Summary
Objectives : We survey recent developments in medical Information Extraction (IE) as reported in the literature from the past three years. Our focus is on the fundamental methodological paradigm shift from standard Machine Learning (ML) techniques to Deep Neural Networks (DNNs). We describe applications of this new paradigm concentrating on two basic IE tasks, named entity recognition and relation extraction, for two selected semantic classes—diseases and drugs (or medications)—and relations between them.
Methods : For the time period from 2017 to early 2020, we searched for relevant publications from three major scientific communities: medicine and medical informatics, natural language processing, as well as neural networks and artificial intelligence.
Results : In the past decade, the field of Natural Language Processing (NLP) has undergone a profound methodological shift from symbolic to distributed representations based on the paradigm of Deep Learning (DL). Meanwhile, this trend is, although with some delay, also reflected in the medical NLP community. In the reporting period, overwhelming experimental evidence has been gathered, as illustrated in this survey for medical IE, that DL-based approaches outperform non-DL ones by often large margins. Still, small-sized and access-limited corpora create intrinsic problems for data-greedy DL as do special linguistic phenomena of medical sublanguages that have to be overcome by adaptive learning strategies.
Conclusions : The paradigm shift from (feature-engineered) ML to DNNs changes the fundamental methodological rules of the game for medical NLP. This change is by no means restricted to medical IE but should also deeply influence other areas of medical informatics, either NLP- or non-NLP-based.
Keywords: Neural networks, deep learning, natural language processing, information extraction, named entity recognition, relation extraction
1 Introduction
The past decade has seen a truly revolutionary paradigm shift for Natural Language Processing (NLP) as a result of which Deep Learning (DL) (for a technical introduction, cf. 1 ; for comprehensive surveys, cf. 2 and 3 ) became the dominating mind-set of researchers and developers in this field (for surveys, cf. 4 5 ). Yet, DL is by no means a new computational paradigm. Rather it can be seen as the most recent offspring of neural computation in the evolution of computer science (cf. the historical background provided by Schmidhuber 6 ). But unlike in previous attempts, it now turns out to be extremely robust and effective for adequately dealing with the contents of unstructured visual 7 , audio/speech 8 , and textual data 9 .
The success of Deep Neural Networks (DNNs) has many roots. Perhaps the most important methodological reason is that, with DNNs, manual feature selection or (semi-)automated feature engineering is abandoned. This time-consuming tuning step was at the same time mandatory and highly influential on the performance of earlier generations of ML systems in NLP based on Markov Models (MMs), Conditional Random Fields (CRFs), Support Vector Machines (SVMs), etc. In a DL system, however, the relevant features (and their relative contribution to a classification decision) are automatically computed as a result of thousands of iterative training cycles.
The ultimate reason for the success behind DNNs is a pragmatic criterion though: system performance . Compared with results in biomedical Information Extraction (IE), obtained in previous years with standard ML methods, DL approaches changed profoundly the rules of the game. In a landslide manner, for the same task and domain, performance figures jumped up to unprecedented levels so far and DL systems consistently outperformed by large margins non-DL state-of-the-art (SOTA) systems for different tasks. Section 3 provides ample evidence for this claim and features the new SOTA results with a deeper look at IE, a major application class of medical NLP (for alternative surveys, cf. 10 11 12 ).
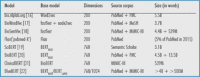
Despite specialized hardware at disposal now, training DNNs still requires tremendous computational resources and processing time. Luckily, for general NLP, huge collections of language models (so-called embeddings ) have already been trained on huge corpora (comprised of hundreds of millions of Web-scraped documents, including newspaper and Wikipedia articles) so that these pre-compiled model resources can be readily reused when dealing with general-purpose language. But medical (and biological) language mirrors special-purpose language characteristics and comprises a large variety of sublanguages of its own. This becomes obvious in Section 3 where we deal with scholarly scientific writing (with documents typically taken from PubMed). Here, differences to general language are mostly due to the use of a highly specialized technical vocabulary (covered by numerous terminologies, such as MeSH, SNOMED-CT, or ICD). Even more challenging are clinical notes and reports (with documents often taken from the MIMIC 1 (Medical Information Mart for Intensive Care) clinical database) which typically exhibit syntactically ill-formed, telegraphic language with lots of acronyms and abbreviations as an additional layer of complexity (cf. the seminal descriptive work distinguishing both these sublanguage types by Friedman et al. 13 ). Newman-Griffis and Fosler-Lussier 14 investigated different sublanguage patterns for the many varieties of clinical reports (pathology reports, discharge summaries, nurse and Intensive Care Unit notes, etc.), while Nunez and Carenini 15 discussed the portability of embeddings across various fields of medicine reflecting characteristic sublanguage use patterns. These constraints have motivated the medical NLP community to adapt embeddings originally trained on general language to the medical language. Table 1 lists those medically informed embeddings, many of which are the basis for the IE applications discussed in Section 3.
Table 1. An Overview of Common Embeddings—Biomedical Language Models.
|
Our survey emphasizes the fundamental methodological paradigm shift of current NLP research from symbolic to distributed representations as the basis of DL. It thus complements earlier contributions to the International Medical Informatics Association (IMIA) Yearbook of Medical Informatics which focused exclusively on the role of social media documents 23 , had a balanced view on the relevance of both Electronic Health Records (EHRs) and social media posts 24 , or dealt with the importance of shared tasks for the progress in medical NLP 25 . The last two Yearbook surveys of the NLP section most closely related to medical IE were published in 2015 26 and 2008 27 . The survey by Velupillai et al. 28 dealt with opportunities and challenges of medical NLP for health outcomes research, with particular emphasis on evaluation criteria and protocols.
We also refer readers to alternative surveys of DL as applied to medical and clinical tasks. Wu et al. 29 reviewed literature for works using DL for a broader view of clinical NLP, whereas Xiao et al. 30 and Shickel et al. 31 performed systematic reviews on the applications of DL to several kinds of EHR data, not only text. Miotto et al. 32 and Esteva et al. 33 further extended that scope to include clinical imaging and genomic data beyond the scope of classical EHRs. From an even broader perspective of the huge amounts of biomedical data, Ching et al. 34 examined various applications of DL to a variety of biomedical problems—patient classification, fundamental biological processes, and treatment of patients—and discussed the unique challenges that biomedical data pose for DL methods. In the same vein, Rajkomar et al. 35 used the entire EHR, including clinical free-text notes, for clinical predictive modeling based on DL (targeted, e.g ., at the prediction of in-hospital mortality or patient’s final discharge diagnoses). They also demonstrated that DL methods outperformed traditional statistical prediction models.
2 Design and Goals of this Survey
In this survey, we concentrated on publications within the time window from 2017 to early 2020 and screened the contributions from three major scientific communities involved in medical IE:
Medicine and medical informatics are covered by PubMed;
Natural language processing is covered by the ACL Anthology, the digital library of the Association for Computational Linguistics;
Neural networks are covered by the major conference series of the neural network community (Neural Information Processing Systems (NIPS/NeurIPS)) whereas the artificial intelligence community gets in via the Association for the Advancement of Artificial Intelligence (AAAI) Digital Library which keeps the records from the AAAI and IJCAI conferences.
We also included health-related publications from the digital libraries of the Association for Computing Machinery (ACM) and the Institute of Electrical and Electronics Engineers (IEEE). When necessary, we also refered to e-preprint archives such as arXiv.org, since they have become a new, increasingly important distribution channel for the most recent research results in computer science (yet, in that state typically without peer review) and thus foreshadow future directions of research.
We searched these literature repositories with a free-text query that can be approximated as follows: (information extraction OR text mining OR named entity recognition OR relation extraction) AND (deep learning OR neural network) AND (medic* OR clinic* OR health)
For this setting, we found approximately 1,000 unique publications, screened them for relevance, and, finally, included roughly 100 into this survey.
3 Deep Neural Networks for Medical Information Extraction
In this section, we introduce applications of DNNs to medical NLP for two different tasks, Named Entity Recognition (NER) and Relation Extraction (REX). The focus of our discussion relies on studies dealing with English as reference language since the vast majority of benchmark and reference data sets are in English 2 . After a brief description of each task, we summarize the current SOTA in tables which generalize often subtle distinctions in experimental design and workflows. Our main goal is to show the diversity of major benchmark datasets, DL approaches, and embeddings being used. For these tables, we extracted all symbolic ( e.g ., corpus or DL approach) and numerical information ( e.g ., about annotation metadata, performance scores) directly from the cited papers.
The assessment of different systems for the same task is centered around their performance on gold data in evaluation experiments. We refrain from highlighting minor differences in the reported scores because of different datasets being used for evaluation, changing volumes of metadata, and sometimes even the genres they contain. Hence, from a strict methodological perspective, the reported results have to be interpreted with utmost caution for two main reasons 37 . First, the choice of pre-processing steps, such as tokenization, inclusion/exclusion of punctuation marks, stop word removal, morphological normalization/lemmatization/stemming, n-gram variability, entity blinding strategies, and, second, the calibration of training methods (split bias, pooling techniques, hyperparameter selection (dropout rate, window size, etc.)) have a strong impact on the way a chosen embedding type and DL model finally performs, even within the same experimental setting. However, the data we report give valuable comparative information of the SOTA, though with fuzzy edges. This situation might be remedied by a recently proposed common evaluation framework for biomedical NLP, the BLUE (Biomedical Language Understanding Evaluation) benchmark 3 22 , which consists of five different biomedical NLP tasks (including NER and REX) with ten corpora (including BC5CDR, DDI, and i2b2 that also occur in the tables below), or the one proposed by Chauhan et al. 37 4 enabling a more lucid comparison of various training methodologies, pre-processing, modeling techniques, and evaluation metrics.
For the tables provided in the next subsections, we used the F 1 score as the main ordering criterion for the cited studies (from highest to lowest) 5 . We usually had to select among a large variety of experimental conditions (with different scores). The final choices we made were led by the criterion to favor comparability among all studies. This means that higher (and lower) outcomes may have been reported in the cited studies for varying experimental conditions. Still, the top-ranked system(s) in each of the following tables defines the current SOTA for a particular application.
3.1 Named Entity Recognition
The task of Named Entity Recognition (NER) is to identify crucial medical named entities ( i.e., spans of concrete mentions of semantic types such as diseases or drugs and their attributes) in running text. For a recent survey of DL-based approaches and architectures underlying NER as a generic NLP application, see 38 .
3.1.1 Diseases
A primary target of NER in the medical field is the automatic identification of diseases in scientific articles and clinical reports. For instance, textual occurrences of disease mentions ( e.g., “Diabetes II” or “cerebral inflammation” ) are mapped to a common semantic type, Disease 6 . The crucial role of recognizing diseases in medical discourse is also emphasized by a number of surveys dealing with the recognition of special diseases. For instance, Sheikhalishahi et al. 40 discussed NLP methods targeted at chronic diseases and found that shallow ML and rule-based approaches (as opposed to more sophisticated DL-based ones) prevail. Koleck et al. 41 summarized the use of NLP to analyze symptom information documented in EHR free-text narratives as an indication of diseases and similar to the previous survey found little coverage of DL methods in this application area as well. Savova et al. 42 reviewed the current state of clinical NLP with respect to oncology and cancer phenotyping from EHR. Datta et al. 43 focused on an even more specialized use case—the lexical representation required for the extraction of cancer information from EHR notes in a frame-semantic format.
The research summarized in Table 2 is strictly focused on Disease recognition and, for reasons of comparability, based on the use of shared data sets and metadata (gold annotations). Two benchmarks are prominently featured, BC5CDR 44 and NCBI 45 7 . BC5CDR is a corpus made of 1,500 PubMed articles, with 4,409 annotated chemicals, 5,818 diseases, and 3,116 chemical-disease interactions, created for the BioCreative V Chemical and Disease Mention Recognition Task 44 . As an alternative, the NCBI Disease Dataset 45 consists of a collection of 793 PubMed abstracts annotated with 6,892 disease mentions which are mapped to 790 unique disease concepts (thus, this corpus can also be used for grounding experiments).
Table 2. Medical Named Entity Recognition: Diseases. Benchmark Datasets from BC5CDR 44 and NCBI 45 .

|
The current top performance for Disease recognition comes close to 90% F 1 8 . Lee et al. 20 use a Transformer model with in-domain training (BioBERT), but also (attention-based) BiLSTMs which perform strongly in the range of 88–89% F 1 score. For the choice of embeddings being used, self-trained ones might be a better choice than pre-trained ones, e.g., those provided by bio.nlplab.org 16 . The incorporation of (large) dictionaries does not provide a competitive advantage in the experiments reported here. Though multi-task learning and transfer learning seem reasonable choices ( 39 46 and 47 , respectively) to combat the sparsity of datasets, they generally do not boost systems to the top ranks.
Interesting though are differences for the same approach on different evaluation data sets. For the second-best system by Sachan et al. 47 , F 1 scores differ for BC5CDR and NCBI by 2.0 (for the third-best 46 by 2.7) percentage points, whereas for the best non-DL approach by Lou et al. 48 , this difference amounts to remarkable 4.1 percentage points. This hints at a strong dependence of the results of the same system set-up on the specific corpus these results have been worked out and, thus, limits generalizability. On the other hand, corpora obviously cannot be blamed for intrinsic analytical hardness since cross-rankings occurs: the system by Lee et al. 20 gets the over-all highest F 1 score for NCBI but underperforms for BC5CDR, whereas for the tagger used by Sachan et al. 47 the ranking is reversed—their system performs better on BC5CDR than on NCBI (differences are in the range of 2 percentage points). The most stable system in this respect is the one by Zhao et al. 39 . Finally, the distance between the best- and second-best-performing DL systems ( 20 and 47 , respectively) and their best non-DL counterpart 48 amounts to 7.6 percentage points (for NCBI) and 3.1 percentage points (for BC5CDR), respectively.
3.1.2 Medication
The second major medical named entity type we here discuss is related to medication information. NER complexity is increased for this task since it is split into several subtasks, including the recognition of drug names ( Drug ), frequency ( Dr-Freq ) and (manner or) route of drug administration ( Dr-Route ), dosage ( Dr-Dose ), duration of administration ( Dr-Dur ), and adverse drug events ( Dr-ADE ). These subtypes are highly relevant in the context of medication information and are backed up by an international standard, the HL7 Fast Healthcare Interoperability Resources (FHIR) 9 . Tables 3 and 4 provide an overview of the SOTA on this topic.
Table 3. Medical Named Entity Recognition: Drugs. Benchmark Datasets: n2c2 56 ; i2b2 2009 57 ; MADE 1.0 59 ; DDI 60 .

|
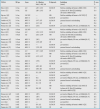
Table 4. Medical Named Entity Recognition: Medication Attributes. Benchmark Datasets: n2c2 56 ; i2b2 2009 57 ; MADE 1.0 59 ; DDI 60 .
|
For medication information, four gold standards had a great impact on the field in the past years. The most recent one came out of the 2018 n2c2 Shared Task on Adverse Drug Events and Medication Extraction in Electronic Health Records 56 , a successor of the 2009 i2b2 Medication Challenge 57 , now with a focus on Adverse Drug Events (ADEs). It includes 505 discharge summaries (303 in the training set and 202 in the test set), which originate from the MIMIC-III clinical care database 58 . The corpus contains nine types of clinical concepts (including drug name), eight attributes (reason, ADE, frequency, strength, duration, route, form, and dosage – from which we chose five for comparison), and 83,869 concept annotations. Relations between drugs and the eight attributes were also annotated and summed up to 59,810 relation annotations (see Section 3.2.1). The third corpus, MADE 1.0 59 , formed the basis for the 2018 Challenge for Extracting Medication, Indication, and Adverse Drug Events (ADEs) from Electronic Health Record (EHR) Notes and consists of 1,092 de-identified EHR notes from 21 cancer patients. Each note was annotated with medication information (drug name, dosage, route, frequency, duration), ADEs, indication (symptom as reason for drug administration), other signs and symptoms, severity (of disease/symptom), and relations among those entities, resulting in 79,000 mention annotations. Finally, the DDI corpus 60 , originally developed for the Drug-Drug Interaction (DDI) Extraction 2013 Challenge 61 , is composed of 792 texts selected from the (semi-structured) DrugBank database 10 and other 233 (unstructured) MEDLINE abstracts, summing up 1,025 documents. This fine-grained corpus has been annotated with a total of 18,502 pharmacological substances and 5,028 drug-drug interactions 11 . Hence, the medication NER task not only comes with a higher entity type complexity but also with text genres different from the disease recognition task—while the former puts emphasis on clinical reports, the latter focuses on scholarly writing.
Except for route and ADE, all top scores for NER were achieved on the n2c2 corpus. For drug names, the current SOTA exceeds 95% F 1 score established by Wei et al. 62 . As to the subtypes, their system also compares favorably to alternative architectures by a large F 1 margin ranging from 8.6 percentage points (for duration) down to 1.0 (for drug name). For route, the distance to the best system is marginal (around 1 percentage point) 12 , whereas for ADE it is huge (more than 10 percentage points, a strong outlier). Overall, frequency, route, and dosage recognition reach outstanding F 1 scores in the range of 95 up to 97%, while for duration information top F 1 scores drop remarkably by at least 10 to 20 percentage points. Still, the recognition of ADEs seems to be the hardest task, with the best system by Wunnava et al. 67 peaking at around 64% F 1 on MADE 1.0 data (here the top performing system by Wei et al. 62 plummets down to 53% F 1 ). Interestingly, ADEs are verbally the least constrained type of natural language utterance compared with all the other entity types considered here.
In terms of DL methodology, BiLSTM-CRFs are the dominating approach. Yet, the type of embeddings used by different DL systems varies a lot ranging from pre-trained Word2vec embeddings and those self-trained on MIMIC-III (for the top performers) to GloVe embeddings pre-trained on CommonCrawl, Wikipedia, EHR notes, and PubMed. There seems to be no generalizable winner for either choice of embeddings given the current state of evaluations, but self-training on medical raw data, such as MIMIC-III, challenge data sets, or, more advisable, using the now available BioSentVec 18 and BlueBERT 22 embeddings pre-trained on MIMIC-III, might be advantageous.
Studies in which the same system configuration was tested on different corpora are still lacking so that corpus effects are unknown (unlike for diseases; see Table 2 ). Yet, there is one interesting though not so surprising observation: Unanue et al. 65 explored the two slices of the DDI corpus, with a span of F 1 scores of more than 16 percentage points. This obviously witnesses the influence of a priori (lack of) structure—DrugBank data is considerably more structured than MEDLINE free texts and, thus, the former gets much higher scores than the latter.
Comparing DL approaches vs. non-DL ones (a CRF architecture) on the same corpus (MADE 1.0), we found that for the core entity type (Drug), the recognition performance differs by almost 3 percentage points, for frequency, route and dose marginally by less than 1, yet for duration and ADE it amounts to roughly 5 and 12 percentage points, respectively—consistently in favor of Deep Neural Networks (DNNs).
3.2 Relation Extraction
Once named entities have been identified, a follow-up question emerges: does some sort of semantic relation hold among these entities? We surveyed this Relation Extraction (REX) task with reference to results that have been achieved for information related to medication attributes and drug-drug interaction.
3.2.1 Medication-Attribute Relations
In Section 3.1.2, we already dealt with single named entity types typically associated with medication information, namely drug names and administration frequency, duration, dosage, route, and ADE, yet in isolated form only. In this subsection, we are concerned with making the close associative ties between Drugs and typical conceptual attributes, such as Frequency , Duration , Dosage , Route , ADE , and Reason (for prescription), explicit. Hence, the recognition of the respective named entity types (Drugs, Dr-Freq , Dr-Dur , Dr-Dose , Dr-Route , Dr-ADE , and Dr-Reason ) turns out to be a good starting point for solving this REX task. Not surprisingly, the benchmarks for this task are a subset of the ones in Tables 3 and 4 depicting the results for medication-related NER. Table 5 provides an overview of the experimental results for finding medication-attribute relations in medical, in effect, clinical, documents.
Table 5. Medical Relation Extraction: Medication-Attribute Relations (including ADEs). Benchmark Datasets: n2c2 56 ; MADE 1.0 59 .

|
The overall results from medication-focused NER are mostly confirmed for the REX task. The n2c2 corpus is the reference dataset for top performance. The group who achieved top F 1 scores for the medication NER problem also performed best for the medication-attribute REX task 62 , with extraordinary figures for Frequency , Route , and Dosage relations (in the upper 98% F 1 range), a superior one for the Duration relation (93% F 1 ), and good ones on the (hard to deal with) Adverse and Reason relations (85% F 1 ). Still, the distances to the second-best system for the same corpus (n2c2) are not so pronounced in most cases, ranging by 1 percentage point (for Frequency , Route , Dosage, and Duration ), yet increased up to 3 (for Adverse ) and 7 (for Reason ) percentage points.
For the MADE 1.0 corpus, a similar picture emerges. From a lower offset (typically around 3 F 1 percentage points compared with n2c2), differences between the best and second-best systems were on the order of (negligible) 1 percentage point for Frequency , Route, and Dosage , yet increased by roughly 3, 5, and 7 percentage points for Reason , Duration , and Adverse events , respectively. Yet, in 4 out of 6 cases ( Frequency , Dosage , Duration , and Adverse events ) non-DL systems (CRFs, SVMs) outperformed their DL counterparts with small margins (in the range of (again, negligible) 1 percentage point) for Frequency and Dosage , yet with higher ones for Duration and Adverse events (5 and 7 percentage points, respectively). In cases where the DL approach ranked higher than a non-DL one, differences ranged between 1 and 3 percentage points (for Route and Reason , respectively). Thus, the MADE 1.0 corpus constitutes a benchmark where well-engineered standard ML classifiers can still play a competitive role. However, we did not find this pattern of partial supremacy of non-DL approaches for the n2c2 benchmark.
The top performers for the medication attribute REX task 62 employed a joint learning approach based on CNN-RNN (thus diverging from the most successful architectures for medication NER; see Tables 3 and 4 ) and rule-based post-processing that outperformed a simple CNN-RNN. Summarizing, the CNN-RNN approach seems more favorable than an (attention-based) BiLSTM, with preferences for self-trained in-domain embeddings.
3.2.2 Drug-Drug Interaction
The second type of medication-focused relation we consider here are drug-drug interactions as featured in the DDI challenge (for surveys on the impact of DL on recent research on drug-drug interactions, cf. 82 83 , for a survey on drug-drug interaction combining progress in data and text mining from EHRs, scientific papers, and clinical reports but lacking in-depth coverage of DL methods, cf. 84 , for the NLP-focused recognition of ADEs also lacking awareness of DL contributions to this topic, cf. 85 ). Four main types of relations between drugs are considered: pharmacokinetic Mechanism , drug Effect , recommendation or Advice regarding a drug interaction, and Interaction between drugs without providing any additional information. Overall, the DDI corpus on which these evaluations were run is divided into 730 documents taken from DrugBank and 175 abstracts from MEDLINE and contains 4,999 relation annotations (4,020 train, 979 test).
Recognition rates for these relations (cf. Table 6 ) are considerably lower than for the medication-related attributes when linked to drugs (cf. Table 5 ). The best systems peak at 85% F 1 score for Advice (a distance of more than 13 percentage points to the top recognition results for medication-attributes), they slip to 78% 13 and 77% for Mechanism and Effect , respectively, and plummet to 59% for Interaction 14 . Differences between the first and second-ranked systems are typically small, yet become larger on subsequent ranks (roughly between 3 to 4 percentage points relative to the top-ranked system). As with medication attributes, drug-drug interactions can also be recognized in a competitive way by CNN-RNN architectures, but attention-based LSTMs perform also considerably well. Again, self-trained embeddings using in-domain corpora seem to be advantageous for this relation class. Reflecting the drop in performance, one may conclude that drug-drug interactions constitute a markedly harder task than the conceptually much closer medication-attribute relations.
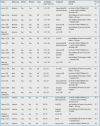
Table 6. Medical Relation Extraction: Drug-Drug Interaction. Benchmark Dataset: DDI 60 .
|
Finally, Table 6 most drastically supports our claim that DL approaches outperform non-DL ones. The difference between both approaches amounts to 5 percentage points for Mechanism , 7 for Effect and Interaction , and 8 for Advice .
4 Conclusions
We have presented various forms of empirical evidence that (with one exception only) Deep Learning-based neural networks outperformed non-DL, feature engineered, approaches for several information extraction tasks. However, despite their success, Deep Neural Networks and their embedding models have their shortcomings as well.
One of the most problematic issues is their dependence on huge amounts of training data: SOTA embedding models are currently trained on hundreds of billions of tokens 89 . This magnitude of data volume is out of reach for any training effort in the medical/clinical domain 90 . Also, embeddings are very vulnerable to malicious attacks or adversarial examples—small changes at the input level may result in severe misclassification 5 . Another well-known problem relates to the instability of word embeddings. Word embeddings depend on their random initialization and the processing order of the underlying examples and therefore they do not necessarily converge on exactly the same embeddings even after several thousands of training iterations 91 92 . Finally, although DL is celebrated for not requiring manual feature engineering, the effects of proper hyperparameter tuning on DNNs 93 remain an issue for DL 94 . Apart from these intrinsic problems, Kalyan and Sangeetha 95 and Khattak et al. 96 refer to extrinsic drawbacks of neural networks, such as opaque encodings (resulting in lacking interpretability) or limited transferability of large models (hindering knowledge distillation for smaller models).
Still, the sparsity of corpora and special linguistic phenomena of the medical (clinical) sublanguage(s) create intrinsic problems for data-greedy DL approaches that have to be overcome by special learning strategies for neural systems, such as transfer learning or domain adaptation. Research on adapting general language models to medical language constraints is just in its beginning. Yet, there is no simple solution to this problem. Wang et al. 97 evaluated Word2vec embeddings trained on private clinical notes, PMC, Wikipedia, and the Google News corpus both qualitatively and quantitatively and showed that the ones trained on Electronic Health Record data performed better on most of the tested scenarios. However, they also found that word embeddings trained on biomedical domain corpora do not necessarily have better performance than those trained on general domain corpora for any downstream biomedical NLP task (other experimental evidence of the effects of in- and out-of-domain corpora and further parameters, such as corpus size, on word embedding performance is reported by Lai et al., 98 ).
While this survey focused on the application domain of medical IE to demonstrate the outstanding role of DL for medical Natural Language Processing, one might be tempted to generalize this trend to other applications as well. There is, indeed, plenty of evidence in the literature that other application fields, such as question answering (and the closely related area of machine reading), summarization, machine translation, and speech processing, reveal the same pattern. However, for text categorization (in the sense of mapping free text to some pre-defined medical category system, such as ICD, SNOMED, or MeSH) this preference is less obvious, since traditional Machine Learning or rule-based models still play an important role here and, more often than for the IE application scenario, show competitive performance against DL approaches. Whether this exception will persist or will be swept away by future research remains an open issue.
Acknowledgments
The first author was partially funded by the German Bundesministerium für Bildung und Forschung (BMBF) under grant no. 01ZZ1803G. We thank all six reviewers for their insightful and helpful comments.
Footnotes
https://mimic.physionet.org/
Wu et al. [ 29 , Table 3(b)] found that 71% of the corpora they screened were English, 20% Chinese, 2% Spanish, Japanese or Finnish and all other languages ranked below 1%. For a survey on medical NLP dealing explicitly with languages other than English, see 36 .
https://github.com/ncbi-nlp/BLUE_Benchmark
https://github.com/geetickachauhan/relation-extraction
We disregard here the common distinction between strict and partial matching; numbers given in the tables typically reflect the strongest condition, i.e., strict (complete) match between system prediction and gold standard data.
Even more ambitious is the task of linking (or grounding) textual mentions and semantic types to unique identifiers of a given terminology or ontology (such as SNOMED-CT, ICD, or the Human Disease Ontology, https://www.ebi.ac.uk/ols/ontologies/doid), an issue we will not elaborate on in this survey, cf. e.g., 39 .
Concrete numbers in the column “Number of Mentions,” indicating the number of named entity mentions (possibly split into training, development, and test set, if provided), may slightly differ for the same corpus because of data cleansing (e.g., removal of duplicates), different pre-processing (e.g., tokenization), and other version issues.
Beltagy et al. 19 report an F1 score of 90% on the BC5CDR corpus, but it remains unclear whether this result refers to the type Disease , Drug , or both of them.
See, e.g., the HL7 FHIR Medication Statement at https://www.hl7.org/fhir/medicationstatement.html#MedicationStatement
https://www.drugbank.ca/
The DDI corpus is actively maintained and enhanced leading to a large number of versions. Hence, comparisons based on DDI have to be carried out very carefully.
Interestingly, the transfer learning approach advocated by Gligic et al. 63 performs well for some medication NER tasks, but fails to deliver competitive results for the medication relation task (cf. Table 5 ).
Xu et al. 86 even reach slightly more than 79% F 1 score for Mechanism (using UMLS-based concept embeddings with a Bi-LSTM approach), but substantially fall below the results for the other three relation types in comparison with all the systems mentioned in Table 6 .
Dewi et al. 87 and Sun et al. 88 report on 86.3% and 84.5% F 1 scores, respectively, for the overall relation classification task both using a multi-layered CNN architecture, yet unfortunately fail to provide details on each of the four single relations under scrutiny here. Both results exceed the overall result of the best-performing system depicted in Table 6 76 (77.3%) by a large margin of 9 and 7 percentage points, respectively.
References
- 1.Goodfellow I J, Bengio Y, Courville A C. MIT Press; 2016. Deep Learning.
- 2.Alom M Z, Taha T M, Yakopcic C, Westberg S, Sidike P, Nasrin M S et al. A state-of-the-art survey on deep learning theory and architectures. Electronics. 2019;8(03):292. [Google Scholar]
- 3.Pouyanfar S, Sadiq S, Yan Y, Tian H, Tao Y, Presa Reyes M Eet al. A survey on deep learning: algorithms, techniques, and applications ACM Computing Surveys 2018510592(92:1–92:36) [Google Scholar]
- 4.Goldberg Y.Neural Network Methods for Natural Language Processing. Number 37 in Synthesis Lectures on Human Language Technologies. Morgan & Claypool; 2017.
- 5.Belinkov Y, Glass J R. Analysis methods in neural language processing: a survey. Transactions of the Association for Computational Linguistics. 2019;7:49–72. [Google Scholar]
- 6.Schmidhuber H J. Deep learning in neural networks: an overview. Neural Networks. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Hohman F M, Kahng M, Pienta R, Chau D H. Visual analytics in deep learning: an interrogative survey for the next frontiers. IEEE Trans Vis Comput Graph. 2019;24(08):2674–93. doi: 10.1109/TVCG.2018.2843369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nassif A B, Shahin I, Attili I, Azzeh M, Shaalan K. Speech recognition using deep neural networks: a systematic review. IEEE Access. 2019;7:19143–65. [Google Scholar]
- 9.Young T, Hazarika D, Poria S, Cambria E. Recent trends in deep learning based natural language processing. IEEE Computational Intelligence Magazine. 2018;13(03):55–75. [Google Scholar]
- 10.Spasić I, Uzuner Ö, Zhou L. Emerging clinical applications of text analytics. Int J Med Inform. 2020;134:103974. doi: 10.1016/j.ijmedinf.2019.103974. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Wang L, Rastegar-Mojarad M A, Moon S, Shen F, Afzal N et al. Clinical information extraction applications: a literature review. J Biomed Inform. 2018;77:34–49. doi: 10.1016/j.jbi.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreimeyer K, Foster M, Pandey A, Arya N, Halford G, Jones S Fet al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review J Biomed Inform 201773(Supplement C):14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. J Biomed Inform. 2002;35(04):222–35. doi: 10.1016/s1532-0464(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 14.Newman-Griffis D, Fosler-Lussier E. Writing habits and telltale neighbors: analyzing clinical concept usage patterns with sublanguage embeddings. In: Proceedings of the 10 th International Workshop on Health Text Mining and Information Analysis LOUHI@EMNLP-IJCNLP 2019. p. 146–56.
- 15.Nunez J J, Carenini G. Comparing the intrinsic performance of clinical concept embeddings by their field of medicine. In: Proceedings of the 10 th International Workshop on Health Text Mining and Information Analysis LOUHI@EMNLP-IJCNLP 2019. p. 11–7.
- 16.Pyysalo S, Ginter F, Moen H, Salakoski T, Ananiadou S. Distributional semantics resources for biomedical text processing. In: Proceedings of the 5 th International Symposium on Languages in Biology and Medicine, LMB 2013. p. 39–43
- 17.Zhang Y, Chen Q, Yang Z, Lin H, Lu Z. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Scientific Data. 2019;6:52. doi: 10.1038/s41597-019-0055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Q, Peng Y, Lu Z. BioSentVec : creating sentence embeddings for biomedical texts. In: Proceedings of the 7 th IEEE International Conference on Healthcare Informatics, ICHI 2019.
- 19.Beltagy I, Lo K, Cohan A. SciBert : a pretrained language model for scientific text. In: Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing & 9 th International Joint Conference on Natural Language Processing EMNLP-IJCNLP 2019. p. 3615–20.
- 20.Lee J, Yoon W, Kim S, Kim D, Kim S, So C H et al. BioBert : a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(04):1234–40. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alsentzer E, Murphy J R, William Boag W, Weng W H, Jin D, Naumann Tet al. Publicly available clinical Bert embeddings. In: Proceedings of the 2 nd Workshop on Clinical Natural Language Processing ClinicalNLP @ NAACL-HLT 2019. p. 72–8.
- 22.Peng Y, Yan S, Zhiyong Lu Z. Transfer learning in biomedical natural language processing: an evaluation of Bert and ELMo on ten benchmarking datasets. In: Proceedings of the 18 th SIGBioMed Workshop on Biomedical Natural Language Processing and Shared Task BioNLP @ ACL 2019. p. 58–65.
- 23.Conway M, Hu M, Chapman W W. Recent advances in using natural language processing to address public health research questions using social media and consumer-generated data. Yearb Med Inform. 2019;28:208–17. doi: 10.1055/s-0039-1677918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Hernandez G, Sarker A, O’Connor K, Savova G K. Capturing the patient’s perspective: a review of advances in natural language processing of health-related text. Yearb Med Inform. 2017;26:214–27. doi: 10.15265/IY-2017-029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Filannino M, Uzuner Ö. Advancing the state of the art in clinical natural language processing through shared tasks. Yearb Med Inform. 2018;27:184–92. doi: 10.1055/s-0038-1667079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velupillai S, Mowery D L, South B R, Kvist M, Dalianis H. Recent advances in clinical natural language processing in support of semantic analysis. Yearb Med Inform. 2015;24:183–93. doi: 10.15265/IY-2015-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meystre S M, Savova G K, Kipper-Schuler K C, Hurdle J F. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008;17:128–44. [PubMed] [Google Scholar]
- 28.Velupillai S, Suominen H, Liakata M, Roberts A, Shah A D, Morley K I et al. Using clinical natural language processing for health outcomes research: overview and actionable suggestions for future advances. J Biomed Inform. 2018;88:11–9. doi: 10.1016/j.jbi.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S, Roberts K, Datta S, Du J, Ji Z, Si Y et al. Deep learning in clinical natural language processing: a methodical review. J Am Med Inform Assoc. 2020;27(03):457–70. doi: 10.1093/jamia/ocz200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao C, Choi E, Sun J. Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review. J Am Med Inform Assoc. 2018;25(10):1419–28. doi: 10.1093/jamia/ocy068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shickel B, Tighe P J, Bihorac A, Rashidi P. Deep EHR: a survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J Biomed Health Inform. 2018;22(05):1589–604. doi: 10.1109/JBHI.2017.2767063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miotto R, Wang F, Wang S, Jiang X, Dudley J T. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2017;19(06):1236–46. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esteva A, Robicquet A, Ramsundar B, Kuleshov V, De Pristo M, Chou K et al. A guide to deep learning in healthcare. Nat Med. 2019;25(01):24–9. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 34.Ching T, Himmelstein D S, Beaulieu-Jones B K, Kalinin A A, Do B T, Way G P et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15(141):2.0170387E7. doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajkomar A, Oren E, Chen K, Dai A M, Hajaj N, Hardt M et al. Scalable and accurate deep learning for electronic health records. NPJ Digit Med. 2018;1:18. doi: 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Névéol A, Dalianis H, Velupillai S, Savova G K, Zweigenbaum P. Clinical natural language processing in languages other than English: opportunities and challenges. J Biomed Semantics. 2018;9(01):12. doi: 10.1186/s13326-018-0179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chauhan G, McDermott M BA, Szolovits P. REflex: flexible framework for relation extraction in multiple domains. In: Proceedings of the 18 th SIGBioMed Workshop on Biomedical Natural Language Processing and Shared Task BioNLP @ ACL 2019. p. 30–47.
- 38.Li J, Sun A, Han J, Li C.A survey on deep learning for named entity recognition. IEEE Transactions on Knowledge and Data Engineering, page [Early Access]; 2020. Available at:https://arxiv.org/pdf/1812.09449.pdf
- 39.Zhao S, Liu T, Zhao S, Wang F. A neural multi-task learning framework to jointly model medical named entity recognition and normalization. In: Proceedings of the 33 rd AAAI Conference on Artificial Intelligence 2019. p. 817–24.
- 40.Sheikhalishahi S, Miotto R, Dudley J T, Lavelli A, Fabio Rinaldi F, Osmani V. Natural language processing of clinical notes on chronic diseases: systematic review. JMIR Med Inform. 2019;7(02):e12239. doi: 10.2196/12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koleck T A, Dreisbach C, Bourne P E, Bakken S. Natural language processing of symptoms documented in free-text narratives of electronic health records: a systematic review. J Am Med Inform Assoc. 2019;26(04):364–79. doi: 10.1093/jamia/ocy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savova G K, Danciu I, Alamudun F, Miller T A, Lin C, Bitterman D S et al. Use of natural language processing to extract clinical cancer phenotypes from electronic medical records. Cancer Res. 2019;79(21):5463–70. doi: 10.1158/0008-5472.CAN-19-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Datta S, Bernstam E V, Roberts K. A frame semantic overview of NLP-based information extraction for cancer-related EHR notes. J Biomed Inform. 2019;100:103301. doi: 10.1016/j.jbi.2019.103301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Sun Y, Johnson R J, Sciaky D, Wei C H, Leaman Ret al. BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database (Oxford) 2016;2016:baw068. [DOI] [PMC free article] [PubMed]
- 45.Doğan R I, Robert Leaman R, Lu Z. NCBI Disease Corpus: a resource for disease name recognition and concept normalization. J Biomed Inform. 2014;47:1–10. doi: 10.1016/j.jbi.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Zhang Y, Ren X, Zhang Y, Žitnik M, Shang J et al. Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics. 2019;35(10):1745–52. doi: 10.1093/bioinformatics/bty869. [DOI] [PubMed] [Google Scholar]
- 47.Sachan D S, Xie P, Sachan M, Xing E P.Effective use of bidirectional language modeling for transfer learning in biomedical named entity recognition. In: Proceedings of the [2nd] Conference on Machine Learning for Healthcare 2018. p. 383–402.
- 48.Lou Y, Zhang Y, Qian T, Li F, Xiong S, Ji D. A transition-based joint model for disease named entity recognition and normalization. Bioinformatics. 2017;33(15):2363–71. doi: 10.1093/bioinformatics/btx172. [DOI] [PubMed] [Google Scholar]
- 49.Xu K, Yang Z, Kang P, Wang Q, Liu W. Document-level attention-based BiLSTM-CRF incorporating disease dictionary for disease named entity recognition. Comput Biol Med. 2019;108:122–32. doi: 10.1016/j.compbiomed.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Mikolov T, Sutskever I, Chen K, Corrado G S, Dean J. Distributed representations of words and phrases and their compositionality. In: Proceedings of the 27 th Annual Conference on Neural Information Processing Systems NIPS 2013. p. 3111–9.
- 51.Hong S K, Lee J G. DTranNER: biomedical named entity recognition with deep learning-based label-label transition model. BMC Bioinformatics. 2020;21(01):53. doi: 10.1186/s12859-020-3393-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peters M E, Neumann M, Iyyer M, Gardner M, Clark C T, Lee Ket al. Deep contextualized word representations. In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL-HLT 2018; 1: Long Papers. p. 2227–37.
- 53.Pennington J, Socher R, Manning C D.GloVe: Global Vectors for word representation. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, EMNLP 2014. p. 1532–43.
- 54.Collobert R, Weston J, Bottou L, Karlen M, Kavukcuoglu K, Kuksa P P. Natural language processing (almost) from scratch. Journal of Machine Learning Research. 2011;12(76):2493–537. [Google Scholar]
- 55.He K, Zhang X, Ren S, Sun J.Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. In: Proceedings of the 2015 IEEE International Conference on Computer Vision, ICCV 2015. p. 1026–34.
- 56.Henry S, Buchan K, Filannino M, Stubbs A, Uzuner Ö. 2018 n2c2 Shared Task on Adverse Drug Events and Medication Extraction in Electronic Health Records. J Am Med Inform Assoc. 2020;27(01):3–12. doi: 10.1093/jamia/ocz166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uzuner Ö, Solt I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010;17(05):514–8. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson A EW, Pollard T J, Shen L, Lehman L WH, Feng M, Ghassemi M M et al. Mimic-III, a freely accessible critical care database. Scientific Data. 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jagannatha A, Liu F, Liu W, Yu H. Overview of the First Natural Language Processing Challenge for Extracting Medication, Indication, and Adverse Drug Events from Electronic Health Record Notes (Made 1) Drug Saf. 2019;42(01):99–111. doi: 10.1007/s40264-018-0762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herrero-Zazo M, Segura-Bedmar I, Martínez P, Declerck T. The DDI corpus: an annotated corpus with pharmacological substances and drug-drug interactions. J Biomed Inform. 2013;46(05):914–20. doi: 10.1016/j.jbi.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 61.Segura-Bedmar I, Martínez P, Herrero-Zazo M. SemEval-2013 Task 9: Extraction of Drug-Drug Interactions from Biomedical Texts (DDIExtraction 2013). In: Proceedings of the 7 th International Workshop on Semantic Evaluation, SemEval@NAACL-HLT 2013. p. 341–50.
- 62.Wei Q, Ji Z, Li Z, Du J, Wang J, Xu J et al. A study of deep learning approaches for medication and adverse drug event extraction from clinical text. J Am Med Inform Assoc. 2019;27(01):13–21. doi: 10.1093/jamia/ocz063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gligic L, Kormilitzin A, Goldberg P, Nevado-Holgado A. Named entity recognition in electronic health records using transfer learning bootstrapped neural networks. Neural Netw. 2020;121:132–9. doi: 10.1016/j.neunet.2019.08.032. [DOI] [PubMed] [Google Scholar]
- 64.Zeng D, Sun C, Lin L, Liu B. LSTM-CRF for drug-named entity recognition. Entropy. 2017;19(06):283. [Google Scholar]
- 65.Unanue I J, Borzeshi E Z, Piccardi M. Recurrent neural networks with specialized word embeddings for health-domain named-entity recognition. J Biomed Inform. 2017;76:102–9. doi: 10.1016/j.jbi.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 66.Li F, Liu W, Yu H. Extraction of information related to adverse drug events from electronic health record notes: design of an end-to-end model based on deep learning. JMIR Med Inform. 2018;6(04):e121594. doi: 10.2196/12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wunnava S, Qin X, Kakar T, Sen C, Rundensteiner E A, Kong X. Adverse drug event detection from electronic health records using hierarchical recurrent neural networks with dual-level embedding. Drug Saf. 2019;42(01):113–22. doi: 10.1007/s40264-018-0765-9. [DOI] [PubMed] [Google Scholar]
- 68.Jagannatha A N, Yu H.Bidirectional RNN for medical event detection in electronic health records. In: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL-HLT 2016. p. 473–82. [DOI] [PMC free article] [PubMed]
- 69.Jagannatha A N, Yu H.Structured prediction models for RNN based sequence labeling in clinical text. In: Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, EMNLP 2016. p. 856–65. [DOI] [PMC free article] [PubMed]
- 70.Dandala B, Joopudi V, Devarakonda M V. Adverse drug events detection in clinical notes by jointly modeling entities and relations using neural networks. Drug Saf. 2019;42(01):135–46. doi: 10.1007/s40264-018-0764-x. [DOI] [PubMed] [Google Scholar]
- 71.Tao C, Filannino M, Uzuner Ö. Prescription extraction using CRFs and word embeddings. J Biomed Inform. 2017;72:60–6. doi: 10.1016/j.jbi.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chapman A B, Peterson K S, Alba P R, Du Vall S L, Patterson O V. Detecting adverse drug events with rapidly trained classification models. Drug Saf. 2019;42(01):147–56. doi: 10.1007/s40264-018-0763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang X, Bian J, Gong Y, Hogan W R, Wu Y. MADEx: a system for detecting medications, adverse drug events, and their relations from clinical notes. Drug Saf. 2019;42(01):123–33. doi: 10.1007/s40264-018-0761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Christopoulou F, Tran T T, Sahu S K, Miwa M, Ananiadou S. Adverse drug events and medication relation extraction in electronic health records with ensemble deep learning methods. J Am Med Inform Assoc. 2020;27(01):39–46. doi: 10.1093/jamia/ocz101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun X, Dong K, Ma L, Sutcliffe R FE, He F, Chen S et al. Drug-drug interaction extraction via recurrent hybrid convolutional neural networks with an improved focal loss. Entropy. 2019;21(01):37. doi: 10.3390/e21010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng W, Lin H, Luo L, Zhao Z, Li Z, Zhang Y et al. An attention-based effective neural model for drug-drug interactions extraction. BMC Bioinformatics. 2017;18:445. doi: 10.1186/s12859-017-1855-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang W, Yang X, Yang C, Guo X W, Zhang X, Wu C.Dependency-based long short term memory network for drug-drug interaction extraction BMC Bioinformatics 201718(Supplement 16):578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mikolov T, Chen K, Corrado G S, Dean J.Efficient estimation of word representations in vector space. In: Proceedings of the 1st International Conference on Learning Representations, ICLR 2013.
- 79.Lim S, Lee K, Kang J. Drug drug interaction extraction from the literature using a recursive neural network. PLoS One. 2018;13(01):e0190926. doi: 10.1371/journal.pone.0190926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, Zheng W, Lin H, Wang J, Yang Z, Dumontier M. Drug-drug interaction extraction via hierarchical RNNs on sequence and shortest dependency paths. Bioinformatics. 2018;34(05):828–35. doi: 10.1093/bioinformatics/btx659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raihani A, Laachfoubi N. Extracting drug-drug interactions from biomedical text using a feature-based kernel approach. Journal of Theoretical and Applied Information Technology. 2016;92(01):109–20. [Google Scholar]
- 82.Zhang T, Leng J, Liu Y. Deep learning for drug-drug interaction extraction from the literature: a review. Brief Bioinform. 2019:bbz087. doi: 10.1093/bib/bbz087. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Y, Lin H, Yang Z, Wang J, Su Y, Xu B et al. Neural network-based approaches for biomedical relation classification: a review. J Biomed Inform. 2019;99:103294. doi: 10.1016/j.jbi.2019.103294. [DOI] [PubMed] [Google Scholar]
- 84.Vilar S, Friedman C, Hripcsak G M. Detection of drug-drug interactions through data mining studies using clinical sources, scientific literature and social media. Brief Bioinform. 2018;19(05):863–77. doi: 10.1093/bib/bbx010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luo Y, Thompson W K, Herr T M, Zeng Z, Berendsen M A, Jonnalagadda S R et al. Natural language processing for EHR-based pharmacovigilance: a structured review. Drug Saf. 2017;40(11):1075–89. doi: 10.1007/s40264-017-0558-6. [DOI] [PubMed] [Google Scholar]
- 86.Xu B, Shi X, Zhao Z, Zheng W. Leveraging biomedical resources in Bi-LSTM for drug-drug interaction extraction. IEEE Access. 2018;6:33432–9. [Google Scholar]
- 87.Dewi I N, Dong S, Hu J.Drug-drug interaction relation extraction with deep convolutional neural networks. In: Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2017. p. 1795–802.
- 88.Sun X, Ma L, Du X, Feng J, Dong K.Deep convolution neural networks for drug-drug interaction extraction. In: Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2018. p.1662–8.
- 89.Grave E, Bojanowski P, Gupta P, Joulin A, Mikolov T.Learning word vectors for 157 languages. In: Proceedings of the 11th International Conference on Language Resources and Evaluation, LREC 2018. p. 3483–7.
- 90.Spasić I, Nenadić G. Clinical text data in machine learning: systematic review. JMIR Med Inform. 2020;8(03):e17984. doi: 10.2196/17984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hellrich J, Hahn U.Bad company: neighborhoods in neural embedding spaces considered harmful. In: Proceedings of the 26th International Conference on Computational Linguistics: Technical Papers, COLING 2016. p. 2785–96.
- 92.Wendlandt L, Kummerfeld J K, Mihalcea R.Factors influencing the surprising instability of word embeddings. In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL-HLT 2018(1): Long Papers. p. 2092–102.
- 93.Diaz GI Fokoue-Nkoutche A, Nannicini G, Samulowitz H.An effective algorithm for hyperparameter optimization of neural networks IBM Journal of Research and Development 201761(4-5):9 [Google Scholar]
- 94.Chiu B, Crichton G KO, Korhonen A, Pyysalo S.How to train good word embeddings for biomedical NLP. In: Proceedings of the 15th Workshop on Biomedical Natural Language Processing BioNLP @ ACL 2016. p. 166–74.
- 95.Kalyan K S, Sangeetha S. SECNLP : a survey of embeddings in clinical natural language processing. J Biomed Inform. 2020;101:103323. doi: 10.1016/j.jbi.2019.103323. [DOI] [PubMed] [Google Scholar]
- 96.Khattak F K, Jeblee S, Pou-Prom C, Abdalla M, Meaney C, Rudzicz F. A survey of word embeddings for clinical text. J Biomed Inform. 2019;4:100057. doi: 10.1016/j.yjbinx.2019.100057. [DOI] [PubMed] [Google Scholar]
- 97.Wang Y, Liu S, Afzal N, Rastegar-Mojarad M A, Wang L, Shen F et al. A comparison of word embeddings for the biomedical natural language processing. J Biomed Inform. 2018;87:12–20. doi: 10.1016/j.jbi.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lai S, Liu K, He S, Zhao J. How to generate a good word embedding. IEEE Intelligent Systems. 2016;31(06):5–14. [Google Scholar]


