Abstract
Purpose:
Cancer immunoediting shapes tumor progression by the selection of tumor cell variants that can evade immune recognition. Given the immune evasion and intra-tumor heterogeneity characteristic of gliomas, we hypothesized that CD8+ T-cells mediate immunoediting in these tumors.
Experimental design:
We developed retrovirus-induced PDGF+Pten−/− murine gliomas and evaluated glioma progression and tumor immunogenicity in the absence of CD8+ T-cells by depleting this immune cell population. Furthermore, we characterized the genomic alterations present in gliomas that developed in the presence and absence of CD8+ T-cells.
Results:
Upon transplantation, gliomas that developed in the absence of CD8+ T-cells engrafted poorly in recipients with intact immunity but engrafted well in those with CD8+ T-cell depletion. In contrast, gliomas that developed under pressure from CD8+ T-cells were able to fully engraft in both CD8+ T-cell-depleted mice and immunocompetent mice. Remarkably, gliomas developed in the absence of CD8+ T-cells exhibited increased aneuploidy, MAPK pathway signaling, gene fusions, and macrophage/microglial infiltration, and showed a proinflammatory phenotype. MAPK activation correlated with macrophage/microglia recruitment in this model and in the human disease.
Conclusions:
Our studies indicate that, in these tumor models, CD8+ T-cells influence glioma oncogenic pathways, tumor genotype, and immunogenicity. This suggests immunoediting of immunogenic tumor clones through their negative selection by CD8+ T-cells during glioma formation.
Keywords: CD8+ T-cells, MAPK, immunoediting, cancer evolution, glioma
Introduction
Glioblastoma (GBM) is the most common and malignant brain cancer in adults, with a median overall survival of approximately 21 months (1). This tumor develops multiple mechanisms of immune suppression including induction of T-cell anergy, exhaustion, negative regulation, apoptosis, and sequestration in the bone marrow (2,3). As a consequence, the efficacy of immunotherapy is hindered.
Cancer immunoediting has been proposed as a mechanism where the adaptive and innate immunity sculpts the immunogenicity of developing tumors, selecting for tumor cell variants that can evade immune surveillance (4-6). This understudied mechanism could help explain why many tumors do not respond to immunotherapies. If anti-tumoral immunity is capable of recognizing some tumor cell variants and not others, cancer immunoediting likely influences the evolutionary path of the cancer. Cancer immunoediting relies on tumor genomic heterogeneity—a hallmark of GBM (7). We recently described the disappearance of immunogenic glioma cell variants following PD-1 blockade in recurrent GBM patients who responded to treatment (8). This suggests that immunoediting may occur not only during formation of these gliomas, but also in the setting of immunotherapy. In spite of these and other clinical observations that suggest immunoediting in gliomas, this phenomenon has not been experimentally investigated in this disease.
We hypothesized that CD8+ T-cells subject gliomas to immunoediting. By depleting this immune cell population during development of transgenic murine gliomas, we investigated how these immune effectors influence tumor immunogenicity and progression. Upon transplantation, gliomas developed in the absence of CD8+ T-cells engrafted poorly in recipients with intact immunity, but readily grafted in hosts with CD8+ T-cell depletion. On the other hand, gliomas that developed in the presence of CD8+ T-cells engrafted well in immunocompetent mice. This implies a more immunogenic profile of gliomas that develop without the pressure exerted by CD8+ T-cells. Moreover, these gliomas exhibited increased activation of MAPK signaling, mitoses, aneuploidy, gene fusions, and macrophage/microglia infiltration. Our results reveal that CD8+ T-cells mediate immunoediting in gliomas, influencing the selection of genomic alterations and modulating the tumor microenvironment during gliomagenesis, leading to a non-immunogenic phenotype observed at the end of tumor development.
Materials and Methods
Generation and transplantation of PDGF+Pten−/− gliomas
PDGF+Pten−/− gliomas were generated by intracranial injection of a PDGF-IRES-Cre retrovirus into the subcortical white matter of adult mice with the Ptenlox/lox genotype as previously described (C57BL/6 background, Jackson Lab Stock No: 006440)(9). Gliomas generated in hosts with intact immunity can grow in syngeneic immune-competent recipients. Transplantation was done by extracting the gliomas that developed in the absence or presence of CD8+ T-cells that were dissociated with 2.5% TrypLE in PBS upon which they were cultured with DMEM (Gibco) + 0.5% fetal bovine serum (Gibco), 1% N2 (Gibco), 1% penicillin/streptomycin (Gibco), 10ng/mL PDGF-AA (PeproTech), and 10ng/mL FGF (Basal FGF PDGF) (PeproTech) for up to a week prior to transplant. Animals were anesthetized with ketamine (115 mg/kg) and xylazine (17 mg/kg), a skin incision was made in the middle of the head, and a burr hole was made with a drill. PDGF+Pten−/− glioma cells were then intracranially implanted through a stereotactic injection using a 10 μL Hamilton syringe (Hamilton, Reno, Nevada, USA) with a 30-gauge needle while animals were mounted on a stereotactic apparatus (Harvard Apparatus; Holliston, MA). For survival analysis, animals losing ≥ 30% of their body weight or having trouble ambulating, feeding, or grooming were euthanized by CO2 administration followed by cervical dislocation. After conclusion of survival analysis, animals were sacrificed. Brains were harvested, sectioned, and stained by immunohistochemistry (IHC), and hematoxylin & eosin (H&E). All the animal procedures were performed and approved in compliance with institutional guidelines under the Institutional Animal Care and Use Committee of Columbia University.
CD8+ T-cell depletion
Animals were treated with either anti-mouse CD8α (BE0004-1, BioXCell) or anti-rat IgG2a isotype control, anti-trinitrophenol (BE0089, BioXCell). The antibody concentration given to each mouse was 2 mg/mL diluted in PBS at a final volume of 100 μL administered via intraperitoneal injection one week prior to glioma induction and continued twice per week in order to achieve consistent CD8+ T-cell depletion. Depletion was monitored at three time points starting from intracranial virus injection, 3 weeks apart.
RNA extraction, sequencing and gene expression analysis
After 7 tumor growth/days, gliomas treated either with the anti-mouse CD8α (8 mice) or the anti-rat IgG2a isotype control (8 mice) were dissected from the brain of the animals. RNA extraction was performed from the tumor bulk using the RNeasy Mini kit (Qiagen). Paired-end transcriptome reads were processed using STAR(10) aligner on the basis of the Ensembl GRCm38 mouse genome assembly with default parameters. Normalized gene expression values were calculated by featureCounts(11) as RPKM. ssGSEA was performed using the R package GSVA(12).
Immunohistochemistry
Formalin-fixed, paraffin-embedded (FFPE) material from animals was deparaffinized with xylene and antigen retrieval was performed with 10mM sodium citrate buffer (pH=6). IHC was performed with a 1:150 diluted rat monoclonal antibody against MHC-I (AB15680) (Abcam; Cambridge, UK), 1:150 diluted rabbit polyclonal antibody against MHC-II (AB180779) (Abcam; Cambridge, UK), 1:2000 diluted rabbit monoclonal antibody against CD11b (AB133357) (Abcam; Cambridge, UK), 1:3000 diluted rabbit monoclonal antibody against Iba1 (AB178846) (Abcam; Cambridge, UK), 1:250 diluted rabbit monoclonal antibody against p38 (4511) (Cell Signaling; Danvers, MA), and 1:250 diluted rabbit monoclonal antibody against pERK (4370) (Cell Signaling; Danvers, MA). Counterstaining was performed on the same material with hematoxylin (Abcam; Cambridge, UK). IHC staining was performed on a Leica Bond-Max automatic immunostainer (Bannockburn, IL). Images were acquired using a standard light microscope (Leica DM2000 LED, Leica Microsystems, Wetzlar, Germany) mounted with a digital microscope camera (Leica DFC450 C, Leica Microsystems, Wetzlar, Germany).
Machine learning of IHC quantification was used by setting parameters to identify positivity and subtracting counter-stained/negative background using ImageJ software (available from the National Institute of Health). The substrate optical density and intensity was determined by quantifying the positive nuclei or cytoplasm (dependent on particular antibody assessed) paired with the intensity of the staining in neoplastic tumor cell regions of the brain at 4x light microscopic magnification in order to infer immunoreactivity index.
Bioanalysis of aneuploidy
Mutational burden was called by analysis of germline and somatic variation by Strelka2 Small Variant Caller (Illumina; San Diego, CA). The germline identification employed a tiered haplotype model adaptively selected between assembly and alignment-based haplotyping at each variant locus. The aneuploidy score from somatic copy number alterations was calculated by calling the presence or absence of amplifications or deletions as described previously (13). Aneuploidy score as a comparison between two groups was called by copy number detection, implemented in the software package CNVkit, using both targeted reads and non-specifically captured off-target reads to infer copy number evenly across the genome (14). STAR-Fusion was used to identify candidate fusion events (10). The impact of the fusion event on coding regions was explored by invoking the ‘examine_coding_effect’ parameter. Only the candidates that affected the coding regions were retained for fusion load analysis. Copy number variance events were counted and the percentage of the genome affected by such events was calculated as described (13).
Calculation of Shannon index
For each tumor, the Shannon diversity was estimated using the command ‘entropy.empirical’ from the ‘entropy’ R package. This was calculated on the basis of the estimated prevalence of different immune cell compartments found in the tumor. The Shannon diversity score, H, followed the formula H = −Σpi × log(pi).
Prediction of neoantigen binders
Novel 9–11mer peptides that could arise from identified non-silent mutations or gene fusions present in the tumor samples were determined. We used the pVAC-Seq54 pipeline with the NetMHCcons55 binding strength predictor to identify neoantigens (15,16). The predicted IC50 inhibitory concentration binding affinities and rank percentage scores were calculated for all peptides that bound to each of the tumor’s HLA alleles. Using established thresholds, predicted binders were considered to be those peptides that had a predicted binding affinity < 500 nM.
Statistical analysis
All statistical analysis was conducted using GraphPad Prism 8 (GraphPad Software; San Diego CA) and R version 3.1.2 (R Core Team; Vienna, Austria). Numerical data was reported as mean ± SEM. Mann Whitney U test or unpaired Student’s t test was used for two group comparisons. ANOVA was used for more than two groups. Kaplan-Meier survival curves were generated, and a log-rank test was utilized to compare survival distribution. Mutational and gene fusion counts were analyzed using a binomial model for count data with over-dispersion. All reported q values were calculated by statistical adjustment of p values using the Benjamini-Hochberg constant. The Pearson coefficient of correlation (r) was calculated by the linear regression of the data points according to the goodness of fit where significance was determined at which point the slope was significantly non-zero. In silico analysis was performed with 161 cataloged, human GBM samples provided by The Cancer Genome Atlas (TCGA) with mRNA expression data available (RNAseq V2 RSEM).
Results
Gliomas that develop in the absence of CD8+ T-cells are more immunogenic, and show more malignant histologic features.
We investigated the effects of CD8+ T-cells on glioma development using a transgenic PDGF+Pten−/− murine glioma model. In this model, gliomas are induced de novo by retroviral vector-based expression of PDGF and Cre recombinase, which leads to loss of the Pten tumor suppressor gene (in Ptenlox/lox mice) and over-expression of PDGF. Resultant gliomas accumulate genetic alterations that resemble those of the human disease (17). CD8+ T-cell-mediated immunoediting would require glioma infiltration by these lymphocytes. In order to investigate this hypothesis, we evaluated the presence of CD4+ and CD8+ T-cells at different stages of glioma progression. At 21 days post-virus injection (d.p.i) for glioma induction, which is an early phase of glioma development that precedes the accumulation of the genetic alterations and histological features of high-grade gliomas (17), there was both CD4+ and CD8+ T-cell infiltration in the glioma (Supplementary Fig. S1A). We then evaluated T-cell infiltration at 35 d.p.i., a period in which this model exhibits the genetic alterations and histology of high-grade gliomas (17). Given that these gliomas exhibit considerable growth within weeks, we normalized T-cell infiltration by the average glioma size (estimated by bioluminescence) for each time point (Supplementary Fig. S1B/C). This analysis revealed a decline in the fraction of CD4+ as well as CD8+ T-cells in the glioma at 35 d.p.i. compared to that exhibited at 21 d.p.i. (p=0.0189 [CD4+], p=0.0026 [CD8+], t-test; Supplementary Fig. S1D).
To investigate the effects of CD8+ T-cells on glioma development and immunogenicity, we depleted this immune cell population by administration of an anti-CD8+ antibody starting one week prior to retroviral injection, and throughout glioma development. Additionally, we injected an isotype control antibody to another cohort of mice (intact immunity). Then, we induced gliomas in these hosts treated with and without the anti-CD8+ T-cell antibody. As a second experiment, we isolated and generated primary cultures of these tumors, and subsequently transplanted these glioma cells into recipients with or without CD8+ T-cell depletion (Fig. 1A shows a schematic of both experiments). CD8+ T-cell depletion was confirmed by flow cytometry analysis of splenocytes (Fig. 1B). The absence of CD8+ T-cells during glioma development had no effect on glioma growth, as mice treated with the anti-CD8+ antibody had comparable survival to mice treated with the isotype control antibody (p=0.7563, Log-rank test; Fig. 1C). This is consistent with the results of the transgenic PDGFB+RCAS-Stat3−/− glioma model, in which CD8+ T-cells had no effect on overall survival in tumor-bearing mice (Rao G et al. unpublished data).
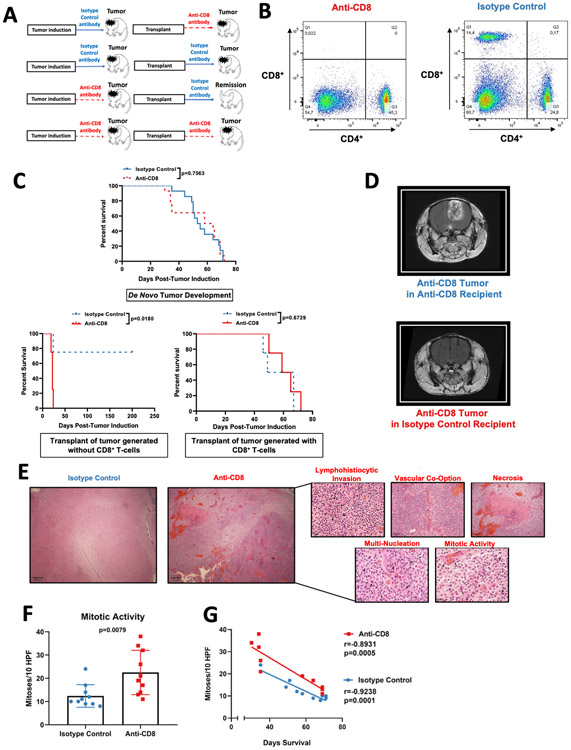
Figure 1: Gliomas that develop in the absence of CD8+ T-cells are more immunogenic yet display increased oncogenic features.
(A) Scheme of the experimental design to investigate glioma immunoediting. Mice were treated with either Isotype control antibody or anti-CD8+ T cell antibody prior to the injection of the PDGF+-IRES-Cre retrovirus, and throughout tumor development. Tumors were harvested and used to create primary cultures. These cell lines were intracranially implanted into recipients treated with either anti-CD8+ antibody or Isotype control antibody prior tumor implantation, and until death (B) Representative flow cytometry dot-plots of CD4+ and CD8+ T cells in the spleen after administration of anti-CD8+ antibody and the isotype control antibody. (C) Kaplan-Meier plots of animals treated with anti-CD8+ and isotype control antibodies prior to and during de novo tumor formation (n=28) (top). Kaplan-Meier plot of animals that received the transplant of glioma originated in a host treated with anti-CD8+ antibody (depleted of CD8+ T-cells) (n=8) (bottom left). Kaplan-Meier plot of animals that received the transplant of glioma originated in a host treated with the Isotype control antibody (immunocompetent host for tumor the de novo tumor formation) (n=8) (bottom right) (D) Representative MRI of mice implanted with gliomas generated in the absence of CD8+ T-cell. (E) Histopathological micrographs with H&E staining in tumors from mice treated with isotype control and anti-CD8+ antibodies. (F) Bar plot showing mitotic figures identified per 10 high-powered fields (HPFs) compared between tumors treated with the anti-CD8+ and isotype control antibody (n=20). (G) Scatter plot showing the correlation between mitotic activity and survival from mice treated with isotype control and anti-CD8+ antibodies. p=0.7563 and p=0.0180 in (C), Log rank test. Data in (F) are shown as mean ± SEM, unpaired student´s t-test. Points represent individual mice. r=−0.8931, p=0.0005 (anti-CD8+); r=−0.9238, p=0.0001 (isotype control) in (G), Pearson correlation coefficient.
Implantation of glioma cells that were originated in mice with CD8+ T-cell depletion into recipients treated with the anti-CD8 antibody (which have CD8+ T-cell depletion) led to growth of gliomas in all cases. Implantation of the same glioma cells developed in mice with CD8+ T-cell depletion into recipients treated with the isotype control antibody (intact immunity) led to engraftment in only 25% of the mice, with the remaining animals surviving past 220 days without any evidence of glioma or neurological symptoms (p=0.018, Log-rank test compared to; Fig. 1C bottom left, and 1D). In contrast, transplantation of a tumor generated in a mouse host with intact CD8+ T-cells (treated with isotype antibody), led to growth and similar survival for recipients that had intact CD8+ T-cells (isotype antibody treatment) or hosts with CD8+ T-cell depletion (Anti-CD8 antibody treatment) (Fig. 1C bottom right).
Interestingly, gliomas that developed in the absence of CD8+ T-cells exhibited a distinct histology relative to those that developed in the presence of CD8+ T-cells (Fig. 1E). In the former group, H&E staining revealed increased lymphohistiocytic infiltrates in addition to a higher extent of necrosis. Vasculature in these gliomas was co-opted by neoplastic cells that were dysmorphic and multi-nucleated. We observed more mitotic activity in gliomas that developed in the absence of CD8+ T-cells (mean mitoses per high power field (HPF): 22.5) compared to gliomas that developed in the presence of CD8+ T-cells (mean mitoses per HPF: 12.4; p=0.0079, t-test; Fig. 1F). Furthermore, mitotic activity showed an inverse correlation with survival (r=−0.8931, p=0.0005 [anti-CD8]; r=−0.9238, p=0.0001 [isotype control], Pearso´s r; Fig. 1G).
In accordance to the cancer immunoediting paradigm, these results show that gliomas developed without CD8+ T-cells display increased immunogenicity compared to tumors developed under the CD8+ T-cell pressure. Along with these findings, these immunogenic tumors exhibited more malignant histological characteristics than the less immunogenic tumors.
Gliomas that develop in the absence of CD8+ T-cells have more chromosomal deletions and gene fusions.
We investigated whether the absence of CD8+ T-cells during glioma development has an effect on the tumor genome. We performed paired somatic and germline exome sequencing, as well as paired-end bulk RNA-seq, of gliomas induced de novo that developed in the presence or absence of CD8+ T-cells. Overall, there were only a few non-synonymous mutations detected, mostly in TP53, consistent with our previous characterization of this glioma model (17). There was no change in the number of point mutations between these gliomas (p=0.7047, t-test; Fig. 2A). Yet gene fusions occurred more frequently in the gliomas that developed in the absence of CD8+ T-cells. Specifically, the mean fusion count of gliomas that developed in the absence of CD8+ T-cells was 2.5 versus 0.5 in the presence of CD8+ T-cells (p=0.0309, Mann Whitney U test; Fig. 2B). In the group of gliomas that developed in the absence of CD8+ T-cells, one glioma showed two neoantigens derived from two separate gene fusion events with predicted high binding affinity to MHC-1 (H2-Kb and H2-Db).
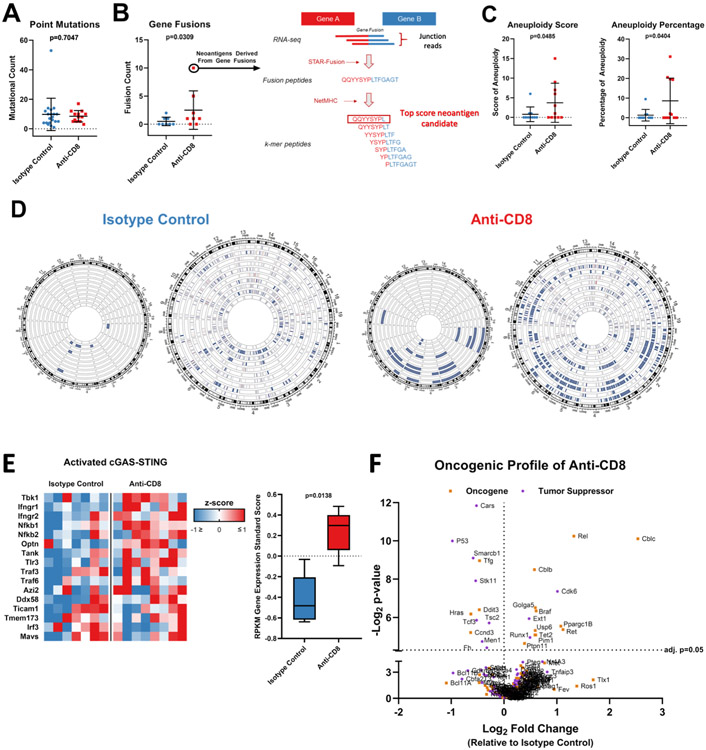
Figure 2: Loss of CD8+ T-cells during glioma formation is associated with increase in tumor genomic instability.
(A) Bee swarm plots showing the number of point mutations (n=33) and (B) gene fusions (n=16) compared between tumors from animals treated with anti-CD8+ and isotype control antibodies. Predicted neoantigens derived from gene fusions with high binding affinity to H2-Kb and H2-Db are shown via schematic representation of the neoantigen prediction pipeline in the right. (C) Bee swarm plots showing the aneuploidy score (n=21) (left) and percentage of aneuploidy across the genome (n=21) (right) compared between tumors from animals treated with anti-CD8+ and isotype control antibodies. (D) Circos plot showing the chromosomal losses and amplifications compared between tumors from animals treated with anti-CD8+ and isotype control antibodies (n=21; p=0.034). Each wheel represents a track of a tumor for the copy number variation analysis. Blue represents deletion while red represents amplification. The left schematic of each panel depicts only deep copy number variations while the right also depicts shallow variations. (E) Heatmap and box plot of the gene expression of the cGAS-STING signaling. The RPKM z-score of genes involved in cGAS-STING signaling is compared between tumors developed in the absence of CD8+ T-cells and the isotype control (n=15). (F) Volcano plots show the relative expression of all oncogenes and tumor suppressors as plotted by their RPKM log2 fold change versus adjusted p-value= 0.05 by differential expression analysis in each group. Data in (A-C, E) are shown as mean ± SEM, p values in (A and C) by student´s t test, p value in (B) by Mann Whitney U test. Points in (A-C) represent individual mice. p=0.0138 in (E), two-way ANOVA.
We assigned an aneuploidy score to each tumor as determined by Davoli et al. (13). Gliomas that developed in the absence of CD8+ T-cells exhibited increased aneuploidy relative to those that developed in the presence of CD8+ T-cells. The mean aneuploidy score of gliomas that developed in the absence of CD8+ T-cells was 3.7, while it was 0.8 in the presence of CD8+ T-cells (p=0.0485, t-test; Fig. 2C). We next used a complementary aneuploidy score calculation to infer somatic copy number alterations evenly across the genome as previously described (18). This analysis revealed that gliomas that developed in the absence of CD8+ T-cells had aneuploidy in 8.587% of the genome while that of the isotype control was only in 1.281% (p=0.0404, Mann Whitney U test). Copy number variation events were mapped in the genome. The increase in aneuploidy exhibited by gliomas that developed in the absence of CD8+ T-cells primarily consisted of chromosomal losses (Fig. 2D). As hyperploidy has been reported to drive innate immune activation via cGAS-STING signaling (19), we evaluated the gene expression associated with cGAS-STING (Tbk1) activation by RNA-seq (20). This analysis revealed increased expression of Tbk1, as well as evidence-based associated genes in gliomas that developed in the absence of CD8+ T-cells (p=0.0138; ANOVA; Fig. 2E).
In line with the malignant histological features revealed in gliomas developed in the absence of CD8+ T-cells, oncogenes and tumor suppressor genes were examined by their relative expression between gliomas that developed in the presence versus absence of CD8+ T-cells. We found that the gliomas that developed in the absence of CD8+ T-cells exhibited differential expression of both oncogenes and tumor suppressors relative to gliomas that developed in the setting of intact immunity (Fig. 2F).
Jointly, our results indicate that CD8+ T-cells influence expression of certain oncogenes and tumor suppressor genes, in addition to reducing aneuploidy and gene fusions. In contrast, the absence of. CD8+ T-cells during glioma formation is associated with these genomic alterations and the increased gene expression of the STING pathway.
ERK and p38 signaling is activated in gliomas that develop in the absence of CD8+ T-cells.
Serine/threonine kinase BRAF, as well as phosphatase PTPN11, are upstream activators of the MAPK signaling pathway that have oncogenic properties (21). Both genes are involved in the activation of ERK. We recently described the over-representation of activating mutations in these genes in recurrent GBM patients who responded to PD-1 blockade (8). Given that these mutations were associated with susceptibility to immunotherapy, we hypothesized that murine gliomas that developed in the absence of CD8+ T-cells may exhibit an overexpression and activation of MAPK/ERK. Murine gliomas did not have mutations in these genes, yet RNA-seq analysis revealed that both Braf (p=0.0232, t-test) and Ptpn11 (p=0.0256, t-test) were over-expressed in gliomas that developed in the absence of CD8+ T-cells (Fig. 3A). The oncogenic MAPK signaling pathway has distinct branches that include the ERK, p38, and JNK effectors. We next investigated the expression of other known activators of these signaling cascades. Nras is an activator of the ERK cascade and was overexpressed in these gliomas (p=0.0483, t-test). Traf6, an activator of the p38 cascade, was also overexpressed (p=0.0483, t-test; Fig. 3A). There were no differences in expression of known activators of the JNK cascade (data not shown). Given the overexpression of these activators of MAPK signaling in gliomas that developed in the absence of CD8+ T-cells, we then investigated the functional activity of the ERK and p38 cascades. Gliomas that developed in the absence of CD8+ T-cells had elevated activation of these cascades, as demonstrated by immunoreactivity for phosphorylation of ERK (pERK) and p38 (p-p38). pERK in gliomas that developed in the absence of CD8+ T-cells revealed intense diffuse positivity with a mean immunoreactivity score of 25.91, as compared to gliomas that developed in the presence of CD8+ T-cells that had a mean immunoreactivity score of 13.41 (p=0.0229, t-test; Fig. 3B). Phospho-p38 expression in gliomas that developed in the absence of CD8+ T-cells demonstrated scattered focal positivity with a mean immunoreactivity score of 14.50, as compared to gliomas that developed in the presence of CD8+ T-cells that had a mean immunoreactivity score of 9.41 (p=0.0285, t-test). To determine if this signaling was unique to the PDGF+Pten−/− murine glioma model, we evaluated the phosphorylation of the ERK and p38 cascades in a PDGFB+RCAS-Stat3−/− transgenic glioma model, in which CD8+ T-cells were depleted by germline homozygous knock-out of the Cd8a gene (22). In this model, pERK had a mean immunoreactivity score of 95.39 and 73.76 in the gliomas from either the CD8−/− or the CD8+/+ background, respectively (p=0.4848, t-test; Fig. 3C). Similar to the PDGF+Pten−/− murine glioma model, PDGFB+RCAS-Stat3+/+ gliomas demonstrated similar findings in p-p38. More specifically, gliomas in the CD8−/− background revealed scattered to moderate p-p38 positivity with a mean immunoreactivity score of 71.56, as compared to a mean immunoreactivity score of 9.18 in the CD8+/+ gliomas (p=0.0022, t-test). We next investigated whether the expression of upstream regulators of the MAPK pathway correlated with phosphorylation of effectors of this pathway in the PDGF+Pten−/− murine glioma model. Braf (r=0.7276; p=0.0408, Pearso´s r) and Ptpn11 (r=0.8499; p=0.0075, Pearso´s r) expression by RNA-seq correlated with pERK in gliomas that developed in the absence of CD8+ T-cells (Supplementary Fig. S2A/B). Nras expression did not correlate with pERK in either gliomas (Supplementary Fig. S2C). Traf6 showed a statistical trend in expression with p-p38 in gliomas that developed in the absence of CD8+ T-cells (r=0.6496; p=0.0813, Pearso´s r) (Supplementary Fig. S2D).
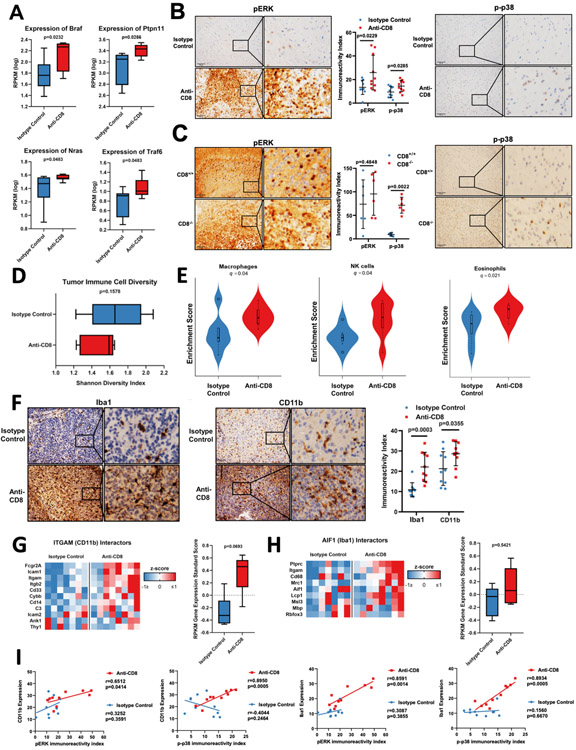
Figure 3: MAPK pathway (ERK and p38) activation is increased gliomas that develop in the absence of CD8+ T-cells and is correlated with increased macrophage/microglial infiltration.
(A) Box plots showing the Braf, Ptpn11, Nras, and Traf6 (n=16) expression compared between tumors developed from mice treated with anti-CD8 and isotype control antibodies by analysis of RNA-seq data in a Pten−/− transgenic murine glioma model. (B) Representative micrographs showing the pERK (left) and p-p38 (right) immunoreactivity index compared between groups. Bee swarm plot showing the quantification of pERK and p-p38 (n=20) (middle). (C) pERK immunoreactivity index compared between tumors from CD8−/− and CD8+/+ mice by IHC in a Stat3−/− transgenic murine glioma model. Bee swarm plot showing the quantification of pERK and p-p38 (n=16) expression. p-p38 compared groups. (D) Box plot showing the shannon diversity index of immune cell populations in tumors developed in the absence of CD8+ T-cells compared to the isotype control (n=20). (E) Violin plots showing the macrophage, NK cell, and eosinophil (n=20) gene expression signature between groups by CIBERSORT of RNA-seq data. (F) Representative micrographs showing the Iba1 (left) and CD11b (middle) immunoreactivity index compared between groups by IHC. Quantification of CD11b and Iba1 (n=20) IHC expression (right). (G) RNA-seq based expression analysis of genes of proteins known to interact with ITGAM (CD11b) (H) and of genes of proteins known to interact with AIF1 (Iba1) in tumors developed in the absence of CD8+ T-cells versus the isotype control (n=15). (I) Correlation between CD11b expression and pERK (n=10). Correlation between CD11b expression and p-p38 (n=10). Correlation between Iba1 expression and pERK (n=10). Correlation between Iba1 expression and p-p38 (n=10). Data are shown as mean ± SEM, p values in A, D, G, and H by unpaired student´s t-test. p values in B, C, and F by unpaired student´s t-test. For B, C, F, and G, points represent individual mice. q values by statistical adjustment of p values using the Benjamini-Hochberg constant. r and p values in (I) by Pearson correlation coefficient. Points represent individual mice.
Together, these results indicate that the overexpression and activation of the p38 and ERK cascade of the MAPK signaling pathway, as oncogenic features that are suppressed by CD8+ T-cells during the progression of gliomas. Our results also confirm the integrity of the MAPK signaling cascade in these gliomas, as expression of upstream members of the pathway are associated with phosphorylation of the downstream effectors, which is indicative of activation.
Macrophage/microglial infiltration increases in gliomas that develop in the absence of CD8+ T-cells.
We hypothesized that the immunogenicity of gliomas that developed in the absence of CD8+ T-cells relates to a distinct immune microenvironment. We determined the Shannon diversity index between the immune cell populations within glioma groups. Gliomas that developed in the absence of CD8+ T-cells had a collective index ranging from 1.228 to 1.652 while those that developed with intact immunity had an index ranging from 1.223 to 2.081 (p=0.1578, t-test; Fig. 3D), indicating that the diversity of glioma infiltrating immune cell populations between both groups of mice is similar. We then used CIBERSORT to estimate the immune cell populations in the tumors by RNA-seq, as previously done (23). This analysis showed an increase in the expression signature of natural killer (NK) cells (q=0.04), macrophages (q=0.04), and eosinophils (q=0.021) in gliomas that developed while undergoing treatment with anti-CD8+ antibody (Fig. 3E).
IHC staining confirmed that macrophage/microglia infiltration in gliomas was enhanced in the absence of CD8+ T-cells. Iba1, a macrophage/microglia marker, showed higher expression with a mean immunoreactivity score of 22.12 in gliomas that developed in the absence of CD8+ T-cells, relative to a mean immunoreactivity score of 11.22 in gliomas that developed in the presence of CD8+ T-cells (p=0.0003, t-test; Fig. 3F). Additionally, we evaluated the integrin subunit CD11b highly expressed on monocytes, granulocytes, and/or tissue resident macrophages, which has been recently reported to be implicated with pro-inflammatory polarization of tumor-associated macrophages (24,25). This marker revealed diffuse positivity with a mean immunoreactivity score of 27.87 in gliomas that developed in the absence of CD8+ T-cells, as compared to a mean immunoreactivity score of 20.79 in gliomas that developed in the presence of CD8+ T-cells (p=0.0355, t-test; Fig. 3F). To strengthen this point, genes coding for proteins known to interact with ITGAM (CD11b) (26) showed a trend for elevated mRNA expression in gliomas that developed in the absence of CD8+ T-cells (p=0.0693, ANOVA; Fig. 3G). Genes coding for proteins known to interact with AIF1 (Iba1) (26) were no different between the groups of mice (p=0.5421, ANOVA; Fig. 3H).
These results suggest increased infiltration of cells from innate immunity, including macrophages/microglia, with strong expression of CD11b in gliomas that developed in the absence of CD8+ T-cells.
MAPK signaling correlates with macrophage infiltration in gliomas that develop in the absence of CD8+ T-cells.
Since we encountered robust yet variable macrophage infiltration and MAPK pathway activation in the gliomas that developed in the absence of CD8+ T-cells, we investigated whether this signaling correlated with the degree of immune infiltration. CD11b expression highly correlated with pERK in gliomas that developed in the absence of CD8+ T-cells (r=0.6512; p=0.0414, Pearso´s r) but not in those gliomas that developed in the presence of CD8+ T-cells (r=0.3252; p=0.3591, Pearso´s r; Fig. 3I). CD11b also correlated with p-p38 in gliomas that developed in the absence of CD8+ T-cells (r=0.8950; p=0.0005, Pearso´s r) but not in gliomas developed with CD8+ T-cells present (r=0.4044; p=0.2464, Pearso´s r). Similarly, Iba1 expression correlated with pERK in gliomas that developed in the absence of CD8+ T-cells (r=0.8591; p=0.0014, Pearso´s r) but not in gliomas that developed in the presence of CD8+ T-cells (r=0.3087; p=0.3855, Pearso´s r). Iba1 also correlated with p-p38 in gliomas that developed in the absence of CD8+ T-cells (r=0.8934; p=0.0005, Pearso´s r) but not in gliomas with intact immunity (r=0.1560; p=0.6670, Pearso´s r).
We then investigated the relationship between MAPK signaling and macrophage/microglial infiltration in human GBM. Messenger RNA (mRNA) expression analysis of 161 GBM patients in TCGA revealed a correlation between ITGAM (CD11b) and MAPK13 (p38delta), an activator of the p38 cascade (r=0.6194; p<0.0001, Pearso´s r; Supplementary Fig. S3A). mRNA expression analysis of GBM patients in TCGA revealed a correlation between AIF1 (Iba1) and RRAS, an activator of the ERK cascade (r=0.6141; p<0.0001, Pearso´s r; Supplementary Fig. S3B). AIF1 gene expression also correlated with MAPK13 (r=0.6008; p<0.0001, Pearso´s r: Supplementary Fig. S3C), and MAP3K8, an activator of the ERK cascade (r=0.8159; p<0.0001, Pearso´s r; Supplementary Fig. S3D). These results suggest that MAPK activation and macrophage/microglial recruitment are highly interrelated in both murine gliomas and human GBM patients. Furthermore, this association appears to be strengthened by the absence of CD8+ T-cells during glioma development.
Gliomas that develop in the absence of CD8+ T-cells have a pro-inflammatory macrophage gene signature.
Given that macrophage/microglial infiltration was robust in gliomas that developed in the absence of CD8+ T-cells, we explored whether this immune cell infiltration was associated with an inflammatory milieu. Inflammatory chemokines and cytokines released during the elimination phase of cancer immunoediting (4) were increased in gliomas that developed in the presence of CD8+ T-cells relative to the CD8+ T-cell-depleted background (p=0.0106, ANOVA; Fig. 4A). However, gene set enrichment analysis (GSEA) employing the tumor inflammation signature, a gene expression profile indicative of an inflamed tumor microenvironment (TME) developed by Ayers et al (27), revealed that gliomas treated with the anti-CD8+ antibody had an active inflammatory TME compared to gliomas developed under the presence of CD8+ T-cells (p=0.033, Kolmogorov-Smirnov test; Fig. 4B).
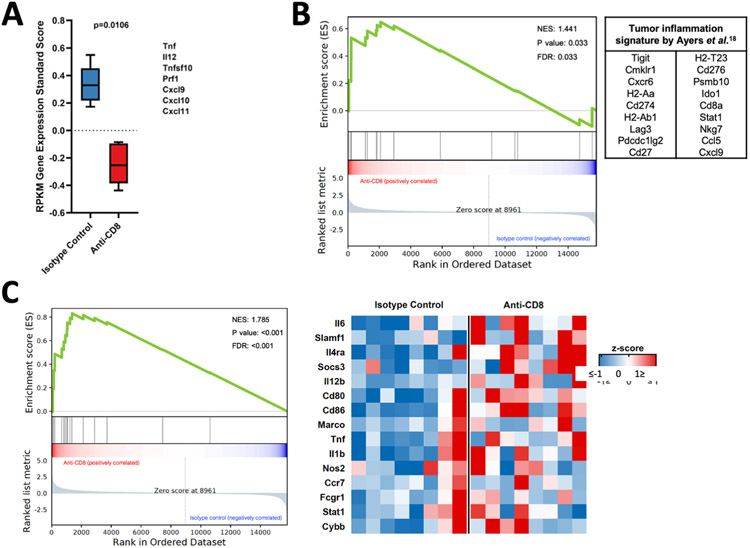
Figure 4: Macrophages/microglia in tumors that develop in the absence of CD8+ T-cells have an enriched pro-inflammatory phenotype.
(A) Box plot showing RPKM gene expression standardized score. (B) GSEA employing the tumor inflammation signature between animals treated with anti-CD8+ and isotype control antibodies. (C) GSEA (left) and heatmap (right) evaluating the expression signature of pro-inflammatory phenotype in macrophages/microglia in tumors developed in the absence of CD8+ T-cells and the isotype control (left, NES=1.785; FDR<0.001), (n=15). Data in (A) is shown as mean ± SEM, p=0.0106 by 2-way ANOVA test. p= 0.033 in (B) and p<0.001 in (C), two-sided Kolmogorov-Smirnov test.
As macrophages can exist either in a non-polarized state or along a continuum between pro- and anti-inflammatory (28), we evaluated such phenotypic changes in these gliomas. GSEA revealed an increased pro-inflammatory macrophage phenotype by a gene signature reported to contribute to such polarization (29) in gliomas that developed in the absence of CD8+ T-cells, with a normalized enrichment score (NES) of 1.785 compared to those that developed in the presence of CD8+ T-cells (p<0.001, Kolmogorov-Smirnov test; Fig. 4C). A gene signature associated with an anti-inflammatory macrophage phenotype (29,30) did not reveal significant differences in gliomas developed in a CD8+ T-cell-depleted background (NES 1.2, p=0.232, data not shown).
Together, these results indicate that gliomas that develop in the absence of CD8+ T-cells have an inflamed tumor microenvironment, including pro-inflammatory macrophage/microglial polarization.
Discussion
Cancer immunoediting shapes tumor evolution by eliminating immunogenic tumor cell variants that can evade immune surveillance, while others are selected for resistance to immune recognition (6). Particularly, CD8+ T-cell recognition of tumor antigens drives the immunological sculpting of a developing cancer (4). Whereas this concept has previously been suggested in GBM (31-33), our study directly investigated the effects of CD8+ T-cell-dependent immunoediting in the development of gliomas. We generated experimental evidence of immunoediting by CD8+ T-cell in a transgenic murine glioma model, as depletion of this immune cell population had a profound effect on tumor genomic evolution, attributable to selection of immunogenic tumor clones by CD8+ T-cells. While we found strong differences between gliomas that developed with or without this immune cell population, we also found important variation of these features within the group of gliomas that developed in the absence of CD8+ T-cells. Recent evidence has shown that glioma genomic evolution is to some extent stochastic (34), i.e. is a neutral evolutionary process, with genomic events that can lead to immune recognition or evasion (32). Surprisingly, whereas depletion of CD8+ T-cells during glioma development had profound effects on tumor genotype, phenotype, immunogenicity and microenvironment, depletion of these immune effector cells did not lead to shorter survival. CD8+ T-cell depletion did not alter survival of other transgenic glioma models in which tumors develop de novo (Rao G et al., submitted). Previous studies also showed that, whereas immunosuppression can increase the incidence of cancer, it usually does not affect survival upon cancer diagnosis (35). These observations in the face of tumor progression are termed the “Hellstrom paradox” (36,37). How this paradoxical situation manifests remains unresolved. De novo tumor formation and the influence of host immunity have been previously explored (38,39). For instance, Urban et al., described that UV-light related immunosuppression in mice influences grafting of skin tumors upon transplantation, suggesting immune selection (38). Yet our study is different, as we show that depletion of CD8+ T-cells during gliomagenesis leads to tumors that exhibit more immunogenicity.
We sought to explore genomic differences as a result of tumorigenesis in the absence of CD8+ T-cells. Low mutational burden is a common explanation for the relative lack of immunogenicity in GBM (40). We found no change in the number of point mutations as a result of the absence of T-cells. Our results imply that CD8+ T-cells do not protect against point mutations or influence this facet of immunogenicity to an appreciable extent. Yet, it is important to consider that in this particular glioma model, mice may not survive long enough for the development and selection of mutations other than those with a strong advantage for cell growth (i.e. Trp53). On the other hand, we observed an increase in the frequency of glioma gene fusions in the absence of CD8+ T-cells. Gene fusions are known to be strong drivers of neoplasia as well as the formation of neoantigens from junctional epitopes (41,42). Bioinformatic modeling predicted these gene fusions to have high binding affinity to MHC-I (H2-Kb and H2-Db). Whereas the antigenic properties predicted warrant experimental validation, this finding is interesting as junctional epitopes from gene fusions are a potential source for development of neoantigen-based vaccination. Among other genomic alterations, increased levels of aneuploidy were also observed in gliomas developed in the absence of CD8+ T-cells. Previous research associated aneuploidy with immune evasion in cancer (13). Meanwhile, we show that gliomas developed in the absence of CD8+ T-cells have both, high aneuploidy and a more immunogenic phenotype, reinforcing the biological importance of negative immunoselection of cancer cells harboring somatic copy number alterations during cancer development (19). Chromosomal instability drives innate immune activation via cGAS-STING in cancer (20), and in support of this, we showed upregulation of genes related to this pathway in gliomas that developed in the absence of CD8+ T-cells (38). Considering that the aneuploidy content in human gliomas is relatively low compared to other cancers (43), our results imply that CD8+ T-cells might be negatively selecting against high aneuploidy glioma cells during tumor development that manifest as low aneuploidy tumors at late tumoral stages as observed in the murine gliomas that developed under the CD8+ T-cell pressure.
We recently reported the presence of MAPK activating mutations in BRAF and PTPN11 in recurrent GBM patients who responded to PD-1 blockade (44). Given the implication of these genes and MAPK in response to immunotherapy, we explored how this pathway is affected by the presence of CD8+ T-cells during glioma development. Whereas these genes were not mutated in the absence of CD8+ T-cells, we did find them over-expressed in this setting, along with several other known activators of ERK and p38 signaling (44). In PDGFB+RCAS-Stat3−/− gliomas, a separate transgenic murine glioma model, the p38 cascade was also more phosphorylatyed/activated when tumors developed in the absence of CD8+ T-cells. We also observed that in the setting of CD8+ T-cell depletion, MAPK activation in gliomas was associated with robust tumor infiltration with CD11b+ and Iba1+ macrophages/microglia. Moreover, in lung cancer, activating mutations of KRAS (which leads to MAPK signaling) are associated with robust tumor infiltration by macrophages (45). MAPK activation has been extensively studied as a driver of innate immunity with effector pro-inflammatory polarization (46), yet these results suggest that activation of MAPK signaling in tumors can influence cancer microenvironment and immunogenicity. These observations of active MAPK signaling in gliomas developed in the absence of CD8+ T-cells add to our recent discovery of MAPK activating mutations as molecular features associated with glioblastoma response to immunotherapy (44). Collectively, this evidence implicates MAPK in immunoediting during glioma development and in susceptibility to immunotherapy, yet the causal relationship and mechanism that drives this association remains unknown.
Our mouse glioma model recapitulates loss of Pten function, a genetic alteration that has been associated with an immunosuppressive microenvironment, and lack of response to immunotherapy, including poor response to PD-1 blockade in recurrent GBM patients (8,47,48). Indeed, PTEN deficiency was recently shown. to promote macrophage infiltration through lysil oxidase secretion promoting tumor progression (49). Whereas our transgenic model is generated through Pten deletion, we observed increased infiltration of macrophages/microglia and a pro-inflammatory phenotype in gliomas that developed in the absence of CD8+ T-cells. Additionally, we encountered a strong relationship between MAPK signaling and macrophage/microglial recruitment in these gliomas. mRNA expression analysis of human GBM patients in silico revealed a strong correlation between levels of known activators of the MAPK signaling pathway and macrophage markers, asserting the biological significance of these findings in the human disease. Furthermore, MAPK is known to promote a pro-inflammatory innate immune response (46). Consistent with this, we established an association between MAPK pathway activation and a pro-inflammatory macrophage recruitment in gliomas. Whereas we observed that CD8+ T-cells dampen MAPK signaling in gliomas, the effect of MAPK signaling on pro-inflammatory microenvironment in the context of glioma immunoediting remains to be elucidated.
In sum, our study suggests that glioma immunoediting involves CD8+ T-cell-dependent selection of tumor clones, leading to immune evasion by the time tumors reach a clinical stage. Cancer immunoediting mediated by CD8+ T-cells simultaneously protects against certain evolutionary hallmarks of cancer and selects for tumor cell variants that are capable of immune evasion. Understanding the interplay between glioma progression and anti-tumoral immunity is essential for the development of effective immunotherapy for this disease. Manipulation of tumor evolution towards a more immunogenic path may be explored by surpassing the CD8+ T-cell-mediated immune evasion in this disease.
Supplementary Material
Statement of translational relevance.
Immune evasion renders gliomas resistant to several types of immunotherapy. As part of the cancer immunoediting process, many cancer cells are eliminated before they develop into a clinically evident malignancy, while other tumor cells prevail evading immune recognition. In this context, therapeutic intervention often fails for gliomas because tumor cell variants that resist immune surveillance get selected by the immune system. The elucidation of how cancer immunoediting leads to immune evasion in gliomas may allow optimization of immunotherapeutic approaches to induce sustained clinical responses in these patients. Our results demonstrate immune evasion in murine gliomas can be attributed to selection pressure by CD8+ T-cells during the development and progression of these tumors.
Acknowledgments
This work was funded by 1R01NS110703–01A1 (AMS), 5DP5OD021356–05 (AMS), P50CA221747 SPORE for Translational Approaches to Brain Cancer (AMS), developmental funds from the Robert H. Lurie Cancer Center Support Grant #P30CA060553 (AMS), Institutional grant from the Lou and Jean Malnati Brain Tumor Institute (AMS), and the Matthew Larson Foundation Iron Matt Research Award (AMS). Histology services were provided by the Northwestern University Mouse Histology and Phenotyping Laboratory, which is supported by NCI P30-CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center.
Footnotes
Conflict of interest
The authors have declared that no conflict of interest exists.
References
- 1.Stupp R, Taillibert S, Kanner A, Read W, Steinberg DM, Lhermitte B, et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA 2017;318(23):2306–16 doi 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nduom EK, Weller M, Heimberger AB. Immunosuppressive mechanisms in glioblastoma. Neuro Oncol 2015;17 Suppl 7:vii9–vii14 doi 10.1093/neuonc/nov151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chongsathidkiet P, Jackson C, Koyama S, Loebel F, Cui X, Farber SH, et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med 2018;24(9):1459–68 doi 10.1038/s41591-018-0135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012;482(7385):400–4 doi 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 2011;331(6024):1565–70 doi 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 6.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 2002;3(11):991–8 doi 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 7.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344(6190):1396–401 doi 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, Chu T, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med 2019;25(3):462–9 doi 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei L, Sonabend AM, Guarnieri P, Soderquist C, Ludwig T, Rosenfeld S, et al. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS One 2011;6(5):e20041 doi 10.1371/journal.pone.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2013;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 12.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC bioinformatics 2013;14(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355(6322) doi 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol 2016;12(4):e1004873 doi 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, et al. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med 2016;8(1):11 doi 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karosiene E, Lundegaard C, Lund O, Nielsen M. NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 2012;64(3):177–86 doi 10.1007/s00251-011-0579-8. [DOI] [PubMed] [Google Scholar]
- 17.Sonabend AM, Bansal M, Guarnieri P, Lei L, Amendolara B, Soderquist C, et al. The transcriptional regulatory network of proneural glioma determines the genetic alterations selected during tumor progression. Cancer Res 2014;74(5):1440–51 doi 10.1158/0008-5472.CAN-13-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. Plos Comput Biol 2016;12(4) doi ARTN e1004873 10.1371/journal.pcbi.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, et al. An immunosurveillance mechanism controls cancer cell ploidy. Science 2012;337(6102):1678–84 doi 10.1126/science.1224922. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 2012;5(214):ra20 doi 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med 2018;24(7):954–60 doi 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 22.Kong LY, Wu AS, Doucette T, Wei J, Priebe W, Fuller GN, et al. Intratumoral mediated immunosuppression is prognostic in genetically engineered murine models of glioma and correlates to immunotherapeutic responses. Clin Cancer Res 2010;16(23):5722–33 doi 10.1158/1078-0432.CCR-10-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12(5):453–7 doi 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid MC, Khan SQ, Kaneda MM, Pathria P, Shepard R, Louis TL, et al. Integrin CD11b activation drives anti-tumor innate immunity. Nat Commun 2018;9(1):5379 doi 10.1038/s41467-018-07387-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panni RZ, Herndon JM, Zuo C, Hegde S, Hogg GD, Knolhoff BL, et al. Agonism of CD11b reprograms innate immunity to sensitize pancreatic cancer to immunotherapies. Sci Transl Med 2019;11(499) doi 10.1126/scitranslmed.aau9240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doorn KJ, Breve JJ, Drukarch B, Boddeke HW, Huitinga I, Lucassen PJ, et al. Brain region-specific gene expression profiles in freshly isolated rat microglia. Front Cell Neurosci 2015;9:84 doi 10.3389/fncel.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127(8):2930–40 doi 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabrusiewicz K, Rodriguez B, Wei J, Hashimoto Y, Healy LM, Maiti SN, et al. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight 2016;1(2) doi 10.1172/jci.insight.85841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gensel JC, Kopper TJ, Zhang B, Orr MB, Bailey WM. Predictive screening of M1 and M2 macrophages reveals the immunomodulatory effectiveness of post spinal cord injury azithromycin treatment. Sci Rep 2017;7:40144 doi 10.1038/srep40144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 2014;6:13 doi 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Appolloni I, Alessandrini F, Ceresa D, Marubbi D, Gambini E, Reverberi D, et al. Progression from low- to high-grade in a glioblastoma model reveals the pivotal role of immunoediting. Cancer Lett 2019;442:213–21 doi 10.1016/j.canlet.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Arrieta VA, Cacho-Díaz B, Zhao J, Rabadan R, Chen L, Sonabend AM. The possibility of cancer immune editing in gliomas. A critical review. Oncoimmunology 2018;7(7):e1445458 doi 10.1080/2162402X.2018.1445458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn GP, Fecci PE, Curry WT. Cancer immunoediting in malignant glioma. Neurosurgery 2012;71(2):201–22; discussion 22–3 doi 10.1227/NEU.0b013e31824f840d. [DOI] [PubMed] [Google Scholar]
- 34.Barthel FP, Johnson KC, Varn FS, Moskalik AD, Tanner G, Kocakavuk E, et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature 2019;576(7785):112–20 doi 10.1038/s41586-019-1775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A 1998;95(13):7556–61 doi 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 2001;410(6832):1107–11 doi 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 37.Hellstrom I, Hellstrom KE, Pierce GE, Yang JP. Cellular and humoral immunity to different types of human neoplasms. Nature 1968;220(5174):1352–4. [DOI] [PubMed] [Google Scholar]
- 38.Urban JL, Burton RC, Holland JM, Kripke ML, Schreiber H. Mechanisms of syngeneic tumor rejection. Susceptibility of host-selected progressor variants to various immunological effector cells. J Exp Med 1982;155(2):557–73 doi 10.1084/jem.155.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature 2007;450(7171):903–7 doi 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 40.Hodges TR, Ott M, Xiu J, Gatalica Z, Swensen J, Zhou S, et al. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro Oncol 2017;19(8):1047–57 doi 10.1093/neuonc/nox026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desrichard A, Snyder A, Chan TA. Cancer Neoantigens and Applications for Immunotherapy. Clin Cancer Res 2016;22(4):807–12 doi 10.1158/1078-0432.CCR-14-3175. [DOI] [PubMed] [Google Scholar]
- 42.Yang W, Lee KW, Srivastava RM, Kuo F, Krishna C, Chowell D, et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat Med 2019;25(5):767–75 doi 10.1038/s41591-019-0434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018;33(4):676–89.e3 doi 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, Chu T, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med 2019. doi 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang SH, Mirabolfathinejad SG, Katta H, Cumpian AM, Gong L, Caetano MS, et al. T helper 17 cells play a critical pathogenic role in lung cancer. Proc Natl Acad Sci U S A 2014;111(15):5664–9 doi 10.1073/pnas.1319051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol 2013;13(9):679–92 doi 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 47.Cascone T, McKenzie JA, Mbofung RM, Punt S, Wang Z, Xu C, et al. Increased Tumor Glycolysis Characterizes Immune Resistance to Adoptive T Cell Therapy. Cell Metab 2018;27(5):977–87 e4 doi 10.1016/j.cmet.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov 2016;6(2):202–16 doi 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen P, Zhao D, Li J, Liang X, Chang A, Henry VK, et al. Symbiotic Macrophage-Glioma Cell Interactions Reveal Synthetic Lethality in PTEN-Null Glioma. Cancer Cell 2019;35(6):868–84.e6 doi 10.1016/j.ccell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.