Abstract
Metabolic pathways leading to the synthesis, uptake, and usage of the nonessential amino acid serine are frequently amplified in cancer. Serine encounters diverse fates in cancer cells, including being charged onto tRNAs for protein synthesis, providing head groups for sphingolipid and phospholipid synthesis, and serving as a precursor for cellular glycine and one-carbon units, which are necessary for nucleotide synthesis and methionine cycle reloading. This review will focus on the participation of serine and glycine in the mitochondrial one-carbon (SGOC) pathway during cancer progression, with an emphasis on the genetic and epigenetic determinants that drive SGOC gene expression. We will discuss recently elucidated roles for SGOC metabolism in nucleotide synthesis, redox balance, mitochondrial function, and epigenetic modifications. Finally, therapeutic considerations for targeting SGOC metabolism in the clinic will be discussed.
Keywords: serine, glycine, one-carbon, metabolism, cancer, mitochondria
1.1. Introduction
The field of cancer biology has appreciated altered metabolism as an essential hallmark of malignant transformation (Hanahan and Weinberg, 2011; Pavlova and Thompson, 2016). A century ago, the German biochemist Otto Warburg observed that cells of cancerous origin displayed an increased uptake of glucose that was fermented to lactic acid rather than oxidized in the mitochondria (Warburg et al., 1924; Warburg et al., 1927). To account for the reduced pyruvate entry into the mitochondrial tricarboxylic acid (TCA) cycle for ATP generation, cancer cells must obtain massive amounts of glucose to maintain energy balance. Moreover, the preference of cancer cells to express the M2 isoform of pyruvate kinase (PKM2) — which can be posttranslationally or allosterically modified to adjust its catalytic activity — is thought to provide additional benefit — namely, providing biosynthetic precursors — by shunting glucose flux to pathways such as pentose phosphate pathway and serine biosynthesis in response to growth factor stimulation and cellular nutrient status (Anastasiou et al., 2011; Chaneton et al., 2012; Ye et al., 2012). Indeed, genomic profiling has revealed that many cancers, particularly breast and melanoma, harbor amplifications and upregulate expression of the rate-limiting enzyme in serine biosynthesis, 3-phosphoglycerate dehydrogenase PHGDH (Locasale et al., 2011; Possemato et al., 2011). Rapidly proliferating cells such as cancer cells also rapidly consume serine, obtained either through synthesis or import from the extracellular milieu. Thus, understanding the myriad metabolic fates of serine is of key interest to the cancer biology field. Serine plays important roles in macromolecule synthesis, either through charging transfer RNAs (tRNAs) for protein synthesis, serving as a precursor to amino acids such as cysteine and glycine, providing head groups for sphingolipid and phospholipid synthesis, or donating 1C units for nucleotide synthesis. We are now beginning to appreciate that, in cancer cells, the mitochondrial serine, glycine, and one-carbon (SGOC) pathway exquisitely catabolizes serine to generate other ―by-products‖ such as NAD(P)H for redox defense and functional mitochondrial tRNAs. Could there be other cellular processes affected by enhanced mitochondrial SGOC pathway activity in cancer? A few outstanding reviews about the SGOC pathway have discussed the biochemistry, compartmentalization, biological regulation, research history, and disease relevance of this pathway (Ducker and Rabinowitz, 2017; Locasale, 2013; Tibbetts and Appling, 2010; Yang and Vousden, 2016). In this review, we will focus on the recent progress in elucidating the role of the mitochondrial branch of this pathway, which is universally upregulated in many cancer types during cancer progression, particularly the connection between this pathway to epigenetic regulations.
2.1. The major cellular one-carbon species
Eukaryotic cells utilize the vitamin B9 derivative tetrahydrofolate (THF) as a carrier for cellular one-carbon (1C) units. Attachment of 1C units to either the N5 or N10 position of THF relies on the biochemical action of 1C metabolic enzymes coupled with oxidation and reduction reactions. Furthermore, the polyglutamylation of THF molecules is thought to facilitate the retention of 1C units in specific compartments—e.g. either the cytosol or mitochondria—by reducing 1C unit affinity for intercompartmental folate transporters and increasing their affinity for compartment-specific folate transformation enzymes (Ducker and Rabinowitz, 2017; Kim et al., 1996). The universal importance of folates in supporting cell proliferation has been long appreciated since a landmark study by Sydney Farber demonstrated that inhibitors of folate transformation enzymes could induce regression of acute lymphoblastic leukemia (ALL) (Farber et al., 1947; Farber and Diamond, 1948). The related compounds methotrexate and pemetrexed are now widely used in the clinic as chemotherapeutics targeting folate metabolism, although like in many other targeted therapies, resistance mechanisms often emerge in tumors (Bertino et al., 1996; Guo et al., 1999).
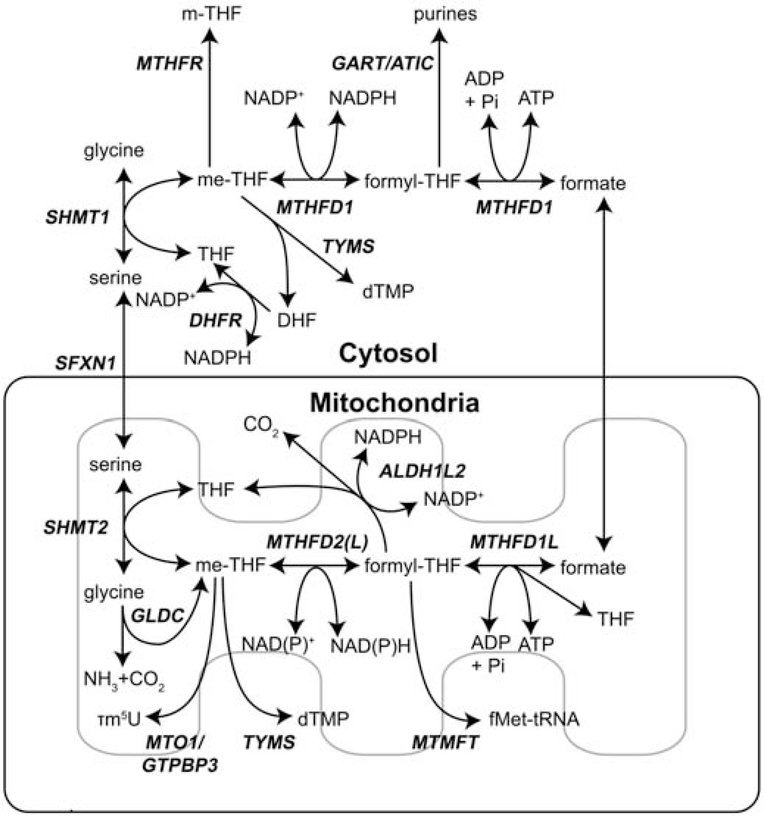
While various metabolites can donate methyl groups to THF, serine and glycine are the major sources of 1C units. Serine catabolism initiated by serine hydroxymethyltransferase (SHMT) transfers the γ-carbon amino acid side chain to THF, forming glycine and 5,10-methylene-THF (me-THF) (Figure 1). The cytosolic (SHMT1) and mitochondrial (SHMT2) isoforms perform the same reactions. Each isoform carries a different burden of total one-carbon flux in cancer cells and will be discussed later. Glycine generated by this reaction can also be catabolized to me-THF in the mitochondria, although the relative contribution of glycine cleavage to total me-THF pools may depend on the expression of the glycine decarboxylase complex (GLDC) and the ability of cells to clear or re-assimilate the toxic ammonia cations generated (Spinelli et al., 2017). Importantly, me-THF generated in both the cytosol and mitochondria functions as 1C donor for thymidylate pyrimidine nucleotide synthesis through the action of thymidylate synthetase (TYMS) (Anderson et al., 2011).
Figure 1. Cytosolic and mitochondrial folate-mediated serine, glycine, and one-carbon cycle.
Serine, and to a lesser extent glycine, provides 1C precursors for biosynthesis in the cytosol and mitochondria. Flux through either branch also maintains redox homeostasis through NAD(P)H consumption or generation. Important enzymes are highlighted in bold and italicized.
me-THF generated from the action of SHMT can be further oxidized to 10-formyl-THF (formyl-THF) by cytosolic methylene-tetrahydrofolate dehydrogenase 1 (MTHFD1) or mitochondrial methylene-tetrahydrofolate dehydrogenase 2 (MTHFD2) and methylene-tetrahydrofolate dehydrogenase 2-like (MTHFD2L). MTHFD1 interconverts me-THF and formyl-THF using NADPH/NADP+, while MTHFD2 and MTHFD2L have recently been reported to utilize both NADPH/NADP+ and NADH/NAD+ (Shin et al., 2017). The primary anabolic role for 10-formyl-THF is supporting cytosolic purine nucleotide synthesis, specifically in the two steps catalyzed by the trifunctional enzyme phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase (GART) and the bifunctional enzyme 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC) (Figure 1).
Cytosolic MTHFD1 and mitochondrial MTHFD1L perform the release of the final transformation product of folate-mediated 1C metabolism, formate, while concomitantly generating ATP. This liberated 1C unit can pass freely across the mitochondrial membrane, with the major direction of exchange being from the mitochondria to the cytosol. In the next sections, we will discuss several hypotheses that attempt to explain why cancer cells preferentially perform oxidative transformation of 1C units in the mitochondria and reductive assimilation of 1C units in the cytosol.
2.2. Directionality of the cytosolic and mitochondrial one-carbon folate cycle
Isotope tracing studies discriminate usage of the cytosolic vs. mitochondrial branches
Initial studies performed in yeast and knockout mice supported the idea that cytosolic 1C units could be derived from mitochondrial 1C metabolism (MacFarlane et al., 2008; Pasternack et al., 1992; Piper et al., 2000). Subsequent studies using deuterium atoms-labeled serine ([2,3,3-2H]serine) elucidated the relative contribution of cytosolic and mitochondrial branches to the total cellular 1C pool (Ducker et al., 2016). me-THF generated directly from cytosolic serine catabolism is labeled with two deuteriums (2H) such that incorporation of the 1C unit into thymidylate (dTTP) produces dTTP mass heavy by 2 (M+2). In contrast, me-THF derived from mitochondrial serine catabolism retains only one labeled 2H following oxidation in the mitochondria to formate, transport across the membrane, and reduction back to me-THF in the cytosol. Therefore, dTTP synthesized from me-THF derived from mitochondrial 1C metabolism is M+1. Using dTTP as a readout, Ducker et al. observed in many proliferating cancer cell lines that most cellular 1C units originate in the mitochondria. In comparison, MTHFD2 expression was not detected in neither proliferating fibroblast cells nor proliferating liver cells, though it is induced upon T cell activation and early embryo development, suggesting that not all normal cells utilize the mitochondrial pathway for proliferation (Nilsson et al., 2014). Intriguingly, tumor cells display plasticity such that deletion of mitochondrial 1C pathway enzymes causes a switch to serine catabolism in the cytosolic 1C pathway. This way, cancer cells retain the ability to generate glycine and 1C units for nucleotide synthesis. In contrast, Shmt2 deficient mice are embryonic lethal, the fibroblast and liver cells from the mice display mitochondrial respiration defects and slower growth rate, suggesting that SHMT2 is indispensable for normal development (Tani et al., 2019; Tani et al., 2018).
A separate study by Lewis et al. employing A549 lung cancer cells used an ectopic expression system of either mutant isocitrate dehydrogenase 1 (IDH1) or 2 (IDH2) to track NADPH production in either compartment of 1C metabolism (Lewis et al., 2014). As described above, the conversion of me-THF to formyl-THF by either MTHFD1 in the cytosol or MTHFD2/MTHFD2L in the mitochondria can generate NADPH. Likewise, this NADPH can be consumed by mutant IDH1 in the cytosol or mutant IDH2 in the mitochondria to generate the R enantiomer of 2-hydroxyglutarate (R-2HG) from alpha-ketoglutarate (Dang et al., 2009; Ward et al., 2010). The amount of deuterium-labeled 2HG can be used as a measure of compartmentalized NADPH flux through the SGOC pathway. When cultured in media containing [2,3,3-2H]serine, M+1 2HG was observed only in cells expressing mutant IDH2 (mitochondria) but not mutant IDH1 (cytosol), suggesting that the mitochondrial 1C metabolism proceeds in the direction whereby NADPH is produced (oxidative), but in the cytosol the 1C pathway may go in reverse direction (reductive) to generate formyl-THF for purine biosynthesis, possibly due to the high NADPH levels generated from pentose phosphate pathway activity..
Serine and glycine cleavage in the mitochondria
Many cancer types display enhanced serine biosynthesis and import of serine from the extracellular milieu, as evidenced by their high expression of de novo serine synthesis enzymes phosphoglycerate dehydrogenase (PHGDH), phosphoserine aminotransferase (PSAT1), and phosphoserine phosphatase (PSPH) and serine transporters solute carrier family 1 member 4 and 5 (SLC1A4 and SLC1A5). As outlined above, much of this serine is shuttled to the mitochondria for catabolism to glycine and 1C units. A meta-analysis study revealed that enzymes involved in the mitochondrial 1C pathway are among the most frequently overexpressed genes in cancer (Nilsson et al., 2014). Recently, a CRISPR-CAS9 screen performed in serine-starved cell lines with loss of cytosolic 1C activity identified sideroflexin 1 (SFXN1) as a key mitochondrial transporter of serine (Kory et al., 2018). Although it is unknown whether serine synthesized de novo encounters different fates from serine transported from the extracellular space, studies suggest that, at least in some cells, there exists coordination between cytoplasmic serine synthesis and mitochondrial 1C unit fate. To start, the expression of the rate limiting enzymes of serine biosynthesis, PHGDH, and mitochondrial serine catabolism, SHMT2, are positively correlated with each other in breast cancer and neuroblastoma (Ye et al., 2014a). In addition, PHGDH activity suppresses SHMT1 activity to promote serine incorporation into nucleotides through the mitochondrial 1C pathway (Pacold et al., 2016).
The role of glycine in cancer cells appears to be somewhat more convoluted. An initial profiling of cancer cell lines described a correlation between glycine consumption and cancer cell proliferation rates (Jain et al., 2012). Later studies revealed that serine, rather than glycine, was the most rapidly consumed amino acid supporting proliferation, and that some cells might switch to glycine only when serine is depleted (Labuschagne et al., 2014). It was even observed that, in some cells, high levels of glycine could impair cell proliferation, presumably through mechanisms such as product inhibition of the SHMT2 reaction or glycine-to-serine conversion through the reverse action of SHMT1 leading to ―wasting‖ of cellular 1C pools, thereby impairing nucleotide synthesis (Fan et al., 2014; Labuschagne et al., 2014; Pacold et al., 2016).
Given the complex roles of glycine—dependent on the relative availability of serine, glycine, and 1C units that drive the rate and direction of the SHMT1/2 reactions—effective glycine clearance through export or cleavage (through GLDC) may be essential to support optimal cell proliferation. The hypoxic tumor microenvironment of brain tumors drives expression of SHMT2, a HIF-target gene (Kim et al., 2015; Ye et al., 2014a). Kim et al. observed that, in a subset of these cells, particularly those located in ischemic regions of gliomas, GLDC is required to prevent toxic accumulation of the glycine-derived metabolites aminoacetone and methylglyoxal. In non-small cell lung cancer, GLDC is enriched in tumor initiating cells (TICs) and drives tumorigenesis by linking glucose to serine/glycine flux with increased synthesis/accumulation of pyrimidine nucleotides (Zhang et al., 2012). Consistent with this finding, a recent study demonstrated that GLDC expression at least partly contributes to the maintenance of TICs derived from human lung cancer patients by coupling 1C flux to the methionine cycle and histone methylation (Wang et al., 2019). In a following section, we will discuss further evidence connecting SGOC metabolism to epigenetic modifications on DNA and histones. In sum, the role of glycine in supporting cancer cell proliferation is likely to be both environment-specific and cell-type specific.
2.3. Beyond cytosolic anabolism: redox balance, mitochondrial function, and metabolic control of epigenetics
The above discussion leads us to the general belief that the preferred route of serine utilization is catabolism in the mitochondria followed by export of the 1C units to support cytosolic anabolic reactions. The reason for this may be compartment-specific NAD(P)H/NAD(P) ratios that favor 1C unit oxidation in the mitochondria and 1C unit reduction in the cytosol. Intriguingly, recent studies have highlighted the concept that SGOC metabolism can actively modulate redox balance in a compartment-specific manner (Fan et al., 2014; Yang et al., 2020; Ye et al., 2014a).
Mitochondrial 1C metabolism generates NAD(P)H
NADPH serves as a crucial molecule providing reducing power for biosynthesis as well as buffering against redox stress in rapidly proliferating or detached cells (Pavlova and Thompson, 2016; Schafer et al., 2009). Isotope tracer studies and quantitative flux analyses revealed that 1C metabolism can generate a significant fraction of total cellular NADPH. In particular, Fan et al. determined that while most cytosolic NADPH is devoted to biosynthesis, the complete oxidation of serine in the mitochondria was required to generate NADPH to suppress reactive oxygen species (ROS) and maintain the ratio of reduced to oxidized glutathione (Fan et al., 2014). These two functions are accomplished by the conversion of me-THF to formyl-THF by MTHFD2/MTHFD2L and a side reaction catalyzed by ALDH1L2 converting formyl-THF to CO2. Circulating metastatic melanoma cells particularly upregulate ALDH1L2, perhaps reflecting a preference towards diverting pathway flux towards NADPH-mediated ROS suppression rather than cytosolic 1C units for cell anabolism (Piskounova et al., 2015). A recent study demonstrated that mitochondrial serine catabolism can also generate a substantial portion of NADH in cultured cells and mice, particularly in the pancreas, spleen, and tumors (Yang et al., 2020). When cell respiration was impaired, other pathways generating NADH (such as the TCA cycle) shut down while mitochondrial serine catabolism persisted, generating toxic amounts of NADH that impeded cell proliferation. Paradoxically, inhibition of either SHMT2 or MTHFD2 normalized intracellular NADH/NAD+ ratios and boosted HCT116 colon cancer cell proliferation in respiration-impaired conditions (treatment with metformin).
Interplay between 1C metabolism and mitochondrial function
For cells under hypoxic stress, upregulation of the mitochondrial 1C pathway provided further benefit to combat ROS generated from an impaired electron transport chain (ETC) (Ye et al., 2014a). Subsequent studies reported that ETC dysfunction could impair mitochondrial catabolism of serine to formate (Bao et al., 2016; Meiser et al., 2016). In particular, treatment of cells with inhibitors of various ETC complexes induced a dependency on high extracellular/cytosolic serine levels to drive serine catabolism through the cytosolic 1C pathway (Bao et al., 2016; Maddocks et al., 2013). Importantly, the growth of cells with impaired ETC under serine starvation could be rescued by addition of formate and hypoxanthine, a precursor to purine nucleotides (Bao et al., 2016). Since xanthine dehydrogenase (XDH) converts hypoxanthine and NAD+ to xanthine and NADH, the rescue effect of hypoxanthine may be through restoring NADH levels (Sato et al., 1995).
Recent work has estimated that, in some cancer cells, mitochondrial formate is produced in excess beyond the 1C demand for biosynthesis (Meiser et al., 2018; Meiser et al., 2016). Although it is still unclear why these cells display this property, one possibility is that heightened serine catabolism is an alternative route to generate mitochondrial NADH without upregulating glucose flux to the TCA cycle. In turn, a functional ETC sustains mitochondrial oxidative power to recycle NADH back to NAD+, thereby ensuring adequate cofactors for the me-THF to formyl-THF reaction catalyzed by MTHFD2/MTHFD2L. It is worth noting that a major function of the ETC is to also support aspartate synthesis, and aspartate and 1C units are both required for nucleotide synthesis (Birsoy et al., 2015; Sullivan et al., 2015). Thus, electron shuttling through the ETC and mitochondrial 1C metabolism may cooperate, through redox regulation, to sense and balance anabolic demand. Indeed, Chen et al. have recently elucidated an analogous relationship in the cytosol whereby the pentose phosphate pathway supports the activity of the cytosolic 1C enzyme dihydrofolate reductase (DHFR) through NADPH (Chen et al., 2019b). We believe that the ability of mitochondrial 1C metabolism to utilize both NAD+ and NADP+ may provide cancer cells the metabolic flexibility to sustain proliferation even under stress conditions: when the ETC is active, the mitochondrial 1C pathway can utilize sufficient NAD+ to generate 1C for purine biosynthesis; under hypoxia or starvation conditions, although the NAD+/NADH ratio decreases, elevated mitochondrial ROS leads to an increased NADP+/NADPH ratio, which can also drive the 1C pathway forward.
Emerging evidence suggests that a reciprocal relationship may indeed exist; namely, mitochondrial serine catabolism supports ETC function by supporting mitochondrial protein translation. While profiling various CRISPR-CAS9 deletion mutants in human colon cancer HCT116 cells, Morscher et al. observed that loss of SHMT2 increased extracellular media acidification due to impaired mitochondrial respiratory chain activity (Morscher et al., 2018). They determined this defect was caused by the loss of ETC protein translation stemming from a deficiency in SHMT2-derived me-THF, which was necessary to generate 5-taurinomethyl modifications at the wobble position of mitochondrial tRNAs. Minton et al. arrived at a similar result in a CRISPR-CAS9 screen for metabolic enzymes that are essential for growth of Jurkat leukemic T cells in low glucose (Minton et al., 2018). SHMT2 was the highest scoring non-ETC protein in the screen, and loss of SHMT2 also resulted in loss of ETC protein translation. Interestingly, Minton et al. determined that the crucial 1C species was mitochondrial formyl-THF for maintaining formylmethionyl-tRNA pools, rather than the upstream metabolite me-THF. Together, this pair of studies lays groundbreaking work for investigating the precise biochemical reactions that link mitochondrial 1C metabolism with mitochondrial translation and respiration.
Coupling the folate cycle to the methionine cycle and cellular methylation
S-adenosyl methionine (SAM), the universal methyl donor for DNA and histone methylation, is generated from the ligation of ATP to methionine through the action of methionine adenosyltransferase (MAT). Early studies using whole-organism infusion methods of isotopically labeled serine suggested that serine catabolism in the folate cycle could contribute to the re-methylation of homocysteine back to methionine (Davis et al., 2004; Gregory et al., 2000). This occurs through methylenetetrahydrofolate reductase (MTHFR)-mediated reduction of me-THF to 5-methyltetrahydrofolate (m-THF), followed by 1C transfer from m-THF to homocysteine catalyzed by methionine synthase (MS). Several groups have reported a positive association between folic acid levels and tumor methylation, and folate supplementation could reverse hypomethylation (Kim et al., 2001; Pufulete et al., 2005; Wasson et al., 2006; Xue et al., 2017). Thus, the discovery of upregulated serine synthetic and serine catabolic flux in many tumors raised the exciting possibility that tumor cells channel serine-derived 1C units through the folate cycle to methylate chromatin.
However, isotope tracing studies in cultured cells failed to identify a substantial contribution of 1C’s from serine to the methyl-group on SAM (Ducker et al., 2016; Maddocks et al., 2016; Pike et al., 2010). Only in the absence of medium methionine could serine contribute 1C units for the re-methylation of homocysteine to methionine (Maddocks et al., 2016). Surprisingly, Maddocks et al. found that in cells grown in full media, serine supported SAM levels through the synthesis of ATP, a substrate for MAT. Taken together, these results suggest that methionine, rather than serine, may be the key amino acid substrate for SAM synthesis and chromatin control in physiological contexts (Dai et al., 2018; Mentch et al., 2015). A diminished contribution of folate-mediated 1C metabolism to SAM may also explain why other clinical studies have failed to see impressive effects of dietary folic acid supplementation on global methylation (Cravo et al., 1998; Figueiredo et al., 2009; Jung et al., 2011), since dysregulation of methionine metabolism—a common event in cancer cells—would predominately influence methylation dynamics (Hoffman and Erbe, 1976). To this end, Gao et al. recently reported therapeutic synergism between dietary methionine restriction and standards of care such as radiation therapy and chemotherapy in tumor-bearing mice, although it is still unclear if and how this occurs through modulating methylation (Gao et al., 2019).
A pair of studies investigating autochthonous mouse models of pancreatic cancer and prostate cancer have attempted to link loss of the tumor suppressors LKB1 and PKCλ/ι, respectively, to increased SGOC flux, DNA methylation, and gene expression changes (Kottakis et al., 2016; Reina-Campos et al., 2019). Green et al. recently reported a mechanism whereby MTHFD2 sustains renal cell carcinoma by controlling global N6-methylated adenosine (m6A) levels on mRNA, enhancing the translation of tumor-promoting factors such as HIF-2α (Green et al., 2019). Intriguingly, work by Koufaris and Nilsson suggests that the regulation of RNA metabolism and translation by MTHFD2 might occur independently of its metabolic functions (Koufaris and Nilsson, 2018). In the absence of isotope tracing experiments clearly showing labeling onto SAM from the third position carbon of serine, the likeliest explanation for these observations may be increased ATP synthesis. Alternatively, indirect mechanisms might explain observed increases in SAM, RNA, DNA, and histone methylation, such as changes in metabolite abundances that either inhibit demethylation (e.g. 2HG production) or reduce antagonistic acetylation (e.g. decreased acetyl-CoA) (Shyh-Chang et al., 2013; Sivanand et al., 2018; Ye et al., 2018). In this model, upregulated one-carbon metabolism may be just a single feature associated with broader metabolic dysregulation, following loss of tumor suppressor function, that fuels an altered epigenetic cancer cell state.
2.4. Genetic and epigenetic regulation of SGOC metabolism
The above studies in genetically engineered mouse models illustrate the broad dysregulation of SGOC metabolism driven by activation of oncogenes or loss of tumor suppressor activity. It is now appreciated that epigenetic factors, such as post-translational modification and subcellular localization, can also regulate the expression and function of enzymes involved in 1C metabolism.
Important transcription factors
The metabolic role of the MYC oncogene has been extensively studied, particularly with regards to regulating glucose, glutamine, nucleotide, and fatty acid metabolism (Stine et al., 2015). MYC also controls SGOC metabolism, as the key enzymes SHMT1, SHMT2, MTHFD1, and MTHFD2 contain E-Box binding motifs for MYC (Haggerty et al., 2003; Ju et al., 2019; Mao et al., 2003; Nikiforov et al., 2002; Pikman et al., 2016). Functional evidence furthermore suggests that in cancer, MYC may at least in part exert its cancer cell fitness-promoting functions through SGOC metabolism. Ye et al. demonstrated cooperation between MYC and HIF1α to drive SHMT2 expression under hypoxia (Ye et al., 2014a). c-Myc is frequently overexpressed in breast cancer, and mitochondrial 1C pathway genes were found to be upregulated in breast cancer cells grown under hypoxia and as mammospheres (Samanta et al., 2016). Finally, either glucose or glutamine starvation triggers the c-Myc isoform-mediated upregulation of enzymes involved in serine synthesis (Sun et al., 2015), as well as those involved in serine catabolism such as SHMT2, further supporting the notion that products of mitochondrial 1C metabolism protect against stresses such as nutrient starvation.
A key transcription factor for the adaptation to cellular stress, activating transcription factor 4 (ATF4), upregulates all the three enzymes in serine biosynthesis, PHGDH, PSAT1 and PSPH, upon amino acid starvation (Ye et al., 2012). ATF4 can also activate expression of downstream SGOC metabolism genes such as SHMT2 and MTHFD2 (Ben-Sahra et al., 2016; DeNicola et al., 2015). ATF4 itself is the transcriptional target of factors such as NRF2 (Ye et al., 2014b). DeNicola et al. showed that in non-small cell lung cancer, NRF2 promotes ATF4-dependent expression of 1C metabolism genes such as SHMT2 to supply substrates for glutathione and nucleotide synthesis (DeNicola et al., 2015). Given the central role of NRF2 in cellular oxidative stress responses and recent observations that the lung tumor microenvironment may select for pathways that protect cells from high oxygen tension (Alvarez et al., 2017; Rojo de la Vega et al., 2018), it is perhaps not a coincidence that the NRF2-KEAP1 axis is frequently dysregulated in lung cancer. Ben-Sahra et al. also reported mTORC1-mediated stimulation of ATF4 activity resulting in coordinated MTHFD2 transcription and purine synthesis, demonstrating a link between growth factor signaling and mitochondrial 1C metabolism. Thus, the ability of ATF4 to transcriptionally activate SGOC metabolism is crucial for integrating inputs during both stress and growth conditions. ATF4 is translationally upregulated by the integrated stress response (ISR) (Harding et al., 2003; Lu et al., 2004), which can also be induced by mitochondrial stress (Quiros et al., 2017; Tyynismaa et al., 2010), raising the question of whether ATF4-dependent mitochondrial 1C pathway reprogramming can play a key role in the cellular adaptation to stress through crosstalk between the ISR and mitochondrial stress response (Melber and Haynes, 2018).
Regulation by post-transcriptional mechanisms
Sole transcriptional output is not the best metric for understanding the activity of an enzyme or pathway. Interactions with other enzymes, particularly those leading to post-translational modifications, may be particularly important for function and localization. Indeed, functional SHMT1 and SHMT2 undergo tetramerization (Giardina et al., 2015). A proteomics screen for interactors of the sirtuin family member SIRT5 elucidated a mechanism whereby SIRT5-mediated desuccinylation of lysine 280 on SHMT2 activates tetramerization and enzymatic activity (Yang et al., 2018). In glioblastoma, elevated levels of IDH3α led to colocalization and interaction with SHMT1, thus regulating a variety of 1C-related processes such as pyrimidine synthesis and DNA methylation (May et al., 2019). Of note, a role for SHMT1 in scaffolding and localizing de novo thymidylate synthesis enzymes to the nuclear lamina has been reported (Anderson et al., 2012).
With the rapid technological advances being made in proximity proteomics (Chen and Perrimon, 2017), in the near future the field will likely gain rich insights into the identity and assembly of various 1C metabolism protein complexes, particularly for the mitochondrial isoforms that are most frequently upregulated in cancer. These investigations may also address some of the proposed “moonlighting‖” functions of 1C metabolism enzymes outside of their native cellular compartments. For example, it was reported that MTHFD2 co-immunoprecipitates with nuclear proteins (Koufaris and Nilsson, 2018) involved in RNA processing and protein translation as well as co-localizes with DNA replication sites in the nucleus (Gustafsson Sheppard et al., 2015), highlighting the potential for nuclear functions of this mitochondrial 1C pathway protein.
Summary of SGOC metabolism by cancer type
For the convenience of the reader, we present in Table 1 a compendium of clinical and functional evidence from the literature describing the involvement of SGOC enzymes in different cancer types.
Table 1.
The reprogramming of serine, glycine and one-carbon pathway across cancer types.
| Study | Enzyme(s) | Expression up or down | Cancer Type(s) | Regulated by | Function/Phenotype/Major conclusion |
|---|---|---|---|---|---|
| (Nilsson et al., 2012) | SHMT1, SHMT2 | Up | Acute myeloid leukemia (murine) | λ-MYC | Dispensable for tumor initiation and progression |
| (Pikman et al., 2016) | MTHFD2 | Up | Acute myeloid leukemia (human, murine) | MYC | Knockdown decreased proliferation, induced differentiation, impaired colony formation, and suppressed TCA cycle |
| (Leivonen et al., 2011) | SHMT2 | Up | Breast (human) | miR-193b | Repressed by miR-193b to restrain ER+ growth |
| (Jain et al., 2012) | SHMT1, SHMT2 | N/A | Breast (human) | N/A | SHMT2 but not SHMT1 correlates with mortality |
| (Lehtinen et al., 2013) | MTHFD2 | Up | Breast (human) | N/A | Knockdown reduced vimentin expression, migration, invasion, and fraction of CD44hi cells |
| (Ye et al., 2014a) | SHMT2 | Up | Breast, Neuroblastoma (human) | HIF1α, N-Myc | Induced by hypoxia to maintain redox balance through NADPH generation |
| (Yin, 2015) | SHMT2 | Up | Breast (human) | N/A | Expression correlated with tumor grade |
| (Samanta et al., 2016) | SHMT2 | Up | Breast (human) | HIF1α, HIF2α | Induced by hypoxia and mammosphere culture |
| (Bernhardt et al., 2017) | SHMT2 | Up | Breast (human) | N/A | High expression correlates with poor patient survival |
| (Chen et al., 2019a) | SHMT2 | Up | Breast (human) | EIF2A, ATF4 | Involvement in integrated stress response following paclitaxel treatment |
| (Li et al., 2020) | SHMT2, MTHFD2, MTHFD1L | Up | Breast (human) | MYC (for MTHFD2 and MTHFD1L) | Upregulated in aggressive, highly metastatic subclones of triple-negative breast cancer cells; SHMT2 knockdown reduced metastatic growth |
| (Miyo et al., 2017) | SHMT2, MTHFD2, ALDH1L2 | Up | Colorectal (human) | N/A | High expression of all 3 enzymes correlated with poor patient survival |
| (Agarwal et al., 2019) | MTHFD1L | Up | Colorectal (human) | N/A | Supports proliferation, colony formation, invasion and migration |
| (Ju et al., 2019) | MTHFD2 | Up | Colorectal (human) | c-Myc | Knockdown disturbed redox balance and accelerated cell death under hypoxia and anchorage independent growth |
| (Tanner et al., 2017) | SHMT1, SHMT2 | Up | Ewing sarcoma (human) | EWS/FLI | High expression correlated with poor patient survival |
| (Kim et al., 2015) | GLDC, SHMT2 | Up | Glioma (human) | N/A | SHMT2 suppresses PKM2 activity and decreases oxygen consumption; GLDC clears excess glycine, providing survival benefit under hypoxia |
| (Wang et al., 2017) | SHMT2 | Up | Glioma (human) | N/A | Expression correlated with tumor grade |
| (Woo et al., 2016) | SHMT2, GLDC | Up | Hepatocellular carcinoma (human) | N/A | SHMT2 is necessary but not sufficient for liver tumorigenesis |
| (Wu et al., 2016b) | SHMT2 | Up | Hepatocellular carcinoma (human) | miR-615-5p | Promotes proliferation and migration |
| (Paone et al., 2014) | SHMT1, SHMT2 | Up | Lung cancer (human) | N/A | SHMT1 knockdown leads to misincorporation of uracil in genomic DNA, cell cycle arrest, apoptosis |
| (Wu et al., 2016a) | SHMT1 | Up | Lung adenocarcinoma (human) | miR-198 | Knockdown leads to less proliferation, increased apoptosis, cell-cycle arrest |
| (Zhang et al., 2012) | GLDC | Up | Non-small cell lung cancer (human) | N/A | Necessary and sufficient for lung tumorigenicity |
| (DeNicola et al., 2015) | SHMT2 | Up | Non-small cell lung cancer (human) | NRF2, ATF4 | Links serine biosynthesis to nucleotide and glutathione synthesis |
| (Piskounova et al., 2015) | ALDH1L2 | Up | Melanoma (human, murine) | N/A | Necessary for NADPH-mediate ROS suppression in metastatic cells |
| (Kottakis et al., 2016) | GLDC, SHMT1, SHMT2, | Up | Pancreatic adenocarcinoma (murine) | LKB1, mTOR | Links serine biosynthesis to DNA methylation |
| (Reina-Campos et al., 2019) | MTHFD2 | Up | Neuroendocrine prostate cancer (murine) | PKCλ/ι, mTOR, ATF4 | Links serine biosynthesis to DNA methylation and neuroendocrine progenitor cell differentiation |
| (Lin et al., 2018) | MTHFD2 | Up | Renal cell carcinoma | N/A | Knockdown decreased cell proliferation, invasion, and migration |
| (Green et al., 2019) | MTHFD2 | Up | Renal cell carcinoma | HIF-2α | Forms positive feedforward loop with HIF-2α to control RNA methylation and reprogram metabolism |
| (Nilsson et al., 2014) | MTHFD2 | Up | Pan-cancer (human) | N/A | Consistently elevated in many cancers, correlates with poor survival in breast cancer, knockdown causes cell death in most cancer cell lines |
2.5. Therapeutic targeting of reprogrammed SGOC metabolism
A number of groups have now reported on the efficacy and mechanism of pharmacological inhibitors against SGOC enzymes. We have compiled the subset of drugs that have been tested in live cells or animals, and their corresponding enzymatic targets (Table 2):
Table 2.
Drugs targeting one-carbon metabolism in cancer.
| Name/Commercial name | Target(s) | Mechanism of action and effect in cells | Status | Cancer Type | Refs. |
|---|---|---|---|---|---|
| SHIN1 | SHMT1/2 | Competitive inhibitor; reduces proliferation by inhibiting glycine and me-THF generation | Preclinical | Various human cancer cell lines | (Ducker et al., 2017) |
| AGF347 | SHMT1/2, GART, ATIC | NA; reduces proliferation by inhibiting glycine and me-THF generation | Preclinical | NSCLC, colon, pancreatic | (Dekhne et al., 2019) |
| 2.12 | SHMT1/2 | Competitive inhibitor; induces apoptosis, cell cycle arrest, and uracil incorporation into DNA | Preclinical | Lung cancer | (Marani et al., 2016) |
| LY345899 | MTHFD2 | Competitive inhibitor; induces apoptosis through reduced mitochondrial NADP(H) generation | Preclinical | Colorectal | (Gustafsson et al., 2017; Ju et al., 2019) |
| Carolacton | MTHFD1/2 | Inhibition of both substrate and cofactor binding in active site; inhibits cell proliferation by an unknown mechanism | Preclinical | Colon, endocervical cancer cell lines | (Fu et al., 2017) |
| LY231514/MTA/Pemetrexed | TYMS, DHFR, GART, ATIC | Competitive inhibitor; inhibits cell proliferation by limiting thymidylate for DNA synthesis | Approved | Various solid and hematological tumors | (Chattopadhyay et al., 2007; Shih et al., 1997) |
| Amethopterin/MTX/Methotrexate | TYMS, DHFR | Competitive inhibitor (folate analog); induces cell death by depleting THF levels | Approved | Various solid and hematological tumors | (Kremer, 2004) |
| 5-FU | TYMS | Suicide complex agent; prevents DNA synthesis by blocking conversion of dUMP to dTMP | Approved | Various solid and hematological tumors | (Danenberg, 1977) |
The initial clinical success of the first generation of drugs targeting 1C metabolism, particularly antifolates (methotrexate and pemetrexed), is complicated by the observation of acquired resistance in many solid tumors. Proposed mechanisms of resistance include reduced transport/increased export by folate carriers, impaired polyglutamylation, or increased levels of DHFR (Walling, 2006). Moreover, clinical studies noted broad toxicities in highly proliferative, non-transformed cells such as gut epithelial and immune cells. Thus, the key question arises: is 1C metabolism the right pathway to drug for cancer treatment? The answer may lie in future studies investigating the newer generation of drugs selectively targeting mitochondrial folate enzymes such as SHMT2 and MTHFD2. In preclinical models, the current generation of inhibitors selectively inhibit the generation of products of mitochondrial 1C metabolism such as me-THF and glycine, although inhibitory activity against cytosolic folate (SHMT1) and purine biosynthesis enzymes (GART and ATIC) were also reported (Dekhne et al., 2019; Ducker et al., 2017). Inhibition of mitochondrial 1C metabolism might be therapeutically exploited in certain tumors, such as diffuse large B-cell lymphoma, that exhibit impaired import of exogenous glycine (Ducker et al., 2017). It is also possible that targeting mitochondrial 1C metabolism may restrict tumor growth through cancer cell-autonomous effects beyond restriction of building blocks for biosynthesis, such as a reduced ability to combat oxidative stress and an altered epigenetic landscape. In turn, more rigorous investigations in vivo will help elucidate the contributions of 1C metabolism to various stages of cancer progression, including tumor initiation and metastasis. Regardless, these studies must take into careful consideration cancer cell non-autonomous effects as well, such as modulating the metabolism and function of other cell types in the tumor microenvironment such as immune cells, in which activation of SGOC enzymes is crucial to their differentiation and effector functions (Ma et al., 2017; Ron-Harel et al., 2018).
3.1. Conclusion
Recent work has highlighted roles for serine, glycine and one-carbon metabolism beyond serving as precursors to protein synthesis, providing 1C precursors for nucleotide synthesis, as well as providing head groups for sphingolipid and phospholipid synthesis. In the mitochondria, this pathway may also toggle the ratio of NAD(P) to NAD(P)H to maintain redox homeostasis and provide essential precursors for mitochondrial protein translation. While several studies have flirted with the concept of direct regulation by presenting an association between reprogrammed SGOC metabolism and histone/DNA methylation in cancer cells, the key piece of evidence missing is significant labeling data showing transfer of the third position carbon of serine onto the methyl group of SAM and onto methyl groups present on histones or DNA. More work needs to be done to clarify these findings, and the answer may likely be dependent on cell type or environmental context.
Further investigations are also needed in understanding the requirements and sensitivities of cancer cells to mitochondrial 1C enzyme inhibition in vivo. The first barrier to this goal is developing specific inhibitors that affect only mitochondrial 1C enzymes, as the most well-characterized inhibitors to-date have residual activity against the cytosolic isozymes or enzymes involved in purine biosynthesis. Second, the majority of genetic and pharmacologic experiments to date have been performed in xenograft models in immunodeficient mice, which may present an overestimation of the therapeutic efficacy of targeting this pathway due to the fact that SGOC pathway is also essential for immune cells’ proliferation. This warrants the need to study mitochondrial 1C metabolic interventions in alternative systems such as 3D organoids or humanized mice, to more faithfully recapitulate human tumor biology. Nonetheless, it is reasonable to hope that a therapeutic window might be achieved to selectively target cancer cells, perhaps in specific cancer types or a subset of cells in a tumor that exhibit the highest flux through the mitochondrial 1C pathway.
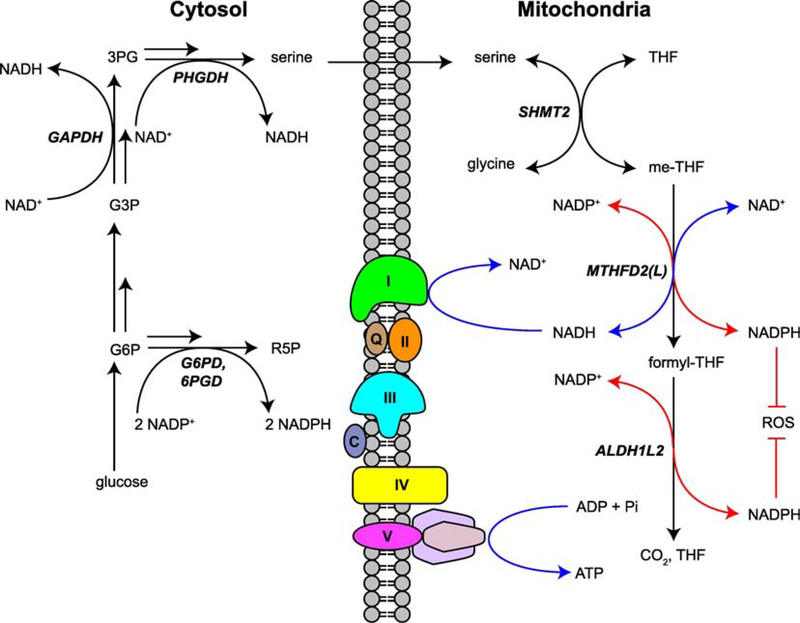
Figure 2. Mitochondrial serine catabolism contributes to shifting NADH and NADPH levels.
Serine catabolism through the mitochondrial folate cycle contributes substantial amounts of NADPH and NADH, particularly under stress conditions such as respiration deficiency. NADH production may be intimately tied to energy production through coupling of MTHFD2(L)-derived NADH to the electron transport chain, which produces ATP. Electron transport chain complexes I, II, III, IV, V are denoted by their respective Roman numerals, and the mobile electron carriers ubiquinone and cytochrome C by Q and C, respectively. Only the inner membrane is shown. NADPH produced by either MTHFD2(L) or ALDH1L2 may be crucial to controlling ROS accumulation. An analogy to cytoplasmic pathways that produce NADH and NADPH from glucose (either through glycolysis or shunt pathways) is provided.
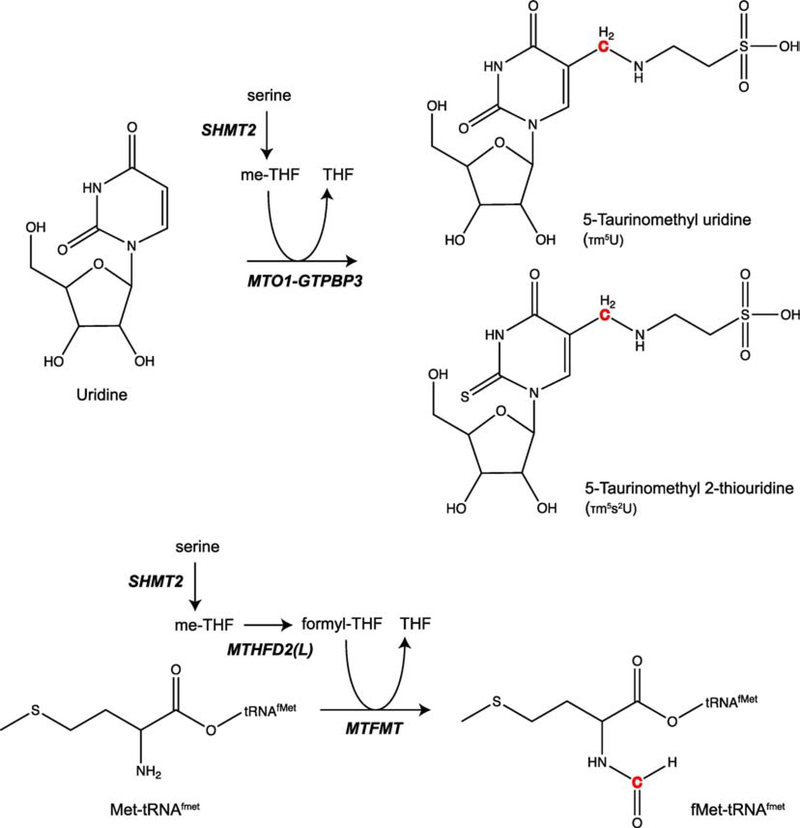
Figure 3. Mechanism of tRNA modification from products of either SHMT2 or MTHFD2(L) activity.
5,10-methylene-THF (me-THF) produced by SHMT2 is used as a substrate by the enzyme complex of MTO1 and GTPBP3 to generate the 5-taurinomethyl modification on uridine, producing either 5-taurinomethyl uridine or its derivative 5-taurionmethyl 2-thiouridine. 10-formyl-THF (formyl-THF) produced by MTHFD2(L), downstream of SHMT2 production of me-THF, is used as a substrate by MTMFT to formylate methionine on formyl-methionine tRNAs (tRNAMet), the tRNAs used to initiate mitochondrial protein translation. The 1C unit contribution to each respective modification from either me-THF or formyl-THF is highlighted in red.
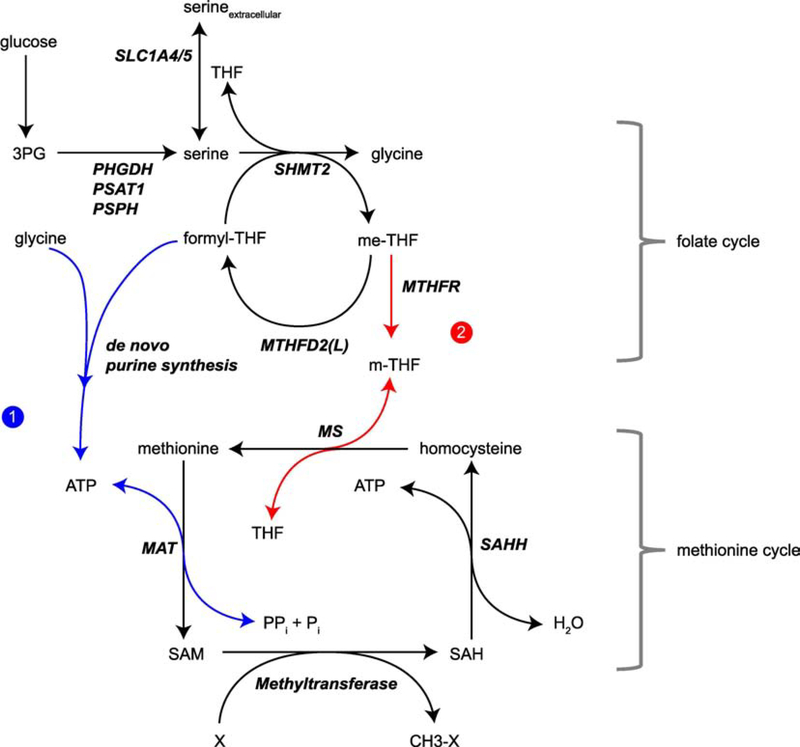
Figure 4. The folate cycle is coupled to the methionine cycle through distinct mechanisms.
The products of serine, either transported into the cell or synthesized from glucose de novo, contributes to SAM generation in two ways. Under normal culture conditions with sufficient methionine, the serine catabolism products glycine and formyl-THF support SAM synthesis through ATP (pathway 1, in blue). In the absence of methionine, serine becomes the crucial 1C donor for the re-methylation of methionine from homocysteine (pathway 2, in red).
Highlights.
Elevated mitochondrial serine, glycine, one-carbon (SGOC) metabolism is a feature of many cancers
In addition to providing substrates and precursors for biosynthesis, mitochondrial SGOC metabolism may help cancer cells maintain redox homeostasis and mitochondrial function
There exists considerable controversy in the link between SGOC metabolic control of epigenetics
Targeting mitochondrial SGOC is an attractive direction for cancer therapy
Acknowledgments
Financial Support: This work was supported by a NIH T32 Training Grant (CA009302-40) to A.M.L., and a NIH R00 Grant (CA184239) and a Mary Kay Foundation Innovative Cancer Research Award (017-37) to J.Y.
Abbreviations used
- tRNA
transfer ribonucleic acid
- SGOC
serine glycine and one-carbon
- TCA
tricarboxylic acid
- ATP
adenosine triphosphate
- PKM2
pyruvate kinase M2 isoform
- PHGDH
3-phosphoglycerate dehydrogenase
- THF
tetrahydrofolate
- 1C
one-carbon
- ALL
acute lymphoblastic leukemia
- SHMT1/2
serine hydroxymethyltransferase 1/2
- me-THF
5,10-methylene-THF
- GLDC
glycine decarboxylase complex
- TYMS
thymidylate synthetase
- formyl-THF
10-formyl-THF
- MTHFD1/2
methylene-tetrahydrofolate dehydrogenase 1/2
- MTHFD1/2L
methylene-tetrahydrofolate dehydrogenase 1/2-like
- NAD(P)(H)
nicotinamide adenine dinucleotide (phosphate
- GART
trifunctional enzyme phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase
- ATIC
bifunctional enzyme 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
- M+n
mass heavy by n atomic units
- dTTP
deoxythymidine triphosphate
- IDH1/2/3
isocitrate dehydrogenase 1/2/3
- PSAT1
phosphoserine aminotransferase
- R-2HG
R enantiomer of 2-hydroxyglutarate
- PSPH
phosphoserine phosphatase
- SLC1A4/5
solute carrier family 1 member 4/5
- CRISPR-CAS9
clustered regularly interspaced short palindromic repeats – CRISPR-associated protein 9 gene editing system
- SFXN1
sideroflexin 1
- HIF1/2
hypoxia-inducible factor 1/2
- TIC
tumor initiating cell
- DNA
deoxyribonucleic acid
- ROS
reactive oxygen species
- ALDH1L2
aldehyde dehydrogenase 1 family member L2
- CO2
carbon dioxide
- ETC
electron transport chain
- OXPHOS
oxidative phosphorylation
- DHFR
dihydrofolate reductase
- SAM
S-adenosyl methionine
- MAT
methionine adenosyltransferase
- MTHFR
methylenetetrahydrofolate reductase
- m-THF
5-methyltetrahydrofolate
- MS
methionine synthase
- LKB1
liver kinase B1
- PKCλ/ι
atypical protein kinase C lambda/iota
- m6A
N6-methylated adenosine
- ATF4
activating transcription factor 4
- NRF2
nuclear factor erythroid 2-related transcription factor 2
- KEAP1
kelch-like ECH-associated protein 1
- mTORC1
mammalian target of rapamycin complex 1
- SIRT5
silent mating type information regulation 2 homolog 5
- EIF2A
eukaryotic initiation factor 2
- EWS/FLI
RNA-binding protein EWS/Frind leukemia integration 1 transcription factor fusion protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Works Cited
- Agarwal S, Behring M, Hale K, Al Diffalha S, Wang K, Manne U, and Varambally S (2019). MTHFD1L, A Folate Cycle Enzyme, Is Involved in Progression of Colorectal Cancer. Transl Oncol 12, 1461–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S, Sabatini DM, Birsoy K, and Possemato R (2017). NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature 551, 639–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, et al. (2011). Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334, 1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DD, Quintero CM, and Stover PJ (2011). Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria. Proc Natl Acad Sci U S A 108, 15163–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DD, Woeller CF, Chiang EP, Shane B, and Stover PJ (2012). Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J Biol Chem 287, 7051–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et al. (2016). Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, and Manning BD (2016). mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt S, Bayerlova M, Vetter M, Wachter A, Mitra D, Hanf V, Lantzsch T, Uleer C, Peschel S, John J, et al. (2017). Proteomic profiling of breast cancer metabolism identifies SHMT2 and ASCT2 as prognostic factors. Breast Cancer Res 19, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertino JR, Goker E, Gorlick R, Li WW, and Banerjee D (1996). Resistance mechanisms to methotrexate in tumors. Stem Cells 14, 5–9. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, and Sabatini DM (2015). An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, et al. (2012). Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491, 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Moran RG, and Goldman ID (2007). Pemetrexed: biochemical and cellular pharmacology, mechanisms, and clinical applications. Mol Cancer Ther 6, 404–417. [DOI] [PubMed] [Google Scholar]
- Chen CL, and Perrimon N (2017). Proximity-dependent labeling methods for proteomic profiling in living cells. Wiley Interdiscip Rev Dev Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, He J, Zhou J, Xiao Z, Ding N, Duan Y, Li W, and Sun LQ (2019a). EIF2A promotes cell survival during paclitaxel treatment in vitro and in vivo. J Cell Mol Med 23, 6060–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang Z, Hoshino A, Zheng HD, Morley M, Arany Z, and Rabinowitz JD (2019b). NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab 1, 404–415. [PMC free article] [PubMed] [Google Scholar]
- Cravo ML, Pinto AG, Chaves P, Cruz JA, Lage P, Nobre Leitao C, and Costa Mira F (1998). Effect of folate supplementation on DNA methylation of rectal mucosa in patients with colonic adenomas: correlation with nutrient intake. Clin Nutr 17, 45–49. [DOI] [PubMed] [Google Scholar]
- Dai Z, Mentch SJ, Gao X, Nichenametla SN, and Locasale JW (2018). Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat Commun 9, 1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danenberg PV (1977). Thymidylate synthetase - a target enzyme in cancer chemotherapy. Biochim Biophys Acta 473, 73–92. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. (2009). Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SR, Stacpoole PW, Williamson J, Kick LS, Quinlivan EP, Coats BS, Shane B, Bailey LB, and Gregory JF 3rd (2004). Tracer-derived total and folate-dependent homocysteine remethylation and synthesis rates in humans indicate that serine is the main one-carbon donor. Am J Physiol Endocrinol Metab 286, E272–279. [DOI] [PubMed] [Google Scholar]
- Dekhne AS, Shah K, Ducker GS, Katinas JM, Wong-Roushar J, Nayeen MJ, Doshi A, Ning C, Bao X, Fruhauf J, et al. (2019). Novel Pyrrolo[3,2-d]pyrimidine Compounds Target Mitochondrial and Cytosolic One-carbon Metabolism with Broad-spectrum Antitumor Efficacy. Mol Cancer Ther 18, 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, et al. (2015). NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 47, 1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, and Rabinowitz JD (2016). Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab 23, 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Ghergurovich JM, Mainolfi N, Suri V, Jeong SK, Hsin-Jung Li S, Friedman A, Manfredi MG, Gitai Z, Kim H, et al. (2017). Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A 114, 11404–11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, and Rabinowitz JD (2017). One-Carbon Metabolism in Health and Disease. Cell Metab 25, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, and Rabinowitz JD (2014). Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber S, Cutler EC, Hawkins JW, Harrison JH, Peirce EC 2nd, and Lenz GG (1947). The Action of Pteroylglutamic Conjugates on Man. Science 106, 619–621. [DOI] [PubMed] [Google Scholar]
- Farber S, and Diamond LK (1948). Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med 238, 787–793. [DOI] [PubMed] [Google Scholar]
- Figueiredo JC, Grau MV, Haile RW, Sandler RS, Summers RW, Bresalier RS, Burke CA, McKeown-Eyssen GE, and Baron JA (2009). Folic acid and risk of prostate cancer: results from a randomized clinical trial. J Natl Cancer Inst 101, 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Sikandar A, Donner J, Zaburannyi N, Herrmann J, Reck M, Wagner-Dobler I, Koehnke J, and Muller R (2017). The natural product carolacton inhibits folate-dependent C1 metabolism by targeting FolD/MTHFD. Nat Commun 8, 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Sanderson SM, Dai Z, Reid MA, Cooper DE, Lu M, Richie JP Jr., Ciccarella A, Calcagnotto A, Mikhael PG, et al. (2019). Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 572, 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina G, Brunotti P, Fiascarelli A, Cicalini A, Costa MG, Buckle AM, di Salvo ML, Giorgi A, Marani M, Paone A, et al. (2015). How pyridoxal 5′-phosphate differentially regulates human cytosolic and mitochondrial serine hydroxymethyltransferase oligomeric state. FEBS J 282, 1225–1241. [DOI] [PubMed] [Google Scholar]
- Green NH, Galvan DL, Badal SS, Chang BH, LeBleu VS, Long J, Jonasch E, and Danesh FR (2019). MTHFD2 links RNA methylation to metabolic reprogramming in renal cell carcinoma. Oncogene 38, 6211–6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory JF 3rd, Cuskelly GJ, Shane B, Toth JP, Baumgartner TG, and Stacpoole PW (2000). Primed, constant infusion with [2H3]serine allows in vivo kinetic measurement of serine turnover, homocysteine remethylation, and transsulfuration processes in human one-carbon metabolism. Am J Clin Nutr 72, 1535–1541. [DOI] [PubMed] [Google Scholar]
- Guo W, Healey JH, Meyers PA, Ladanyi M, Huvos AG, Bertino JR, and Gorlick R (1999). Mechanisms of methotrexate resistance in osteosarcoma. Clin Cancer Res 5, 621–627. [PubMed] [Google Scholar]
- Gustafsson R, Jemth AS, Gustafsson NM, Farnegardh K, Loseva O, Wiita E, Bonagas N, Dahllund L, Llona-Minguez S, Haggblad M, et al. (2017). Crystal Structure of the Emerging Cancer Target MTHFD2 in Complex with a Substrate-Based Inhibitor. Cancer Res 77, 937–948. [DOI] [PubMed] [Google Scholar]
- Gustafsson Sheppard N, Jarl L, Mahadessian D, Strittmatter L, Schmidt A, Madhusudan N, Tegner J, Lundberg EK, Asplund A, Jain M, et al. (2015). The folate-coupled enzyme MTHFD2 is a nuclear protein and promotes cell proliferation. Sci Rep 5, 15029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty TJ, Zeller KI, Osthus RC, Wonsey DR, and Dang CV (2003). A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc Natl Acad Sci U S A 100, 5313–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. (2003). An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11, 619–633. [DOI] [PubMed] [Google Scholar]
- Hoffman RM, and Erbe RW (1976). High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc Natl Acad Sci U S A 73, 1523–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, and Mootha VK (2012). Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336, 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju HQ, Lu YX, Chen DL, Zuo ZX, Liu ZX, Wu QN, Mo HY, Wang ZX, Wang DS, Pu HY, et al. (2019). Modulation of Redox Homeostasis by Inhibition of MTHFD2 in Colorectal Cancer: Mechanisms and Therapeutic Implications. J Natl Cancer Inst 111, 584–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung AY, Smulders Y, Verhoef P, Kok FJ, Blom H, Kok RM, Kampman E, and Durga J (2011). No effect of folic acid supplementation on global DNA methylation in men and women with moderately elevated homocysteine. PLoS One 6, e24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, Cantor JR, et al. (2015). SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature 520, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Huang T, Schirch D, and Schirch V (1996). Properties of tetrahydropteroylpentaglutamate bound to 10-formyltetrahydrofolate dehydrogenase. Biochemistry 35, 15772–15783. [DOI] [PubMed] [Google Scholar]
- Kim YI, Baik HW, Fawaz K, Knox T, Lee YM, Norton R, Libby E, and Mason JB (2001). Effects of folate supplementation on two provisional molecular markers of colon cancer: a prospective, randomized trial. Am J Gastroenterol 96, 184–195. [DOI] [PubMed] [Google Scholar]
- Kory N, Wyant GA, Prakash G, Uit de Bos J, Bottanelli F, Pacold ME, Chan SH, Lewis CA, Wang T, Keys HR, et al. (2018). SFXN1 is a mitochondrial serine transporter required for one-carbon metabolism. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottakis F, Nicolay BN, Roumane A, Karnik R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T, et al. (2016). LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature 539, 390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koufaris C, and Nilsson R (2018). Protein interaction and functional data indicate MTHFD2 involvement in RNA processing and translation. Cancer Metab 6, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JM (2004). Toward a better understanding of methotrexate. Arthritis Rheum 50, 1370–1382. [DOI] [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, and Maddocks OD (2014). Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep 7, 1248–1258. [DOI] [PubMed] [Google Scholar]
- Lehtinen L, Ketola K, Makela R, Mpindi JP, Viitala M, Kallioniemi O, and Iljin K (2013). High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget 4, 48–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivonen SK, Rokka A, Ostling P, Kohonen P, Corthals GL, Kallioniemi O, and Perala M (2011). Identification of miR-193b targets in breast cancer cells and systems biological analysis of their functional impact. Mol Cell Proteomics 10, M110 005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, and Metallo CM (2014). Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell 55, 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li AM, Ducker GS, Li Y, Seoane JA, Xiao Y, Melemenidis S, Zhou Y, Liu L, Vanharanta S, Graves EE, et al. (2020). Metabolic Profiling Reveals a Dependency of Human Metastatic Breast Cancer on Mitochondrial Serine and One-Carbon Unit Metabolism. Mol Cancer Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Huang B, Wang H, Liu X, Hong Y, Qiu S, and Zheng J (2018). MTHFD2 Overexpression Predicts Poor Prognosis in Renal Cell Carcinoma and is Associated with Cell Proliferation and Vimentin-Modulated Migration and Invasion. Cell Physiol Biochem 51, 991–1000. [DOI] [PubMed] [Google Scholar]
- Locasale JW (2013). Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. (2011). Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 43, 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, and Ron D (2004). Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167, 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, Mainolfi N, Suri V, Guak H, Balmer ML, et al. (2017). Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell Metab 25, 345–357. [DOI] [PubMed] [Google Scholar]
- MacFarlane AJ, Liu X, Perry CA, Flodby P, Allen RH, Stabler SP, and Stover PJ (2008). Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J Biol Chem 283, 25846–25853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, and Vousden KH (2013). Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Labuschagne CF, Adams PD, and Vousden KH (2016). Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol Cell 61, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao DY, Watson JD, Yan PS, Barsyte-Lovejoy D, Khosravi F, Wong WW, Farnham PJ, Huang TH, and Penn LZ (2003). Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr Biol 13, 882–886. [DOI] [PubMed] [Google Scholar]
- Marani M, Paone A, Fiascarelli A, Macone A, Gargano M, Rinaldo S, Giardina G, Pontecorvi V, Koes D, McDermott L, et al. (2016). A pyrazolopyran derivative preferentially inhibits the activity of human cytosolic serine hydroxymethyltransferase and induces cell death in lung cancer cells. Oncotarget 7, 4570–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JL, Kouri FM, Hurley LA, Liu J, Tommasini-Ghelfi S, Ji Y, Gao P, Calvert AE, Lee A, Chandel NS, et al. (2019). IDH3alpha regulates one-carbon metabolism in glioblastoma. Sci Adv 5, eaat0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser J, Schuster A, Pietzke M, Vande Voorde J, Athineos D, Oizel K, Burgos-Barragan G, Wit N, Dhayade S, Morton JP, et al. (2018). Increased formate overflow is a hallmark of oxidative cancer. Nat Commun 9, 1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser J, Tumanov S, Maddocks O, Labuschagne CF, Athineos D, Van Den Broek N, Mackay GM, Gottlieb E, Blyth K, Vousden K, et al. (2016). Serine one-carbon catabolism with formate overflow. Sci Adv 2, e1601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melber A, and Haynes CM (2018). UPR(mt) regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res 28, 281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gomez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, et al. (2015). Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab 22, 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton DR, Nam M, McLaughlin DJ, Shin J, Bayraktar EC, Alvarez SW, Sviderskiy VO, Papagiannakopoulos T, Sabatini DM, Birsoy K, et al. (2018). Serine Catabolism by SHMT2 Is Required for Proper Mitochondrial Translation Initiation and Maintenance of Formylmethionyl-tRNAs. Mol Cell 69, 610–621 e615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyo M, Konno M, Colvin H, Nishida N, Koseki J, Kawamoto K, Tsunekuni K, Nishimura J, Hata T, Takemasa I, et al. (2017). The importance of mitochondrial folate enzymes in human colorectal cancer. Oncol Rep 37, 417–425. [DOI] [PubMed] [Google Scholar]
- Morscher RJ, Ducker GS, Li SH, Mayer JA, Gitai Z, Sperl W, and Rabinowitz JD (2018). Mitochondrial translation requires folate-dependent tRNA methylation. Nature 554, 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, and Cole MD (2002). A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol 22, 5793–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson LM, Forshell TZ, Rimpi S, Kreutzer C, Pretsch W, Bornkamm GW, and Nilsson JA (2012). Mouse genetics suggests cell-context dependency for Myc-regulated metabolic enzymes during tumorigenesis. PLoS Genet 8, e1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, and Mootha VK (2014). Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun 5, 3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacold ME, Brimacombe KR, Chan SH, Rohde JM, Lewis CA, Swier LJ, Possemato R, Chen WW, Sullivan LB, Fiske BP, et al. (2016). A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol 12, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paone A, Marani M, Fiascarelli A, Rinaldo S, Giardina G, Contestabile R, Paiardini A, and Cutruzzola F (2014). SHMT1 knockdown induces apoptosis in lung cancer cells by causing uracil misincorporation. Cell Death Dis 5, e1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternack LB, Laude DA Jr., and Appling DR (1992). 13C NMR detection of folate-mediated serine and glycine synthesis in vivo in Saccharomyces cerevisiae. Biochemistry 31, 8713–8719. [DOI] [PubMed] [Google Scholar]
- Pavlova NN, and Thompson CB (2016). The Emerging Hallmarks of Cancer Metabolism. Cell Metab 23, 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike ST, Rajendra R, Artzt K, and Appling DR (2010). Mitochondrial C1-tetrahydrofolate synthase (MTHFD1L) supports the flow of mitochondrial one-carbon units into the methyl cycle in embryos. J Biol Chem 285, 4612–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikman Y, Puissant A, Alexe G, Furman A, Chen LM, Frumm SM, Ross L, Fenouille N, Bassil CF, Lewis CA, et al. (2016). Targeting MTHFD2 in acute myeloid leukemia. J Exp Med 213, 1285–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper MD, Hong SP, Ball GE, and Dawes IW (2000). Regulation of the balance of one-carbon metabolism in Saccharomyces cerevisiae. J Biol Chem 275, 30987–30995. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, Leitch AM, Johnson TM, DeBerardinis RJ, and Morrison SJ (2015). Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 527, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. (2011). Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufulete M, Al-Ghnaniem R, Khushal A, Appleby P, Harris N, Gout S, Emery PW, and Sanders TA (2005). Effect of folic acid supplementation on genomic DNA methylation in patients with colorectal adenoma. Gut 54, 648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, and Auwerx J (2017). Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol 216, 2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-Campos M, Linares JF, Duran A, Cordes T, L’Hermitte A, Badur MG, Bhangoo MS, Thorson PK, Richards A, Rooslid T, et al. (2019). Increased Serine and One-Carbon Pathway Metabolism by PKClambda/iota Deficiency Promotes Neuroendocrine Prostate Cancer. Cancer Cell 35, 385–400 e389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo de la Vega M, Chapman E, and Zhang DD (2018). NRF2 and the Hallmarks of Cancer. Cancer Cell 34, 21–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron-Harel N, Notarangelo G, Ghergurovich JM, Paulo JA, Sage PT, Santos D, Satterstrom FK, Gygi SP, Rabinowitz JD, Sharpe AH, et al. (2018). Defective respiration and one-carbon metabolism contribute to impaired naive T cell activation in aged mice. Proc Natl Acad Sci U S A 115, 13347–13352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D, Park Y, Andrabi SA, Shelton LM, Gilkes DM, and Semenza GL (2016). PHGDH Expression Is Required for Mitochondrial Redox Homeostasis, Breast Cancer Stem Cell Maintenance, and Lung Metastasis. Cancer Res 76, 4430–4442. [DOI] [PubMed] [Google Scholar]
- Sato A, Nishino T, Noda K, Amaya Y, and Nishino T (1995). The structure of chicken liver xanthine dehydrogenase. cDNA cloning and the domain structure. J Biol Chem 270, 2818–2826. [DOI] [PubMed] [Google Scholar]
- Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, and Brugge JS (2009). Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 461, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C, Chen VJ, Gossett LS, Gates SB, MacKellar WC, Habeck LL, Shackelford KA, Mendelsohn LG, Soose DJ, Patel VF, et al. (1997). LY231514, a pyrrolo[2,3d]pyrimidine-based antifolate that inhibits multiple folate-requiring enzymes. Cancer Res 57, 1116–1123. [PubMed] [Google Scholar]
- Shin M, Momb J, and Appling DR (2017). Human mitochondrial MTHFD2 is a dual redox cofactor-specific methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase. Cancer Metab 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. (2013). Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 339, 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanand S, Viney I, and Wellen KE (2018). Spatiotemporal Control of Acetyl-CoA Metabolism in Chromatin Regulation. Trends Biochem Sci 43, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli JB, Yoon H, Ringel AE, Jeanfavre S, Clish CB, and Haigis MC (2017). Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science 358, 941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine ZE, Walton ZE, Altman BJ, Hsieh AL, and Dang CV (2015). MYC, Metabolism, and Cancer. Cancer Discov 5, 1024–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, and Vander Heiden MG (2015). Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 162, 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Song L, Wan Q, Wu G, Li X, Wang Y, Wang J, Liu Z, Zhong X, He X, et al. (2015). cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res 25, 429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Mito T, Velagapudi V, Ishikawa K, Umehara M, Nakada K, Suomalainen A, and Hayashi JI (2019). Disruption of the mouse Shmt2 gene confers embryonic anaemia via foetal liver-specific metabolomic disorders. Sci Rep 9, 16054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Ohnishi S, Shitara H, Mito T, Yamaguchi M, Yonekawa H, Hashizume O, Ishikawa K, Nakada K, and Hayashi JI (2018). Mice deficient in the Shmt2 gene have mitochondrial respiration defects and are embryonic lethal. Sci Rep 8, 425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner JM, Bensard C, Wei P, Krah NM, Schell JC, Gardiner J, Schiffman J, Lessnick SL, and Rutter J (2017). EWS/FLI is a Master Regulator of Metabolic Reprogramming in Ewing Sarcoma. Mol Cancer Res 15, 1517–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts AS, and Appling DR (2010). Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr 30, 57–81. [DOI] [PubMed] [Google Scholar]
- Tyynismaa H, Carroll CJ, Raimundo N, Ahola-Erkkila S, Wenz T, Ruhanen H, Guse K, Hemminki A, Peltola-Mjosund KE, Tulkki V, et al. (2010). Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet 19, 3948–3958. [DOI] [PubMed] [Google Scholar]
- Walling J (2006). From methotrexate to pemetrexed and beyond. A review of the pharmacodynamic and clinical properties of antifolates. Invest New Drugs 24, 37–77. [DOI] [PubMed] [Google Scholar]
- Wang B, Wang W, Zhu Z, Zhang X, Tang F, Wang D, Liu X, Yan X, and Zhuang H (2017). Mitochondrial serine hydroxymethyltransferase 2 is a potential diagnostic and prognostic biomarker for human glioma. Clin Neurol Neurosurg 154, 28–33. [DOI] [PubMed] [Google Scholar]
- Wang Z, Yip LY, Lee JHJ, Wu Z, Chew HY, Chong PKW, Teo CC, Ang HY, Peh KLE, Yuan J, et al. (2019). Methionine is a metabolic dependency of tumor-initiating cells. Nat Med 25, 825–837. [DOI] [PubMed] [Google Scholar]
- Warburg O, Posener K, and Negelein E (1924). On the metabolism of carcinoma cells. Biochem Z 152, 309–344. [Google Scholar]
- Warburg O, Wind F, and Negelein E (1927). The metabolism of tumors in the body. J Gen Physiol 8, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. (2010). The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson GR, McGlynn AP, McNulty H, O’Reilly SL, McKelvey-Martin VJ, McKerr G, Strain JJ, Scott J, and Downes CS (2006). Global DNA and p53 region-specific hypomethylation in human colonic cells is induced by folate depletion and reversed by folate supplementation. J Nutr 136, 2748–2753. [DOI] [PubMed] [Google Scholar]
- Woo CC, Chen WC, Teo XQ, Radda GK, and Lee PT (2016). Downregulating serine hydroxymethyltransferase 2 (SHMT2) suppresses tumorigenesis in human hepatocellular carcinoma. Oncotarget 7, 53005–53017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhang G, Li P, Chen S, Zhang F, Li J, Jiang C, Chen X, Wang Y, Du Y, et al. (2016a). miR-198 targets SHMT1 to inhibit cell proliferation and enhance cell apoptosis in lung adenocarcinoma. Tumour Biol 37, 5193–5202. [DOI] [PubMed] [Google Scholar]
- Wu X, Deng L, Tang D, Ying G, Yao X, Liu F, and Liang G (2016b). miR-615–5p prevents proliferation and migration through negatively regulating serine hydromethyltransferase 2 (SHMT2) in hepatocellular carcinoma. Tumour Biol 37, 6813–6821. [DOI] [PubMed] [Google Scholar]
- Xue G, Lu CJ, Pan SJ, Zhang YL, Miao H, Shan S, Zhu XT, and Zhang Y (2017). DNA hypomethylation of CBS promoter induced by folate deficiency is a potential noninvasive circulating biomarker for colorectal adenocarcinomas. Oncotarget 8, 51387–51401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Garcia Canaveras JC, Chen Z, Wang L, Liang L, Jang C, Mayr JA, Zhang Z, Ghergurovich JM, Zhan L, et al. (2020). Serine Catabolism Feeds NADH when Respiration Is Impaired. Cell Metab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, and Vousden KH (2016). Serine and one-carbon metabolism in cancer. Nat Rev Cancer 16, 650–662. [DOI] [PubMed] [Google Scholar]
- Yang X, Wang Z, Li X, Liu B, Liu M, Liu L, Chen S, Ren M, Wang Y, Yu M, et al. (2018). SHMT2 Desuccinylation by SIRT5 Drives Cancer Cell Proliferation. Cancer Res 78, 372–386. [DOI] [PubMed] [Google Scholar]
- Ye D, Guan KL, and Xiong Y (2018). Metabolism, Activity, and Targeting of D- and L-2-Hydroxyglutarates. Trends Cancer 4, 151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, et al. (2014a). Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov 4, 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, and Thompson CB (2012). Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci U S A 109, 6904–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Mimura J, Okada T, Sato H, Liu T, Maruyama A, Ohyama C, and Itoh K (2014b). Nrf2- and ATF4-dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome inhibition. Mol Cell Biol 34, 3421–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K (2015). Positive correlation between expression level of mitochondrial serine hydroxymethyltransferase and breast cancer grade. Onco Targets Ther 8, 1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al. (2012). Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 148, 259–272. [DOI] [PubMed] [Google Scholar]