Summary/Abstract
The split inteins from the DnaE cyanobacterial family are efficient and versatile tools for protein engineering and chemical biology applications. Their ultrafast splicing kinetics allow the efficient production of native proteins from two separate polypeptides both in vitro and in cells. They can also be used to generate proteins with C-terminal thioesters for downstream applications. In this chapter, we describe a method based on a genetically fused version of the DnaE intein Npu for the preparation of doubly modified proteins through recombinant expression. In particular, we provide protocols for the recombinant production of modified ubiquitin through amber suppression where 1) fused Npu is used as a traceless purification tag, or 2) as a protein engineering tool to introduce C-terminal modifications for subsequent attachment to other proteins of interest. Our purification protocol allows for quick and facile separation of truncated products and eliminates the need for engineering protease cleavage sites. Our approach can be easily adapted to different proteins and applications where the simultaneous presence of internal and C-terminal modifications is desirable.
Keywords: Intein, Ubiquitin, Amber Suppression, Unnatural Amino Acid Incorporation, Protein Modification, Tagless Protein Purification
1. Introduction
Inteins are a group of autoprocessing domains that can excise themselves from their host polypeptide chain while concurrently building a native peptide bond that connects the flanking host residues. This process, called intein splicing, does not require energy input in the form of ATP and relies instead on the correct folding of the intein and a series of steps that involve strategically positioned cysteine or serine residues [1–4]. There are two types of inteins found in nature: contiguous versions where the intein domain represents one continuous polypeptide chain [5,6], and split versions where the intein domain is separated into two polypeptides flanked by the N- or C-terminal segments of the host protein (also called extein) [7–9]. Both the contiguous and split inteins have become valuable tools for protein engineering including the purification, modification, cyclization and ligation of proteins and peptides of various lengths [1,10]. Historically, these applications have been performed with contiguous inteins such as the robust gyrase A intein from Mycobacterium xenopi (Mxe GyrA) [11–13]. For example, commercial kits for affinity column purification are available where the protein of interest is fused to an engineered version of GyrA and a chitin binding domain [14]. After expression, the construct is bound to a chitin resin column and on column cleavage is performed with a thiol reagent such as dithiothreitol (DTT), resulting in the release of pure full-length protein of interest. Similarly, the GyrA intein has been a reliable and useful tool for the generation of C-terminal thioesters and their downstream use in expressed protein ligation applications [11,13]. The use of genetically fused split inteins from the cyanobacterial DnaE split intein family in lieu of GyrA, however, offers several advantages [7]. First, Npu and other members of this family exhibit ultrafast kinetics and high splicing efficiency, properties that can increase the yield of the purified or modified protein and reduce the time required to handle aggregation prone samples. Second, engineered versions of Npu can work efficiently at high temperatures and high concentrations of urea enabling the purification and modification of proteins that require denaturing conditions [15,16].
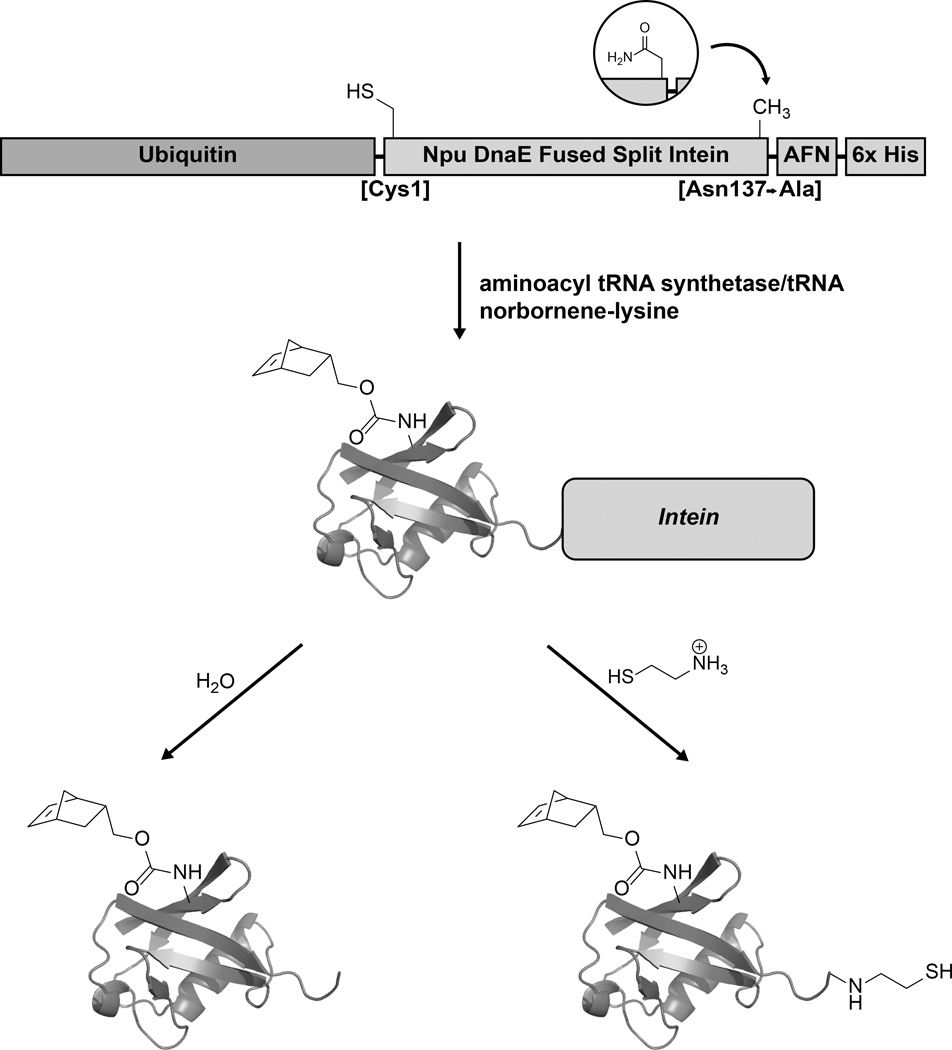
In this chapter, we describe a generalizable method to harness the beneficial properties of genetically fused Npu and to introduce modifications at multiple sites in a single protein of interest, in this case the 76-residue protein ubiquitin. Our construct starts with ubiquitin, followed by fused Npu, a small linker and a hexahistidine purification tag (Figure 1). The hexahistidine tag allows the separation of the full-length protein from any truncations, while the intein subsequently removes the histidine tag in a traceless and efficient manner without the need of proteases. In the intein portion of the construct, the last residue of Npu – typically an asparagine residue important for the splicing mechanism [7] – is mutated to alanine (Figure 2). This mutation biases the intein reactivity toward N-terminal cleavage at the junction between ubiquitin and Npu [1]. Cleavage can be promoted by water molecules at slightly basic pH resulting in the release of the protein of interest, or by appropriate thiol reagents leading to the modification of the C-terminus. The water-mediated cleavage reaction can be exploited to efficiently purify ubiquitin produced by amber suppression [17] and to incorporate the unnatural amino acid (UAA) norborne-lysine at position 6 of the ubiquitin sequence [18]. The thiolysis reaction, on the other hand, enables the introduction of a C-terminal thioester that can be used in protein ligation applications and in the modification of the C-terminus with various reagents. In this chapter, we illustrate this process using the reagent aminoethanethiol. This generates a ubiquitin construct that can be attached to other proteins of interest through an asymmetric disulfide linkage [19]. More importantly, the two strategies can be combined to produce ubiquitin containing both an internal and a C-terminal modification [20]. We have also successfully used this strategy to produce isotopically labeled and modified ubiquitin proteins [20], further illustrating the utility and efficiency of the protocol described below.
Figure 1.
The fused split intein Npu can be used as a traceless purification tag for the recombinant production of modified proteins through amber suppression and/or as a tool to introduce C-terminal thioesters for downstream protein ligation. In our construct, ubiquitin is genetically fused to Npu, followed by a small linker (AFN) and a hexahistidine tag for affinity purification. The last residue of Npu, N137, is mutated to alanine to promote intein cleavage at the ubiquitin-Npu junction.
Figure 2.
Sequence of the K6X-Ubiquitin-NpuDnaE-AFN-His6 protein used as an example in this protocol. The position of the norbornene lysine amino acid (X) is underlined, the ubiquitin sequence is shown in black, the NpuDnaE intein is in bold characters, while the AFN-His6 sequence at the C-terminus is shown in italic font.
Our protocol starts with the optimization of protein expression and the incorporation of the unnatural amino acid norbornene lysine through amber suppression on a small scale. Following a scale-up of the protein expression, cell lysis and affinity purification, we split the protocol into two parts. The first part describes the intein hydrolysis protocol used to generate the native ubiquitin C-terminus, while the second part describes the intein thiolysis steps necessary to modify the protein with C-terminal aminoethanethiol moiety. We also present a preparative-scale reverse-phase purification protocol applicable to both intein cleavage reactions and end with a refolding protocol for the modified ubiquitin construct.
2. Materials
All solutions and media are prepared in ultrapure deionized water (purified to a resistivity of 18.2 MΩ·cm at 25 °C) using analytical grade reagents unless indicated otherwise in the text. All materials used in the preparation of gels and purification columns are reagent grade unless otherwise stated.
2.1. Plasmid Constructs
The pTXB1-Ubiquitin-NpuDnaE-AFN-His6 plasmid used herein was provided as a generous gift by the lab of Dr. Tom Μuir [7]. The plasmid contains an ampicillin resistance marker and expression of the ubiquitin-fused split intein construct Ubiquitin-NpuDnaE-AFN-H6 (Figure 2) is placed under control of the T7 promoter. Store at −20 °C until use.
Primers to replace the codon for lysine 6 in the ubiquitin sequence with the amber codon (TAG).
The pEVOL-tRNApyl/pylRSY306A,Y384F plasmid used herein was provided as a generous gift by the lab of Dr. Carsten Schultz [18]. The plasmid contains a chloramphenicol resistance marker and expression of the tRNA synthetase/tRNA pair is placed under control of the pBAD promoter. The encoded tRNA synthetase can efficiently incorporate lysine variants carrying large head groups such as the norbornene-lysine UAA used here. Store at −20 °C until use.
2.2. Protein Expression
Ampicillin stock solution: 100 mg in 1 mL water. Filter with a 0.22 μm syringe-tip filter. Store at −20 °C until use.
Chloramphenicol stock solution: 35 mg in 1 mL ethanol. Filter with a 0.22 μm syringe-tip filter. Store at −20 °C until use.
IPTG stock solution: 500 mM in water. Filter with a 0.22 μm syringe-tip filter. Store at −20 °C until use.
Arabinose stock solution: 20% w/v in water. Filter with a 0.22 μm syringe-tip filter. Store at −20 °C until use.
BL21 (DE3) competent cells. Store as received at −80 °C until use.
LB Agar Plates: 40 g Miller LB Agar (see Note 1) in 1 L water. Autoclave and when the temperature of the media has dropped below 65 °C, add 1 mL of ampicillin stock solution for a final concentration of 100 μg/mL and 1 mL of chloramphenicol stock solution for a final concentration of 35 μg/mL. Pour media into sterile petri dishes and allow them to cool to room temperature before use. Store unused plates at 4 °C.
LB media: 25 g Miller LB (see Note 2) in 1 L water. Autoclave and allow media to cool to room temperature before use. Store unused media at 4 °C.
Norbornene lysine: 0.1 M in 0.5 M sodium hydroxide. See synthesis protocol below.
12% Tris-glycine gels for SDS-PAGE
Thiol-free SDS-PAGE loading buffer: 2% w/v sodium dodecyl sulfate (SDS), 10% v/v glycerol, 12.5 mM ethylenediaminetetraacetic acid (EDTA), and 0.02% w/v bromophenol blue in 50 mM Tris-HCl, pH 6.8.
2.3. Protein Purification
2.3.1. Ni-NTA Affinity Purification
1 M tris(2-carboxyethyl)phosphine (TCEP): 280 mg in 530 μL of 5 M sodium hydroxide and 470 μL of 1 M unbuffered Tris base.
Lysis buffer: 50 mM sodium phosphate (monobasic), 300 mM sodium chloride, and 5 mM imidazole (see Note 3). Adjust to a final pH of 7.5. Add 1 protease inhibitor tablet per 50 mL buffer immediately before use (see Note 4).
Wash buffer 1: 50 mM sodium phosphate (monobasic), 300 mM sodium chloride, and 20 mM imidazole. Adjust to a final pH of 7.5.
Wash buffer 2: 50 mM sodium phosphate (monobasic), 300 mM sodium chloride, and 50 mM imidazole. Adjust to a final pH of 7.5.
Elution buffer: 50 mM sodium phosphate (monobasic), 300 mM sodium chloride, and 250 mM imidazole. Adjust to a final pH of 7.5.
Nickel (II) nitrilotriacetic acid (Ni-NTA) resin (see Note 5).
150 mL aluminum beaker.
Sonicator: 1/8″ diameter probe tip, power 125 W, frequency 20 kHz.
2.3.2. High Performance Liquid Chromatography (HPLC) Purification
XBridge Peptide BEH C18 300Å, 5 μm, 4.6 mm x 150 mm analytical HPLC column (Waters).
XBridge Peptide BEH C18 300Å, 5 μm, 10 mm x 250 mm preparative-scale HPLC column (Waters).
HPLC solvent A: 0.1% TFA in ultrapure or HPLC grade water.
HPLC solvent B: 0.1% TFA in HPLC grade acetonitrile.
Trifluoroacetic acid (TFA).
2.3.3. Modification of the C-terminus
2-aminoethanethiol stock solution: 1 M in 50 mM sodium phosphate and 300 mM sodium chloride. Adjust to a final pH of 7.5.
2.4. Synthesis of norbornene lysine
Triphosgene: 4.8 g (1 equivalent), CAS 32315-10-9.
5-Norbornene-2-methanol: 2 g (1 equivalent), CAS 95-12-5.
Boc-L-Lysine-OH: 4.8 g (1.2 equivalent), CAS 13734-28-6.
Anhydrous tetrahydrofuran (THF): 40 mL.
1.0 M NaOH/THF: 2 to 1 v/v. Combine 60 mL of 1.0 M sodium hydroxide (NaOH) and 30 mL of anhydrous THF.
Ethyl acetate.
Hexanes.
N,N-Dimethyl formamide (DMF): 90 mL.
Anhydrous sodium sulfate.
1 M HCl solution: 83.3 mL of 12 N HCl and deionized water up to 1 L total volume.
50 mM HCl solution: 10 mL of 1 M HCl solution to 190 mL of deionized water.
Saturated NaCl solution.
60% formic acid in chloroform: 6 to 4 v/v. Combine 15 mL of formic acid and 10 mL of chloroform.
Schlenk line with argon gas and high vacuum valve.
Magnetic stirrer and stir bars.
Rotary evaporator.
Separatory funnel.
Glass column for flash chromatography.
Silica gel powder: CAS 7631-86-9.
2.5. Ubiquitin Refolding
Denaturing buffer: 6 M guanidium chloride, 10 mM Tris-HCl, and 10 mM KCl. Adjust to a final pH of 7.5.
Refolding buffer: 10 mM Tris-HCl and 10 mM KCl. Adjust to a final pH of 7.5.
Dialysis cassette or dialysis membrane, 3,500 MWCO, 3 mL capacity.
3. Methods
Our methodology section is organized in the following way. First, in Section 3.1 we outline the synthesis of norbornene-lysine. While we use this as an example modification, the subsequent steps in our protocol are compatible with the incorporation of other UAAs provided that the appropriate matching tRNA synthetase plasmid is used. Then, in Section 3.2 we provide protocols for the expression and purification of the Ub-NpuDnaE-His6 construct on a small and large scale, respectively. We also provide details on how to optimize the incorporation of the UAA. Section 3.3 outlines the protocols for intein cleavage through hydrolysis and thiolysis, while section 3.4 describes the preparative HPLC purification of the generated constructs. We end with Section 3.5 which presents the refolding protocol for the doubly modified final ubiquitin construct. We should note that Sections 3.2, 3.3 and 3.4 are general and applicable to any other protein construct of interest.
3.1. Synthesis of norbornene-lysine.
Since to our knowledge norbornene lysine is not commercially available, we provide our synthesis protocol, based on the published literature [21]. All synthetic steps should be performed in a hood equipped with standard synthesis equipment (see Section 2.4). Care should be taken when handling all reagents. All applicable material safety data sheets should be consulted and followed before proceeding with the protocol.
Dissolve triphosgene (4.8 g) in dry THF (0.5 M, 35 mL) while stirring and add 5-norbornene-2-methanol (2 g) dropwise for 1 hour to the solution at 0 °C under argon atmosphere.
Stir the reaction mixture for 6 hours.
Remove all volatile components under reduced pressure using a rotary evaporator and dry under high vacuum for an additional 1 hour.
Add dry THF to the oily intermediate to prepare a 5 mL solution at 3.5 M.
Dissolve Boc-L-Lys-OH (4.8 g) in 1.0 M NaOH/THF (2:1 v/v, 90 mL, 0.3 M) and slowly add the intermediate solution from the previous step to the solution at 0 °C.
Warm the reaction mixture up to room temperature and stir for 16 hours.
Add ethyl acetate (100 mL) to the mixture and acidify the aqueous layer to around pH 4 with 1M HCl solution.
Collect the organic layer and extract the aqueous layer with additional ethyl acetate (100 mL each) three times.
Combine all organic layer fractions and wash them with saturated NaCl solution (400 mL).
Dry the organic solution with anhydrous sodium sulfate, and remove it by filtration.
Evaporate all volatile components under reduced pressure in a rotary evaporator.
Purify the crude product by silica gel flash column chromatography (ethyl acetate/hexanes, gradient: 1:2 to 2:1).
Remove all volatile components by rotary evaporation and dissolve the residue in 60% formic acid in chloroform (6:4 v/v, 90 mL, 0.2 M).
Stir the reaction mixture for 24 h at room temperature.
Add DMF (90 mL) to the reaction mixture and remove all volatile components under reduced pressure.
Add 50 mM HCl (200 mL) to the residue and lyophilize the solution to obtain the pure HCl salt of norbornene-lysine.
3.2. Ub-NpuDnaE-His6 expression
This section describes the expression and purification protocol for the ubiquitin-intein construct and the incorporation of norbornene-lysine at position 6 of the ubiquitin sequence. We refer to this construct as K6X-Ubiquitin-NpuDnaE-AFN-His6. We describe cloning and transformation steps (Section 3.2.1), a protocol for the optimization of protein expression on a small scale (Section 3.2.2) and the large-scale expression and purification of the modified construct (Section 3.2.3).
3.2.1. Cloning and Transformation
The first steps of the protocol involve the cloning of the UAA incorporation site into the plasmid of interest. This is followed by the double transformation of the ubiquitin-intein and tRNA synthetase plasmids into competent cells for protein expression.
Pick the desired site for unnatural amino acid incorporation in the protein plasmid sequence. This site will be replaced with the TAG amber stop codon. In our case, we choose to replace lysine 6 in ubiquitin with the unnatural amino acid norbornene lysine.
Using the template plasmid (pTXB1-Ubiquitin-NpuDnaE-AFN-His6) and appropriate primers, introduce the TAG mutation by site-directed mutagenesis (see Note 6). We denote the modified plasmid as pTXB1-K6X-Ubiquitin-NpuDnaE-AFN-His6.
Sequence the modified plasmid and store at −20 °C.
Retrieve a 50 μL aliquot of the BL21 (DE3) competent cells, the pTXB1-K6X-Ubiquitin-NpuDnaE-AFN-His6 plasmid stock, and the pEVOL-tRNApyl/pylRSY306A, Y384F plasmid stock and thaw them on ice.
Using sterile technique, add 1.5 μL (50 – 100 ng of DNA) of each plasmid stock to the BL21 (DE3) competent cells (see Note 7).
Incubate the cells and plasmids on ice for 20 minutes.
Heat-shock the cells at 42 °C for 40 seconds.
Incubate the cells on ice for 5 minutes.
Using sterile technique, add 150 μL of antibiotic-free, sterile LB media to the transformation mixture. Gently mix the cells by pipetting. Do not vortex.
Incubate the cells at 37 °C for 1.5 hours in an incubator at 220 rpm.
Plate 200 μL of the transformation mixture on LB agar containing chloramphenicol and ampicillin.
Incubate the plate at 37 °C. Colonies are generally observed after 10–15 hours (see Note 8). Once colonies are visible, proceed to step 3.2.2. Colonies should remain viable for a week if stored at 4 °C but are best used immediately.
3.2.2. Optimization of Protein Expression
Here, we provide a protocol to optimize protein expression when the incorporation of unnatural amino acids by amber suppression is desired. This protocol is general and can be adapted for the optimization of UAA incorporation for other proteins or for other UAAs. The optimal expression conditions (temperature, time, antibiotic) may differ for other systems.
Pick a single colony from the plate prepared in Section 3.2.1 and inoculate 5 mL of autoclaved LB media containing 5 μL of the stock solutions of ampicillin (final 100 μg/mL) and chloramphenicol (final 35 μg/mL) in a 15 mL culture tube under sterile conditions.
Incubate the preculture for 12 – 16 hours at 37 °C while shaking at 220 rpm. If desired, cell stocks can be prepared after incubation (see Note 9).
Prepare two 15 mL culture tubes (one for norbornene lysine incorporation and one for a negative control) containing 5 mL of autoclaved LB and antibiotic as described in step 1 above. Inoculate each tube with 50 μL of the overnight preculture.
Incubate the cultures at 37 °C while shaking at 220 rpm until the OD600 reaches 0.2 – 0.4. This should take 2.5 to 4 hours.
Add 50 μL of the 0.1 M norbornene lysine stock solution to one of the cultures for a final concentration of 1 mM norbornene lysine. Add 50 μL of 0.5 M NaOH to the other tube to serve as a control.
Incubate the cultures for 30 minutes to 1 hour until an OD600 of 0.4 – 0.6 has been reached.
Take a 250 μL aliquot from each culture for analysis by SDS-PAGE. To prepare each SDS-PAGE sample, measure the OD600 of each culture, pellet the cells by centrifugation at 4 °C and 3,000 x g, and decant the supernatant. Resuspend the cells in 10 μL of SDS loading buffer per 0.1 units of OD600. This aliquot will serve as the “Time 0” timepoint for characterizing the expression of the ubiquitin-fused split intein construct. Store on ice or at −20 °C until analysis by SDS-PAGE.
Add 5 μL of IPTG stock and 5 μL of arabinose stock to each culture tube for a final concentration of 0.5 mM and 0.02% (w/v), respectively. The addition of IPTG and arabinose will induce the expression of the ubiquitin-fused split intein construct and the aminoacyl tRNA synthetase/tRNA pair.
Take aliquots from both cultures at defined time points and prepare them for SDS-PAGE analysis as described in step 7 above. For the K6X-Ubiquitin-NpuDnaE-AFN-His6 construct, we typically take time points at 0, 1, 2, 3, and 4 hours after the addition of IPTG. After all time points have been taken, the cultures can be discarded as biohazardous waste.
Heat the SDS-PAGE samples at 85 °C for 5 minutes in thiol-free SDS PAGE loading buffer (see Note 10).
Analyze the samples on a 12% SDS-PAGE Tris-glycine gel.
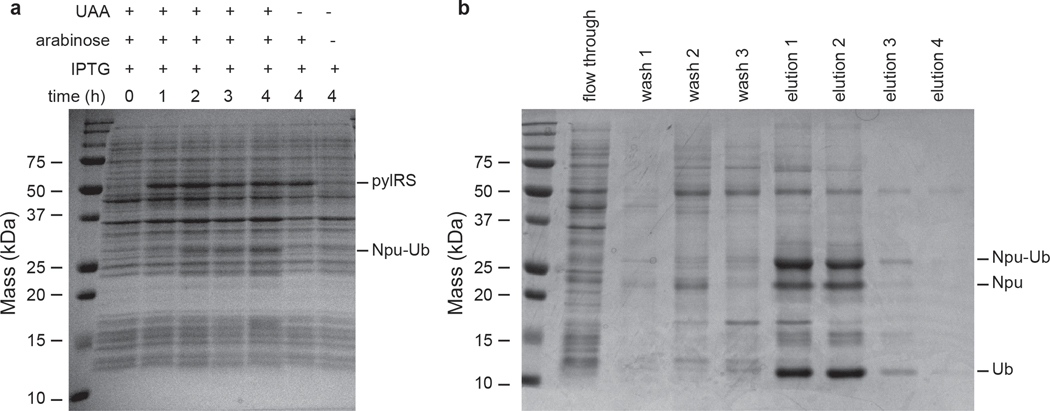
Choose the best time course for scaling up protein production (Figure 3a). The aminoacyl tRNA synthetase bands should appear in both samples, but the desired ubiquitin-fused split intein band should only be present in the samples containing norbornene lysine. For optimal expression, determine the time point when the ubiquitin-fused split intein band is maximized and in vivo hydrolysis is minimized (see Note 11).
Figure 3.
(a) Test expression of ubiquitin-Npu containing norbornene lysine at position 6 of the ubiquitin sequence, a modification introduced genetically through amber suppression. Expression of the construct is expected only when the unnatural amino acid (UAA), arabinose and IPTG are added to the culture media. In our case, the optimal expression conditions are 4 hours and 37 °C. (b) Ni-NTA column purification of the ubiquitin-Npu construct analyzed by SDS-PAGE gel electrophoresis. Intein hydrolysis should not be visible in the wash fractions and should only be initiated upon elution. In this gel image, some hydrolysis has already taken place in the elution fractions.
3.2.3. Large-Scale Protein Expression and Purification
Once protein expression has been optimized on a small scale (Section 3.2.2), large scale cultures (1L or more) can be prepared. In this section, we provide a method to scale the culture growth and the purification steps that need to be performed before intein cleavage. This includes cell lysis and affinity column purification of the His-tagged ubiquitin-intein construct. These steps remove truncation products that may be generated due to premature translation termination at the stop TAG codon site. It is important to perform these steps as fast as possible while keeping the protein sample at 4 °C to avoid premature intein cleavage during purification.
Start with a frozen cell stock or a freshly transformed plate as described in Section 3.2.1 (see Note 12). Inoculate 10 mL of autoclaved LB media containing chloramphenicol and ampicillin in a sterile 50 mL culture tube and grow cells overnight (12 – 16 hours) at 37 °C while shaking at 220 rpm.
Add 10 mL of saturated preculture to 1 L of sterile LB media containing chloramphenicol and ampicillin in a 4 L Erlenmeyer flask.
Incubate the cultures at 37 °C while shaking at 220 rpm until OD600 reaches 0.2 – 0.4. This should take 2.5 to 4 hours.
Add 10 mL of the 0.1 M norbornene lysine stock solution to the culture for a final concentration of 1 mM norbornene lysine.
Incubate the cultures for 30 minutes to 1 hour until an OD600 of 0.4 – 0.6 has been reached.
Take a 250 μL aliquot from each culture for analysis by SDS-PAGE as described in step 1 of Section 3.2.2.
Add 1 mL of 0.5 M of IPTG (for a final concentration of 0.5 mM) and 20% w/v arabinose (for a final concentration of 0.02% w/v) to the culture.
Allow the protein to express under the conditions and time determined in Section 3.2.2. In our example, we express the protein at 37 °C for 4 hours and shaking at 220 rpm.
Pour the culture into a 1 L Nalgene centrifuge bottle and pellet the cells by centrifugation at 10,000 x g for 30 minutes.
Decant the supernatant and carefully transfer the pellet to a 50 mL centrifuge tube using a plastic scraper. If desired, the pellet can be flash frozen at this point and stored at −80 °C until purification can be resumed.
Add 20 mL of cold lysis buffer and 20 μL of 1 M TCEP stock solution to the pellet. While keeping the sample on ice, resuspend the pellet until the solution becomes homogeneous. If necessary, pass the solution through an 18-gauge syringe needle to break any leftover clumps.
Transfer the suspension into a 150 mL aluminum beaker and place the beaker on ice. Lyse the cells by sonication. We use a cycle set at 65% amplitude with 30 s on/off intervals for 6 minutes (see Note 13).
Clarify the lysate by centrifugation at 30,000 x g for 1 hour at 4 °C.
Prepare the Ni-NTA column by pouring 2 ml of Ni-NTA agarose resin (4 mL of stock solution slurry) into a glass or disposable plastic column outfitted with a filter frit and a flow adaptor.
The Ni-NTA resin is supplied as a suspension in 20% ethanol. Drain the ethanol from the resin and wash with 10 mL (5 column volumes, CV) of water, followed by 10 mL (5 CV) of lysis buffer. To avoid premature intein hydrolysis, perform the Ni-NTA purification in a 4 – 6 °C cold room as quickly as possible.
Pour the supernatant of the centrifuged lysate into the column.
Mix the resin and lysate by pipetting up and down. Transfer the suspension into a 50 mL centrifuge tube and incubate on a rotator for 1 hour at 4 °C to allow the protein to bind to the resin.
Return the suspension back to the column to perform the wash and elution steps. An aliquot from each sample should be collected for SDS-PAGE analysis. Drain and collect the flow-through.
- Perform the following washes:
- 10 mL (5 CV) of lysis buffer – collect the eluent as Wash 1
- 10 mL (5 CV) of wash buffer 1 – collect the eluent as Wash 2
- 5 ml (2.5 CV) of wash buffer 2 – collect the eluent as Wash 3
Elute the protein with 3 mL of elution buffer (1.5 CV). Repeat four times and collect all fractions as Elution 1, 2, 3, and 4 (see Note 14).
Analyze each fraction by SDS-PAGE and combine the fractions that contain the desired protein (Figure 3b).
3.3. Intein Cleavage
The following portion of the protocol is split into two discrete sections. If the generation of the native C-terminal carboxylic acid is required, follow the instructions in Section 3.3.1. In this case, the intein is used as a purification tag. When the generation of C-terminal thioesters is desired, follow the protocol in Section 3.3.2. In our case, we modify the C-terminus of ubiquitin with an aminoethanethiol moiety that can be used to attach ubiquitin to other proteins of interest. In both cases, ubiquitin also contains the unnatural amino acid norbornene lysine at position 6 as introduced above.
3.3.1. Generation of the Native C-terminus via Hydrolysis
Add 1 M TCEP stock to the pooled fractions for a final TCEP concentration of 10 mM. Verify that the pH is ~ 7.5 using 2–3 μL of the pooled sample and a pH indicator strip.
Incubate the sample overnight (12 – 16 hours) at room temperature to allow intein hydrolysis to occur (see Note 15).
Prepare a small amount of a 1:1 solution of the hydrolysis reaction and HPLC solvent A (50–100 μL total volume). Centrifuge to remove any precipitate and load the sample onto an analytical HPLC column. For the ubiquitin sample described here, we use the analytical column described in Section 2.3.2 with a flow rate of 1 mL/min, a solvent gradient of 30% to 70% B for 30 min, and UV absorbance detection at 214 nm.
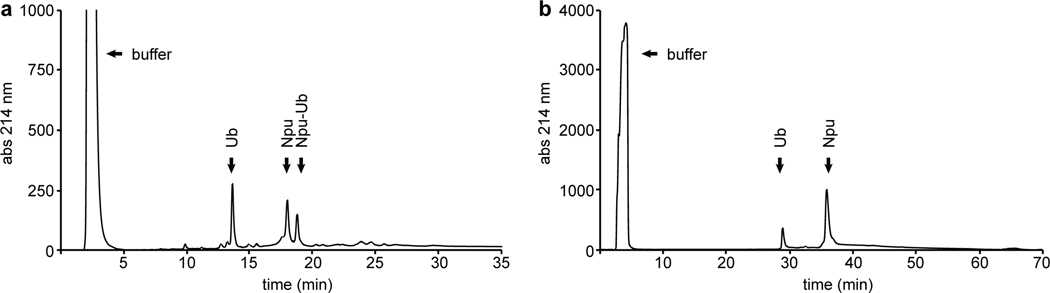
The hydrolysis is complete if the ubiquitin-intein peak has disappeared and two separate peaks are visible, one corresponding to the hydrolyzed ubiquitin and the other to the intein (Figure 4). Confirm the identity of the protein of interest by electrospray mass spectrometry (ESI-MS).
Figure 4.
Analysis and purification of the intein cleavage by reverse-phase HPLC. (a) Analytical HPLC chromatogram of an intermediate stage where a fraction of the ubiquitin-intein construct is still intact, and another fraction has hydrolyzed to form ubiquitin and Npu. The chromatogram was collected using a 30 – 70% HPLC Solvent B gradient over 30 minutes with a flow rate of 1 mL/min. (b) Preparative scale reverse-phase HPLC purification of the fully hydrolyzed ubiquitin-intein construct. Here only two peaks are visible, one corresponding to ubiquitin and the other one to the intein. Fractions containing ubiquitin were collected and lyophilized. Purification was performed with a 5 – 70% HPLC Solvent B gradient over 60 min with a flow rate of 4 mL/min.
3.3.2. C-Terminal Modification via Thiolysis
Add 1 M 2-aminoethanethiol solution and 1 M TCEP solution to the combined Ni-NTA fractions to a final concentration of 100 mM and 10 mM, respectively. Verify that the pH is ~ 7.5 using 2–3 μL of the pooled sample and a pH indicator strip.
Incubate the sample overnight (12 – 16 hours) at room temperature to allow the reaction to complete (see Note 15).
Prepare a small amount of 1:1 solution of the hydrolysis reaction and HPLC solvent A (50–100 μL total volume). Centrifuge to remove any precipitate and load the sample onto an analytical HPLC column. For the ubiquitin sample described here, we use the analytical column described in Section 2.3.2 with a flow rate of 1 mL/min, a solvent gradient of 30% to 70% B for 30 min, and UV absorbance detection at 214 nm.
The reaction is complete if the ubiquitin-intein peak has disappeared and two separate peaks are visible, one corresponding to the modified ubiquitin and the other to the intein (Figure 4). Confirm the identity of the protein of interest by electrospray mass spectrometry (ESI-MS).
3.4. Preparative-Scale HPLC Purification
The reaction products from the intein cleavage can be easily separated by reverse-phase C18 HPLC chromatography as described below. This purification protocol is compatible with any protein construct that can be lyophilized and refolded.
Take the reaction mixture from 3.3.1 or 3.3.2 and adjust the pH to ~4 by the dropwise addition of concentrated HPLC grade TFA (see Note 16).
Centrifuge the sample to remove any precipitate (see Note 17), filter through a 0.45 μL HPLC certified syringe-tip filter (e.g. GHP Acrodiscs from Pall Corporation) and load the sample onto a preparative-scale HPLC column. For the ubiquitin samples described here, we use the preparative column described in section 2.3.2 with a flow rate of 4 mL/min, a solvent gradient of 30% to 70% B for 60 min, and UV absorbance detection at 214 nm. Under these conditions, ubiquitin elutes at ~ 25 minutes and the intein elutes at ~ 30 minutes (Figure 4b).
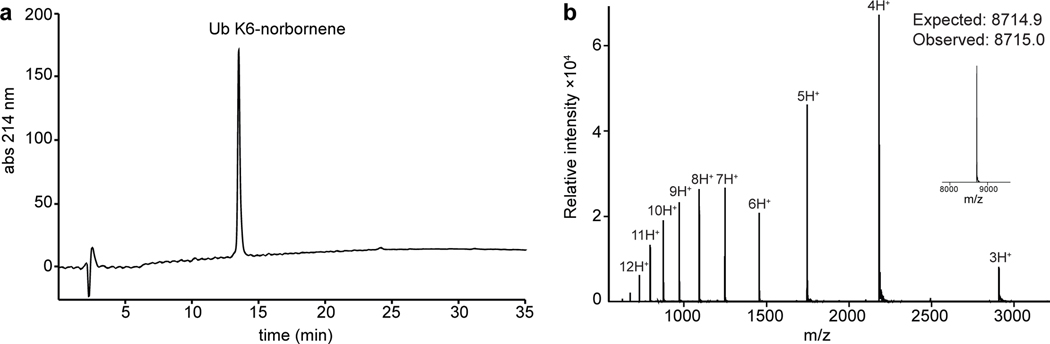
Collect the desired fractions and verify purity by analytical HPLC and ESI-MS (Figure 5). Combine the pure fractions.
Flash-freeze the sample by dipping the tube into liquid nitrogen and lyophilize. Lyophilized samples can be stored at −80 °C indefinitely. Typical yields for the doubly modified ubiquitin construct described here are ~ 5 mg/1 L of culture (see Note 18 and Note 19).
Figure 5.
Analysis of the purified ubiquitin construct containing norbornene lysine at position 6 of the sequence. (a) Analytical reverse-phase HPLC chromatogram depicting a single ubiquitin peak. The chromatogram was collected with a 30 – 70% HPLC Solvent B gradient over 30 minutes with a flow rate of 1 mL/min. (b) ESI-MS analysis of the purified protein. Calculated expected mass is 8714.9, experimentally determined mass is 8715.0.
3.5. Protein Refolding
Ubiquitin can be easily refolded by dialysis. We have found that the final ubiquitin fold is not perturbed by the presence of the UAA or the C-terminal modification [20]. If working with other proteins, follow their respective refolding steps and, if possible, confirm that the final structure is not perturbed, e.g. by circular dichroism (CD), nuclear magnetic resonance (NMR) spectroscopy, X-ray crystallography, or other suitable biophysical methods.
Dissolve the lyophilized ubiquitin into 6M guanidium chloride denaturing buffer and adjust the concentration to 1 mg/mL. In this case, we use 1 mL of protein solution.
Centrifuge the sample to remove any precipitation and load the supernatant into a dialysis cassette.
Place the cassette into 1 L of refolding buffer at 4 °C and gently stir the solution by magnetic stir bar for 4 – 6 hours. Replace with fresh refolding buffer and continue dialysis overnight.
Carefully remove the solution from the dialysis cassette, centrifuge to remove precipitation and measure the absorbance at A280 to determine the concentration of protein.
4. Notes
Any common LB agar recipe will likely work in this case. The Miller LB agar recipe that we use contains 10 g tryptone, 5 g yeast extract, 10 g sodium chloride, and 15 g agar per liter.
Any common LB recipe will likely work in this case. The Miller LB agar recipe that we use contains 10 g tryptone, 5 g yeast extract, and 10 g sodium chloride per liter.
A stock solution of 4X buffer will be useful in preparing the lysis buffer, wash buffer 1, wash buffer 2, and elution buffer. To make the 4X stock, prepare a solution containing 200 mM sodium phosphate (monobasic) and 1.2 M sodium chloride. Adjust to a final pH of 7.5.
We use cOmplete™ Protease Inhibitor Cocktail EDTA-free tablets (Roche) since they contain a combination of multiple protease inhibitors for serine and cysteine proteases but do not inhibit metallo- or aspartic proteases.
We use HisPur™ Ni-NTA resin (Thermo Fisher Scientific) and we follow the purification protocols suggested by the manufacturer. If another vendor is used, the protocol should be adapted according to the manufacturer instructions.
Any site-directed mutagenesis protocol or vendor kit should work. Follow the instructions provided by the vendor.
Other competent cell lines can be used for expression provided that they do not carry chloramphenicol resistant plasmids.
The appearance of colonies and cell culture growth may be slower than usual due to the use of two antibiotics.
Working near a flame, take 250 μL of cell culture and add 250 μL of autoclaved 50% glycerol solution. Flash-freeze in liquid nitrogen and store at −80 °C for future protein expression.
If SDS-PAGE gel loading buffer with thiols (dithiothreitol or β-mercaptoethanol) is used, the gel analysis may show misleading premature intein cleavage.
If substantial in vivo hydrolysis is observed, protein expression may be performed at lower temperature. In our case, we found that the aminoacyl tRNA synthetase does not express well below 30 °C, which affects the efficiency of UAA incorporation. To improve expression of the aminoacyl tRNA synthetase, the final concentration of arabinose may also be increased up to a final concentration of 0.2% w/v.
To inoculate from a transformed plate, use a sterilized glass pipette to pick a colony from the plate and dip the pipette into the autoclaved LB media. To inoculate from a frozen cell stock, thaw the stock on ice and use a sterile pipette tip to transfer 2 – 5 μL of cell stock to the LB media.
If premature hydrolysis is an issue, a French press may be used for lysis.
If C-terminal modification is desired, we recommend moving to step 3.3.2 as soon as possible. We collect the Ni-NTA elution fractions quickly while working at 4 °C, take a small aliquot for gel analysis and add 2-aminoethanthiol and TCEP immediately. As soon as the thiol is added to the elution fractions, they can be moved to room temperature. This is necessary to prevent premature hydrolysis of the protein before the thiolysis reaction can take place.
Since ubiquitin is a stable protein, we routinely incubate overnight. However, the incubation times can be shortened if more unstable proteins are being purified. In this case, we recommend following the hydrolysis or thiolysis reaction by SDS-PAGE or analytical HPLC in real time. If the protein of interest requires denaturants, we also recommend using the fused Cfa intein instead of Npu [15]. On the other hand, if the reaction is not complete after 12 – 16 hours of incubation time, we recommend adding fresh TCEP solution (10 mM) and EDTA to a final concentration of 0.2 mM. To speed up hydrolysis, the pH can also be raised to 8.0 and the sample can be incubated at 37 °C for a few hours.
If the protein of interest contains cysteines or C-terminal thioesters, incubate the sample with freshly added TCEP (10 mM) for 30 min at room temperature before the addition of TFA. This prevents undesired disulfide formation and ensures that only monomeric protein is loaded on the column.
It is common to observe a significant amount of precipitation at this step. We found that the precipitate contains the intein while ubiquitin remains in solution. Should the protein of interest also precipitate at low pH, the precipitate can be redissolved in 6 M unbuffered guanidinium HCl and loaded onto the preparative HPLC column directly.
Based on our experience purifying and modifying different proteins with the protocol described here, several factors can affect the final yield. The most significant factor is the efficiency of the UAA incorporation. For example, higher yields can be obtained with the orthogonal tyrosyl tRNA/tRNA synthetase pair that can incorporate p-benzoyl-L-phenylalanine by amber suppression [22,20]. Second, the expression temperature can be an important factor in preventing in vivo hydrolysis. If possible, we recommend expressing the protein of interest-split intein fusions at 18 °C overnight. Third, ubiquitin contains two glycine residues at the C-terminus and the flexibility imparted by those residues should aid the hydrolysis reaction. Thus, ubiquitin might be more prone to in vivo hydrolysis compared to most proteins.
Finally, we note that reverse-phase HPLC purification and refolding are not required for this protocol. If native conditions are to be maintained, the removal of the intein can be performed by reverse Ni-NTA purification, ion exchange or size-exclusion. The removal of leftover reagents such as imidazole and 2-aminoethanethiol can also be performed by dialysis.
5. References
- 1.Shah NH, Muir TW (2014) Inteins: nature’s gift to protein chemists. Chem Sci 5 (2):446–461. doi: 10.1039/c3sc52951g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volkmann G, Mootz HD (2013) Recent progress in intein research: from mechanism to directed evolution and applications. Cell Mol Life Sci 70 (7):1185–1206. doi: 10.1007/s00018-012-1120-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klabunde T, Sharma S, Telenti A, Jacobs WR, Sacchettini JC (1998) Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat Struct Biol 5 (1):31–36. doi:DOI 10.1038/nsb0198-31 [DOI] [PubMed] [Google Scholar]
- 4.Liu ZH, Frutos S, Bick MJ, Vila-Perello M, Debelouchina GT, Darst SA, Muir TW (2014) Structure of the branched intermediate in protein splicing. P Natl Acad Sci USA 111 (23):8422–8427. doi: 10.1073/pnas.1402942111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirata R, Ohsumi Y, Nakano A, Kawasaki H, Suzuki K, Anraku Y (1990) Molecular-Structure of a Gene, Vma1, Encoding the Catalytic Subunit of H+-Translocating Adenosine-Triphosphatase from Vacuolar Membranes of Saccharomyces-Cerevisiae. Journal of Biological Chemistry 265 (12):6726–6733 [PubMed] [Google Scholar]
- 6.Kane PM, Yamashiro CT, Wolczyk DF, Neff N, Goebl M, Stevens TH (1990) Protein Splicing Converts the Yeast Tfp1 Gene-Product to the 69-Kd Subunit of the Vacuolar H+-Adenosine Triphosphatase. Science 250 (4981):651–657. doi:DOI 10.1126/science.2146742 [DOI] [PubMed] [Google Scholar]
- 7.Shah NH, Dann GP, Vila-Perello M, Liu ZH, Muir TW (2012) Ultrafast Protein Splicing is Common among Cyanobacterial Split Inteins: Implications for Protein Engineering. Journal of the American Chemical Society 134 (28):11338–11341. doi: 10.1021/ja303226x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, Hu ZM, Liu XQ (1998) Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. P Natl Acad Sci USA 95 (16):9226–9231. doi:DOI 10.1073/pnas.95.16.9226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvajal-Vallejos P, Pallisse R, Mootz HD, Schmidt SR (2012) Unprecedented Rates and Efficiencies Revealed for New Natural Split Inteins from Metagenomic Sources. Journal of Biological Chemistry 287 (34):28686–28696. doi: 10.1074/jbc.M112.372680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debelouchina GT, Muir TW (2017) A molecular engineering toolbox for the structural biologist. Q Rev Biophys 50:e7. doi: 10.1017/S0033583517000051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Southworth MW, Amaya K, Evans TC, Xu MQ, Perler FB (1999) Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques 27 (1):110-+. doi:DOI 10.2144/99271st04 [DOI] [PubMed] [Google Scholar]
- 12.Batjargal S, Walters CR, Petersson EJ (2015) Inteins as Traceless Purification Tags for Unnatural Amino Acid Proteins. Journal of the American Chemical Society 137 (5):1734–1737. doi: 10.1021/ja5103019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frutos S, Goger M, Giovani B, Cowburn D, Muir TW (2010) Branched intermediate formation stimulates peptide bond cleavage in protein splicing. Nature Chemical Biology 6 (7):527–533. doi: 10.1038/Nchembio.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chong SR, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, Hirvonen CA, Pelletier JJ, Paulus H, Xu MQ (1997) Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192 (2):271–281. doi:Doi 10.1016/S0378-1119(97)00105-4 [DOI] [PubMed] [Google Scholar]
- 15.Stevens AJ, Brown ZZ, Shah NH, Sekar G, Cowburn D, Muir TW (2016) Design of a Split Intein with Exceptional Protein Splicing Activity. Journal of the American Chemical Society 138 (7):2162–2165. doi: 10.1021/jacs.5b13528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens AJ, Sekar G, Shah NH, Mostafavi AZ, Cowburn D, Muir TW (2017) A promiscuous split intein with expanded protein engineering applications. P Natl Acad Sci USA 114 (32):8538–8543. doi: 10.1073/pnas.1701083114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Brock A, Herberich B, Schultz PG (2001) Expanding the genetic code of Escherichia coli. Science 292 (5516):498–500. doi: 10.1126/science.1060077 [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann JE, Plass T, Nikic I, Aramburu IV, Koehler C, Gillandt H, Lemke EA, Schultz C (2015) Highly Stable trans-Cyclooctene Amino Acids for Live-Cell Labeling. Chem-Eur J 21 (35):12266–12270. doi: 10.1002/chem.201501647 [DOI] [PubMed] [Google Scholar]
- 19.Chatterjee C, McGinty RK, Fierz B, Muir TW (2010) Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nature Chemical Biology 6 (4):267–269. doi: 10.1038/Nchembio.315 [DOI] [PubMed] [Google Scholar]
- 20.Debelouchina GT, Gerecht K, Muir TW (2017) Ubiquitin utilizes an acidic surface patch to alter chromatin structure. Nat Chem Biol 13 (1):105–110. doi: 10.1038/nchembio.2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang K, Davis L, Torres-Kolbus J, Chou CJ, Deiters A, Chin JW (2012) Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat Chem 4 (4):298–304. doi: 10.1038/nchem.1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin JW, Martin AB, King DS, Wang L, Schultz PG (2002) Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. P Natl Acad Sci USA 99 (17):11020–11024. doi: 10.1073/pnas.172226299 [DOI] [PMC free article] [PubMed] [Google Scholar]