Abstract
Background
SARS-CoV-2, an emerging strain of coronavirus, has affected millions of people from all the continents of world and received worldwide attention. This emerging health crisis calls for the urgent development of specific therapeutics against COVID-19 to potentially reduce the burden of this emerging pandemic.
Purpose
This study aims to evaluate the anti-viral efficacy of natural bioactive entities against COVID-19 via molecular docking and molecular dynamics simulation.
Methods
A library of 27 caffeic-acid derivatives was screened against 5 proteins of SARS-CoV-2 by using Molegro Virtual Docker 7 to obtain the binding energies and interactions between compounds and SARS-CoV-2 proteins. ADME properties and toxicity profiles were investigated via www.swissadme.ch web tools and Toxtree respectively. Molecular dynamics simulation was performed to determine the stability of the lead-protein interactions.
Results
Our obtained results has uncovered khainaoside C, 6-O-Caffeoylarbutin, khainaoside B, khainaoside C and vitexfolin A as potent modulators of COVID-19 possessing more binding energies than nelfinavir against COVID-19 Mpro, Nsp15, SARS-CoV-2 spike S2 subunit, spike open state and closed state structure respectively. While Calceolarioside B was identified as pan inhibitor, showing strong molecular interactions with all proteins except SARS-CoV-2 spike glycoprotein closed state. The results are supported by 20 ns molecular dynamics simulations of the best complexes.
Conclusion
This study will hopefully pave a way for development of phytonutrients-based antiviral therapeutic for treatment or prevention of COVID-19 and further studies are recommended to evaluate the antiviral effects of these phytochemicals against SARS-CoV-2 in in vitro and in vivo models.
Keywords: COVID-19, SARS-CoV-2, Bioactive phytochemicals, Caffeic acid derivative
Graphical abstract
Introduction
A severe respiratory coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) has emerged as a pandemic at the end of 2019 (Du et al., 2020; Zhu et al., 2020). By July 2020, SARS-CoV-2 has affected more than 200 territories, infecting more than 13 million individuals and causing more than 0.5 million deaths (WHO, Situation Report–177). COVID-19 has marked the history with third life-threatening coronavirus epidemic into the human population during 21st century (Guo et al., 2020). By 7th January, 2020, Chinese scientists released the sequenced SARS-CoV-2 genome for the identification and development of potential candidates against COVID-19 by computational methods and other therapeutic techniques (Lu et al., 2020).
The SARS-CoV-2 is a β-coronavirus, enveloped and positive sense-RNA virus, with ~30 kb genome (Wu et al., 2020). SARS-CoV-2 genome possesses a complex organization encoding various structural as well as non-structural proteins (Nsps) (Kim et al., 2020a). Majority part of viral genome (replicase ORF1ab encompassing Nsps) is translated into two overlapping polyproteins known as pp1a and pp1ab (Amoretti et al., 2002). These polypeptides also codes for ~306 amino acid long main protease (Mpro) (Liu and Wang, 2020), which digests polypeptides at various conserved sites yielding 16 functional viral Nsps possessing multiple enzymatic activities especially in viral replication (Amoretti et al., 2002; Kim et al., 2020a). One of such enigmatic protein, Nsp15, is an endoribonuclease known to be indispensable for protein interference during innate immune response (Kim et al., 2020a). Due to functional importance of Mpro and Nsp15 in viral replication and survival, both could be potential therapeutic drug targets to combat COVID-19.
In addition to Nsps, SARS-CoV-2 genome also consists of structural protein encoding genes including S (spike) gene, E gene (viral envelop protein), and N (nucleo-capsid protein) gene (Khan et al., 2020). Viral spike proteins possess strong affinity with the human ACE2 (angiotensin-converting enzyme 2) receptor by which virus fuses with target membrane to gain entry into human cells (Hussain et al., 2020). SARS-CoV-2 fusion potential and ACE2 affinity is much greater as compared to SARS-CoV, suggesting that the SARS-CoV-2 fusion machinery is a novel target for coronavirus fusion inhibitors. S protein binds ACE2 receptor via its S1 subunit, while its S2 subunit interacts to form fusion core, which brings viral and target cell membranes into proximity for efficient fusion and subsequent infection (Xia et al., 2020). Thus, SARS-CoV-2 S2 subunit could be a potential target for coronavirus fusion inhibitors. Moreover, S1 trimeric crowns removal or opening is expected to be essential for exposure of receptor binding domain (RBD) to ACE2 receptor and S2 conformational changes which enable binding and membrane fusion (Walls et al., 2020b) suggesting that SARS-CoV-2 spike ectodomain structure and SARS-CoV-2 spike closed state glycoprotein structure might be novel therapeutic targets to develop anti-COVID-19 drugs.
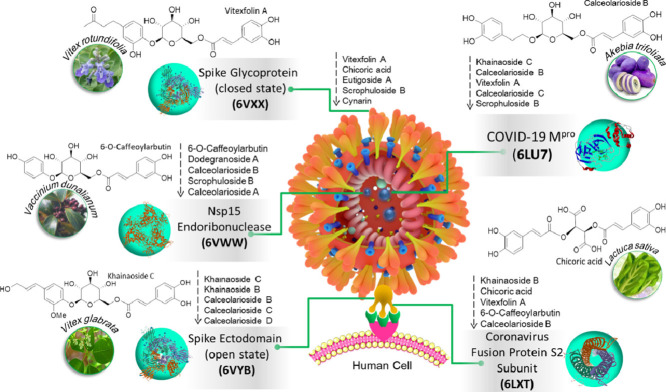
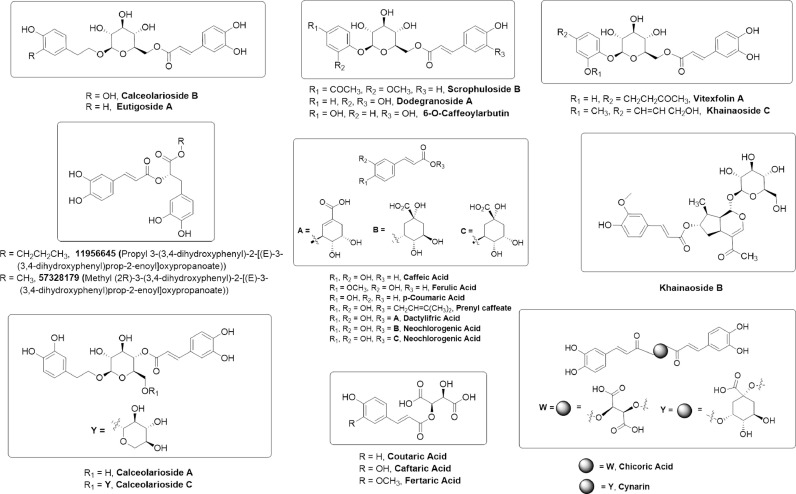
Growing evidences have established the worth of polyphenols as lead compounds for drug discovery against various human diseases (Dos Santos et al., 2018). Recent studies reported that polyphenol have potential to combat with COVID-19 (Adem et al., 2020). Caffeic acids are one of the abundant plant-based polyphenols possessing 2 phenolic hydroxyl moieties and commonly found in coffee, fruits and vegetables (Magnani et al., 2014). Caffeic acids have been reported for their potent virucidal activity against herpes simplex virus (Langland et al., 2018), SFTS (severe fever with thrombocytopenia syndrome) virus (Ogawa et al., 2018), and influenza virus (Utsunomiya et al., 2014). Based upon these results, we have screened a library of caffeic acid derivatives (CAFDs) (Fig. 1 ) for the identification of novel natural anti-COVID-19 compounds against various SARS-CoV-2 drug targets including COVID-19 Mpro (6LU7), SARS-CoV-2 S2 subunit (6LXT), Nsp15 endoribonuclease (6VWW), SARS-CoV-2 spike ectodomain open state structure (6VYB), and SARS-CoV-2 spike closed state glycoprotein structure (6VXX). Our results present in silico-based identification of khainaoside C, 6-O-Caffeoylarbutin, khainaoside B, khainaoside C and vitexfolin A as potent modulators of COVID-19 Mpro, Nsp15, coronavirus fusion protein, spike open state and closed state structure respectively. Our findings will provide valuable data for exploration and development of caffeic acid-derivatives as lead structures, novel therapeutic and prophylaxis agents against COVID-19 in the near future.
Fig. 1.
Chemical structures of caffeic acid and its derivatives.
Methods
To obtain binding interactions between CAFDs and binding pockets of 5 different proteins of SARS-CoV-2, five independent docking analyses were performed by using Molegro Virtual Docker (MVD) software in a computer cluster system provided by EXPER, model-FQC-01266 running Intel Core i3–2100 CPU @3.10GHz Processor, 64 BİT, 4 GB RAM, 1TB hard disk, and NVIDIA GeForce GT 630 Graphic card. The crystal structures of the following SARS-CoV-2 proteins were retrieved from the protein data bank web site (http://www.rcsb.org/pdb): SARS-CoV-2 Mpro (PDB ID: 6LU7: Resolution 2.16 Å) (Jin et al., 2020), Nsp15 endoribonuclease (PDB ID: 6VWW) (Kim et al., 2020b), coronavirus fusion protein (PDB ID: 6LXT) (Xia et al., 2020), SARS-CoV-2 spike ectodomain (PDB ID: 6VYB) and spike glycoprotein (open state) (PDB ID: 6VXX) (Walls et al., 2020a). The selected cavity of 6LU7 is the binding site of natural inhibitor N3. Nelfinavir, utilized for the cure of the HIV (human immunodeficiency virus), was utilized as a positive control. For other targets, the docking cavities of target proteins were selected according to the amino acids involved in the activity of protein. The docking x y z positions of the proteins were identified as-10.87 15 68.21 for 6LU7 (radius 15 Å) −68.51 29.06 29 for 6VWW (radius 14 Å), -20 19 -25 for 6LXT (radius 22 Å), 217 195 265 for 6VXX (radius 24 Å) and 231 185 168 for 6VYB (radius 21 Å). MolDock Score was selected at the scoring function and the search algorithm. After docking, energy minimization and H-bond optimizations were performed. The docking simulation was repeated for each ligand 20 times. The top binding scores were utilized for further analysis. Also, Discovery Studio Visualizer 2020 was used for in-depth analysis of docking results.
The chemical structures of selected compounds were received at 3D SDF conformer from the PubChem site. PubChem CIDs of examined compounds are given respectively; Khainaoside C (44606078), Calceolarioside B (5273567), Vitexfolin A(10458788), Calceolarioside C (45360240), Scrophuloside B (11712581), Cynarin (CYN) (5281769), Eutigoside A (10026568), Calceolarioside D (14015431), Robustaside D (38358972), Chicoric acid (CHA) (5281764), Robustaside E (50994836), Dodegranoside A (44513070), 6-O-Caffeoylarbutin (15689808), Khainaoside B (44606238), Calceolarioside A (5273566), Propyl 3-(3.4-dihydroxyphenyl)-2-[(E)-3-(3.4-dihydroxyphenyl)prop-2-enoyl]oxypropanoate (11956645), Dactylifric acid (6124136), Neochlorogenic acid (NCHL) (5280633), Fertaric acid (FTA) (22298372), Prenyl caffeate acid (5281790), Caftaric acid (CFT) (6440397), Chlorogenic acid (CHL) (1794427), Coutaric acid (CTA) (57517924), Caffeic acid (689043), p-Coumaric acid (637542), Ferulic acid (445858). 3D SDF structures were prepared with ChemBio3D or MarvinSketch for simple molecules.
ADME and toxicity prediction
In silico ADME analysis was conducted to investigate physicochemical properties of potent hits, such as water solubility, lipophilicity and pharmacokinetics by using following website http://www.swissadme.ch (Daina et al., 2017). Absorption (% ABS) of potent hits from intestine was evaluated by: % ABS = 109 × (0.345xTPSA). Toxicity analysis was performed using offline software Toxtree 3.1 application (Zhao et al., 2002).
Molecular dynamics study
The structures of the best-docked complex for each protein are selected for in-depth molecular dynamics simulation (MDS) study for a period of 20 ns. NAMD software was utilized to conduct the MDS with CHARMM 36 force field (Huang and MacKerell, 2013; Phillips et al., 2005). VMD is used to prepare the complexes for the MDS (Humphrey et al., 1996). Complexes are subjected to equilibration using the CHARMM GUI web server after that a production run for 20 ns is performed on Shaheen supercomputer of King Abdullah University of Science and Technology (KAUST) under the project number k1482 (Jo et al., 2008). The equilibration is done on the protein-small molecule solvated in the TIP3P water model and 0.154 M NaCl solution at 310 K temperature and pH 7 (Mark and Nilsson, 2001). VMD is utilized in trajectories analysis, while the Chimera software of UCSF is used for cluster analysis (Mark and Nilsson, 2001; Pettersen et al., 2004). After trajectory clustering, the five most populous clusters are represented by a conformation and tested for its binding to the protein. AutoDock Vina software is used in the binding energy calculations using 40 Å × 40 Å × 40 Å box dimensions (Morris et al., 2009; Trott and Olson, 2010).
Results
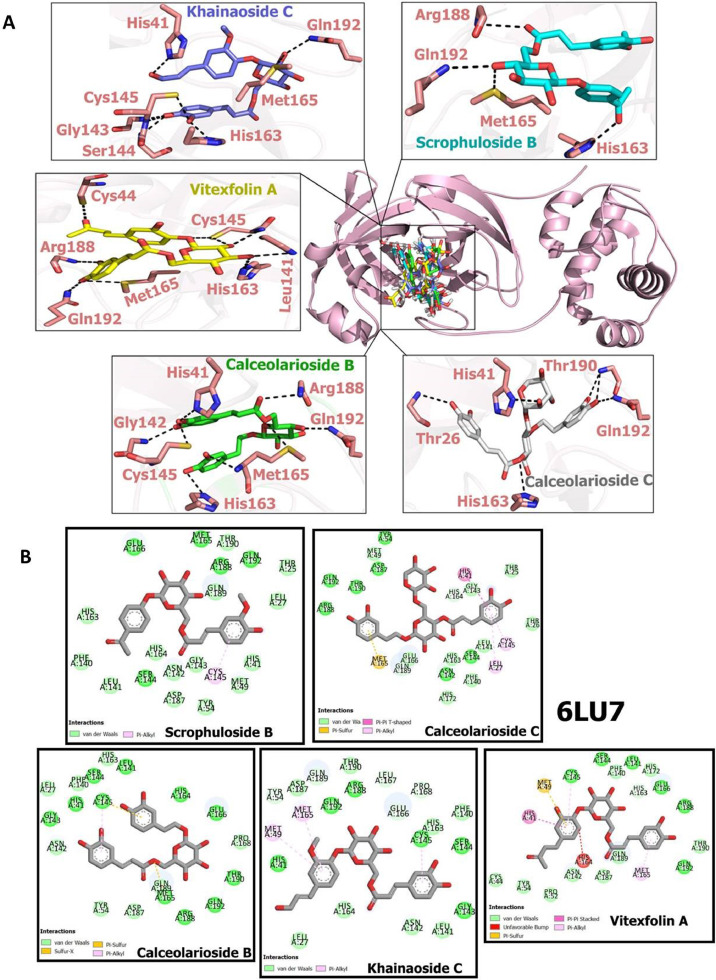
The MolDock Scores obtained from the docking studies of CAFDs and 6LU7 are summarized in Table 1 . Based on these in-silico results, khainaoside C, calceolarioside B, vitexfolin A, calceolarioside C and scrophuloside B exhibited best binding potential with COVID-19 virus Mpro having MolDock scores of -191.5990, -191.2950, -186.2820, -178.5540 and -177.799 respectively. Khainaoside C showed the highest binding affinity at the active site. It forms hydrogen bond interactions with His 41, His 163, Met 165, Arg 188, Gln 192, Glu 166, Cys 145, Gly 143, Ser 144, Leu 141 (Fig. 2 A). Calceolarioside B binds to SARS-CoV-2 Mpro by forming hydrogen bonds with His 41, His 163, His 164, Met 165, Asp 187, Gln 189, Thr 190, Arg 188, Gln 192, Glu 166, Cys 145, Gly 143, Ser 144, Leu 141 (Fig. 2A). Vitexfolin A and calceolarioside C also showed good binding affinities to the active sites of COVID-19 virus Mpro via interacting with Met 49, Tyr 54, Cys 44, His 163, His 164, Arg 188, Gln 192, Glu 166, Cys 145, Ser 144, Leu 141, Phe 140 (Fig. 2A) and His 41, Tyr 54, Thr 26, His 163, His 164, Met 165, Asp 187, Gln 189, Thr 190, Arg 188, Gln 192, Glu 166, Asn 142, Cys 145, Ser 144, Leu 141 respectively (Fig. 2A). According to the obtained results, khainaoside C and calceolarioside B affinities with COVID-19 virus Mpro and their scores are significantly higher than Nelfinavir. Ligand plot in Fig. 2B represents residual wise van der Waals interactions, pi-alkyl interactions, pi-pi interactions and pi-sulfur interactions of potent hits with Mpro.
Table 1.
Results of the docking of CAFDs on the crystal structure of COVID-19 virus Mpro (6LU7)
| Ligand | MolDock Score | Protein-Ligand Interactions | Internal Ligand Interactions | H-Bond |
|---|---|---|---|---|
| Nelfinavir | -147.3800 | -171.698 | 22.284 | -6.8731 |
| Khaınaosıde C | -191.599 | -200.025 | 8.42644 | -27.1197 |
| Calceolarioside B | -191.295 | -192.449 | 1.15387 | -29.335 |
| Vitexfolin A | -186.282 | -195.849 | 9.56729 | -27.845 |
| Calceolarioside C | -178.554 | -230.49 | 51.9361 | -26.4726 |
| Scrophuloside B | -177.799 | -174.45 | -3.34965 | -13.1931 |
| Cynarin | -177.1841 | -207.737 | 30.5529 | -20.6951 |
| Eutıgosıde A | -167.291 | -178.443 | 11.1524 | -19.147 |
| Calceolarioside D | -165.78 | -173.156 | 7.37642 | -19.0178 |
| Robustaside D | -164.794 | -181.854 | 17.0598 | -31.946 |
| Chicoric acid | -162.6854 | -178.756 | 16.0706 | -18.3829 |
| Robustaside E | -161.535 | -177.951 | 16.4157 | -24.393 |
| Dodegranoside A | -157.933 | -192.633 | 34.6997 | -32.2933 |
| 6-O-Caffeoylarbutin | -156.602 | -166.442 | 9.84068 | -17.4251 |
| Khaınaosıde B | -155.284 | -185.867 | 30.5834 | -21.4445 |
| Calceolarioside A | -149.67 | -174.029 | 24.3592 | -24.5915 |
| Propyl 3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2- | -148.144 | -166.589 | 18.4446 | -11.8087 |
| Methyl (2R)-3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxypropanoate | -145.999 | -165.943 | 19.944 | -10.591 |
| Dactylifric acid, | -134.2368 | -156.667 | 22.4302 | -19.4056 |
| Neochlorogenic acid | -130.0202 | -159.793 | 29.7728 | -21.9994 |
| Fertaric acid | -127.1668 | -144.398 | 17.2312 | -12.0719 |
| Prenyl caffeate acid | -122.69655 | -130.602 | 7.90545 | -5.00251 |
| Caftaric acid | -121.9308 | -145.024 | 23.0932 | -12.1007 |
| Chlorogenic acid | -119.6166 | -147.503 | 27.8864 | -14.8102 |
| Coutaric acid | -119.1713 | -136.215 | 17.0437 | -10.6761 |
| Caffeic acid | -91.5046 | -100.307 | 8.80237 | -12.4985 |
| p-Coumaric acid | -90.0385 | -98.9344 | 8.89594 | -6.95915 |
| Ferulic | -78.5495 | -83.6803 | 5.13082 | -4.9909 |
Fig. 2.
Docking poses of caffeic acid derivatives with COVID-19 virus Mpro(A) Hydrogen bonding interactions of khaınaosıde C, scrophuloside B, vitexfolin A, calceolarioside B and calceolarioside C with amino acid residues of COVID-19 virus Mpro, (B) 2D view of interaction types of khaınaosıde C, scrophuloside B, vitexfolin A, calceolarioside B and calceolarioside C with surrounding amino acids of COVID-19 virus Mpro.
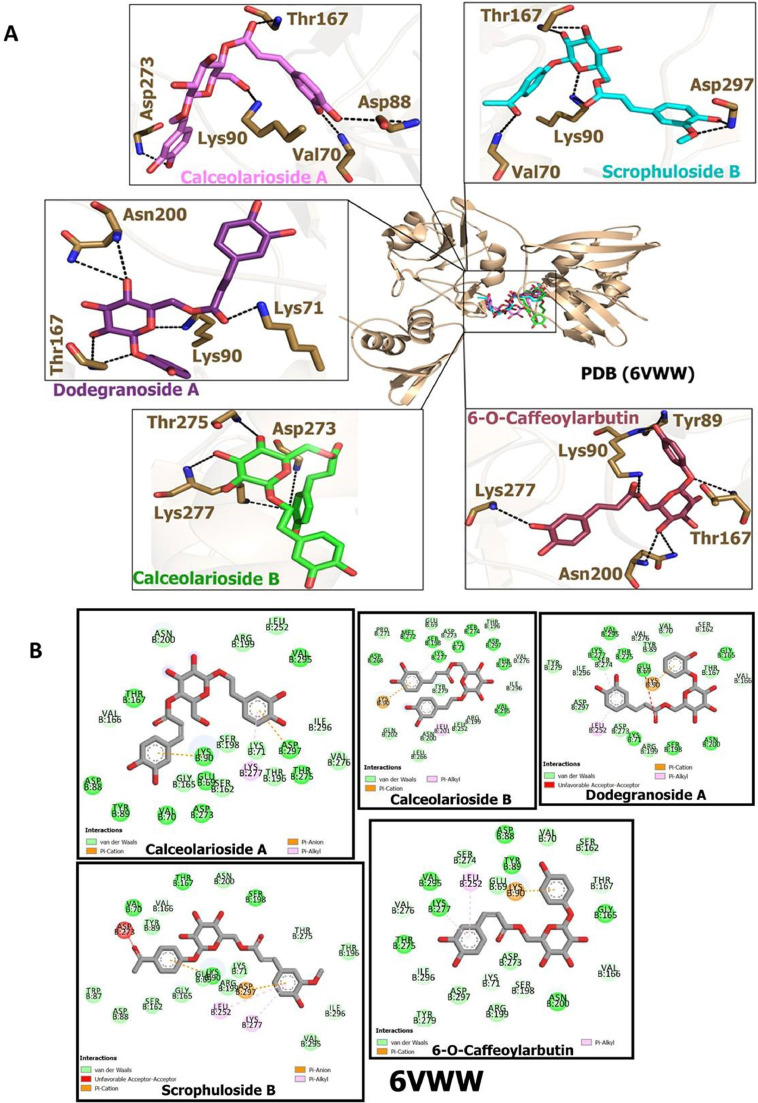
The MolDock Scores obtained from the docking studies of CAFDs and 6VWW are provided in Table 2 . 6-O-Caffeoylarbutin, dodegranoside A, calceolarioside B, scrophuloside B and calceolarioside A possess significantly good binding potential to Nsp15 endoribonuclease with MolDock Scores -171.541, -168.82, -164.77, -163.023 and -157.557 respectively as compared to Nelfinavir which possess MolDock score of -148.413 The interactions of these compounds with amino acid residues of target protein are shown in Fig. 3 A. Ligand plot representing residual wise van der Waals interaction, pi-alkyl interactions, pi-cation and pi-anion interactions are presented in Fig. 3B.
Table 2.
Results of the docking of CAFDs on the crystal structure of Nsp15 endoribonuclease (6VWW).
| Ligand | MolDock Score | Protein-Ligand Interactions | Internal Ligand Interactions | H-Bond |
|---|---|---|---|---|
| Nelfinavir | -148.413 | -176.918 | 28.5042 | -6.24452 |
| 6-O-Caffeoylarbutin | -171.541 | -192.585 | 21.0444 | -21.747 |
| Dodegranoside A | -168.82 | -189.321 | 20.5011 | -20.0539 |
| Calceolarioside B | -164.77 | -178.002 | 13.2327 | -16.1947 |
| Scrophuloside B | -163.023 | -188.728 | 25.7045 | -17.7 |
| Calceolarioside A | -157.557 | -191.866 | 34.3089 | -22.6471 |
| Calceolarioside D | -157.531 | -176.714 | 19.1827 | -13.5793 |
| Robustaside D | -152.212 | -166.649 | 14.4366 | -13.1528 |
| Vitexfolin A | -151.664 | -174.554 | 22.8901 | -14.0706 |
| Eutıgosıde A | -150.42 | -173.682 | 23.2626 | -14.3851 |
| Robustaside E | -150.175 | -181.359 | 31.1841 | -20.7424 |
| Chicoric acid | -149.662 | -165.153 | 15.4907 | -9.06476 |
| Propyl 3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2- | -148.42 | -160.41 | 11.9901 | -11.2875 |
| Khaınaosıde B | -146.216 | -159.431 | 13.2149 | -18.838 |
| Khaınaosıde C | -144.449 | -169.295 | 24.8469 | -14.338 |
| Calceolarioside C | -143.258 | -190.29 | 47.0315 | -26.9485 |
| Methyl (2R)-3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxypropanoate | -136.959 | -151.082 | 14.1226 | -7.66985 |
| Cynarin | -136.239 | -168.638 | 32.3994 | -9.84515 |
| Dactylifric acid | -124.395 | -135.853 | 11.4586 | -11.8072 |
| Chlorogenic acid | -117.22 | -146.224 | 29.004 | -19.2892 |
| Fertaric acid | -115.811 | -131.072 | 15.2609 | -10.3567 |
| Neochlorogenic acid (NCHL) | -114.913 | -140.215 | 25.3017 | -13.8222 |
| Prenyl caffeate acid | -113.339 | -118.376 | 5.03646 | -9.94435 |
| Coutaric acid | -112.946 | -123.273 | 10.3278 | -9.20319 |
| Caftaric acid | -110.286 | -130.278 | 19.9917 | -13.6044 |
| p-Coumaric acid | -90.4353 | -98.5634 | 8.12807 | -7.41811 |
| Caffeic acid | -86.1121 | -92.4587 | 6.34661 | -7.61903 |
| Ferulic acid | -78.5357 | -83.0275 | 4.49178 | -4.65339 |
Fig. 3.
Docking poses of caffeic acid derivatives with COVID-19 virus Nsp15 endoribonuclease (A) Hydrogen bonding interactions of 6-O-Caffeoylarbutin, calceolarioside B, dodegranoside A, calceolarioside A and scrophuloside B with amino acid residues of virus Nsp15 endoribonuclease (B) 2D view of interaction types of 6-O-Caffeoylarbutin, calceolarioside B, dodegranoside A, calceolarioside A and scrophuloside B with surrounding amino acids of COVID-19 virus Nsp15 endoribonuclease.
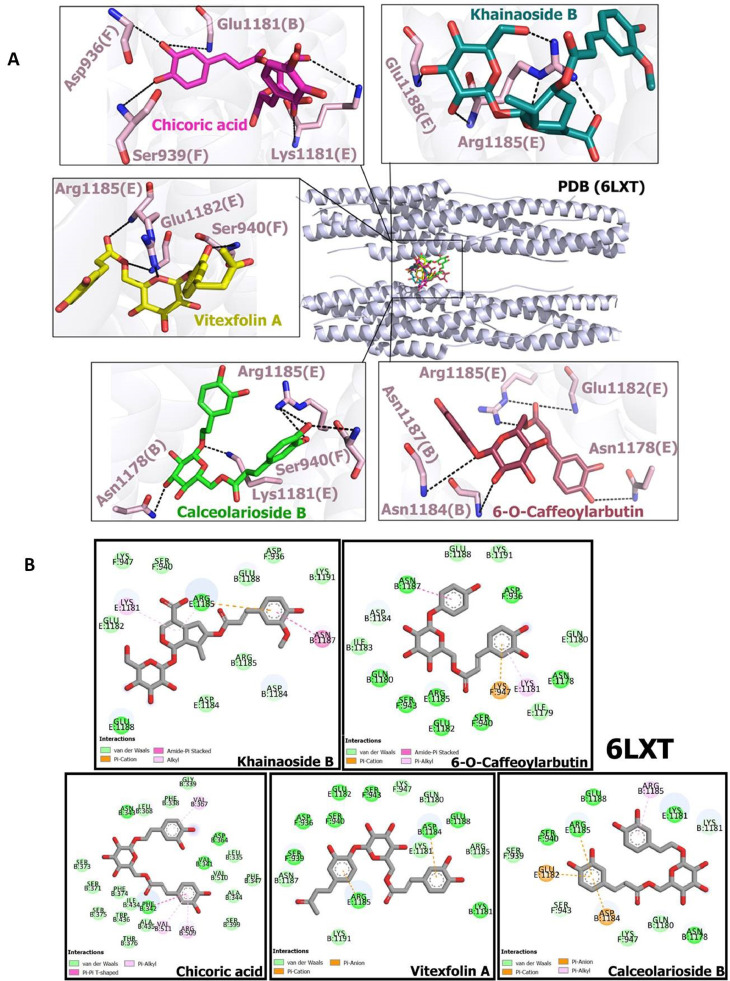
Table 3 presents MolDock Score, interactions and H-Bonds obtained from the docking studies of CAFDs with 6LXT. The results show highest binding potential of khaınaosıde B (-150.44), followed by chicoric acid (-150.017), vitexfolin A (-149.558) 6-O-Caffeoylarbutin (-146.12) and calceolarioside B (-141.587) as compared to Nelfinavir (-130.687). The binding interactions of potent compounds with S2 subunit of fusion protein are shown in Fig. 4 A. Fig. 4B shows residual wise van der Waals interactions, pi-alkyl interactions, pi-pi interactions, amide-pi-stacked interactions and pi-anion interactions of potent hits with 6LXT.
Table 3.
Results of the docking of CAFDs on the crystal structure of post fusion core of SARS-CoV-2 S2 subunit (6LXT).
| Ligand | MolDock Score | Protein-Ligand Interactions | Internal Ligand Interactions | H-Bond |
|---|---|---|---|---|
| Nelfinavir | -130.687 | -164.232 | 33.5452 | -9.3432 |
| Khaınaosıde B | -150.44 | -161.896 | 11.4557 | -8.36351 |
| Chicoric acid | -150.017 | -168.62 | 18.603 | -11.1833 |
| Vitexfolin A | -149.558 | -185.217 | 35.6583 | -20.1769 |
| 6-O-Caffeoylarbutin | -146.12 | -164.641 | 18.5207 | -16.9512 |
| Calceolarioside B | -141.587 | -172.326 | 30.7394 | -11.9822 |
| Khaınaosıde C | -139.261 | -147.331 | 8.06987 | -11.3859 |
| Calceolarioside D | -138.843 | -151.697 | 12.8542 | -8.86834 |
| Calceolarioside A | -135.155 | -170.652 | 35.4964 | -13.9423 |
| Scrophuloside B | -134.911 | -150.3 | 15.3891 | -11.735 |
| Robustaside D | -132.712 | -148.2 | 15.488 | -8.16426 |
| Calceolarioside C | -132.021 | -171.948 | 39.9268 | -21.5846 |
| Cynarin | -129.89 | -173.962 | 44.0712 | -15.3668 |
| Eutıgosıde A | -129.684 | -148.713 | 19.0289 | -10.3091 |
| Dodegranoside A | -129.625 | -153.199 | 23.5741 | -8.37051 |
| Robustaside E | -128.164 | -158.032 | 29.8676 | -14.1894 |
| Propyl 3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2- | -125.955 | -141.065 | 15.1099 | -5 |
| Methyl (2R)-3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxypropanoate | -120.097 | -139.184 | 19.087 | -10.5192 |
| Neochlorogenic acid | -112.361 | -138.544 | 26.183 | -15.9721 |
| Fertaric acid | -106.658 | -131.958 | 25.3 | -9.22242 |
| Dactylifric acid | -106.238 | -127.635 | 21.3977 | -12.3803 |
| Caftaric acid | -104.253 | -120.779 | 16.5262 | -10.7807 |
| Coutaric acid | -103.181 | -114.849 | 11.6674 | -9.25386 |
| Chlorogenic acid | -101.663 | -127.123 | 25.4594 | -12.0677 |
| Prenyl caffeate acid | -96.4395 | -106.955 | 10.5154 | -7.95977 |
| p-Coumaric acid | -77.652 | -84.3751 | 6.72302 | -8.07627 |
| Caffeic acid | -70.7393 | -77.8201 | 7.08078 | -8.17656 |
| Ferulic | -68.4726 | -74.5274 | 6.05481 | -5.1854 |
Fig. 4.
Docking poses of caffeic acid derivatives with COVID-19 virus fusion protein S2 subunit (A) Hydrogen bonding interactions of khaınaosıde B, chicoric acid, vitexfolin A, 6-O-Caffeoylarbutin, and calceolarioside B with amino acid residues of fusion protein S2 subunit (B) 2D view of interaction types of khaınaosıde B, chicoric acid, vitexfolin A, 6-O-Caffeoylarbutin, and calceolarioside B with surrounding amino acids of COVID-19 virus fusion protein S2 subunit.
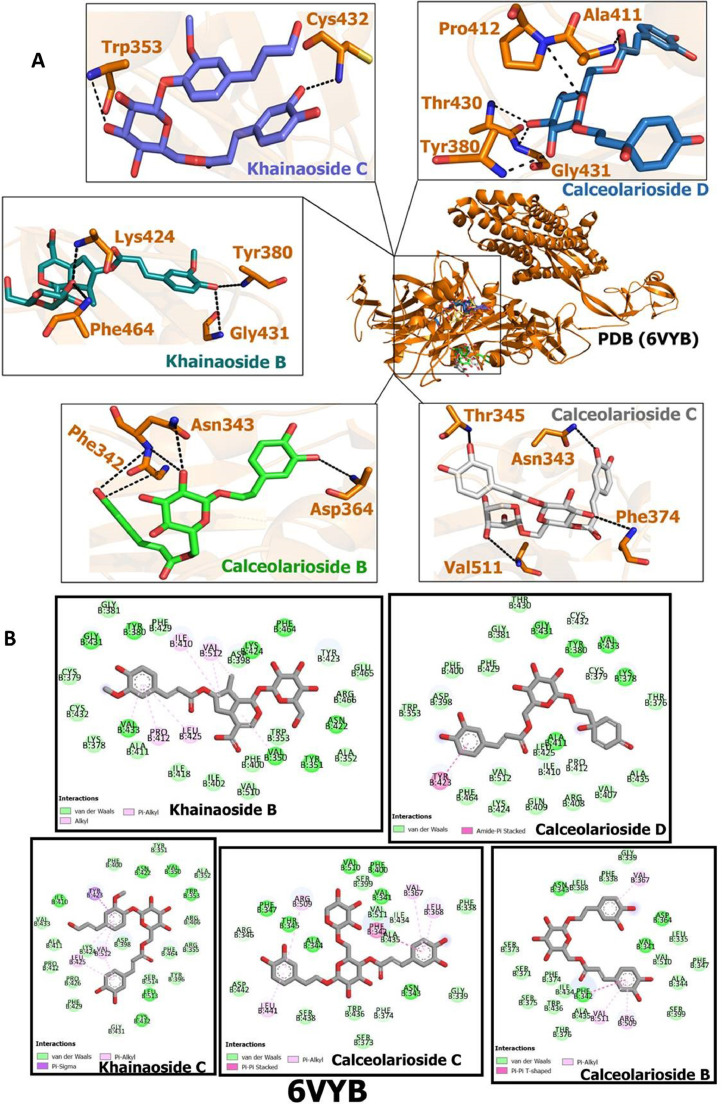
Table 4 shows the obtained results from the docking studies of CAFDs with crystal structure 6VYB. The results show highest binding potential of khaınaosıde C (-166.448), followed by khaınaosıde B (-165.435), and calceolarioside B (-153.135), calceolarioside C (-151.284), and calceolarioside D (-149.841) with comparison to reference drug Nelfinavir (-148.747). The amino acid residues of spike protein participating in interaction with these compounds are presented in Fig. 5 A. CAFDs interact with target protein via van der Waals interactions, pi-alkyl interactions, pi-pi interactions, amide-pi-stacked interactions and pi-sigma interactions (Fig. 5B).
Table 4.
Results of the docking of CAFDs on the crystal structure of SARS-CoV-2 spike ectodomain structure (open state) (6VYB).
| Ligand Name | MolDock Score | Protein-Ligand Interactions | Internal Ligand Interactions | H-Bond |
|---|---|---|---|---|
| Nelfinavir | -148.747 | -176.89 | 28.1432 | -2.49896 |
| Khaınaosıde C | -166.448 | -161.803 | -4.64496 | -8.71808 |
| Khaınaosıde B | -165.435 | -193.843 | 28.408 | -9.1829 |
| Calceolarioside B | -153.135 | -177.198 | 24.0631 | -11.6422 |
| Calceolarioside C | -151.284 | -203.133 | 51.8485 | -15.5073 |
| Calceolarioside D | -149.841 | -171.435 | 21.5938 | -6.20592 |
| Eutıgosıde A | -144.298 | -150.757 | 6.45875 | -8.94751 |
| Dodegranoside A | -142.831 | -161.943 | 19.1118 | -6.51745 |
| Vitexfolin A | -142.604 | -173.882 | 31.278 | -12.4266 |
| Chicoric acid | -141.566 | -164.796 | 23.2302 | -6.62961 |
| Propyl 3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2- | -139.154 | -149.85 | 10.6965 | -6.13396 |
| 6-O-Caffeoylarbutin | -137.225 | -160.96 | 23.7351 | -9.99943 |
| Cynarin | -133.964 | -180.052 | 46.0882 | -3.61595 |
| Robustaside D | -133.688 | -139.895 | 6.20761 | -7.9848 |
| Calceolarioside A | -133.035 | -170.659 | 37.6243 | -13.4663 |
| Robustaside E | -132.278 | -154.031 | 21.7532 | -4.63369 |
| Methyl (2R)-3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxypropanoate | -129.816 | -140.552 | 10.7361 | -3.44702 |
| Scrophuloside B | -129.274 | -159.233 | 29.959 | -5.75544 |
| Neochlorogenic acid | -116.183 | -137.163 | 20.9802 | -8.85752 |
| Dactylifric acid | -114.666 | -128.192 | 13.5263 | -5.90671 |
| Fertaric acid | -111.217 | -123.587 | 12.3705 | -2.53938 |
| Caftaric acid | -109.539 | -124.99 | 15.4504 | -9.02851 |
| Prenyl caffeate acid | -109.296 | -118.722 | 9.42623 | -3.47987 |
| Chlorogenic acid | -108.993 | -132.887 | 23.8942 | -8.6096 |
| Coutaric acid | -103.861 | -113.724 | 9.86261 | -2.97925 |
| p-Coumaric acid | -87.9961 | -96.8163 | 8.82013 | -4.60597 |
| Caffeic acid | -85.3883 | -91.7692 | 6.38084 | -7.06512 |
| Ferulic | -83.9251 | -89.969 | 6.04391 | -2.71179 |
Fig. 5.
Docking poses of caffeic acid derivatives with SARS-CoV-2 spike ectodomain (A) Hydrogen bonding interactions of khaınaosıde C, khaınaosıde B, calceolarioside B, calceolarioside C, calceolarioside D with amino acid residues of SARS-CoV-2 spike ectodomain (open state) (B) 2D view of interaction types of khaınaosıde C, khaınaosıde B, calceolarioside B, calceolarioside C, calceolarioside D with surrounding amino acids of SARS-CoV-2 spike ectodomain (open state).
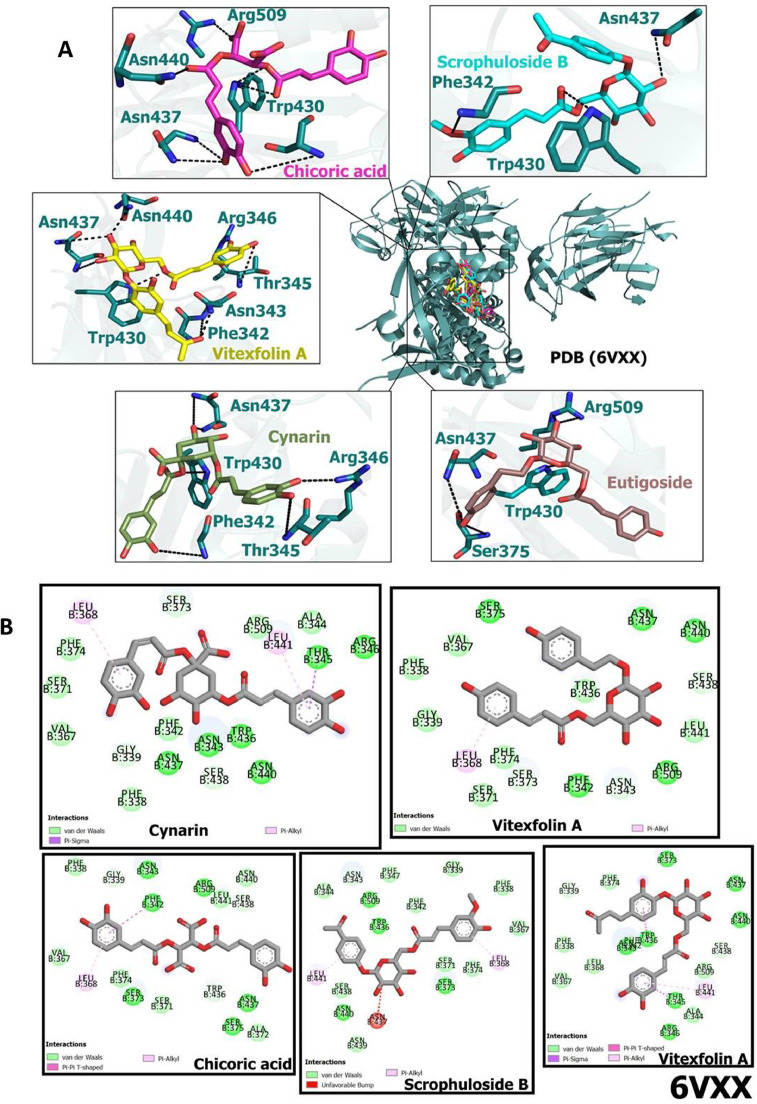
The binding energies obtained from the docking analysis of 6VXX with ligands are presented in Table 5 . Vitexfolin A, chicoric acid, eutıgosıde A exhibited the best potential against spike glycoproteins of SARS-CoV-2. According to in silico results, 7 of the compounds have a better affinity to bind with spike glycoproteins than Nelfinavir (Table 5). The ligand-protein interactions are shown in Fig. 6 A. Amino acid residues of 6VXX interact with CAFDs via van der Waals interaction, pi-alkyl interactions, pi-pi interactions and pi-sigma interactions are (Fig. 6B).
Table 5.
Results of the docking of CAFDs on the crystal structure of SARS-CoV-2 spike glycoprotein (closed state) (6VXX).
| Ligand Name | MolDock score | Protein-ligand interactions | Internal ligand interactions | H-Bond |
|---|---|---|---|---|
| Nelfinavir | -133.655 | -160.117 | 26.4619 | -7.62916 |
| Vitexfolin A | -158.443 | -177.854 | 19.4111 | -15.769 |
| Chicoric acid | -141.781 | -166.842 | 25.0606 | -7.55396 |
| Eutıgosıde A | -137.834 | -167.189 | 29.3553 | -11.5249 |
| Scrophuloside B | -136.54 | -169.158 | 32.6181 | -10.2562 |
| Cynarin | -136.457 | -176.339 | 39.8825 | -11.3919 |
| Calceolarioside D | -135.748 | -145.865 | 10.1171 | -7.33425 |
| Khaınaosıde B | -134.98 | -141.167 | 6.18667 | -14.3158 |
| Robustaside D | -130.586 | -154.38 | 23.7941 | -12.9145 |
| Robustaside E | -126.92 | -126.404 | -0.5159 | -14.472 |
| Khaınaosıde C | -126.895 | -124.812 | -2.0832 | -10.5752 |
| Propyl 3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2- | -125.712 | -153.822 | 28.1099 | -11.792 |
| Calceolarioside A | -123.256 | -152.379 | 29.1231 | -13.9537 |
| Calceolarioside B | -122.232 | -149.1 | 26.8684 | -13.6966 |
| 6-O-Caffeoylarbutin | -121.921 | -116.367 | -5.5538 | -8.70194 |
| Dodegranoside A | -119.728 | -144.496 | 24.7679 | -9.47601 |
| Methyl (2R)-3-(3,4-dihydroxyphenyl)-2-[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxypropanoate | -118.965 | -140.467 | 21.502 | -14.4652 |
| Calceolarioside C | -116.478 | -176.585 | 60.1073 | -21.2389 |
| Dactylifric acid, | -111.795 | -132.804 | 21.0096 | -3.32111 |
| Caftaric acid | -106.167 | -121.402 | 15.2351 | -12.1219 |
| Coutaric acid | -105.763 | -118.798 | 13.0344 | -11.9847 |
| Fertaric acid | -104.402 | -115.523 | 11.1211 | -9.61248 |
| Neochlorogenic acid | -102.203 | -130.877 | 28.6741 | -9.99063 |
| Prenyl caffeate acid | -100.523 | -107.187 | 6.66386 | -2.5 |
| Chlorogenic acid | -92.1219 | -117.747 | 25.6247 | -17.3432 |
| Ferulic | -78.2151 | -82.4019 | 4.18678 | -0.5326 |
| Caffeic acid | -75.6669 | -83.0829 | 7.41603 | -0.37606 |
| p-Coumaric acid | -73.8693 | -82.9404 | 9.07116 | -6.05487 |
Fig. 6.
Docking poses of caffeic acid derivatives with SARS-CoV-2 spike glycoprotein (closed state) (A) Hydrogen bonding interactions of scrophuloside B, chicoric acid, vitexfolin A, cynarin and eutıgosıde A with amino acid residues of SARS-CoV-2 spike glycoprotein (closed state) (B) 2D view of interaction types of scrophuloside B, chicoric acid, vitexfolin A, cynarin and eutıgosıde A with surrounding amino acids of spike glycoprotein (closed state).
Among all the screened compounds, only calceolarioside B possess good binding affinities with four out of five selected targets of SARS-CoV-2 (COVID-19 virus Mpro, Fusion S2 subunit, spike ectodomain (open state), and Nsp15 endoribonuclease) while vitexfolin A exhibits good binding efficacy against three out of five selected proteins of SARS-CoV-2 (COVID-19 virus Mpro, Fusion S2 subunit and spike protein). Based upon these findings, calceolarioside B could be regarded as pan inhibitor of SARS-CoV-2 proteins.
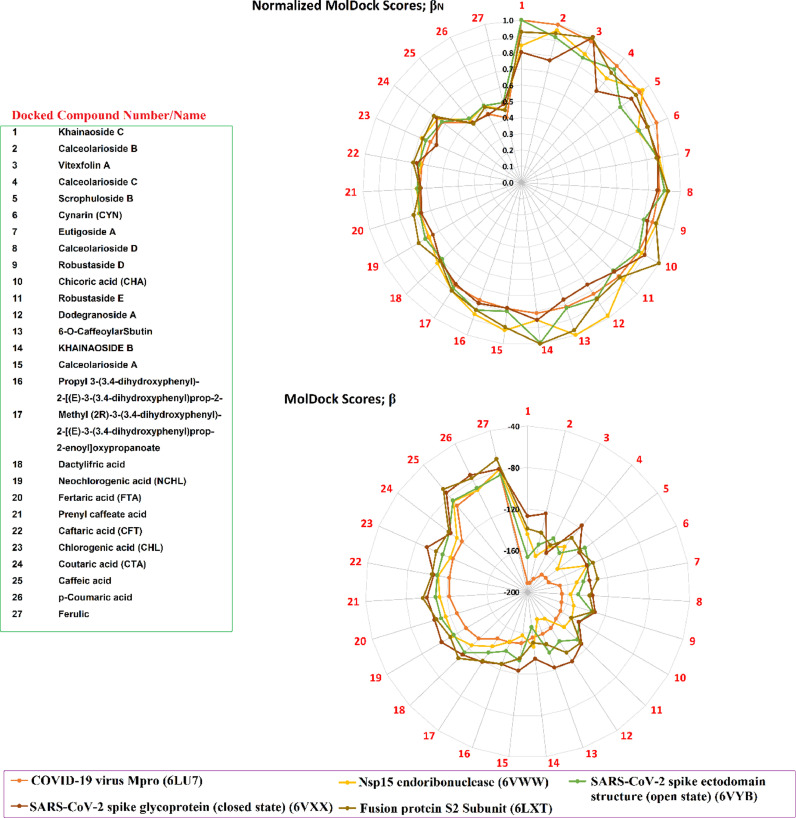
Relative assessment of MolDock scores were performed with (β), of CAFDs against SARS-CoV-2 proteins encoded by PDB IDs as 6LU7, 6VWW, 6TLX, 6VYB, and 6VXX (Fig. 7 ). We normalized all MolDock scores using equation as described below
Where; βN, βi, βmax represent the normalized MolDock score, MolDock score of a compound for any drug target protein and maximum MolDock score among all compounds of any drug target protein.
Fig. 7.
MolDock Score comparison among 27 compounds versus active sites SARS-CoV-2 6LU7, 6VWW, 6LXT, 6VYB, and 6VXX.
ADME profiling of potent caffeic acid derivatives
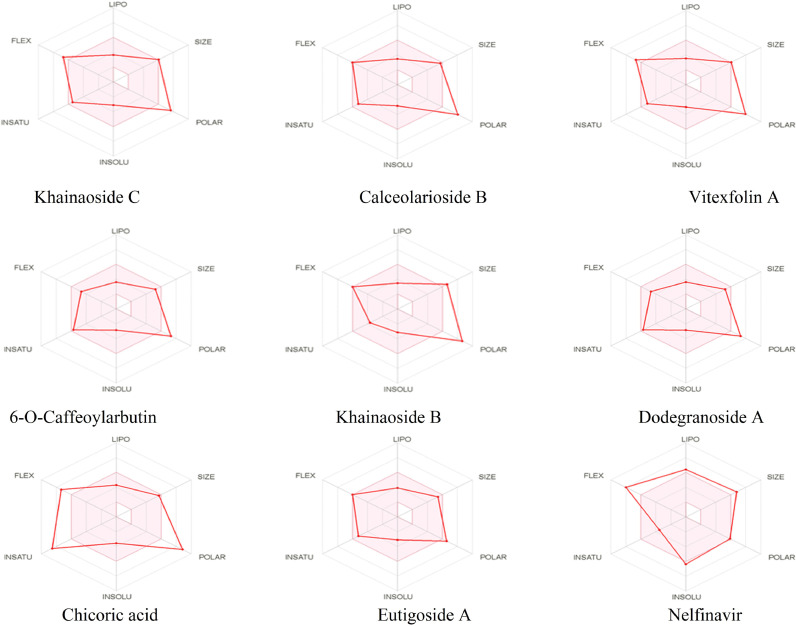
The eight compounds with high-binding affinity against COVID-19 were analysed for ADME properties using SwissADME web tool. The results of eight compounds with high activity potential are presented in Fig. 8 . Eutigoside A meets all criteria for oral use. Physicochemical, pharmacokinetics and drug-likeness properties of potent hits are presented in Table 6 . All of the compounds are water soluble. They do not cross the blood brain barrier, do not interact with interaction of main enzymes of Cytochromes P450, and have P-gp substrate properties. Toxicity assessment according to the chemical structure was carried out using Toxtree software.
Fig. 8.
Bioavailability Radar related to physicochemical properties of molecules (Criterias: Lipophilicity: - 0.7 < XLOGP3 < ++5.0, Size: 150 MW 500 g/mol, Polarity: 20 < TPSA < 130 Å2, Insolubility: 0 < log S < 6, Insaturation, Flexibility: 0.25 < rotatable bonds < 9).
Table 6.
ADME profiling of potent caffeic acid derivatives.
| Compound name | Ghose | TPSA | Absorption (% ABS) | Water solubilityLog S (ESOL) | BBB permeant | P-gp substrate | CYP isoform interact |
|---|---|---|---|---|---|---|---|
| Khaınaosıde C | No | 175.37 | 48.49 | -3.12- Soluble | No | Yes | No |
| Calceolarioside B | Yes | 186.37 | 44.70 | -2.85- Soluble | No | Yes | No |
| Vitexfolin A | No | 183.21 | 45.79 | -3.01- Soluble | No | Yes | No |
| 6-O-Caffeoylarbutin | Yes | 166.14 | 51.68 | -2.86- Soluble | No | Yes | No |
| Khaınaosıde B | No | 201.67 | 39.42 | -3.14- Soluble | No | Yes | No |
| Dodegranoside A | Yes | 166.14 | 51.68 | -2.86- Soluble | No | Yes | No |
| Chicoric acid | Yes | 208.12 | 37.19 | -3.58- Soluble | No | Yes | No |
| Eutıgosıde A | Yes | 145.91 | 58.66 | -3.11- Soluble | No | Yes | No |
| Nelfinavir | No | 127.20 | 65.18 | -6.36-Poorly soluble | No | Yes | 1A2, 2C19, 3A4 |
Molecular dynamics simulation
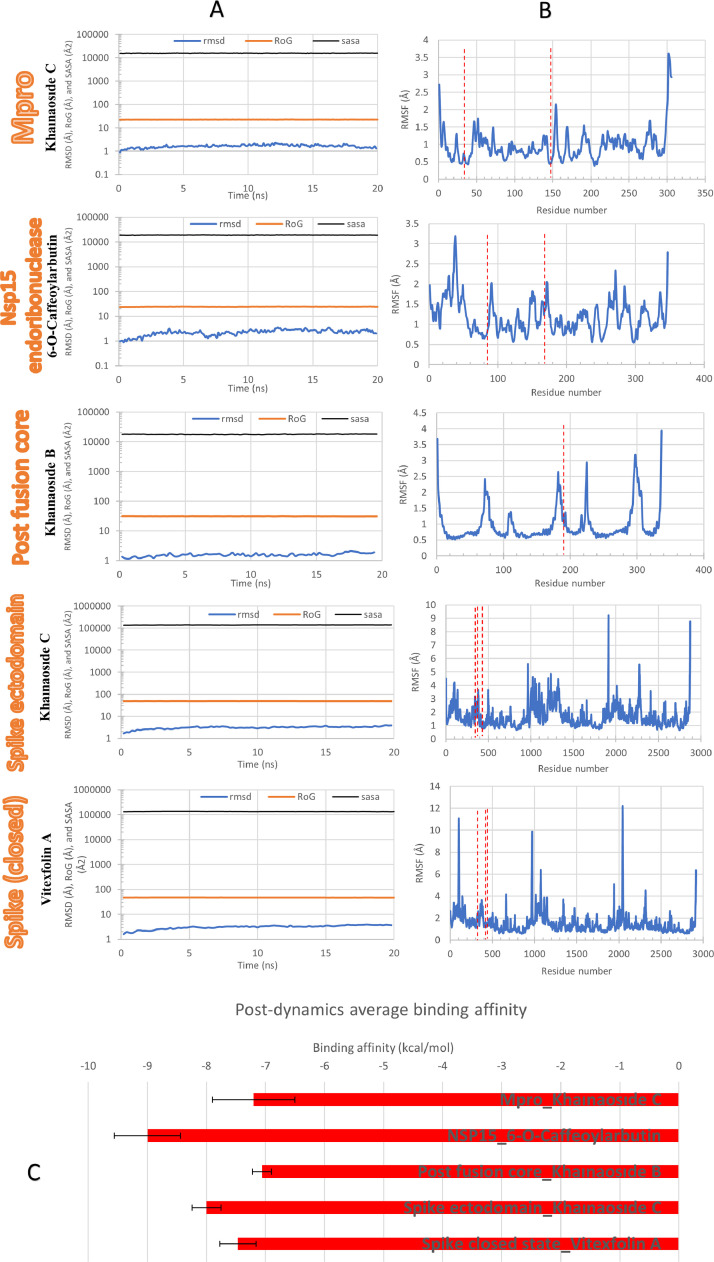
Fig. 9A shows the Root Mean Square Deviation (RMSD) in Å, Radius of Gyration (RoG) in Å, and Surface Accessible Surface Area (SASA) in Å2. The values of the three parameters indicate the equilibration of the systems during the 20 ns MDS. On the other hand, Fig. 9B shows the stability of the residues during the simulation through per residue Root Mean Square Fluctuation (RMSF) in Å. Dashed-red lines indicate the positions of the active site residues. Fig. 9C shows the average binding energies obtained for the representative conformation for each cluster after the MDS run for 20 ns on the protein-ligand complexes.
Fig. 9.
Post-dynamics analysis: (A) Root Mean Square Deviation (RMSD) (blue line), Radius of Gyration (RoG) (orange line), and surface accessible surface area (SASA) (gray line) for the best complex for each viral protein during 20 ns MDS run. (B) Per-residue Root Mean Square Fluctuation (RMSF) for the same complexes with dashed lines represents the active residues. (C) Post-dynamics average binding energies (in kcal/mol) calculated for the complexes using AutoDock Vina. Error bars represent the standard deviations.
Discussion
Polyphenols possess various beneficial properties against viral diseases such as modulation of immune system (Jasso-Miranda et al., 2019), inhibiting viral replication (Lalani and Poh, 2020) and reduction of viral uptake by target membrane (Vazquez-Calvo et al., 2017). CAFDs are polyphenolic compounds which possess prominent antiviral activity especially against hepatitis B virus (HBV) (Zhang et al., 2014) and HIV (Pluymers et al., 2000). Keeping in view the potential of CAFDs against viral diseases (Fig. 1), 27 CAFDs were docked with 5 proteins of SARS-CoV-2 to evaluate their binding energies against these targets. Out of 27 CAFDs, 19 of them showed greater MD score (ranging between -191 to -159, Table 1) against COVID-19 virus Mpro as compared to nelfinavir (-147) (Adem et al., 2020). 11 out of 27 CAFDs possess greater MD score against Nsp15 endoribonuclease (Table 2) and SARS-CoV-2 fusion S2 subunit as compared to reference drug (Table 3), while 5 were found to be good binder of spike ectodomain (open state) (Table 4) and 7 exhibits good binding interaction with spike glycoprotein (closed state) as compared to Nelfinavir (Table 5). From chemical point of view, the binding potential of potent CAFDs i.e., khainaoside C, scrophuloside B, vitexfolin A, calceolarioside B & C with SARS-CoV-2 proteins can largely be attributed to the presence of hydroxyl and carbonyl groups. These groups specifically –OH group serves as excellent candidate for hydrogen bonding and exhibits tendency to form strong interactions with amino acid residues of target protein. As reported in Fig. 9, the 20 ns MDS runs were enough for the protein-ligand complexes. RMSD for each system show equilibration, while SASA and RoG both show stabilities during the MDS. The per-residues RMSF profiles in Fig. 9B indicate the stability of the binding since the interacting residues (H41, C149 in Mpro, K90, T167 in NSP15, R185 in the post-fusion core, N343, Y380, G431 in spike ectodomain, and F342, T430, N437 in spike closed state) have low RMSF values in all the complexes. Additionally, the average binding energies (Fig. 9C) calculated for the different clusters during the MDS runs show that the binding is stable. Hence, the MDS study supports previous docking scores values.
Most of the CAFDs possessing good binding energies with various proteins of SARS-CoV-2 have been isolated from traditionally used medicinal plants such as: khainaoside B and khainaoside C are isolated from Thai medicinal plant Vitex glabrata which is traditionally used to support lactation (Luecha et al., 2009) and possess anti-diabetic potential (Somtimuang et al., 2018). Vitexfolin A is an important constituent of Vitex rotundifolia and possesses strong analgesic effect (Okuyama et al., 1998). Scrophuloside B has been identified in traditional Chinese medicinal plants including Picrorhiza scrophulariiflora (Wang et al., 2013) and Radix scrophulariae (Jing et al., 2011). Calceolarioside B has been isolated from Fraxinus sieboldiana and Forsythia suspensea plants both of which possess anti-inflammatory and anti-viral properties (Kim et al., 2002; Wang et al., 2009). While some of potent CAFDs are found in vegetables and fruits such as: Lettuce (Lactuca sativa), a widely consumed leafy vegetable, is found to be enriched with various CAFDs such as chicoric acid and chlorogenic acid (Abu-Reidah et al., 2013). Chicoric acid has been long known as potent anti-viral agent against HIV (Lin et al., 1999) and its mechanism of action involves deactivation of HIV-1 integrase, increased T-lymphoblastoid viability, and downregulation of reverse transcriptase of HIV-1 (Peng et al., 2019). 6′-O-caffeoylarbutin is abundantly found in Vaccinium dunalianumas. V. dunalianumas is a commonly cultivated blueberry species in China and its dried leaf buds as herbal tea while its leaves are used as folk medicine (Li et al., 2016; Luo et al., 2015). Sunflower (Helianthus annuus) sprouts (nutritious sunflower lettuce) found to be enriched with cynarin (Sun et al., 2012), recommending these foods as beneficial food choice for COVID-19 patients.
Among all these potent compounds, calceolarioside B is identified as pan-inhibitor of SARS-CoV-2 having potential to target SARS-CoV-2 Mpro, Nsp15, coronavirus fusion protein, as well as spike ectodomain which brought excitement about calceolarioside B's potential against COVID-19. Interestingly, calceolarioside B is also an important constituent of Akebia trifoliata fruit which is popularized in Asia due to its nutritional values as well as delicious taste and also used as dietary supplement for its various health benefits including anti-microbial and anti-inflammation (Wang et al., 2019), suggesting that Akebia trifoliata fruit could potentially help COVID-19 patients to fight this disease. Calceolarioside B has potential to inhibit gp4l transmembrane glycoproteins of HIV (human immunodeficiency virus) (Kim et al., 2002). These glycoproteins play critical role in virus cell fusion with target membrane to enable viral entry into the host cell (Buzon et al., 2010). As calceolarioside B is identified as potent binder of SARS-CoV-2 spike glycoprotein during our study, the results from both of these studies propose calceolarioside B as viral and target membrane fusion inhibitor further strengthening its role against COVID-19.
In addition, calceolarioside B exhibits good anti-RSV (respiratory syncytial virus) effects (Dong et al., 2017) which recommend that calceolarioside B rich foods could be a potential alternative approach for the prevention and treatment of COVID-19. In addition to its anti-viral properties, it has potential to inhibit IL-6 production to exert it anti-inflammatory properties (Jin et al., 2014). COVID-19 patients suffer with severe inflammatory response in the later stages of infection, thus, dual anti-viral/anti-inflammatory properties of this compound makes it ideal candidate for drug development for prevention and treatment of COVID-19. As coronaviruses mutations make it difficult to develop vaccine, so does the single-target drugs. Single-target drugs might encounter low efficacy as the virus mutates. Thus, complex diseases like COVID-19 are more likely to be alleviated or healed though simultaneous modulation of multiple targets. Based on the above discussion, in our personal opinion, efficacy of CAFDs-enriched foods against COVID-19 as well as safety suggests that their adoption in daily diets might help prevent the onset of COVID-19 in an alternative and nonpharmacological approach.
Conclusively, this study provides scientific basis for the possible utilization of CAFDs as drug leads to develop anti-COVID-19 therapeutics. Since there is an urgent and timely need to find out effective and specific anti-viral treatment for COVID-19, this study will hopefully lay the foundation to work forward on small scale studies for the determination of 1) efficacy of CAFDs in reducing viral load and shortening the infectious period, 2) optimal dosing regimen based on impact on viremia, 3) impact on antibody production, inflammatory signaling and oxidative stress in COVID-19 patients. It seems that CAFDs have better absorption and good safety profiles as many studies pointed out the better absorption of CAFDs and CAFDs are important constituent of dietary foods. However, it would be worthwhile to conduct pharmacological studies to determine whether the bioavailability of CAFDs is good enough or it is better to administer in combinations. Once we understand the best way to deal with the CAFDs, it will be a reasonable starting point for therapeutic interventions based on the pharmacokinetic and pharmacodynamic studies on CAFDs. Despite excitement about CAFD's potential from antiviral and anti-inflammatory researches, there is an urgent and timely need to fill the room of knowledge for validation of CAFDs potential against COVID-19.
Acknowledgment
Shaheen supercomputer of King Abdullah University of Science and Technology (KAUST) is used to perform the MDS study (under the project number k1482). We thank Dr. Bahaa Mostafa for using his computational power unit during the analysis of the MDS data.
References
- Abu-Reidah I.M., Contreras M.M., Arraez-Roman D., Segura-Carretero A., Fernandez-Gutierrez A. Reversed-phase ultra-high-performance liquid chromatography coupled to electrospray ionization-quadrupole-time-of-flight mass spectrometry as a powerful tool for metabolic profiling of vegetables: Lactuca sativa as an example of its application. J. Chromatogr. A. 2013;1313:212–227. doi: 10.1016/j.chroma.2013.07.020. [DOI] [PubMed] [Google Scholar]
- Adem, S., Eyupoglua, V., Sarfraz, I., Rasul, A., 2020. Identification of potent COVID-19 main protease (Mpro) inhibitors from natural polyphenols: an in silico strategy unveils a hope against CORONA. [DOI] [PMC free article] [PubMed]
- Amoretti M., Amsler C., Bonomi G., Bouchta A., Bowe P., Carraro C., Cesar C.L., Charlton M., Collier M.J., Doser M., Filippini V., Fine K.S., Fontana A., Fujiwara M.C., Funakoshi R., Genova P., Hangst J.S., Hayano R.S., Holzscheiter M.H., Jorgensen L.V., Lagomarsino V., Landua R., Lindelof D., Lodi Rizzini E., Macri M., Madsen N., Manuzio G., Marchesotti M., Montagna P., Pruys H., Regenfus C., Riedler P., Rochet J., Rotondi A., Rouleau G., Testera G., Variola A., Watson T.L., van der Werf D.P., Collaboration A. Production and detection of cold antihydrogen atoms. Nature. 2002;419:456–459. doi: 10.1038/nature01096. [DOI] [PubMed] [Google Scholar]
- Buzon V., Natrajan G., Schibli D., Campelo F., Kozlov M.M., Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Lu X., Tong X., Dong Y., Tang L., Liu M. Forsythiae fructus: a review on its phytochemistry, quality control, pharmacology and pharmacokinetics. molecules. 2017;22 doi: 10.3390/molecules22091466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos C.N., Menezes R., Stewart D. Polyphenols as new leads in drug discovery: biological activity and mechanisms. Curr. Pharm. Des. 2018;24:2041–2042. doi: 10.2174/138161282419180924094610. [DOI] [PubMed] [Google Scholar]
- Du R.H., Liang L.R., Yang C.Q., Wang W., Cao T.Z., Li M., Guo G.Y., Du J., Zheng C.L., Zhu Q., Hu M., Li X.Y., Peng P., Shi H.Z. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur. Respir. J. 2020 doi: 10.1183/13993003.00524-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Military Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics 14. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., Aziz B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J.Med. Virol. 2020 doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasso-Miranda C., Herrera-Camacho I., Flores-Mendoza L.K., Dominguez F., Vallejo-Ruiz V., Sanchez-Burgos G.G., Pando-Robles V., Santos-Lopez G., Reyes-Leyva J. Antiviral and immunomodulatory effects of polyphenols on macrophages infected with dengue virus serotypes 2 and 3 enhanced or not with antibodies. Infect. Drug Resist. 2019;12:1833–1852. doi: 10.2147/IDR.S210890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H.G., Kim A.R., Ko H.J., Lee S.K., Woo E.R. Three new lignan glycosides with IL-6 inhibitory activity from Akebia quinata. Chem. Pharm. Bull. 2014;62:288–293. doi: 10.1248/cpb.c13-00668. [DOI] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jing J., Chan C.O., Xu L., Jin D., Cao X., Mok D.K., Parekh H.S., Chen S. Development of an in-line HPLC fingerprint ion-trap mass spectrometric method for identification and quality control of Radix Scrophulariae. J. Pharm. Biomed. Anal. 2011;56:830–835. doi: 10.1016/j.jpba.2011.07.032. [DOI] [PubMed] [Google Scholar]
- Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Khan, M.F., Khan, M.A., Khan, Z.A., Ahamad, T., Ansari, W.A., 2020. Identification of dietary molecules as therapeutic agents to combat COVID-19 using molecular docking studies.
- Kim H.J., Yu Y.G., Park H., Lee Y.S. HIV gp41 binding phenolic components from Fraxinus sieboldiana var. Angustata. Planta Med. 2002;68:1034–1036. doi: 10.1055/s-2002-35665. [DOI] [PubMed] [Google Scholar]
- Kim, Y., Jedrzejczak, R., Maltseva, N.I., Endres, M., Godzik, A., Michalskaa, K., Joachimiak, A., 2020a. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. [DOI] [PMC free article] [PubMed]
- Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci.: A Publ. Protein Soc. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani S., Poh C.L. Flavonoids as antiviral agents for enterovirus A71 (EV-A71) Viruses. 2020;12 doi: 10.3390/v12020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langland J., Jacobs B., Wagner C.E., Ruiz G., Cahill T.M. Antiviral activity of metal chelates of caffeic acid and similar compounds towards herpes simplex, VSV-Ebola pseudotyped and vaccinia viruses. Antiviral Res. 2018;160:143–150. doi: 10.1016/j.antiviral.2018.10.021. [DOI] [PubMed] [Google Scholar]
- Li Z.J., Shen X.Y., Hou C.L. Fungal endophytes of South China blueberry (Vaccinium dunalianum var. urophyllum) Lett. Appl. Microbiol. 2016;63:482–487. doi: 10.1111/lam.12673. [DOI] [PubMed] [Google Scholar]
- Lin Z., Neamati N., Zhao H., Kiryu Y., Turpin J.A., Aberham C., Strebel K., Kohn K., Witvrouw M., Pannecouque C., Debyser Z., De Clercq E., Rice W.G., Pommier Y., Burke T.R., Jr. Chicoric acid analogues as HIV-1 integrase inhibitors. J. Med. Chem. 1999;42:1401–1414. doi: 10.1021/jm980531m. [DOI] [PubMed] [Google Scholar]
- Liu X., Wang X.J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J. Genet. Genom. = Yi chuan xue bao. 2020;47:119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecha P., Umehara K., Miyase T., Noguchi H. Antiestrogenic constituents of the Thai medicinal plants Capparis flavicans and Vitex glabrata. J. Nat. Prod. 2009;72:1954–1959. doi: 10.1021/np9006298. [DOI] [PubMed] [Google Scholar]
- Luo X.L., Li N., Xu M., Zhu H.T., He P., Ding Y., Zhao P., Zhang Y.J. HPLC simultaneous determination of arbutin, chlorogenic acid and 6′-O-caffeoylarbutin in different parts of Vaccinium dunalianum Wight. Nat. Prod. Res. 2015;29:1963–1965. doi: 10.1080/14786419.2015.1013472. [DOI] [PubMed] [Google Scholar]
- Magnani C., Isaac V., Corrêa M.A., Salgado H. Caffeic acid: a review of its potential use in medications and cosmetics. Anal. Methods. 2014;6:3203. [Google Scholar]
- Mark P., Nilsson L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. 2001;105:9954–9960. [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Shirasago Y., Ando S., Shimojima M., Saijo M., Fukasawa M. Caffeic acid, a coffee-related organic acid, inhibits infection by severe fever with thrombocytopenia syndrome virus in vitro. J. Infect. Chemother.: Off. J. Jpn Soc. Chemother. 2018;24:597–601. doi: 10.1016/j.jiac.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Okuyama E., Fujimori S., Yamazaki M., Deyama T. Pharmacologically active components of viticis fructus (Vitex rotundifolia). II. The components having analgesic effects. Chem. Pharm. Bull. 1998;46:655–662. doi: 10.1248/cpb.46.655. [DOI] [PubMed] [Google Scholar]
- Peng Y., Sun Q., Park Y. The bioactive effects of chicoric acid as a functional food ingredient. J. Med. Food. 2019;22:645–652. doi: 10.1089/jmf.2018.0211. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kale L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluymers W., Neamati N., Pannecouque C., Fikkert V., Marchand C., Burke T.R., Jr., Pommier Y., Schols D., De Clercq E., Debyser Z., Witvrouw M. Viral entry as the primary target for the anti-HIV activity of chicoric acid and its tetra-acetyl esters. Mol. Pharmacol. 2000;58:641–648. doi: 10.1124/mol.58.3.641. [DOI] [PubMed] [Google Scholar]
- Somtimuang C., Olatunji O.J., Ovatlarnporn C. Evaluation of in vitro alpha-amylase and alpha-glucosidase inhibitory potentials of 14 medicinal plants constituted in Thai folk antidiabetic formularies. Chem. Biodivers. 2018;15 doi: 10.1002/cbdv.201800025. [DOI] [PubMed] [Google Scholar]
- Sun Z., Chen J., Ma J., Jiang Y., Wang M., Ren G., Chen F. Cynarin-rich sunflower (Helianthus annuus) sprouts possess both antiglycative and antioxidant activities. J. Agric. Food Chem. 2012;60:3260–3265. doi: 10.1021/jf300737y. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya H., Ichinose M., Ikeda K., Uozaki M., Morishita J., Kuwahara T., Koyama A.H., Yamasaki H. Inhibition by caffeic acid of the influenza A virus multiplication in vitro. Int. J. Mol. Med. 2014;34:1020–1024. doi: 10.3892/ijmm.2014.1859. [DOI] [PubMed] [Google Scholar]
- Vazquez-Calvo A., Jimenez de Oya N., Martin-Acebes M.A., Garcia-Moruno E., Saiz J.C. Antiviral properties of the natural polyphenols delphinidin and epigallocatechin gallate against the Flaviviruses West Nile Virus, Zika Virus, and Dengue Virus. Front. Microbiol. 2017;8:1314. doi: 10.3389/fmicb.2017.01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.N., Ma Z.Q., Liu Y., Guo Y.Z., Gu Z.W. New phenylethanoid glycosides from the fruits of forsythia suspense (thunb.) vahl. Molecules. 2009;14:1324–1331. doi: 10.3390/molecules14031324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhao W., Choomuenwai V., Andrews K.T., Quinn R.J., Feng Y. Chemical investigation of an antimalarial Chinese medicinal herb Picrorhiza scrophulariiflora. Bioorg. Med. Chem. Lett. 2013;23:5915–5918. doi: 10.1016/j.bmcl.2013.08.077. [DOI] [PubMed] [Google Scholar]
- Wang X., Yu N., Peng H., Hu Z., Sun Y., Zhu X., Jiang L., Xiong H. The profiling of bioactives in Akebia trifoliata pericarp and metabolites, bioavailability and in vivo anti-inflammatory activities in DSS-induced colitis mice. Food & Funct. 2019;10:3977–3991. doi: 10.1039/c9fo00393b. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.L., Dai L.H., Wu Y.H., Yu X.P., Zhang Y.Y., Guan R.F., Liu T., Zhao J. Evaluation of hepatocyteprotective and anti-hepatitis B virus properties of Cichoric acid from Cichorium intybus leaves in cell culture. Biol. Pharm. Bull. 2014;37:1214–1220. doi: 10.1248/bpb.b14-00137. [DOI] [PubMed] [Google Scholar]
- Zhao Y.H., Abraham M.H., Le J., Hersey A., Luscombe C.N., Beck G., Sherborne B., Cooper I. Rate-limited steps of human oral absorption and QSAR studies. Pharm. Res. 2002;19:1446–1457. doi: 10.1023/a:1020444330011. [DOI] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus, I., Research, T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]