (in press) doi: 10.1111/nph.16098. [DOI] [PubMed] [Google Scholar]
- Hao M, Luo J, Zhang L, et al. 2013. Production of hexaploid triticale by a synthetic hexaploid wheat–rye hybrid method. Euphytica 193: 347–357. [Google Scholar]
- Harvey PH, Pagel MD. 1991. The comparative method in evolutionary biology. Oxford: Oxford University Press. [Google Scholar]
- Heiberger RM. 2015. HH: statistical analysis and data display: Heiberger and Holland. R package version 3.1-23. http://CRAN.R-project.org/package=HH [Google Scholar]
- Hewitt GM. 1999. Post-glacial re-colonization of European biota. Biological Journal of the Linnean Society 68: 87–112. [Google Scholar]
- Hijmans R, Cameron S, Parra J, Jones P, Jarvis A. 2005. Very high resolution interpolated climate surfaces for global land areas. International Journal of Climatology 25: 1965–1978. [Google Scholar]
- Hijmans RJ, Gavrilenko T, Stephenson S, Bamberg J, Salas A, Spooner DM. 2007. Geographical and environmental range expansion through polyploidy in wild potatoes (Solanum section Petota). Global Ecology and Biogeography 16: 485–495. [Google Scholar]
- Husband BC, Sabara HA. 2004. Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae). New Phytologist 161: 703–713. [DOI] [PubMed] [Google Scholar]
- Husband BC, Schemske DW. 1998. Cytotype distribution at a diploid–tetraploid contact zone in Chamerion (Epilobium) angustifolium (Onagraceae). American Journal of Botany 85: 1688–1694. [PubMed] [Google Scholar]
- Husband BC, Schemske DW. 2000. Ecological mechanisms of reproductive isolation between diploid and tetraploid Chamerion angustifolium. Journal of Ecology 88: 689–701. [Google Scholar]
- Husband BC, Schemske DW, Burton TL, Goodwillie C. 2002. Pollen competition as a unilateral reproductive barrier between sympatric Chamerion angustifolium. Proceedings of the Royal Society B: Biological Sciences 269: 2565–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husband BC, Baldwin SJ, Sabara HA. 2016. Direct vs. indirect effects of whole-genome duplication on prezygotic isolation in Chamerion angustifolium: implications for rapid speciation. American Journal of Botany 103: 1259–1271. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Kadereit JW. 2015. The geography of hybrid speciation in plants. Taxon 64: 673–687. [Google Scholar]
- Keeler KH, Davis GA. 1999. Comparison of common cytotypes of Andropogon gerardii (Andropogoneae, Poaceae). American Journal of Botany 86: 974–979 [PubMed] [Google Scholar]
- Kellogg EA. 2016. Has the connection between polyploidy and diversification actually been tested? Current Opinion in Plant Biology 30: 25–32. [DOI] [PubMed] [Google Scholar]
- Kirchheimer B, Schinkel CCF, Dellinger AS, et al. 2016. A matter of scale: apparent niche differentiation of diploid and tetraploid plants may depend on extent and grain of analysis. Journal of Biogeography 43: 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolář F, Štech M, Trávníček P, et al. 2009. Towards resolving the Knautia arvensis agg. (Dipsacaceae) puzzle: primary and secondary contact zones and ploidy segregation at landscape and microgeographic scales. Annals of Botany 103: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolář F, Lučanová M, Záveská E, et al. 2016. Ecological segregation does not drive the intricate parapatric distribution of diploid and tetraploid cytotypes of the Arabidopsis arenosa group (Brassicaceae). Biological Journal of the Linnean Society 119: 673–688. [Google Scholar]
- Kraaijeveld K. 2010. Genome size and species diversification. Evolutionary Biology 37: 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejčíková J, Sudová R, Oberlander K, Dreyer L, Suda J. 2013. The spatio-ecological segregation of different cytotypes of Oxalis obtusa (Oxalidaceae) in contact zones. South African Journal of Botany 88: 62–68. [Google Scholar]
- Krpač VT. 2000. Taksonomija I horologija na rodot Veronica L. (Scrophulariaceae) vo Skopskata kotlina. Master Thesis, Ss. Cyril and Methodius University in Skopje, Republic of Macedonia (FYROM). [Google Scholar]
- Kutnjak D, Kuttner M, Niketić M, Dullinger S, Schönswetter P, Frajman B. 2014. Escaping to the summits: phylogeography and predicted range dynamics of Cerastium dinaricum, an endangered high mountain plant endemic to the western Balkan Peninsula. Molecular Phylogenetics and Evolution 78: 365–374. [DOI] [PubMed] [Google Scholar]
- Laliberté E, Legendre P, Shipley B.2014. https://cran.r-project.org/web/packages/FD/FD.pdf FD: Measuring functional diversity from multiple traits, and other tools for functional ecology. R package version 1.0-12.
- Legendre P, Anderson MJ. 1999. Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecological Monographs 69: 1–24. [Google Scholar]
- Leitch IJ, Bennett MD. 2004. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society 82: 651–663. [Google Scholar]
- Levin DA. 1975. Minority cytotype exclusion in local plant populations. Taxon 24: 35–43. [Google Scholar]
- Levin DA. 2000. The origin, expansion, and demise of plant species. Oxford: Oxford University Press. [Google Scholar]
- Levin D. 2002. The role of chromosomal change in plant evolution. Oxford: Oxford University Press. [Google Scholar]
- Levin DA, Soltis DE. 2018. Factors promoting polyploid persistence and diversification and limiting diploid speciation during the K–Pg interlude. Current Opinion in Plant Biology 42: 1–7. [DOI] [PubMed] [Google Scholar]
- Li WD, Biswas DK, Xu H, et al. 2009. Photosynthetic responses to chromosome doubling in relation to leaf anatomy in Lonicera japonica subjected to water stress. Functional Plant Biology 36: 783–792. [DOI] [PubMed] [Google Scholar]
- Liu S, Chen S, Chen Y, Guan Z, Yin D, Chen F. 2011. In vitro induced tetraploid of Dendranthema nankingense (Nakai) Tzvel. shows an improved level of abiotic stress tolerance. Scientia Horticulturae 127: 411–419. [Google Scholar]
- Lord J, Westoby M, Leishman M. 1995. Seed size and phylogeny in six temperate floras: constraints, niche conservatism, and adaptation. The American Naturalist 146: 349–364. [Google Scholar]
- Losos JB. 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecology Letters 11: 995–1003. [DOI] [PubMed] [Google Scholar]
- Loureiro J, Rodriguez E, Dolezel J, Santos C. 2007. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Annals of Botany 100: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumaret R, Guillerm JL, Delay J, Loutfi AAL, Izco J, Jay M. 1987. Polyploidy and habitat differentiation in Dactylis glomerata L. from Galicia (Spain). Oecologia 73: 436–446. [DOI] [PubMed] [Google Scholar]
- Lumaret R, Barrientos E. 1990. Phylogenetic relationships and gene flow between sympatric diploid and tetraploid plants of Dactylis glomerata (Gramineae). Plant Systematics and Evolution 169: 81–96. [Google Scholar]
- Luttikhuizen PC, Stift M, Kuperus P, Van Tienderen PH. 2007. Genetic diversity in diploid vs. tetraploid Rorippa amphibia (Brassicaceae). Molecular Ecology 16: 3544–3553. [DOI] [PubMed] [Google Scholar]
- Lysak MA, Dolezel J. 1998. Estimation of nuclear DNA content in Sesleria (Poaceae). Caryologia 51: 123–132. [Google Scholar]
- Maceira NO, Jacquard P, Lumaret R. 1993. Competition between diploid and derivative autotetraploid Dactylis glomerata L. from Galicia. Implications for the establishment of novel polyploid populations. New Phytologist 124: 321–328. [DOI] [PubMed] [Google Scholar]
- Maherali H, Walden AE, Husband BC. 2009. Genome duplication and the evolution of physiological responses to water stress. New Phytologist 184: 721–731. [DOI] [PubMed] [Google Scholar]
- Mairal M, Šurinová M, Castro S, Münzbergová Z. 2018. Unmasking cryptic biodiversity in polyploids: origin and diversification of Aster amellus aggregate. Annals of Botany 122:1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T, Münzbergová Z. 2006. Distribution and ecology of cytotypes of the Aster amellus aggregates in the Czech Republic. Annals of Botany 98: 845–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzaneda AJ, Rey PJ, Bastida JM, Weiss-Lehman C, Raskin E, Mitchell-Olds T. 2012. Environmental aridity is associated with cytotype segregation and polyploidy occurrence in Brachypodium distachyon (Poaceae). New Phytologist 193: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant BD, Soltis DE, Soltis PS. 2016. Patterns of abiotic niche shifts in allopolyploids relative to their progenitors. New Phytologist 212: 708–718. [DOI] [PubMed] [Google Scholar]
- Martin SL, Husband BC. 2009. Influence of phylogeny and ploidy on species ranges of North American angiosperms. Journal of Ecology 97: 913–922. [Google Scholar]
- Martínez-Ortega MM, Delgado L, Albach DC, Elena-Rosselló JA, Rico E. 2004. Species boundaries and phylogeographic patterns in cryptic taxa inferred from AFLP markers: Veronica subgen. Pentasepalae (Scrophulariaceae) in the western Mediterranean. Systematic Botany 29: 965–986. [Google Scholar]
- Mayrose I, Zhan SH, Rothfels CJ, et al. 2011. Recently formed polyploid plants diversify at lower rates. Science 333: 1257–1257. [DOI] [PubMed] [Google Scholar]
- McAllister CA, Miller AJ. 2016. Single nucleotide polymorphism discovery via genotyping by sequencing to assess population genetic structure and recurrent polyploidization in Andropogon gerardii. American Journal of Botany 103: 1314–1325. [DOI] [PubMed] [Google Scholar]
- McAllister C, Blaine R, Kron P, et al. 2015. Environmental correlates of cytotype distribution in Andropogon gerardii (Poaceae). American Journal of Botany 102: 92–102. [DOI] [PubMed] [Google Scholar]
- McHale NA. 1983. Environmental induction of high frequency 2n pollen formation in diploid Solanum. Canadian Journal of Genetics and Cytology 25: 609–615. [Google Scholar]
- Meimberg H, Rice KJ, Milan NF, Njoku CC, McKay JK. 2009. Multiple origins promote the ecological amplitude of allopolyploid Aegilops (Poaceae). American Journal of Botany 96: 1262–1273. [DOI] [PubMed] [Google Scholar]
- Meudt HM, Rojas-Andrés BM, Prebble JM, Low E, Garnock-Jones PJ, Albach DC. 2015. Is genome downsizing associated with diversification in polyploid lineages of Veronica? Botanical Journal of the Linnean Society 178: 243–266. [Google Scholar]
- Muñoz-Pajares AJ, Perfectti F, Loureiro J, et al. 2018. Niche differences may explain the geographic distribution of cytotypes in Erysimum mediohispanicum. Plant Biology 20: 139–147. [DOI] [PubMed] [Google Scholar]
- Narum SR. 2006. Beyond Bonferroni: less conservative analyses for conservation genetics. Conservation Genetics 7: 783–787. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, et al. 2013. https://cran.r-project.org/package=vegan vegan: community ecology package. R package version 2.0-7.
- Otto S, Whitton J. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Padilla-García N, Rojas-Andrés BM, López-González N, et al. 2018. The challenge of species delimitation in the diploid–polyploid complex Veronica subsection Pentasepalae. Molecular Phylogenetics and Evolution 119: 196–209. [DOI] [PubMed] [Google Scholar]
- Pandit MK, White SM, Pocock MJO. 2014. The contrasting effects of genome size, chromosome number and ploidy level on plant invasiveness: a global analysis. New Phytologist 203: 697–703. [DOI] [PubMed] [Google Scholar]
- Parisod C, Holderegger R, Brochmann C. 2010. Evolutionary consequences of autopolyploidy. New Phytologist 186: 5–17. [DOI] [PubMed] [Google Scholar]
- Paule J, Sharbel TF, Dobeš C. 2011. Apomictic and sexual lineages of the Potentilla argentea L. group (Rosaceae): cytotype and molecular genetic differentiation. Taxon 60: 721–732. [Google Scholar]
- Paule J, Wagner ND, Weising K, Zizka G. 2017. Ecological range shift in the polyploid members of the South American genus Fosterella (Bromeliaceae). Annals of Botany 120: 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paun O, Bateman RM, Fay MF, et al. 2011. Altered gene expression and ecological divergence in sibling allopolyploids of Dactylorhiza (Orchidaceae). BMC Evolutionary Biology 11: 113. doi: 10.1186/1471-2148-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peev D. 1972. New taxa and ploidy levels of some Bulgarian Veronica species. Doklady Bolgarskoi Akademii Nauk 25: 811–814. [Google Scholar]
- Pockman WT, Sperry JS. 1997. Freezing-induced xylem cavitation and the northern limit of Larrea tridentata. Oecologia 109: 19–27. [DOI] [PubMed] [Google Scholar]
- Ramsey J. 2007. Unreduced gametes and neopolyploids in natural populations of Achillea borealis (Asteraceae). Heredity 98: 143–150. [DOI] [PubMed] [Google Scholar]
- Ramsey J. 2011. Polyploidy and ecological adaptation in wild yarrow. Proceedings of the National Academy of Sciences, USA 108: 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Ramsey TS. 2014. Ecological studies of polyploidy in the 100 years following its discovery. Philosophical Transactions of the Royal Society B: Biological Sciences 369: 1–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annual Review of Ecology and Systematics 29: 467–501. [Google Scholar]
- R Development Core Team 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; Available from: http://www.R-project.org/ [Google Scholar]
- Rice A, Šmarda P, Novosolov M, et al. 2019. The global biogeography of polyploid plants. Nature Ecology & Evolution 3: 265–273. [DOI] [PubMed] [Google Scholar]
- Rodriguez DJ. 1996. A model for the establishment of polyploidy in plants. The American Naturalist 147: 33–46. [Google Scholar]
- Rojas-Andrés BM, Martínez-Ortega MM. 2016. Taxonomic revision of Veronica subsection Pentasepalae (Plantaginaceae sensu APG III). Phytotaxa 285: 1–100. [Google Scholar]
- Rojas-Andrés BM, Albach DC, Martínez-Ortega MM. 2015. Exploring the intricate evolutionary history of the diploid–polyploid complex Veronica subsection Pentasepalae (Plantaginaceae). Botanical Journal of the Linnean Society 179: 670–692. [Google Scholar]
- Scalone R, Albach DC. 2014. Cytological evidence for gametophytic self-incompatibility in the genus Veronica. Turkish Journal of Botany 38: 197–201. [Google Scholar]
- Scalone R, Kolf M., Albach DC. 2013. Mating system variation in Veronica (Plantaginaceae): inferences from pollen/ovule ratios and other reproductive traits. Nordic Journal of Botany 31: 372–384. [Google Scholar]
- Scheerer H. 1937. Experimentelle und zytologische Untersuchungen innerhalb der Veronica-Gruppe Pentasepala. Flora 131: 287–323. [Google Scholar]
- Scheerer H. 1949. Zur Polyploidie und Genetik der Veronica-Gruppe Pentasepala. Planta 37: 293–298. [Google Scholar]
- Segraves KA, Anneberg TJ. 2016. Species interactions and plant polyploidy. American Journal of Botany 103: 1326–1335. [DOI] [PubMed] [Google Scholar]
- Shaffer JP. 1995. Multiple hypothesis testing. Annual Reviews of Psychology 46: 561–584. [Google Scholar]
- Schranz ME, Mohammadin S, Edger PP. 2012. Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Current Opinion in Plant Biology 15: 147–153 [DOI] [PubMed] [Google Scholar]
- Signorell A, Aho K, Alfons A, et al. 2017. https://cran.r-project.org/package=DescTools DescTools: tools for descriptive statistics. R package version 0.99.22.
- Simonin KA, Roddy AB. 2018. Genome downsizing, physiological novelty, and the global dominance of flowering plants. PLoS Biology 16: e2003706. doi: 10.1371/journal.pbio.2003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. 1999. Polyploidy: recurrent formation and genome evolution. Trends in Ecology & Evolution 14: 348–352. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Segovia-Salcedo MC, Jordon-Thaden I, et al. 2014. Are polyploids really evolutionary dead-ends (again)? A critical reappraisal of Mayrose et al. (2011). New Phytologist 202: 1105–1117. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Marchant DB, Van de Peer Y, Soltis DE. 2015. Polyploidy and genome evolution in plants. Current Opinion in Genetics and Development 35: 119–125. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. 1950. Variation and evolution in plants. Boston, MA: Springer. [Google Scholar]
- Stebbins GL. 1984. Polyploidy and the distribution of the artic–alpine flora: new evidence and a new approach. Botanica Helvetica 94: 1–13. [Google Scholar]
- Stebbins GL. 1985. Polyploidy, hybridization, and the invasion of new habitats. Annals of the Missouri Botanical Garden 72: 824–832. [Google Scholar]
- Strid A. 1986. Veronica austriaca. In: Löve A, Chromosome Number Reports XCIII. Taxon; 35: 897–903. [Google Scholar]
- Surina B, Schönswetter P, Schneeweiss GM. 2011. Quaternary range dynamics of ecologically divergent species (Edraianthus serpyllifolius and E. tenuifolius, Campanulaceae) within the Balkan refugium. Journal of Biogeography 38: 1381–1393. [Google Scholar]
- Temsch E, Greilhuber J, Krisai R. 2010. Genome size in liverworts. Preslia 82: 63–80. [Google Scholar]
- Theodoridis S, Randin C, Broennimann O, Patsiou T, Conti E. 2013. Divergent and narrower climatic niches characterize polyploid species of European primroses in Primula sect. Aleuritia. Journal of Biogeography 40: 1278–1289. [Google Scholar]
- Thiers B. 2017. continuously updated. Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium http://sweetgum.nybg.org/ih/ (Accessed 15 January 2017).
- Thompson JD. 2005. Plant evolution in the Mediterranean. Oxford: Oxford University Press. [Google Scholar]
- Thompson JD, Lumaret R. 1992. The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends in Ecology & Evolution 7: 302–307. [DOI] [PubMed] [Google Scholar]
- Thompson JN, Merg KF. 2008. Evolution of polyploidy and the diversification of plant–pollinator interactions. Ecology 89: 2197–2206. [DOI] [PubMed] [Google Scholar]
- Thompson KA, Husband BC, Maherali H. 2014. Climatic niche differences between diploid and tetraploid cytotypes of Chamerion angustifolium (Onagraceae). American Journal of Botany 101: 1868–1875. [DOI] [PubMed] [Google Scholar]
- Vandenberghe J, French HM, Gorbunov A, et al. 2014. The Last Permafrost Maximum (LPM) map of the Northern Hemisphere: permafrost extent and mean annual air temperatures, 25–17 ka BP. Boreas 43: 652–666. [Google Scholar]
- Visger CJ, Germain-Aubrey CC, Patel M, Sessa EB, Soltis PS, Soltis DE. 2016. Niche divergence between diploid and autotetraploid Tolmiea. American Journal of Botany 103: 1396–1406. [DOI] [PubMed] [Google Scholar]
- Visser V, Molofsky J. 2015. Ecological niche differentiation of polyploidization is not supported by environmental differences among species in a cosmopolitan grass genus. American Journal of Botany 102: 36–49. [DOI] [PubMed] [Google Scholar]
- Watzl B. 1910. Veronica prostrata L., teucrium L. und austriaca L. Nebst einem Anhang über deren nächste Verwandte. Abhandlungen der Kaiserlich-Koniglichen Zoologisch-Botanischen Gesellschaft in Wien 5: 1–94. [Google Scholar]
- Wefferling KM, Castro S, Loureiro J, Castro M, Tavares D, Hoot SB. 2017. Cytogeography of the subalpine marsh marigold polyploid complex (Caltha leptosepala s.l., Ranunculaceae). American Journal of Botany 104: 271–285. [DOI] [PubMed] [Google Scholar]
- Wendel JF. 2000. Genome evolution in polyploids. Plant Molecular Biology 42: 225–249 [PubMed] [Google Scholar]
- Wendel JF. 2015. The wondrous cycles of polyploidy in plants. American Journal of Botany 102: 1753–1756. [DOI] [PubMed] [Google Scholar]
- Zuur AF, Ieno EN, Elphick CS. 2010. A protocol for data exploration to avoid common statistical problems. Methods in Ecology and Evolution 1: 3–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.