Abstract
Genetic variations in the 18S ribosomal DNA (18S), 28S ribosomal DNA (28S), second internal transcribed spacer of ribosomal DNA (ITS2), and mitochondrial cytochrome c oxidase subunit 1 (cox1) of Neoschoengastia gallinarum collected from subtropical China were examined. First, a portion of the 18S (p18S), a portion of the 28S (p28S), and the complete ITS2 were separately amplified from individual mites and sequenced. The lengths of the sequences of p18S, p28S, and ITS2 were found to be 1379 bp, 3465~3468 bp, and 200 bp, respectively. The intraspecific sequence variation was 0~0.1% for p28S and 0~1.6% for ITS2, though no variation was observed for p18S, suggesting conservation of rDNA sequences. Second, a portion of the mitochondrial cox1 gene (pcox1) of N. gallinarum was analyzed. The length of the pcox1 sequence is 460 bp, and two distinct groups were observed in N. gallinarum. All pcox1 sequences in group I were identical, and there was only one nucleotide transition observed in group II; however, 7.0~7.2% variations between the two groups were observed, suggesting that two genotypes of N. gallinarum: genotype I and genotype II. Phylogenetic analyses based on pcox1 sequences indicated that N. gallinarum isolates (genotype I or genotype II) clustered into one branch; according to cox1 sequence analysis of Trombiculidae, Walchia hayashii is the closest species. The present study shows that ITS2 rDNA sequence can act as marker for the identification of N. gallinarum samples. Furthermore, analysis of the mitochondrial pcox1 sequence suggests the existence of two genotypes, which has implications for further studies of the ecology and population genetic structures of N. gallinarum.
Keywords: Neoschoengastia gallinarum, Genetic variation, rDNA, cox1, Genotype
Introduction
Neoschoengastia gallinarum (Arthropoda: Arachnida: Acarina: Trombiculidae), a common ectoparasite, was first discovered in Taiwan and named by Hatori (Sugimoto 1936). The life cycle of chigger mites involves incomplete metamorphosis and includes four stages: egg, larva, nymph, and adult (Jeu 1961). The larvae of N. gallinarum feed on the blood of many species of birds, including poultry, primarily parasitizing under the wing and the medial side of the thigh, and these larvae develop into nymphs after falling off the host. Poultry are at a high risk for N. gallinarum infection, and chickens have strong tissue responses following parasitization. The primary clinical symptoms of N. gallinarum infection are anemia, emaciation, mental retardation, and even death when infestation by a large population occurs (Jeu 1956, 1957). N. gallinarum is one of the most common and highly pathogenic ectoparasites in free-range broilers in China, causing mostly whole-group infections and repeated infections. At present, the prevention and treatment of this disease mainly rely on chemical drugs. However, as N. gallinarum is very resistant to chemical drugs, new types of agents, such as botanical drugs, are currently a hot topic of research against this N. gallinarum (Wang et al. 2018).
Currently, the most frequently used sequences for investigating mite phylogeny are ribosomal DNA (rDNA) and mitochondrial genes (mtDNA). rDNA is a type of moderately repetitive sequence that exists in chromosomes as tandem multiple copies. Different regions of rDNA exhibit different levels of evolution, which can be used for various levels of classification. Eukaryotic 18S rDNA and 28S rDNA are genes that encode ribosomal subunits and are highly conserved during evolution; thus, sequence differences between homologous species are small and are generally considered suitable for high-order classification (Hillis and Dixon 1991). Due to the low selection pressure and rapid evolutionary variability, the internal transcribed spacer (ITS) sequence is more suitable for studying phylogenetic relationships at low levels, such as species and population (Lin et al. 2007a, b; Zhao et al. 2012). Mitochondrial DNA follows a matrilineal inheritance pattern, and the primary structure of mtDNA differs significantly between and within species, with the advantages of evolutionary autonomy and simplicity (Cheng et al. 2016; Jin et al. 2019). Among mitochondrial protein-encoding genes, the evolutionary rate of the cox1 gene is three times that of other protein-encoding genes; this gene exhibits few insertions and deletions, and the sequences at both ends are relatively well conserved and are among the most widely used molecular markers (Medina and Walsh 2000; Gao et al. 2017).
The objectives of the present study were to examine sequence variations in rDNA and the mt cox1 gene among N. gallinarum isolates in Guangdong and Fujian provinces, subtropical China. Phylogenetic relationships among 14 Trombiculidae species were also reconstructed based on the pcox1 sequences.
Materials and methods
Samples and DNA extraction
N. gallinarum samples were collected from yellow-feathered broilers from six different chicken farms in Yunfu city, Guangdong Province, and Putian city, Fujian Province, China (Table 1). The mites were obtained from skin with crateriform lesions using fine scissors. The samples were washed in normal saline and identified primarily based on morphological characteristics with optical microscopy and electron microscopy. The parasites were then fixed in 70% (v/v) ethanol and stored at − 20 °C until use. Total genomic DNA was extracted from individual samples by sodium dodecyl sulfate/proteinase K treatment, column-purified (Wizard™ DNA Clean-Up System, Promega, USA) and eluted with 50 μL of H2O according to the manufacturer’s recommendations.
Table 1.
Geographical origins and GenBank accession numbers of N. gallinarum samples
| Sample codes | Geographical origin | Accession number | Sample codes | Geographical origin | Accession number | ||
|---|---|---|---|---|---|---|---|
| pcox1 | ITS2 | pcox1 | ITS2 | ||||
| NGY1 | Yunfu | MK423976 | MK643333 | NGF1 | Putian | MK423976 | MK423981 |
| NGY2 | Yunfu | MK423976 | MK423981 | NGF2 | Putian | MK423977 | MK643334 |
| NGY3 | Yunfu | MK423976 | MK423981 | NGF3 | Putian | MK423977 | MK423981 |
| NGY4 | Yunfu | MK423977 | MK423980 | NGF4 | Putian | MK423976 | MK423981 |
| NGY5 | Yunfu | MK423977 | MK643334 | NGF5 | Putian | MK423978 | MK423982 |
| NGY6 | Yunfu | MK423976 | MK423980 | NGF6 | Putian | MK423977 | MK423980 |
| NGY7 | Yunfu | MK423978 | MK643334 | NGF7 | Putian | MK423976 | MK423979 |
| NGY8 | Yunfu | MK423978 | MK423981 | NGF8 | Putian | MK423976 | MK423980 |
| NGY9 | Yunfu | MK423977 | MK423981 | NGF9 | Putian | MK423976 | MK423980 |
| NGY10 | Yunfu | MK423977 | MK643334 | NGF10 | Putian | MK423978 | MK643333 |
| NG1 | Yunfu | MK423976 | MK423981 | NGFA1 | Putian | MK423977 | MK643334 |
| NG2 | Yunfu | MK423977 | MK423982 | NGFA2 | Putian | MK423976 | MK643335 |
| NG3 | Yunfu | MK423978 | MK423979 | NGFA3 | Putian | MK423976 | MK643333 |
| NG4 | Yunfu | MK423976 | MK423980 | NGFA4 | Putian | MK423977 | MK423981 |
| NG5 | Yunfu | MK423976 | MK423982 | NGFA5 | Putian | MK423978 | MK423980 |
| NGD11 | Yunfu | MK423978 | MK643333 | NGFA6 | Putian | MK423977 | MK423982 |
| NGD12 | Yunfu | MK423977 | MK423981 | NGFA7 | Putian | MK423977 | MK643333 |
| NGD13 | Yunfu | MK423976 | MK423979 | NGFA8 | Putian | MK423978 | MK423982 |
| NGD14 | Yunfu | MK423977 | MK423981 | NGFA9 | Putian | MK423977 | MK423979 |
| NGD15 | Yunfu | MK423977 | MK423982 | NGA10 | Putian | MK423977 | MK423982 |
PCR amplification
Based on partial sequences of the 18S rDNA and 28S rDNA of Eutrombicula splendens available in GenBank (accession nos. KY922159 and KY922031, respectively), primers (Table 2) to amplify N. gallinarum p18S and p28S were designed based on the conserved regions. The complete ITS2 and pcox1 were amplified by PCR with primers designed based on analysis of the corresponding sequences amplified by universal primers (Hedin 2001; Hedin and Maddison 2001; Ji et al. 2010) (Table 2). These primers were synthesized with a Biosearch Model 8700 DNA synthesizer (Biosearch, USA). PCR (50 μL) was performed in a thermocycler (Biometra, German) in a reaction containing 2.5 mM MgCl2, 0.4 mM each dNTP, 5 μL of 10× LA Taq buffer, 0.5 mM each primer, 2.5 U rTaq polymerase (Takara, Japan), and 2 μL of DNA sample. Except for p28S, all amplifications were carried out under the following conditions: initial denaturation at 94 °C for 5 min; 35 cycles of 94 °C for 30 s (denaturation), 55 °C for 30 s min (annealing), and 72 °C for 30 s ~ 2 min (extension) according to the product length; and a final extension at 72 °C for 5 min. Long-PCR amplification was used for p28S amplification under the following conditions: 94 °C for 5 min (initial denaturation); followed by 10 cycles of 94 °C for 30 s (denaturation), 53 °C for 30 s (annealing), 68 °C for 4 min (extension), and 94 °C for 5 min (initial denaturation); 25 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 4 min; and a final extension at 72 °C for 7 min. Samples without DNA (no-DNA controls) were included in each amplification run, and in no case were amplicons detected in the no-DNA controls (data not shown). Each amplicon (3 μL) was examined by agarose (1.0%) gel electrophoresis, stained with ethidium bromide and photographed using a gel documentation system (UVItec, UK).
Table 2.
Primers used to amplify p18S rDNA, p28S rDNA, ITS2 rDNA, and mitochondrial pcox1
| Name of primer | Sequence (5′–3′) | Reference |
|---|---|---|
| For pcox1 | ||
| COI-F4 | CGTAACAGCTCATGCTTTCATT | This study |
| COI-R4 | AGTGGTGTTGAAGTTTCGATCT | This study |
| For ITS2 | ||
| 5.8s-F2 | CACGCCGAGCACTCGACATT | This study |
| 28s-R2 | GATCCTTCGCTCGCCGTTACT | This study |
| For p18S | ||
| 18-f1 | GGCTCATTAAATCAGTTACGGTT | This study |
| 18-R1 | ATTCCTCGTTCATGGGCAAT | This study |
| For p28S | ||
| 28-f1 | GAAAGCAAAAGCGATTCCCT | This study |
| 28-R1 | CGACCTTTGACAACTACGAAC | This study |
Purification, cloning, and sequencing of PCR products
For p18S, p28S, ITS2, and pcox1 regions, the amplification products were separated by 1.0% agarose gel electrophoresis; the gels were then stained with ethidium bromide for DNA visualization under UV light. The PCR products were excised from the agarose gel and purified using spin columns (Wizard PCR-Prep DNA Purification System, Promega, USA), after which the purified PCR products were ligated to the pGEM-T easy vector (Promega, USA) according to the manufacturer’s recommendations. The recombinant plasmid was then transformed into Escherichia coli JM109 competent cells (Promega, USA), and positive transformants containing recombinant plasmids were selected by PCR amplification. Cell cultures with confirmed recombinant plasmids were sent to Shanghai Sangon Biological Engineering Biotechnology Company for sequencing using an ABI 377 automated DNA sequencer (with BigDye terminator chemistry).
Sequence analyses
Sequences of p18S, p28S, ITS2, and pcox1 were separately aligned using the computer program ClustalX 1.83 (Thompson et al. 1997). Pairwise comparisons of the level of sequence difference (D) were conducted among and within species using the formula D = 1-(M/L), where M is the number of alignment positions at which the two sequences have a base in common and L is the total number of alignment positions over which the two sequences are compared (Chilton et al. 1995).
Phylogenetic analyses
To study phylogenetic relationships among Trombiculidae species, 14 published pcox1 sequences of chigger mites from the family Trombiculidae were selected, and the Amblyomma americanum (Acarina: Ixodidae, GenBank: DQ168131) pcox1 sequence was used as the outgroup. Sequences of pcox1 with consensus lengths (460 bp) were aligned using the MAFFT 7.263 program (Katoh and Standley 2013), and comparison of the results was performed using MEGA5.1 software (Tamura et al. 2011). The distance method was applied to calculate the genetic distance between samples. Based on the Kimura 2-parameter model, a phylogenetic tree was constructed by the neighbor-joining (NJ) method. Bootstrap support for NJ trees was calculated using 1000 bootstrap replicates.
Results
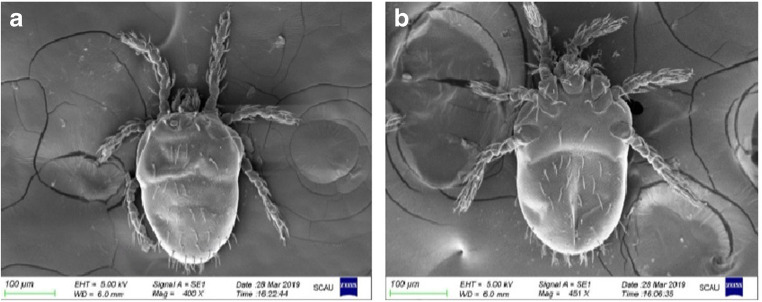
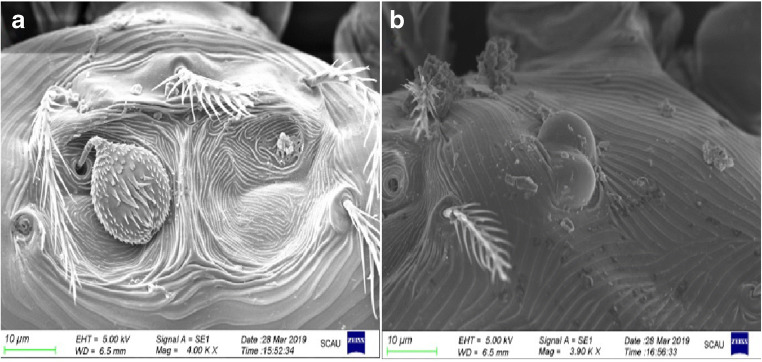
Combined with optical and electronic microscope observations, living mites were observed to be orange-red, oval, with usually 46 back setae and 40 abdomen setae (Fig. 1). The thigh seta, knee seta, dorsal tibia seta, lateral tibia seta, and abdominal tibia seta were all branched, and the cheliceral sheath setae were bare (data not shown). Under the electronic microscope, the back plate was very small and trapezoidal, and the front half had many engraved points; the front edge was almost straight, and the middle of the rear edge was slightly recessed forward and hairy ball-shaped; most of the expansion had many small spines and was covered by skin folds (Fig. 2a). The eyespots were 2 × 2, with the front ones larger than the back ones; only two eyespots in the right half of the mite were photographed (Fig. 2b). All mite samples collected from chickens from different areas of China were identified as N. gallinarum by using the reported identification formula, and the results showed that the findings were consistent with previous reports (Jeu 1959; Li et al. 1997). In this study, there were no obvious differences found between individual mites.
Fig. 1.
General morphology observation of N. gallinarum. a Back side × 400. b Ventral side × 451
Fig. 2.
Scanning electron microscopy observation of N. gallinarum. a Back plate × 4000. b Right eyespot × 3900
Genomic DNA was extracted from 10 individual mites from Yunfu city and Putian city, China. The 1379-bp p18S rDNA region and 3465~3468-bp p28S rDNA region of N. gallinarum samples from subtropical China were amplified using universal PCR primers for 18S rDNA and 28S rDNA (Table 2). The amplified p18S rDNA regions of the N. gallinarum samples from different geographical locations were 100% identical, and no sequence polymorphism was detected. The sequence was deposited in GenBank under accession no. MK400440. Comparison with the p18S rDNA of E. splendens (GenBank KY922159) revealed rather low interspecific sequence variation (0.6%) caused by only 8 nucleotide substitutions. The p28S rDNA sequences of N. gallinarum were deposited in GenBank under accession nos. MK418676–83. A slightly higher level of intraspecific sequence divergence (0.1%) was caused by 12 point mutations; the maximum-likelihood estimation of the transition/transversion bias (R) was 0.65, and 3 insertions or deletions were detected in the N. gallinarum 28S rDNA sequence. Comparison with the p28S rDNA of E. splendens (GenBank KY922031) revealed interspecific sequence variation (3.3~3.4%). The average levels of bases A, T, C, and G in the p18S sequence were calculated to be 26.69%, 24.80%, 21.10%, and 27.41%, respectively, with slightly higher A + T content (51.49%) than C + G (48.51%) content. The average levels of bases A, T, C, and G in the p28S sequence were determined to be 25.30%, 24.75%, 21.15%, and 28.79%, respectively, with slightly higher A + T content (50.06%) than C + G (49.94%) content.
Genomic DNA was extracted from each of 40 individual mites from Yunfu city and Putian city, China. PCR amplification with universal primers that annealed to the 5.8S and 28S rDNA regions (Table 2) resulted in a 371-bp fragment that included complete ITS2 sequences and partial 5.8S and 28S rDNA sequences. The ITS2 sequences were deposited in GenBank under accession nos. MK423979–83 and MK643333–35. Mutual comparison of the sequences revealed 7 point mutations (1.6% sequence divergence) in the 200-bp ITS2 region, and the maximum-likelihood estimation of transition/transversion bias (R) was 5.73. The average levels of bases A, T, C, and G in the ITS2 sequence were 20.00%, 39.50%, 17.50%, and 23.00%, respectively, and the A + T content (59.50%) of these samples was distinctly higher than the C + G (40.50%) content. The ITS2 sequences were aligned in NCBI, and no significant similarity was observed in the middle ITS2 region. Only the 5.8S and 28S regions at both ends exhibited significant sequence similarity to close species.Using a pair of universal primers for Acarina (Hedin 2001; Hedin and Maddison 2001), a fragment of the 1065-bp pcox1 region was amplified from genomic DNA extracted from each of 40 individual mites from Yunfu city and Putian city, China. After sequencing (data not shown), specific primers (Table 2) were designed for the N. gallinarum pcox1 sequence, and a 460-bp fragment was amplified. The pcox1 sequences of N. gallinarum were deposited in GenBank under accession nos. MK423976–78. Mutual comparison of the sequences revealed 2 distinctly different groups, suggesting the existence of 2 genotypes existed: genotype I and genotype II. All gene sequences in the genotype I group were identical, and there were two sequences with only one nucleotide transition in the genotype II group (Fig. 3). Sequence variation between the 2 genotypes was found to be 7.0~7.2%. The observed variations were due to 32 polymorphic sites. The maximum-likelihood estimation of transition/transversion bias (R) was 5.73. There was no insertion or deletion, and the transition rate was significantly higher than the transversion rate. Compared with the sequence from Blankaartia acuscutellaris (GenBank: KY930738), the interspecific sequence variation was calculated to be 20.0~21.5%. The base compositions of the two genotypes were similar, with average levels of A, T, C, and G in the pcox1 sequence of 35.43%, 33.77%, 18.70%, and 12.10%, respectively, and a distinctly higher A + T content (69.20%) than C + G (30.80%) content.
Fig. 3.
Alignment of three representative pcox1 sequences of N. gallinarum. Sequence NGY5 belongs to genotype I, and sequences NGY1 and NGY7 belong to genotype II; dark gray region: only 1 nucleotide transition (T-C) in genotype II
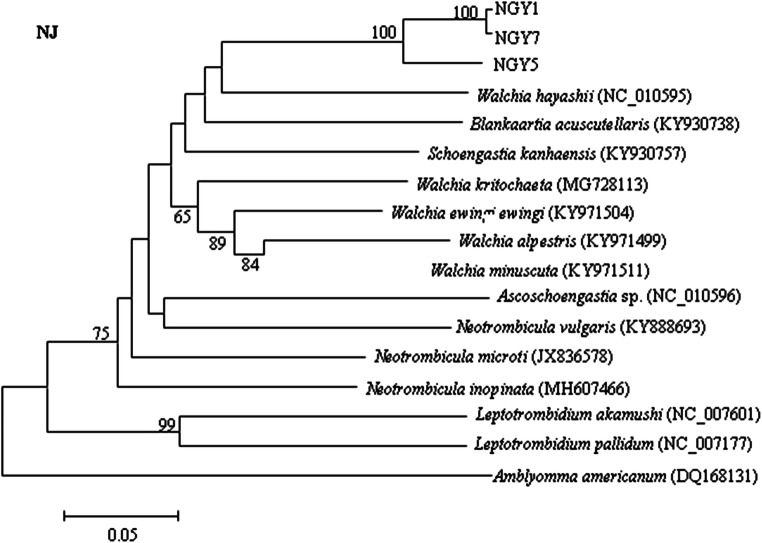
Based on pcox1 sequences, the phylogenetic relationships among 14 Trombiculidae species were reconstructed (Fig. 4). The topological structure of the phylogenetic tree showed the representative isolates NGY1 and NGY7 (genotype II) of N. gallinarum to be located in the same large branch and clustered together with the representative isolate NGY5 (genotype I); the probability value was 100. The closest relative to N. gallinarum was Walchia hayashii (NC_010595), followed by B. acuscutellaris (KY930738). Leptotrombidium akamushi (NC_007601), and Leptotrombidium pallidum (NC_007177) clustered together in a single branch and were found to be most distantly related to the other chigger mites in the family Trombiculidae.
Fig. 4.
Phylogenetic relationships among Trombiculidae species inferred by neighbor-joining (NJ) analysis based on mitochondrial pcox1 sequences and using Amblyomma americanum as an outgroup. Probability values below 50 are hidden (NGY1: MK423976, NGY5: MK4239777, NGY7: MK423978)
Discussion
In the present study, N. gallinarum samples were identified primarily based on larval morphological characteristics (Li et al. 1997). Although morphological identification is indispensable for the classification of parasites, it sometimes requires a high degree of experience with specific instruments and procedures. Therefore, molecular biological methods play an important role in the identification of mites.
Sequence alignment of the two populations of N. gallinarum from different geographical locations showed that the sequence variability of 18S rDNA and 28S rDNA was much lower than that of ITS2 and cox1; the p18S rDNA sequence was more highly conserved than the p28S rDNA sequence with no intraspecific difference. These sequence differences were small, and they are thus not suitable for low-level phylogenetic analysis. Few differences between the intraspecific sequences of p28S rDNA were detected, and though some interspecific differences were observed and they may be used as genetic markers for phylogenetic analysis at the genus level or higher. Various studies have demonstrated that rDNA ITS2 provides genetic markers for accurate identification of a number of parasite groups (Lin et al. 2007a, b; Zhao et al. 2012). ITS2 is relatively conserved within N. gallinarum, is highly variable among other chigger mite species, and can be used as a species marker. Analysis of N. gallinarum pcox1 sequences revealed two distinct genotypes, with a ratio of approximately 1:1. There were no obvious morphological differences found between mites of the two different genotypes by optical or electronic microscopy. Based on sequence alignment, some identical ITS2 sequences between the two N. gallinarum cox1 genotypes indicated that the N. gallinarum ITS2 sequence is more highly conserved than the cox1 sequence, and this sequence variation was not associated with the geographic location. The occurrence of different genotypes may be due to ecological and geographic factors, such as host conditions, climatic conditions, and geographical isolation. In the present study, sequence alignment showed that the two genotypes are present in the populations from different geographic locations, different chicken breeds, and different individual chickens, suggesting that genotype prevalence is not associated with host species or individual; mutual invasion between populations from different geographical locations may cause different genotypes to be detected at multiple locations. A phylogenetic tree of the species in the family Trombiculidae was constructed based on the pcox1 sequence. The results showed that N. gallinarum is more closely related to W. hayashii than other chigger mites, but the p value was low. It can be speculated that the chigger mite mt cox1 gene has a high evolutionary rate, which indirectly indicates that this chigger mite exhibits strong adaptability to the environment. To fully understand the relationships among different cox1 genotypes, further sample collection from different areas and further biological and molecular studies are needed.
This is the first report of the p18S, p28S, ITS2, and pcox1 sequences of N. gallinarum, which fills a gap in the knowledge of the molecular biology of this species. The results indicate two genotypes of N. gallinarum, as based on cox1 variation, and that the cox1 sequence is a suitable genetic marker for interspecies and intraspecies phylogenetic analyses and exploration of new genotypes. Compared with cox1, ITS2 has lower intraspecies variation and high interspecies variation, and it is thus suitable for species identification in the order Acarina. Currently, research on the molecular aspects of the family Trombiculidae is limited, so this subject needs to be further explored via comprehensive phylogenetic analyses of chigger mites.
Funding information
This work was supported by the Key Research and Development Program of Guangdong Province (2019B020218004) and the National Key Research and Development Program of China (Grant No. 2016YFD0501300).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
The experiments of this study were conducted following regulations of the Laboratory Animal Center of South China Agricultural University and approved by the Laboratory Animal Ethics Committee of South China Agricultural University.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ya-Biao Weng, Email: ybweng@scau.edu.cn.
Zhi-Jian Tan, Email: 16306888@qq.com.
References
- Cheng T, Liu GH, Song HQ, Lin RQ, Zhu XQ. The complete mitochondrial genome of the dwarf tapeworm Hymenolepis nana--a neglected zoonotic helminth. Parasitol Res. 2016;115(3):1253–1262. doi: 10.1007/s00436-015-4862-8. [DOI] [PubMed] [Google Scholar]
- Chilton NB, Gasser RB, Beveridge I. Differences in a ribosomal DNA sequence of morphologically indistinguishable species within the Hypodontus macropi complex (Nematoda, Strongyloidea) Int J Parasitol. 1995;25(5):647–651. doi: 10.1016/0020-7519(94)00171-J. [DOI] [PubMed] [Google Scholar]
- Gao DZ, Liu GH, Wang GL, Zhu XQ, Wang CR. Sequence variation in three mitochondrial genes among Gasterophilus intestinalis isolates from two distinct regions in China. Mitochondrial DNA A DNA. Mapp Seq Anal. 2017;28(1):37–40. doi: 10.3109/19401736.2015.1106529. [DOI] [PubMed] [Google Scholar]
- Hedin MC. Molecular insights into species phylogeny, biogeography and morphological stasis in the ancient spider genus Hypochilus (Araneae, Hypochilidae) Mol Phylogenet Evol. 2001;18(2):238–251. doi: 10.1006/mpev.2000.0882. [DOI] [PubMed] [Google Scholar]
- Hedin MC, Maddison WP. A combined molecular approach to phylogeny of the jumping spider subfamily Dendryphantinae (Araneae: Salticidae) Mol Phylogenet Evol. 2001;18(3):386–403. doi: 10.1006/mpev.2000.0883. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991;66(4):411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- Jeu MH. Studies on Tsutsugamushi part VI. Preliminary investigation of chiggers in Nanjing area. Acta Entomol Sin. 1956;4:433–448. [Google Scholar]
- Jeu MH. Studies on Tsutsugamushi part IX. A preliminary study on chiggers infesting chicken in China (Acariformes, Trombiculidae) Acta Entomol Sin. 1957;4:449–466. [Google Scholar]
- Jeu MH. Studies on Tsutsugamushi part XX. Morphology of the nymph and adult of chigger mite, Neoschongastia galinarum Hatori, 1920 (Acariformes, Trombiculidae) Acta Entomol Sin. 1959;9:167–177. [Google Scholar]
- Jeu MH. Experimental studies on the hibernation of chigger mites (Acariformes, Trombiculidea). (studies on Tsutsugamushi part XVIII) Acta Entomol Sin. 1961;1:485–492. [Google Scholar]
- Ji YJ, Zhang DX, He LJ. Evolutionary conservation and versatility of a new set of primers for amplifying the ribosomal internal transcribed spacer regions in insects and other invertebrates. Mol Ecol Resour. 2010;3(4):581–585. doi: 10.1046/j.1471-8286.2003.00519.x. [DOI] [Google Scholar]
- Jin YC, Li XY, Liu JH, Zhu XQ, Liu GH. Comparative analysis of mitochondrial DNA datasets indicates that Toxascaris leonina represents a species complex. Parasit Vectors. 2019;12(1):194. doi: 10.1186/s13071-019-3447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. Mafft multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JC, Wang DQ, Chen XB (1997) Trombiculid mites of China, studies on vector and pathogen of tsutsugamushi disease Guangdong Sci Technol Press: 215–224 (In Chinese)
- Lin RQ, Dong SJ, Nie K, Wang CR, Song HQ, Li AX, Huang WY, Zhu XQ. Sequence analysis of the first internal transcribed spacer of rDNA supports the existence of the intermediate Fasciola between F. hepatica and F. gigantica in mainland China. Parasitol Res. 2007;101(3):813–817. doi: 10.1007/s00436-007-0512-0. [DOI] [PubMed] [Google Scholar]
- Lin RQ, Zhu XQ, Wei DX, Deng Y, Liu W, Song HQ, Li AX, Lun ZR. Characterization of Oesophagostomum spp. from pigs in China by PCR-based approaches using genetic markers in the internal transcribed spacers of ribosomal DNA. Parasitol Res. 2007;101(2):351–356. doi: 10.1007/s00436-007-0498-7. [DOI] [PubMed] [Google Scholar]
- Medina M, Walsh PJ. Molecular systematics of the order anaspidea based on mitochondrial DNA sequence (12S, 16Sand COI) Mol Phylogenet Evol. 2000;15(1):41–58. doi: 10.1006/mpev.1999.0736. [DOI] [PubMed] [Google Scholar]
- Sugimoto M. On the Formosan chicken mite, Neoschngastia gallinarum (Hatori, 1920) J Soi Trop Agr Formosa. 1936;8:241–253. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mega5: molecular evolutionary genetics analysis using maximum-likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgin DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LM, Wang ZX, Weng YB, Lin RQ. The killing test in vitro of Chinese traditional medicine extracts and chemical drugs against Neoschngastia gallinarum. Poult Husb Dis Control. 2018;10:41–42. [Google Scholar]
- Zhao GH, Li J, Mo XH, Li XY, Lin RQ, Zou FC, Weng YB, Song HQ, Zhu XQ. The second transcribed spacer rDNA sequence: an effective genetic marker for inter-species phylogenetic analysis of trematodes in the order Strigeata. Parasitol Res. 2012;111(4):1467–1472. doi: 10.1007/s00436-012-2981-z. [DOI] [PubMed] [Google Scholar]