Abstract
Background -
Genetic factors that influence kidney traits have been understudied for low frequency and ancestry-specific variants.
Methods -
This study used imputed whole genome sequencing from the Trans-Omics for Precision Medicine project to identify novel loci for estimated glomerular filtration rate (eGFR) and urine albumin to creatinine ratio (ACR) in up to 12,207 Hispanics/Latinos. Replication was performed in the Women’s Health Initiative and the UK Biobank when variants were available.
Results -
Two low frequency intronic variants were associated with eGFR (rs58720902 at AQR, minor allele frequency [MAF] = 0.01, P=1.6 × 10−8) or ACR (rs527493184 at ZBTB16, MAF=0.002, P=1.1 × 10−8). An additional variant at PRNT (rs2422935, MAF=0.54, P=2.89 × 10−8) was significantly associated with eGFR in meta-analysis with replication samples. We also identified two known loci for ACR (BCL2L11 rs116907128, P=5.6 × 10−8 and HBB rs344, P=9.3 × 10−11) and validated eight loci for ACR previously identified in the UK Biobank.
Conclusions -
Our study shows gains in gene discovery when using dense imputation from multi-ethnic whole genome sequencing data in admixed Hispanics/Latinos. It also highlights limitations in genetic research of kidney traits, including the lack of suitable replication samples for variants that are more common in non-European ancestry and those at low frequency in populations.
Keywords: kidney, genetic polymorphism, genetic variation, genome-wide association scan, ancestry
Journal Subject Terms: Genetic, Association Studies; Genetics; Epidemiology; Race and Ethnicity
Introduction
Urine albumin to creatinine ratio (ACR) and decreased estimated glomerular filtration rate (eGFR) reflect different dimensions of chronic kidney disease (CKD), i.e., kidney damage and reduced kidney function, respectively. Hispanics/Latinos have increased aged-adjusted prevalence of CKD demonstrated by increased ACR and/or decreased eGFR compared to non-Hispanic U.S. whites based on recent data from the Hispanic Community Health Study / Study of Latinos (HCHS/SOL)1. In HCHS/SOL, the prevalence of albuminuria and reduced eGFR was 14% and 3%, respectively, among individuals who were on average 41 year-old.
Genome wide association studies (GWAS) have uncovered novel loci for ACR and eGFR, although the number of identified loci for ACR is modest compared to other complex traits. Few studies have included Hispanics/Latinos for these CKD traits. In our recent work using GWAS approaches, we identified significant associations of ACR at the CUBN and the HBB genes, the later related to associations for the sickle cell variant rs334, which is present in Hispanics with African admixture2. Additional research using admixture mapping identified an Amerindian variant at BCL2L11 associated with ACR in HCHS/SOL Hispanics/Latinos3. For eGFR, 93 loci have been recently described in multi-ethnic GWAS meta-analyses that included approximately 23,000 Hispanics/Latinos4. Interestingly, there has been little overlap in loci identified for ACR and eGFR in both Hispanics/Latinos and in studies of individuals of European ancestry5.
Prior GWAS have assessed imputed genotypes using references from the 1000 Genome Project. The NHLBI Trans-Omics for Precision Medicine (TOPMed) project recently generated deep-coverage (mean depth 30x) whole genome sequencing (WGS) on over 50,000 individuals from multi-ethnic studies, including 7.5% Hispanic/Latinos. This resource provides a large reference of common and low frequency genetic variants in diverse populations for high quality imputation in diverse populations. We used the TOPMed WGS haplotypes for a dense imputation of single nucleotide variants (SNVs) and small deletion/insertions (indels) in the HCHS/SOL study6. This study reports findings from GWAS of eGFR and ACR in Hispanics/Latinos using this data. We attempted to validate associations from a recently published GWAS of ACR in the UK Biobank white British, which identified 32 novel loci that have not yet been validated7.
Methods
HCHS/SOL8 genotype and phenotype data are publicly available at the Database of Genotypes and Phenotypes (dbGaP) and can be accessed at https://www.ncbi.nlm.nih.gov/gap, study accession phs000810. The freeze 5b TOPMed data used for imputation of WGS data is available at the dbGap, study accession phs001395. In order to minimize the possibility of unintentionally sharing information that can be used to re-identify private information in this single study, summary data of this study is available from the corresponding author upon reasonable request.
The study was approved by the institutional review boards (IRBs) at each field center, where all participants gave written informed consent, and by the Non-Biomedical IRB at the University of North Carolina at Chapel Hill.
Methods are included in the Supplemental Material.
Results
Data were available in 12,207 participants for eGFR and 11,688 for ACR. The mean age of participants was 46.1 years (standard deviation 13.9), 58.7% were women, 20.0% had diabetes and 28.0% had hypertension treated with medications. Mean eGFR was 96.6 (standard deviation 18.9) ml/min/1.73 m2 and median ACR was 6.5 (interquartile 4.5–12.2) mg/g creatinine.
GWAS results
GWAS of eGFR and ACR showed little evidence for genomic inflation (λ=1,005 and λ=1.00, respectively). Quantile-quantile plots are shown in Supplemental Figure 1 and Manhattan plots for eGFR and ACR are shown in Supplemental Figure 2. GWAS of eGFR identified fifteen loci at P<10−7 (Table 1), including a significant association for a low frequency intronic SNV at the AQR gene (rs58720902, minor allele frequency [MAF]= 0.01, P=1.6 × 10−8)(Figure 1A). Most of the SNVs/indels shown in Table 1 were low frequency variants and showed a large effect on eGFR. These variants were more commonly seen in non-European ancestry populations. For ACR, we identified twelve loci at P<10−7 including two loci previously identified in admixture mapping (BCL2L11 rs116907128, P=3.5 × 10−8) and in a GWAS in HCHS/SOL (HBB rs344, P=8.4 × 10−11), in addition to a novel loci at ZBTB16 (P=1.1 × 10−8) (Table 2, Figure 1B). The HBB rs344 was also nominally associated with eGFR (P=5.0 × 10−3).
Table 1.
Main findings for association with eGFR at p<10−7 using TOPMed reference imputation in the HCHS/SOL (n=12,207)
| Ch | Position (hg38) |
SNPID | Coded allele |
Non-coded | AF coded | beta | P | Nearby Gene |
Function | 1000 Genome Project allele frequencies | Replication WHI AA (n=8,224) | Replication WHI HA (n=3,549) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFR | AMR | EUR | Beta (SE), P | Beta (SE), P | ||||||||||
| 2 | 191304743 | rs566396416 | A | G | 0.002 | −12.801 (2.523) | 3.8×10−7 | MYO1B | intronic | NA | NA | NA | NA | NA |
| 5 | 113371933 | rs17379925 | T | C | 0.02 | 3.159 (0.645) | 9.0 ×10−7 | MCC | intronic | 0.005 | 0.02 | 0.05 | NA | 1.135 (1.143), 0.32 |
| 5 | 171905787 | rs77109276 | G | A | 0.01 | −4.179 (0.847) | 8.2 × 10−7 | FBXW11 | intronic | NA | 0.006 | 0.03 | NA | 2.600 (1.408), 0.06 |
| 6 | 34250388 | rs10080749 | G | A | 0.33 | −0.987 (0.200) | 8.0 × 10−7 | C6orf1 | intergenic | 0.52 | 0.37 | 0.12 | NA | − 0.249 (0.357), 0.49 |
| 6 | 54013111 | NA | AT | A | 0.001 | −20.946 (4.233) | 7.5 × 10−7 | MLIP | intergenic | NA | NA | NA | NA | NA |
| 7 | 128864667 | rs537479423 | T | C | 0.002 | −12.396 (2.282) | 5.6 × 10−8 | ATP6V1F | intronic | 0.01 | 0.003 | NA | NA | NA |
| 7 | 151442610 | rs145127841 | A | G | 0.006 | −6.862 (1.3890) | 7.8 × 10−7 | CRYGN | intergenic | 0.03 | 0.006 | NA | NA | NA |
| 10 | 25871827 | rs150486305 | T | G | 0.003 | 9.716 (1.934) | 5.0 × 10−7 | LOC101929073 | intergenic | NA | 0.001 | 0.001 | NA | NA |
| 11 | 57930162 | NA | T | C | 0.001 | −17.786 (2.283) | 6.0 × 10−8 | OR9Q1 | intergenic | NA | NA | NA | NA | NA |
| 11 | 58498757 | NA | G | A | 0.001 | −17.028 (3.223) | 1.3 × 10−7 | OR5B21 | intergenic | NA | NA | NA | NA | NA |
| 13 | 88310365 | rs530730032 | G | A | 0.002 | −12.862 (2.555) | 4.8 × 10−7 | LINC00433 | intergenic | 0.005 | NA | NA | NA | NA |
| 13 | 112853280 | rs560559296 | G | A | 0.001 | −14.559 (2.730) | 9.6 × 10−8 | ATP11A | intronic | NA | 0.001 | 0.005 | NA | NA |
| 15 | 32721018 | rs11857586 | A | T | 0.02 | 3.527 (0.706) | 5.9 × 10−7 | GREM1 | intronic | 0.15 | 0.01 | 0.002 | −0.865 (0.498), 0.08 | −0.401 (1.229), 0.74 |
| 15 | 34926038 | rs58720902 | T | G | 0.01 | 4.791 (0.847) | 1.6 × 10−8 | AQR | intronic | 0.11 | 0.01 | 0.00 | 0.086 (0.523), 0.86 | 1.97 (6.49), 0.03 |
| 20 | 4739670 | rs2422935 | A | G | 0.54 | −0.965 (0.188) | 3.0 × 10−7 | PRNT | ncRNA intronic | 0.53 | 0.54 | 0.55 | −0.346 (0.278), 0.21 | −0.877 (0.314), 0.005 |
Abbreviations: AF, allele frequency; Chr, chromosome; P, p-value; AFR, African; AMR, Admixed Americans; EUR, European; AA, African American; HA, Hispanics; SE, standard deviation
Figure 1.
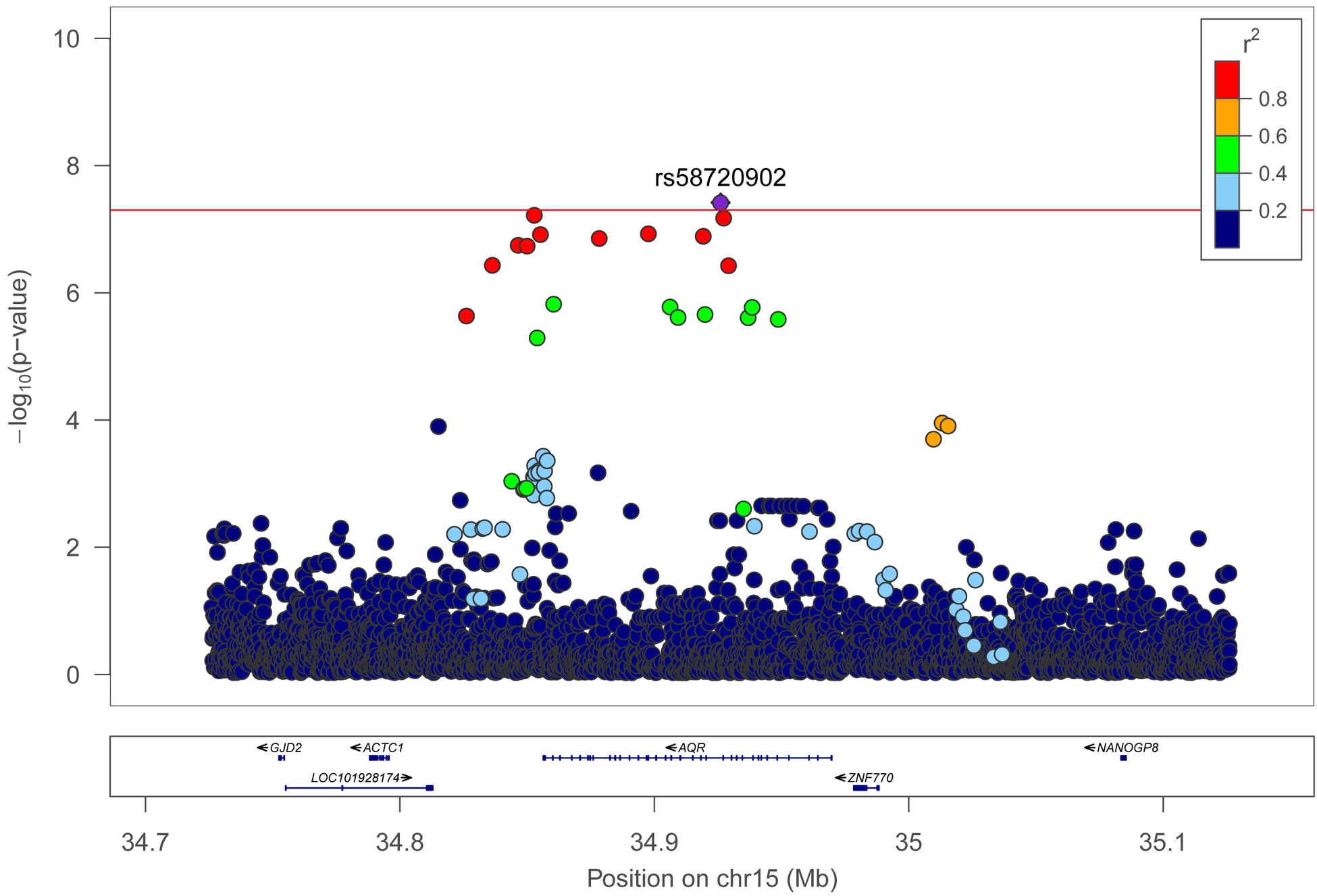

Regional plot for associations at the AQR gene on chromosome 15 for eGFR (A), and association at the ZBTB16 gene for ACR (B) in the HCHS/SOL discovery samples. X-axis shows the chromosome position and underlying genes in the region. The y-axis is the –log (p-values). Each dot is a SNV and the color indicates linkage disequilibrium (r2) with the best variant (in purple). Red horizontal line is the genome-wide association threshold.
Table 2.
Main findings for association with ACR at p<10−7 using TOPMed reference imputation in the HCHS/SOL (n=11,688)
| Chr | Position (hg38) | SNPID | Coded allele |
Non-coded | AF coded | beta | P | Nearby Gene | Function | 1000 Genome Project allele frequencies | Replication UK Biobank P | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AFR | AMR | EUR | |||||||||||
| 2 | 27346004 | rs10205592 | C | T | 0.56 | −0.075 (0.014) | 1.1×10−7 | GTF3C2 | intronic | 0.87 | 0.51 | 0.41 | NA |
| 2 | 108390209 | rs13021399 | T | A | 0.56 | −0.081 (0.015) | 9.9×10−8 | SULT1C4 | intergenic | 0.02 | 0.53 | 0.26 | 0.36 |
| 2 | 108897145 | rs3827760 | G | A | 0.29 | 0.097 (0.019) | 1.6×10−7 | EDAR | missense | 0.003 | 0.39 | 0.01 | 0.10 |
| 2 | 109624640 | rs919942 | A | C | 0.63 | 0.071 (0.014) | 8.2×10−7 | SOWAHC | intergenic | 0.33 | 0.69 | 0.54 | 0.52 |
| 2 | 111121122 | rs116907128 | A | C | 0.14 | 0.120 (0.022) | 3.5×10−8 | BCL2L11 | UTR5 | 0.002 | 0.17 | 0.001 | NA |
| 3 | 196146539 | rs540878340 | A | G | 0.002 | 0.914 (0.176) | 2.0 × 10−7 | LINC00885 | ncRNA_intronic | 0.02 | 0.001 | NA | NA |
| 11 | 5227002 | rs334 | A | T | 0.01 | 0.530 (0.082) | 8.4 ×10−11 | HBB | missense | NA | NA | ||
| 11 | 103974051 | rs7103465 | A | G | 0.03 | 0.248 (0.049) | 3.7 × 10−7 | PDGFD | intronic | 0.11 | 0.02 | 0.02 | 0.41 |
| 11 | 114155245 | rs527493184 | A | G | 0.002 | 1.029 (0.180) | 1.1 × 10−8 | ZBTB16 | intronic | 0.01 | 0.003 | NA | NA |
| 16 | 86734887 | rs11117207 | C | T | 0.08 | −0.128 (0.025) | 3.6 × 10−7 | FOXL1 | intergenic | 0.27 | 0.08 | 0.04 | 0.62 |
| 17 | 54416610 | rs115573116 | T | C | 0.002 | 0.756 (0.145) | 1.7 × 10−7 | TOM1L1 | intergenic | 0.03 | 0.001 | NA | NA |
| 22 | 16823805 | rs1032642268 | A | G | 0.002 | 0.873 (0.176) | 7.1 × 10−7 | XKR3 | intergenic | NA | NA | NA | NA |
Abbreviations: AF, allele frequency; Chr, chromosome; P, p-value; 1kg, 1000 Genome Project; AA, African American; HA, Hispanics; SE, standard deviation; UKB, UK Biobank; AFR, African; AMR, Admixed Americans; EUR, European
Replication of HCHS/SOL GWAS findings
We attempted replication of the SNVs independently associated with eGFR at P<10−7 in the Women’s Health Initiative (WHI) African Americans and Hispanics/Latinos guided by the allele frequency in ancestry specific datasets9, 10. Most of the low frequency/rare SNVs were not available for replication given the studies were imputed to the 1000 Genome Project reference panels. The significantly associated eGFR SNV at the AQR locus nominally replicated in WHI Hispanics (MAF=0.01, P=0.03). This SNV was more common in African ancestry (MAF=0.08) but the association was not significant (P=0.86) in WHI African Americans. Although the direction of effect was concordant among discovery and replication samples, there was significant heterogeneity in meta-analysis (P= 1.4×10−5) (Table 1). The SNV at the PRNT gene, rs2422935, was nominally associated with eGFR in WHI Hispanics/Latinos but not in African Americans. However, the effect estimates were concordant in direction among HCHS/SOL, WHI Hispanics/Latinos and African Americans. In meta-analysis across HCHS/SOL and WHI samples, the association reached genome-wide significance (P=1.4 × 10−8, P for heterogeneity=0.18). Among remaining SNVs present in at least one replication sample, meta-analyses of discovery and replication samples showed significant heterogeneity (P<0.05) and P-values were higher than those observed in the discovery sample.
For ACR, because there is no available data in Hispanics or individuals of African ancestry, we attempted replication for SNVs at P<10−7 using summary statistics from UK Biobank white British7. Five SNVs were available including two that were rare in the UK Biobank white British dataset. None of the SNVs replicated at nominal level and just three SNVs had concordant direction of effect between HCHS/SOL and the UK Biobank data.
Secondary analyses showed no differences in effect estimates within diabetes-strata for SNVs that replicated for eGFR or ACR (Supplemental Table 1).
Validation of previously reported ACR loci identified in the UK Biobank
We next examined the association for 46 loci recently reported in the UK Biobank by Haas et al7 and additional 32 loci reported by Teumer et al11 (both included the UK Biobank data), which have not been validated in independent studies. Replication criteria consider a nominal association (P<0.05) and consistency in direction of effects between our data and the UK Biobank. Six SNVs (five at novel loci) described by Haas et al. replicated: SNX17 (P=1.7 × 10−7), HOTTIP (P=0.001); WIPF3 (p=0.005); CUBN (P=0.05); C10orf11 (P=0.002) and ACTN1 (P=0.002) (Table 3). The most significant SNV at the known CUBN locus in our data was rs144250387 (P=1.9 × 10−6). Only three loci replicated from Teumer et al (KCNK5, rs1544935, P=0.003; OAF, rs508205, P=0.02; DPEP1 rs2460448, P=0.04) (Supplemental Table 2).
Table 3.
Replication of associated SNVs from the UK Biobank for ACR in Hispanics/Latinos of the HCHS/SOL study.
| SNV | CHR | POS (hg38) | Coded allele | Other allele | Coded allele frequency | Coded Allele count |
N | BETA | SE | P | Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs12032996 | 1 | 33454985 | A | G | 0.19 | 4380 | 11688 | −0.001 | 0.018 | 0.97 | ZSCAN20 |
| rs10157710 | 1 | 47496019 | T | C | 0.70 | 16357 | 11688 | 0.017 | 0.015 | 0.27 | FOXD2 |
| rs11162351 | 1 | 77479047 | G | C | 0.32 | 7576 | 11688 | −0.001 | 0.015 | 0.95 | AK5 |
| rs11264327 | 1 | 155122631 | A | G | 0.51 | 12027 | 11688 | −0.011 | 0.014 | 0.44 | EFNA1 |
| rs12727104 | 1 | 171454028 | A | G | 0.17 | 3973 | 11688 | −0.005 | 0.019 | 0.78 | PRRC2C |
| rs12727980 | 1 | 200289967 | T | C | 0.58 | 13624 | 11688 | 0.018 | 0.014 | 0.21 | LINC00862 |
| rs4665972 | 2 | 27375230 | C | T | 0.67 | 15758 | 11688 | −0.078 | 0.015 | 1.7 × 10−7 | SNX17 |
| rs6750228 | 2 | 51084986 | A | T | 0.06 | 1462 | 11688 | 0.040 | 0.030 | 0.18 | NRXN1 |
| rs13394343 | 2 | 85527219 | A | C | 0.40 | 9464 | 11688 | 0.004 | 0.014 | 0.78 | LOC100630918 |
| rs10207567 | 2 | 202850250 | C | G | 0.77 | 18089 | 11688 | 0.025 | 0.016 | 0.13 | ICA1L |
| rs1047891 | 2 | 210675783 | A | C | 0.31 | 7307 | 11688 | −0.024 | 0.015 | 0.11 | CPS1 |
| rs35483183 | 2 | 227011971 | A | G | 0.07 | 1698 | 11688 | 0.037 | 0.027 | 0.17 | COL4A4 |
| rs6768627 | 3 | 46853886 | T | C | 0.10 | 2388 | 11688 | 0.014 | 0.023 | 0.55 | MYL3 |
| rs112607182 | 3 | 170309619 | T | C | 0.03 | 654 | 11688 | 0.046 | 0.046 | 0.32 | PRKCI |
| rs3805382 | 4 | 55605384 | G | A | 0.40 | 9288 | 11688 | 0.014 | 0.015 | 0.34 | NMU |
| rs7654754 | 4 | 76488642 | A | G | 0.48 | 11204 | 11688 | −0.003 | 0.014 | 0.84 | SHROOM3 |
| rs6535594 | 4 | 148211605 | A | G | 0.57 | 13421 | 11688 | 0.024 | 0.014 | 0.08 | NR3C2 |
| rs702634 | 5 | 53975590 | A | G | 0.79 | 18513 | 11688 | 0.022 | 0.017 | 0.21 | ARL15 |
| rs7731168 | 5 | 65000644 | C | G | 0.20 | 4761 | 11688 | 0.019 | 0.017 | 0.27 | CWC27 |
| rs4410790 | 7 | 17244953 | C | T | 0.38 | 8913 | 11688 | −0.006 | 0.015 | 0.66 | AHR |
| rs2023844 | 7 | 27203619 | A | G | 0.88 | 20613 | 11688 | 0.072 | 0.022 | 0.00095 | HOTTIP |
| rs17158386 | 7 | 29765745 | A | G | 0.15 | 3583 | 11688 | 0.057 | 0.020 | 0.003 | WIPF3 |
| rs55798132 | 8 | 2808621 | A | G | 0.008 | 193 | 11687 | −0.076 | 0.078 | 0.33 | LOC101927815 |
| rs28601761 | 8 | 125487789 | G | C | 0.32 | 7553 | 11687 | −0.012 | 0.015 | 0.40 | TRIB1 |
| rs45551835 | 10 | 16890385 | A | G | 0.017 | 405 | 11688 | 0.149 | 0.054 | 0.005 | CUBN |
| rs144360241 | 10 | 16925418 | C | T | 0.005 | 122 | 11688 | 0.087 | 0.109 | 0.43 | CUBN |
| rs1276720 | 10 | 16929427 | T | C | 0.57 | 13251 | 11688 | 0.014 | 0.014 | 0.33 | CUBN |
| rs10995311 | 10 | 62805174 | G | C | 0.27 | 6208 | 11688 | 0.001 | 0.016 | 0.94 | ADO |
| rs67339103 | 10 | 76133928 | A | G | 0.30 | 7106 | 11688 | 0.047 | 0.015 | 0.002 | C10orf11 |
| rs17368443 | 11 | 10275289 | C | G | 0.04 | 1027 | 11688 | 0.043 | 0.035 | 0.22 | SBF2 |
| rs1124694 | 11 | 11077129 | G | A | 0.30 | 6916 | 11688 | 0.001 | 0.015 | 0.96 | GALNT18 |
| rs2601006 | 12 | 69585737 | T | C | 0.44 | 10299 | 11688 | 0.011 | 0.014 | 0.42 | CCT2 |
| rs4288924 | 14 | 68835682 | A | G | 0.56 | 12993 | 11688 | −0.044 | 0.014 | 0.002 | ACTN1 |
| rs8035855 | 15 | 41785763 | A | G | 0.60 | 14121 | 11688 | 0.004 | 0.014 | 0.80 | MAPKBP1 |
| rs1145074 | 15 | 45411626 | A | T | 0.55 | 12930 | 11688 | −0.011 | 0.015 | 0.47 | SPATA5L1 |
| rs146311723 | 15 | 63512308 | C | T | 0.096 | 2261 | 11688 | −0.014 | 0.023 | 0.55 | USP3 |
| rs2472297 | 15 | 74735539 | T | C | 0.095 | 2232 | 11688 | 0.003 | 0.024 | 0.89 | CYP1A1 |
| rs2338796 | 17 | 39399374 | G | A | 0.28 | 6471 | 11688 | −0.027 | 0.015 | 0.08 | FBXL20 |
| rs35572189 | 17 | 81451999 | A | G | 0.35 | 8126 | 11688 | −0.004 | 0.014 | 0.78 | BAHCC1 |
| rs784257 | 18 | 55729968 | C | T | 0.90 | 20951 | 11688 | −0.025 | 0.024 | 0.29 | TCF4 |
| rs838142 | 19 | 48748894 | G | A | 0.40 | 9448 | 11688 | −0.003 | 0.014 | 0.84 | FUT1 |
SNVs rs183131780 (MIR548AR), rs35924503 (SPHKAP), rs189107782 (FRG1), rs144994089 (AQP7) and rs141640975 (CUBN) were not available.
Discussion
In this genetic study of Hispanics/Latinos using multi-ethnic dense imputed WGS genotypes, we identified two novel loci for eGFR, a novel locus for ACR and replicated additional eight ACR loci identified in GWAS using the UK Biobank white British samples. Overall, the imputation of TOPMed SNVs allowed for identification of several associations for low frequency variants and those that are more common in non-European ancestry. However, our results also underscore the limitations of current genetic studies, including the lack of suitable replication samples for variants that are more common in non-European ancestry or are low frequency.
For eGFR, the AQR locus finding was driven by an intronic variant that is rare in European and Admixed Americans but it is a common variant in African Americans (Table 1). The association replicated in WHI Hispanics but not in African Americans. However, there was significant heterogeneity in meta-analysis, with a larger effect on eGFR in Hispanics/Latinos than African Americans, suggesting potential differences by ancestry background. A recent study identified a SNV 1 kb downstream of AQR (rs3743121) associated with type 2 diabetes in East Asians, although the sample size was very small12. Experimental knockdown of AQR in immortalized cells (HepG2) showed improved glucose uptake and insulin sensitivity with additional effects on glycogen synthesis and gluconeogenesis12. In our data, there was no difference in effect estimates by diabetes status at the AQR locus (Supplemental Table 1). However, the identified SNV was associated with a protective effect on eGFR, which mechanisms may include improvement in glucose metabolism. Further studies are needed to validate the association in Hispanics/Latinos. At the PRNT locus, an intronic variant was significantly associated with decreased eGFR in the combined discovery and replication samples. This variant is common across all populations and there is little knowledge on the function of the gene and its relation to kidney traits. The AQR and PRNT SNVs had little evidence for any regulatory function in in silico analysis.
Our GWAS of ACR identified a new locus at ZBTB16 driven by a rare variant that was not available in the UK Biobank for replication. We confirmed associations at the HBB gene (rs344 related to hemoglobin S or sickle cell trait) and the BCL2L11 gene, which we have been previously reported in this cohort2. We have shown that rs344 is associated with eGFR variation in our data, although at modest P-values. We also replicated eight loci initially reported in the UK Biobank for white British, including 5 that were novel: SNX17 (intronic), HOTTIP (ncRNA intronic), WIPF3 (intergenic), C10orf11 (intronic) and ACTN1 (intergenic)7. At the SNX17 locus, the SNV is an expression quantitative trait loci for SNX17 in GTEx muscle skeletal tissue. This gene has no known function related to kidney traits. HOTTIP produces a long RNA in antisense to the HOXA gene cluster, and regulates expression of HOXA genes. This locus has been identified in GWAS of blood pressure in individuals of African ancestry and in Hispanics/Latinos13, 14, but its relation to kidney disease is unknown. The intergeneic SNV at WIPF3 is an expression quantitative trait loci for WIPF3 in GTEx left ventricle. At least six SNVs identified in the UK Biobank were rare in our data and did not replicate in our study: rs189107782 (FRG1), rs55798132 (LOC101927815), rs144994089 (AQP7), rs45551835, rs144360241 and rs141640975 (CUBN). Three additional loci from a recent GWAS of ACR that included the UK Biobank replicated at modest p-values11. Overall, the number of ACR loci that we were able to validate from these previous studies was small.
Both albuminuria and eGFR are independently associated with cardiovascular mortality and progression to end-stage kidney disease15–17. Understanding their genetic determinants may offer opportunities for more targeted interventions to reduce these outcomes. Most GWAS studies of eGFR and ACR have included large number of individuals of European ancestry, and findings are driven by European populations. This may explain the lack of replication of some of the ACR findings from the UK Biobank, although Hispanics/Latinos have European ancestry admixture. Trans-ethnic studies with large samples of diverse populations and studies within a single diverse population such as this report are still needed to better characterize disease risk across and within populations. We and others have already shown that the study of admixed populations can identify population specific SNVs2, 18 or loci driven by SNVs with a higher allele frequency in one population14. In addition, we have successfully fine-mapped loci to SNVs with evidence of functionality. For example, the SNV at the BCL2L11 locus, rs116907128, identified in our study of Hispanics/Latinos, is located within the promoter region of the gene in a region enriched for regulatory markers (DNAse I hypersensitive sites in human kidney cells and histone modification binding sites) which are strong evidence for its regulatory function. This locus replicated in recent analyses of the UK Biobank for ACR. However, the most significant SNVs at the region were either a low frequency SNV (rs183131780)7 or an intronic variant to ACOXL (rs2880119)11, and none of these variants showed evidence for functional regulation of nearby genes.
In summary, our GWAS of Hispanics/Latinos using multi-ethnic WGS imputed genotypes identified novel loci for eGFR and replicated published associations for ACR. This study provides evidence for gains in gene discovery and for identifying variants with regulatory function in a study of Hispanics/Latinos and when using dense imputed variant panels.
Supplementary Material
Acknowledgments:
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. The authors thank the staff and participants of HCHS/SOL for their important contributions. Investigators website - http://www.cscc.unc.edu/hchs/. This manuscript has been reviewed by the HCHS/SOL Publications Committee for scientific content and consistency of data interpretation with previous HCHS/SOL publications.
Trans-Omics in Precision Medicine (TOPMed) program imputation panel (version 5b) is supported by the National Heart, Lung and Blood Institute (NHLBI); see www.nhlbiwgs.org. TOPMed study investigators contributed data to the reference panel, which can be accessed through the Michigan Imputation Server; see https://imputationserver.sph.umich.edu. The panel was constructed and implemented by the TOPMed Informatics Research Center at the University of Michigan (3R01HL-117626-02S1; contract HHSN268201800002I). The TOPMed Data Coordinating Center (3R01HL-120393-02S1; contract HHSN268201800001I) provided additional data management, sample identity checks, and overall program coordination and support. We gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
Sources of Funding: The baseline examination of HCHS/SOL was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236), and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contributed to the first phase of HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research (NIDCR), National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at Washington University was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Additional analysis support was provided by the NHLBI grant HL123677-01 (NF). Genotyping efforts were supported by NHLBI HSN 26220/20054C, NCATS CTSI grant UL1TR000124, and NIDDK Diabetes Research Center (DRC) grant DK063491. This study is supported by the following grants: National Institutes of Health DK117445, MD012765 and HL140385 to NF.
Nonstandard Abbreviations and Acronyms:
- ACR
urine albumin-to-creatinine ratio
- HCHS/SOL
Hispanic Community Health Study/Study of Latinos
- SNV
single nucleotide variant
- WHI
Women’s Health Initiative
Footnotes
Disclosures: None
References:
- 1.Ricardo AC, Flessner MF, Eckfeldt JH, Eggers PW, Franceschini N, Go AS, Gotman NM, Kramer HJ, Kusek JW, Loehr LR, et al. Prevalence and Correlates of CKD in Hispanics/Latinos in the United States. Clin J Am Soc Nephrol. 2015;10:1757–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kramer HJ, Stilp AM, Laurie CC, Reiner AP, Lash J, Daviglus ML, Rosas SE, Ricardo AC, Tayo BO, Flessner MF, et al. African Ancestry-Specific Alleles and Kidney Disease Risk in Hispanics/Latinos. J Am Soc Nephrol. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown LA, Sofer T, Stilp AM, Baier LJ, Kramer HJ, Masindova I, Levy D, Hanson RL, Moncrieft AE, Redline S, et al. Admixture Mapping Identifies an Amerindian Ancestry Locus Associated with Albuminuria in Hispanics in the United States. J Am Soc Nephrol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris AP, Le TH, Wu H, Akbarov A, van der Most PJ, Hemani G, Smith GD, Mahajan A, Gaulton KJ, Nadkarni GN, et al. Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat Commun. 2019;10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genome Catalog. https://www.ebi.ac.uk/gwas/. assessed July 2019.
- 6.Kowalski MH, Qian H, Hou Z, Rosen JD, Tapia AL, Shan Y, Jain D, Argos M, Arnett DK, Avery C, et al. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 2019;15:e1008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas ME, Aragam KG, Emdin CA, Bick AG, International Consortium for Blood P, Hemani G, Davey Smith G and Kathiresan S. Genetic Association of Albuminuria with Cardiometabolic Disease and Blood Pressure. Am J Hum Genet. 2018;103:461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sorlie PD, Aviles-Santa LM, Wassertheil-Smoller S, Kaplan RC, Daviglus ML, Giachello AL, Schneiderman N, Raij L, Talavera G, Allison M, et al. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol. 2010;20:629–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control Clin Trials. 1998;19:61–109. [DOI] [PubMed] [Google Scholar]
- 10.Anderson GL, Manson J, Wallace R, Lund B, Hall D, Davis S, Shumaker S, Wang CY, Stein E and Prentice RL. Implementation of the Women’s Health Initiative study design. Annals of epidemiology. 2003;13:S5–17. [DOI] [PubMed] [Google Scholar]
- 11.Teumer A, Li Y, Ghasemi S, Prins BP, Wuttke M, Hermle T, Giri A, Sieber KB, Qiu C, Kirsten H, et al. Genome-wide association meta-analyses and fine-mapping elucidate pathways influencing albuminuria. Nat Commun. 2019;10:4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song C, Yan H, Wang H, Zhang Y, Cao H, Wan Y, Kong L, Chen S, Xu H, Pan B, et al. AQR is a novel type 2 diabetes-associated gene that regulates signaling pathways critical for glucose metabolism. J Genet Genomics. 2018;45:111–120. [DOI] [PubMed] [Google Scholar]
- 13.Franceschini N, Carty CL, Lu Y, Tao R, Sung YJ, Manichaikul A, Haessler J, Fornage M, Schwander K, Zubair N, et al. Variant Discovery and Fine Mapping of Genetic Loci Associated with Blood Pressure Traits in Hispanics and African Americans. PLoS One. 2016;11:e0164132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang J, Le TH, Edwards DRV, Tayo BO, Gaulton KJ, Smith JA, Lu Y, Jensen RA, Chen G, Yanek LR, et al. Single-trait and multi-trait genome-wide association analyses identify novel loci for blood pressure in African-ancestry populations. PLoS Genet. 2017;13:e1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerstein HC, Mann JF, Yi Q, Zinman B, Dinneen SF, Hoogwerf B, Halle JP, Young J, Rashkow A, Joyce C, et al. Albuminuria and risk of cardiovascular events, death, and heart failure in diabetic and nondiabetic individuals. JAMA. 2001;286:421–6. [DOI] [PubMed] [Google Scholar]
- 16.Astor BC, Matsushita K, Gansevoort RT, van der Velde M, Woodward M, Levey AS, Jong PE, Coresh J, Chronic Kidney Disease Prognosis C, Astor BC, et al. Lower estimated glomerular filtration rate and higher albuminuria are associated with mortality and end-stage renal disease. A collaborative meta-analysis of kidney disease population cohorts. Kidney Int. 2011;79:1331–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattix HJ, Hsu CY, Shaykevich S and Curhan G. Use of the albumin/creatinine ratio to detect microalbuminuria: implications of sex and race. J Am Soc Nephrol. 2002;13:1034–9. [DOI] [PubMed] [Google Scholar]
- 18.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


