Abstract
During the COVID-19 pandemic, SARS-CoV-2 infected millions of people and claimed hundreds of thousands of lives. Virus entry into cells depends on the receptor binding domain (RBD) of the SARS-CoV-2 spike protein (S). Although there is no vaccine, it is likely that antibodies will be essential for protection. However, little is known about the human antibody response to SARS-CoV-21–5. Here we report on 149 COVID-19 convalescent individuals. Plasmas collected an average of 39 days after the onset of symptoms had variable half-maximal pseudovirus neutralizing titers: less than 1:50 in 33% and below 1:1000 in 79%, while only 1% showed titers >1:5000. Antibody sequencing revealed expanded clones of RBD-specific memory B cells expressing closely related antibodies in different individuals. Despite low plasma titers, antibodies to three distinct epitopes on RBD neutralized at half-maximal inhibitory concentrations (IC50s) as low as single digit ng/mL. Thus, most convalescent plasmas obtained from individuals who recover from COVID-19 do not contain high levels of neutralizing activity. Nevertheless, rare but recurring RBD-specific antibodies with potent antiviral activity were found in all individuals tested, suggesting that a vaccine designed to elicit such antibodies could be broadly effective.
Between April 1 and May 8, 2020, 157 eligible participants enrolled in the study. Of these, 111 (70.7%) were individuals diagnosed with SARS-CoV-2 infection by RT-PCR (cases), and 46 (29.3%) were close contacts of individuals diagnosed with SARS-CoV-2 infection (contacts). While inclusion criteria allowed for enrollment of asymptomatic participants, 8 contacts that did not develop symptoms were excluded from further analyses. The 149 cases and contacts were free of symptoms suggestive of COVID-19 for at least 14 days at the time of sample collection. Participant demographics and clinical characteristics are shown in SI Tables 1 and 2 and Extended Data Fig. 1a. Only one individual who tested positive for SARS-CoV-2 infection by RT-PCR remained asymptomatic. The other 148 participants reported symptoms suggestive of COVID-19 with an average onset of approximately 39 days (range 17 to 67 days) before sample collection. In this cohort, symptoms lasted for an average of 12 days (0–35 days), and 11 (7%) of the participants were hospitalized. The most common symptoms were fever (83.9%), fatigue (71.1%), cough (62.4%) and myalgia (61.7%) while baseline comorbidities were infrequent (10.7%) (SI Tables 1 and 2). There were no significant differences in duration or severity (see Methods) of symptoms, or in time from onset of symptoms to sample collection between genders or between cases and contacts. There was no age difference between females and males in our cohort (Extended Data Fig. 1).
Plasma samples were tested for binding to the SARS-CoV-2 RBD and trimeric spike (S) proteins by a validated ELISA using anti-IgG or -IgM secondary antibodies for detection (Fig. 1, SI Table 1 and Extended Data Figs. 2 and 3)6,7. Eight independent negative controls and the positive control plasma sample from participant 21 (COV21) were included for normalization of the area under the curve in all experiments (AUC). Overall, 78% and 70% of the plasma samples tested showed anti-RBD and anti-S IgG AUCs that were at least 2 standard deviations above the control (Fig. 1 a, b). In contrast, only 15% and 34% of the plasma samples showed IgM responses to anti-RBD and anti-S that were at least 2 standard deviations above control, respectively (Fig. 1 c, d). There was no positive correlation between anti-RBD or -S IgG or IgM levels and duration of symptoms or the timing of sample collection relative to onset of symptoms (Fig. 1e, and Extended Data Figs. 3 a–c and 3 g–j). On the contrary, as might be expected, anti-RBD IgM titers were negatively correlated with duration of symptoms and the timing of sample collection (Fig. 1e and Extended Data Fig. 3h). Anti-RBD IgG levels were modestly correlated to age, and the severity of symptoms including hospitalization (Fig. 1 f, g and Extended Data Fig. 3k). Interestingly, females had lower anti-RBD and -S IgG titers than males (Fig. 1h, Extended Data Fig. 2f).
Figure 1. Plasma antibodies against SARS-CoV-2.
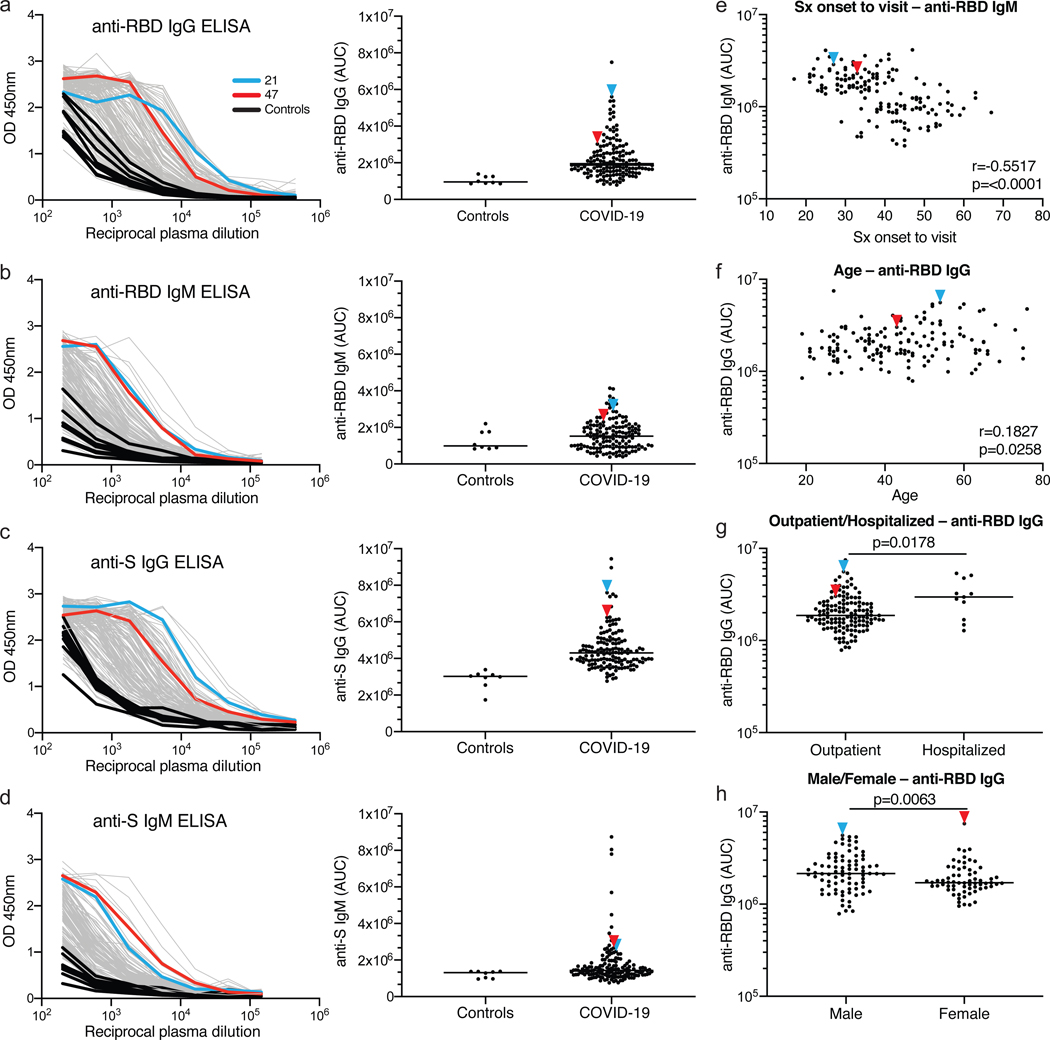
a-d, Graphs show results of ELISAs measuring plasma reactivity to RBD (a, b) and S protein (c, d). Left shows optical density units at 450 nm (OD, Y axis) and reciprocal plasma dilutions (X axis). Negative controls in black; individuals 21, and 47 in blue and red lines and arrowheads, respectively. Right shows normalized area under the curve (AUC) for 8 controls and each of 149 individuals in the cohort. e, Symptom (Sx) onset to time of sample collection in days (X axis) plotted against normalized AUC for IgM binding to RBD (Y axis); r=0.5517 and p=<0.0001. f, Participant age in years (X axis) plotted against normalized AUC for IgG binding to RBD (Y axis); r=0.1827 and p=0.0258. The r and p values for the correlations in e and f were determined by two-tailed Spearman’s. g, Normalized AUC of anti-RBD IgG ELISA for outpatients (n=138) and hospitalized (n=11) individuals; p=0.0178. h, Normalized AUC of anti-RBD IgG ELISA for males (n=83) and females (n=66); p=0.0063. For g and h horizontal bars indicate median values. Statistical significance was determined using two-tailed Mann-Whitney U test.
To measure the neutralizing activity in convalescent plasmas we used HIV-1-based virions carrying a nanoluc luciferase reporter that were pseudotyped with the SARS-CoV-2 spike (SARS-CoV-2 pseudovirus, see Methods, Fig. 2 and Extended Data Fig. 4). Negative (historical) and positive (COV21) controls were included in all experiments. The overall level of neutralizing activity in the cohort, as measured by the half-maximal neutralizing titer (NT50) was generally low, with 33% less than 50 and 79% below 1,000 (Fig. 2 a, b). The geometric mean NT50 was 121 (arithmetic mean = 714), and only 2 individuals reached NT50s above 5,000 (Fig. 2 a, b and SI Table 1).
Figure 2. Neutralization of SARS-CoV-2 pseudovirus by plasma.
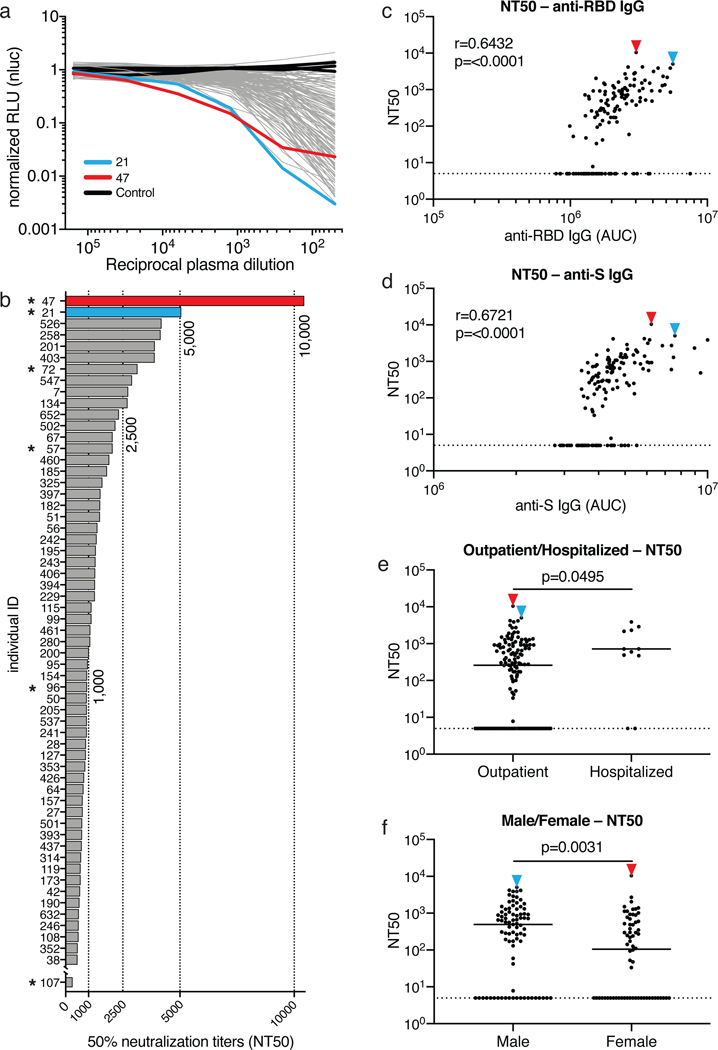
a, Graph shows normalized relative luminescence values (RLU, Y axis) in cell lysates of 293TACE2 cells 48 hours after infection with nanoluc-expressing SARS-CoV-2 pseudovirus in the presence of increasing concentrations of plasma (X axis) derived from 149 participants (grey, except individuals 21 and 47 in blue and red lines, bars and arrowheads, respectively) and 3 negative controls (black lines). Mean of duplicates; representative of two independent experiments. b, Ranked average half-maximal inhibitory plasma neutralizing titer (NT50) for the 59 of 149 individuals with NT50s >500 and individual 107. Asterisks indicate donors from which antibody sequences were derived. c, Normalized AUC for anti-RBD IgG ELISA (X axis) plotted against NT50 (Y axis); r=0.6432, p=<0.0001. d, Normalized AUC for anti-S IgG ELISA (X axis) plotted against NT50 (Y axis); r=0.6721, p=<0.0001. The r and p values for the correlations in c and d were determined by two-tailed Spearman’s. e, NT50 for outpatients (n=138) and hospitalized (n=11) individuals; p=0.0495. f, NT50 for males (n=83) and females (n=66) in the cohort; p=0.0031. Statistical significance in e and f was determined using two-tailed Mann-Whitney U test and horizontal bars indicate median values. Dotted lines in c to f (NT50=5) represents lower limit of detection (LLOD). Samples with neutralizing titers below 1:50 were plotted at LLOD.
Notably, levels of anti-RBD- and -S IgG antibodies correlated strongly with NT50 (Fig. 2 c, d). Neutralizing activity also correlated with age, duration of symptoms and symptom severity (Extended Data Fig. 5). Consistent with this observation, hospitalized individuals with longer symptom duration showed slightly higher average levels of neutralizing activity than non-hospitalized individuals (p=0.0495, Fig. 2e). Finally, we observed a significant difference in neutralizing activity between males and females (p=0.0031, Fig. 2f). The difference between males and females was consistent with higher anti-RBD and -S IgG titers in males, and could not be attributed to age, severity, timing of sample collection relative to onset of symptoms or duration of symptoms (Fig. 1h, Extended Data Fig. 1 a–d and 2f).
To determine the nature of the antibodies elicited by SARS-CoV-2 infection we used flow cytometry to isolate individual B lymphocytes with receptors that bound to RBD from the blood of 6 selected individuals including the 2 top and 4 high to intermediate neutralizers (Fig. 3). The frequency of antigen-specific B cells, identified by their ability to bind to both Phycoerythrin (PE)- and AF647-labeled RBD, ranged from 0.07 to 0.005% of all circulating B cells in COVID-19 convalescents but they were undetectable in pre-COVID-19 controls (Fig. 3a and Extended Data Fig. 6). We obtained 534 paired IgG heavy and light chain (IGH and IGL) sequences by reverse transcription and subsequent PCR from individual RBD-binding B cells from the 6 convalescent individuals (see Methods and SI Table 3). When compared to the human antibody repertoire, several IGHV and IGLV genes were significantly over-represented (Extended Data Fig. 7). The average number of V genes nucleotide mutations for IGH and IGL was 4.2 and 2.8, respectively (Extended Data Fig. 8), which is lower than in antibodies cloned from individuals suffering from chronic infections such as Hepatitis B or HIV-1, and similar to antibodies derived from primary malaria infection or non-antigen-enriched circulating IgG memory cells8–11. Among other antibody features, IGH CDR3 length was indistinguishable from the reported norm and hydrophobicity was below average (Extended Data Fig. 8)12.
Figure 3. Anti-SARS-CoV-2 RBD antibodies.
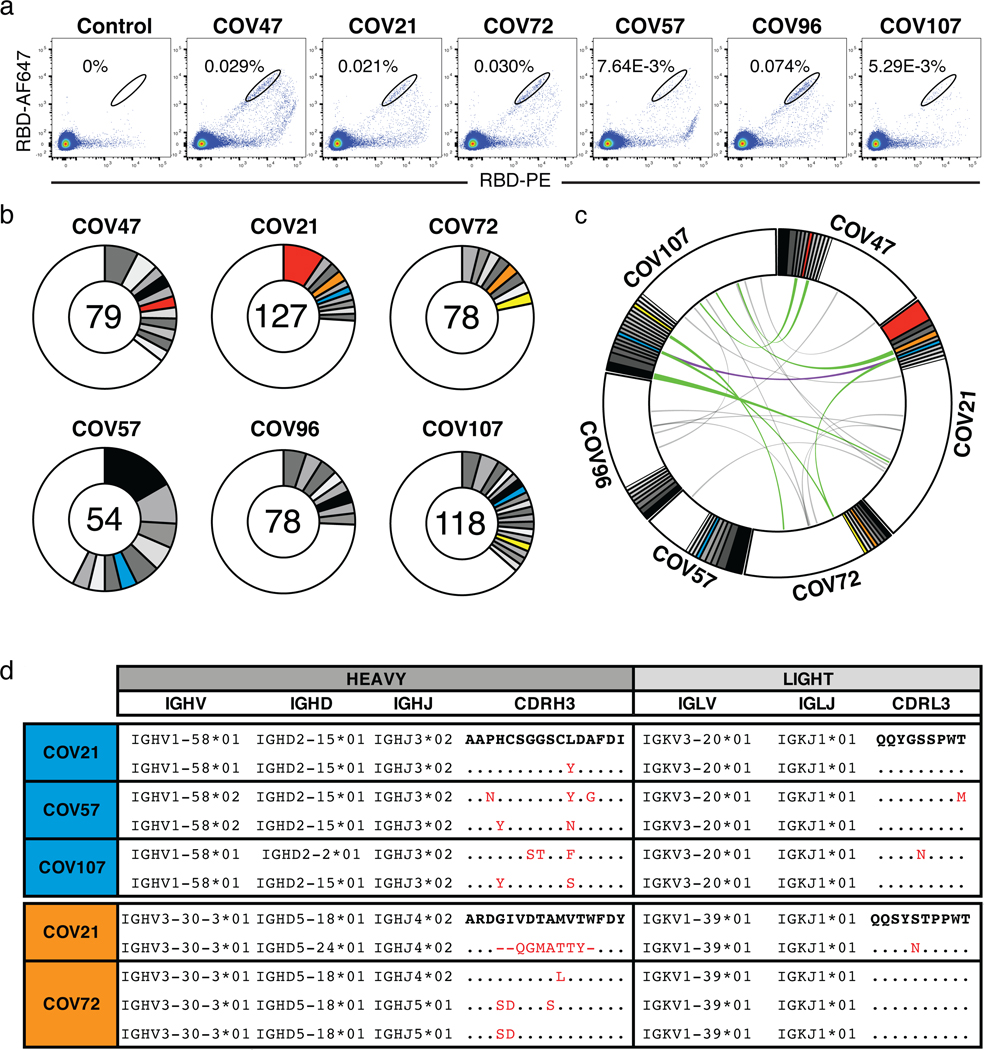
a. Representative flow cytometry plots showing dual AF647- and PE-RBD binding B cells in control and 6 study individuals (for gating strategy see Extended Data Fig. 6). Percentages of antigen specific B cells are indicated. Control is a healthy sample obtained before COVID-19. b, Pie charts depicting the distribution of antibody sequences from 6 individuals. The number in the inner circle indicates the number of sequences analyzed for the individual denoted above the circle. White indicates sequences isolated only once, and grey or colored pie slices are proportional to the number of clonally related sequences. Red, blue, orange and yellow pie slices indicate clones that share the same IGHV and IGLV genes. c, Circos plot shows sequences from all 6 individuals with clonal relationships depicted as in b. Interconnecting lines indicate the relationship between antibodies that share V and J gene segment sequences at both IGH and IGL. Purple, green and gray lines connect related clones, clones and singles, and singles to each other, respectively. d, Sample sequence alignment for antibodies originating from different individuals that display highly similar IGH V(D)J and IGL VJ sequences including CDR3s. Amino acid differences in CDR3s to the bolded reference sequence above are indicated in red and dots represent identities.
As is the case with other human pathogens, there were expanded clones of viral antigen binding B cells in all COVID-19 individuals tested (see Methods and Fig. 3 b,c). Overall, 32.2% of the recovered IGH and IGL sequences were from clonally expanded B cells (range 21.8–57.4% across individuals, Fig. 3b). Antibodies that shared specific combinations of IGHV and IGLV genes in different individuals comprised 14% of all the clonal sequences (colored pie slices in Fig. 3 b,c). Remarkably, the amino acid sequences of some antibodies found in different individuals were nearly identical (Fig. 3 e,d). For example, antibodies expressed by clonally expanded B cells with IGHV1-58/IGKV3-20 and IGHV3-30-3/IGKV1-39 found repeatedly in different individuals had amino acid sequence identities of up to 99% and 92%, respectively (Fig. 3d and SI Table 4). We conclude that the IgG memory response to the SARS-CoV-2 RBD is rich in recurrent and clonally expanded antibody sequences.
To examine the binding properties of anti-SARS-CoV-2 antibodies, we expressed 94 representative antibodies, 67 from clones and 27 from singlets (SI Table 5). ELISA assays showed that 95% (89 out of 94) of the antibodies tested including clonal and unique sequences bound to the SARS-CoV-2 RBD with an average half-maximal effective concentration (EC50) of 6.9 ng/mL (Fig. 4a and Extended Data Fig. 9a). A fraction of these (7 out of 77 that were tested, or 9%) cross-reacted with the RBD of SARS-CoV with EC50s below 1μg/mL (Extended Data Fig. 9b and c). No significant cross-reactivity was noted to the RBDs of MERS, HCoV-OC43, HCoV-229E or HCoV-NL63.
Figure 4. Anti-SARS-CoV-2 RBD antibody reactivity.
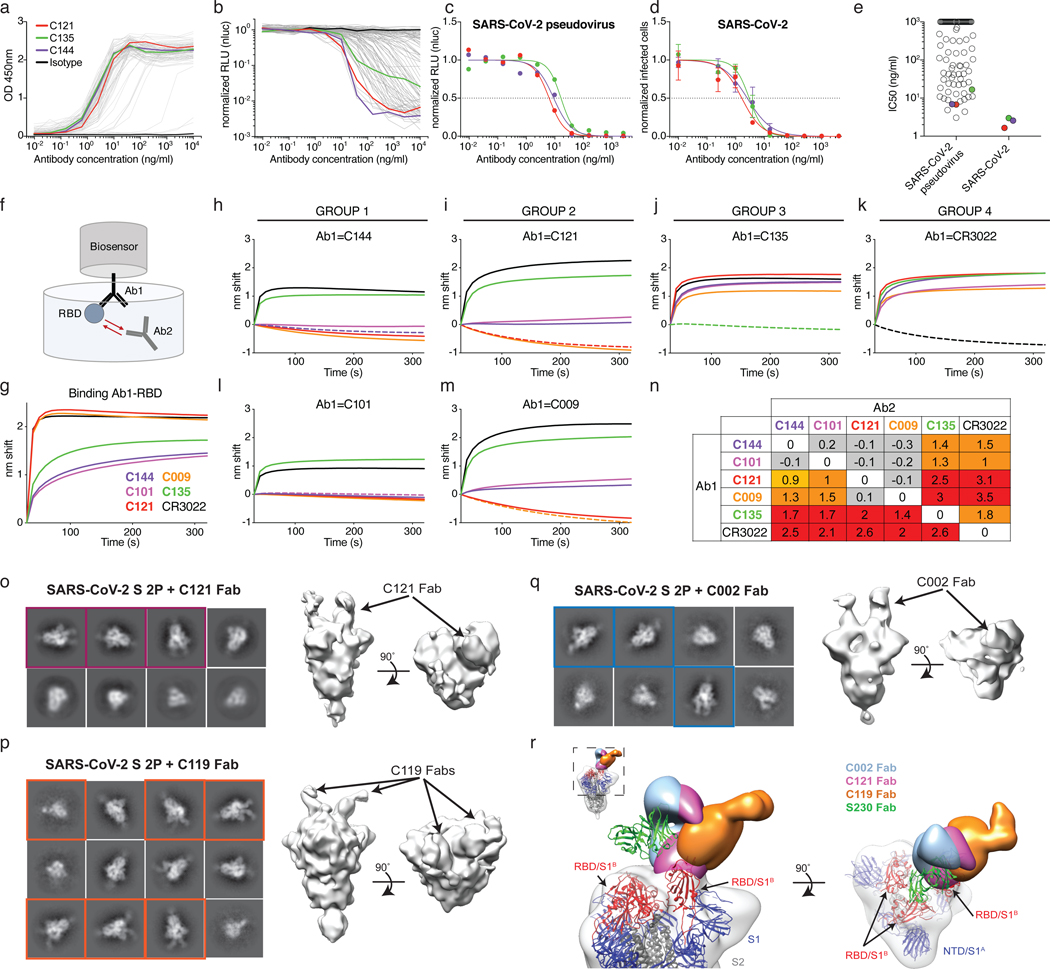
a, Graph shows results of ELISA assays measuring monoclonal antibody binding to RBD. Optical density units at 450 nm (OD, Y axis) vs. antibody concentrations (X axis); 94 samples and 1 isotype control. C121, C135 C144 and isotype control in red, green, purple, and black respectively, in all panels. b, Graph shows normalized relative luminescence values (RLU, Y axis) in cell lysates of 293TACE2 cells 48 hours after infection with SARS-CoV-2 pseudovirus in the presence of increasing concentrations of monoclonal antibodies (X axis). 89 samples and 1 isotype control. c, Normalized RLU for SARS-CoV-2 pseudovirus neutralization (Y axis) vs. titration of monoclonal antibodies C121, C135 and C144. d, SARS-CoV-2 real virus neutralization assay. Normalized infected cells (Y axis, determined by dividing the amount of infection per well by the average of control wells infected in the absence of antibodies) vs. titration of monoclonal antibodies C121, C135 and C144. a to d show a representative of two independent experiments. In b and c is mean of duplicates and in d is mean with standard deviation of triplicates. e, IC50s for antibodies assayed in b and d, the average value of at least two experiments is shown. Samples with IC50s above 1μg/ml were plotted at 1μg/ml; n=89 (pseudovirus) and n=3 (virus), respectively. f, Diagrammatic representation of biolayer interferometry experiment. g, Graph shows binding of C144, C101, C121, C009, C135, and CR302213,14 to RBD. h-n, Secondary antibody binding to preformed IgG-RBD complexes (Ab1). The table displays the shift in nanometers after second antibody (Ab2) binding to the antigen in the presence of the first antibody (Ab1). Values are normalized by the subtraction of the autologous antibody control. Representative of two experiments. o-q, Representative 2D-class averages and 3D reconstructed volumes for SARS-CoV-S 2P trimers complexed with C002, C119, and C121 Fabs. 2D class averages with observable Fab density are boxed. r,
Overlay of S-Fab complexes with fully-occupied C002 (blue), C121 (magenta) and C119 (orange) Fabs. The SARS-CoV-2 S model from PDB 6VYB was fit into the density and the SARS-CoV mAb S230 (PDB 6NB6) is shown as a reference (green ribbon).
To determine whether the monoclonal antibodies have neutralizing activity, we tested them against the SARS-CoV-2 pseudovirus (Fig. 4 and SI Table 6). Among 89 RBD binding antibodies tested, we found 52 that neutralized SARS-CoV-2 pseudovirus with nanogram per milliliter half-maximal inhibitory concentrations (IC50s) ranging from 3 to 709 (Fig. 4 b,c and e, SI Table 6). A subset of the most potent of these antibodies were also tested against authentic SARS-CoV-2 and neutralized with IC50s of less than 5 ng/ml (Fig. 4 d,e). Only two of the antibodies which cross-reacted with the RBD of SARS-CoV showed significant neutralizing activity against SARS-CoV pseudovirus (Extended Data Fig. 9d and e).
Potent neutralizing antibodies were found in individuals irrespective of their plasma NT50s. For example, C121, C144, and C135 with IC50s of 1.64, 2.55 and 2.98 ng/mL against authentic SARS-CoV-2, respectively, were obtained from individuals COV107, COV47, and COV72 whose plasma NT50 values were of 297, 10,433 and 3,138, respectively (Figs. 2b and 4). Finally, clones of antibodies with shared IGHV and IGLV genes were among the best neutralizers, e.g., antibody C002 composed of IGHV3-30/IGKV1-39 is shared by the 2 donors with the best plasma neutralizing activity (red pie slice in Fig. 3b and Fig. 4). We conclude that even individuals with modest plasma neutralizing activity harbor rare IgG memory B cells that produce potent SARS-CoV-2 neutralizing antibodies.
To determine whether human anti-SARS-CoV-2 monoclonal antibodies with neutralizing activity can bind to distinct domains on the RBD, we performed bilayer interferometry experiments in which a preformed antibody-RBD immune complex was exposed to a second monoclonal. The antibodies tested comprised 3 groups, all of which differ in their binding properties from CR3022, an antibody that neutralizes SARS-CoV and binds to, but does not neutralize SARS-CoV-213,14. Representatives of each of the 3 groups include: C144 and C101 in Group 1; C121 and C009 in Group 2; C135 in Group 3. All of these antibodies can bind after CR3022. Groups 1 and 2 also bind after Group 3, and Groups 1 and 2 differ in that Group 1 can bind after Group 2 but not vice versa (Fig. 4 f–n). We conclude that similar to SARS-CoV, there are multiple distinct neutralizing epitopes on the RBD of SARS-CoV-2.
To further define the binding characteristics of Groups 1 and 2 antibodies, we imaged SARS-CoV-2 S–Fab complexes by negative stain electron microscopy (nsEM) using C002 (Group 1, an IGHV3-30/IGKV1-39 antibody, which is clonally expanded in 2 donors), C119 and C121 (both in Group 2) Fabs (Fig. 4 f–r and Extended Data Fig. 10). Consistent with the conformational flexibility of the RBD, 2D class averages showed heterogeneity in both occupancy and orientations of bound Fabs for both groups (Fig. 4o–q). The low resolution of NS EM reconstructions precludes detailed binding interpretations, but the results are consistent with Fabs from both groups being able to recognize “up” and “down” states of the RBD, as previously described for some antibodies targeting this epitope15,16. The 3D reconstructions are also consistent with competition measurements indicating that Groups 1 and 2 antibodies bind a RBD epitope distinct from antibody CR3022 (Fig. 4f–n) and with a single-particle cryo-EM structure of a C105-S complex (https://www.biorxiv.org/content/10.1101/2020.05.28.121533v1.full.pdf). In addition, the structures suggest that the antibodies bind the RBD with different angles of approach, with Group 1 antibodies more similar to the approach angle of the SARS-CoV antibody S23017 (Fig. 4r).
Human monoclonal antibodies with neutralizing activity against pathogens ranging from viruses to parasites have been obtained from naturally infected individuals by single cell antibody cloning. Several have been shown to be effective in protection and therapy in model organisms and in early phase clinical studies, but only one antiviral monoclonal is currently in clinical use18. Antibodies are relatively expensive and more difficult to produce than small molecule drugs. However, they differ from drugs in that they can engage the host immune system through their constant domains that bind to Fc gamma receptors on host immune cells19. These interactions can enhance immunity and help clear the pathogen or infected cells, but they can also lead to disease enhancement during Dengue20 and possibly coronavirus infections21. This problem has impeded Dengue vaccine development but would not interfere with the clinical use of potent neutralizing antibodies that can be modified to prevent Fc gamma receptor interactions and remain protective against viral pathogens22.
Antibodies are essential elements of most vaccines and will likely be crucial component of an effective vaccine against SARS-CoV-223–25. Recurrent antibodies have been observed in other infectious diseases and vaccinal responses11,26–29. The observation that plasma neutralizing activity is low in most convalescent individuals, but that recurrent anti-SARS-CoV-2 RBD antibodies with potent neutralizing activity can be found in individuals with unexceptional plasma neutralizing activity suggests that humans are intrinsically capable of generating anti-RBD antibodies that potently neutralize SARS-CoV-2. Thus, vaccines that selectively and efficiently induce antibodies targeting the SARS-CoV-2 RBD may be especially effective.
Methods
Study participants.
Study participants were recruited at the Rockefeller University Hospital in New York from April 1 through May 8, 2020. Eligible participants were adults aged 18–76 years who were either diagnosed with SARS-CoV-2 infection by RT-PCR and were free of symptoms of COVID-19 for at least 14 days (cases), or who were close contacts (e.g., household, co-workers, members of same religious community) with someone who had been diagnosed with SARS-CoV-2 infection by RT-PCR and were free of symptoms suggestive of COVID-19 for at least 14 days (contacts). Exclusion criteria included presence of symptoms suggestive of active SARS-CoV-2 infection, or hemoglobin < 12 g/dL for males and < 11 g/dL for females.
Most study participants were residents of the Greater New York City tri-state region and were enrolled sequentially according to eligibility criteria. Participants were first interviewed by phone to collect information on their clinical presentation, and subsequently presented to the Rockefeller University Hospital for a single blood sample collection. Participants were asked to rate the highest severity of their symptoms on a numeric rating scale ranging from 0 to 10. The score was adapted from the pain scale chart, where 0 was the lack of symptoms, 4 were distressing symptoms (e.g. fatigue, myalgia, fever, cough, shortness of breath) that interfered with daily living activities, 7 were disabling symptoms that prevented the performance of daily living activities, and 10 was unimaginable/unspeakable discomfort (in this case, distress due to shortness of breath). All participants provided written informed consent before participation in the study and the study was conducted in accordance with Good Clinical Practice and clinical data collection and management was with software iRIS by iMedRIS. The study was performed in compliance with all relevant ethical regulations and the protocol for human subject studies was approved by the Institutional Review Board (IRB) of the Rockefeller University.
Blood samples processing and storage.
Peripheral Blood Mononuclear Cells (PBMCs) were obtained by gradient centrifugation and stored in liquid nitrogen in the presence of FCS and DMSO. Heparinized plasma and serum samples were aliquoted and stored at −20°C or less. Prior to experiments, aliquots of plasma samples were heat-inactivated (56C for 1 hour) and then stored at 4C.
Cloning, expression and purification of recombinant coronavirus proteins.
Codon-optimized nucleotide sequences encoding the SARS-CoV-2 S ectodomain (residues 16–1206) and receptor binding domain (RBD; residues 331–524) were synthesized and subcloned into the mammalian expression pTwist-CMV BetaGlobin vector by Twist Bioscience Technologies based on an early SARS-CoV-2 sequence isolate (GenBank MN985325.1). The SARS-CoV-2 RBD construct included an N-terminal human IL-2 signal peptide and dual C-terminal tags ((GGGGS)2-HHHHHHHH (octa-histidine), and GLNDIFEAQKIEWHE (AviTag)). In addition, the corresponding S1B or receptor binding domains for SARS-CoV (residues 318–510; GenBank AAP13441.1), MERS-CoV (residues 367–588; GenBank JX869059.2), HCoV-NL63 (residues 481–614; GenBank AAS58177.1), HCoV-OC43 (residues 324–632; GenBank AAT84362.1), and HCoV-229E (residues 286–434; GenBank AAK32191.1) were synthesized with the same N- and C-terminal extensions as the SARS-CoV-2 RBD construct and subcloned into the mammalian expression pTwist-CMV BetaGlobin vector (Twist Bioscience Technologies). The SARS-CoV-2 S ectodomain was modified as previously described4. Briefly, the S ectodomain construct included an N-terminal mu-phosphatase signal peptide, 2P stabilizing mutations (K986P and V987P), mutations to remove the S1/S2 furin cleavage site (682RRAR685 to GSAS), a C-terminal extension (IKGSG-RENLYFQG (TEV protease site), GGGSG-YIPEAPRDGQAYVRKDGEWVLLSTFL (foldon trimerization motif), G-HHHHHHHH (octa-histidine tag), and GLNDIFEAQKIEWHE (AviTag)). The SARS-CoV-2 S 2P ectodomain and RBD constructs were produced by transient transfection of 500 mL of Expi293F cells (Thermo Fisher) and purified from clarified transfected cell supernatants four days post-transfection using Ni2+-NTA affinity chromatography (GE Life Sciences). Affinity-purified proteins were concentrated and further purified by size-exclusion chromatography (SEC) using a Superdex200 16/60 column (GE Life Sciences) running in 1x TBS (20 mM Tris-HCl pH 8.0, 150 mM NaCl, and 0.02% NaN3). Peak fractions were analyzed by SDS- PAGE, and fractions corresponding to soluble S 2P trimers or monomeric RBD proteins were pooled and stored at 4°C.
ELISAs.
Validated ELISAs6,7 to evaluate antibodies binding to SARS-CoV-2 RBD and trimeric spike proteins, and to SARS-CoV RBD, were performed by coating of high binding 96 half well plates (Corning #3690) with 50 μL per well of a 1μg/mL protein solution in PBS overnight at 4°C. Plates were washed 6 times with washing buffer (1xPBS with 0.05% Tween 20 (Sigma-Aldrich)) and incubated with 170 μL per well blocking buffer (1xPBS with 2% BSA and 0.05% Tween20 (Sigma)) for 1 hour at room temperature (RT). Immediately after blocking, monoclonal antibodies or plasma samples were added in PBS and incubated for 1 hr at RT. Plasma samples were assayed at a 1:200 starting dilution and seven additional 3-fold serial dilutions. Monoclonal antibodies were tested at 10μg/ml starting concentration and 10 additional 4-fold serial dilutions. Plates were washed 6 times with washing buffer and then incubated with anti-human IgG or IgM secondary antibody conjugated to horseradish peroxidase (HRP) (Jackson Immuno Research 109-036-088 and 109-035-129) in blocking buffer at a 1:5000 dilution. Plates were developed by addition of the HRP substrate, TMB (ThermoFisher) for 10 minutes, then the developing reaction was stopped by adding 50μl 1M H2SO4 and absorbance was measured at 450nm with an ELISA microplate reader (FluoStar Omega, BMG Labtech) with Omega and Omega MARS software for analysis. For plasma samples, a positive control (plasma from patient COV21, diluted 200-fold in PBS) and negative control historical plasma samples was added in duplicate to every assay plate for validation. The average of its signal was used for normalization of all the other values on the same plate with Excel software prior to calculating the area under the curve using Prism 8 (GraphPad). For monoclonal antibodies, the half-maximal effective concentration (EC50) was determined using 4-parameter nonlinear regression (GraphPad Prism).
293TACE2 cells.
For constitutive expression of ACE2 in 293T cells, a cDNA encoding ACE2, carrying two inactivating mutations in the catalytic site (H374N & H378N), was inserted into CSIB 3’ to the SFFV promoter30. 293TACE2 cells were generated by transduction with CSIB based virus followed by selection with 5 μg/ml Blasticidin.
SARS-CoV-2 and SARS-CoV pseudotyped reporter viruses.
A plasmid expressing a C-terminally truncated SARS-CoV-2 S protein (pSARS-CoV2-Strunc) was generated by insertion of a human-codon optimized cDNA encoding SARS-CoV-2 S lacking the C-terminal 19 codons (Geneart) into pCR3.1. The S ORF was taken from “Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1” (NC_045512). For expression of full-length SARS-CoV S protein, “Human SARS coronavirus Spike glycoprotein Gene ORF cDNA clone expression plasmid (Codon Optimized)” (here referred to as pSARS-CoV-S) was obtained from SinoBiological (Cat: VG40150-G-N). An env-inactivated HIV-1 reporter construct (pNL4-3ΔEnv-nanoluc) was generated from pNL4-331 by introducing a 940 bp deletion 3’ to the vpu stop-codon, resulting in a frameshift in env. The human codon-optimized nanoluc Luciferase reporter gene (Nluc, Promega) was inserted in place of nucleotides 1–100 of the nef-gene. To generate pseudotyped viral stocks, 293T cells were transfected with pNL4-3ΔEnv-nanoluc and pSARS-CoV2-Strunc or pSARS-CoV-S using polyethyleneimine. Co-transfection of pNL4-3ΔEnv-nanoluc and S-expression plasmids leads to production of HIV-1-based virions carrying either the SARS-CoV-2 or SARS-CoV spike protein on the surface. Eight hours after transfection, cells were washed twice with PBS and fresh media was added. Supernatants containing virions were harvested 48 hours post transfection, filtered and stored at −80°C. Infectivity of virions was determined by titration on 293TACE2 cells. See also https://www.biorxiv.org/content/10.1101/2020.06.08.140871v1.
Pseudotyped virus neutralization assay.
Five-fold serially diluted plasma from COVID-19 convalescent individuals and healthy donors or four-fold serially diluted monoclonal antibodies were incubated with the SARS-CoV-2 or SARS-CoV pseudotyped virus for 1 hour at 37°C degrees. The mixture was subsequently incubated with 293TACE2 cells for 48 hours after which cells were washed twice with PBS and lysed with Luciferase Cell Culture Lysis 5x reagent (Promega). Nanoluc Luciferase activity in lysates was measured using the Nano-Glo Luciferase Assay System (Promega) with Modulus II Microplate Reader User interface (TURNER BioSystems). Relative luminescence units obtained were normalized to those derived from cells infected with SARS-CoV-2 or SARS-CoV pseudotyped virus in the absence of plasma or monoclonal antibodies. The half-maximal inhibitory concentration for plasma (NT50) or monoclonal antibodies (IC50) was determined using 4-parameter nonlinear regression (GraphPad Prism).
Cell lines, virus and virus titration.
VeroE6 kidney epithelial cells (Chlorocebus sabaeus; ATCC) and Huh-7.5 hepatoma cells (H. sapiens; Dr. Charles Rice, Laboratory of Virology and Infectious Disease, The Rockefeller University) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 1% nonessential amino acids (NEAA) and 10% fetal bovine serum (FBS) at 37°C and 5% CO2. All cell lines have been tested negative for contamination with mycoplasma and were obtained from the ATCC (with the exception for Huh-7.5). SARS-CoV-2, strain USA-WA1/2020, was obtained from BEI Resources and amplified in VeroE6 cells at 33°C. Viral titers were measured on Huh-7.5 cells by standard plaque assay (PA). Briefly, 500 μL of serial 10-fold virus dilutions in Opti-MEM were used to infect 400,000 cells seeded the day prior in a 6-well plate format. After 90 min adsorption, the virus inoculum was removed, and cells were overlayed with DMEM containing 10% FBS with 1.2% microcrystalline cellulose (Avicel). Cells were incubated for five days at 33°C, followed by fixation with 3.5% formaldehyde and crystal violet staining for plaque enumeration. All experiments were performed in a biosafety level 3 laboratory.
Microscopy-based neutralization assay of authentic SARS-CoV-2.
The day prior to infection VeroE6 cells were seeded at 12,500 cells/well into 96-well plates. Antibodies were serially diluted in BA-1, mixed with a constant amount of SARS-CoV-2 (grown in VeroE6) and incubated for 60 min at 37°C. The antibody-virus-mix was then directly applied to VeroE6 cells (MOI of ~0.1 PFU/cell). Cells were fixed 18 hours post infection by adding an equal volume of 7% formaldehyde to the wells, followed by permeabilization with 0.1% Triton X-100 for 10 min. After extensive washing, cells were incubated for 1 hour at room temperature with blocking solution of 5% goat serum in PBS (catalog no. 005–000-121; Jackson ImmunoResearch). A rabbit polyclonal anti-SARS-CoV-2 nucleocapsid antibody (catalog no. GTX135357; GeneTex) was added to the cells at 1:500 dilution in blocking solution and incubated at 4°C overnight. A goat anti-rabbit AlexaFluor 594 (catalog no. A-11012; Life Technologies) at a dilution of 1:2,000 was used as a secondary antibody. Nuclei were stained with Hoechst 33342 (catalog no. 62249; Thermo Scientific) at a 1:1,000 dilution. Images were acquired with a fluorescence microscope and analyzed using ImageXpress Micro XLS and MetaXpress software (Molecular Devices, Sunnyvale, CA). All statistical analyses were done using Prism 8 software (GraphPad).
Biotinylation of viral protein for use in flow cytometry.
Purified and Avi-tagged SARS-CoV-2 RBD was biotinylated using the Biotin-Protein Ligase-BIRA kit according to manufacturer’s instructions (Avidity). Ovalbumin (Sigma, A5503-1G) was biotinylated using the EZ-Link Sulfo-NHS-LC-Biotinylation kit according to the manufacturer’s instructions (Thermo Scientific). Biotinylated Ovalbumin was conjugated to streptavidin-BV711 (BD biosciences, 563262) and RBD to streptavidin-PE (BD biosciences, 554061) and streptavidin-Alexa Fluor 647 (AF647, Biolegend, 405237) respectively32.
Single cell sorting by flow cytometry.
PBMCs were enriched for B cells by negative selection using a pan B cell isolation kit according to the manufacturer’s instructions (Miltenyi Biotec, 130-101-638). The enriched B cells were incubated in FACS buffer (1 X Phosphate-buffered Saline (PBS), 2% calf serum, 1 mM EDTA) with the following anti-human antibodies (all at 1:200 dilution): anti-CD20-PECy7 (BD Biosciences, 335793), anti-CD3-APC-eFluro 780 (Invitrogen, 47-0037-41), anti-CD8-APC-eFluro 780 (Invitrogen, 47-0086-42), anti-CD16-APC-eFluro 780 (Invitrogen, 47-0168-41), anti-CD14-APC-eFluro 780 (Invitrogen, 47-0149-42), as well as Zombie NIR (BioLegend, 423105), and fluorophore-labeled RBD and Ovalbumin for 30 minutes on ice32. Single CD3−CD8−CD16−CD20+Ova−RBD-PE+RBD-AF647+ B cells were sorted into individual wells of 96-well plates containing 4 μl of lysis buffer (0.5 X PBS, 10mM DTT, 3000 units/mL RNasin Ribonuclease Inhibitors (Promega, N2615) per well using a FACS Aria III and FACSDiva software (Becton Dickinson) for acquisition and FlowJo for analysis. The sorted cells were frozen on dry ice, and then stored at −80°C or immediately used for subsequent RNA reverse transcription. Although cells were not stained for IgG expression, they are memory B cells based on the fact that they are CD20+ (a marker absent in plasmablasts) and they express IgG (since antibodies were amplified from these cells using IgG-specific primers).
Antibody sequencing, cloning and expression.
Antibodies were identified and sequenced as described previously27,33,34. Briefly, RNA from single cells was reverse-transcribed (SuperScript III Reverse Transcriptase, Invitrogen, 18080–044) and the cDNA stored at −20°C or used for subsequent amplification of the variable IGH, IGL and IGK genes by nested PCR and Sanger sequencing33. Anti-Zika virus monoclonal antibody Z02127 was used as isotype control. Sequence analysis was with MacVector. Amplicons from the first PCR reaction were used as templates for Sequence- and Ligation-Independent Cloning (SLIC) into antibody expression vectors. Recombinant monoclonal antibodies and Fabs were produced and purified as previously described35,36.
Biolayer interferometry.
BLI assays were performed on the Octet Red instrument (ForteBio) at 30°C with shaking at 1,000 r.p.m. Epitope binding assays were performed with protein A biosensor (ForteBio 18–5010), following the manufacturer’s protocol “classical sandwich assay”. (1) Sensor check: sensors immersed 30 sec in buffer alone (buffer ForteBio 18–1105). (2) Capture 1st Ab: sensors immersed 10min with Ab1 at 40 μg/mL. (3) Baseline: sensors immersed 30 sec in buffer alone. (4) Blocking: sensors immersed 5 min with IgG isotype control at 50 μg/mL. (6) Antigen association: sensors immersed 5 min with RBD at 100 μg/mL. (7) Baseline: sensors immersed 30 sec in buffer alone. (8) Association Ab2: sensors immersed 5 min with Ab2 at 40 μg/mL. Curve fitting was performed using the Fortebio Octet Data analysis software (ForteBio).
Computational analyses of antibody sequences.
Antibody sequences were trimmed based on quality and annotated using Igblastn v1.14.037 with IMGT domain delineation system. Annotation was performed systematically using Change-O toolkit v.0.4.538. Heavy and light chains derived from the same cell were paired, and clonotypes were assigned based on their V and J genes using in-house R and Perl scripts (Fig. 3 b,c). All scripts and the data used to process antibody sequences are publicly available on GitHub (https://github.com/stratust/igpipeline).
The frequency distributions of human V genes in anti-SARS-CoV-2 antibodies from this study was compared to Sequence Read Archive SRP01097039. The V(D)J assignments were done using IMGT/High V-Quest and the frequencies of heavy and light chain V genes were calculated for 14 and 13 individuals, respectively, using sequences with unique CDR3s. The two-tailed t test with unequal variances was used to determine statistical significance (Extended Data Fig. 7). Nucleotide somatic hypermutation and CDR3 length were determined using in-house R and Perl scripts. For somatic hypermutations, IGHV and IGLV nucleotide sequences were aligned against their closest germlines using Igblastn and the number of differences were considered nucleotide mutations. The average mutations for V genes was calculated by dividing the sum of all nucleotide mutations across all patients by the number of sequences used for the analysis. To calculate the GRAVY scores of hydrophobicity40 we used Guy H.R. Hydrophobicity scale based on free energy of transfer (kcal/mole)41 implemented by the R package Peptides available in the Comprehensive R Archive Network repository (https://journal.r-project.org/archive/2015/RJ-2015-001/RJ-2015-001.pdf). We used 533 heavy chain CDR3 amino acid sequences from this study (sequence COV047_P4_IgG_51-P1369 lacks CDR3 amino acid sequence) and 22,654,256 IGH CDR3 sequences from the public database of memory B-cell receptor sequences42. The Shapiro-Wilk test was used to determine whether the GRAVY scores are normally distributed. The GRAVY scores from all 533 IGH CDR3 amino acid sequences from this study were used to perform the test and 5000 GRAVY scores of the sequences from the public database were randomly selected. The Shapiro-Wilk p-values were 6.896 × 10−3 and 2.217 × 10−6 for sequences from this study and the public database, respectively, indicating the data are not normally distributed. Therefore, we used the Wilcoxon non-parametric test to compare the samples, which indicated a difference in hydrophobicity distribution (p = 5 × 10−6; Extended Data Fig. 8).
Negative-stain EM Data Collection and Processing
Purified Fabs (C002, C119, and C121) were complexed with SARS-CoV-2 S trimer at a 2-fold molar excess for 1 min and diluted to 40 μg/mL in TBS immediately before adding 3 μL to a freshly-glow discharged ultrathin, 400 mesh carbon-coated copper grid (Ted Pella, Inc.). Samples were blotted after a 1 min incubation period and stained with 1% uranyl formate for an additional minute before imaging. Micrographs were recorded on a Thermo Fisher Talos Arctica transmission electron microscope operating at 200 keV using a K3 direct electron detector (Gatan, Inc) and SerialEM automated image acquisition software43. Images were acquired at a nominal magnification of 28,000x (1.44 Å/pixel size) and a −1.5 to −2.0 μm defocus range. Images were processed in cryoSPARC, and reference-free particle picking was completed using a gaussian blob picker44. Reference-free 2D class averages and ab initio volumes were generated in cryoSPARC, and subsequently 3D-classified to identify classes of S-Fab complexes, that were then homogenously refined. Figures were prepared using UCSF Chimera45. The resolutions of the final single particle reconstructions were ~17–20 Å calculated using a gold-standard FSC (0.143 cutoff) and ~24–28 Å using a 0.5 cutoff.
Extended Data
Extended Data Figure 1. Clinical correlates.
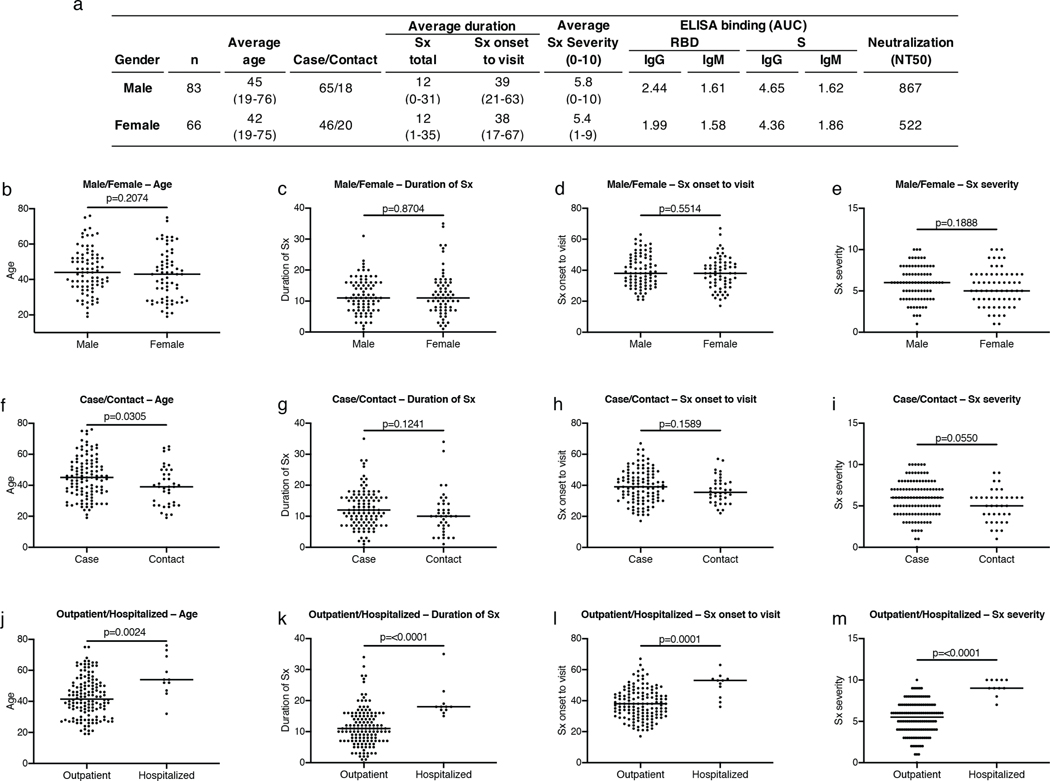
a, Summary of the cohort’s characteristics. b, Age distribution (Y axis) for all males (n=83) and females (n=66) in the cohort; p=0.2074. c, Duration of symptoms in days (Y axis) for all males (n=83) and females (n=66) in the cohort; p=0.8704. d, Time between symptom onset and plasma collection (Y axis) for all males (n=83) and females (n=66) in the cohort; p=0.5514. e, Subjective symptom severity on a scale of 0–10 (Y axis) for all males (n=83) and females (n=66) in the cohort; p=0.1888. f, Age distribution (Y axis) for all cases (n=111) and contacts (n=38) in the cohort; p=0.0305. g, Duration of symptoms in days (Y axis) for all cases (n=111) and contacts (n=38) in the cohort; p=0.1241. h, Time between symptom onset and plasma collection in days (Y axis) for all cases (n=111) and contacts (n=38) in the cohort; p=0.1589. i, Symptom severity (Y axis) for all cases (n=111) and contacts (n=38) in the cohort; p=0.0550. j, Age distribution (Y axis) for all outpatient (n=138) and hospitalized (n=11) participants; p=0.0024. k, Duration of symptoms in days (Y axis) for all outpatient (n=138) and hospitalized (n=11) participants in the cohort; p=<0.0001. l, Time between symptom onset and plasma collection in days (Y axis) for all outpatient (n=138) and hospitalized (n=11) participants in the cohort; p=0.0001. m, Symptom severity (Y axis) for all outpatient (n=138) and hospitalized (n=11) participants in the cohort; p=<0.0001. Horizontal bars indicate median values. Statistical significance was determined using two-tailed Mann-Whitney U test.
Extended Data Figure 2. Clinical correlates of plasma antibody titers.
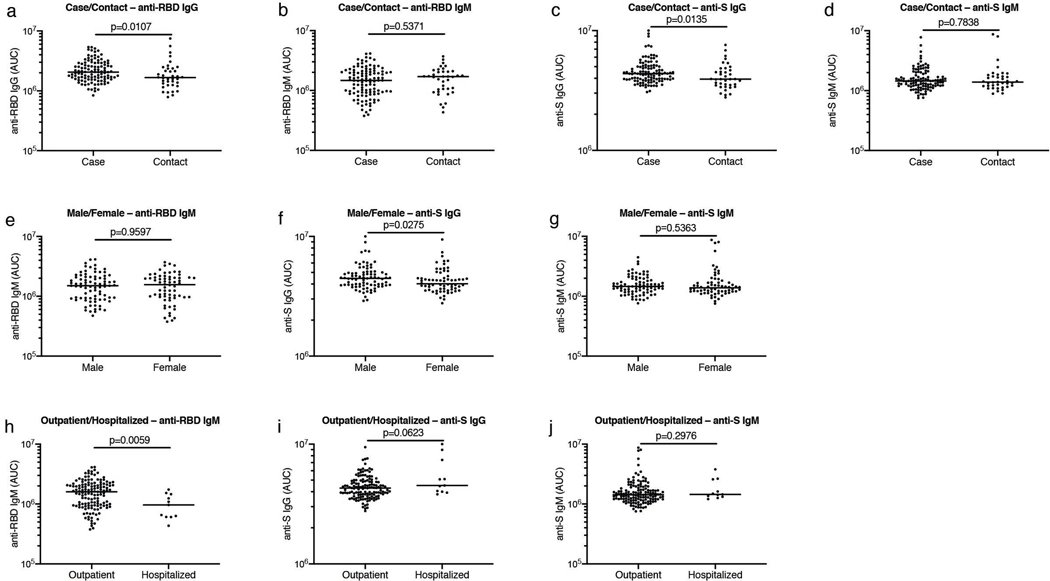
a, Normalized AUC for IgG anti-RBD (Y axis) for all cases (n=111) and contacts (n=38) in the cohort; p=0.0107. b, Normalized AUC for IgM anti-RBD (Y axis) for all cases (n=111) and contacts (n=38) in the cohort; p=0.5371. c, Normalized AUC for IgG anti-S (Y axis) for all cases (n=111) and contacts (n=38) in the cohort; p=0.0135. d, Normalized AUC for IgM anti-S (Y axis) for all cases (n=111) and contacts (n=38) in the cohort; p=0.7838. e, Normalized AUC for IgM anti-RBD (Y axis) for all males (n=83) and females (n=66) in the cohort; p=0.9597. f, Normalized AUC for IgG anti-S (Y axis) for all males (n=83) and females (n=66) in the cohort; p=0.0275. g, Normalized AUC for IgM anti-S (Y axis) for all males (n=83) and females (n=66) in the cohort; p=0.5363. h, Normalized AUC for IgM anti-RBD (Y axis) for all outpatient (=138) and hospitalized (n=11) participants in the cohort; p=0.0059. i, Normalized AUC for IgG anti-S (Y axis) for all outpatient (=138) and hospitalized (=11) participants in the cohort; p=0.0623. j, Normalized AUC for IgM anti-S (Y axis) for all outpatient (=138) and hospitalized (=11) participants in the cohort; p=0.2976. Horizontal bars indicate median values. Statistical significance was determined using two-tailed Mann-Whitney U test.
Extended Data Figure 3. Additional clinical correlates of plasma antibody titers.
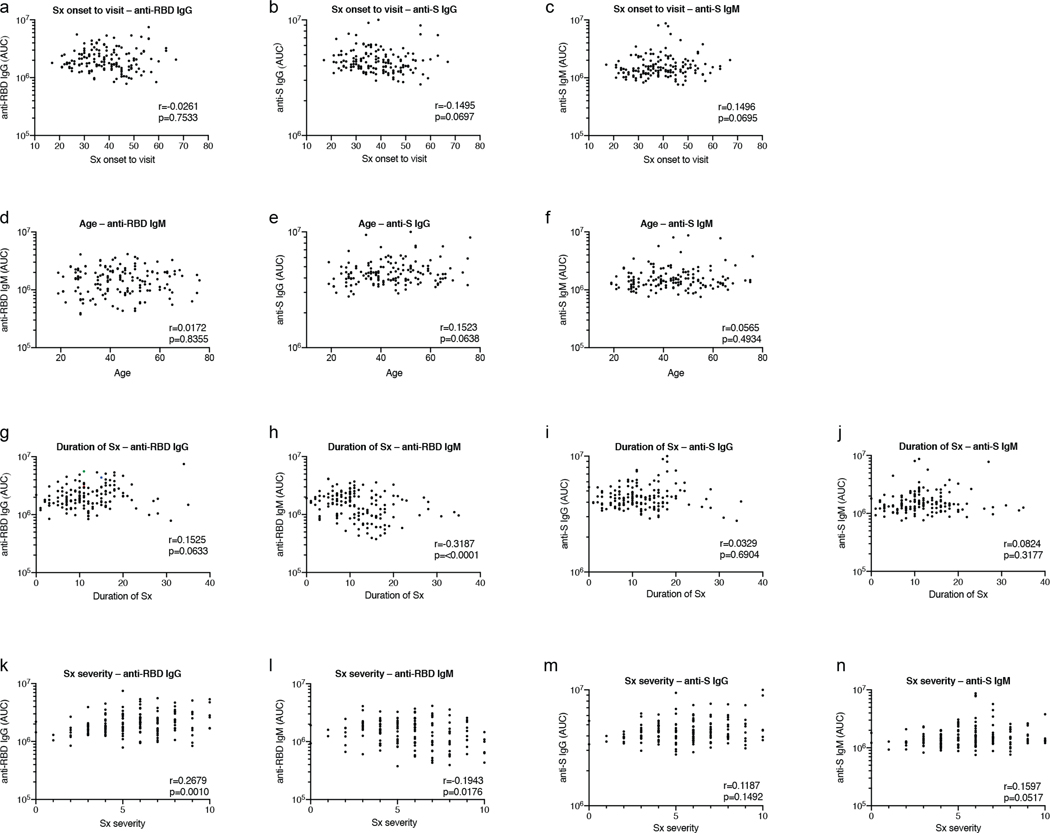
a, Time between symptom onset and plasma collection in days (X axis) plotted against normalized AUC for IgG anti-RBD (Y axis); r=−0.0261 p=0.7533. b, Time between symptom onset and plasma collection in days (X axis) plotted against normalized AUC for IgG anti-S (Y axis); r=−0.1495 p=0.0697. c, Time between symptom onset and plasma collection in days (X axis) plotted against normalized AUC for IgM anti-S (Y axis); r=0.1496 p=0.0695. d, Age (X axis) plotted against AUC for IgM anti-RBD (Y axis); r=0.0172 p=0.8355. e, Age (X axis) plotted against normalized AUC for IgG anti-S (Y axis); r=0.1523 p=0.0638. f, Age (X axis) plotted against normalized AUC for IgM anti-S (Y axis); r=0.0565 p=0.4934. g, Duration of symptoms in days (X axis) plotted against normalized AUC for IgG anti-RBD (Y axis); r=0.1525, p=0.0633. h, Duration of symptoms in days (X axis) plotted against normalized AUC for IgM anti-RBD (Y axis); r=−0.3187, p=<0.0001. i, Duration of symptoms in days (X axis) plotted against normalized AUC for IgG anti-S (Y axis); r=0.0329, p=0.6904. j, Duration of symptoms in days (X axis) plotted against normalized AUC for IgM anti-S (Y axis); r=0.0824, p=0.3177. k, Severity of symptoms (X axis) plotted against normalized AUC for IgG anti-RBD (Y axis); r=0.2679 p=0.0010. l, Severity of symptoms (X axis) plotted against normalized AUC for IgM anti-RBD (Y axis); r=−0.1943 p=0.0176. m, Severity of symptoms (X axis) plotted against normalized AUC for IgG anti-S (Y axis); r=0.1187 p=0.1492. n, Severity of symptoms (X axis) plotted against normalized AUC for IgM anti-S (Y axis); r=0.1597 p=0.0517. All correlations were analyzed by two-tailed Spearman’s.
Extended Data Figure 4. Diagrammatic representation of the SARS-CoV-2 pseudovirus luciferase assay.

a, Co-transfection of pNL4-3ΔEnv-nanoluc and pSARS-CoV-2 spike vectors into 293T cells (ATCC) leads to production of SARS-CoV-2 Spike-pseudotyped HIV-1 particles (SARS-CoV-2 pseudovirus) carrying the Nanoluc gene. b, SARS-CoV-2 pseudovirus is incubated for 1 h at 37°C with plasma or monoclonal antibody dilutions. The virus-antibody mixture is used to infect ACE2-expressing 293T cells, which will express nanoluc Luciferase upon infection. c, Relative luminescence units (RLU) reads from lysates of ACE2-expressing 293T cells infected with increasing amounts of SARS-CoV-2 pseudovirus. Error bars represent standard deviation of triplicates, two experiments.
Extended Data Figure 5. Clinical correlates of neutralization.
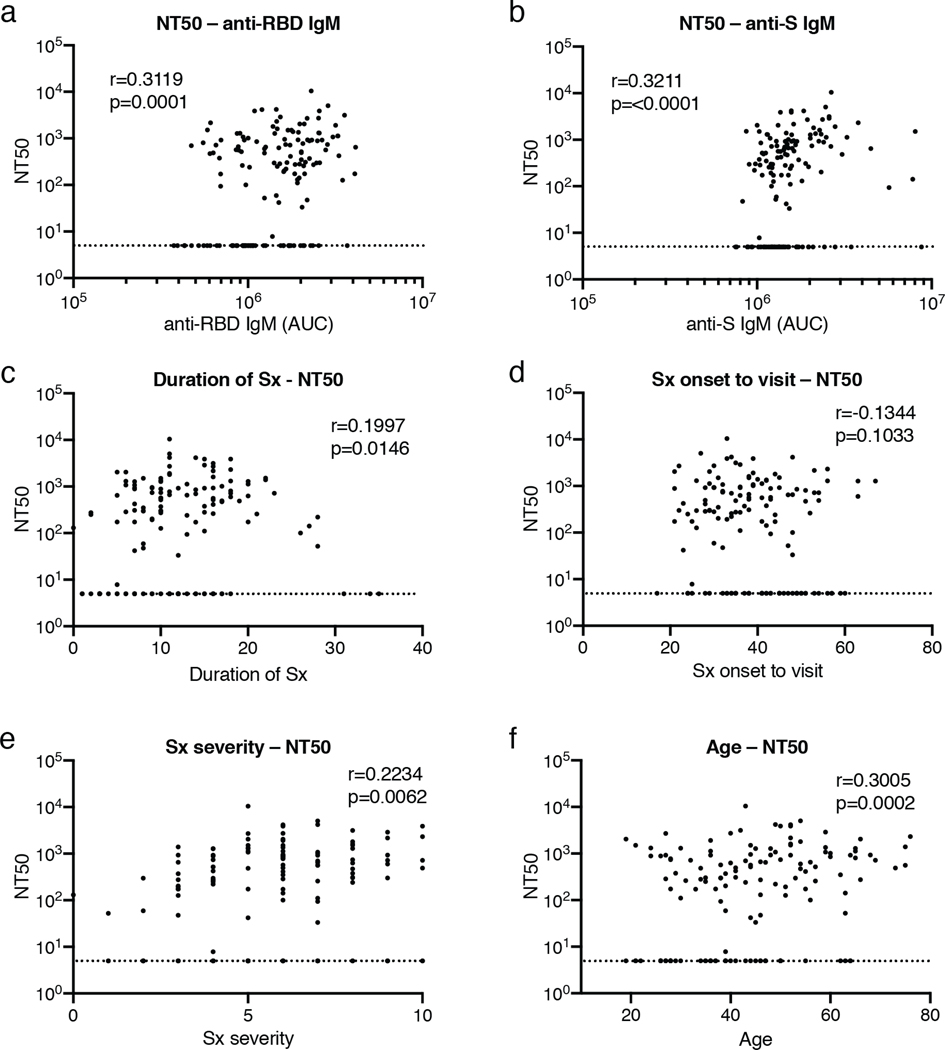
a, Normalized AUC for anti-RBD IgM (X axis) plotted against NT50 (Y axis); r=0.3119, p=0.0001. b, Normalized AUC for anti-S IgM (X axis) plotted against NT50 (Y axis); r=0.3211, p=<0.0001. c, Duration of symptoms in days (X axis) plotted against NT50 (Y axis); r=0.1997, p=0.0146. d, Time between symptom onset and plasma collection in days (X axis) plotted against NT50 (Y axis); r=−0.1344, p=0.1033. e, Symptom severity (X axis) plotted against NT50 (Y axis); r=0.2234, p=0.0062. f, Age (X axis) plotted against NT50 (Y axis); r=0.3005, p=0.0002. All correlations were analyzed by two-tailed Spearman’s. Dotted line (NT50=5) represents lower limit of detection (LLOD) of pseudovirus neutralization assay. Samples with neutralizing titers below 1:50 were plotted at LLOD.
Extended Data Figure 6. Flow cytometry.
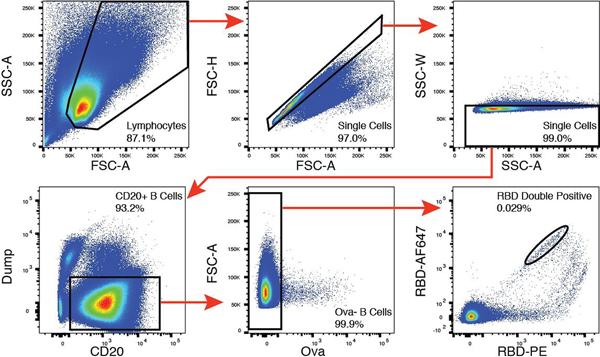
Gating strategy used for cell sorting. Gating was on singlets that were CD20+ and CD3−CD8−CD16−Ova−. Sorted cells were RBD-PE+ and RBD-AF647+.
Extended Data Figure 7. Frequency distributions of human V genes.
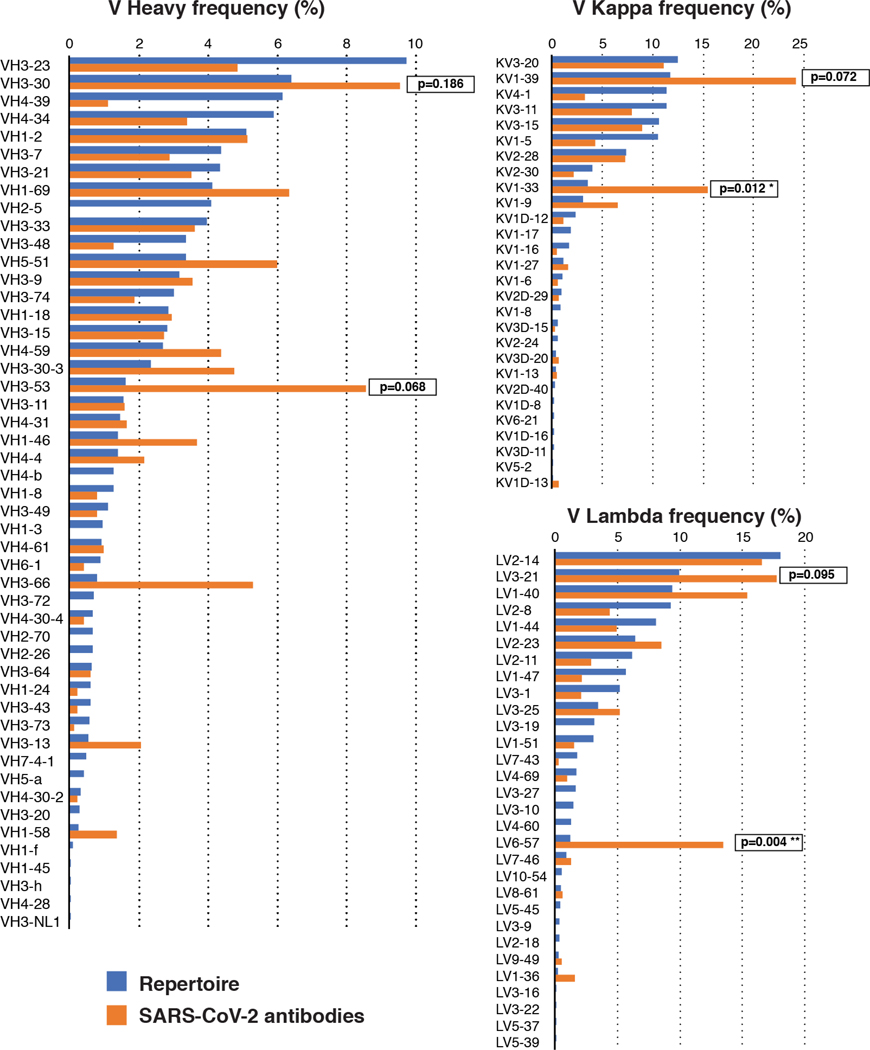
The two-tailed t test with unequal variance was used to compare the frequency distributions of human V genes of anti-SARS-CoV-2 antibodies from this study to Sequence Read Archive SRP01097039.
Extended Data Figure 8. Analysis of antibody somatic hypermutation and CDR3 length.
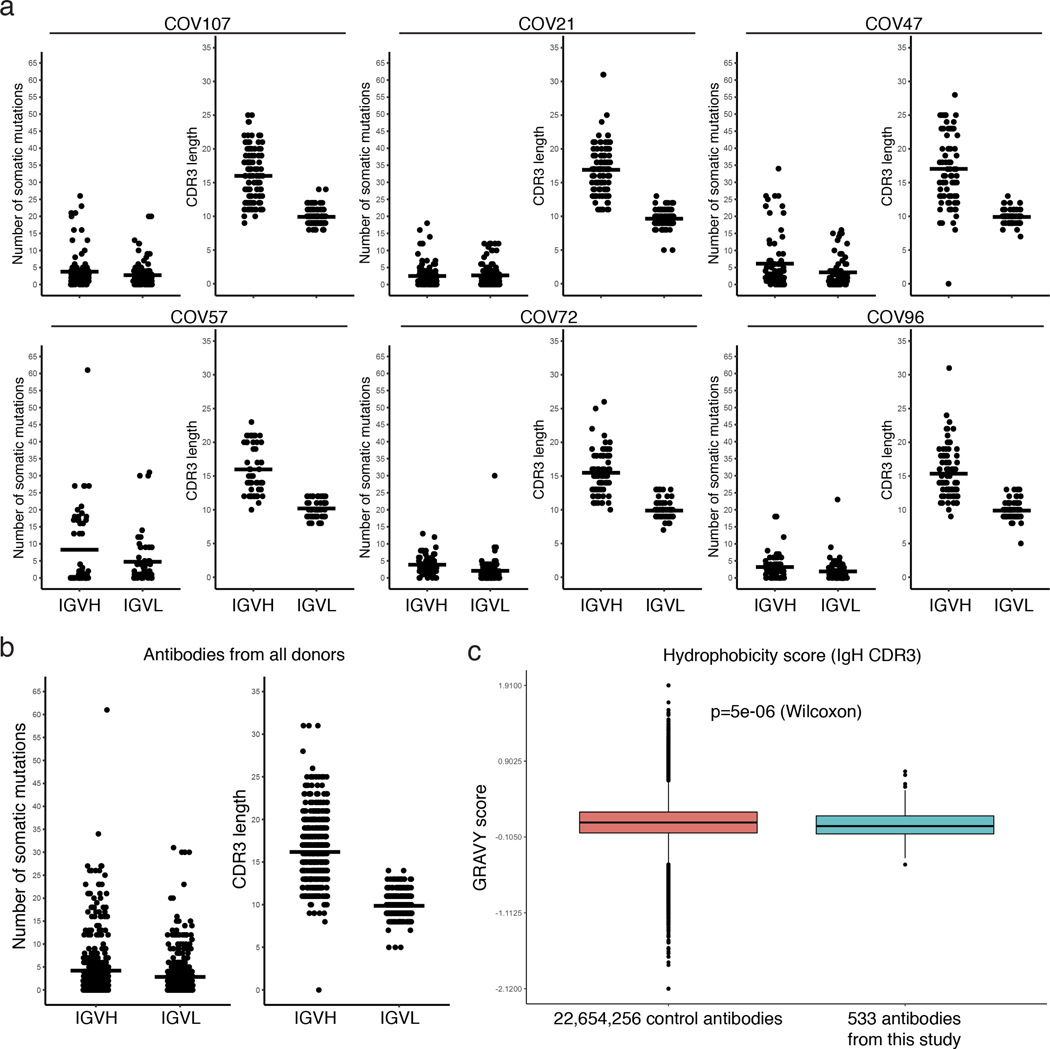
a, For each individual, the number of somatic nucleotide mutations (Y axis) at the IGVH and IGVL are shown on the left panel, and the amino acid length of the CDR3s (Y axis) are shown on the right panel. The horizontal bars indicate the mean. The number of antibody sequences (IGVH and IGVL) evaluated in each participant are n=118 (COV107), n=127 (COV21), n=79 (COV47), n=54 (COV57), n=78 (COV72), n=78 (COV96). b, same as in a but for all antibodies combined (n=534 for both IGVH and IGVL). c, Distribution of the hydrophobicity GRAVY scores at the IGH CDR3 in antibody sequences from this study compared to a public database (see Methods for statistical analysis). The box limits are at the lower and upper quartiles, the center line indicates the median, the whiskers are 1.5x interquartile range and the dots represent outliers.
Extended Data Figure 9. Binding of the monoclonal antibodies to the RBD of SARS-CoV-2 and cross-reactivity to SARS-CoV.
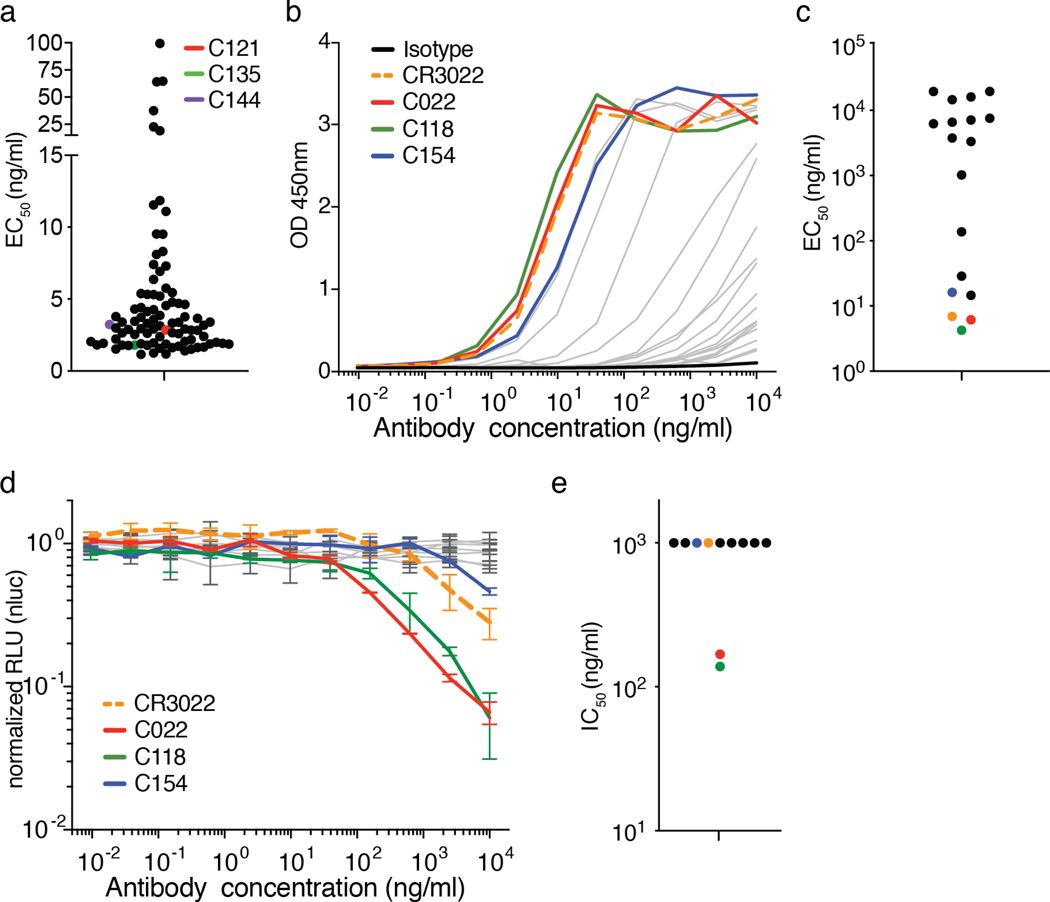
a, EC50 values for binding to the RBD of SARS-CoV-2. Average of two or more experiments; n=89. b and c, Binding curves and EC50 values (average of two experiments) for binding to the RBD of SARS-CoV; n=20 and n=17 (excluding isotype and CR3022), respectively. d and e, SARS-CoV pseudovirus neutralization curves and IC50 values. Shown in d are the standard deviations of duplicates for one representative experiment and in e is the average of two experiments (n=10, excluding CR3022). Samples with IC50s above 1μg/ml were plotted at 1μg/ml.
Extended Data Figure 10. Biolayer interferometry experiment.
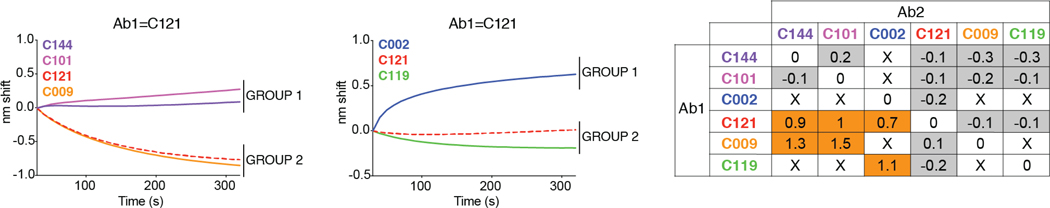
Binding of antibodies C144, C101, C002, C121, C009, C119. Graphs show secondary antibody binding to preformed C121 IgG-RBD complexes. The table displays the shift in nanometers after second antibody (Ab2) binding to the antigen in the presence of the first antibody (Ab1). Values are normalized by the subtraction of the autologous antibody control.
Supplementary Material
Acknowledgements:
We thank all study participants who devoted time to our research; Drs. Barry Coller and Sarah Schlesinger, the Rockefeller University Hospital Clinical Research Support Office and nursing staff. Dr. Joseph L. DeRisi for facilitating interactions with the Chan Zuckerberg BioHub. All members of the M.C.N. laboratory for helpful discussions, Drs. Amelia Escolano, Gaëlle Breton and Bernardo Reis, and Maša Jankovic for laboratory support, and Dr. Jost Vielmetter and the Protein Expression Center in the Beckman Institute at Caltech. This work was supported by NIH grant P01-AI138398-S1 (M.C.N., C.M.R., P.J.B.) and 2U19AI111825 (M.C.N. and C.M.R).; the Caltech Merkin Institute for Translational Research and P50 AI150464 (P.J.B.), George Mason University Fast Grant (D.F.R. and P.J.B.) and the European ATAC consortium EC 101003650 (D.F.R.); 3 R01-AI091707-10S1 to C.M.R.; R37-AI64003 to P.D.B.; R01AI78788 to T.H.; The G. Harold and Leila Y. Mathers Charitable Foundation to C.M.R.. Electron microscopy was performed in the Caltech Beckman Institute Resource Center for Transmission Electron Microscpy (Drs. Songye Chen and Andrey Malyutin, Directors). C.G. was supported by the Robert S. Wennett Post-Doctoral Fellowship, in part by the National Center for Advancing Translational Sciences (National Institutes of Health Clinical and Translational Science Award program, grant UL1 TR001866), and by the Shapiro-Silverberg Fund for the Advancement of Translational Research. P.D.B. and M.C.N. are Howard Hughes Medical Institute Investigators.
Footnotes
Competing financial interest: In connection with this work The Rockefeller University has filed a provisional patent application on which D.F.R. and M.C.N. are inventors.
Data availability statement: Data are provided in SI Tables 1, 3, 4, 5 and 6; and in Figure 3 has associated raw sequencing data deposited at Github (https://github.com/stratust/igpipeline). This study also includes A Public Database of Memory and Naive B-Cell Receptor Sequences (https://datadryad.org/stash/dataset/doi:10.5061/dryad.35ks2), PDB datasets 6VYB and 6NB6 and Sequence Read Archive SRP010970.
Code availability statement: Computer code to process the antibody sequences is available at GitHub (https://github.com/stratust/igpipeline).
References
- 1.Graham RL, Donaldson EF & Baric RS A decade after SARS: strategies for controlling emerging coronaviruses. Nat Rev Microbiol 11, 836–848, doi: 10.1038/nrmicro3143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gralinski LE & Baric RS Molecular pathology of emerging coronavirus infections. J Pathol 235, 185–195, doi: 10.1002/path.4454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280 e278, doi: 10.1016/j.cell.2020.02.052 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walls AC et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281–292 e286, doi: 10.1016/j.cell.2020.02.058 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang S, Hillyer C & Du L Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol, doi: 10.1016/j.it.2020.03.007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amanat F et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med, doi: 10.1038/s41591-020-0913-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grifoni A et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell, doi: 10.1016/j.cell.2020.05.015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheid JF et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640, doi: 10.1038/nature07930 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Tiller T et al. Autoreactivity in human IgG+ memory B cells. Immunity 26, 205–213, doi: 10.1016/j.immuni.2007.01.009 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murugan R et al. Clonal selection drives protective memory B cell responses in controlled human malaria infection. Sci Immunol 3, doi: 10.1126/sciimmunol.aap8029 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Wang Q et al. A Combination of Human Broadly Neutralizing Antibodies against Hepatitis B Virus HBsAg with Distinct Epitopes Suppresses Escape Mutations. Cell Host Microbe, doi: 10.1016/j.chom.2020.05.010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briney B, Inderbitzin A, Joyce C & Burton DR Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 566, 393–397, doi: 10.1038/s41586-019-0879-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ter Meulen J et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med 3, e237, doi: 10.1371/journal.pmed.0030237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan M et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633, doi: 10.1126/science.abb7269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls AC et al. Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell 176, 1026–1039 e1015, doi: 10.1016/j.cell.2018.12.028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinto D et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature, doi: 10.1038/s41586-020-2349-y (2020). [DOI] [PubMed] [Google Scholar]
- 17.Zhu Z et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A 104, 12123–12128, doi: 10.1073/pnas.0701000104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salazar G, Zhang N, Fu TM & An Z Antibody therapies for the prevention and treatment of viral infections. NPJ Vaccines 2, 19, doi: 10.1038/s41541-017-0019-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bournazos S & Ravetch JV Anti-retroviral antibody FcgammaR-mediated effector functions. Immunol Rev 275, 285–295, doi: 10.1111/imr.12482 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feinberg MB & Ahmed R Advancing dengue vaccine development. Science 358, 865–866, doi: 10.1126/science.aaq0215 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki A & Yang Y The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol, doi: 10.1038/s41577-020-0321-6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Rompay KKA et al. A combination of two human monoclonal antibodies limits fetal damage by Zika virus in macaques. Proc Natl Acad Sci U S A 117, 7981–7989, doi: 10.1073/pnas.2000414117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plotkin SA Correlates of protection induced by vaccination. Clin Vaccine Immunol 17, 1055–1065, doi: 10.1128/CVI.00131-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science, doi: 10.1126/science.abc6284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandrashekar A et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science, doi: 10.1126/science.abc4776 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheid JF et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637, doi: 10.1126/science.1207227 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robbiani DF et al. Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico. Cell 169, 597–609 e511, doi: 10.1016/j.cell.2017.04.024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrhardt SA et al. Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV. Nat Med 25, 1589–1600, doi: 10.1038/s41591-019-0602-4 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Pappas L et al. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 516, 418–422, doi: 10.1038/nature13764 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Kane M et al. Identification of Interferon-Stimulated Genes with Antiretroviral Activity. Cell Host Microbe 20, 392–405, doi: 10.1016/j.chom.2016.08.005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adachi A et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59, 284–291 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z et al. Isolation of single HIV-1 Envelope specific B cells and antibody cloning from immunized rhesus macaques. J Immunol Methods 478, 112734, doi: 10.1016/j.jim.2019.112734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiller T et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329, 112–124, doi: 10.1016/j.jim.2007.09.017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Boehmer L et al. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat Protoc 11, 1908–1923, doi: 10.1038/nprot.2016.102 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Klein F et al. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J Exp Med 211, 2361–2372, doi: 10.1084/jem.20141050 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoofs T et al. Broad and Potent Neutralizing Antibodies Recognize the Silent Face of the HIV Envelope. Immunity 50, 1513–1529 e1519, doi: 10.1016/j.immuni.2019.04.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J, Ma N, Madden TL & Ostell JM IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res 41, W34–40, doi: 10.1093/nar/gkt382 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta NT et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358, doi: 10.1093/bioinformatics/btv359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubelt F et al. Onset of immune senescence defined by unbiased pyrosequencing of human immunoglobulin mRNA repertoires. PLoS One 7, e49774, doi: 10.1371/journal.pone.0049774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyte J & Doolittle RF A simple method for displaying the hydropathic character of a protein. J Mol Biol 157, 105–132, doi: 10.1016/0022-2836(82)90515-0 (1982). [DOI] [PubMed] [Google Scholar]
- 41.Guy HR Amino acid side-chain partition energies and distribution of residues in soluble proteins. Biophys J 47, 61–70, doi: 10.1016/S0006-3495(85)83877-7 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeWitt WS et al. A Public Database of Memory and Naive B-Cell Receptor Sequences. PLoS One 11, e0160853, doi: 10.1371/journal.pone.0160853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51, doi: 10.1016/j.jsb.2005.07.007 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296, doi: 10.1038/nmeth.4169 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Goddard TD, Huang CC & Ferrin TE Visualizing density maps with UCSF Chimera. J Struct Biol 157, 281–287, doi: 10.1016/j.jsb.2006.06.010 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


