ABSTRACT
While conventional nutrition research has yielded biomarkers such as doubly labeled water for energy metabolism and 24-h urinary nitrogen for protein intake, a critical need exists for additional, equally robust biomarkers that allow for objective assessment of specific food intake and dietary exposure. Recent advances in high-throughput MS combined with improved metabolomics techniques and bioinformatic tools provide new opportunities for dietary biomarker development. In September 2018, the NIH organized a 2-d workshop to engage nutrition and omics researchers and explore the potential of multiomics approaches in nutritional biomarker research. The current Perspective summarizes key gaps and challenges identified, as well as the recommendations from the workshop that could serve as a guide for scientists interested in dietary biomarkers research. Topics addressed included study designs for biomarker development, analytical and bioinformatic considerations, and integration of dietary biomarkers with other omics techniques. Several clear needs were identified, including larger controlled feeding studies, testing a variety of foods and dietary patterns across diverse populations, improved reporting standards to support study replication, more chemical standards covering a broader range of food constituents and human metabolites, standardized approaches for biomarker validation, comprehensive and accessible food composition databases, a common ontology for dietary biomarker literature, and methodologic work on statistical procedures for intake biomarker discovery. Multidisciplinary research teams with appropriate expertise are critical to moving forward the field of dietary biomarkers and producing robust, reproducible biomarkers that can be used in public health and clinical research.
Keywords: dietary biomarkers, dietary intervention studies, diet, nutrition, metabolomics
Introduction
Prevailing dietary intake assessment methods (e.g., FFQ) rely heavily on self-reported dietary recall and have a variety of systematic and random measurement errors. A systematic underreporting of dietary intake, especially of total calories and absolute amounts of macronutrients, in weight-loss trials has been well documented (1). This problem is further exacerbated by the increasing prevalence of “ready-to-eat meals” in the Western diet, with incomplete ingredient lists and inability of the participants to complete the cumbersome and complicated dietary questionnaires. In addition, imperfect or incomplete food composition databases can lead to inaccuracies when food intake data are converted to the corresponding nutrient intake data. Finally, differences in individual metabolism, due to genetics or the gut microbiome, add complexity to intake measurements. Ideally, self-reported dietary intake information should be independently validated against a biologic or chemical marker that provides an accurate measure of the dietary intake and exposure. For example, candidate biomarkers, such as alkyl resorcinols for measuring wheat and rye intake, are beginning to be used in epidemiologic studies (2). However, such objective markers of intake are limited to few nutrients and do not exist for most foods and dietary patterns.
Recent advances in high-throughput MS and NMR spectroscopy combined with improved metabolomic, genomic, and metagenomic techniques are now making it possible to identify new and improved dietary biomarkers. Several studies have demonstrated the feasibility of this combined multiomic approach (3–5). To explore the potential of multiomics approaches in dietary biomarker development and to identify related challenges and approaches to address them, the NIH organized a workshop on “Omics Approaches to Nutritional Biomarkers” from 26 to 27 September 2018 in Bethesda, MD. This workshop engaged nutrition and omics researchers from the United States, Canada, and several countries in Europe, all of whom participated in scientific presentations and focused breakout sessions to discuss various aspects of dietary biomarker development.
Table 1 presents a summary of the challenges and the resulting recommendations from the workshop. Each challenge is discussed in greater detail below. The recommendations are intended to serve as a guide for scientists wishing to identify, develop, validate, or use dietary biomarkers in their research programs.
TABLE 1.
Strategies and approaches for advancing dietary biomarker development1
| Challenges | Recommendations/resources needed |
|---|---|
| Define dietary biomarkers and their utility in nutrition research | |
| Multiple dietary biomarker definitions in use | Adopt a universally accepted biomarker classification scheme with a well-developed ontology for use by the nutritional epidemiology and dietary biomarker community |
| Lack of publicly available comprehensive databases on dietary biomarkers | Develop or expand well-curated, publicly available international databases on dietary biomarkers such as Exposome-Explorer and Phenol-Explorer for prioritization of candidate biomarkers |
| Lack of comprehensive food composition databases | Develop and maintain comprehensive food composition databases |
| Approaches to studying biomarkers | |
| Studies are often conducted with no clear regard for human heterogeneity | Capture information on host factors (e.g., genetics, gut microbiome, behavioral and cultural practices) that may help to explain heterogeneity in dietary biomarker measures |
| Current feeding studies are “siloed” and often single studies conducted for a shorter duration, involving smaller sample sizes | Conduct larger CFSs, testing a variety of foods and dietary patterns across diverse populations to identify universal candidate biomarkers |
| Shortage of appropriately collected specimen repositories for dietary biomarker development | Collect a variety of biospecimens (e.g., fecal samples, blood cells, saliva, toenails, hair) as part of feeding studies to discover and validate both short- and long-term dietary biomarkers |
| Leverage existing biospecimen repositories from feeding studies and prospective cohorts to validate dietary biomarkers | |
| Encourage long-term storage of biospecimens from completed feeding studies for dietary biomarker development studies | |
| Lack of standardized specimen collection and processing protocols for omics analysis | Implement well-standardized specimen collection and processing protocols to ensure reproducibility, comparability, and generalizability across studies |
| Cumbersome sampling procedures and lack of integration of advanced devices for sample collection | Develop new sampling techniques for efficient collection and wider acceptance and improved adherence in large studies (e.g., dried blood spots) and adopt wearables and smartphone devices that allow for continuous metabolite monitoring |
| Analytical and statistical considerations of biomarker development | |
| Metabolite coverage and reproducibility | Encourage sharing of spectral data and chemical databases of biologically feasible structures of metabolites |
| Support internationally co-ordinated efforts for providing resources on food constituent libraries and biomarker data from various laboratories | |
| Facilitate distribution of relevant metabolite standards (e.g., Food Compound Exchange) | |
| Shortage of strategies to evaluate variation within and between laboratories | Develop standardized approaches for evaluating laboratory variation and normalizing for drift and differences across laboratories |
| Shortage of statistical methods for handling measurement error and applying to dietary exposure assessment | Conduct methodologic work on statistical procedures for intake biomarker discovery and disease application |
| Sharing sensitive metadata across laboratories is difficult | Establish secure portals accessible via cloud computing and portability environments for sharing metadata |
| Lack of minimum reporting standards for statistical analytic pipeline/workflow for nutritional metabolomics studies | Establish minimum reporting standards to support study replication |
| Dietary biomarker discovery and validation | |
| Dietary biomarker development is lengthy with no clear validation criteria | Adopt a universal dietary biomarker validation strategy that is accepted by the nutrition research community |
| Untargeted metabolomics produces multiple metabolites with no quantitative measures | Develop targeted and quantitative assays for validation studies after initial biomarker identification |
| Areas where more data are needed | |
| Lack of comprehensive food composition databases | Create and maintain truly comprehensive food composition databases by expanding existing databases, such as FooDB, in terms of chemical coverage and breadth of human food intake |
| Integrate more fully the various food composition databases using shared links, common identifiers, and common ontologies | |
| Extend food composition databases to archive experimentally acquired or accurately predicted referential MS/MS and/or NMR spectra data to facilitate food or dietary biomarker identification | |
| Lack of concerted efforts and community resources necessary for dietary biomarker development | Support international efforts to prepare, acquire, or synthesize authentic food-specific compounds and their MS/MS and/or NMR spectra and enable access via open-source databases (e.g., GNPS, MoNA, FooDB, HMDB, the Metabolomics Workbench, and MetaboLights) |
| Support international efforts to prepare, acquire, or synthesize authentic gut-derived, liver-derived, or similarly biotransformed food compounds and their MS/MS and/or NMR spectra. Facilitate access, via open-source databases such as GNPS, MoNA, FooDB, HMDB, the Metabolomics Workbench, and MetaboLights | |
| Improve algorithms and open-access software to more accurately predict metabolic biotransformation products (mimicking liver, microbial, or promiscuous biotransformations) to facilitate in silico metabolomics | |
| Improve algorithms and open-source software to more accurately predict MS/MS spectra (at multiple collision energies and on different platforms), NMR spectra, collisional cross-section data (for IMS data), and GC or HPLC retention times of small molecules | |
| Specificity is a challenge for dietary biomarker development | Use combinations of biomarkers from either a single study or pooled data from several feeding studies to increase marker specificity |
| Develop reference ranges for biomarkers across different populations and age ranges (children compared with adults) | |
| Integration of dietary biomarkers with other omics techniques | |
| Neither genomics nor metabolomics tools alone provide complete understanding of how dietary components are metabolized | Integrate other omics methods in dietary biomarker analysis with a view to understanding the impact of individual variation and personalized responses |
| Identify and further explore the effect of SNPs on dietary biomarker measures | |
| Improve tools (databases, software, statistical methods) to facilitate the integration of genomics, metagenomics, proteomics, and metabolomics data in nutritional studies | |
| Lack of systematically collected catalogs of SNPs | Continuously update databases or catalogs of SNPs, genes, and gene signatures that alter the metabolism, presence, or abundance of known and potential dietary biomarkers |
| Other critical elements | |
| Lack of concerted efforts for biomarker development | Foster collaboration among multidisciplinary researchers |
| Encourage public–private partnerships for collecting and sharing the data on dietary biomarkers that would not be otherwise freely available | |
| Train early career scientists in dietary biomarker development | |
| Lack of common ontology for dietary biomarker literature | Support standard ontology efforts through development of newer and broader algorithms for electronically mining the literature |
| Convene taskforces for developing common data elements for dietary biomarker research | |
1CFS, controlled feeding study; GNPS, Global Natural Products Social Molecular Networking; HMDB, Human Metabolome Database; IMS, Ion Mobility Separation; MoNA, MassBank of North America; SNP, single-nucleotide polymorphism.
Dietary Biomarker Definitions and Their Utility in Nutrition Research
A dietary biomarker enables an objective measure of either dietary intake, its impact on host physiology or its modification of disease risk (6). Following a broader paradigm for biomarker utility, diet-related biomarkers are typically classified into 3 groups: 1) exposure biomarkers, 2) susceptibility markers, and 3) outcome biomarkers. An exposure biomarker provides an objective measure of dietary intake of a particular food or nutrient (7). A susceptibility biomarker provides information about resilience or susceptibility to effects caused by food components, such as susceptibility to iron overload from meat consumption. In contrast, an outcome biomarker is used to assess how physiologic and clinical outcomes are affected by nutrient exposures (8). In addition to this “classical” set of biomarkers, several other dietary biomarker classification schemes have also emerged in the field of nutrition, depending on how a biomarker changes in relation to intake and length of exposure (6, 9).
No single classification scheme covers all the aspects of dietary biomarker functions and features (9). The same compound may be classified in different categories depending on the purpose of use. For example, total plasma homocysteine concentrations indicate folate status and serve as a marker of nutrient status and a biomarker of treatment response to folate supplementation (10). Most of these biomarker classification schemes assume a unidirectional interaction, with specific dietary components affecting physiologic systems. However, it is increasingly recognized that the relationship between dietary components and physiologic systems is bidirectional. In fact, dietary components affect the host's physiology, which in turn has an impact on how these dietary substances are metabolized (Figure 1). Moreover, the food-host metabolic interaction is embedded in a broader cultural and environmental system that influences the type and extent of food exposure and affects the metabolic end products (i.e., biomarkers) detectable in human biospecimens.
FIGURE 1.
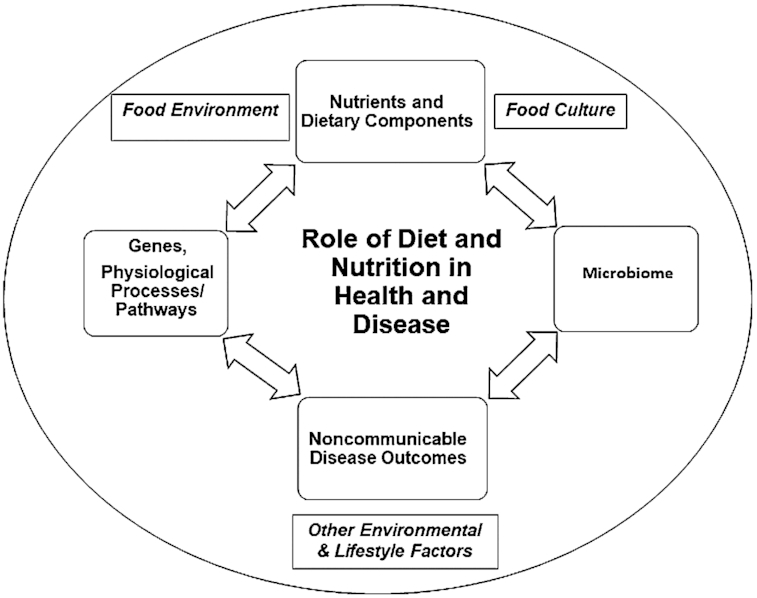
Bidirectional interaction between dietary components and physiologic systems embedded in food consumption driven by food environments and further influenced by cultural and lifestyle factors. Consumption of nutrients such as fatty acids, amino acids, vitamins, trace elements, and bioactive compounds has an impact on host physiology, affecting both the health status and susceptibility to disease. Metabolism of dietary components is also influenced by the genetic make of an individual. In addition, dietary components may directly affect gut microbiota composition and function, which may exacerbate metabolic and physiologic outcomes, further influencing disease susceptibility. Host physiology and altered susceptibility to disease in turn affect how these dietary substances are metabolized.
The application of metabolomics allows a better characterization of this bidirectional relationship between diet and physiology, enabling the measurement of both nutrient and nonnutrient metabolites that could serve as candidate biomarkers (11). However, nonnutrient markers are not well integrated in the current paradigm of biomarker classifications, and no common biomarker ontology can address all these classifications. In recognition of these challenges, Gao et al. (9) developed a detailed dietary biomarker classification framework that integrates both nutrient and nonnutrient markers from food components.
Under this new classification scheme, exposure bio-markers (which may be single biomarkers or combinations of multiple biomarkers) are further classified into food component intake biomarkers (FCIBs), biomarkers of food intake (BFIs), and dietary pattern biomarkers (DPBs). FCIBs are typically metabolites of chemicals present in different foods and include both nutrients and nonnutrients. BFIs, on the other hand, are associated with a given food type or food group and mostly consist of nonnutrients, such as proline betaine for citrus fruit consumption. DPBs are used to distinguish between different dietary regimens such as Mediterranean, Western, or Nordic diet patterns. DPBs can include both FCIB and BFI markers from a variety of foods found in a specific dietary pattern. This new type of diet-related biomarker classification scheme appears to offer both breadth and flexibility as it allows the same markers to be used for a variety of different purposes (9).
Currently, only a few reliable intake biomarkers are known. These include 24-h urinary nitrogen for protein intake, doubly labeled water (DLW) for total energy expenditure measurements, and 24-h urinary sodium and potassium for sodium/potassium intake. Unfortunately, methods such as DLW analysis are very expensive, while 24-h urinary nitrogen or urinary sodium and potassium measurements are too cumbersome for regular participant compliance to be used in large studies (12).
One approach that appears to be particularly promising for finding biomarkers of macronutrient intake is the use of isotope ratio mass spectrometry (IRMS) (13). Naturally occurring differences in the stable isotope ratios (SIRs) of lighter elements among foods such as carbon (13C compared with 12C, measured as δ13C) and nitrogen (15N compared with 14N, measured as δ15N) are reliably incorporated into tissues and can be measured by IRMS. One of the advantages of this method is that SIRs are very stable and can be measured in a variety of biologic specimens, including blood, hair, and toenails. Biomarkers for macronutrient food components such as carbohydrates and protein have been explored using SIR analysis (14–16).
Another approach is MS-based metabolomics, which is opening the door to measuring both micronutrient and nonnutrient biomarkers to reliably predict food intake. For example, the Phenol-Explorer database contains information about >500 nonnutrient plant polyphenols that are specific for particular foods or food groups (17). Using standard metabolomic methods and the Phenol-Explorer database to annotate polyphenol metabolites in urine, it was possible to measure >80 polyphenol metabolites in 24-h urine samples and to identify good predictors of intake from some of their main food sources such as citrus fruit, coffee, tea, and wine as estimated with 24-h dietary records (18). More recently, using a targeted assay for 34 dietary polyphenols measured in urine, it was possible to study variations of urinary excretion according to geographic variations of the diet in 4 different countries, enabling the identification of those phenolic compounds most strongly associated with intake of 110 plant-derived food groups (19). A recent study used a nonnutrient biomarker alkylresorcinol metabolite in plasma for whole-grain consumption to demonstrate its protective effect on the risk for ischemic stroke, revealing its potential clinical utility (2). These examples illustrate that metabolomics, when combined with the right kinds of databases, can be used to identify some useful dietary biomarkers.
Approaches to Studying Biomarkers
Study designs
Dietary intake biomarker development is best approached as an iterative process, involving a well-integrated methodologic strategy from biomarker discovery through validation. Biomarker development should also rely on sufficiently robust study designs to identify candidate biomarkers that subsequently can be successfully validated (20). While controlled feeding studies (CFSs) are particularly informative for both biomarker discovery and validation, other study designs may be used to capture the characteristics of dietary variation and identify candidate dietary biomarkers for a wide diversity of foods.
Cross-sectional studies are routinely used for initial dietary biomarker exploration for capturing the continuous distribution of dietary constituents in the habitual diets, including food groups, or of dietary patterns (7). Key challenges of using cross-sectional studies to discover dietary biomarkers lie in the limitations of common dietary assessment instruments used, such as dietary recalls, food diaries, and FFQs (21). Additional challenges relate to measurement errors in dietary self-reporting (22), the inadequacies of food-composition tables, and the limited generalizability of diet-biomarker associations to other populations. Most reported candidate dietary biomarkers arise from foods that are routinely consumed and potentially more accurately recalled by participants (23, 24). In contrast, foods that are consumed infrequently are often difficult to capture with a recall or an FFQ and typically will result in the sporadic appearance of measurable biomarkers in blood or urine. In such cases, cross-sectional studies may be ineffective to identify such biomarkers, unless they have unusually long half-lives. These biomarkers may be less easily identified and would likely be among the more lipophilic metabolites (7). Such biomarkers may not effectively replace traditional self-report dietary assessment methods of longer-term exposure, but their integration with dietary intake data may provide a more accurate assessment of exposure.
In contrast to cross-sectional studies, which are often used for dietary biomarker discovery, CFSs are primarily used to evaluate the effects of diet on biologic and physiologic processes in humans. Nonetheless, feeding known amounts of specific foods or nutrients to study participants also provides an opportunity to evaluate biomarkers of dietary exposure (25). Typically, CFSs use the same standardized menus for all participants, thereby reducing the variation in nutrient intake and the variance introduced by food type, as well as by the handling, storage, preparation, and processing of the food. These studies permit the testing of several factors, such as the magnitude of consumption and duration of feeding (e.g., short term compared with long term), and can provide rich data on biomarker nutrikinetic and nutridynamic properties, similar to drug metabolism (26, 27). CFSs also allow the assessment of metabolite variability due to host physiology, the type of intervention (e.g., dietary component, food, dietary pattern), the biomarker performance, and when and how often the samples should be collected.
In CFSs, dietary constituents or foods may be administered at the same dose to all participants (28), various doses to provide a range of exposures (29), or doses based on body weight (30). These diets also depart from participants’ habitual intake and consequently need adequate duration for biomarker equilibration. An alternative to a set-menu CFS is a variable-menu CFS that preserves the normal variation in nutrient and food consumption at the individual level in the study population. This approach requires individualized menu plans for each participant that mimic their habitual food intake as estimated by using a 4-d food record and adjusted for energy requirements, on the basis of calibrated energy estimates and standard energy estimating equations (31).
To date, most CFSs have been conducted for shorter durations, with small sample sizes and a limited capacity to capture interpersonal heterogeneity. In addition, these studies are often expensive and laborious to conduct, thereby necessitating several methodologic compromises (limiting the sample size, reducing the study duration, etc.) that may potentially affect the final study results. There is no clear consensus on the choice of feeding study designs or sample sizes needed for dietary biomarker development and validation. Recently, there have been attempts to combine a variety of study designs such as crossover, controlled feeding, and cross-sectional studies for biomarker explorations from discovery phase to testing them in free-living populations on habitual diets (32, 33). The final design depends, in large part, on the specific questions being addressed.
When substantial information is available on certain biomarkers, there may be no need to start from the beginning of the biomarker discovery process. Small, short-term feeding studies that yield candidate biomarkers may be followed by studies that characterize biomarker time and dose response. Likewise, validation and testing of biomarker performance may be done in separate cohorts, and the process may be repeated with necessary corrections, until an optimal biomarker performance is achieved.
The replication of initial biomarker studies in different populations is often necessary to generalize the results, to accommodate population heterogeneity, and to properly account for food choice diversity and dietary patterns. Ideally, the first validation study should be conducted in a similar population to the initial discovery cohort, favoring repeated measures to minimize intraperson variation in biomarker measures. Existing large cohorts, such as the Women's Health Initiative, the Framingham Health Study, and the Nurses’ Health Study, can be leveraged for large validation studies. However, it is important to recognize the limitations of the dietary assessment methods and the biosampling protocols used in these types of studies. Collaborative multicenter feeding studies using habitual diet feeding study designs such as the one employed by the Nutrition and Physical Activity Association Study provide an excellent opportunity for recruiting diverse populations and exploring several nutritional factors and candidate biomarkers (34). Citizen science projects, such as the American Gut Project (35), may also be useful to validate candidate biomarkers because they rapidly generate large sample sizes, involve broad national and international participation, and help to capture biomarkers of more prolonged exposure to a particular diet.
Criteria such as high sensitivity and high specificity for the dietary intake of interest are fundamental to good biomarkers that can be quantified in terms of the area under the receiver operating characteristic curve (AUROC). In addition, a potential food or diet intake biomarker should be able to explain a sizable fraction of the feeding study variation in the given diet. The AUROCs allow setting a cutoff value for a given biomarker, ranging from 0.5 with random association to 1.0 with strong association between the biomarker and dietary consumption, and rely on the continuous performance of the biomarker on a binary outcome (36). The specific cut point to be met in applying these criteria may depend on the context of the feeding study. It may also have to be adjusted to reflect the accuracy of estimated intake in the feeding study (e.g., accuracy of food composition databases), as well as the study duration and other aspects of the feeding study design. Investigators proposing novel biomarkers need to provide convincing evidence of a close correspondence between the actual intake and the biomarker estimated intake, rather than simply demonstrating a positive correlation between the two. While the AUROCs are useful for dietary biomarker research, their utility highly depends on the availability of good gold-standard markers with which they can be compared for their classification (36).
Biologic sampling
Regardless of the study design, careful consideration of the types of biologic samples collected and analyzed is fundamental to ensuring meaningful outcomes. In terms of dietary biomarkers, urine appears to provide better metabolite coverage compared with plasma due to the relative lack of interfering proteins and the fact that many dietary biomarkers are in higher concentrations in urine (11). Several factors affect the choice of biospecimen matrix for dietary biomarkers. Different specimens may yield different candidate biomarkers due to their unique physiologic origins or their different duration of exposure. Specimens such as saliva and sweat may provide insight on short-term dietary exposures, whereas RBCs better capture medium-term exposure (11, 37, 38), while toenails (39) and hair appear to be promising matrices for long-term exposures (40, 41). Assessment of biomarkers in more than 1 matrix (both plasma and urine) will also provide information on the distribution and dynamic range of biomarkers in the system and their half-lives (11).
Archived biospecimens from well-conducted dietary interventions are potentially very useful resources for biomarker discovery and validation. However, precise information on the stability of certain dietary biomarkers upon storage and sample handling practices, especially for multisite studies is needed. It is important to determine potential confounders that contribute to metabolite variability. Currently, there are several large repositories for plasma and serum (42). Unfortunately, there are not many cohort studies with repositories containing urine samples. While urine is often the preferred biomatrix for dietary biomarker studies, it is also important to remember that many useful dietary biomarkers have been identified in plasma, although sample collection requires trained phlebotomists (7). Indeed, the dietary metabolites with the strongest food correlations in population studies tend to replicate in both blood and urine and predict habitual diets (24). For this reason, it is still useful to expand dietary biomarker studies in blood-based (plasma) specimens. Overall, the lack of appropriately collected, publicly accessible repositories of specimens from intervention and cross-sectional studies represents a continuing impediment to dietary biomarker discovery and development. Certainly, encouraging the long-term storage of biospecimens from completed feeding studies will expedite the biomarker discovery and development process.
There is a critical need to standardize specimen collection and sample processing protocols to ensure greater reproducibility, comparability, and generalizability across studies (43). For instance, Lloyd et al. (44) conducted a systematic validation of biomarkers of habitual citrus fruit intake and demonstrated that both spot and overnight fasting urine samples provide a good correlation with FFQ data. Garcia-Perez et al. (43) explored the timing of urine collection and compared the quantification of biomarkers in spot urine with 24-h urine samples.
Although repeated samples are highly desirable for accurate quantitative measurement, single samples may also be reasonably informative, especially for frequently consumed foods (45). This is especially true for biomarkers showing good reproducibility over time, which are well suited for prospective studies involving larger cohorts (45, 46). Furthermore, sample handling (standing time, storage temperature, and freeze/thaw cycles) affects many metabolites, which obviously affects the robustness of any identified metabolite biomarkers. However, if samples are handled consistently, then biomarker-outcome associations (e.g., in nested case-control studies) can still perform well. Nonetheless, standardized sample handling practices should be encouraged.
The development of new sampling techniques is also becoming important to enable more efficient, cost-effective sample collection and better coverage in larger cohort studies. This is particularly important for studies with geographically isolated cohorts (47). For example, dried blood spots are proving to be an inexpensive method for sample collection and storage; they require minimal specialized equipment and offer several advantages, including convenient transportation (48). Novel gastrointestinal tract sampling methods are emerging that may identify novel dietary biomarkers related to intake and food microbial metabolism (49). Advances in wearable technology that can continuously monitor metabolites or allow intermittent sampling will likely complement and expedite the biomarker development process.
Analytical and Statistical Considerations in Biomarker Development
Analytical and technological issues
High-throughput, untargeted metabolomics approaches have revolutionized dietary biomarker development, allowing unbiased interrogation of both nutrients and nonnutrients. Several analytical methods, including NMR, MS combined with LC, and GC, have been used for dietary biomarker research. These methods differ in their sensitivity, sample processing requirements, and metabolite coverage. NMR is a robust, relatively unbiased, inherently quantitative method that allows novel metabolite identification and requires little sample processing. However, NMR suffers from low sensitivity, enabling detection and/or quantification of 30–100 different, more abundant metabolites in a given biologic sample. GC-MS is ideal for detecting a variety of nutrients (amino acids, sugars, organic acids, steroids, fatty acids, and volatile metabolite analysis), and it is sufficiently sensitive to detect ≤300 different chemicals in certain biomatrices. However, GC-MS requires extensive sample workup and sample derivatization, making it more time-consuming and more difficult to quantify compounds than NMR. High-resolution LC-MS is a highly sensitive technique that allows the detection of ≤10,000 features and the identification of between 400 and 1500 different chemicals depending on the platform and method (targeted compared with untargeted). LC-MS is particularly suitable for detecting nonnutrient metabolites that occur in very low concentrations. As a result, it is gaining popularity as the preferred platform in both metabolomics and dietary biomarker studies. One of the limitations of LC-MS is that no single LC system can cover all metabolite classes. Hydrophilic interaction chromatography typically must be used to separate more polar metabolites, reversed-phase chromatography must be used to analyze neutral and nonpolar metabolites, and chemical derivatization may be required to detect lower-abundance metabolites. While obtaining relative quantitation of compounds using LC-MS is straightforward and typical in metabolomics, determining the absolute concentration of compounds is more difficult and requires expensive isotopically labeled standards as well as multipoint calibration curves. Ideally, data from a variety of metabolomics platforms should be interrogated for discovering and/or quantifying candidate dietary biomarkers of specific dietary exposures.
Interlaboratory reproducibility of untargeted LC-MS metabolomics data is another challenge and is heavily influenced by the instrument type and design. Nevertheless, Cajka et al. (50) have recently shown that 9 different mass spectrometers can give rise to nearly identical results for identical biologic samples if detection saturation is avoided. Metabolite identification, coverage, and reproducibility are also influenced by experimental conditions that include sample processing, storage, mode of detection, instrument run, and choice of data reduction methods used. No standardized and universally accepted protocols and pipelines exist for untargeted LC-MS–based metabolomics. Furthermore, LC-MS methods from individual laboratories are not freely shared among the community, further reducing confidence and reproducibility. The variability can be minimized by adopting appropriate quality control measures such as proper blanks, controls, and standards in the experimental runs. There are now efforts within the metabolomics community to develop such standards and protocols to improve reliability and reduce variability within and across studies. It is also important to adopt the use of pooled reference samples (such as standard reference materials) to adjust for instrumental differences and batch variations over time. To improve the quality of data processing and metabolite identification, there is a critical need for sharing the raw data, including quality control measures and blanks for data processing. Interlaboratory comparison of assays, with appropriate standards, should be encouraged to ensure cross-validation of assays.
Data analysis and metabolite identification for biomarker discovery
While thousands of “features” can be detected on untargeted LC-MS–based metabolomics platforms, the actual identification of metabolites continues to be a major challenge. Raw data from MS instruments must undergo several processing steps before they can be statistically analyzed and compared. These steps involve the removal of adducts, peak identification and peak alignment, spectral deconvolution, compound identification (via matching to an MS/MS spectrum), and multivariate statistical analysis. Many software tools, including commercial packages, offer a wide range of excellent features, but they all differ in their algorithms for picking MS peaks (51–53). As a result, there is only a 50–70% overlap between the MS peaks detected by different packages, from the same raw data files using identical or near-identical settings (54). Clearly, more standardization of the data analysis pipelines and peak picking algorithms is needed.
Compound identification, which is the next step after spectral alignment and peak detection, typically involves the comparison of MS/MS spectral features with well-curated MS databases. However, there is considerable diversity in the types of MS instruments and the types of MS spectra that can be collected on these instruments. As a result, it is often difficult to find a comprehensive MS database that fits with the type of MS spectra being collected other than the instrument-specific database provided by the vendor. Because the vendor-specific databases are often costly or do not cover the compounds of interest, there is a growing need for comprehensive, open-access MS databases that provide MS spectra for multiple platforms and that support broad metabolite identification activities. One such database is the MassBank of North America (MoNA) (55). MoNA is an open-access MS database that actively harvests and displays a large portion of the public MS/MS fragment spectral data for metabolites into a single, web-accessible resource containing >130,000 experimental MS/MS spectral records from authentic compounds (including many food compounds) and nearly 140,000 predicted spectra generated for lipids.
Several other public databases also contain large, freely available collections of reference metabolite MS/MS spectral data covering multiple MS platforms (56–58). However, these databases are also populated with a large fraction (sometimes >80%) of predicted spectra commonly generated using programs such as MetFrag (59), CFM-ID (60), or Mass Frontier (61). While still useful, these predicted MS spectra are not as accurate or as correct as experimentally collected MS spectra. Indeed, the dearth of experimental MS spectra collected for authentic compounds continues to be a major challenge in metabolite annotation. Recently, an effort known as the Food Compound Exchange has been launched to help address this problem (62). This community-driven concept, which was sponsored by the Food Biomarker Alliance (63), allows researchers from around the world to freely share metabolite standards and experimentally collected metabolite and food constituent spectra in a collaborative manner. Efforts such as the Food Compound Exchange should help create key community resources to expedite biomarker discovery (62). Clearly, more support is needed for these types of community-driven, bottom-up efforts.
Dietary biomarker discovery
After the candidate metabolites have been identified (either through targeted or untargeted metabolomic approaches), the next challenge is to identify the most useful or important biomarkers from the collection of identified metabolites. Biomarker discovery and biomarker assessment are often aided by the availability of specialized statistical software packages. These packages typically use multivariate statistics and feature selection and/or machine learning to identify 1 or more compounds that maximize the sensitivity and/or specificity of the biomarker or biomarker panel for the dietary exposure on a receiver operating characteristic (ROC) curve. While ROC curves mainly enable the stratification of individuals based on their consumption of dietary components, it may be difficult to identify individuals with sporadic consumption. Maximizing the AUROC by selecting the right chemical or the right combination of chemicals is often a central goal of biomarker identification or biomarker discovery. Several freely available webservers and software packages such as MetaboAnalyst and Galaxy have emerged over the past decade to provide not only comprehensive web-based tools for routine metabolomic data analysis and functional interpretation but also tools for metabolomic-based biomarker discovery suites (64, 65). However, advanced statistical tools are essential for validating dietary biomarkers, integrating multiomics data in nutritional studies, correcting measurement errors in self-reported dietary reports (66), and generating disease-diet biomarker regression models (67). There is a clear need for greater standardization, including standardized reporting (or minimum reporting standards), for the statistical analysis of nutritional metabolomics data. Similarly, standardized processes for evaluating study reliability, data normalization, handling multiple testing effects, performing study replications, and cross-validation or external validation are also needed. In this regard, it would be particularly useful for the community to have statistical code repositories to foster greater uniformity and greater levels of reproducibility.
Dietary Biomarker Validation
LC-MS–based dietary biomarker discovery can provide hundreds of candidate biomarkers, but these biomarkers need to be thoroughly validated to be meaningfully used in large cohort studies. The goal of validation is to ensure that newly discovered biomarkers can reliably and reproducibly predict dietary intake of food components. Biomarker validation requires analytical and biologic testing of the performance of the biomarkers. It also requires an assessment of their specificity to food components and their robustness in larger cohorts. While several concepts exist regarding the validation of biomarkers, there are no universally accepted validation criteria for dietary intake biomarkers. Dragsted et al. (20) have recently devised an 8-step validation process that systematically assesses candidate biomarker plausibility, dose response, time response, robustness, reliability, stability, analytical performance, and reproducibility. Each criterion is important for establishing overall biomarker validity but may be evaluated in a different order, depending on the status of the candidate dietary biomarker.
Standardized dietary biomarker validation criteria allow the grading of markers based on their performance. Biomarker plausibility evaluates the credibility of the association between the biomarker and its food components. Plausibility can be based on a variety of sources of evidence, including research literature or in silico analysis of predictable biomarkers from compounds in the existing food composition databases and/or experimental data from metabolomics. Biomarker kinetics (including dose response and temporal response to a single acute exposure) can be used to determine the suitability of the biomarker over heterogeneous food intake distributions and variable biomarker half-lives. Time-related response to multiple exposures (e.g., medium- or longer-term feeding studies) may yield information on the distribution pattern of the biomarker across biologic tissues (RBCs, hair, nails, etc.). The robustness and reliability criteria are used to determine how the biomarker behaves in a mixed meal or as part of a normal diet in the real world among diverse populations (i.e., generalizability) and how it performs in comparison with other known biomarkers or other gold standards. Analytical performance criteria are used to determine the biomarker performance in both qualitative and quantitative terms, using known chemical standards ensuring a higher level of confidence in the biomarker performance. Cross-validation of the biomarker across laboratories confirms the reproducibility of the biomarker against food intake and completes the entire validation process.
This 8-step view of biomarker validation covers the entire spectrum of biomarker development from discovery through validation, using similar strategies and analytical platforms as they move from one step to another, depending on the purpose of the biomarkers. When there is substantial information available on a biomarker or a set of biomarkers, some of these validation steps may be eliminated to make more significant strides toward validation. Initial studies can employ small-scale acute feeding studies, followed by other studies enabling characterization of other elements such as dose and temporal response relationships or the testing of the candidate biomarker performance in separate cohorts. Wider acceptance and adoption of such a systematic approach by the research community will expedite dietary biomarker research, bridge the gaps between discovery and validation, and turn biomarker development into a tractable process.
Areas Where More Data Are Needed
The paucity of validated dietary intake biomarkers represents a fundamental challenge for food and nutrition research, and it highlights the need to acquire more data about the chemical compounds found in food and their fate after ingestion. Some are metabolically inert, and the biomarker compound found in blood, urine, or feces is identical to the compound found in a specific food (i.e., proline betaine for citrus consumption). In other cases, the consumed nutrients or nonnutrients are metabolically transformed by endogenous processes or the gut microbiota. This leads to chemical by-products that are very different from the ones originally ingested in the food (e.g., microbial product equol from daidzein after soy consumption). Therefore, to develop a large set of robust, specific, and fully validated food-specific biomarkers, it will be necessary to do 2 things: 1) acquire more data about the chemical constituents found in food (the “food metabolome”) and 2) acquire more data about the way that these chemical constituents are biologically transformed in the human body.
More than 150 food composition databases exist; however, most of these databases contain a relatively small number (10–100) of nonunique compounds for a vast number of foods. For instance, the USDA nutrient composition database (68) contains chemical data for nearly 250,000 different foods, but it only lists an average of 50 chemical compounds in each food item. While this information is useful for general nutritional assessment, it is not useful for identifying potential food-specific biomarkers.
More recently, a small number of online, electronic databases have emerged with more detailed chemical composition data for a smaller number of “raw” or mildly processed foods (Table 2) (57, 58, 69–72). However, their utility in the nutrition research community is limited by their lack of visibility and their lack of standardization or integration with each other. Another issue relates to the fact that these databases are still relatively incomplete. Most raw foods contain >10,000 different compounds, yet the average compound coverage in even the most comprehensive food composition database is <1000 compounds per food item. Indeed, untargeted analyses of hundreds of different foods by the Dorrestein laboratory at the University of California at San Diego (UCSD) has found that <5% of the detected MS peaks in any given food item can be assigned using these databases (73) (PC Dorrestein, UCSD, personal communication, 2018). This highlights an even more serious problem with today's food composition databases; that is, they do not have sufficient authentic reference NMR or MS/MS spectra to permit broad and accurate compound identification. The availability of more authentic reference spectra would permit identification of more food-specific compounds in both foods and in human biofluids or excreta. Fewer than 1000 food-derived compounds have had their NMR or MS/MS spectra experimentally collected and deposited into food-specific databases (72). The lack of authentic chemical standards for food constituents and the lack of authentic referential spectra are the 2 most serious data-related issues hampering the identification, discovery, or validation of food-specific biomarkers.
TABLE 2.
Food metabolite databases1
| Database | Description | Unique features | Reference |
|---|---|---|---|
| GNPS | Global Natural Products Social Molecular Networking: database of raw, processed, or identified MS/MS data | Food-specific data and includes MS/MS spectra from a large (>3500) number of different foods | (57) |
| HMDB | Human Metabolome Database on small-molecule metabolites found in the human body | A variety of endogenous metabolites with >1000 biotransformation products; data on 3056 metabolites linked to 2192 SNPs with 6777 specific metabolite-SNP interactions; data on 2901 metabolites that vary with physiology and data on 5498 metabolites that vary with pathophysiologic conditions | (58) |
| PhytoHub | Plant-based metabolite database on phytochemicals present in foods commonly ingested with human diets | Plant metabolites with 578 biotransformation products | (69) |
| Exposome-Explorer | Biomarkers of exposure to environmental risk factors for diseases | Data on 145 dietary biomarkers including their concentrations in various populations, type of biospecimens analyzed, the analytical techniques used, their reproducibility over time, and correlations with food intake | (70) |
| Phenol-Explorer | First comprehensive database on polyphenol content in foods | Plant polyphenol metabolites with 375 biotransformation products | (71) |
| FooDB | Database on food constituents, chemistry, and biology | Data and referential MS and NMR spectra on >26,000 food chemicals found in >720 raw or lightly processed foods | (72) |
SNP, single-nucleotide polymorphism.
To find better food-specific biomarkers, it is important to know more about the way that these chemical constituents are biologically transformed in vivo. The fact that gut microbial activity influences the presence/abundance of certain food-specific metabolites adds another layer of complexity to food-specific biomarker identification. Indeed, the interindividual variation due to differences in host genetics and the gut microbiome suggests that some degree of personalization may be required to properly interpret a number of food-specific biomarkers.
While steady progress is being made to identify food-specific, liver-specific, and other tissue-derived and microbially derived biomarkers, many challenges still exist. Just as with food constituents mentioned above, there is a profound shortage of authentic chemical standards and authentic reference NMR or MS/MS spectra for these important compounds. Fewer than 200 of these compounds appear to exist in chemical or spectral libraries, yet they probably number in the tens of thousands in human biofluids or excreta (72).
Because biotransformed compounds are difficult to isolate and expensive to synthesize via classical organic synthetic chemistry, there are 2 emerging approaches to address these problems. One approach is to enzymatically synthesize these compounds, while the other approach is to computationally generate them (in silico metabolomics). The biosynthetic approach involves adding purified precursors to an artificial gut (74), to homogenized fecal material (75), or to isolated liver microsomes (76) and allowing the selected biomatrix to perform the work. The limitation of this approach is that substantial effort is required to purify the products from each biomatrix and to collect the required MS/MS or NMR spectra. Furthermore, as highlighted earlier, there are relatively few precursor molecules (<1000) available to feed such a biosynthetic pipeline. So, while the experimental approach will likely generate many novel and authentic compounds, it is unlikely to generate enough compounds to cover >10–20% of the desired chemical space.
The in silico approach involves using computational approaches to generate metabolite structure by modeling biotransformation reactions (phase I, phase II, and microbial reactions) on a known set of food constituent precursors. Several commercial programs effectively model these biotransformation processes, as well as a new freeware tool such as BioTransformer (77). Once the compound structures are computationally generated, it is possible to identify them in real samples by matching the observed MS/MS spectra using tools such as CSI-FingerID (78), molecular networking approaches via Global Natural Products Social Molecular Networking (57), or through the comparison of observed MS/MS spectra with predicted MS/MS spectra via CFM-ID (60). The advantages of this in silico approach are that it is fast, inexpensive, and not limited by the availability of physical compounds. The disadvantages are that the predictions are not sufficiently accurate, and no authentic compounds or authentic spectra are generated.
Biomarker measurement should be sensitive enough to capture dietary exposure information and should fall within the dynamic range of measurable limits commonly found in a population. However, dynamic ranges for most biomarkers are not currently known. In addition, from the personalized nutrition and health perspective, ranges may differ depending on physiologic status and vary among adults and children. Capturing this variation is important to understand response compared with nonresponse to a dietary exposure. To ensure sensitivity, concentration ranges (for different age groups) for each biomarker should be well defined (79, 80). Developing and establishing reference ranges across different populations, including children and adults, for a variety of dietary markers is helpful before planning larger studies.
Another area where more data are needed concerns the half-life of putative dietary biomarkers. How fast a dietary compound is absorbed and how long it stays in the system before elimination can affect the timing of sampling and the utility of the biomarker. For example, food components with faster absorption and elimination kinetics have a very narrow window for sampling (e.g., proline betaine for citrus fruits) (44). Similarly, some biomarkers from microbial metabolism (e.g., urolithin) can be detected only 30–45 h after the intake of ellagitannin (81, 82). Metabolites with very short half-lives can provide information only about recent dietary intake, and timing of sampling contributes to variability in measurement and the estimate of intra- and interpersonal variation (83). Depending on study objectives, it may be desirable to combine the metabolites with short half-lives with self-reported dietary intake measurements or with additional select biomarkers with sufficiently longer half-lives (e.g., lipophilic metabolites) to reduce intraindividual variation (7, 84). Comprehensive knowledge of the half-life of metabolites will certainly enhance biomarker identification approaches and expedite biomarker development. To this end, high-throughput methods for evaluating half-lives of metabolites (i.e., biomarkers) are needed to help advance the field.
Integration of Dietary Biomarkers with Other Omics Techniques
Dietary biomarkers are primarily small molecules derived from either the food itself or the digestion and biotransformation of specific food-derived compounds. However, the abundance and the type of potential dietary biomarkers can be significantly altered by physiologic parameters, which can contribute to significant interindividual variability.
Gut microbial metabolism also plays a vital role in determining which circulating metabolites may be present. This has become apparent in relation to several classes of phytochemicals, including the Brassica-derived glucosinolates and flavonoids present in a variety of plant foods (85, 86). A well-known example is the bacterial conversion of the soy isoflavone daidzein to equol, which, due to interindividual differences in gut microbial community composition, occurs in only a subset of individuals upon soy consumption (87, 88). The ability to characterize the gut microbiome and its functional capacity (via 16S rRNA gene sequencing and metagenomics, respectively) has helped to explain the variation in production of some putative dietary biomarkers. However, the fact that so many compounds (both endo-genous and food derived) are affected by microbial metabolism suggests that these effects must be considered in selecting reliable dietary biomarkers (89).
Host genetics also has an important role in determining both the type and abundance of certain dietary biomarkers. Of note is the impact of single-nucleotide polymorphisms (SNPs) on both nutrient metabolism and dietary preferences affecting the metabolites that can be detected and potential biomarkers. Metabolome-wide association studies (MWASs) or genome-wide association studies with metabolomics have linked metabolite levels to many human SNPs (90–92). So far, these studies have identified thousands of SNPs and thousands of metabolites that appear to covary. Some of these SNPs are known to account for ≤60% of the variability of circulating levels of certain metabolites (93).
Given the significant effects of genetics on metabolite levels, it is essential that anyone conducting dietary biomarker studies carefully consider genetic data when selecting or identifying potential dietary biomarkers. The dietary biomarker community has 2 options: 1) use previously collected MWAS data to exclude certain metabolites as potential dietary biomarkers (due to their strong genetic control) or 2) use genetic/SNP data to adjust or recalibrate dietary biomarker data to work for specific individuals. Both approaches are feasible; however, over the short term, it is likely that the use of pre-existing MWAS data to exclude or disqualify proposed dietary biomarkers will be the easiest and most cost-effective approach.
A number of interesting applications of genetics to dietary biomarkers are also starting to emerge. Some of the most fascinating ones may lie with the impact of SNPs on dietary preferences. Individuals who have adverse reactions to certain foods are unlikely to consume them and therefore should not have nutrient markers for those foods. On the other hand, individuals who have cravings for certain foods will likely have an abundance of markers for those foods. Several examples of how SNPs affect dietary preferences (and therefore dietary biomarker levels) are described in Table 3 (94–106).
TABLE 3.
Gene variants and dietary intake association1
| Closest gene | Reference SNP cluster ID | Dietary intake association | Reference |
|---|---|---|---|
| MCM6(intron) | rs182549 | Lactose intolerance | (94) |
| MCM6 (intron) | rs4988235 | Lactose intolerance | (94) |
| MCM6 | rs3754686 | Proxy for milk intake | (95) |
| ALDH2 | rs671 | Alcohol intolerance | (96) |
| ADH1B | rs1229984 | Alcohol aversion | (97) |
| ADH1C | rs698 | Alcohol dependence | (98) |
| KLB | rs11940694 | Increased alcohol consumption | (99) |
| TAS2R38 | rs713598 | Brassica vegetable and coffee aversion | (100) |
| TAS2R38 | rs1726866 | Brassica vegetable and coffee aversion | (100) |
| TAS2R38 | rs10246939 | Brassica vegetable and coffee aversion | (100, 101) |
| OR10A2 | rs72921001 | Cilantro/coriander aversion | (102) |
| CYP1A1 | rs2472297 | Increased coffee consumption | (103) |
| CYP1A1 | rs2470893 | Increased coffee consumption | (103) |
| AHR | rs6968865 | Increased coffee consumption | (103) |
| FGF21 | rs838133 and rs838145 | Sweet tooth (candy preference); increased carbohydrate and lower fat consumption | (104–106) |
| RARB | rs7619139 | Increased carbohydrate consumption | (104) |
| DRAM1 | rs77694286 | Increased protein consumption | (104) |
| FTO | rs1421085 | Increased protein consumption | (104) |
1SNP, single-nucleotide polymorphism.
Overall, the existing evidence strongly suggests that genomics, microbiome analysis, and metagenomics can play a role in the detection, identification, validation, and quantification of many known and putative dietary biomarkers. Therefore, the use of other omics (i.e., nonmetabolomic) techniques in dietary biomarker analysis can serve to complement the metabolomic information that is normally collected for dietary biomarker studies.
Pathways to Precision Nutrition
Simply stated, precision nutrition is the nutritional analog of precision medicine. More specifically, it is nutrition or dietary guidance designed to optimize health, facilitate disease prevention, and enhance therapeutic benefit through molecular (metabolomic, genomic, proteomic, metagenomic) profiling at the level of the individual. Precision nutrition approaches require a keen understanding of how genetic-metabotype-diet interactions affect dietary biomarker levels and determine nutrient status. There are classical examples wherein genetic variation (i.e., SNPs) influences metabolic differences by influencing dietary requirements and responses to different diets. For example, dietary choline deficiency produces liver or muscle dysfunction in most men and postmenopausal women. Fortunately, most premenopausal women are actually protected against choline deficiency, because of the hormonal induction of phosphatidyl ethanolamine-N-methyltransferase (PEMT), an enzyme that enables endogenous synthesis of choline (107). However, a SNP in PEMT (rs12325817) prevents induction by estrogen, making a subset of these women susceptible to choline deficiency, which illustrates how polymorphisms in enzymes, in critical metabolic pathways, can impair nutrient metabolism (108). Unfortunately, these kinds of diet-related SNPs can only be confirmed by challenging individuals with differential diet regimens (low and high).
From this example of differential choline metabolism, it is apparent that precision nutrition approaches require data on both dietary intake and SNPs. Yet, to date, no catalogs of SNPs can inform dietitians or other clinicians about specific nutrient requirements that might serve as the basis for practicing precision nutrition. Therefore, there is a critical need for catalogs of gene signatures that alter the metabolism of nutrients. These SNPs need to be confirmed for whether they can predict changes in a biomarker's relationship to an individual's nutrient status.
Systematic integration of SNP data together with the broader metabotype-based biomarkers will certainly advance precision nutrition efforts. The metabotype-based personalized nutrition approach uses a broader metabolic phenotype that characterizes biologic diversity between and within individuals. For example, comprehensive metabolite and/or lipidomic profiles may provide insights in relation to the response or not to dietary challenges.
Precision nutrition efforts are also emerging through integrated studies of the microbiome and metabolome. In particular, Zeevi et al. (109) showed how a machine-learning algorithm that integrates metabolomic data, dietary habits, physiologic measurements, physical activity, and gut microbiota can predict personalized postprandial glycemic response to complex (regular) meals. This result was further validated in a separate cohort of 100 test subjects and then again in a blinded randomized controlled dietary intervention of 26 individuals. Implementing these molecularly informed custom diets led to significantly lower postprandial glycemic responses and consistent alterations in the gut microbiota of these test subjects. This is an excellent example of a well-conducted, carefully validated biomarker study. It also demonstrates the remarkable potential of precision nutrition and shows how customized dietary guidance can be computationally designed to optimize health and enhance therapeutic benefit through comprehensive, multiomic molecular profiling.
Conclusions
It is evident that there are gaps and challenges to establishing nutrient or food-specific biomarkers. There are also some compelling ideas and novel resources that are starting to emerge that may help to address these challenges. Clearly, more human feeding studies, with well-chosen designs, are needed to fuel the dietary intake biomarker development process.
In addition, it is important to appreciate that dietary biomarker development is a multidisciplinary enterprise and benefits from engaging several collaborative efforts. Fostering collaborations among analytical or natural product chemists, omics (metabolomics, genomics, proteomics, metagenomics) specialists, physicians, dietitians and nutritionists, statisticians, epidemiologists, and bioinformaticians is critical for advancing the field. Chemists are needed to measure, synthesize, or isolate the appropriate chemical standards and to collect the relevant referential spectra. Omics specialists are needed to perform large-scale omics studies to discover or validate the appropriate biomarkers. Physicians, dietitians, nutritionists, and epidemiologists are needed to design the diets or dietary interventions, assemble the cohorts, collect the samples, and acquire the metadata. Statisticians need to be involved at various levels of biomarker development to help with biomarker discovery and validation, assist with data modeling, and account for measurement error. Bioinformaticians are needed to consolidate or integrate the data, develop data exchange standards, create ontologies, and bring some order to this very diverse array of data types. Finally, dedicated study participants are needed to generate the specimens.
Interestingly, such a multifaceted collaboration aimed at discovering food-based biomarkers has recently been undertaken by several countries in the European Union (and Canada) under the Joint Programming Initiative, a Healthy Diet for a Healthy Life. Over the past 4 y the initiative, called FoodBAll or the Food Biomarker Alliance, has generated a wealth of data on dietary biomarkers (63). In particular, FoodBAll brought chemists, omics (metabolomics, transcriptomics, genomics) scientists, dietitians, clinicians, statisticians, and bioinformaticians together to work, collaborate, and create much-needed resources. The result has been a number of useful tools, databases, chemical libraries, white papers, guidelines, and other resources that are starting to form the basis for using dietary biomarkers in nutritional epidemiology (63). This effort has stimulated a keen interest by many other scientific groups and communities around the world to extend and expand these promising ideas and resources. More support for these kinds of concerted and co-ordinated activities is essential to advance dietary biomarker research and to establish precision nutrition as an integral part of the drive toward precision health.
Acknowledgments
The authors’ responsibilities were as follows—PM: has been the primary lead for organizing the workshop; JWL and DSW: chaired the workshop; PM, JWL, and DSW: have taken the primary responsibility of writing the manuscript and for the final content; other authors have actively participated in planning the workshop, presenting their work and sharing their opinions on dietary biomarker development; and all authors: read and approved the final manuscript.
Notes
Perspective articles allow authors to take a position on a topic of current major importance or controversy in the field of nutrition. As such, these articles could include statements based on author opinions or point of view. Opinions expressed in Perspective articles are those of the author and are not attributable to the funder(s) or the sponsor(s) or the publisher, Editor, or Editorial Board of Advances in Nutrition. Individuals with different positions on the topic of a Perspective are invited to submit their comments in the form of a Perspectives article or in a Letter to the Editor.
The workshop was supported by the funds from National Institute of Diabetes and Digestive and Kidney Diseases.
Disclaimer: Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Author disclosures: PM, JWL, DSW, DB, DNC, DD, YD-F, PCD, LOD, JD, LCD, JTD, NJE, OF, REG, FBH, RWK, DMK, MRL, ARL, CJL, SCM, HLN, DMO, JMO, SKO, MP, RP, DR, NR, HMR, SAR, SS, AS, and PRS, no conflicts of interest. SHZ has an equity interest in SNP therapeutics, a company that identifies people with certain genetic polymorphisms that are associated with health problems and develops medical foods to treat these problems. He has had grant funding from Balchem Company, a company that makes choline for diet supplements and animal feed. SHZ is on advisory boards for Baxter, Procter and Gamble, and Abbott, all companies with an interest in choline relative to their products. He received an honorarium for speaking from Nutrapharma, a company that makes diet supplements.
Abbreviations used: AUROC, area under the receiver operating characteristic curve; BFI, biomarker of food intake; CFS, controlled feeding study; DLW, doubly labeled water; DPB, dietary pattern biomarker; FCIB, food component intake biomarker; FoodBAll, Food Biomarker Alliance; IRMS, isotope ratio mass spectrometry; MoNA, MassBank of North America; MWAS, metabolome-wide association study; PEMT, phosphatidyl ethanolamine-N-methyltransferase; ROC, receiver operating characteristic; SIR, stable isotope ratio; SNP, single-nucleotide polymorphism.
References
- 1. Freedman LS, Commins JM, Moler JE, Arab L, Baer DJ, Kipnis V, Midthune D, Moshfegh AJ, Neuhouser ML, Prentice RL et al.. Pooled results from 5 validation studies of dietary self-report instruments using recovery biomarkers for energy and protein intake. Am J Epidemiol. 2014;180(2):172–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun T, Zhang Y, Huang H, Wang X, Zhou L, Li S, Huang S, Xie C, Wen Y, Zhu Y et al.. Plasma alkylresorcinol metabolite, a biomarker of whole-grain wheat and rye intake, and risk of ischemic stroke: a case-control study. Am J Clin Nutr. 2019;109(2):1–7. [DOI] [PubMed] [Google Scholar]
- 3. Brennan L. The nutritional metabolomics crossroads: how to ensure success for dietary biomarkers. Am J Clin Nutr. 2017;105(2):293–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheung W, Keski-Rahkonen P, Assi N, Ferrari P, Freisling H, Rinaldi S, Slimani N, Zamora-Ros R, Rundle M, Frost G et al.. A metabolomic study of biomarkers of meat and fish intake. Am J Clin Nutr. 2017;105(3):600–8. [DOI] [PubMed] [Google Scholar]
- 5. Guasch-Ferre M, Bhupathiraju SN, Hu FB. Use of metabolomics in improving assessment of dietary intake. Clin Chem. 2018;64(1):82–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Corella D, Ordovas JM.. Biomarkers: background, classification and guidelines for applications in nutritional epidemiology. Nutr Hosp. 2015;31 Suppl 3:177–88. [DOI] [PubMed] [Google Scholar]
- 7. Scalbert A, Rothwell JA, Keski-Rahkonen P, Neveu V. The Food Metabolome and Dietary Biomarkers. Boca Raton (FL): CRC Press; 2017. [Google Scholar]
- 8. Biesalski HK, Dragsted LO, Elmadfa I, Grossklaus R, Muller M, Schrenk D, Walter P, Weber P. Bioactive compounds: definition and assessment of activity. Nutrition. 2009;25(11–12):1202–5. [DOI] [PubMed] [Google Scholar]
- 9. Gao Q, Pratico G, Scalbert A, Vergeres G, Kolehmainen M, Manach C, Brennan L, Afman LA, Wishart DS, Andres-Lacueva C et al.. A scheme for a flexible classification of dietary and health biomarkers. Genes Nutr. 2017;12:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sobczynska-Malefora A, Harrington DJ.. Laboratory assessment of folate (vitamin B9) status. J Clin Pathol. 2018;71(11):949–56. [DOI] [PubMed] [Google Scholar]
- 11. Scalbert A, Brennan L, Manach C, Andres-Lacueva C, Dragsted LO, Draper J, Rappaport SM, van der Hooft JJ, Wishart DS. The food metabolome: a window over dietary exposure. Am J Clin Nutr. 2014;99(6):1286–308. [DOI] [PubMed] [Google Scholar]
- 12. Thomas DM, Watts K, Friedman S, Schoeller DA. Modelling the metabolism: allometric relationships between total daily energy expenditure, body mass, and height. Eur J Clin Nutr. 2019;73(5):763–9. [DOI] [PubMed] [Google Scholar]
- 13. O'Brien DM. Stable isotope ratios as biomarkers of diet for health research. Annu Rev Nutr. 2015;35:565–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choy K, Nash SH, Kristal AR, Hopkins S, Boyer BB, O'Brien DM. The carbon isotope ratio of alanine in red blood cells is a new candidate biomarker of sugar-sweetened beverage intake. J Nutr. 2013;143(6):878–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yeung EH, Saudek CD, Jahren AH, Kao WH, Islas M, Kraft R, Coresh J, Anderson CA. Evaluation of a novel isotope biomarker for dietary consumption of sweets. Am J Epidemiol. 2010;172(9):1045–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yun HY, Lampe JW, Tinker LF, Neuhouser ML, Beresford SAA, Niles KR, Mossavar-Rahmani Y, Snetselaar LG, Van Horn L, Prentice RL et al.. Serum nitrogen and carbon stable isotope ratios meet biomarker criteria for fish and animal protein intake in a controlled feeding study of a Women's Health Initiative Cohort. J Nutr. 2018;148(12):1931–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Neveu V, Perez-Jimenez J, Vos F, Crespy V, du Chaffaut L, Mennen L, Knox C, Eisner R, Cruz J, Wishart D et al.. Phenol-Explorer: an online comprehensive database on polyphenol contents in foods. Database (Oxford). 2010;2010:bap024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edmands WM, Ferrari P, Rothwell JA, Rinaldi S, Slimani N, Barupal DK, Biessy C, Jenab M, Clavel-Chapelon F, Fagherazzi G et al.. Polyphenol metabolome in human urine and its association with intake of polyphenol-rich foods across European countries. Am J Clin Nutr. 2015;102(4):905–13. [DOI] [PubMed] [Google Scholar]
- 19. Zamora-Ros R, Achaintre D, Rothwell JA, Rinaldi S, Assi N, Ferrari P, Leitzmann M, Boutron-Ruault MC, Fagherazzi G, Auffret A et al.. Urinary excretions of 34 dietary polyphenols and their associations with lifestyle factors in the EPIC cohort study. Sci Rep. 2016;6:26905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dragsted LO, Gao Q, Scalbert A, Vergeres G, Kolehmainen M, Manach C, Brennan L, Afman LA, Wishart DS, Andres Lacueva C et al.. Validation of biomarkers of food intake-critical assessment of candidate biomarkers. Genes Nutr. 2018;13:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Illner AK, Freisling H, Boeing H, Huybrechts I, Crispim SP, Slimani N. Review and evaluation of innovative technologies for measuring diet in nutritional epidemiology. Int J Epidemiol. 2012;41(4):1187–203. [DOI] [PubMed] [Google Scholar]
- 22. Neuhouser ML, Tinker L, Shaw PA, Schoeller D, Bingham SA, Horn LV, Beresford SA, Caan B, Thomson C, Satterfield S et al.. Use of recovery biomarkers to calibrate nutrient consumption self-reports in the Women's Health Initiative. Am J Epidemiol. 2008;167(10):1247–59. [DOI] [PubMed] [Google Scholar]
- 23. Lloyd AJ, Beckmann M, Haldar S, Seal C, Brandt K, Draper J. Data-driven strategy for the discovery of potential urinary biomarkers of habitual dietary exposure. Am J Clin Nutr. 2013;97(2):377–89. [DOI] [PubMed] [Google Scholar]
- 24. Playdon MC, Sampson JN, Cross AJ, Sinha R, Guertin KA, Moy KA, Rothman N, Irwin ML, Mayne ST, Stolzenberg-Solomon R et al.. Comparing metabolite profiles of habitual diet in serum and urine. Am J Clin Nutr. 2016;104(3):776–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tasevska N, Runswick SA, Welch AA, McTaggart A, Bingham SA. Urinary sugars biomarker relates better to extrinsic than to intrinsic sugars intake in a metabolic study with volunteers consuming their normal diet. Eur J Clin Nutr. 2009;63(5):653–9. [DOI] [PubMed] [Google Scholar]
- 26. Serrano JC, Jove M, Gonzalo H, Pamplona R, Portero-Otin M. Nutridynamics: mechanism(s) of action of bioactive compounds and their effects. Int J Food Sci Nutr. 2015;66 Suppl 1:S22–30. [DOI] [PubMed] [Google Scholar]
- 27. van Duynhoven JPM, van Velzena EJJ, Westerhuis JA, Foltz M, Jacobs DM, Smilde AK. Nutrikinetics: concept, technologies, applications, perspectives. Trends Food Sci Tech. 2012;26(1):4–13. [Google Scholar]
- 28. Lampe JW, King IB, Li S, Grate MT, Barale KV, Chen C, Feng Z, Potter JD. Brassica vegetables increase and apiaceous vegetables decrease cytochrome P450 1A2 activity in humans: changes in caffeine metabolite ratios in response to controlled vegetable diets. Carcinogenesis. 2000;21(6):1157–62. [PubMed] [Google Scholar]
- 29. McEvoy CT, Wallace IR, Hamill LL, Hunter SJ, Neville CE, Patterson CC, Woodside JV, Young IS, McKinley MC. Increasing fruit and vegetable intake has no dose-response effect on conventional cardiovascular risk factors in overweight adults at high risk of developing cardiovascular disease. J Nutr. 2015;145(7):1464–71. [DOI] [PubMed] [Google Scholar]
- 30. Navarro SL, Schwarz Y, Song X, Wang CY, Chen C, Trudo SP, Kristal AR, Kratz M, Eaton DL, Lampe JW. Cruciferous vegetables have variable effects on biomarkers of systemic inflammation in a randomized controlled trial in healthy young adults. J Nutr. 2014;144(11):1850–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lampe JW, Huang Y, Neuhouser ML, Tinker LF, Song X, Schoeller DA, Kim S, Raftery D, Di C, Zheng C et al.. Dietary biomarker evaluation in a controlled feeding study in women from the Women's Health Initiative cohort. Am J Clin Nutr. 2017;105(2):466–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heinzmann SS, Brown IJ, Chan Q, Bictash M, Dumas ME, Kochhar S, Stamler J, Holmes E, Elliott P, Nicholson JK. Metabolic profiling strategy for discovery of nutritional biomarkers: proline betaine as a marker of citrus consumption. Am J Clin Nutr. 2010;92(2):436–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Munger LH, Trimigno A, Picone G, Freiburghaus C, Pimentel G, Burton KJ, Pralong FP, Vionnet N, Capozzi F, Badertscher R et al.. Identification of urinary food intake biomarkers for milk, cheese, and soy-based drink by untargeted GC-MS and NMR in healthy humans. J Proteome Res. 2017;16(9):3321–35. [DOI] [PubMed] [Google Scholar]
- 34. Tasevska N, Midthune D, Tinker LF, Potischman N, Lampe JW, Neuhouser ML, Beasley JM, Van Horn L, Prentice RL, Kipnis V. Use of a urinary sugars biomarker to assess measurement error in self-reported sugars intake in the Nutrition and Physical Activity Assessment Study (NPAAS). Cancer Epidemiol Biomarkers Prev. 2014;23(12):2874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y et al.. American Gut: an Open Platform for Citizen Science Microbiome Research. (2018 May-Jun) mSystems 3,e00031-1829795809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Raghavan R, Ashour FS, Bailey R. A review of cutoffs for nutritional biomarkers. Adv Nutr. 2016;7(1):112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burri BJ, Neidlinger TR, Clifford AJ. Serum carotenoid depletion follows first-order kinetics in healthy adult women fed naturally low carotenoid diets. J Nutr. 2001;131(8):2096–100. [DOI] [PubMed] [Google Scholar]
- 38. Davidson EA, Pickens CA, Fenton JI. Increasing dietary EPA and DHA influence estimated fatty acid desaturase activity in systemic organs which is reflected in the red blood cell in mice. Int J Food Sci Nutr. 2018;69(2):183–91. [DOI] [PubMed] [Google Scholar]
- 39. Filippini T, Ferrari A, Michalke B, Grill P, Vescovi L, Salvia C, Malagoli C, Malavolti M, Sieri S, Krogh V et al.. Toenail selenium as an indicator of environmental exposure: a cross-sectional study. Mol Med Rep. 2017;15(5):3405–12. [DOI] [PubMed] [Google Scholar]
- 40. Le Marchand L, Yonemori K, White KK, Franke AA, Wilkens LR, Turesky RJ. Dose validation of PhIP hair level as a biomarker of heterocyclic aromatic amines exposure: a feeding study. Carcinogenesis. 2016;37(7):685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Valenzuela LO, O'Grady SP, Enright LE, Murtaugh M, Sweeney C, Ehleringer JR. Evaluation of childhood nutrition by dietary survey and stable isotope analyses of hair and breath. Am J Hum Biol. 2018;30(3):e23103. [DOI] [PubMed] [Google Scholar]
- 42. Prentice RL, Willett WC, Greenwald P, Alberts D, Bernstein L, Boyd NF, Byers T, Clinton SK, Fraser G, Freedman L et al.. Nutrition and physical activity and chronic disease prevention: research strategies and recommendations. J Natl Cancer Inst. 2004;96(17):1276–87. [DOI] [PubMed] [Google Scholar]
- 43. Garcia-Perez I, Posma JM, Chambers ES, Nicholson JK, Mathers JC, Beckmann M, Draper J, Holmes E, Frost G. An analytical pipeline for quantitative characterization of dietary intake: application to assess grape intake. J Agric Food Chem. 2016;64(11):2423–31. [DOI] [PubMed] [Google Scholar]
- 44. Lloyd AJ, Beckmann M, Fave G, Mathers JC, Draper J. Proline betaine and its biotransformation products in fasting urine samples are potential biomarkers of habitual citrus fruit consumption. Br J Nutr. 2011;106(6):812–24. [DOI] [PubMed] [Google Scholar]
- 45. Kotsopoulos J, Tworoger SS, Campos H, Chung FL, Clevenger CV, Franke AA, Mantzoros CS, Ricchiuti V, Willett WC, Hankinson SE et al.. Reproducibility of plasma and urine biomarkers among premenopausal and postmenopausal women from the Nurses’ Health Studies. Cancer Epidemiol Biomarkers Prev. 2010;19(4):938–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Townsend MK, Clish CB, Kraft P, Wu C, Souza AL, Deik AA, Tworoger SS, Wolpin BM. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin Chem. 2013;59(11):1657–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fave G, Beckmann M, Lloyd AJ, Zhou S, Harold G, Lin W, Tailliart K, Xie L, Draper J, Mathers JC. Development and validation of a standardized protocol to monitor human dietary exposure by metabolite fingerprinting of urine samples. Metabolomics. 2011;7(4):469–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jacob M, Malkawi A, Albast N, Al Bougha S, Lopata A, Dasouki M, Abdel Rahman AM. A targeted metabolomics approach for clinical diagnosis of inborn errors of metabolism. Anal Chim Acta. 2018;1025:141–53. [DOI] [PubMed] [Google Scholar]
- 49. Elliott DRF, Walker AW, O'Donovan M, Parkhill J, Fitzgerald RC. A non-endoscopic device to sample the oesophageal microbiota: a case-control study. Lancet Gastroenterol Hepatol. 2017;2(1):32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cajka T, Smilowitz JT, Fiehn O. Validating quantitative untargeted lipidomics across nine liquid chromatography-high-resolution mass spectrometry platforms. Anal Chem. 2017;89(22):12360–8. [DOI] [PubMed] [Google Scholar]
- 51. Domingo-Almenara X, Montenegro-Burke JR, Ivanisevic J, Thomas A, Sidibe J, Teav T, Guijas C, Aisporna AE, Rinehart D, Hoang L et al.. XCMS-MRM and METLIN-MRM: a cloud library and public resource for targeted analysis of small molecules. Nat Methods. 2018;15(9):681–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12(6):523–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rafiei A, Sleno L.. Comparison of peak-picking workflows for untargeted liquid chromatography/high-resolution mass spectrometry metabolomics data analysis. Rapid Commun Mass Spectrom. 2015;29(1):119–27. [DOI] [PubMed] [Google Scholar]
- 55. Fiehn O. MassBank of North America. [Internet].2018. Available from: mona.fiehnlab.ucdavis.edu. [Google Scholar]
- 56. Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30(9):826–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T et al.. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34(8):828–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, Sajed T, Johnson D, Li C, Karu N et al.. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ruttkies C, Schymanski EL, Wolf S, Hollender J, Neumann S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J Cheminform. 2016;8:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Allen F, Pon A, Wilson M, Greiner R, Wishart D. CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res. 2014;42(Web Server issue):W94–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mass Frontier 8.0 Spectral Interpretation Software. Waltham (MA):Thermo Fisher Scientific; 2018. [Google Scholar]
- 62. Manach C, Weinert C, Wishart D. Food Compound Exchange [Internet].2018. Available from: http://foodcomex.org/. [Google Scholar]
- 63. FoodBAll FBA. [Internet]. Available from: http://foodmetabolome.org/. [Google Scholar]
- 64. Chong J, Xia J.. MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics. 2018;34(24):4313–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kirpich AS, Ibarra M, Moskalenko O, Fear JM, Gerken J, Mi X, Ashrafi A, Morse AM, McIntyre LM. SECIMTools: a suite of metabolomics data analysis tools. BMC Bioinformatics. 2018;19(1):151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Prentice RL, Huang Y, Tinker LF, Beresford SA, Lampe JW, Neuhouser ML. Statistical aspects of the use of biomarkers in nutritional epidemiology research. Stat Biosci. 2009;1(1):112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Prentice RL, Pettinger M, Neuhouser ML, Tinker LF, Huang Y, Zheng C, Manson JE, Mossavar-Rahmani Y, Anderson GL, Lampe JW. Application of blood concentration biomarkers in nutritional epidemiology: example of carotenoid and tocopherol intake in relation to chronic disease risk. Am J Clin Nutr. 2019;109(4):1189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gebhardt SEC, Howe RL, Haytowitz JC, Pehrsson DB, Lemar PR, Holcomb LE, Stup GT, Thomas MA, Exler RG, Showell J et al.. USDA national nutrient database for standard reference, release 19, Beltsville (MD): USDA; 2006. [Google Scholar]
- 69. Manach C. Phytohub. [Internet] 2016. Available from: http://phytohub.eu/. [Google Scholar]
- 70. Neveu V, Moussy A, Rouaix H, Wedekind R, Pon A, Knox C, Wishart DS, Scalbert A. Exposome-Explorer: a manually-curated database on biomarkers of exposure to dietary and environmental factors. Nucleic Acids Res. 2017;45(D1):D979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rothwell JA, Urpi-Sarda M, Boto-Ordonez M, Llorach R, Farran-Codina A, Barupal DK, Neveu V, Manach C, Andres-Lacueva C, Scalbert A. Systematic analysis of the polyphenol metabolome using the Phenol-Explorer database. Mol Nutr Food Res. 2016;60(1):203–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wishart DS. FooDB: the food composition database. [Internet] 2018. Available from: http://foodb.ca. [Google Scholar]
- 73. da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proc Natl Acad Sci USA. 2015;112(41):12549–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Alander M, De Smet I, Nollet L, Verstraete W, von Wright A, Mattila-Sandholm T. The effect of probiotic strains on the microbiota of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME). Int J Food Microbiol. 1999;46(1):71–9. [DOI] [PubMed] [Google Scholar]
- 75. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA et al.. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551(7682):648–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zuniga A, Li L.. Ultra-high performance liquid chromatography tandem mass spectrometry for comprehensive analysis of urinary acylcarnitines. Anal Chim Acta. 2011;689(1):77–84. [DOI] [PubMed] [Google Scholar]
- 77. Djoumbou-Feunang Y, Fiamoncini J, Gil-de-la-Fuente A, Greiner R, Manach C, Wishart DS. BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J Cheminform. 2019;11(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Duhrkop K, Shen H, Meusel M, Rousu J, Bocker S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc Natl Acad Sci USA. 2015;112(41):12580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Achaintre D, Bulete A, Cren-Olive C, Li L, Rinaldi S, Scalbert A. Differential isotope labeling of 38 dietary polyphenols and their quantification in urine by liquid chromatography electrospray ionization tandem mass spectrometry. Anal Chem. 2016;88(5):2637–44. [DOI] [PubMed] [Google Scholar]
- 80. Achaintre D, Gicquiau A, Li L, Rinaldi S, Scalbert A. Quantification of 38 dietary polyphenols in plasma by differential isotope labelling and liquid chromatography electrospray ionization tandem mass spectrometry. J Chromatogr A. 2018;1558:50–8. [DOI] [PubMed] [Google Scholar]
- 81. Cerda B, Periago P, Espin JC, Tomas-Barberan FA. Identification of urolithin a as a metabolite produced by human colon microflora from ellagic acid and related compounds. J Agric Food Chem. 2005;53(14):5571–6. [DOI] [PubMed] [Google Scholar]
- 82. Piwowarski JP, Granica S, Stefanska J, Kiss AK. Differences in metabolism of ellagitannins by human gut microbiota ex vivo cultures. J Nat Prod. 2016;79(12):3022–30. [DOI] [PubMed] [Google Scholar]
- 83. National Research Council Human Biomonitoring for Environmental Chemicals. Washington (DC): National Academies Press; 2006. [Google Scholar]
- 84. Radtke J, Linseisen J, Wolfram G. Fasting plasma concentrations of selected flavonoids as markers of their ordinary dietary intake. Eur J Nutr. 2002;41(5):203–9. [DOI] [PubMed] [Google Scholar]
- 85. Cassidy A, Minihane AM.. The role of metabolism (and the microbiome) in defining the clinical efficacy of dietary flavonoids. Am J Clin Nutr. 2017;105(1):10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Narbad A, Rossiter JT.. Gut glucosinolate metabolism and isothiocyanate production. Mol Nutr Food Res. 2018;62(18):e1700991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Atkinson C, Berman S, Humbert O, Lampe JW. In vitro incubation of human feces with daidzein and antibiotics suggests interindividual differences in the bacteria responsible for equol production. J Nutr. 2004;134(3):596–9. [DOI] [PubMed] [Google Scholar]
- 88. Decroos K, Vanhemmens S, Cattoir S, Boon N, Verstraete W. Isolation and characterisation of an equol-producing mixed microbial culture from a human faecal sample and its activity under gastrointestinal conditions. Arch Microbiol. 2005;183(1):45–55. [DOI] [PubMed] [Google Scholar]
- 89. Hullar MA, Lancaster SM, Li F, Tseng E, Beer K, Atkinson C, Wahala K, Copeland WK, Randolph TW, Newton KM et al.. Enterolignan-producing phenotypes are associated with increased gut microbial diversity and altered composition in premenopausal women in the United States. Cancer Epidemiol Biomarkers Prev. 2015;24(3):546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kastenmuller G, Raffler J, Gieger C, Suhre K. Genetics of human metabolism: an update. Hum Mol Genet. 2015;24(R1):R93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP et al.. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yet I, Menni C, Shin SY, Mangino M, Soranzo N, Adamski J, Suhre K, Spector TD, Kastenmuller G, Bell JT. Genetic influences on metabolite levels: a comparison across metabolomic platforms. PLoS One. 2016;11(4):e0153672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rhee EP, Yang Q, Yu B, Liu X, Cheng S, Deik A, Pierce KA, Bullock K, Ho JE, Levy D et al.. An exome array study of the plasma metabolome. Nat Commun. 2016;7:12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ingram CJ, Mulcare CA, Itan Y, Thomas MG, Swallow DM. Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet. 2009;124(6):579–91. [DOI] [PubMed] [Google Scholar]
- 95. Smith CE, Coltell O, Sorli JV, Estruch R, Martinez-Gonzalez MA, Salas-Salvado J, Fito M, Aros F, Dashti HS, Lai CQ et al.. Associations of the MCM6-rs3754686 proxy for milk intake in Mediterranean and American populations with cardiovascular biomarkers, disease and mortality: Mendelian randomization. Sci Rep. 2016;6:33188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Li D, Zhao H, Gelernter J. Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol-induced medical diseases in Asians. Hum Genet. 2012;131(5):725–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D et al.. Genome-wide association study of alcohol dependence: significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry. 2014;19(1):41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Li D, Zhao H, Gelernter J. Further clarification of the contribution of the ADH1C gene to vulnerability of alcoholism and selected liver diseases. Hum Genet. 2012;131(8):1361–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Schumann G, Liu C, O'Reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S et al.. KLB is associated with alcohol drinking, and its gene product beta-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci USA. 2016;113(50):14372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Calancie L, Keyserling TC, Taillie LS, Robasky K, Patterson C, Ammerman AS, Schisler JC. TAS2R38 predisposition to bitter taste associated with differential changes in vegetable intake in response to a community-based dietary intervention. G3 (Bethesda). 2018;8(6):2107–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mikolajczyk-Stecyna J, Malinowska AM, Chmurzynska A. TAS2R38 and CA6 genetic polymorphisms, frequency of bitter food intake, and blood biomarkers among elderly woman. Appetite. 2017;116:57–64. [DOI] [PubMed] [Google Scholar]
- 102. Eriksson N, Wu S, Do CB, Kiefer AK, Tung JY, Mountain JL, Hinds DA, Francke U. A genetic variant near olfactory receptor genes influences cilantro preference. Flavour. 2012;1:22. [Google Scholar]
- 103. Coffee and Caffeine Genetics Consortium; Cornelis MC, Byrne EM, Esko T, Nalls MA, Ganna A, Paynter N, Monda KL, Amin N. Genome-wide meta-analysis identifies six novel loci associated with habitual coffee consumption. Mol Psychiatry. 2015;20(5):647–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Merino J, Dashti HS, Li SX, Sarnowski C, Justice AE, Graff M, Papoutsakis C, Smith CE, Dedoussis GV, Lemaitre RN et al.. Genome-wide meta-analysis of macronutrient intake of 91,114 European ancestry participants from the cohorts for heart and aging research in genomic epidemiology consortium. Mol Psychiatry. 2019;24(12):1920–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Tanaka T, Ngwa JS, van Rooij FJ, Zillikens MC, Wojczynski MK, Frazier-Wood AC, Houston DK, Kanoni S, Lemaitre RN, Luan J et al.. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr. 2013;97(6):1395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. von Holstein-Rathlou S, BonDurant LD, Peltekian L, Naber MC, Yin TC, Claflin KE, Urizar AI, Madsen AN, Ratner C, Holst B et al.. FGF21 mediates endocrine control of simple sugar intake and sweet taste preference by the liver. Cell Metab. 2016;23(2):335–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zeisel SH. Nutritional genomics: defining the dietary requirement and effects of choline. J Nutr. 2011;141(3):531–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Resseguie ME, da Costa KA, Galanko JA, Patel M, Davis IJ, Zeisel SH. Aberrant estrogen regulation of PEMT results in choline deficiency-associated liver dysfunction. J Biol Chem. 2011;286(2):1649–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M et al.. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–94. [DOI] [PubMed] [Google Scholar]


