Abstract
Activation of oncogenic KRAS is the most common driving event in lung adenocarcinoma development. Despite the existing rationale for targeting activated KRAS and its downstream effectors, the failure of clinical trials to date indicates that the mechanism of KRAS-driven malignancy remains poorly understood. Here we report that histone deacetylase 10 (HDAC10) might function as a putative tumor suppressor in mice carrying a spontaneously activated oncogenic Kras allele. Hdac10 deletion accelerated KRAS-driven early-onset lung adenocarcinomas, increased macrophage infiltration in the tumor microenvironment, and shortened survival time in mice. Highly tumorigenic and stem-like lung adenocarcinoma (LUAD) cells were increased in Hdac10-deleted tumors compared to Hdac10 wild-type tumors. HDAC10 regulated the stem-like properties of KRAS-expressing tumor cells by targeting SOX9. Expression of SOX9 was significantly increased in Hdac10-deleted tumor cells and depletion of SOX9 in Hdac10 knockout (KO) LUAD cells inhibited growth of tumor spheres. The genes associated with TGF-β pathway were enriched in Hdac10 KO tumor cells, and activation of TGF-β signaling contributed to SOX9 induction in Hdac10 KO LUAD cells. Overall, our study evaluates the functions and mechanisms of action of HDAC10 in lung carcinogenesis which will inform the rationale for targeting its related regulatory signaling as an anticancer strategy.
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide(1). Approximately 40% of human lung cancers are adenocarcinomas, of which more than half have a known oncogenic driver mutation. Oncogenic KRAS activation is the most common driving event in lung adenocarcinoma. Up to 30% of patients with lung adenocarcinoma have a KRAS mutation(2). Although many major downstream signaling pathways and co-effectors of KRAS have been intensely studied over the last 30 years, these discoveries have not yet been translated into an effective targeted therapy(3).
Histone deacetylases (HDACs) reversibly modulates chromatin structure and gene expression by removing acetyl groups from histone and non-histone proteins. Dysregulated HDAC expression and/or function often contribute to tumorigenesis by disrupting acetylation homeostasis in cells(4). Several preclinical studies have indicated that the combined inhibition of HDACs with Ras signaling, such as MAPK and PI3K, could produce synergistic effects in KRAS-mutant cancer cells(5). HDAC inhibitors (HDACi) can also restore tumor cell sensitivity to EGFR-tyrosine kinase inhibitors (TKIs) treatment, and induce cell death in KRAS-mutant non-small cell lung cancer (NSCLC) cells(6, 7). While broad-spectrum HDACi have been pre-clinically or clinically used for cancer treatment, the precise function of individual HDACs as a central mediator of proliferation and tumorigenic capacity remains a conundrum(4). Although HDAC knockdown (KD) or inhibition in a variety of cancer cells induces cell cycle arrest and apoptosis, in some cancers, genetic inactivation of HDACs in vivo might have tumorigenic effects(8, 9). For example, a frameshift mutation of HDAC2 is present in many human cancer cells and the loss of HDAC2 expression confers resistance to HDACi treatment(8). Also, SIRT6 was found to be a tumor suppressor that regulates aerobic glycolysis and ribosome metabolism in cancer cells. Sirt6 deficiency induced transformation of immortalized MEFs and promoted the tumorigenesis of colorectal cancer in ApcMin mice(9). Thus, there is an urgent need to systematically dissect the role of individual HDACs in different cancer types at different stages of tumorigenesis.
In humans, there are 18 HDACs grouped into four classes. HDAC10 is a class IIb HDAC that possesses one catalytic domain and one additional leucine-rich incomplete catalytic domain. HDAC10 deacetylates histones in vitro and represses transcription when tethered to a target promoter(10, 11). HDAC10 transcriptionally represses the expression of DUB3 deubiquitinating enzyme by forming a protein complex with nuclear receptor co-repressor 2 (NCOR2) and aberrant expression of DUB3 confers BET inhibitor resistance in cancer cells by increasing BRD4 expression(12). In addition to histone and non-histone deacetylase activities, a recent study demonstrated that HDAC10 also functions as a polyamine deacetylase (PDAC), which can remove an acetyl group from cationic polyamines(13). Defining the enzymatic activity and substrate specificity of HDAC10 will help to better understand the role of HDAC10 in human cancers.
HDAC10 has been reported to be the strongest predictor of poor prognosis for lung cancer patients. Low expression of HDAC10 is associated with short survival time of patients with non-small cell lung cancer (NSCLC)(14), indicating a potential role of HDAC10 in lung cancer development. In our previous study, we showed that the depletion of HDAC10 widely suppresses cell proliferation in a panel of human lung cancer cell lines by inhibiting mitotic entry, which is related with the loss of cyclin A2(15). In another study, it was revealed that HDAC10 knockdown can induce cell cycle arrest and apoptosis in lung cancer cells through regulating the phosphorylation of AKT(16). To further determine the role of HDAC10 in tumorigenesis in vivo and further support the relevance of HDAC10 in human lung cancer development, we used Hdac10 genetic knockout (KO) C57BL/6 mice to study the effect of HDAC10 on KRAS-driven lung cancer formation. We found that HDAC10 plays different roles in a cell context dependent manner. In established tumor cells, HDAC10 is critical for unchecked cell growth, so transient depletion or inhibition of HDAC10 suppresses tumor cell growth(15). In contrast, the deletion of Hdac10 in mice accelerated oncogenic KRAS-driven lung tumorigenesis. HDAC10 might function as a tumor suppressor by regulating cancer stem-like cells (CSCs). Lung adenocarcinoma (LUAD) cells isolated from Hdac10 KO tumor tissues exhibited highly tumorigenic and stem-like properties. Moreover, activation of TGFβ signaling and upregulation of SOX9 in Hdac10 KO LUAD cells were required for HDAC10-mediated CSCs regulation.
Materials and Methods
Mouse strains
Hdac10 KO mice, obtained from Taconic Biosciences (Model TF1813), were generated from ES cells using the Cre-loxP method. The KrasLA1 mouse, which carries latent, oncogenic alleles of Kras, has been reported previously(17). This mouse model recapitulates spontaneous KRAS oncogenic activation in vivo and causes early onset multifocal lung adenocarcinoma in the mouse. To characterize the role of HDAC10 in KRAS-driven lung tumorigenesis, the Hdac10 KO mouse was bred with the KrasLA mouse to obtain littermate Hdac10 KO homozygous, heterozygous and Hdac10 WT mice, all of which carry KrasLA1alleles. The mice were sacrificed at different ages, and the effect of Hdac10 deletion on tumor cell growth was detected by examination of the number of visible nodules and histopathological analysis. All animal studies have been approved by the George Washington University Institutional Animal Care and Use Committee.
LUAD isolation and tumor sphere assay
Primary mouse lung adenocarcinoma (LUAD) cells were isolated as previously described(18). Briefly, Hdac10−/−, KrasLA1 and Hdac10+/+, KrasLA1mice bearing lung tumors were euthanized at 20–30 weeks. Dissected lung tumors from each mouse were pooled and dissociated in Collagenase (0.5 μg/mL) and DNase solution (0.2 μg/mL) for 50 min at 37°C. The dissociated cells were filtered through a 70 μm strainer and grown in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2. The fibroblasts were removed by different sensitivity with trypsin digestion and the LUAD cells were authenticated by detecting the lung epithelial cell marker EpCAM, lineage marker SFTPC (surfactant associated protein C), and SCGB1A1 (secretoglobin family 1A member 1). Moreover, the expression of the mutant Kras allele in LUAD cells were validated by RT-PCR as previously described(17). Additional details on LUAD and the tumor sphere assay can be found in Supplementary Materials and Methods.
Cell lines, plasmids, antibodies and xenograft mouse models
HEK 293T and NSCLC cell lines H157, A549, H441, H23, H358, H1299, PC9, H1975, H322, H596, H522, H292 and H661 were purchased from ATCC and came with comprehensive authentication and quality controls. All cell lines were confirmed regularly to be free of mycoplasma contaminations. Cells were grown in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2. Lentivirus-transduced cells were selected for puromycin or neomycin resistance, and the stably expressing cells were used at passages P5 to P10. Details of plasmid constructions, sources of antibodies and xenograft mouse models are provided in Supplementary Materials and Methods.
Immunoblotting, immunostaining and flow cytometry analyses
Western blotting, immunostaining, and flow cytometry analyses are described in Supplementary Materials and Methods.
RNA sequencing and Pathway Analysis
Comparative RNA sequencing was performed by PrimBio Research Institute to detect HDAC10 downstream pathways. Pathway analysis was performed using Ingenuity Pathway Analysis (IPA) and gene set enrichment analysis (GSEA)(19, 20). See Supplementary Materials and Methods for details.
Real-time RT-PCR
Real-time RT-PCR was performed as previously described(15). See Supplementary Methods for details of gene expression analysis. PCR primer sequences are shown in Table S1.
Statistics
Statistical analyses were carried out using R and GraphPad Prism 8. The normality of continuous variables was analyzed by Normal probability plots and Shapiro-Wilk test. Normally distributed data were compared by un-pooled two sample t-test; non-normally distributed data were analyzed by nonparametric Wilcoxon rank-sum test. Ordinary one-way ANOVA followed by Tukey test was used for multiple comparisons. Survival data were analyzed by non-parametric methods. Survival curves were plotted by Kaplan–Meier method and log-rank test was applied to compare survival curves between groups. p < 0.05 was considered statistically significant.
Results
Deletion of Hdac10 promotes KRAS-driven lung tumorigenesis
Our previous study has revealed the role of HDAC10 in cell cycle regulation by using human non-small cell lung cancer (NSCLC) cell lines (15); however, the effect of HDAC10 on primary lung cancer initiation and progression is not yet known. In this study, we investigate whether germline deletion of Hdac10 can regulate cancer formation in KRAS-driven lung cancer mouse model. The Hdac10 KO mouse, which has a whole body in-frame deletion of the catalytic domain of HDAC10, was generated by the Cre-loxP method (Supplementary Fig. S1A). The deletion of Hdac10 exons 4–8 in the mouse genome was validated by tail-biopsy genotyping and real-time RT-PCR (Supplementary Figs. S1B and S1C). Loss of expression of the HDAC10 full length protein in different mouse tissues was also confirmed by Western blot (Supplementary Fig. S1D). Although the catalytic activity of HDAC10 is abolished, Hdac10 KO mice are viable and fertile without any developmental defects. Hdac10 KO mice have similar body weight to wild-type (WT) mice (Supplementary Fig. S1E). The deletion of Hdac10 alone in mice did not cause known spontaneous carcinogenesis.
To characterize the role of HDAC10 in KRAS-induced lung cancer development, we employed the KrasLA1 mouse model, in which the latent G12D mutant Kras gene can become spontaneously activated by sporadic recombination and causes 100% of animals to develop multifocal lung adenocarcinoma(17). Hdac10 KO mice were crossed with KrasLA1 mice to generate littermate Hdac10 KO homozygous (Hdac10−/−, KrasLA1), heterozygous (Hdac10+/−, KrasLA1) and WT (Hdac10+/+, KrasLA1) mice. The effect of Hdac10 deletion on lung cancer development was determined by measuring the number and size of visible lesions together with histological analysis. The results indicated that deletion of Hdac10 can cooperate with the Kras mutation to accelerate tumor development in the lung (Figs. 1A–C). Compared with Hdac10 WT littermates, both Hdac10 KO homozygous and heterozygous mice displayed a significant increase in the total number of tumor nodules and tumor burden (Figs. 1A and 1C). The loss of HDAC10 predominantly caused an increase in the number of small nodules (<1 mm) instead of large nodules (>1 mm) at 8 weeks of age (Fig. 1B).
Figure 1. The role of Hdac10 deletion in KrasG12D-driven lung tumorigenesis.
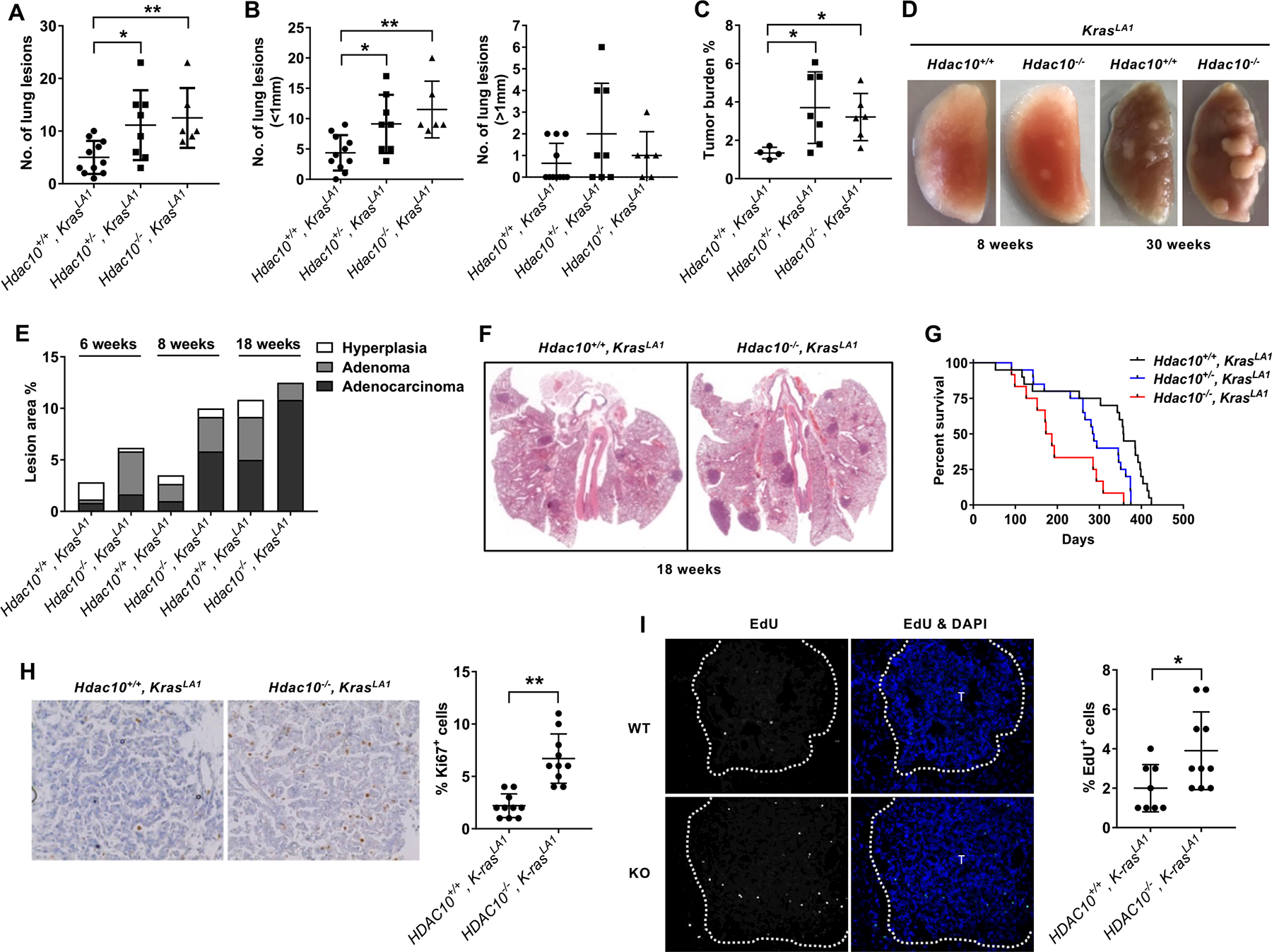
(A-B) The number of tumor nodules was counted on the lung surface of mice at 8 weeks of age. (A) The total number of tumor nodules on the lungs of mice with different genotypes. (B) The number of tumor nodules with a diameter less than 1 mm (left panel) or greater than 1 mm (right panel). (C) Tumor burden of mice at 8 weeks of age is quantified on the lung H&E cross-section as the ratio of total tumor area to total lung area. (D) Gross morphology of the left lobes of the lungs isolated from littermate Hdac10 WT and KO KrasLA1 mice. (E) Histopathological analysis of tumors in the lungs of Hdac10+/+, KrasLA1 and Hdac10−/−, KrasLA1 mice at 6, 8 and 18 weeks of age. (F) Hematoxylin and eosin staining of the lungs of Hdac10+/+, KrasLA1 and Hdac10−/−, KrasLA1 mice at 18 weeks of age. (G) The Kaplan–Meier survival of mice with different genotypes. (H) Immunohistochemistry of Ki67, counterstained with hematoxylin (nucleus), is illustrated (left panel). Ki67 positive cells were counted and graphed (right panel). (I) Representative images showing that Hdac10 KO mice exhibited more EdU positive cells than control mice (left). The right panel is the quantification of EdU-positive cell percentage. *, P < 0.05 or **, P < 0.01.
The malignant progression of lung tumors was graded by histopathological analysis according to the recommendation of the mouse models of human cancers consortium (21). In the mouse pulmonary pathologic criteria, hyperplasia, which is not included in the human classifications, is the initial stages of carcinogenesis and will further progress to adenoma. The proliferative lesions of the lung will develop, ranging from hyperplasia, to adenoma to adenocarcinomas (21). Representative hematoxylin and eosin staining of the lung tumors with hyperplasia, adenoma, and adenocarcinoma are indicated in Supplementary Fig. S2A. Hdac10 KO mice had a higher incidence of adenoma at 6 weeks and exhibited increased conversion of adenoma to adenocarcinoma after 8 weeks, suggesting that the deletion of Hdac10 promotes tumor initiation and progression through a series of morphological stages from mild hyperplasia and adenoma to overt lung adenocarcinoma (Figs. 1D–F). Furthermore, this pro-tumor effect led to a significant reduction in survival in Hdac10 KO mice. The median survival time for Hdac10 KO mice was 180 days, while the median survival time for Hdac10 heterozygous and WT mice are 285.5 and 357.5 days (Fig. 1G). In addition, the decrease in survival time occurs in both male and female Hdac10 KO mice (Supplementary Fig. S2B). Together, these data support that oncogenic driver mutations and the deletion of Hdac10 synergistically promote tumor growth and contribute to the poor prognosis of lung adenocarcinoma.
Tumor growth can be reflected by the ability of cells to proliferate. To further investigate the effect of Hdac10 deletion in KRAS-induced tumor growth, the cell proliferation in tumors was examined by immunohistochemistry with anti-Ki-67 antibody and EdU incorporation assay. As shown in Fig. 1H, immunohistochemical analysis of Ki-67 indicate that the number of Ki-67 positive cells in tumor lesions was markedly increased in Hdac10 KO mice compared with WT control mice. Furthermore, the deletion of Hdac10 also increased the percentage of EdU-labeled cells (Fig. 1I). Consistent with the promotion of KRAS-driven tumor growth by Hdac10 deletion, tumors in Hdac10 KO mice exhibit increased cell proliferation compared with Hdac10 WT tumors. Cell apoptosis in tumor tissues was detected using TUNEL assay based on the detection of DNA strand breaks. There were no significant differences in apoptosis between Hdac10 WT and KO tumors (Supplementary Fig. S2C).
The absence of HDAC10 induces massive recruitment of macrophages
To understand the molecular consequences of Hdac10 deletion in vivo, tumors were micro-dissected, and Hdac10 WT and KO KrasLA1mice (Hdac10+/+, KrasLA1 and Hdac10−/−, KrasLA1) and a comparative gene expression analysis was performed by RNA sequencing, followed by Ingenuity Pathway Analysis (IPA). As shown in Fig. 2A, differentially expressed genes were categorized by function, and a significant increase in agranulocyte and granulocyte adhesion and diapedesis indicate that Hdac10 deletion might modulate lung inflammation along with carcinogenesis.
Figure 2. The absence of HDAC10 induces massive macrophage infiltration into the tumor.
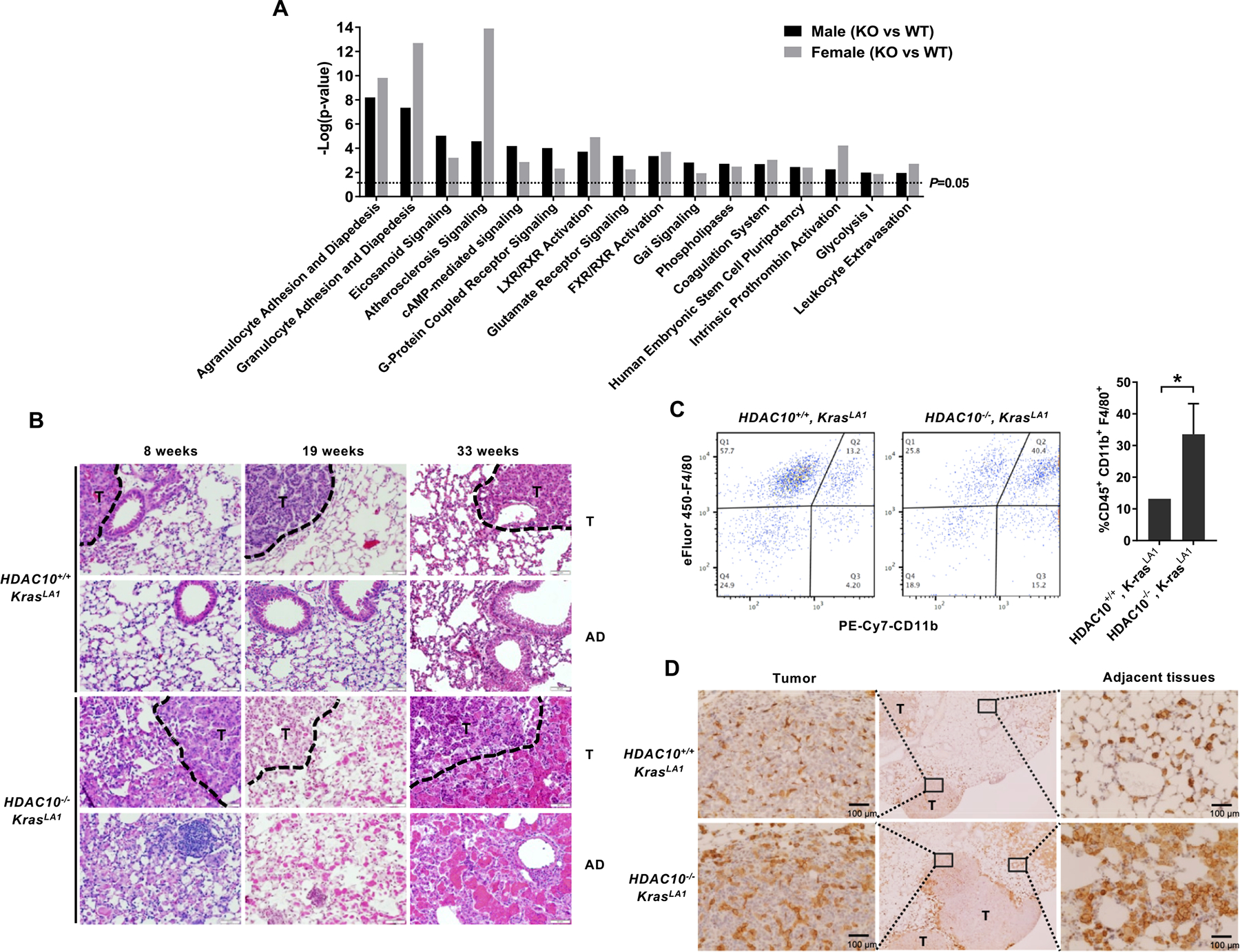
(A) Ingenuity Pathway Analysis of RNA sequencing data. Notable gene sets are listed with normalized enrichment scores for each group according to RNA sequencing data. (B) Representative H&E histology of lung sections from Hdac10 KO and WT KrasLA1 mice. Tumor adjacent (AD) and tumor (T) areas are indicated. (C) Representative flow cytometric analysis of macrophage populations from Hdac10+/+, KrasLA1 and Hdac10−/−, KrasLA1 mice (left panel) and the quantification of macrophage population among two groups (n=3). The cells were gated on live CD45+ single cells and subsequently CD11b+, F4/80+ macrophages were selected. (D) Immunohistochemistry of F4/80+ macrophages in lung sections. Tumor (T) and tumor adjacent areas are indicated.
The lung inflammation was investigated by standard histopathological analysis on H&E stained sections. The loss of HDAC10 in KrasLA1mice led to dramatically increased inflammatory response by enhancing macrophage recruitment into alveoli and within tumor foci (Fig. 2B). The high density of macrophage (CD45+ F4/80+ CD11b+) infiltration in tumors was validated by flow cytometry analysis (Fig. 2C). The immunohistochemical analysis of murine macrophage marker F4/80 indicated a significant increase in macrophage recruitment in Hdac10 KO tumors, predominantly in the area adjacent to and within tumors (Fig. 2D). This massive macrophage recruitment occurred with high incidence in the lung of Hdac10 KO mice at different ages (Supplementary Table. S2). Since KrasG12D activation has been reported to cause accumulation of alveolar macrophages in lung tumors(22), Hdac10 deletion can accelerate both KRAS-driven early onset lung adenocarcinoma and macrophage infiltration in the tumor microenvironment. The high density of macrophage infiltration in the lungs of Hdac10 KO mice can cause changes in size, color and structural rigidity of lung parenchymal tissue, resulting in lobar consolidation and decreased elastic recoil. Moreover, we found MAC-2 (Galectin 3) positive tumor-associated macrophages (TAMs) were highly infiltrated into the tumor microenvironment in Hdac10-deleted tumors (Supplementary Fig. S2D). MAC-2 is up-regulated in M2-type macrophages and is considered as a marker of M2 skewing (23, 24). M2 macrophages have been reported to exert a tumor-promoting activity by secreting a series of anti-inflammatory molecules (25). Hdac10 deletion not only accelerates KRAS-driven lung tumorigenesis, but also increases tumor-associated inflammation in the tumor microenvironment.
HDAC10 modulates stem-like properties in cancer cells
Given that the deletion of HDAC10 can enhance macrophage recruitment into the tumor microenvironment, we questioned whether HDAC10 could regulate tumor intrinsic malignancy in addition to the extrinsic regulation of the tumor microenvironment. The Ingenuity Pathway Analysis (IPA) of RNA sequencing data indicated that genes associated with embryonic stem cell pluripotency was increased in both male and female Hdac10 KO mice compared to WT mice (Fig. 2A). To determine whether HDAC10 can regulate the pluripotency of tumor cells, primary lung adenocarcinoma (LUAD) cells were isolated from Hdac10 WT and KO tumor tissues, and RT-PCR was performed to validate the activation of oncogenic KRAS in the primary cultured lung cancer cells. The expression of the mutant Kras allele can be detected in both tumor tissues and LUAD cells, but not in normal adjacent lung tissues (Supplementary Fig. S3A). The LUAD cells expressed alveolar type II cell lineage marker SFTPC (surfactant associated protein C), but not Clara cell marker SCGB1A1 (secretoglobin family 1A member 1), indicating that type II cells are the origin of LUAD cells (Supplementary Fig. S3B).
Next, the effect of Hdac10 deletion on tumor cell pluripotency was validated by the 3D tumor sphere assay. When the LUAD cells were cultured in an ultra-low-attachment plate at low cell density, Hdac10 KO LUAD cells gave rise to persistently proliferating tumor spheres, compared to Hdac10 WT LUAD cells (Figs. 3A and 3B). Overexpression of HDAC10 in the KO tumor cells reduced the sphere formation efficiency (Fig. 3C). These data indicate that HDAC10 suppresses the proliferative capacity of primary LUAD cells in 3D sphere culture. Furthermore, we established a xenograft tumor model to confirm the tumor initiating capacity (TIC) of Hdac10 KO lung tumor cells. We chose nude mice instead of syngeneic C57BL/6 mice because the immunity in syngeneic wild-type mice will negatively influence TIC readouts. Furthermore, it is easier to identify tumors and keep track of tumor size in nude mice. The limiting diluted Hdac10 WT and KO LUAD cells were subcutaneously transplanted into the left and right flank of athymic nude mice, respectively. Hdac10 KO tumor cells have relatively higher tumor incidence than Hdac10 WT tumor cells (Fig. 3D). Tumor weight measurements indicated that Hdac10 KO tumor cells grow much faster than WT tumor cells (Fig. 3E).
Figure 3. HDAC10 regulates tumor pluripotency.
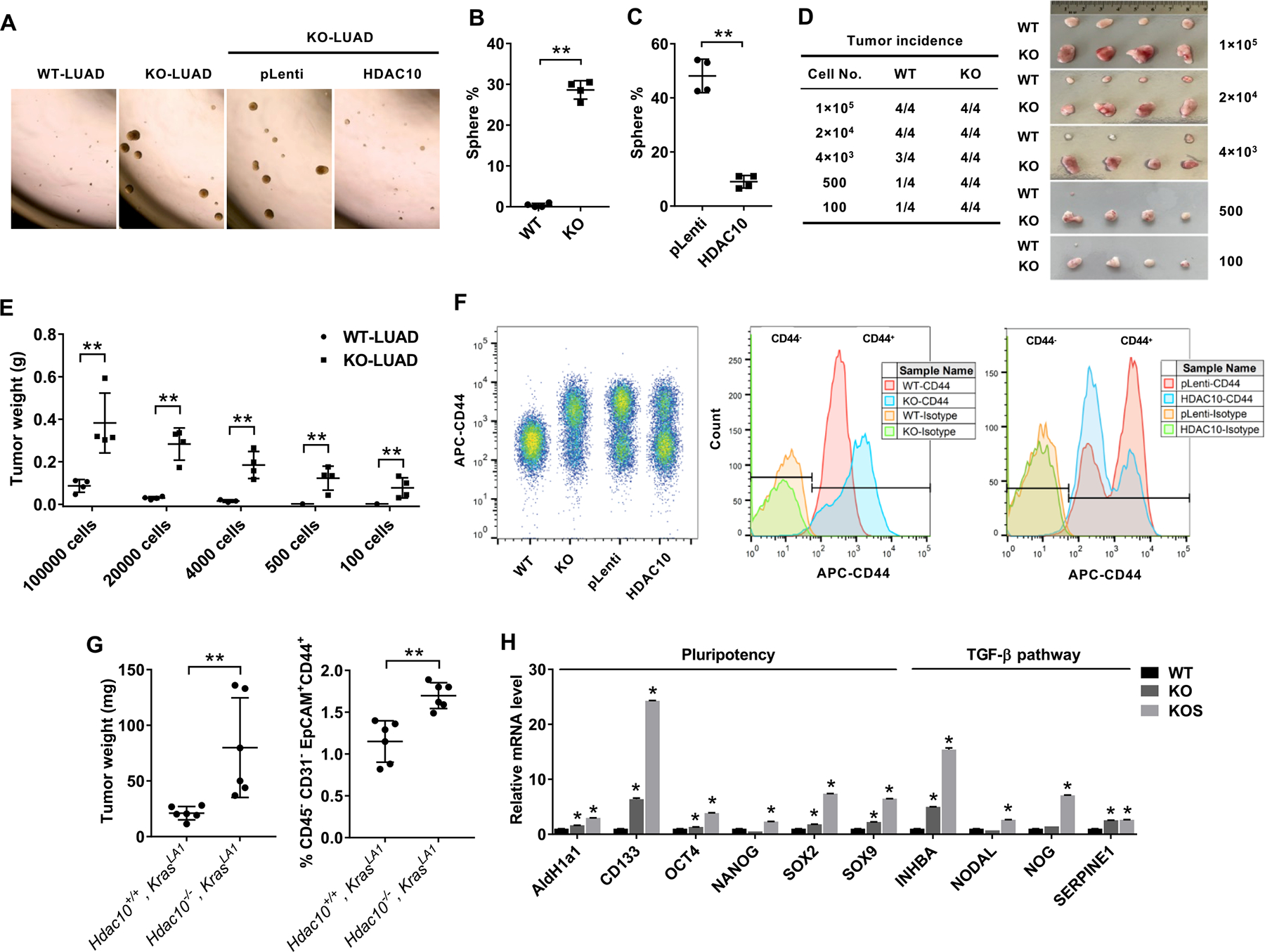
(A) Tumor sphere cultures of mouse LUAD cells 1 week after plating in a 24-well ultra-low attachment plate. (B) Quantification of the percentage of tumor spheres from Hdac10 WT and KO LUAD cells. (C) Quantification of tumor spheres from Hdac10 KO LUAD cells transduced with lentiviral control (pLenti) or Hdac10 expression vectors (HDAC10). (D) Tumor incidence of the in vivo limiting dilution assay. Representative image of tumors (right panel) and tumor incidence (left panel) at 3 weeks were indicated. (E) The tumors of the indicated groups from (D) were weighed. (F) Flow cytometric analysis of CD44 expression in LUAD cells. CD44 expression was increased in Hdac10 KO LUAD cells, whereas overexpression of HDAC10 in Hdac10 KO LUAD cells caused a decrease of CD44high population. (G) Flow cytometry showed a markedly increased CD45−CD31−EpCAM+CD44+ population in lung tumors from Hdac10−/−, KrasLA1mice at 30 weeks (right panel). (Left panel) The weights of total lung tumors from mice were indicated. (H) Results of real-time RT-PCR analysis of the expression of pluripotency transcription factors and TGF-β pathway in Hdac10 WT and KO LUAD, as well as KO LUAD spheres (KOS). *, P < 0.05 or **, P < 0.01.
CD44 has been reported as a cell surface marker for the enrichment of cancer stem cells, and it can promote oncogenic KRAS-induced adenocarcinoma(26). The level of CD44 was highly increased in Hdac10-deleted LUAD cells and there is a larger CD44high cell population in Hdac10 KO LUAD cells (70–80%) compared to WT LUAD cells (less than 10%) (Fig. 3F). Overexpression of HDAC10 caused a significant decrease in CD44 expression in Hdac10 KO LUAD cells and reduce the CD44high cell population from 65% to 34% compared to control vector (pLenti)-transduced cells (Fig. 3F). To further validate that there are more CD44-positive tumor cells in Hdac10 KO lung tumors, the tumor tissues were microdissected from Hdac10−/−, KrasLA1 and Hdac10+/+, KrasLA1 mice at 28–30 weeks, and the percentage of EpCAMpos, CD44pos cells in the CD45neg, CD31neg population were detected by flow cytometry. As shown in Figure 3G, the total tumor weights are higher in Hdac10 KO mice. Meanwhile, there are more CD44-positive cells in Hdac10 KO lung tumors compared with Hdac10 WT tumors.
To further validate that there are more highly tumorigenic or stem-like cells in Hdac10-deleted LUAD cells, the expression of known pluripotency transcription factors was assessed by real-time RT-PCR. The levels of OCT4, SOX2 and SOX9 were increased in Hdac10 KO LUAD cells, and were even higher in the proliferating tumor spheres of Hdac10 KO cells (Fig. 3H). Meanwhile, the levels of ALDH1A1 (aldehyde dehydrogenase 1 family member A1) and CD133, which are expressed in various cancer stem cells, were remarkably upregulated in Hdac10 KO cells (Fig. 3H). TGF-β is a regulator of cancer stemness and metastasis(27). We found that genes associated with TGF-β pathway were increased in Hdac10 KO LUAD cells (Fig. 3H). NOG is an antagonist of bone morphogenetic protein (BMP), a TGF-β superfamily protein. NOG has been reported to support the maintenance of pluripotency of human embryonic stem cells in vitro(28). We found that the NOG mRNA level was increased in Hdac10 KO tumor spheres (Fig. 3H). TGF-β is known to be a strong inducer of plasminogen activator inhibitor type-1 (PAI-1; SERPINE1)(29). SERPINE1 high expression has been linked with poor prognosis in a number of malignancies, including lung adenocarcinoma(30, 31). The mRNA level of SERPINE1 is increased in Hdac10 KO LUAD cells (Fig. 3H). Overall, the highly tumorigenic and stem-like cells are present in primary non-small cell lung cancer (NSCLC) tissues(18). HDAC10 may regulate oncogenic KRAS-driven lung cancer development through targeting the stem-like property of tumor cells.
HDAC10 regulates cancer cell stem-like properties by targeting SOX9
The dysregulation of SOX9, a stem cell transcriptional regulator, has been implicated in various types of cancers. Overexpression of SOX9 is found in more than 50% of lung adenocarcinomas, particularly those with Kras mutations, and a high level of SOX9 is associated with poor prognosis in patients(32, 33). SOX9 can promote the expansion of CSCs and epithelial-to-mesenchymal transition (EMT) by activating SLUG, and SLUG stabilizes SOX9 protein and subsequent tumorigenic function in NSCLC cells(34, 35). Given that the inhibition of HDACs in lung adenocarcinoma cell lines could elevate a SOX9-positive cell population(34), we hypothesize that HDAC10 might regulate cell pluripotency in KRAS-expressing tumors through targeting SOX9.
Western blotting analysis of SOX9 showed that the level of SOX9 was dramatically increased in Hdac10 KO lung tumors compared with Hdac10 WT tumors (Fig. 4A). The level of NOG and oncogenic KRAS (G12D) were also slightly increased, while FOXJ1 (Forkhead Box J1), a marker of a differentiated lineage-ciliated airway epithelial cells, was decreased in Hdac10 KO tumors (Fig. 4A). The upregulation of SOX9 might be associated with the high proliferation rate in lung tumor tissues, as demonstrated by immunohistochemical staining of SOX9 and Ki67 (Fig. 4B). The endogenous level of SOX9 is also significantly increased in isolated Hdac10 KO LUAD cells compared with Hdac10 WT LUAD (Figs. 4C and 4D). The tumor sphere cultured from Hdac10 KO LUAD cells exhibited an even higher level of SOX9 (Figs. 4E and 4F). NOG was also found to be upregulated in tumor sphere cells (Figs. 4E and 4F); however, the level of NOG is only increased in the early-passage (less than 8) Hdac10 KO cells (Fig. 4D), indicating that the induction of NOG in Hdac10 KO cells might be under the control of the tumor microenvironment in vivo.
Figure 4. SOX9 is upregulated in Hdac10-deleted tumor cells.
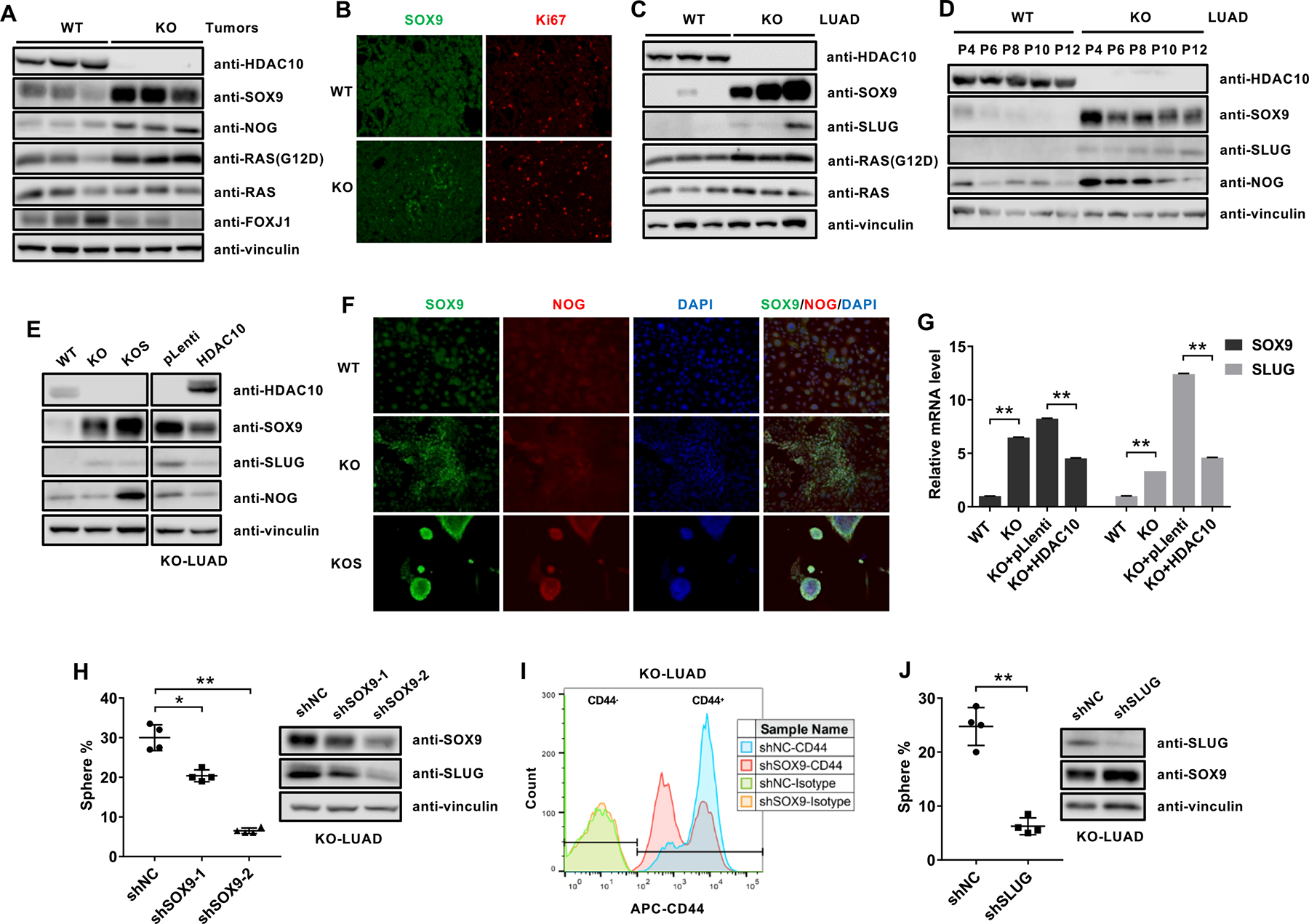
(A) Western analysis of the indicated proteins in Hdac10 WT and KO tumor tissues from mice at 30 weeks. (B) Immunostaining of SOX9 (green) and Ki67 (red) from mice at 30 weeks. (C) Western analysis of the indicated proteins in Hdac10 WT and KO LUAD cells isolated from different mice. (D) Western blot analysis of SOX9, SLUG and NOG expression in Hdac10 WT and KO LUAD cells with indicated passage number. (E) Western blot analysis results for the indicated proteins in Hdac10 WT, KO and KO spheres (KOS) LUAD cells (left panel), or Hdac10 KO LUAD cells transduced with the control vector or vector encoding HDAC10 (right panel). (F) Immunostaining of SOX9 (green), NOG (red) and DAPI (blue) in LUAD cells. (G) Results of real-time RT-PCR analysis of the expression of SOX9 and SLUG. (H) The effect of Sox9 depletion on the growth of tumor spheres in Hdac10 KO LUAD cells. Quantification of tumor spheres (left panel) and Western analysis (right panel) of SOX9 and SLUG in Hdac10 KO LUAD cells transduced with control (shNC) or SOX9 shRNA lentiviral vector (shSOX9–1 and shSOX9–2). (I) Flow cytometric analysis of the effect of SOX9 depletion on CD44 expression in Hdac10 KO LUAD cells. (J) The effect of SLUG depletion on the growth of tumor spheres in Hdac10 KO LUAD cells. *, P < 0.05 or **, P < 0.01.
Since SLUG and SOX9 could function cooperatively to induce and maintain tumor initiating capacity in breast cancer cells(36), we further evaluated the expression of SLUG in cultured LUAD cells. We found that SLUG was also upregulated in Hdac10-deleted LUAD cells (Figs. 4C–4E). The ectopic overexpression of HDAC10 can reduce both the protein and mRNA levels of SOX9 and SLUG in Hdac10 KO LUAD cells (Figs. 4E and 4G). To evaluate the effect of SOX9 on tumor cell pluripotency, endogenous SOX9 was knocked down by transducing SOX9-specific shRNA in Hdac10 KO LUAD cells, which have a high level of SOX9. The result showed that the loss of SOX9 in Hdac10 KO cells can cause a decrease in SLUG expression and inhibit the growth of tumor spheres (Fig. 4H). Meanwhile, knocking down of SOX9 in Hdac10-KO LUAD cells reduced the CD44high cell population from 80% to 43% compared to control shRNA (shNC)-transduced cells (Fig. 4I). Furthermore, depletion of SLUG also caused an inhibition of tumor sphere growth in Hdac10-KO LUAD cells (Fig. 4J). Collectively, these data demonstrate that SOX9 and SLUG, as downstream targets of HDAC10, may play a critical role in HDAC10-related CSC regulation and maintenance.
HDAC10 regulates SOX9 through the TGF-β pathway
Gene Set Enrichment Analysis (GSEA) indicates that genes associated with TGF-β, myogenesis and hypoxia pathways were enriched in Hdac10 KO tumors, whereas genes associated with E2F targets, G2/M checkpoint, interferon and inflammatory response, MYC targets, mitotic spindle and inflammatory response were enriched in Hdac10 WT tumors (Figs. 5A and 5B). The changes in gene expression in Hdac10 KO tumor tissues compared to Hdac10 WT tumor tissues were further confirmed by real-time RT-PCR (Fig. 5C). Consistent with our previous finding that HDAC10 regulates the cell cycle via upregulating cyclin A2 (CCNA2) expression in human lung cancer cell lines(15), the mRNA level of cyclin A2 was decreased in Hdac10 KO tumor tissues (Fig. 5C). TGF-β signaling related genes, such as TGFβ superfamily ligand (NODAL), BMPs antagonist (NOG), SMAD members (SMAD3, SMAD7) and inducible targets of TGFβ signaling (Serpine1, NOCR2), were increased in Hdac10 KO cells (Fig. 5C).
Figure 5. Activation of TGF-β pathway contributes to SOX9 and SLUG induction in Hdac10 KO LUAD cells.
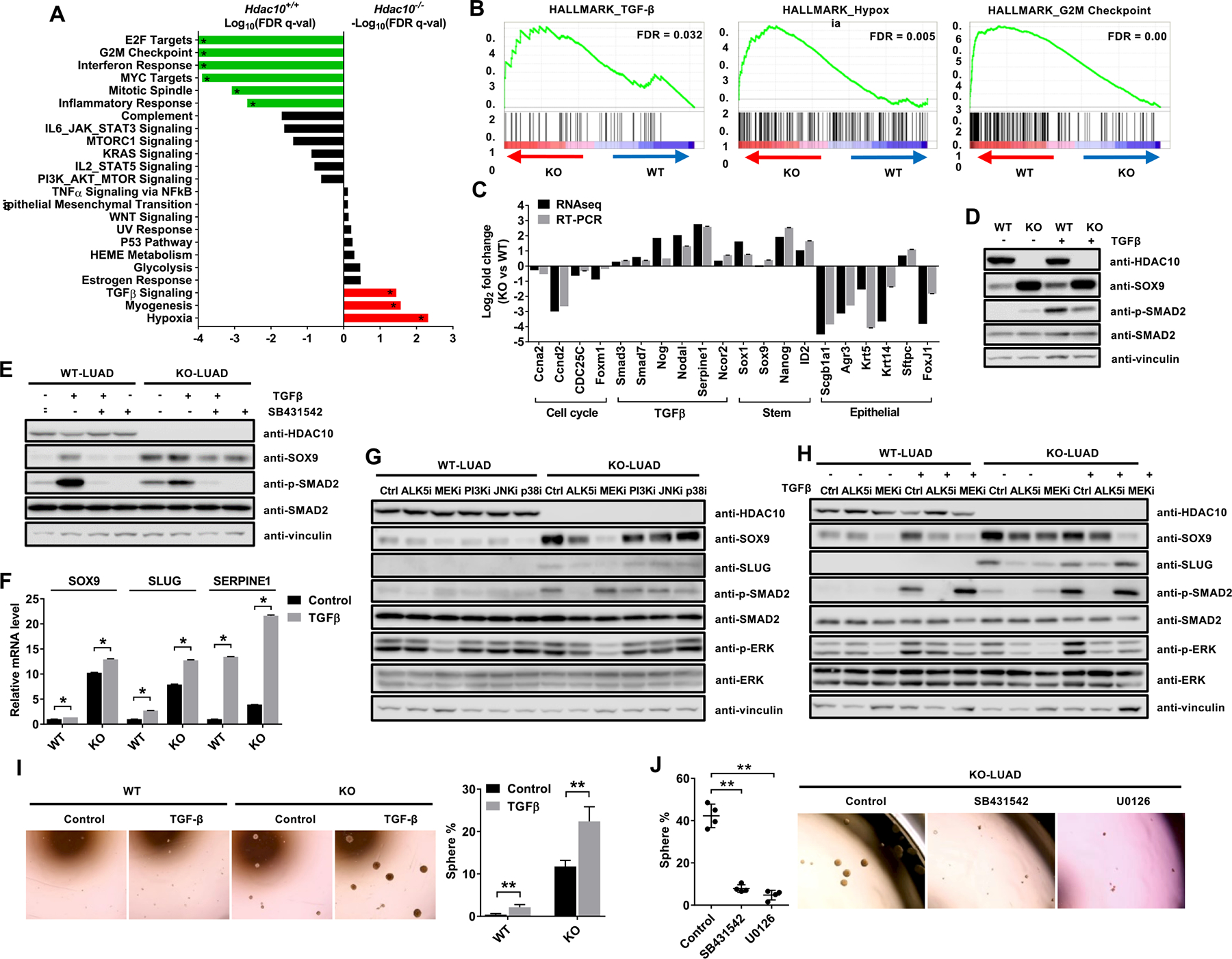
(A) The result of Gene set enrichment analysis (GSEA) of Hallmark Gene Sets. Notable gene sets, which are enriched in either Hdac10 WT (green) or KO (red) tumors, are listed with FDR q-value on a logarithmic scale. (B) GSEA plots showing that genes in TGF-β and hypoxia pathways were upregulated in Hdac10 KO tumors (left and middle panel), and genes associated with the G2M checkpoint were upregulated in Hdac10 WT tumors (right panel). (C) Results of comparative RNA sequence and real-time RT-PCR analysis of gene expression profiles of Hdac10 KO tumor compared with Hdac10 WT tumor. (D) Western blot analysis results for the indicated proteins in Hdac10 WT and KO LUAD cells after treatment with 5 ng/ml TGF-β or solvent control for 3 days. (E) Western analysis of the indicated proteins in LUAD cells treated with 5 ng/ml TGF-β or solvent control together with 10 μM SB431542 for 3 days. (F) Results of real-time RT-PCR analysis of the expression of SOX9, SLUG and SERPINE1 in Hdac10 WT and KO LUAD cells treated with TGF-β or solvent control. (G) Western analysis of the indicated proteins in Hdac10 WT and KO LUAD cells treated with 10 μM SB431542 (ALK5i), 10 μM U0126 (MEKi), 10 μM LY294002 (PI3Ki), 10 μM SP600125 (JNKi), 5 μM SB203580 (p38i) or solvent control for 3 days. (H) Western analysis of the indicated proteins in LUAD cells treated with 5 ng/ml TGF-β or solvent control together with 10 μM SB431542 (ALK5i) or 10 μM U0126 (MEKi) for 3 days. (I) Tumor sphere cultures of LUAD cells treated with TGF-β or solvent control (left panel). Quantification of the percentage of tumor spheres is indicated in the right panel. (J) Tumor sphere assay result of Hdac10 KO LUAD cells treated with SB431542 (ALK5i), U0126 (MEKi) or solvent control. (Left) Quantified percentage of spheres in tumor sphere assay. *, P < 0.05 or **, P < 0.01.
Given that genes associated with the TGF-β pathway were enriched in Hdac10 KO tumor cells and the transcription of SLUG and SOX9 have been reported to be under the control of TGF-β pathway in many cell types(37–40), we next examined whether HDAC10 regulates SOX9 and SLUG expression through the TGF-β pathway. The activation of SMAD2 was evaluated by Western blot using anti-p-SMAD2 antibody. The result indicated that the level of SMAD2 phosphorylation is increased in Hdac10 KO LUAD cells compared to WT LUAD cells (Fig. 5D). TGF-β stimulation could increase SOX9 expression by approximately 2-fold in Hdac10 WT LUAD cells and the induction can be rescued by treatment with SB431542, a selective inhibitor of TGF-β activin receptor-like kinases (ALK5), indicating the specific effect of TGF-β on SOX9 induction (Figs. 5D and 5E). TGF-β exposure can also significantly increase SLUG and SERPINE1 mRNA levels, as has been reported (Fig. 5F).
To determine which signaling events downstream of TGF-β is involved in SOX9 expression regulation, we used SMAD and non-SMAD pathway inhibitors (ALK5 inhibitor: SB431542; MEK inhibitor: U0126; PI3K inhibitor: LY294002; JNK inhibitor: SP600125; p38 MAPK inhibitor: SB203580) to treat Hdac10 WT and KO LUAD cells. Treatment with MEK inhibitor (U0126) resulted in a decrease in ERK phosphorylation, as well as a significant reduction in SOX9 and SLUG levels in Hdac10 KO LUAD (Figs. 5G). Treatment with SB431542, which targets the SMAD pathway, caused a decrease in SMAD2 phosphorylation and reduced SOX9 level in Hdac10 KO LUAD cells (Figs. 5E and 5G). To further evaluate that SOX9 induction in Hdac10 KO cells is dependent on TGF-β signaling, we treated Hdac10 WT and KO LUAD cells with TGF-β or a solvent control together with SB431542 or U0126 (Fig. 5H). In Hdac10 WT LUAD cells, TGF-β stimulation could significantly increase SMAD2 and ERK phosphorylation and upregulate SOX9 expression. SB431542 inhibited SMAD2 signaling induced by TGF-β and downregulated SOX9 level. SOX9 induction by TGF-β could also be rescued by treatment with U0126, a potent inhibitor of MEK, which has no direct effect on the SMAD pathway. U0126 treatment could also reduce SOX9 level in control-treated cells, indicating MEK/ERK signaling is involved in regulating SOX9 basal expression (Fig. 5H). SOX9 has been found to be required for the mesenchymal activation of the ERK/MAPK cascade during the development of the respiratory tract and MEK constitutive activation in mouse enhances Sox9 expression (41). In Hdac10 KO LUAD cells, the endogenous SOX9 level was much higher than in WT LUAD cells and TGF-β treatment could not further upregulate SOX9 expression, which might be due to elevated SOX9 already being saturated in Hdac10 KO LUAD. SB431542 and U0126 treatment could block the activation of SMAD and MEK/ERK signaling, respectively, and reduce SOX9 expression (Fig. 5H). These results indicate that TGF-β1 regulates SOX9 expression via differential activation of ALK5/SMAD2 and MEK/ERK signaling. In addition, consistent with the effect of TGF-β on SOX9 induction, TGF-β stimulation could promote tumor sphere growth in both Hdac10 WT and KO LUAD cells (Fig. 5I). U0126 or SB431542 treatment can inhibit the growth of tumor spheres in Hdac10 KO LUAD cells (Fig. 5J). These findings suggest that both SMAD and non-SMAD TGF-β pathways play a role in SOX9 expression regulation, as well as in tumor cell stemness maintenance.
In addition to SOX9, the Ingenuity Pathway Analysis (IPA) of RNA sequencing data indicates that genes associated with embryonic stem cell (ESCs) pluripotency were enriched in Hdac10 KO tumors, including NODAL (TGF-β superfamily ligand), NOG (antagonist of BMPs), NANOG (ESCs transcription factor) and WNT pathway members, like WNT3, WNT7B, WNT8B and WNT10B (Supplementary Fig. S4A). Upregulated expression of these pluripotency genes in Hdac10 KO LUAD cells was validated by real-time RT-PCR (Supplementary Fig. S4B). To demonstrate whether the expression of these genes is also under the control of TGF-β signaling, we treated Hdac10 WT and KO LUAD cells with TGF-β. TGF-β stimulation could significantly increase the level of SOX9, NODAL, NOG, NANOG, WNT3, WNT7B and WNT10B in Hdac10 WT LUAD cells, but not in Hdac10 KO LUAD cells (Supplementary Fig. S4B). The difference in sensitivity to TGF-β stimulation between Hdac10 WT and KO cells could be a result of either HDAC10’s requirement for TGF-β-mediated induction of pluripotency genes or the pre-existing highly elevated expression levels of these genes in Hdac10 KO LUAD cells. Moreover, treatment with MEK inhibitor (U0126) and ALK5 inhibitor (SB431542) can simultaneously reduce the level of SOX9 in Hdac10 KO LUAD cells (Supplementary Fig. S4C), indicating that the elevated SOX9 transcripts in Hdac10 KO cells is TGF-β pathway-dependent. However, other pluripotency genes cannot consistently be reduced by U0126 and SB431542 treatment (Supplementary Fig. S4C). Together, these results demonstrate that the expression of SOX9 is under the control of TGF-β signaling. The activation of the TGF-β pathway in Hdac10 KO tumor cells upregulates SOX9 expression, and elevated SOX9 level further favors CSCs maintenance and propagation and enhances lung tumorigenesis.
HDAC10 represses SOX9 expression in human lung cancer cells
Our results indicate that HDAC10 might function as a putative tumor suppressor in murine lung cancer development; however, the potential relevance of HDAC10 to human lung cancer is not yet clear. The genetic alterations of HDAC10 in different human lung cancer cohort studies was obtained from cBio Cancer Genomics Portal (42, 43). HDAC10 gene is infrequently altered in lung cancer (less than 3%) and the alteration of HDAC10 is mainly contributed by gene deletion (2% in Lung Adenocarcinoma TCGA cohort) (Supplementary Fig. S5A). In addition, HDAC10 is not genetically altered with KRAS-mutation in lung adenocarcinoma, because genomic alterations of HDAC10 and KRAS are mutually exclusive (Supplementary Fig. S5B).
Then, we examined the level of HDAC10 transcript in human lung adenocarcinoma (LUAD) and normal samples using TCGA and GTEx datasets from GEPIA (44). The expression of HDAC10 is significantly reduced in lung adenocarcinoma compared to matched TCGA normal and GTEx normal lung tissues (Fig. 6A). Expression of HDAC10 in lung adenocarcinoma was further examined in KRAS wild-type and mutant tumors using TCGA LUAD data. The level of HDAC10 is significantly decreased in KRAS mutant tumors compared to KRAS wild-type tumors (Fig. 6B). However, there is no significant correlation between HDAC10 expression and tumor stages in LUAD (Fig. 6C). Kaplan-Meier analysis is used to study the effect of HDAC10 expression on the overall survival of lung adenocarcinoma patients. A survival curve gained from Human Protein Atlas (HPA) shows that low HDAC10 expression correlates with worse survival of lung cancer patients (Fig. 6D). The median survival time for high and low-HDAC10 are 7.178 and 3.778 years, respectively. Furthermore, high SOX9 expression associates with a poor prognosis in human lung cancer patients (Fig. 6D). The median survival time for high and low-SOX9 are 3.279 and 4.186 years, respectively. These results indicate that HDAC10 transcript is decreased in human lung adenocarcinoma and, consistent with an earlier report (14), the low expression of HDAC10 is associated with poor overall survival in human lung cancer patients.
Figure 6. HDAC10 expression in human lung adenocarcinoma and their correlation to patient survival.
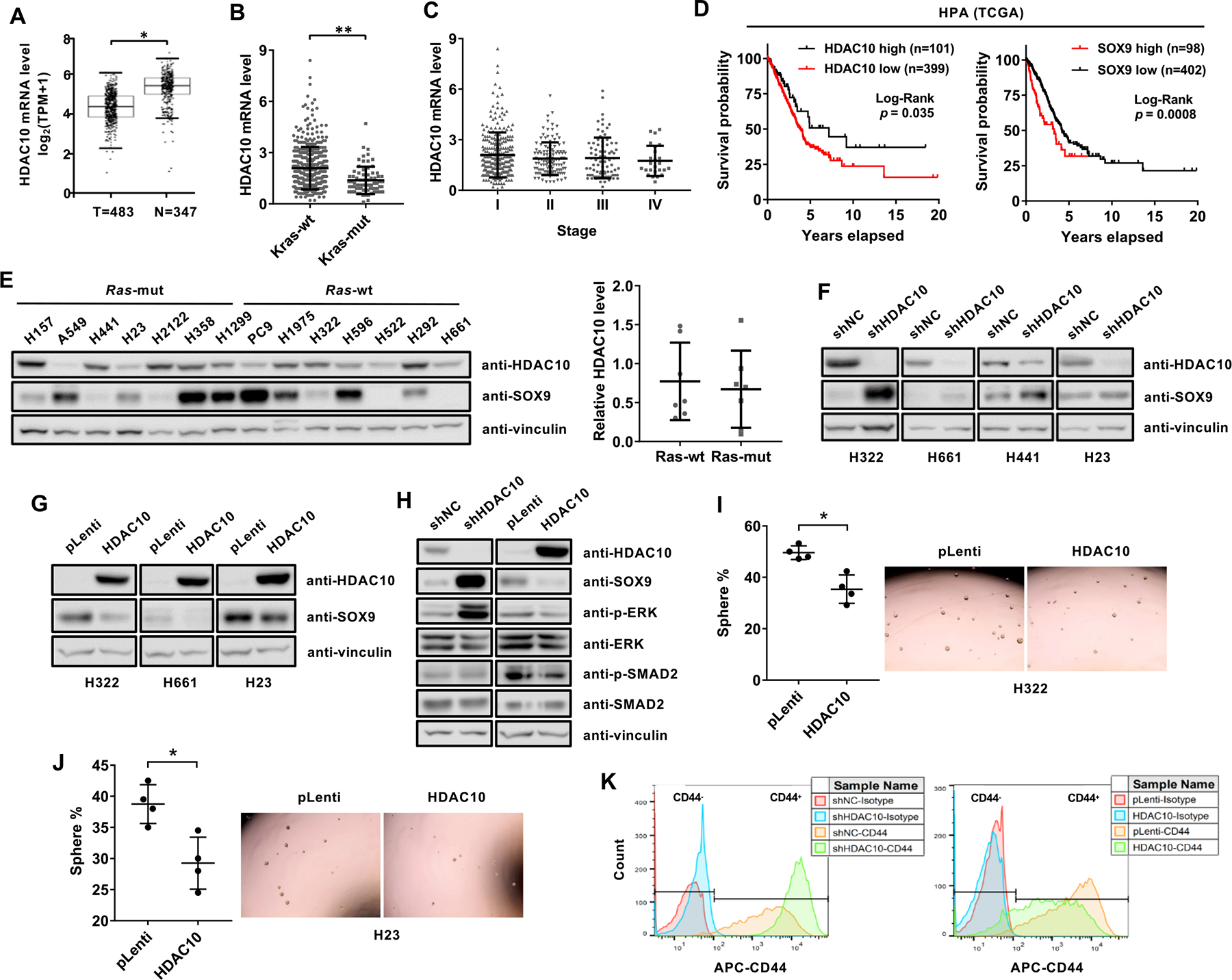
(A) The log2(TPM+1) transformed expression of HDAC10 mRNA in human lung adenocarcinoma and normal tissues from GEPIA. (B) HDAC10 mRNA level in KRAS wild-type (wt) and mutant (mut) tumor tissues using TCGA LUAD dataset. HDAC10 transcript level is indicated as FPKM (Fragments Per Kilobase Million) downloaded from the HPA database. (C) HDAC10 mRNA in different tumor stages from TCGA LUAD data. (D) Kaplan-Meier curves from the HPA database showing the correlation between the expression of HDAC10 (left panel) and SOX9 (right panel) with overall survival of lung adenocarcinoma patients. The cutoff FPKM value of HDAC10 and SOX9 expression are 2.8 and 17, respectively. (E) Western analysis of HDAC10 and SOX9 protein level in a panel of human lung cancer cell lines (left panel) and the semi-quantitative expression of HDAC10 in RAS wild-type (wt) and mutant (mut) cell lines (right panel). (F) Western blot analysis of HDAC10 and SOX9 expression in the indicated cell types expressing control shRNA (shNC) or shRNA targeting HDAC10 (shHDAC10). (G) Western blot analysis of HDAC10 and SOX9 expression in the indicated cell types expressing control vector (pLenti) or HDAC10 expression vector (HDAC10). (H) Western analysis of the indicated proteins in HDAC10 knockdown or HDAC10 overexpressed H322 cells (I) Tumor sphere cultures of H322 cells transduced with control (pLenti) or HDAC10 expression vectors (HDAC10) (right panel) and quantification of tumor spheres (left panel). (J) Tumor sphere cultures of H23 cells (right panel) and quantification of tumor spheres (left panel). (K) Flow cytometric analysis of CD44 expression in HDAC10 knockdown (left panel) or HDAC10 overexpressed H23 cells (right panel). *, P < 0.05 or **, P < 0.01.
To study the effect of HDAC10 on SOX9 expression in human lung cancer cells, the endogenous protein level of HDAC10 and SOX9 were assessed by Western analysis in a panel of human NSCLC lines (Fig. 6E). The negative correlation between HDAC10 and SOX9 in NSCLC lines did not reach statistical significance. Although HDAC10 expression was reduced in KRAS mutant tumors compared to KRAS wild-type tumors in TCGA LUAD data (Fig. 6B), the expression of HDAC10 did not show a signification association with RAS mutation status in NSCLC lines (Fig. 6E). To evaluate the effect of HDAC10 depletion on SOX9 expression, the lentiviral system was used to transduce HDAC10-specific shRNA into H322, H661, H23 and H441 cells. Western blot analysis indicates that HDAC10 knockdown significantly increased SOX9 expression in human lung cancer cell lines (Fig. 6F). Meanwhile, HDAC10 overexpression could inhibit the expression of SOX9 (Fig. 6G) and the effect of HDAC10 on SOX9 expression is not KRAS dependent because HDAC10 could also repress SOX9 transcription in RAS wild-type cells, such as H322 and H661 cells. Consistent with the finding that HDAC10 regulates SOX9 transcription through TGFβ signaling pathway in mouse LUAD cells, HDAC10 knockdown increased the level of p-SMAD2 and p-ERK, while overexpression of HDAC10 decreased the level of p-SMAD2 and p-ERK in H322 cells (Fig. 6H). Moreover, ectopic expression of HDAC10 in H322 and H23 cells can inhibit sphere formation efficiency of cells (Figs. 6I and 6J). Given that CD44 is expressed in H23 cells, overexpression of HDAC10 caused a significant decrease in CD44 expression and knockdown of HDAC10 caused a significant increase in CD44 expression (Fig. 6K). Collectively, these results indicate that HDAC10 might regulate cancer stem-like cells in human lung cancer via targeting SOX9.
Discussion
We investigated the effects of HDAC10 on tumor initiation and progression using a KRAS-driven lung cancer mouse model. A putative tumor suppressor role of HDAC10 was seen in mice carrying spontaneously activated oncogenic alleles of Kras. Hdac10 deletion can accelerate KRAS-driven early onset lung adenocarcinoma and shorten survival time in mice. These results are consistent with human clinical data, in which a reduction in HDAC10 mRNA level was found in lung adenocarcinoma and low HDAC10 expression is associated with shorter survival time in human lung cancer patients(14). In our previous study, we found that transient HDAC10 knockdown in human lung cancer cell lines widely inhibits mitotic entry and suppresses cell proliferation, suggesting that HDAC10 may be required for the growth of tumor cells(15). The tumor suppressor role of HDAC10 in the KrasLA1 mouse model therefore seems contradictory to HDAC10’s effect on cell cycle regulation in established lung cancer cell lines. Conceivably, HDAC10 most likely plays another role in a different cell context. HDAC10 may function in surveillance in vivo, preventing oncogenic KRAS-induced malignant transformation and tumor development. Thus, Hdac10 deletion promoted tumor growth in mice carrying KrasLA1alleles. However, for established malignant cells, HDAC10 is critical for unchecked cell growth, so transient HDAC10 depletion and inhibition suppress tumor cell growth. In addition, the phenotypic discrepancies between gene knockout and knockdown might due to a genetic compensation response or transcriptional adaptation, which are widespread phenomenon in response to gene knockout (45). Although cyclin A2 mRNA level was reduced in Hdac10 KO tumors and LUAD cells, we did not detect a cell cycle defect or reduction of cyclin A2 protein level in Hdac10 KO LUAD compared to WT LUAD cells. Furthermore, although HDAC10 and HDAC6 belong to the same HDAC subclass, HDAC6 is required for RAS-mediated oncogenicity by modulating KRAS acetylation(46). Hdac6-null MEFs are resistant to both oncogenic RAS and ErbB2-induced transformation, and Hdac6-null mice are also resistant to chemical carcinogen-induced skin tumors (47), suggesting that individual HDACs may play different roles in malignant transformation. Thus, a better understanding of the context-dependent effects of individual HDAC enzymes will provide a rationale for targeting HDACs in cancer therapy.
To dissect the mechanism by which HDAC10 regulates tumor initiation, we found that HDAC10 regulates stem-like properties of cancer cells. Cancer stem-like cells (CSCs) have gained increasing attention recently in cancer research. These cells can generate new tumors through their stem cell properties, essentially self-renewal potential and differentiation into multiple cell lineages(48). Highly tumorigenic and stem-like cells have been reported in primary non-small cell lung cancer (NSCLC) tissues(18, 48, 49). CSCs of lung cancer are well recognized by their specific markers, such as CD133, CD44, ABCG2, and ALDH1A1, and exhibits biological characteristics associated with both cancer and stem cells, including spheroid formation, ability to initiate tumor growth and multi-drug resistance(48). Our studies indicate that HDAC10 plays a role in CSCs regulation. First, comparative RNA sequencing data showed that the genes associated with embryonic stem cell pluripotency are enriched in Hdac10 KO tumors. Second, primary cultured LUAD cells isolated from Hdac10 KO tumors exhibited stem-like cell properties. Hdac10 KO LUAD cells could give rise to persistently proliferating tumor spheres and exhibit a greater tumor-initiating capacity in recipient mice. Furthermore, the number of CD44pos cancer cells was significantly upregulated in Hdac10 KO lung tumors versus Hdac10 WT tumors. Lastly, SOX9 and SLUG, stem cell transcriptional regulators, are markedly increased in Hdac10 KO LUAD cells, and knockdown of either SOX9 or SLUG in Hdac10-KO LUAD cells can inhibit its sphere formation efficiency. These results convincingly argue that HDAC10 might regulate cancer stem-like cells in KRAS-expressing tumors via the pluripotency pathway.
SRY (sex determining region Y)-box 9 (SOX9) is a member of the SOX family of transcription factors which possesses the homologous high mobility group (HMG)-box domain. SOX9 function has been widely studied as a key regulator of cartilage, male gonad development as well as lung branching morphogenesis(20). Aberrant expression of SOX9 has been found in many tumor types, including lung cancer(32, 50–52). SOX9 is highly expressed in more than 50% of human lung adenocarcinomas and is associated with poor patient survival(32). SOX9, together with other transcription factors such as SLUG, a member of the Snail family, is involved in regulating CSCs and tumor metastasis(35, 36, 53). Overexpression of SOX9 and SLUG in breast cancer cells promotes the formation of tumor-initiating cells (TICs)(36). SLUG and SOX9 are also co-upregulated in advanced lung cancer and co-expression of SLUG and SOX9 regulates CSCs properties and lung metastasis(35). In our study, the expression of SOX9 is consistently upregulated in Hdac10 KO tumor tissues as well as isolated LUAD cells. Depletion of SOX9 can reduce SLUG expression, inhibit the proliferation of tumor sphere and decrease CD44 expression in Hdac10-KO LUAD cells, indicating that SOX9 unequivocally plays an important role in HDAC10-mediated CSCs regulation.
Given the pivotal role of SOX9 in CSCs regulation, the expression of SOX9 is targeted or modulated by multiple pathways during cancer development. TGF‐β, an essential regulator of cell polarity, growth, differentiation, and lineage specificity in multiple cell types, plays an important role in modulating the cancer stem cell niche(54, 55). We found that TGF-β signaling is activated in Hdac10 KO tumor cells. Compromising TGF-β signaling with either SMAD or non-SMAD pathway inhibitors (SB431542 or U0126) can decrease SOX9 and SLUG levels and inhibit the growth of tumor spheres in Hdac10 KO LUAD cells, indicating the important role of TGF-β pathway in HDAC10–mediated SOX9 and CSCs regulation.
Like HDAC10, other HDAC family members have been reported to regulate the TGF-β pathway in different cell contexts. Several studies have shown that HDAC inhibitor treatment can suppress TGF-β signaling either by directly modulating expression and/or activating TGF-β pathway components, such as SMAD2/3 and TGF-β type II receptor, or by indirectly silencing the expression of TGF-β downstream targets via histone deacetylation (56–59). Although HDAC inhibitors suppress TGF-β signaling, genetic deletion of some HDAC members might enhance the TGF-β pathway in a cell context-dependent manner. Conditional genetic deletion of HDAC1 and/or HDAC2 in ESC lines reduced endothelial-to-hematopoietic transition (EHT) through modulating TGF-β signaling and this defect could be rescued by treatment of SB431542, a selective inhibitor of the receptor ALK5 (60). HDAC3 was reported to repress the TGF-β signaling pathway during early cardiogenesis (61). HDAC3 epigenetically silences TGF-β1 transcription within mesenchymal cells in a deacetylase-independent manner. Deletion of HDAC3 caused cardiac defects by increasing TGF-β1 expression and activating SMAD2/3 phosphorylation. SIRT1 can also inhibit TGF-β pathway and ameliorate renal fibrosis by directly interacting with SMAD3 and modulating SMAD3 deacetylation (62). In our studies, we found that the deletion of Hdac10 activates TGF-β pathway in Kras-mutant tumor cells. However, how the TGF-β pathway is activated in Hdac10 KO tumors remains unclear. One potential mechanism is that the loss of HDAC10 upregulates the expression of NODAL, a TGFβ superfamily ligand, which further activates SMAD and non-SMAD TGF-β pathways in LUAD cells.
Hdac10 deletion not only accelerates KRAS-driven early onset lung adenocarcinoma, but also increases tumor-associated inflammation in the tumor microenvironment, which is characterized by a high density of macrophages in the tumor microenvironment. The predominant macrophage subtypes are MAC-2 positive cells which skew to M2-like macrophages. Macrophages can be polarized into different phenotypes: pro-inflammatory M1 macrophages or anti-inflammatory M2 macrophages. High infiltration of M2 macrophages in the tumor is associated with poor overall survival in NSCLC patients(63). TGF-β is one of the major cytokines secreted by M2 macrophages and M2 polarization has been observed in oncogenic KRAS-induced lung tumorigenesis in mice (64, 65). A recent study showed that TGFβ secreted by tumor-associated macrophages (TAMs) increases SOX9 expression and promotes the epithelial-to-mesenchymal transition of lung cancer cells(64). However, whether the increase of TAMs infiltration in Hdac10-deleted tumors plays a role in the paracrine activation of the TGFβ pathway still needs further investigation.
In summary, we showed that HDAC10 acts as a tumor suppressor in lung cancer development. Hdac10 deficiency in the presence of oncogenic KRAS activation accelerates lung tumorigenesis in mice (Fig. 7). The loss of HDAC10 increased early adenocarcinoma formation and lung macrophage infiltration, and further reduced survival time in mice. LUAD cells isolated from Hdac10 KO tumors exhibited highly tumorigenic and stem-like properties. The expression of CSCs surface marker CD44 and genes associated with stem cell pluripotency are increased in Hdac10 KO tumor cells. The high level of SOX9 in Hdac10 KO LUAD cells is required for the expansion and maintenance of CSCs. Given that the activation of TGF-β signaling contributes to SOX9 induction in Hdac10 KO LUAD cells, HDAC10 might regulate stem-like properties of KRAS-expressing tumor cells through targeting the TGF-β/SOX9 pathway. HDAC inhibitors have been pre-clinically or clinically used to treat cancers; however, inhibition of HDACs’ function might change the expression of specific genes that are beneficial for tumor development. In our study, we elucidate that the loss of HDAC10 promotes KRAS-driven lung tumorigenesis through enhancing CSC properties by SOX9. This finding provides the preclinical rationale for combination treatment of lung cancer by targeting both HDACs activity and pluripotency pathways.
Figure 7. A schematic proposed model of HDAC10 in KRAS-driven lung cancer development.
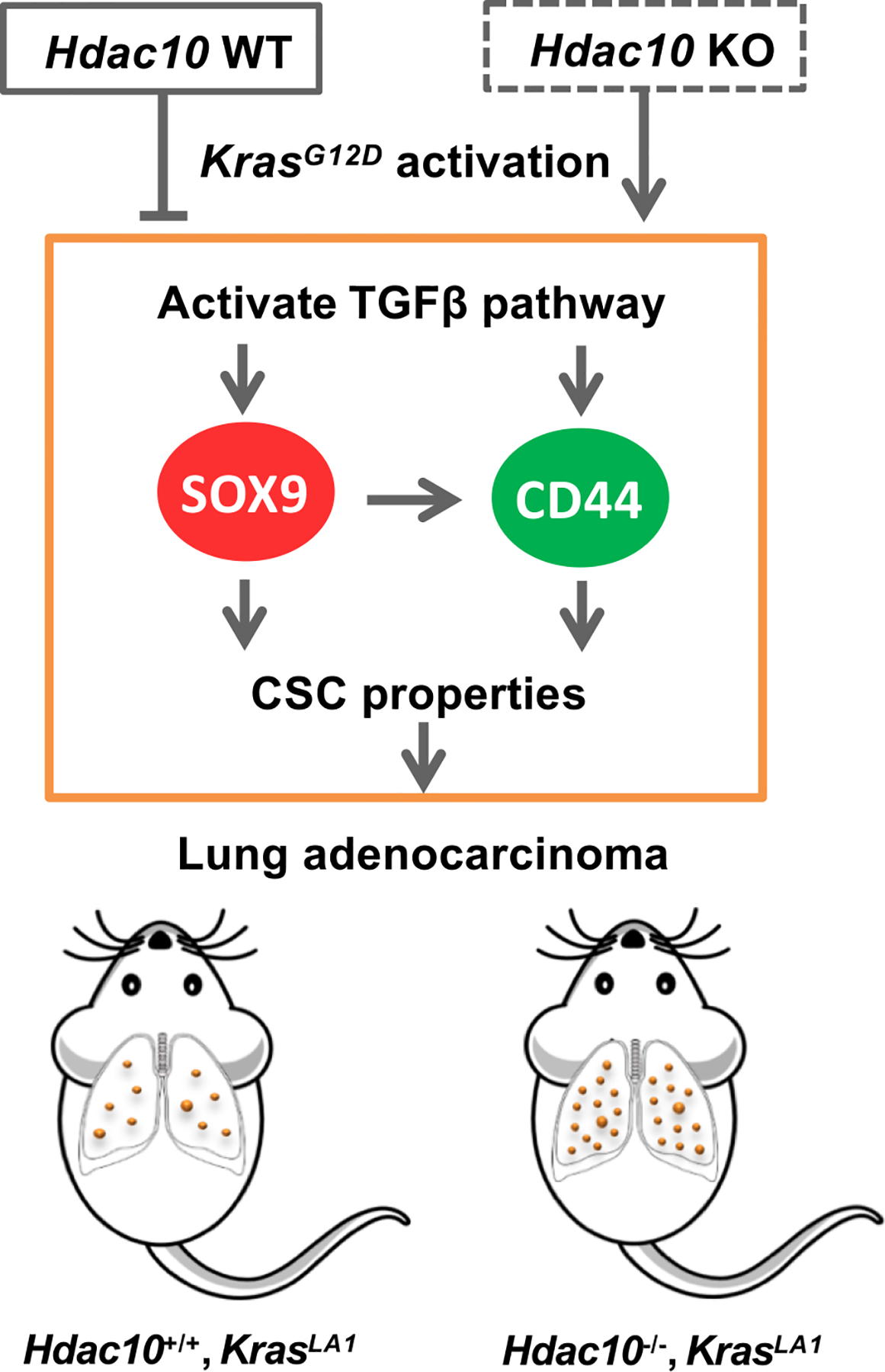
Hdac10 deficiency in the presence of oncogenic KRAS accelerates lung tumorigenesis in mice. The loss of HDAC10 induces SOX9 transcription and upregulates CD44 expression in LUAD cells by activating TGF-β signaling. HDAC10 regulates the stem-like properties of tumor cells through the TGF-β/SOX9 pathway.
Supplementary Material
Significance.
These findings linking HDAC10 and lung tumorigenesis identify potential novel strategies for targeting HDAC10 as a treatment for lung cancer.
Acknowledgements
We thank Lirong Peng, Hongbo Ling, Xiaoyan Zheng, Ya Zhang, plus other members of the Seto Lab, and the Moffitt Cancer Center Pathology Core for support and discussions. We are extremely grateful to Sonali Bahl for editing the manuscript. Funding for this work was awarded to ES (CA187040, CA169210), and to SZ and WP (AI121080 and AI139874), by the National Institutes of Health.
Footnotes
Disclosure of Potential Conflict of Interest: All authors declare that there are no conflicts of interest.
References
- 1.Siegel RL, Miller KD and Jemal A Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7–30 [DOI] [PubMed] [Google Scholar]
- 2.Roman M, Baraibar I, Lopez I, Nadal E, Rolfo C, Vicent S, et al. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol Cancer 2018;17:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox AD, Fesik SW, Kimmelman AC, Luo J and Der CJ Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014;13:828–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y and Seto E HDACs and HDAC Inhibitors in Cancer Development and Therapy. Cold Spring Harb Perspect Med 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada T, Amann JM, Tanimoto A, Taniguchi H, Shukuya T, Timmers C, et al. Histone Deacetylase Inhibition Enhances the Antitumor Activity of a MEK Inhibitor in Lung Cancer Cells Harboring RAS Mutations. Mol Cancer Ther 2018;17:17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greve G, Schiffmann I, Pfeifer D, Pantic M, Schuler J and Lubbert M The pan-HDAC inhibitor panobinostat acts as a sensitizer for erlotinib activity in EGFR-mutated and -wildtype non-small cell lung cancer cells. BMC Cancer 2015;15:947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeannot V, Busser B, Vanwonterghem L, Michallet S, Ferroudj S, Cokol M, et al. Synergistic activity of vorinostat combined with gefitinib but not with sorafenib in mutant KRAS human non-small cell lung cancers and hepatocarcinoma. Onco Targets Ther 2016;9:6843–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ropero S, Fraga MF, Ballestar E, Hamelin R, Yamamoto H, Boix-Chornet M, et al. A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet 2006;38:566–9 [DOI] [PubMed] [Google Scholar]
- 9.Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 2012;151:1185–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong JJ, Liu J, Bertos NR and Yang XJ Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res 2002;30:1114–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao HY, Lee CH, Komarov A, Han CC and Evans RM Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J Biol Chem 2002;277:187–93 [DOI] [PubMed] [Google Scholar]
- 12.Jin X, Yan Y, Wang D, Ding D, Ma T, Ye Z, et al. DUB3 Promotes BET Inhibitor Resistance and Cancer Progression by Deubiquitinating BRD4. Mol Cell 2018;71:592–605 e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hai Y, Shinsky SA, Porter NJ and Christianson DW Histone deacetylase 10 structure and molecular function as a polyamine deacetylase. Nat Commun 2017;8:15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osada H, Tatematsu Y, Saito H, Yatabe Y, Mitsudomi T and Takahashi T Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int J Cancer 2004;112:26–32 [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Peng L and Seto E Histone Deacetylase 10 Regulates the Cell Cycle G2/M Phase Transition via a Novel Let-7-HMGA2-Cyclin A2 Pathway. Mol Cell Biol 2015;35:3547–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Huang Y, Wang Z, Wang HT, Duan B, Ye D, et al. HDAC10 promotes lung cancer proliferation via AKT phosphorylation. Oncotarget 2016;7:59388–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 2001;410:1111–6 [DOI] [PubMed] [Google Scholar]
- 18.Tammela T, Sanchez-Rivera FJ, Cetinbas NM, Wu K, Joshi NS, Helenius K, et al. A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 2017;545:355–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kramer A, Green J, Pollard J Jr. and Tugendreich S Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30:523–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, et al. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res 2004;64:2307–16 [DOI] [PubMed] [Google Scholar]
- 22.Okayama H, Saito M, Oue N, Weiss JM, Stauffer J, Takenoshita S, et al. NOS2 enhances KRAS-induced lung carcinogenesis, inflammation and microRNA-21 expression. Int J Cancer 2013;132:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacKinnon AC, Farnworth SL, Hodkinson PS, Henderson NC, Atkinson KM, Leffler H, et al. Regulation of alternative macrophage activation by galectin-3. J Immunol 2008;180:2650–8 [DOI] [PubMed] [Google Scholar]
- 24.Novak R, Dabelic S and Dumic J Galectin-1 and galectin-3 expression profiles in classically and alternatively activated human macrophages. Biochim Biophys Acta 2012;1820:1383–90 [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Song Y, Du W, Gong L, Chang H and Zou Z Tumor-associated macrophages: an accomplice in solid tumor progression. J Biomed Sci 2019;26:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao P, Damerow MS, Stern P, Liu AH, Sweet-Cordero A, Siziopikou K, et al. CD44 promotes Kras-dependent lung adenocarcinoma. Oncogene 2013;32:5186–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellomo C, Caja L and Moustakas A Transforming growth factor beta as regulator of cancer stemness and metastasis. Br J Cancer 2016;115:761–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaturvedi G, Simone PD, Ain R, Soares MJ and Wolfe MW Noggin maintains pluripotency of human embryonic stem cells grown on Matrigel. Cell Prolif 2009;42:425–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samarakoon R, Higgins CE, Higgins SP and Higgins PJ TGF-beta1-Induced Expression of the Poor Prognosis SERPINE1/PAI-1 Gene Requires EGFR Signaling: A New Target for Anti-EGFR Therapy. J Oncol 2009;2009:342391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 2002;8:816–24 [DOI] [PubMed] [Google Scholar]
- 31.Aviel-Ronen S, Coe BP, Lau SK, da Cunha Santos G, Zhu CQ, Strumpf D, et al. Genomic markers for malignant progression in pulmonary adenocarcinoma with bronchioloalveolar features. Proc Natl Acad Sci U S A 2008;105:10155–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou CH, Ye LP, Ye SX, Li Y, Zhang XY, Xu XY, et al. Clinical significance of SOX9 in human non-small cell lung cancer progression and overall patient survival. J Exp Clin Cancer Res 2012;31:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Capaccione KM, Hong X, Morgan KM, Liu W, Bishop JM, Liu L, et al. Sox9 mediates Notch1-induced mesenchymal features in lung adenocarcinoma. Oncotarget 2014;5:3636–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin SC, Chou YT, Jiang SS, Chang JL, Chung CH, Kao YR, et al. Epigenetic Switch between SOX2 and SOX9 Regulates Cancer Cell Plasticity. Cancer Res 2016;76:7036–48 [DOI] [PubMed] [Google Scholar]
- 35.Luanpitpong S, Li J, Manke A, Brundage K, Ellis E, McLaughlin SL, et al. SLUG is required for SOX9 stabilization and functions to promote cancer stem cells and metastasis in human lung carcinoma. Oncogene 2016;35:2824–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012;148:1015–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi J, Park SY and Joo CK Transforming growth factor-beta1 represses E-cadherin production via slug expression in lens epithelial cells. Invest Ophthalmol Vis Sci 2007;48:2708–18 [DOI] [PubMed] [Google Scholar]
- 38.Brandl M, Seidler B, Haller F, Adamski J, Schmid RM, Saur D, et al. IKK(alpha) controls canonical TGF(ss)-SMAD signaling to regulate genes expressing SNAIL and SLUG during EMT in panc1 cells. J Cell Sci 2010;123:4231–9 [DOI] [PubMed] [Google Scholar]
- 39.Aomatsu K, Arao T, Sugioka K, Matsumoto K, Tamura D, Kudo K, et al. TGF-beta induces sustained upregulation of SNAI1 and SNAI2 through Smad and non-Smad pathways in a human corneal epithelial cell line. Invest Ophthalmol Vis Sci 2011;52:2437–43 [DOI] [PubMed] [Google Scholar]
- 40.Li T, Huang H, Shi G, Zhao L, Li T, Zhang Z, et al. TGF-beta1-SOX9 axis-inducible COL10A1 promotes invasion and metastasis in gastric cancer via epithelial-to-mesenchymal transition. Cell Death Dis 2018;9:849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boucherat O, Nadeau V, Berube-Simard FA, Charron J and Jeannotte L Crucial requirement of ERK/MAPK signaling in respiratory tract development. Development 2014;141:3197–211 [DOI] [PubMed] [Google Scholar]
- 42.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Z, Li C, Kang B, Gao G, Li C and Zhang Z GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98–W102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Brolosy MA and Stainier DYR Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet 2017;13:e1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang MH, Laurent G, Bause AS, Spang R, German N, Haigis MC, et al. HDAC6 and SIRT2 regulate the acetylation state and oncogenic activity of mutant K-RAS. Mol Cancer Res 2013;11:1072–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee YS, Lim KH, Guo X, Kawaguchi Y, Gao Y, Barrientos T, et al. The cytoplasmic deacetylase HDAC6 is required for efficient oncogenic tumorigenesis. Cancer Res 2008;68:7561–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prabavathy D, Swarnalatha Y and Ramadoss N Lung cancer stem cells-origin, characteristics and therapy. Stem Cell Investig 2018;5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zakaria N, Satar NA, Abu Halim NH, Ngalim SH, Yusoff NM, Lin J, et al. Targeting Lung Cancer Stem Cells: Research and Clinical Impacts. Front Oncol 2017;7:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chakravarty G, Moroz K, Makridakis NM, Lloyd SA, Galvez SE, Canavello PR, et al. Prognostic significance of cytoplasmic SOX9 in invasive ductal carcinoma and metastatic breast cancer. Exp Biol Med (Maywood) 2011;236:145–55 [DOI] [PubMed] [Google Scholar]
- 51.Richtig G, Aigelsreiter A, Schwarzenbacher D, Ress AL, Adiprasito JB, Stiegelbauer V, et al. SOX9 is a proliferation and stem cell factor in hepatocellular carcinoma and possess widespread prognostic significance in different cancer types. PLoS One 2017;12:e0187814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruan H, Hu S, Zhang H, Du G, Li X, Li X, et al. Upregulated SOX9 expression indicates worse prognosis in solid tumors: a systematic review and meta-analysis. Oncotarget 2017;8:113163–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 2015;525:256–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakano M, Kikushige Y, Miyawaki K, Kunisaki Y, Mizuno S, Takenaka K, et al. Dedifferentiation process driven by TGF-beta signaling enhances stem cell properties in human colorectal cancer. Oncogene 2019;38:780–93 [DOI] [PubMed] [Google Scholar]
- 55.Woosley AN, Dalton AC, Hussey GS, Howley BV, Mohanty BK, Grelet S, et al. TGFbeta promotes breast cancer stem cell self-renewal through an ILEI/LIFR signaling axis. Oncogene 2019;38:3794–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshikawa M, Hishikawa K, Marumo T and Fujita T Inhibition of histone deacetylase activity suppresses epithelial-to-mesenchymal transition induced by TGF-beta1 in human renal epithelial cells. J Am Soc Nephrol 2007;18:58–65 [DOI] [PubMed] [Google Scholar]
- 57.Halder SK, Cho YJ, Datta A, Anumanthan G, Ham AJ, Carbone DP, et al. Elucidating the mechanism of regulation of transforming growth factor beta Type II receptor expression in human lung cancer cell lines. Neoplasia 2011;13:912–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osada H, Tatematsu Y, Masuda A, Saito T, Sugiyama M, Yanagisawa K, et al. Heterogeneous transforming growth factor (TGF)-beta unresponsiveness and loss of TGF-beta receptor type II expression caused by histone deacetylation in lung cancer cell lines. Cancer Res 2001;61:8331–9 [PubMed] [Google Scholar]
- 59.Kaimori A, Potter JJ, Choti M, Ding Z, Mezey E and Koteish AA Histone deacetylase inhibition suppresses the transforming growth factor beta1-induced epithelial-to-mesenchymal transition in hepatocytes. Hepatology 2010;52:1033–45 [DOI] [PubMed] [Google Scholar]
- 60.Thambyrajah R, Fadlullah MZH, Proffitt M, Patel R, Cowley SM, Kouskoff V, et al. HDAC1 and HDAC2 Modulate TGF-beta Signaling during Endothelial-to-Hematopoietic Transition. Stem Cell Reports 2018;10:1369–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewandowski SL, Janardhan HP and Trivedi CM Histone Deacetylase 3 Coordinates Deacetylase-independent Epigenetic Silencing of Transforming Growth Factor-beta1 (TGF-beta1) to Orchestrate Second Heart Field Development. J Biol Chem 2015;290:27067–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang XZ, Wen D, Zhang M, Xie Q, Ma L, Guan Y, et al. Sirt1 activation ameliorates renal fibrosis by inhibiting the TGF-beta/Smad3 pathway. J Cell Biochem 2014;115:996–1005 [DOI] [PubMed] [Google Scholar]
- 63.Jackute J, Zemaitis M, Pranys D, Sitkauskiene B, Miliauskas S, Vaitkiene S, et al. Distribution of M1 and M2 macrophages in tumor islets and stroma in relation to prognosis of non-small cell lung cancer. BMC Immunol 2018;19:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang S, Che D, Yang F, Chi C, Meng H, Shen J, et al. Tumor-associated macrophages promote tumor metastasis via the TGF-beta/SOX9 axis in non-small cell lung cancer. Oncotarget 2017;8:99801–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Redente EF, Dwyer-Nield LD, Merrick DT, Raina K, Agarwal R, Pao W, et al. Tumor progression stage and anatomical site regulate tumor-associated macrophage and bone marrow-derived monocyte polarization. Am J Pathol 2010;176:2972–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


