Abstract
Glia play a key role in immunosurveillance within the central nervous system (CNS) and can recognize a wide range of pathogen associated molecular patterns (PAMPS) via members of multiple pattern recognition receptor (PRR) families. Of these, the expression of cytosolic/nuclear RNA and DNA sensors by glial cells is of particular interest as their ability to interact with intracellular nucleic acids suggest a critical role in the detection of viral pathogens. The recently discovered DNA sensors cyclic GMP-AMP synthase (cGAS) and interferon gamma inducible protein 16 (IFI16) have been reported to be important for the recognition of DNA pathogens such as herpes virus-1 (HSV-1) in peripheral human cell types, and we have recently demonstrated that human glia express cGAS and its downstream adaptor molecule stimulator of interferon genes (STING). Here, we have demonstrated that human microglial cells functionally express cGAS and exhibit robust constitutive IFI16 expression. While cGAS serves as a significant component in IRF3 activation and IFN-β production by human microglial cells in response to foreign intracellular DNA, IFI16 is not required for such responses. Surprisingly, neither of these sensors mediate effective antiviral responses to HSV-1 in microglia and this may be due, at least in part, to viral suppression of cGAS and/or IFI16 expression. As such, this ability may represent an important HSV immune evasion strategy in glial cells, and approaches that mitigate such suppression might represent a novel strategy to limit HSV-1 associated neuropathology.
Keywords: Microglia, pattern recognition receptors, viral DNA, cGAS, IFI16, HSV-1
INTRODUCTION
It is now recognized that astrocytes and microglia play a critical role in the production of immune mediators that contribute to both protective host defense and disease pathology within the central nervous system (CNS) (Ghoshal et al. 2007; Swarup et al. 2007; Das et al. 2008; Marques et al. 2008; Furr et al. 2010, 2011a; Furr and Marriott 2012; Jiang et al. 2014; Crill et al. 2015; Reinert et al. 2016). The mechanisms by which glia recognize and respond to CNS pathogens are only now becoming apparent with the demonstration that microglia and astrocytes express a wide range of pattern recognition receptors (PRRs) capable of sensing pathogen and damage associated molecular patterns (PAMPs and DAMPs, respectively) (Sterka et al. 2006; Furr et al. 2010, 2011a; Jeffries and Marriott 2017a). Similar to peripheral host cells, activation of glial PRRs initiate signaling cascades that lead to the production of soluble proinflammatory and/or antiviral mediators. Whether such production and release acts in a beneficial or detrimental manner in the CNS during infection is less well understood, and appears to be context dependent (Blank and Prinz 2017).
Of these PRRs, the expression of recently discovered cytosolic/nuclear RNA and DNA sensors by glial cells is of particular interest as their ability to interact with nucleic acids in the intracellular environment suggest an important role in the detection of viral pathogens. Consistent with this notion, we have demonstrated that the RNA sensor, retinoic acid inducible gene I (RIG-I), is important for the detection of vesicular stomatitis virus (VSV) in human glia, and we have shown that such recognition leads to the production of damaging proinflammatory mediators by these cells (Furr et al. 2010). In addition, we have demonstrated that murine glia express DNA dependent activator of interferon regulatory factors (DAI) and showed that this cytosolic DNA sensor contributes to glial inflammatory responses to the neurotropic DNA virus herpes simplex virus-1 (HSV-1) (Furr et al. 2011b; Crill et al. 2015).
More recently, additional DNA sensors such as cyclic GMP-AMP synthase (cGAS), absent in melanoma 2 (AIM2), and interferon γ-inducible protein 16 (IFI16), have been identified in peripheral cell types (Takaoka et al. 2007; Bürckstümmer et al. 2009; Unterholzner et al. 2010; Sun et al. 2013). Of these, cGAS has been the most widely studied and has been shown to recognize relevant CNS pathogens such as HSV-1 and HIV (Gao et al. 2013; Li et al. 2013a). This DNA sensor has been shown to directly bind double stranded DNA and to subsequently produce the secondary messenger 2’3’-cGAMP that then activates the critical downstream adaptor protein stimulator of interferon genes (STING). This activation leads to phosphorylation and nuclear translocation of the transcription factor, interferon regulatory factor 3 (IRF3), which induces the expression of type I interferons such as IFN-β (Sun et al. 2013). However, while we have recently demonstrated that human glia express cGAS and STING (Jeffries and Marriott 2017a), the functional significance of this DNA sensing pathway has not yet been established in these cells.
Here, we demonstrate that cGAS is functionally expressed by human microglial cells and underlies, at least in part, exogenous DNA-mediated cytokine production by this resident CNS cell type. Additionally, we have established that human glia also express the DNA sensor IFI16. However, while cGAS and IFI16 are thought to be important for DNA virus replication restriction in peripheral human cell types (Conrady et al, 2012; Li et al. 2013b; Civril et al. 2013; Zhang et al. 2014; Johnson et al. 2014a; Shu et al. 2014; Ma et al. 2015; Diner et al. 2015a, 2016; Iqbal et al. 2016a; Wang et al. 2017; Merkl et al. 2018a), our data indicates that neither of these sensors mediate effective antiviral responses to HSV-1 in human microglia, perhaps due to an ability of HSV-1 to inhibit their expression in this cell type.
MATERIALS AND METHODS
Source and propagation of glial cells
A human microglia cell line (hμglia) was a kind gift from Dr. Jonathan Karn (Case Western Reserve University). These cells were derived from primary human cells transformed with lentiviral vectors expressing SV40 T antigen and human telomerase reverse transcriptase, and have been classified as microglia due to their microglia-like morphology, migratory and phagocytic activity, presence of the microglial cell surface markers CD11b, TGFβR, and P2RY12, and characteristic microglial RNA expression profile (Garcia-Mesa et al. 2017). These cells were maintained in Dulbecco’s Modified Eagle Medium supplemented with 5% fetal bovine serum (FBS) and penicillin/streptomycin as previously described by our laboratory (Jeffries and Marriott 2017b). In some experiments a second immortalized human microglial cell line, developed by Applied Biological Materials Inc. (ABM; Richmond, Canada), was also employed, and these cells were maintained in PriGrow III media (ABM) with 10% FBS and penicillin/streptomycin. Primary human astrocytes were purchased from ScienCell Research Laboratories (Carlsbad, CA) and were cultured in medium supplied by the vendor. U87-MG, an immortalized human astrocytic cell line, was obtained from the ATCC (HTB-14). Cells were maintained in Eagle’s Minimum Essential Medium supplemented with 10% FBS and penicillin/streptomycin.
In vitro challenge of human microglia and astrocytes with nucleic acid ligands
Synthetic double stranded B-form DNA analog poly (deoxyadenylic-deoxythymidylic) acid sodium salt (Poly(dA:dt)) and G3-ended Y-form short DNA, reported cGAS agonists (Herzner et al. 2015; Jeffries and Marriott 2017b), were purchased from Invivogen (San Diego, CA). These ligands were directly introduced into microglial or astrocytic cells at concentrations of 0.01, 0.1, and/or 1.0 μg/ml using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. At the indicated time points post-transfection, whole cell protein isolates were collected and RNA was isolated for immunoblot analysis and semi-quantitative RT-PCR, respectively.
Preparation of viral stocks and in vitro infection of glial cells
HSV-1 viral stocks were prepared by infecting monolayer cultures of Vero cells (ATCC; CCL-81) with HSV-1 (MacIntyre strain from a patient with encephalitis; ATCC; VR-539) at a MOI of 0.01 and incubated for 48 to 72 hrs, at which time 100% of cells displayed cytopathic effects. Tissue culture flasks were then placed at −80°C for 15 minutes and subsequently warmed to room temperature inside a tissue culture hood. The cell suspension was removed and pulse sonicated (Vibra Cell; Sonics and Materials Inc., Newton, CT) to release intake virions. The sonicated material was centrifuged at 4000 RCF to remove unwanted cell debris and the supernatant mixed with sterile milk for increased stability during freeze/thaw cycles. The stock was aliquoted and viral titers were quantified using a standard plaque assay of serial dilutions on Vero cells at 37°C. The viral titer of the stock solution was 1.2 X 107 PFU/ml. Human astrocytes and microglia were infected with HSV-1 at MOIs of 0.02, 0.2, or 2.0 viral particles to glia and virus was allowed to adsorb for one hour in DMEM in the absence of FBS or antibiotics. Cells were subsequently washed with PBS and cultures were maintained in appropriate growth medium for the indicated times prior to the collection of supernatants, whole cell protein isolates, and/or total RNA.
RNA extraction and semi-quantitative reverse transcription PCR (RT-PCR)
Total RNA was isolated from cultured glial cells using Trizol reagent (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions and quantified using a Nanodrop ND-1000 spectrophotometer. Prior to reverse transcription, RNA was treated with amplification grade DNase (Sigma Aldrich Cat. AMPD1) to remove genomic DNA. All RNA samples were diluted to the same concentration and reverse transcribed in the presence of random hexamers using 200 U of RNase H minus Moloney leukemia virus reverse transcriptase (Promega, Madison, WI) in the buffer supplied by the manufacturer. Semi-quantitative RT-PCR was performed on 16% of the reverse-transcribed cDNA product to assess the relative levels of expression of mRNA-encoding ICP-8, BST2, Viperin, IFITM1, and the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) using the forward and reverse primers shown in Table 1.
Table 1.
PCR primer sequences
| Gene | Forward Primer | Reverse Primer | Size |
|---|---|---|---|
| ICP-8 | GAGCTTCTGGCGTTACTGTC | TATGGTTACCTTGTCCGAGCC | 465bp |
| CXCL10 | TGTACGCTGTACCTGCATCA | CTGTGTGGTCCATCCTTGGAA | 268bp |
| BST2 (Tetherin) | GATGGCCCTAATGGCTTCCC | TAACCGTGTTGCCCCATGAC | 366bp |
| IFITM1 | TCAACATCCACAGCGAGACC | CAAAGGTTGCAGGCTATGGG | 331bp |
| RSAD2 (Viperin) | TGCTGGGAAGCTCTTGAGTG | CATTGCTCACGATGCTCACG | 446bp |
| GAPDH | CCATCACCATCTTCCAGGAGCGAG | CACAGTCTTCTGGGTGGCAGTGAT | 347bp |
Immunoblot analysis
Whole cell protein isolates were collected from microglial and astrocytic cells using Triton lysis buffer (10 mM Tris-HCl pH 10.5, 5 mM MgCl2, and 1% (v/v) Triton X-100) and analyzed by immunoblot analysis. Samples were electrophoresed on a 12% SDS-polyacrylamide gel and transferred to Immobilon-P transfer membranes (Millipore). Membranes were blocked with either 5% milk (cGAS, IFI16, HSV-1 glycoprotein) or 5% BSA (phospho-IRF3) for one hour and then incubated overnight at 4°C with primary antibodies directed against cGAS (Sigma Aldrich), IFI16 (Santa Cruz Biotechnology), pIRF3 (Cell Signaling Technology), HSV-1 glycoprotein g1 (GeneTex), and the housekeeping gene product β-actin (Abcam). Blots were then washed and incubated in the presence of a horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG secondary antibody. Bound enzyme was detected with the Super Signal system (Thermo Fisher Scientific). A plasmid encoding full-length human IFI16 was used to confirm the expression of this protein in human cells (Addgene plasmid cat #35064) (Liao et al. 2011). Immunoblots shown are representative of at least three separate experiments using the Bio-Rad ChemiDoc imaging system and quantification analysis was performed using ImageLab software (Bio-Rad).
Enzyme-linked immunosorbent assay (ELISA)
Specific capture ELISAs were performed to quantify human IL-6 and IFN-β release. The IL-6 ELISA was conducted using a rat anti-human IL-6 capture antibody (BD Pharmingen) and a biotinylated rat anti-human IL-6 detection antibody (BD Pharmingen). The IFN-β ELISA was carried out using a polyclonal rabbit anti-human IFN-β capture antibody (Abcam) and a biotinylated polyclonal rabbit anti-human IFN-β detection antibody (Abcam). Bound antibody was detected using streptavidin-HRP (BD Biosciences) followed by the addition of tetramethylbenzidine (TMB) substrate. H2SO4 was used to stop the reaction and absorbance was measured at 450 nm. Dilutions of recombinant IL-6 and IFN-β (BD Pharmingen, Abcam) were used to generate standard curves and the concentration of each in study samples were determined by extrapolation to the standard curve.
Generation of heterozygous cGAS deletion microglial cell line
To investigate cGAS function, we created a heterozygous deletion microglial cell line that expresses cGAS at reduced levels, cGAS+/−, using CRISPR/Cas9 approaches. A sgRNA targeting cGAS was generated using the CRISPOR algorithm (http://crispor.tefor.net/) and a suitable sgRNA sequence was selected based on minimal off-targeting and proximity to the 5’ end of cGAS (5’ATCTTCTTAAGACAGGGGCACG-3’). The cGAS targeting sgRNA was cloned by BbsI digestion into the px458 plasmid (Addgene plasmid cat #48138) (Ran et al. 2013) that promotes simultaneous expression of cGAS sgRNA, Cas9, and GFP. The hμglia human microglial cell line was transfected at 60% confluency with the cGAS sgRNA-pX458 plasmid (0.5 μg/mL) using Lipofectamine 2000 according to the manufacturer’s instructions and the cells were prepared for fluorescence activated cell sorting (FACS) and clonal isolation at 72 hrs. GFP positive cells were isolated by FACS and seeded at 100, 1000, and 10,000 cells per well in a six well plate in complete growth media and maintained in culture until distinct colonies were visible. Individual cGAS+/− colonies were selected and propagated prior to analysis of cGAS expression by immunoblot analysis and subsequent experimental use.
siRNA transfection
ON-TARGETplus siRNA targeting human IFI16) and non-targeting pool siRNAs were purchased from Dharmacon (Lafayette, CO). Each was transfected into the hμglia human microglial cell line at a concentration of 5 nM using RNAiMAX transfection reagent (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. At 48 hrs, IFI16 protein knockdown was confirmed by immunoblot analysis.
Statistical Analysis
Data is presented as the mean +/− standard error of the mean (SEM). Statistical analyses were performed by one-way analysis of variance (ANOVA) with Bonferroni’s or Tukey’s post hoc tests or Student’s t-test as appropriate using commercially available software (GraphPad Prism, GraphPad Software, La Jolla, CA). In all experiments, results were considered statistically significant when a P-value of less than 0.05 was obtained.
RESULTS
cGAS is functionally expressed by human microglia
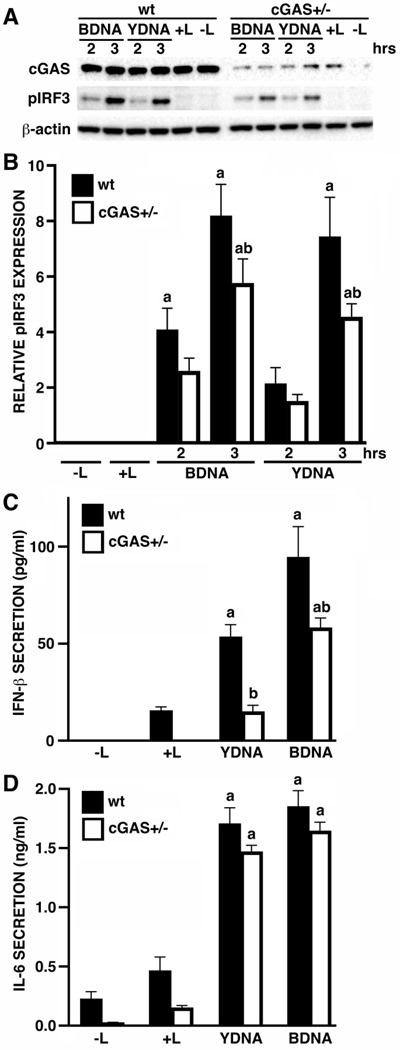
We have previously demonstrated that primary human glia and cell lines can respond to the intracellular introduction of foreign DNA, and showed that these cells express cGAS and its downstream effector molecule STING (Jeffries and Marriott 2017b). Here, we have used CRISPR/Cas9 technology to generate a heterozygous deletion human microglial cell line that expresses cGAS at reduced levels, cGAS+/−, to determine whether this cytosolic DNA sensor is functional in the hμglia human microglial cell line. As shown in Figures 1A and B, intracellular administration of microglia with B- and Y-form DNA, reported ligands for cGAS (Herzner et al. 2015; Jeffries and Marriott 2017b), elicited marked increases in levels of phosphorylated IRF3, and these responses were significantly reduced in cGAS+/− cells.
FIGURE 1:
The DNA sensor cGAS is required for maximal antiviral mediator production by human microglia following intracellular administration of exogenous DNA. Panels A and B: Wildtype (wt) or heterozygous cGAS deletion (cGAS+/−) immortalized human microglia (hμglia) were untreated (-L) or exposed to transfection reagent alone (+L) for 3 hrs, or were challenged with intracellular B-form DNA (BDNA; 0.1 μg/ml) or Y-form DNA (YDNA; 1 μg/ml) for 2 or 3 hours. Whole cell lysates were subsequently collected and analyzed for the expression of cGAS, phosphorylated IRF3 (pIRF3), or the housekeeping gene product β-actin by immunoblot analysis. Relative pIRF3 expression was determined by densitometric analysis and normalized to β-actin (n = 4–6). Panels C and D: wt or cGAS+/− hμglia were untreated (-L) or treated with transfection reagent alone (+L), or exposed to intracellular BDNA or YDNA (0.1 μg/ml and 1 μg/ml, respectively). After 24 hours, cell free supernatants were collected and the concentration of IFN-β (C) and IL-6 (D) was quantified by specific capture ELISA. Results are presented as the mean of four independent experiments +/− SEM (n = 4). The letter a indicates significant differences from cells exposed to transfection reagent alone while b indicates a significant difference from similarly treated wt cells.
Consistent with their ability to induce IRF3 phosphorylation, intracellular administration of either BDNA or YDNA elicited significant increases in the secretion of the type I interferon, IFN-β, and was also able to induce the release of the potent inflammatory cytokine IL-6 by microglial cells (Figures 1C and D). As with phosphorylated IRF3 levels, decreased cGAS expression resulted in a significant reduction in IFN-β secretion following challenge with either BDNA or YDNA (Figures 1C and D). Interestingly, cGAS expression reduction failed to significantly decrease microglial IL-6 secretion, suggesting either that the markedly reduced cGAS levels are sufficient to initiate such expression, or that alternative DNA sensing molecules are responsible for the production of this inflammatory cytokine.
cGAS contributes to antiviral gene expression but does not restrict infectious HSV-1 particle release by infected human microglial cells
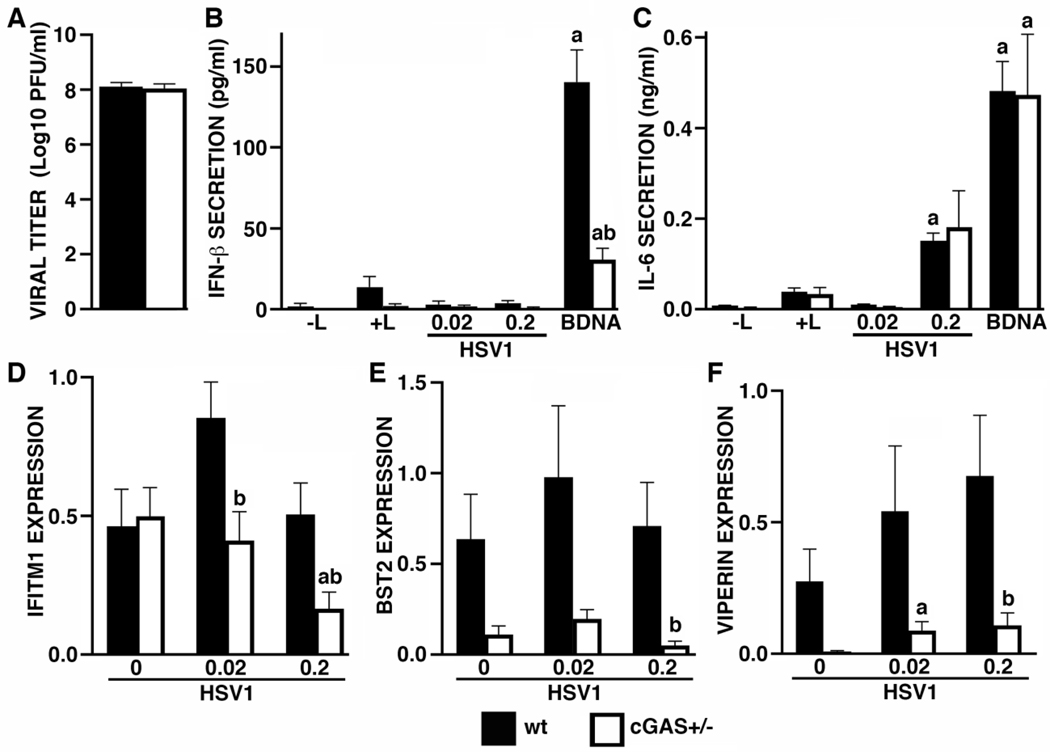
Since cGAS is functional in human microglia, we next investigated the role of this sensor in the antiviral microglial responses to the clinically relevant neurotropic DNA virus, HSV-1. Wild type and cGAS+/− microglia were exposed to HSV-1 at a MOI of 0.2 and the number of infectious HSV-1 particles released by these cells was determined by plaque assay at 24 hours following viral challenge. Surprisingly, reduced cGAS expression failed to elicit significant changes in the level of HSV-1 release (Figure 2A). Such a result could stem from an inability of these cells to secrete detectable levels of IFN-β at 8 hrs (Figure 2B) or 12 hrs (data not shown) following HSV-1 challenge, despite responding to this virus with significant levels of IL-6 release at 8 hrs (Figure 2C) and 12 hrs (data not shown) following infection. Again, cGAS expression reduction significantly reduced transfected BDNA-mediated IFN-β production by microglia, but did not significantly affect either BDNA or HSV-1-induced IL-6 release at either 8 hrs (Figures 2B and C) or 12 hrs (data not shown) following challenge.
FIGURE 2:
The DNA sensor cGAS contributes to antiviral gene expression, but does not restrict the number of infectious HSV-1 particles released by infected human microglia. Panel A: wt or cGAS+/− hμglia were infected with HSV-1 at an MOI of 0.2. At 24 hrs, cell free supernatants were collected and viral titers were determined by plaque assay (n = 7). Panels B and C: wt or cGAS+/− hμglia were transfected with 0.1 μg/mL BDNA or infected with HSV-1 (MOI of 0.02 and 0.2). At 8 hrs, cell free supernatants were collected and the concentrations of IFN-β (B) and IL-6 (C) were quantified by specific capture ELISAs (n = 7). Panels D-F: wt or cGAS+/− hμglia were infected with HSV-1 (MOI of 0.02 and 0.2). After 12 hrs, total RNA was isolated and the level of expression of mRNA encoding IFITM1 (D), BST2 (E), and Viperin (F), was determined by semi-quantitative RT-PCR and levels are reported relative to the expression of the housekeeping gene GAPDH (n = 4–7). In Panels B and C, the letter a indicates significant differences from cells exposed to transfection reagent alone while b indicates a significant difference from similarly treated wt cells. In Panels D-F, the letter a indicates significant differences from uninfected cells while b indicates a significant difference from similarly treated wt cells
Consistent with an absence of significant IFN-β production by HSV-1 challenged human microglial cells, wt cells did not show significant elevations in the expression of mRNA encoding the IFN-stimulated genes IFITM1, BST2, or viperin (Figures 2D–F). Interestingly, cGAS+/− microglia expressed significantly lower levels of IFITM1, BST2, and viperin, mRNA expression following HSV-1 infection than similarly challenged wt cells, and even showed a tendency for lower BST2 and viperin expression in uninfected cells (Figures 2D–F). Together, these data indicate that while cGAS is required, at least in part, for the maintenance of antiviral gene expression by human microglial cells, such cGAS-mediated responses are not sufficient to limit the release of infectious HSV-1 particles by these cells.
Human glia express IFI16 but this DNA sensor does not contribute to microglial responses to HSV-1
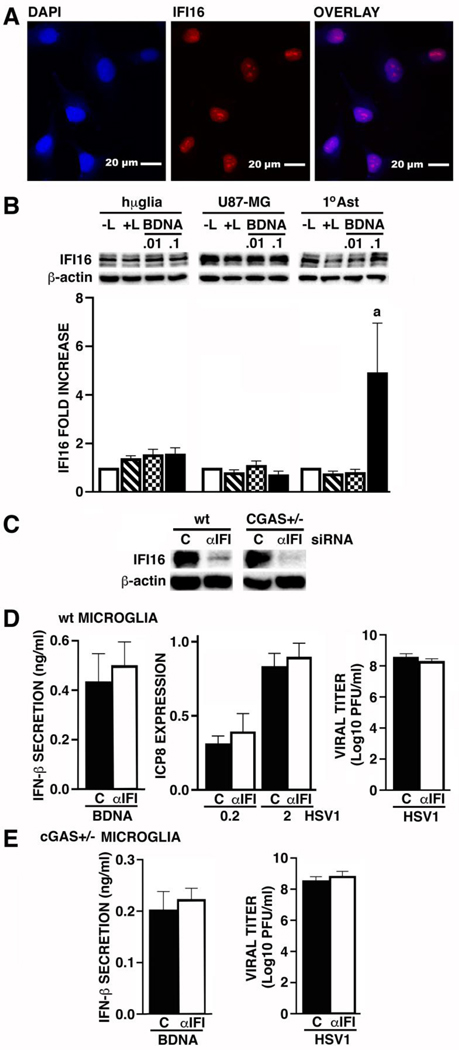
Another intracellular DNA sensor, IFI16, has previously been reported to function as an HSV-1 restriction factor in peripheral human cells (Conrady et al, 2012; Liang et al. 2014; Johnson et al. 2014a; Diner et al. 2015a, 2016; Merkl et al. 2018a). Accordingly, we have determined if human glial cells express IFI16 and whether this molecule mediates, either constitutively or in the reduced expression of cGAS, microglial immune responses to HSV-1 challenge. As shown in Figure 3A, the hμglia human microglial cell line constitutively expresses IFI16 as determined by immunofluorescence microscopy, and this sensor is localized to the nucleus as previously described in other cell types (Roy et al. 2019a). This expression was confirmed by immunoblot analysis with robust constitutive levels of a protein close to the predicted molecular weight of IFI16 (88 KDa), and at an identical size to that seen in whole cell protein isolates from HEK 293T cells transfected with a plasmid vector to express IFI16 (data not shown). Other fainter bands may correspond to the isotypic variants of IFI16 that have been reported to arise due to differential RNA splicing events (Johnstone et al, 1998). Such expression was also confirmed in a commercially available (Applied Biological Materials Inc.) immortalized human microglial cell line (data not shown). Levels of IFI16 protein expression were not increased further following stimulation with intracellular BDNA administration (Figure 3B). Interestingly, while the U87-MG human astrocytic cell line also constitutively expressed robust levels of IFI16 protein that were not increased following BDNA stimulation, primary human astrocytes only showed low levels of IFI16 at rest, but showed marked elevations in response to BDNA transfection (Figure 3B).
FIGURE 3:
Human glia express IFI16, but this DNA sensor does not contribute to BDNA or HSV-1 mediated antiviral mediator production by human microglia, or restrict the number of infectious HSV-1 particles released by infected glia. Panel A: Micrographs show representative nuclear (DAPI), IFI16, and overlaid, immunofluorescence in hμglia human microglial cells (60X objective). Panel B: hμglia, U87-MG astrocytic cells, and primary human astrocytes (1°AST), were transfected with BDNA (0.01 and 0.1 ug/ml). At 24 hrs post-transfection, whole cell lysates were collected and tested for the presence of IFI16 by immunoblot analysis (n = 7–11). Panels C-E: wt or cGAS+/− hμglia were transfected with siRNA targeting IFI16 (αIFI; 5 nM) or scrambled RNA (C) for 48 hrs and IFI16 protein knockdown was confirmed by immunoblot analysis (Panel C; n = 2). After 48 hrs, control (C) or αIFI siRNA treated wt (Panel D) or cGAS+/− (Panel E) hμglia were either transfected with BDNA (0.1 ug/ml) or infected with HSV-1 (MOI of 0.2 and 2), and viral ICP8 mRNA levels relative to GAPDH expression were determined by RT-PCR at 8 hrs following challenge (n = 4), while supernatant IFN-β concentrations (n = 4–5) and viral titers (n = 8) were determined at 24 hrs post-challenge. Results are presented as the mean +/− SEM and no statistically significant differences were observed.
We then utilized siRNA approaches to assess the role of IFI16 in microglial immune responses to HSV1. We confirmed that siRNA targeting IFI16 markedly attenuated the expression of this protein and possible isotypic variants in both wt and cGAS+/− human microglia (Figure 3C) and determined the effect of IFI16 knockdown on microglial responses to intracellular BDNA administration and HSV-1 infection. IFI16 knockdown had no demonstrable effect on BDNA-induced IFN-β production, expression of the HSV-1 gene product ICP8, or infectious HSV-1 particle release, by wt human microglial cells (Figure 3D), and had no effect on IL-6 release (data not shown). Similarly, IFI16 knockdown had no demonstrable effect on BDNA-induced IFN-β production (Figure 3E), infectious HSV-1 particle release (Figure 3E), or IL-6 production (data not shown), by cGAS+/− microglial cells. Together, these data indicate that while human glia express IFI16 this DNA sensor does not contribute to BDNA or HSV-1 mediated antiviral mediator production by human microglia, or restrict the number of infectious HSV-1 particles released by infected cells.
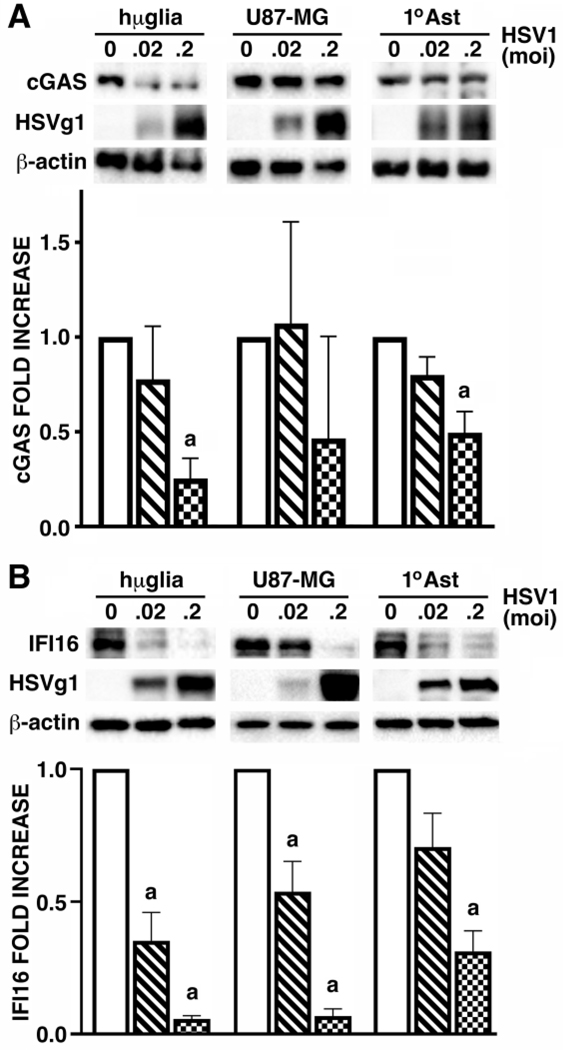
HSV-1 infection down regulates cGAS and IFI16 expression by human glia
To begin to determine the mechanisms underlying the apparent resistance of HSV-1 to cGAS and/or IFI16 mediated antiviral microglial responses we have assessed the effect of this virus on the expression of each of these intracellular DNA sensors by human glia. As shown in Figure 4A, HSV-1 infection significantly decreased cGAS protein levels by human microglial cell and primary human astrocytes in a dose dependent manner, and showed a tendency to reduce cGAS expression in U87-MG astrocytic cells at 24 hours post-challenge. Similarly, HSV-1 infection elicited significant and dose-dependent decreases in IFI16 protein levels in the hμglia human microglial cell line, U87-MG astrocytic cells, and primary human astrocytes (Figure 4B), and a second immortalized human microglial cell line (data not shown). As such, the ability of HSV-1 to downregulate the expression of both of these intracellular DNA sensors may represent an important immune evasion strategy in human glia.
FIGURE 4:
HSV-1 infection down regulates cGAS and IFI16 expression by human glia. hμglia, U87-MG, and primary human astrocytes (1°AST), were untreated or infected with HSV-1 (MOI of 0.02 and 0.2). At 24 hrs, whole cell lysates were collected and analyzed for the expression of cGAS (Panel A; n = 3), IFI16 (Panel B; n = 7–9), viral glycoprotein g1 (HSVg1), and the housekeeping product β-actin, by immunoblot analysis. Protein levels were determined by densitometric analysis relative to β-actin expression, and are shown as fold increases over untreated cells. Results are presented as the mean +/− SEM and the letter a indicates significant differences from uninfected cells.
DISCUSSION
We have previously demonstrated that human glial cells express the DNA sensor cGAS and the downstream adaptor molecule STING (Jeffries and Marriott 2017a). In the present study, we expand upon this work by demonstrating that cGAS is functional in human microglial cells. We show that microglia respond to intracellular administration of either B or Y-form DNA, reported ligands for cGAS in other cell types (Herzner et al. 2015; Jeffries and Marriott 2017b), with increased levels of IRF3 activation and IFN-β secretion. Importantly, we show that these responses are due, in large part, to recognition via cGAS with the demonstration that reduced cGAS expression results in significantly reduced levels of IRF3 activation and IFN-β release in response to either of these ligands, while IL-6 responses are unchanged. These findings are in agreement with previous studies in peripheral cell types showing that cGAS is critical for cytoplasmic dsDNA recognition (Li et al. 2013b; Sun et al. 2013; Civril et al. 2013; Zhang et al. 2013, 2014; Shu et al. 2014; Ma et al. 2015).
Similar to cGAS, IFI16 has also been proposed to directly bind dsDNA and mediate the expression of antiviral mediators such as type I IFNs (Unterholzner et al. 2010). Interestingly, some studies have suggested that IFI16 works in concert with cGAS to initiate host immune responses to viral pathogens (Orzalli et al. 2015; Jønsson et al. 2017; Almine et al. 2017; Liu et al. 2017), while others point to separate and distinct antiviral functions for each (Diner et al. 2016). In the present study, we provide the first demonstration that human glial cells constitutively express the IFI16 protein. Surprisingly, and in contrast to peripheral human cell types that show IFI16-dependent IFN-β production in response to dsDNA ligands (Jønsson et al. 2017; Almine et al. 2017), our studies employing siRNA-mediated IFI16 knockdown indicate that this sensor does not play a significant role in human microglial IFN responses to exogenous DNA administration. Such a finding cannot be explained on the basis of DNA sensing redundancy via cGAS as IFI16 knockdown similarly failed to affect BDNA-induced IFN-β expression in cGAS+/− microglia.
cGAS has previously been demonstrated to be an important PRR for combating numerous infections through direct detection of cytosolic microbial/viral DNA (Gao et al. 2013; Li et al. 2013a; Schoggins et al. 2014; Cox et al. 2015; Watson et al. 2015; Xia et al. 2016; Paijo et al. 2016; Vermeire et al. 2016; Ruangkiattikul et al. 2017; Sun et al. 2017; Cheng et al. 2018). Importantly, this sensor has been shown to be a critical component in the generation of protective immunity in a murine model of acute herpes simplex encephalitis (HSE) (Reinert et al. 2016). This protection is thought to be mediated through the canonical cGAS-STING signaling pathway, which leads to the expression of type I IFNs that act in an autocrine and/or paracrine manner to promote an antiviral state (Reinert et al. 2016). Similarly, IFI16 exhibits antiviral capabilities in peripheral cell types (Conrady et al, 2012; Johnson et al. 2014a; Merkl et al. 2018b; Merkl and Knipe 2019a; Roy et al. 2019b) and has been reported to be an important restriction factor for herpesviruses including HSV-1 in such cells (Dutta et al. 2015; Ansari et al. 2015; Diner et al. 2015b; Iqbal et al. 2016b). However, unlike cGAS, the mechanism of action of IFI16 and the signaling pathways that this intracellular sensor employs are poorly understood.
Given the documented importance of both cGAS and IFI16 as PRRs in the generation of host immune responses to HSV-1 in peripheral cell types, and the present description of constitutive expression of both sensors in glial cells, it is not unreasonable to assume that these sensors could serve a similar function in human microglia during HSV-1 infection. Surprisingly, our studies show that neither cGAS nor IFI16, alone or in concert, appear to significantly impact HSV-1 transcription or the production/release of infectious particles in human microglial cells. While the lack of cGAS and IFI16-mediated antiviral responses to this neuroinvasive HSV-1 clinical isolate may be simply due to host cell type or species dependent differences in sensor function (Kalamvoki and Roizman 2014a; Orzalli et al. 2016), we have determined that HSV-1 infection elicits marked reductions in the expression level of both of these molecules in human microglial cells. Such HSV-1-mediated suppression could explain why siRNA directed against cGAS or IFI16 failed to elicit demonstrable effects in infected cells and suggests a viral immune evasion mechanism. Indeed, these findings are consistent with the previously reported ability of HSV-1 to target the expression and/or the signaling pathways of cGAS and IFI16 in non-CNS cell types (Orzalli et al. 2012; Johnson et al. 2013; Kalamvoki and Roizman 2014b; Christensen et al. 2016; Su and Zheng 2017; Huang et al. 2018; Zhang et al. 2018). Such a strategy could underlie, at least in part, the absence of IFN-β production by HSV-1 infected microglia with little or no induction in the expression of the antiviral interferon-stimulated genes BST2, viperin, and IFITM1, despite retaining an ability to release IL-6. However, it should be noted that IFI16 has been reported to restrict herpesvirus replication via transcriptional regulation rather than effects on IFN responses in other cell types (Johnson et al. 2014b; Merkl et al. 2018c; Merkl and Knipe 2019b; Roy et al. 2019b), and so the downregulation of this molecule could impact microglial responses to HSV-1 via mechanisms other than by inhibiting IFN-β production.
Taken in concert, we have demonstrated that human microglial cells functionally express the cytosolic DNA sensor cGAS and exhibit robust constitutive expression of the nuclear DNA sensor IFI16. While cGAS serves as a significant component in IRF3 activation and IFN-β production by human microglial cells in response to intracellular administration of foreign DNA, IFI16 does not appear to be required for such responses, in contrast to previous reports in other human cell types. Surprisingly, neither of these intracellular DNA sensors mediate effective antiviral responses by human microglial cells to the neurotropic DNA virus HSV-1, and this may be due, at least in part, to an ability of this virus to suppress the expression of cGAS and/or IFI16 in these cells. As such, this ability may represent an important HSV immune evasion strategy in glial cells, and approaches that mitigate such suppression might represent a novel strategy to limit HSV-1 associated neuropathology.
Acknowledgments
Funding: This work was supported by grant NS097840 to IM from the National Institutes of Health.
Footnotes
DECLARATIONS
Ethical Approval and Consent to Participate: All protocols involving animals were approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Charlotte.
Consent for publication: Not applicable.
Availability of data and material: The data used and/or analyzed during the current study will be available from the corresponding author on reasonable request.
Competing interests: The authors declare that they have no competing interests.
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
REFERENCES
- Almine JF, O’Hare CAJ, Dunphy G, et al. (2017) IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat Commun 8:14392 10.1038/ncomms14392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari MA, Dutta S, Veettil MV, et al. (2015) Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-β Responses. PLOS Pathog 11:e1005019. 10.1371/journal.ppat.1005019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank T, Prinz M (2017) Type I interferon pathway in CNS homeostasis and neurological disorders. Glia 65:1397–1406 [DOI] [PubMed] [Google Scholar]
- Bürckstümmer T, Baumann C, Blüml S, et al. (2009) An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol 10:266–272. 10.1038/ni.1702 [DOI] [PubMed] [Google Scholar]
- Cheng W-Y, He X-B, Jia H-J, et al. (2018) The cGas–Sting Signaling Pathway Is Required for the Innate Immune Response Against Ectromelia Virus. Front Immunol 9:1297 10.3389/fimmu.2018.01297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen MH, Jensen SB, Miettinen JJ, et al. (2016) HSV-1 ICP27 targets the TBK1-activated STING signalsome to inhibit virus-induced type I IFN expression. EMBO J 35:1385–99. 10.15252/embj.201593458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, et al. (2013) Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498:332–337. 10.1038/nature12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrady CD, Zheng M, Fitzgerald KA, et al. (2012) Resistance to HSV-1 infection in the epithelium resides with the novel innate sensor, IFI-16. Mucosal Immunol 5:173–83. doi: 10.1038/mi.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DJ, Field RH, Williams DG, et al. (2015) DNA sensors are expressed in astrocytes and microglia in vitro and are upregulated during gliosis in neurodegenerative disease. Glia 63:812–825. 10.1002/glia.22786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crill EK, Furr-Rogers SR, Marriott I (2015) RIG-I is required for VSV-induced cytokine production by murine glia and acts in combination with DAI to initiate responses to HSV-1. Glia 63:2168–80. 10.1002/glia.22883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Mishra MK, Ghosh J, Basu A (2008) Japanese Encephalitis Virus infection induces IL-18 and IL-1β in microglia and astrocytes: Correlation with in vitro cytokine responsiveness of glial cells and subsequent neuronal death. J Neuroimmunol 195:60–72. 10.1016/J.JNEUROIM.2008.01.009 [DOI] [PubMed] [Google Scholar]
- Diner BA, Lum KK, Javitt A, Cristea IM (2015a) Interactions of the Antiviral Factor Interferon Gamma-Inducible Protein 16 (IFI16) Mediate Immune Signaling and Herpes Simplex Virus-1 Immunosuppression. Mol Cell Proteomics 14:2341–2356. 10.1074/mcp.M114.047068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner BA, Lum KK, Javitt A, Cristea IM (2015b) Interactions of the Antiviral Factor Interferon Gamma-Inducible Protein 16 (IFI16) Mediate Immune Signaling and Herpes Simplex Virus-1 Immunosuppression. Mol Cell Proteomics 14:2341–2356. 10.1074/mcp.M114.047068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner BA, Lum KK, Toettcher JE, Cristea IM (2016) Viral DNA Sensors IFI16 and Cyclic GMP-AMP Synthase Possess Distinct Functions in Regulating Viral Gene Expression, Immune Defenses, and Apoptotic Responses during Herpesvirus Infection. MBio 7:. 10.1128/mBio.01553-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Dutta S, Veettil MV, et al. (2015) BRCA1 Regulates IFI16 Mediated Nuclear Innate Sensing of Herpes Viral DNA and Subsequent Induction of the Innate Inflammasome and Interferon-β Responses. PLOS Pathog 11:e1005030. 10.1371/journal.ppat.1005030 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Furr SR, Chauhan VS, Moerdyk-Schauwecker MJ, Marriott I (2011a) A role for DNA-dependent activator of interferon regulatory factor in the recognition of herpes simplex virus type 1 by glial cells. J Neuroinflammation 8:99 10.1186/1742-2094-8-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr SR, Chauhan VS, Moerdyk-Schauwecker MJ, Marriott I (2011b) A role for DNA-dependent activator of interferon regulatory factor in the recognition of herpes simplex virus type 1 by glial cells. J Neuroinflammation 8:99 10.1186/1742-2094-8-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr SR, Marriott I (2012) Viral CNS infections: role of glial pattern recognition receptors in neuroinflammation. Front Microbiol 3:201 10.3389/fmicb.2012.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr SR, Moerdyk-Schauwecker M, Grdzelishvili VZ, Marriott I (2010) RIG-I mediates nonsegmented negative-sense RNA virus-induced inflammatory immune responses of primary human astrocytes. Glia 58:1620–9. 10.1002/glia.21034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu Y-T, et al. (2013) Cyclic GMP-AMP Synthase Is an Innate Immune Sensor of HIV and Other Retroviruses. Science (80- ) 341:903–906. 10.1126/science.1240933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mesa Y, Jay TR, Checkley MA, et al. (2017) Immortalization of primary microglia: a new platform to study HIV regulation in the central nervous system. J Neurovirol 23:47–66. 10.1007/s13365-016-0499-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal A, Das S, Ghosh S, et al. (2007) Proinflammatory mediators released by activated microglia induces neuronal death in Japanese encephalitis. Glia 55:483–496. 10.1002/glia.20474 [DOI] [PubMed] [Google Scholar]
- Herzner A-M, Hagmann CA, Goldeck M, et al. (2015) Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat Immunol 16:1025–1033. 10.1038/ni.3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, You H, Su C, et al. (2018) Herpes Simplex Virus 1 Tegument Protein VP22 Abrogates cGAS/STING-Mediated Antiviral Innate Immunity. J Virol 92:. 10.1128/JVI.00841-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Ansari MA, Kumar B, et al. (2016a) Histone H2B-IFI16 Recognition of Nuclear Herpesviral Genome Induces Cytoplasmic Interferon-β Responses. PLOS Pathog 12:e1005967. 10.1371/journal.ppat.1005967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Ansari MA, Kumar B, et al. (2016b) Histone H2B-IFI16 Recognition of Nuclear Herpesviral Genome Induces Cytoplasmic Interferon-β Responses. PLOS Pathog 12:e1005967. 10.1371/journal.ppat.1005967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries AM, Marriott I (2017a) Human microglia and astrocytes express cGAS-STING viral sensing components. Neurosci Lett 658:. 10.1016/j.neulet.2017.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries AM, Marriott I (2017b) Human microglia and astrocytes express cGAS-STING viral sensing components. Neurosci Lett 658:53–56. 10.1016/j.neulet.2017.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Ye J, Zhu B, et al. (2014) Roles of TLR3 and RIG-I in mediating the inflammatory response in mouse microglia following Japanese encephalitis virus infection. J Immunol Res 2014:787023. 10.1155/2014/787023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Bottero V, Flaherty S, et al. (2014a) IFI16 Restricts HSV-1 Replication by Accumulating on the HSV-1 Genome, Repressing HSV-1 Gene Expression, and Directly or Indirectly Modulating Histone Modifications. PLoS Pathog 10:e1004503. 10.1371/journal.ppat.1004503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Bottero V, Flaherty S, et al. (2014b) IFI16 Restricts HSV-1 Replication by Accumulating on the HSV-1 Genome, Repressing HSV-1 Gene Expression, and Directly or Indirectly Modulating Histone Modifications. PLoS Pathog 10:e1004503. 10.1371/journal.ppat.1004503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KE, Chikoti L, Chandran B (2013) Herpes Simplex Virus 1 Infection Induces Activation and Subsequent Inhibition of the IFI16 and NLRP3 Inflammasomes. J Virol 87:5005–5018. 10.1128/JVI.00082-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW, Kershaw MH, Trapani JA (1998) Isotypic variants of the interferon-inducible transcriptional repressor IFI16 arise through differential mRNA splicing. Biochemistry 37:11924–11931. [DOI] [PubMed] [Google Scholar]
- Jønsson KL, Laustsen A, Krapp C, et al. (2017) IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat Commun 8:14391 10.1038/ncomms14391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki M, Roizman B (2014a) HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci 111:E611–E617. 10.1073/pnas.1323414111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki M, Roizman B (2014b) HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci 111:E611–E617. 10.1073/pnas.1323414111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-D, Wu J, Gao D, et al. (2013a) Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341:1390–4. 10.1126/science.1244040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, et al. (2013b) Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science (80- ). 10.1126/science.1244040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Seo GJ, Choi YJ, et al. (2014) Crosstalk between the cGAS DNA Sensor and Beclin-1 Autophagy Protein Shapes Innate Antimicrobial Immune Responses. Cell Host Microbe 15:228–238. 10.1016/j.chom.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JCC, Lam R, Brazda V, et al. (2011) Interferon-inducible protein 16: Insight into the interaction with tumor suppressor p53. Structure 19:418–429. 10.1016/j.str.2010.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Niu Q, Fan X, et al. (2017) Priming and Activation of Inflammasome by Canarypox Virus Vector ALVAC via the cGAS/IFI16–STING–Type I IFN Pathway and AIM2 Sensor. J Immunol 199:3293–3305. 10.4049/jimmunol.1700698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Li B, Liu S, et al. (2015) Positive Feedback Regulation of Type I IFN Production by the IFN-Inducible DNA Sensor cGAS. J Immunol 194:1545–1554. 10.4049/jimmunol.1402066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques CP, Cheeran MC-J, Palmquist JM, et al. (2008) Prolonged microglial cell activation and lymphocyte infiltration following experimental herpes encephalitis. J Immunol 181:6417–26. 10.4049/jimmunol.181.9.6417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl PE, Knipe DM (2019a) Role for a Filamentous Nuclear Assembly of IFI16, DNA, and Host Factors in Restriction of Herpesviral Infection. MBio 10:1–16. 10.1128/mbio.02621-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl PE, Knipe DM (2019b) Role for a Filamentous Nuclear Assembly of IFI16, DNA, and Host Factors in Restriction of Herpesviral Infection. MBio 10:. 10.1128/mBio.02621-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl PE, Orzalli MH, Knipe DM (2018a) Mechanisms of Host IFI16, PML, and Daxx Protein Restriction of Herpes Simplex Virus 1 Replication. J Virol 92:. 10.1128/JVI.00057-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl PE, Orzalli MH, Knipe DM (2018b) Mechanisms of Host IFI16, PML, and Daxx Protein Restriction of Herpes Simplex Virus 1 Replication. J Virol 92:1–19. 10.1128/jvi.00057-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl PE, Orzalli MH, Knipe DM (2018c) Mechanisms of Host IFI16, PML, and Daxx Protein Restriction of Herpes Simplex Virus 1 Replication. J Virol 92:. 10.1128/JVI.00057-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, Broekema NM, Diner BA, et al. (2015) cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc Natl Acad Sci 112:E1773–E1781. 10.1073/pnas.1424637112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, Broekema NM, Knipe DM (2016) Relative Contributions of Herpes Simplex Virus 1 ICP0 and vhs to Loss of Cellular IFI16 Vary in Different Human Cell Types. J Virol 90:8351–8359. 10.1128/JVI.00939-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, DeLuca NA, Knipe DM (2012) Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci 109:E3008–E3017. 10.1073/pnas.1211302109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paijo J, Döring M, Spanier J, et al. (2016) cGAS Senses Human Cytomegalovirus and Induces Type I Interferon Responses in Human Monocyte-Derived Cells. PLOS Pathog 12:e1005546. 10.1371/journal.ppat.1005546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, et al. (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert LS, Lopušná K, Winther H, et al. (2016) Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defence in the CNS. Nat Commun. 10.1038/ncomms13348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Ghosh A, Kumar B, Chandran B (2019a) IFI16, a nuclear innate immune DNA sensor, mediates epigenetic silencing of herpesvirus genomes by its association with H3K9 methyltransferases SUV39H1 and GLP. Elife 8:. 10.7554/eLife.49500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Ghosh A, Kumar B, Chandran B (2019b) IFI16, a nuclear innate immune DNA sensor, mediates epigenetic silencing of herpesvirus genomes by its association with H3K9 methyltransferases SUV39H1 and GLP. Elife 8:. 10.7554/eLife.49500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruangkiattikul N, Nerlich A, Abdissa K, et al. (2017) cGAS-STING-TBK1-IRF3/7 induced interferon-β contributes to the clearing of non tuberculous mycobacterial infection in mice. Virulence 8:1303–1315. 10.1080/21505594.2017.1321191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, et al. (2014) Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505:691–695. 10.1038/nature12862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu C, Li X, Li P (2014) The mechanism of double-stranded DNA sensing through the cGAS-STING pathway. Cytokine Growth Factor Rev 25:641–648. 10.1016/j.cytogfr.2014.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterka D, Rati DM, Marriott I (2006) Functional expression of NOD2, a novel pattern recognition receptor for bacterial motifs, in primary murine astrocytes. Glia 53:322–330. 10.1002/glia.20286 [DOI] [PubMed] [Google Scholar]
- Su C, Zheng C (2017) Herpes Simplex Virus 1 Abrogates the cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway via Its Virion Host Shutoff Protein, UL41. J Virol 91:. 10.1128/JVI.02414-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Sundström KB, Chew JJ, et al. (2017) Dengue virus activates cGAS through the release of mitochondrial DNA. Sci Rep 7:3594 10.1038/s41598-017-03932-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, et al. (2013) Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–91. 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup V, Ghosh J, Duseja R, et al. (2007) Japanese encephalitis virus infection decrease endogenous IL-10 production: Correlation with microglial activation and neuronal death. Neurosci Lett 420:144–149. 10.1016/J.NEULET.2007.04.071 [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, et al. (2007) DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448:501–505. 10.1038/nature06013 [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, et al. (2010) IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. 10.1038/ni.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeire J, Roesch F, Sauter D, et al. (2016) HIV Triggers a cGAS-Dependent, Vpu- and Vpr-Regulated Type I Interferon Response in CD4 + T Cells. Cell Rep 17:413–424. 10.1016/j.celrep.2016.09.023 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ning X, Gao P, et al. (2017) Inflammasome Activation Triggers Caspase-1-Mediated Cleavage of cGAS to Regulate Responses to DNA Virus Infection. Immunity 46:393–404. 10.1016/j.immuni.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Watson RO, Bell SL, MacDuff DA, et al. (2015) The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host Microbe 17:811–819. 10.1016/j.chom.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Ye B, Wang S, et al. (2016) Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat Immunol 17:369–378. 10.1038/ni.3356 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhao J, Xu S, et al. (2018) Species-Specific Deamidation of cGAS by Herpes Simplex Virus UL37 Protein Facilitates Viral Replication. Cell Host Microbe 24:234–248.e5. 10.1016/j.chom.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, et al. (2013) Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Mol Cell 51:226–235. 10.1016/j.molcel.2013.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wu J, Du F, et al. (2014) The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop. Cell Rep 6:421–430. 10.1016/j.celrep.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]