Abstract
The World Health Organization (WHO) has declared a pandemic caused by a new coronavirus named SARS-CoV-2. The growing demand for commercial kits used for automated extraction of SARS-CoV-2 RNA, a key step before rRT-PCR diagnosis, could cause a shortage of stocks that hinders the rapid processing of samples.
Although the recommendation is to use automated methods for nucleic acid extraction, alternatives are necessary to replace commercial kits. However, these alternatives should be as reliable as automated methods.
This work describes a simple method to detect SARS-CoV-2 from specimens collected in different preservation media. Samples were previously inactivated by heating and precipitating with a PEG/NaCl solution before rRT-PCR assays for Orf1ab, N and S genes. The new method was compared with an automated protocol of nucleic acid extraction. Both procedures showed similar analytical results. Consequently, this simple and inexpensive method is a suitable procedure for laboratory diagnosis of SARS-CoV-2 infection.
Keywords: SARS-CoV-2, rRT-PCR, RNA extraction, COVID-19, PEG/NaCl, Precipitation
1. Introduction
SARS-CoV-2 is a virus that causes the ongoing COVID-19 pandemic, an acute respiratory distress syndrome which represents the most serious problem for global health (Cascella et al., 2020). Most cases are mild, usually with self-limiting symptoms and recovery within two weeks. Severe patients progress rapidly with an acute respiratory distress syndrome and septic shock, eventually ending in multiple organ failure (Wu and McGoogan, 2020).
WHO and the Centers for Disease Control and Prevention (CDC) published reverse transcription-quantitative polymerase chain reactions protocols (rRT-PCR) for SARS-CoV-2 diagnosis. This included an RNA extraction step to purify the viral RNA from a nasopharyngeal swab (CDC, 2020). However, since March 2020, some manufacturers reported the shortage of supplies related to COVID-19 driven by the sudden increase in global demand for RNA extraction kits.
Different approaches avoiding viral nucleic extraction have been tested such as diluting samples and using a heat-processed method. However, heating oropharyngeal swabs is not as sensitive or accurate as RT-qPCR reactions performed on purified samples, detecting 97.4 % of the COVID-19-positive patients with no false positives results (Fomsgaard and Rosenstierne, 2020). SARS-CoV-2 detection by direct rRT-PCR without RNA extraction and inactivating samples at 95 °C for 5 min, was showed from specimens placed in UTM and molecular water, but not from samples in Hanks medium and saline buffer (Merindol et al., 2020). Moreover, SARS-CoV-2 detection without RNA extraction was described by mixing respiratory samples in a 1:1 (v/v) ratio with Sputasol (Oxoid, Basingstoke, England) before adding it directly to the rRT-PCR reaction mix (Wee et al., 2020).
In this research, a simple, inexpensive and reliable alternative method that uses heat inactivation and nucleic acid precipitation was used to process samples placed in different media, without the requirement of using commercial kits. This new procedure showed similar results compared to an automated method. Thus, this new technique is proposed for SARS-CoV-2 laboratory diagnosis.
2. Material and methods
2.1. Nucleic acid purification
One-hundred and four nasopharyngeal swab samples (NPSs), previously extracted using an automated NucliSENS® easyMAG® (bioMérieux) and tested for COVID-19 at the Institute of Public Health of Chile. Ninety-four positive samples, with a Ct range from 12.79 to 39.12, and 10 negative samples were used for this study.
Samples were collected in tubes containing UTM-RT mini transport media (Copan Diagnostics Inc.) (n = 35), PBS 1x solution (n = 46), Hanks medium (n = 13) or DNA/RNA Shield™(Zymo Research) (n = 10). In a Class II Type A2 laminar flow biosafety cabinet (NuAire), 100 μL of a solution 20 % (w/v) Chelex-100 (Sigma Aldrich, USA) prepared in nuclease-free water was added to a 1.5 mL microtube using a P1000 micropipette (range 100−1000 μL). When Chelex-100 was precipitated, the supernatant was removed. Then, 200 μL of a solution 20 % (w/v) PEG 8000 (Sigma Aldrich, USA), 2.5 M NaCl (Ambion, USA) and 200 μL of NPSs were added. Tubes were incubated at 70 °C for 30 min (Thermomixer comfort, Eppendorf) and centrifuged at 13,500 g for 10 min. Supernatant was discarded and 600 μL of 70 % ethanol was added once for pellets washing. Samples were centrifuged at 13,500 g for 5 min and ethanol supernatant was discarded. Pellets were allowed to dry for 2 min at RT and resuspended in 50 μL of nuclease-free water, pipetting up and down using a P1000 to avoid blocking the pipette tip with Chelex-100. Supernatants (5 μL) were used directly for rRT-PCR CoV-2 detection.
2.2. SARS-CoV-2 detection by rRT-PCR
For all samples, TaqMan™ 2019-nCoV Assay Kit v1 (ThermoFisher) was used for viral detection, using duplicates for each, following the manufacturer’s instructions in a QuantStudio 3 Real Time PCR System (ThermoFisher).
3. Results
One-hundred and four NPSs (94 positives and 10 negatives) were extracted with an automated easyMAG® and PEG/NaCl precipitation methods. Then, three viral genes (Orf1ab, N and S) and one internal control gene (RNase P or RNP) were amplified by rRT-PCR.
Viral inactivation at 70 °C for 30 min did not affect rRT-PCR amplification (n = 104). All samples showed a concordant result (94 positive and 10 negative samples) between the easyMAG® extraction and the PEG/NaCl precipitation method. Results of rRT-PCR after easyMAG® extraction were considered as being true. Consequently, the sensibility and specificity for PEG/NaCl method were 100 % and 100 %, respectively. Furthermore, the accuracy was 100 % with each viral gene.
Moreover, the internal RNP gene was successfully amplified in all samples, corroborating extraction effectiveness (data not shown). Thus, the four different media used in this study (UTM, PBS 1x solution, Hanks medium and DNA/RNA Shield™) did not affect analytical results, because all precipitated samples were able to be detected by rRT-PCR using the whole viral panel (Orf1ab, N and S genes) and the internal control gene.
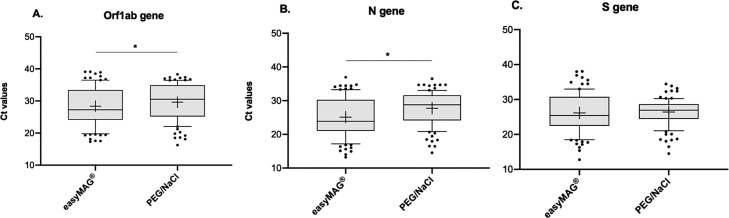
Performance of both easyMAG® and PEG/NaCl extraction methods was evaluated in positive samples (n = 94) by rRT-PCR. The mean of Ct values was compared for each viral gene. Orf1ab and N values were lower when easyMAG® was used and differences between both methods were significant (Wilcoxon test, P < 0.0001). On the other hand, the mean of Ct value for S was lower with easyMAG® extraction and differences were not significant (Wilcoxon test, P = 0.0810) (Fig. 1 and Table 1 ).
Fig. 1.
Comparison of average Ct values between easyMAG® extraction and PEG/NaCl precipitation for A. Orf1ab gene, B. N gene and C. S gene (*p < 0.0001, Wilcoxon test). The line inside the box plot represents the median and the box extends from lower to upper quartiles. Whiskers indicate the 90th and 10th percentiles and dots represent outliers. Mean is shown as “+”.
Table 1.
Descriptive statistics for easyMAG® extraction and PEG/NaCl precipitation. Values corresponding to average Ct for Orf1ab, N and S gene amplified by rRT-PCR.
| easyMAG® |
PEG/NaCl |
|||||
|---|---|---|---|---|---|---|
| Orf1ab | N | S | Orf1ab | N | S | |
| Minimum | 17.31 | 13.23 | 12.79 | 16.26 | 14.55 | 14.51 |
| Maximum | 39.12 | 36.96 | 38.06 | 38.33 | 36.50 | 34.44 |
| Mean | 28.37 | 25.12 | 26.14 | 29.63 | 27.74 | 26.40 |
| Std. Deviation | 5.90 | 5.80 | 5.64 | 5.66 | 4.90 | 3.78 |
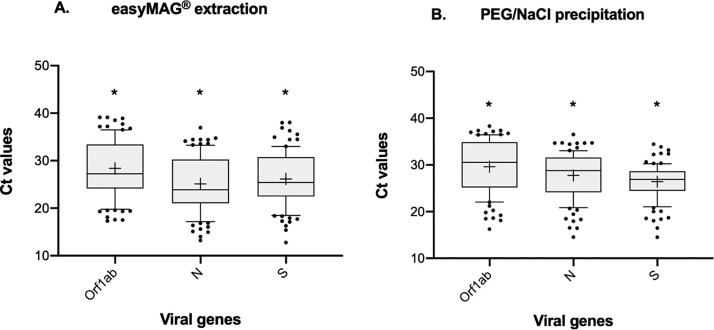
When Ct values from each gene were compared within each extraction method, significant differences were observed (Friedman test, P < 0.0001). The best performance was achieved with the N gene (Ct mean 27.74 ± 4.90) for automated extraction, and with the S gene (Ct mean 26.40 ± 3.78) for the PEG/NaCl precipitation (Fig. 2 ).
Fig. 2.
Comparison of average Ct values for Orf1ab, N and S genes within easyMAG® extraction and PEG/NaCl precipitation (*=p < 0.0001, Dunn’s multiple comparisons test). The line inside the box plot represents the median, and the box extends from lower to upper quartiles. Whiskers indicate the 90th and 10th percentiles and dots represent outliers. Mean is shown as “+”.
4. Discussion
In a pandemic context, it is relevant to obtain rapid and reliable results to diagnose patients and take rapid public health actions. Here, a simple protocol to detect SARS-CoV-2 from NPSs using rRT-PCR after a heat inactivation and a precipitation/concentration step is proposed.
In order to inactivate SARS-CoV-2 present in contaminated objects, a treatment for 3 min at above 75 °C or 5 min at above 65 °C or 20 min at above 60 °C is recommended (Abraham et al., 2020). Moreover, a CoV-2 isolate (USA-WA1/2020), inactivated for 30 min at 65 °C, is being commercialized for different purposes (Bei Resources). In this research, a 30 min incubation step at 70 °C was used to obtain safe samples to be manipulated in the laboratory, without previous RNA extraction using commercial kits. Also, Chelex-100 was incorporated as a protective agent during extraction at high temperatures because this reagent favors chelating groups to bind to cellular components (Lounsbury et al., 2012). Moreover, this resin stabilizes the RNA during the thermal shock and protects it from degradation (Fontaine and Guillot, 2003).
In this study a precipitation step using a PEG/NaCl solution was used, after heat inactivation of NPSs to concentrate samples and remove unwanted inhibitors, before the rRT-PCR. This solution was previously used for selective precipitation of large RNAs (Lounsbury et al., 2012; Nilsen, 2012). Also, it is broadly used for phage precipitation and purification (Yamamoto et al., 1970). Samples collected in different media (UTM, PBS 1x, Hanks and DNA/RNA Shield) were tested using this protocol. All analytical results obtained by this protocol were concordant with those obtained by an easyMAG® procedure. This is an advantage over a direct rRT-PCR without RNA extraction described, where it was possible to detect SARS-CoV-2 only from samples collected in UTM or molecular water, but not from samples collected in phosphate-buffered saline or Hanks medium (Merindol et al., 2020). A protocol that used samples heat-processed for 5 min at 98 °C was reported (Fomsgaard and Rosenstierne, 2020). This protocol allowed detecting only 97.4 % of COVID-19-positive patients in comparison to PEG/NaCl precipitation that detected 100 % of the positive cases.
Performance of viral gene amplification (Orf1ab, N, S) was evaluated and differences in Ct values were slightly better using easyMAG® extraction. However, the three genes were identified in all samples and, remarkably, S gene did not show significant differences in amplification between both extraction methods. It is important to highlight that NPSs input volume used for easyMAG® extraction was 600 μL and for PEG/NaCl precipitation 200 μL were used. Although this protocol used 3 times less volume, it was able to detect samples whose Ct values were at the limit of detection of the rRT-PCR technique.
A direct amplification protocol and its performance against a MagnaPure automated system by rRT-PCR was reported (Fomsgaard and Rosenstierne, 2020). Nevertheless, sensitivity (85 %), specificity (95 %) and accuracy (88 %) were lower than the proposed PEG/NaCl precipitation method. Here, it was obtained 100 % of sensitivity, specificity and accuracy. Another study showed better sensitivity for the N gene than Orf1ab and S, when samples were extracted using a MagnaPure (Iglói et al., 2020). In this study, the best performance was achieved with the N gene for easyMAG® extraction and with the S gene for the PEG/NaCl precipitation.
Ethical statement
Given the important impact that data from this study could have on Public Health, all nasopharyngeal samples from COVID-19 positive patients were unidentified and not considered as Human samples.
CRediT authorship contribution statement
S. Ulloa: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Visualization. C. Bravo: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Visualization. B. Parra: Investigation. E. Ramirez: Methodology. A. Acevedo: Investigation. R. Fasce: Resources. J. Fernandez: Conceptualization, Resources, Supervision.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest associated with this work.
Acknowledgments
We thank all investigators from Molecular Genetics Sub Department and Viral Diseases Sub Department, both from Institute of Public Health of Chile.
References
- Abraham J.P., Plourde B.D., Cheng L. Using heat to kill SARS‐CoV ‐2. Rev. Med. Virol. 2020 doi: 10.1002/rmv.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls. StatPearls Publishing; Treasure Island (FL): 2020. Features, evaluation and treatment coronavirus (COVID-19) [PubMed] [Google Scholar]
- CDC . Centers for Disease Control and Prevention; 2020. Coronavirus Disease 2019 (COVID-19) [WWW Document]https://www.cdc.gov/coronavirus/2019-ncov/lab/index.html (accessed 4.8.20)URL. [Google Scholar]
- Fomsgaard A.S., Rosenstierne M.W. An alternative workflow for molecular detection of SARS-CoV-2 - escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.14.2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine M., Guillot E. Study of 18S rRNA and rDNA stability by real-time RT-PCR in heat-inactivated Cryptosporidium parvum oocysts. FEMS Microbiol. Lett. 2003;226:237–243. doi: 10.1016/S0378-1097(03)00538-X. [DOI] [PubMed] [Google Scholar]
- Iglói Z., Leven M., Abdel-Karem Abou-Nouar Z., Weller B., Matheeussen V., Coppens J., Koopmans M., Molenkamp R. Comparison of commercial realtime reverse transcription PCR assays for the detection of SARS-CoV-2. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury J.A., Coult N., Miranian D.C., Cronk S.M., Haverstick D.M., Kinnon P., Saul D.J., Landers J.P. An enzyme-based DNA preparation method for application to forensic biological samples and degraded stains. Forensic Sci. Int. Genet. 2012;6:607–615. doi: 10.1016/j.fsigen.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Merindol N., Pépin G., Marchand C., Rheault M., Peterson C., Poirier A., Houle C., Germain H., Danylo A. SARS-CoV-2 detection by direct rRT-PCR without RNA extraction. J. Clin. Virol. 2020;128 doi: 10.1016/j.jcv.2020.104423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T.W. Selective precipitation of large RNAs. Cold Spring Harb. Protoc. 2012;2012 doi: 10.1101/pdb.prot072322. [DOI] [PubMed] [Google Scholar]
- Wee S.K., Sivalingam S.P., Yap E.P.H. Rapid direct nucleic acid amplification test without RNA extraction for SARS-CoV-2 using a portable PCR thermocycler. Genes. 2020;11 doi: 10.3390/genes11060664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Yamamoto K.R., Alberts B.M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970;40:734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]