Abstract
Social affiliative behaviors -- engagement in positive (i.e. non-aggressive) social approach and reciprocal social interactions with a conspecific -- comprise a construct within the NIMH RDoC Social Processes Domain. Affiliative behaviors are disrupted in multiple human neurodevelopmental and neuropsychiatric disorders, such as autism, schizophrenia, social phobia, and others. Human genetic studies have strongly implicated synaptic cell adhesion molecules (sCAMs) in several such disorders that involve marked reductions, or other dysregulations, of social affiliative behaviors. Here, we review the literature on the role of sCAMs in social affiliative behaviors. We integrate findings pertaining to synapse structure and morphology, neurotransmission, postsynaptic signaling pathways, and neural circuitry to propose a multi-level model that addresses the impact of a diverse group of sCAMs, including neurexins, neuroligins, protocadherins, immunoglobulin (Ig) superfamily proteins, and leucine rich repeat proteins, as well as their associated scaffolding proteins, including Shanks and others, on social affiliative behaviors. This review finds that the disruption of sCAMs often manifests in changes in social affiliative behaviors, likely through alterations in synaptic maturity, pruning, and specificity leading to excitation / inhibition imbalance in several key regions, namely the medial prefrontal cortex (mPFC), basolateral amygdala (BLA), hippocampus, and ventral tegmental area (VTA). Unraveling the complex network of interacting sCAMs in glutamatergic synapses will be an important strategy for elucidating the mechanisms of social affiliative behaviors and the alteration of these behaviors in many neuropsychiatric and neurodevelopmental disorders.
Keywords: synapse, neurexin, neuroligin, SHANK, cadherin, NMDA, affiliation, behavior
Introduction
Social affiliative behaviors -- engagement in positive (i.e., non-aggressive) social approach and reciprocal social interactions with a conspecific -- comprise a construct within the NIMH Research Domain Criteria (RDoC) Social Processes Domain and are disrupted in multiple human neurodevelopmental and neuropsychiatric disorders, such as autism, schizophrenia and social phobia. Because major disruptions of social affiliative behaviors are common, highly disabling, and refractory to currently-available pharmacological treatments, research into the underlying genetic and molecular basis of social affiliation deficits should be prioritized so that psychiatric treatment can be advanced. In this review, we will examine the neural circuits and molecular mechanisms of social affiliative behaviors, and will focus on the role of synaptic cell adhesion molecules (sCAMs) in these behaviors.
Synapses are highly complex, dynamic structures at the interfaces between neurons and are critical for neuron to neuron communication in the brain. sCAMs are transmembrane or membrane-tethered proteins involved in the maintenance, function, and elimination of synapses. sCAMs can belong to several protein families, including neurexins, neuroligins, cadherins, protocadherins (Pcdhs), immunoglobulin superfamily (IgSF) proteins, and leucine-rich repeat (LRR) proteins. Historically, sCAMs are defined by their combination of repeated adhesion protein domains and their location at the synapse. sCAMs sit on the pre- and/or postsynaptic membrane and bind trans-synaptically to other sCAMs, as well as intracellularly to cytoskeletal elements and scaffolding proteins.
Human genetic studies have strongly implicated sCAMs in several neuropsychiatric and neurodevelopmental disorders involving marked reductions, or other dysregulations, of social affiliative behaviors. Most notably, neurexins and neuroligins have been associated with autism spectrum disorder (ASD), a neurodevelopmental disorder largely defined by reduced or dysregulated social affiliative behaviors (1). Neurexins have been linked to several other disorders associated with social difficulties, including Pitt-Hopkins syndrome, schizophrenia, intellectual disability, attention deficit and hyperactivity disorder (ADHD), and bipolar disorder (2). Additionally, a member of the IgSF, close homologue of L1 (CHL1), has been associated with autism, intellectual disability, and schizophrenia (3-6). A well-studied family of sCAM-associated scaffolding proteins, SHANKs, have been implicated in various neurodevelopmental and psychiatric disorders, including ASD, intellectual disability, schizophrenia, and ADHD (7). Several genes in the cadherin and Pcdh families have also been associated with neuropsychiatric disorders, including epilepsy, intellectual disability, ASD, schizophrenia, and bipolar disorder (8).
This review proposes a mechanistic model for the role of sCAMs in social affiliation behaviors using evidence from rodent models. The NIMH RDoC defines social affiliation as social approach (i.e. initiation of interaction with another) and engagement (i.e. maintenance of interaction beyond initiation) in positive social interactions with other individuals. In this review, we will focus on the role of sCAMs in the three-chamber social approach assay (9, 10) as well as the direct social interaction assay. We do not include studies of social novelty, social memory, aggression, mating, or maternal behaviors, although these are important areas of future focus. By focusing on a key RDoC behavior rather than putative rodent models of complex disorders, our review can approach the mechanistic detail – at the molecular, cellular, circuit, and behavioral levels –necessary to develop new treatment targets for disruptions of this important behavioral domain. This review finds that the disruption of sCAMs often manifests in changes in social affiliative behaviors, along with alterations in dendritic spine morphology and glutamatergic transmission in several key regions, namely the medial prefrontal cortex (mPFC), basolateral amygdala (BLA), ventral tegmental area (VTA), and hippocampus.
Neurexins and neuroligins
Neurexins
The neurexin family of proteins are key organizers of the synapse, with thousands of isoforms and multiple binding partners. Neurexins are present at presynaptic membranes of excitatory and inhibitory synapses (11, 12) (see Figures 2 and 3). They have been shown to be involved in recruitment of synaptic vesicles to the presynaptic membrane for release and in triggering postsynaptic differentiation (11, 13). Neurexins bind GABA-A receptors in addition to other sCAMs, including neuroligins, dystroglycan, LRR transmembrane (LRRTM) neuronal proteins, cerebellins, calsyntenins, latrophilins, and neurexophilin (14-21). In humans and mice, there are three genes encoding classic neurexins: Nrxn1, Nrxn2, and Nrxn3 (22). Neurexin genes in vertebrates have two promoters with the ability to produce either alpha- or beta-neurexin transcripts (22). The Nrxn genes differ in alternative splicing and expression patterns (22, 23) (see Supplementary Table 1).
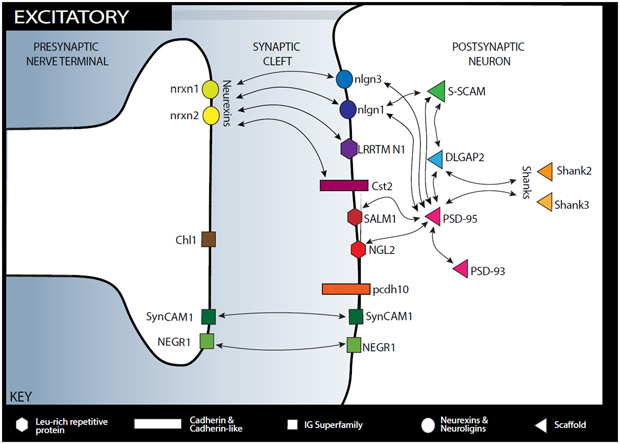
Figure 2. sCAM interaction network in the excitatory synapse.
This figure shows the location of sCAMs discussed in the review within the excitatory synapse as well as their binding partners.
Calsyntenin-2 (Cst2) is a member of the cadherin superfamily and resides on the postsynaptic side of both inhibitory and excitatory synapses.
Cell adhesion molecule L-1 like (Chl1), a member of the immunoglobin superfamily, resides on the presynaptic membrane and is involved in heterophilic binding, though its specific binding partners are unknown.
Disk large associated protein 2 (DLGAP2) is primarily expressed in the postsynaptic side of excitatory synapses, binds DLG and Shank proteins, and may have a role in enrichment of PSD-95 at the plasma membrane.
Leucine rich repeat transmembrane neuronal 1 (LRRTM N1) resides on the postsynaptic side of excitatory synapses and binds trans-synaptically to neurexins.
Netrin-G ligand 2 (ngl-2) is a member of the leucine rich repeat protein, exists on the postsynaptic side of excitatory synapses, and binds to both netrin-G 2 and PSD-95.
Neural growth regulator 1 (NEGR1) is a member of the immunoglobin superfamily and its location on pre- and postsynaptic membranes is dependent on development.
The neurexin family (shown here are nrxn1 and nrxn2) are predominantly presynaptic organizers of the synapse with thousands of isoforms and multiple binding partners.
Neuroligin 1 (nlgn1) resides on the postsynaptic side of excitatory synapses and binds with neurexins and the scaffolding proteins PSD-95 and S-SCAM.
Neuroligin 3 (nlgn3) is on the postsynaptic side of both excitatory and inhibitory synapses and binds trans-synaptically to neurexins.
Postsynaptic density protein 93 (PSD-93) is expressed in postsynaptic neurons in excitatory and inhibitory synapses synapse and serves as a scaffolding protein, binding PSD-95, K+ channels, and NMDA receptors.
Postsynaptic density protein 95 (PSD-95) is expressed in postsynaptic neurons in excitatory synapse and serves as a scaffolding protein, binding many other SCAMs in addition to NMDAR.
Protocadherin 10 (pcdh10), a cadherin-like protein, is a postsynaptic protein that engages in both homophilic and heterophilic binding.
The Shank family (shown here, Shank2 and Shank3) are scaffolding proteins in postsynaptic neurons of excitatory synapses and bind to many synapse proteins as well as glutamatergic receptors directly and indirectly.
Synaptic adhesion like molecule (SALM1) is a postsynaptic molecule in the leucine rich repeat protein family that exists both in excitatory and inhibitory synapses and binds PSD-95.
Synaptic cell adhesion molecule 1 (SynCAM1), a member of the immunoglobin superfamily, is present on both the pre- and postsynaptic sides and is involved in homophilic as well as heterophilic binding.
Synaptic scaffolding molecule (S-SCAM) resides in the postsynaptic neuron in excitatory and inhibitory synapses and connects to glutamatergic receptors (NMDAR and AMPAR) as well as other synaptic proteins.
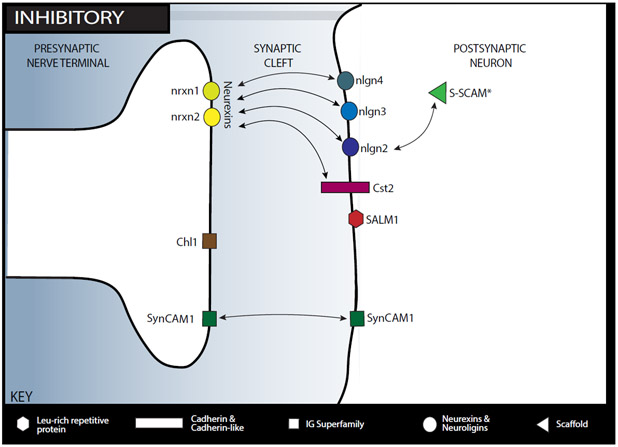
Figure 3. sCAM interaction network in the inhibitory synapse.
This figure shows the location of sCAMs discussed in the review within the inhibitory synapse as well as their binding partners. The following sCAMs are shown:
Calsyntenin-2 (Cst2) is a member of the cadherin superfamily and resides on the postsynaptic side of both inhibitory and excitatory synapses.
Cell adhesion molecule L-1 like (Chl1), a member of the immunoglobin superfamily, resides on the presynaptic membrane and is involved in heterophilic binding, though its specific binding partners are unknown.
The neurexin family (shown here are nrxn1 and nrxn2) are predominantly presynaptic organizers of the synapse with thousands of isoforms and multiple binding partners.
Neuroligin 2 (nlgn2) is on the postsynaptic side of inhibitory synapses and binds with neurexins as well as the scaffolding protein, S-SCAM.
Neuroligin 3 (nlgn3) is on the postsynaptic side of both excitatory and inhibitory synapses and binds trans-synaptically to neurexins.
Neuroligin 4 (nlgn4) is on the postsynaptic side of inhibitory synapses and binds trans-synaptically to neurexins.
Synaptic adhesion like molecule (SALM1) is a postsynaptic molecule in the leucine rich repeat protein family that is present in both excitatory and inhibitory synapses.
Synaptic cell adhesion molecule 1 (SynCAM1), a member of the immunoglobin superfamily, is present on both the pre- and postsynaptic sides and is involved in homophilic as well as heterophilic binding.
Synaptic scaffolding molecule (S-SCAM) is a scaffolding protein in the postsynaptic neuron in excitatory and inhibitory synapses and interacts with select glutamatergic receptors (NMDAR and AMPAR) as well as other synaptic proteins.
The reported effects of deleting Nrxn1 on rodent social affiliation are varied, including decrease (24, 25), increase (26), and no change in the behavior (27, 28). These inconsistencies across studies could be due, in part, to variable expressivity of the mutation related to differing genetic backgrounds of the models (26, 28) and to variation across studies in methods, including behavioral testing procedures, age of the mice at testing, and other differences (see Supplementary Table 2).
In contrast to the variability found with Nrxn1 deletions, knockout of Nrxn2 has been shown consistently to decrease social affiliation (27, 29, 30). Knockout of Nrxn2 also impacted excitatory neurotransmission in layer V of the somatosensory cortex, namely by decreasing the frequency of miniature excitatory postsynaptic currents (mEPSCs) and NMDAR transmission (29). Studies examining the impact of Nrxn3 on social affiliation were not found in our literature search. Overall, while the role of Nrxn1 is more uncertain, it seems clear that Nrxn2 plays a role in social affiliation. More work needs to be done to test the region- and cell-specificity of the role of neurexins in social affiliation, in addition to the potential role of spine changes in the mechanistic pathway linking neurexins to social affiliation behavior.
Neuroligins
The most-studied postsynaptic binding partners for neurexins are neuroligins. Neuroligins facilitate presynaptic neurexin clustering and affect synapse maturation (13, 31). Some work (mostly in overexpression or knockdown models) indicates that neuroligins may influence synaptogenesis, along with spine morphology and number (11, 32-34), though those findings are in conflict with other work examining knockout models (31). In mice, there are four neuroligins, which differ in their cell-type specific expression and binding specificity.
Neuroligin-1 (encoded by Nlgn1) is found at excitatory synapses throughout the brain and, in addition to extracellular interactions with presynaptic neurexins, binds several postsynaptic scaffolding proteins via its cytoplasmic domain, including PSD-95, S-SCAM, Shank1, and Shank3 (35-38). Knockout of Nlgn1 led to decreased social affiliation, decreased long-term potentiation in Hip CA1, and decreased NMDA/AMPA ratio in the striatum (39). Knockin of a deleterious Nlgn1 allele, a P89L substitution originally identified in a proband with ASD, led to decreased social affiliation in a mouse model (40). On the other hand, overexpression of Nlgn1 failed to affect social affiliation, as did knockdown of Nlgn1(41, 42).
Neuroligin-2 (Nlgn2) is present at the postsynaptic membrane of inhibitory synapses throughout the brain (43). Knockout and knockdown of Nlgn2 had no effect on social affiliation behavior (42, 44, 45). Conditional knockout of Nlgn2 in the medial prefrontal cortex (mPFC), however, did lead to decreased social affiliation, accompanied by a decrease in inhibitory transmission, as indicated by a decrease in amplitude and frequency of miniature inhibitory postsynaptic currents (mIPSCs) in the mPFC (46). Conditional, region-specific overexpression of Nlgn2 in the hippocampus did not affect social affiliation (47). Transgenic overexpression of Nlgn2 has led to decreased social affiliation and an increase in frequency of mEPSCs in layer II/III of the prefrontal cortex (48).
Neuroligin-3 (Nlgn3) is found at both excitatory and inhibitory synapses throughout the brain (49). Knockin of the deleterious Nlgn3 allele R451C (a mutation identified in autistic probands) has been reported to decrease social affiliation in some cases (50-52) and to result in no change in others (53-56). One study showed variation in results on social affiliation (no change / decrease) based on the assay used, yet consistent increases in excitatory transmission in hippocampal CA1, as measured by fEPSP, NMDA/AMPA ratio, mEPSC frequency, and NMDAR-dependent LTP (57). Among these studies, researchers tested the impact of genetic background, showing that, for Nlgn3R451C knockin mice, social preference in a 3-chamber assay was eliminated on a 129S2/SvPasCrl background but maintained on a C57BL/6J background (52, 53). In contrast to the knockin R451C models, knockout of Nlgn3 does not consistently affect social affiliation (50, 58, 59). However, one study did find that both global knockout of Nlgn3 and conditional knockout of Nlgn3 in ventral tegmental area (VTA) dopaminergic neurons led to reduced social affiliation (60). Neuroligin 4 (Nlgn4) seems to be limited to glycinergic synapses throughout the brain and central nervous system more generally (61). Thus far, there is one report that knockout of Nlgn4 led to a decrease in social affiliation (62).
Together, current data generally support a role for neuroligins in social affiliation, with some variation in how they impact related neurotransmission. One way Nlgn3 impacts social affiliation is through GluA2-lacking AMPA receptors at inputs onto dopaminergic neurons in the VTA (60), while Nlgn2 and inhibitory transmission are implicated in the mPFC; though, the detailed mechanisms involved still need to be clarified (45, 46, 48).
Cadherins
There are over 100 members of the cadherin superfamily, defined by the presence of one or more approximately 110 amino acid-long cadherin domains in their extracellular portions. Subfamilies of cadherins include the classical cadherins (including type I and type II cadherins), desmosomal cadherins, protocadherins, calsyntenins, and atypical/7-transmembrane cadherins. Cadherins have been implicated in pre- and postsynaptic organization and are likely to play key roles in synapse formation and plasticity as well as in spine maintenance (63, 64). Despite being implicated in neurodevelopmental disorders with social impairments (8), cadherins have received relatively little attention in research related to social behaviors. The present review found research on social affiliation for only two cadherin superfamily genes: calsyntenin-2 (Clstn2) and protocadherin-10 (Pcdh10).
Calsyntenin-2 resides on the postsynaptic side of both inhibitory and excitatory synapses in most areas throughout the brain (65), and Clstn2 knockout mice exhibited decreased social affiliation (66). Protocadherin-10 is present in excitatory synapses and engages in homophilic, and limited heterophilic, trans- and intracellular interactions (67-69). It is expressed throughout the brain, though with some regional specificity that varied depending on whether mRNA or protein expression was analyzed (67, 70, 71). Haploinsufficiency of Pcdh10 in mice decreased social affiliation and led to an increased density of morphologically immature dendritic spines in the BLA (72). Importantly, decreased social affiliation in the Pcdh10 model was rescued by acute, systemic treatment with D-cycloserine, a partial glycine agonist that enhances NMDAR signaling (72).
Thus, though a couple members of the cadherin superfamily have been implicated in social affiliation, there is a vast amount of work still to be done to elucidate the potential roles of the remaining family members. Given the association of many protocadherins with changes in synapse structure and with neuropsychiatric disorders, further study of this diverse subfamily in relation to social affiliation, in particular, should be pursued (73, 74).
Immunoglobulin (Ig) superfamily (IgSF)
Many sCAMs belong to the IgSF, whose members are defined by the presence of extracellular Ig domains similar to those in antibodies, each comprised of a sandwich of two β-sheets. Multiple members of this family have been implicated in synapse formation, maintenance, and plasticity (75). Unfortunately, few IgSF members have been examined in the social affiliation literature as of yet.
Close homologue of L1 (Chl1) is involved in synapse maintenance, via influences on presynaptic organization (85, 86). Chl1 is expressed on the presynaptic membrane of excitatory and inhibitory neurons throughout the brain and is also expressed by some glia (78, 80, 86). Its intracellular binding partners include heat shock cognate 71 kDa protein (Hsc70), synaptosomal nerve-associated protein 25, vesicle-associated membrane protein (VAMP/synaptobrevin), and cysteine string protein, while extracellular interactions with integrins have also been reported (82, 85, 86). Knockout of Chl1 led to a decrease in social affiliation (87).
Synaptic cell adhesion molecule 1 (SynCAM1; a.k.a. Cell Adhesion Molecule 1 [Cadm1]) is involved in the processes of synapse formation pre- and postsynaptically and maintenance via heterophilic transsynaptic binding, as well as in the recruitment of NMDA receptors to the postsynaptic membrane via binding to intracellular effector molecules (76, 77, 79, 81). SynCAM1 is present on both presynaptic and postsynaptic sides of excitatory and inhibitory synapses throughout the brain (76, 83). It is involved in homophilic as well as heterophilic binding with other members of the SynCAM family (specifically with SynCAM2), and also interacts with CASK and syntenin (76, 77). Knockout of the Cadm1 gene (which encodes SynCAM1) led to a decrease in social affiliation (84).
Neural growth regulator 1 (NEGR1, a.k.a. Kilon/IGLON4) is a glycosylphosphatidylinositol (GPI)-linked member of the IgLON family, a subfamily of the IgSF, and is involved in the regulation of synapses, largely active in axons, growth cones, and presynaptic terminals early in development and in spines later in development (88). Negr1 is expressed in multiple regions throughout the brain and binds both homophilically and to heterophilic ligand opioid-binding cell adhesion molecule (OBCAM) (89, 90). It is found on the presynaptic membrane early in development and on the postsynaptic membrane later in development (88). Both knockdown and knockout of Negr1 have been reported to result in reduced social affiliation (91-93). These behavioral changes have been accompanied by decreases in spine density, number of parvalbumin-positive interneurons, neurogenesis, and axon growth in the hippocampus (91-93).
Given the vast number of IgSF sCAMs, there is much more work to be done in testing their relationship to social affiliation. However, current data on the select genes that have been tested (Chl1, Cadm1, Negr1) suggest important roles for members of this family in the regulation of social affiliative behaviors. Work with Negr1, in particular, indicates that spine and synapse formation in the hippocampus is likely involved in behavioral alterations (91-93).
Leucine-rich repeat (LRR) proteins
Leucine-rich repeat (LRR) proteins are defined by LRR domains, comprised of an α-β horseshoe fold, and are known for their role in protein interaction and cell adhesion. Many members of this structurally-defined family have been identified as sCAMs. LRR proteins have been implicated in synapse formation, maturation, maintenance, and plasticity (94, 95). Only a few members of the LRR family have been assessed for effects on social affiliation: netrin-G ligand 2 (NGL-2), synaptic adhesion like molecule (SALM1), and LRR transmembrane neuronal 1 (LRRTM1).
NGL-2 (Lrrc4) is implicated in postsynaptic organization and in synapse formation, differentiation, and maintenance, brain-specific, and expressed in several regions including the cerebellum, cerebral cortex, occipital lobe, frontal lobe, temporal lobe, and putamen, with localization specifically to the postsynaptic side of excitatory synapses (96). NGL-2 binds trans-synaptically to netrin-G2 and intracellularly to PSD-95 and is involved in the recruitment of NMDA receptors (97). Knockout of Lrrc4 led to a decrease in social affiliation, with knockouts demonstrating decreased frequency and amplitude of mEPSCs, AMPAR- and NMDAR-related synaptic activity, and long term potentiation in the hippocampus (98). Treatment of the mice with an NMDAR-agonist, D-cycloserine, recovered social affiliative behavior (98), similar to findings for Pcdh10 haploinsufficient mice noted above (78).
SALM1 (Lrfn2) has been implicated in synapse development and differentiation, along with recruitment of NMDA receptors (99, 100). It is found on the postsynaptic side of excitatory synapses throughout the brain and binds intracellularly to scaffolding proteins PSD95, SAP97, and SAP102 (99, 100). Knockout of Lrfn2 led to a decrease and to no change in social affiliation in separate studies (101, 102). The decrease in social investigation and interaction in the Lrfn2 mutant mice was paired with fewer, more irregular dendritic spines, decreased mEPSC frequency, and an increase in silent synapses in the hippocampus (102).
Finally, LRRTM1 (Lrrtm1) resides on the postsynaptic side of excitatory synapses throughout the brain, binds trans-synaptically to α- and β-neurexins, and is a neurexin ligand involved in both pre- and postsynaptic differentiation (103-105). However, knockout of Lrrtm1 reportedly had no effect on social affiliation (106). From this subsample of LRR proteins, NGL-2 and SALM1 have been implicated in social affiliation, perhaps in part through changes in glutamatergic transmission in the hippocampus, although region-specific knockout and rescue will be needed to test the role of specific circuits.
sCAM-associated scaffolding proteins
Due to limitations in space, the sCAM-associated scaffolding proteins, including the Shank family, are discussed in detail in the Supplemental Information.
Proposed mechanistic model
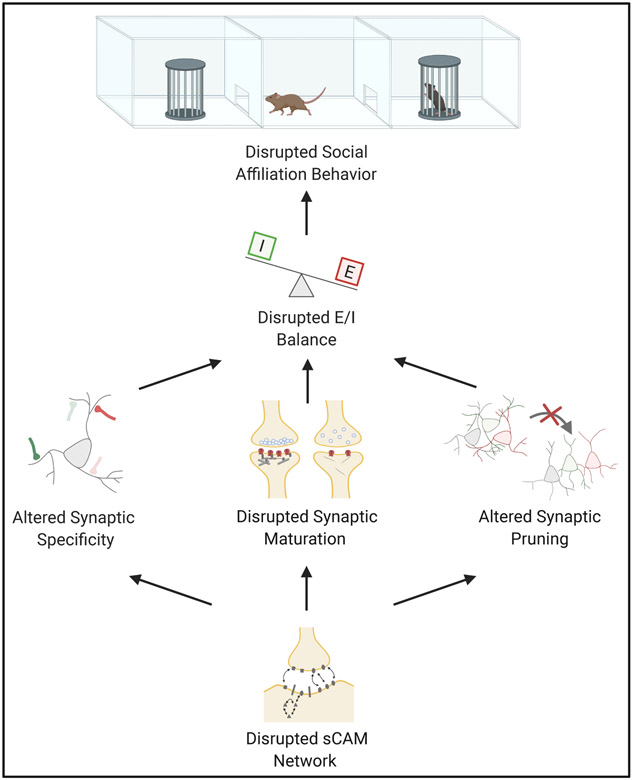
We propose that sCAMs affect social affiliation behavior via one or more of the following mechanisms: altered specificity during synaptogenesis, disrupted synaptic maturation, and/or altered synaptic pruning. According to our model, the net effect of these mechanisms is alteration in excitatory / inhibitory balance in circuits relevant to social affiliative behaviors (circuits including mPFC, BLA, VTA, ACC, and hippocampus; see Figure 1).
Figure 1. Proposed mechanistic model for the role of sCAMs in social affiliative behaviors.
This figure shows our proposed model for how disruption of sCAMs could lead to disrupted social affiliation, namely through altered synaptic specificity, synaptic pruning, and/or synaptic maturation that disrupts the E/I balance of the circuit.
With a single exception (noted below), the following evidence for disrupted synaptic specificity, maturation, and pruning was accompanied by a decrease in social affiliation. Evidence of altered synaptic specificity includes abnormal axon projections, which is seen in the hippocampus of a Negr1 model (91), and abnormal dendritic arborization, which is seen in the ACC and in the striatum of Shank3 models (107, 108). Evidence of disrupted synaptic maturation includes abnormal active zones, vesicle pools, neurotransmitter receptor density, PSD scaffold, synaptic strength, and spine morphology. Abnormal vesicle pools were seen in a transgenic overexpression model of Nlgn2 (48). Stabilization of actin was able to rescue social affiliation deficits and accompanying decreased NMDAR transmission in a Shank3 model, suggesting that compromised actin in PSD scaffolding contributed to the decrease in social affiliation (109). Immature spine morphology was seen with haploinsufficiency of Pcdh10 in the BLA (72), Lrfn2 in the Hip (102), and Shank3 in the ACC (107). A higher number of mature excitatory synapses was demonstrated in the hippocampus in a Lrfn2 model (102).
Evidence of altered synaptic pruning includes altered microglial function, altered arbor maturation, and abnormal density of circuit constructs. Change in density of circuit constructs is seen in Nlgn2 models. Specifically, overexpression of Nlgn2 during early development led to a decrease in the ratio of asymmetric to symmetric synapses, and mPFC-specific knockout of Nlgn2 led to decrease in GAD65-positive synapse density (46, 48). Change in spine density / number, which could indicate either altered synaptic maturation or synaptic pruning, was seen in Pcdh10 (72), Negr1 (93), Lrfn2 (102), Shank2 (110), Shank3 (107, 108, 111), Magi2 (S-SCAM) (112), and Dlgap2 (PSD-95)(113) models. The Pcdh10 model is unique among the other studies in demonstrating an increase in spine density in the BLA (though the spines are also more immature and as such are likely less functional) (72). Models of Negr1, Shank2, and Magi2 demonstrated a decrease of spine density / number in the hippocampus (93, 110, 112). There was also a decrease in spine density in the somatosensory and visual cortex in a Lrfn2 model and in the OFC/mPFC in a Dlgap2 model (though in this case, there was an increase, not a decrease, in social affiliation) (102, 113). Shank 3 models showed a decrease in spine density in the ACC and striatum (107, 108).
Alterations in E/I balance has been a prominent hypothesis for the neurobiology of autism for decades (114, 115), however mechanisms by which alterations in ASD genes might lead to these E/I imbalances in specific circuits, which in turn leads to behavioral phenotypes, remain largely unclear. Moreover, there appears to be heterogeneity of such imbalances (some evidence for excess of excitation, and some for excess of inhibition) and their interpretation (116-118), which may be related to the large number of genes involved in varying pathways in ASD. While there is mixed evidence cited above, the majority of the studies we reviewed point to a decrease in excitation in relation to inhibition, leading to decreased social affiliation. Several studies demonstrate decreased excitatory transmission, including Nrxn2 in the somatosensory cortex (29), Lrrc4 in the hippocampus (98), Shank2 in the hippocampus (110, 119), and Shank3 in the striatum, ACC, VTA, and striatum (60, 107, 108, 111). In a Shank3 model, the pattern of E/I depended on region, with decreased excitation and increased inhibition in the hippocampus and decreased inhibition in the mPFC (120). Additionally, two studies demonstrated a decrease in inhibition, specifically in the mPFC in an mPFC-specific knockout of Nlgn2 (46) and in the hippocampus in a global knockout of Negr1 (92). More specific than general E/I balance, several studies across models and regions demonstrated impaired glutamatergic transmission. Impaired glutamatergic transmission was seen in Nrxn2 model in somatosensory cortex (29), in Lrrc4 in the hippocampus (98), Shank2 in the Hip (121, 122), and Shank3 in the striatum, hippocampus, and PFC (123-126). Interestingly, an age-effect was found in Shank2 mice such that there was an increase in glutamatergic transmission in the hippocampus at P14 and a decrease at P24 (127). These findings in sum point to a general pattern of decreased excitation and glutamatergic transmission associated with decreased social affiliation, though there is variation related to model, age, and region tested that should be further explored.
In terms of defining the social affiliation circuit that may be affected by sCAMs, this review finds strong evidence implicating several brain regions, including the mPFC, BLA, striatum, ACC, and hippocampus. Accompanying decreased social affiliation, in the hippocampus there has been evidence of altered synaptic specificity in Negr1 models (91), altered synaptic maturation in Lrfn2 models (102), and finally changes in spine density / number that could indicate either altered synaptic maturation or pruning in Lrfn2, Shank2, Shank3, and Magi2 models (102, 110, 112, 128). There is altered synaptic specificity, synaptic maturation, and decreased spine density in Shank3 models in the ACC (107, 108). Also in Shank3 mice, altered synaptic specificity and change in spine density were found in the striatum (108, 111). There is evidence of altered synapse maturation based on a increase in spine density and in immature spines in the BLA of Pcdh10 haploinsufficient mice (72). Additionally, conditional regional knockdowns and knockouts of sCAMs decreased social affiliation, including conditional knockdowns / knockouts of Shank3, Nlgn2, and Nlgn3 in the VTA, ACC, and mPFC (46, 60, 107, 129).
Discussion
The overall patterns that we observe in the literature suggest that mutation / dysfunction of sCAM genes alters synaptic maturation, specificity, and pruning and leads to E / I imbalance in several relevant brain regions, including mPFC, BLA, VTA, and hippocampus. Moving forward, study of sCAM gene knockouts on additional genetic backgrounds, more comprehensive studies of mutations in the various sCAMs, and the use of larger sample sizes will help to advance the knowledge base further. Additionally, many of the experiments currently being conducted use global knockouts; while this is a useful starting point, region- and cell-type specific knockouts as well as conditional knockouts at specific developmental time points are needed to provide meaningful information on the role of sCAMs in specific parts of social motivation / approach circuitry across development. Proteomic studies focused on sCAMs of different regions during social affiliation behavior could also provide insight into the circuitry underlying the behavior. Other important areas of future investigation include mechanisms of rescue and whether restoration of function at certain developmental time points or in specific parts of neural circuitry are sufficient to rescue social affiliation overall.
There are also still fundamental gaps in our knowledge base on sCAMs. For some sCAMs, key pieces of basic information (i.e. binding partners, localization within the brain, or localization within the synapse) are still lacking. Advancing our understanding of how each sCAM interacts with glutamatergic receptors and affects spine density, number, and morphology promises to provide greater insight into the mechanisms underlying social affiliation. An additional challenge is studying the role in social affiliation of the many sCAM protein isoforms, with differing binding strengths and specificities, as well as the role of specific binding domains in each protein. By addressing these gaps in future studies, we, as a field, can gain greater insight into synaptic and circuit mechanisms of social affiliation, its disruption in neurodevelopmental and neuropsychiatric disorders, and potential targets for rescue.
Supplementary Material
Acknowledgements
The authors wish to thank the following funding sources: The Autism Spectrum Program of Excellence (ASPE), Research Gift to the University of Pennsylvania; Pennsylvania Department of Health (SAP # 4100042728) (R.T.S.); and 1P50MH096891 (Raquel Gur)– subproject 6773 (E.S.B. and T.A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Institutes of Health. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions. We would like to thank BioRender for the use of their software to create Figure 1.
Financial Disclosures
All authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Südhof TC (2008): Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 455: 903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasem E, Kurihara T, Tabuchi K (2017): Neurexins and neuropsychiatric disorders. Neurosci Res. 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Sakurai K, Migita O, Toru M, Arinami T (2002): An association between a missense polymorphism in the close homologue of L1 (CHL1, CALL) gene and schizophrenia. Mol Psychiatry. 7: 412–415. [DOI] [PubMed] [Google Scholar]
- 4.Shoukier M, Fuchs S, Schwaibold E, Lingen M, Gärtner J, Brockmann K, Zirn B (2013): Microduplication of 3p263 in nonsyndromic intellectual disability indicates an important role of CHL1 for normal cognitive function. Neuropediatrics. 44: 268–271. [DOI] [PubMed] [Google Scholar]
- 5.Salyakina D, Cukier HN, Lee JM, Sacharow S, Nations LD, Ma D, et al. (2011): Copy number variants in extended autism spectrum disorder families reveal candidates potentially involved in autism risk. PLoS One. 6: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li C, Liu C, Zhou B, Hu C, Xu X (2016): Novel microduplication of CHL1 gene in a patient with autism spectrum disorder: A case report and a brief literature review. Mol Cytogenet. 9: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guilmatre A, Huguet G, Delorme R, Bourgeron T (2014): The emerging role of SHANK genes in neuropsychiatric disorders. Dev Neurobiol. 74: 113–122. [DOI] [PubMed] [Google Scholar]
- 8.Redies C, Hertel N, Hübner CA (2012): Cadherins and neuropsychiatric disorders. Brain Res. 1470: 130–144. [DOI] [PubMed] [Google Scholar]
- 9.Sankoorikal GM V, Kaercher KA, Boon CJ, Lee JK, Brodkin ES (2006): A mouse model system for genetic analysis of sociability: C57BL/6J versus BALB/cJ inbred mouse strains. Biol Psychiatry. 59: 415–423. [DOI] [PubMed] [Google Scholar]
- 10.Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, et al. (2004): Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes, Brain Behav. 3: 287–302. [DOI] [PubMed] [Google Scholar]
- 11.Graf ER, Zhang X, Jin S, Linhoff MW, Craig AM (2004): Neurexins Induce Differentiation of GABA and Glutamate Postsynaptic Specializations via Neuroligins. Cell. 119: 1013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC (1992): Neurexins : Synaptic Cell Surface Proteins Related to the cL-Latrotoxin Receptor and Laminin. Science (80-). 257: 50–56. [DOI] [PubMed] [Google Scholar]
- 13.Dean C, Scholl FG, Choih J, Demaria S, Berger J, Isacoff E, Scheiffele P (2003): Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 6: 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boucard AA, Ko J, Südhof TC (2012): High affinity neurexin binding to cell adhesion Gprotein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J Biol Chem. 287: 9399–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC (2005): A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to α- and β-neurexins. Neuron. 48: 229–236. [DOI] [PubMed] [Google Scholar]
- 16.Pettem KL, Yokomaku D, Luo L, Linhoff MW, Prasad T, Connor SA, et al. (2013): The Specific a -Neurexin Interactor Calsyntenin-3 Promotes Excitatory and Inhibitory Synapse Development. Neuron. 80: 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko J, Fuccillo M V, Malenka RC, Su TC (2009): LRRTM2 Functions as a Neurexin Ligand in Promoting Excitatory Synapse Formation. Neuron. 64: 791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joo JY, Lee SJ, Uemura T, Yoshida T, Yasumura M, Watanabe M, Mishina M (2011): Differential interactions of cerebellin precursor protein (Cbln) subtypes and neurexin variants for synapse formation of cortical neurons. Biochem Biophys Res Commun. 406: 627–632. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Atasoy D, Araç D, Yang X, Fucillo MV, Robison AJ, et al. (2010): Neurexins physically and functionally interact with GABA A receptors. Neuron. 66: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugita S, Saito F, Tang J, Satz J, Campbell K, Südhof TC (2001): A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 154: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Missler M, Hammer RE, Südhof TC (2002): Neurexophilin Binding to α-Neurexins. J Biol Chem. 273: 34716–34723. [DOI] [PubMed] [Google Scholar]
- 22.Tabuchi K, Südhof TC (2002): Structure and Evolution of Neurexin Genes : Insight into the Mechanism of Alternative Splicing. Genomics. 79: 849–859. [DOI] [PubMed] [Google Scholar]
- 23.Ullrich B, Ushkaryov YA, Südhof TC (1995): Cartography of neurexins: More than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 14: 497–507. [DOI] [PubMed] [Google Scholar]
- 24.Armstrong EC, Caruso A, Servadio M, Andreae LC, Trezza V, Scattoni L, Fernandes C (2019): Assessing the developmental trajectory of mouse models of neurodevelopmental disorders: social and communication deficits in mice with Neurexin 1α deletion. Genes, Brain Behav. . doi: doi: 10.1111/gbb.12630. [DOI] [PubMed] [Google Scholar]
- 25.Twining RC, Vantrease JE, Love S, Padival M, Rosenkranz JA (2017): An intra-amygdala circuit specifically regulates social fear learning. Nat Neurosci. 20: 458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grayton HM, Missler M, Collier DA, Fernandes C (2013): Altered Social Behaviours in Neurexin 1α Knockout Mice Resemble Core Symptoms in Neurodevelopmental Disorders. PLoS One. 8. doi: 10.1371/journal.pone.0067114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dachtler J, Ivorra JL, Rowland TE, Lever C, John Rodgers R, Clapcote SJ (2015): Heterozygous deletion of α-neurexin I or α-neurexin II results in behaviors relevant to autism and schizophrenia. Behav Neurosci. 129: 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Etherton MR, Blaiss CA, Powell CM, Südhof TC (2009): Mouse neurexin-1 deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci. 106: 17998–18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Born G, Grayton HM, Langhorst H, Dudanova I, Rohlmann A, Woodward BW, et al. (2015): Genetic targeting of NRXN2 in mice unveils role in excitatory cortical synapse function and social behaviors. Front Synaptic Neurosci. 7: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dachtler J, Glasper J, Cohen RN, Ivorra JL, Swiffen DJ, Jackson AJ, et al. (2014): Deletion of α-neurexin II results in autism-related behaviors in mice. Transl Psychiatry. 4: e484–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chanda S, Hale WD, Zhang B, Wernig M, Sudhof TC (2017): Unique versus redundant functions of neuroligin genes in shaping excitatory and inhibitory synapse properties. J Neurosci. 37: 6816–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chih B, Engelman H, Scheiffele P (2005): Control of excitatory and inhibitory synapse formation by neuroligins. Science (80-). 307: 1324–1328. [DOI] [PubMed] [Google Scholar]
- 33.Kwon H, Kozorovitskiy Y, Oh W, Peixoto RT (2013): Cortical synaptogenesis and excitatory synapse number are determined via a Neuroligin-1-dependent intercellular competition. Nat Neurosci. 15: 1667–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheiffele P, Fan J, Choih J, Fetter R, Serafini T (2000): Neuroligin Expressed in Nonneuronal Cells Triggers Presynaptic Development in Contacting Axons. Cell. 101: 657–669. [DOI] [PubMed] [Google Scholar]
- 35.Iida J, Hirabayashi S, Sato Y, Hata Y (2004): Synaptic scaffolding molecule is involved in the synaptic clustering of neuroligin. Mol Cell Neurosci. 27: 497–508. [DOI] [PubMed] [Google Scholar]
- 36.Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, et al. (1997): Binding of Neuroligins to PSD-95. Science (80-). 277: 1511–1516. [DOI] [PubMed] [Google Scholar]
- 37.Song J-Y, Ichtchenko K, Sudhof TC, Brose N (1999): Neuroligin 1 is a postsynaptic celladhesion molecule of excitatory synapses. Proc Natl Acad Sci USA. 96: 1100–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer G, Varoqueaux F, Neeb A, Oschlies M, Brose N (2004): The complexity of PDZ domain-mediated interactions at glutamatergic synapses : a case study on neuroligin. Neuropharmacology. 47: 724–733. [DOI] [PubMed] [Google Scholar]
- 39.Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, et al. (2010): Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 30: 2115–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakanishi M, Nomura J, Ji X, Tamada K, Arai T, Takahashi E, et al. (2017): Functional significance of rare neuroligin 1 variants found in autism. PLoS Genet. 13: 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoy JL, Haeger PA, Constable JR, Arias RJ, McCallum R, Kyweriga M, … & Washbourne P (2014): Neuroligin1 drives synaptic and behavioral maturation through intracellular interactions. J Neurosci. 33: 9364–9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gkogkas CO, Khoutorsky A, Ran I, Rampakakis E, Nevarko T, Weatherill DB, et al. (2013): Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature. 493: 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varoqueaux F, Jamain S, Brose N (2004): Neuroligin 2 is exclusively localized to inhibitory synapses. Eur J Cell Biol. 83: 449–456. [DOI] [PubMed] [Google Scholar]
- 44.Wohr M, Silverman JL, Scattoni ML, Turner SM, Harris MJ, Saxena R, Crawley JN (2013): Developmental delays and reduced pup ultrasonic vocalizations but normal sociability in mice lacking the postsynaptic cell adhesion protein neuroligin2. Behav Brain Res. 251: 50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blundell J, Tabuchi K, Bolliger MF, Blaiss C a, Liu X, Südhof TC, Powell CM (2009): Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes, Brain Behav. 8: 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang J, Xu W, Hsu YT, Yee AX, Chen L, Südhof TC (2015): Conditional neuroligin-2 knockout in adult medial prefrontal cortex links chronic changes in synaptic inhibition to cognitive impairments. Mol Psychiatry. 20: 850–859. [DOI] [PubMed] [Google Scholar]
- 47.Van Zandt M, Weiss E, Almyasheva A, Lipior S, Maisel S, Naegele JR (2019): Adenoassociated viral overexpression of neuroligin 2 in the mouse hippocampus enhances GABAergic synapses and impairs hippocampal-dependent behaviors. Behav Brain Res. 362: 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hines RM, Wu L, Hines DJ, Steenland H, Mansour S, Dahlhaus R, et al. (2008): Synaptic Imbalance, Stereotypies, and Impaired Social Interactions in Mice with Altered Neuroligin 2 Expression. J Neurosci. 28: 6055–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Budreck EC, Scheiffele P (2007): Neuroligin-3 is a neuronal adhesion protein at GABAergic and glutamatergic synapses. Eur J Neurosci. 26: 1738–1748. [DOI] [PubMed] [Google Scholar]
- 50.Tabuchi K, Blundell J, Etherton MR, Hammer RE, Powell CM, Südhof TC (2011): A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science (80-). 318: 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar M, Duda JT, Hwang W-T, Kenworthy C, Ittyerah R, Pickup S, et al. (2014): High Resolution Magnetic Resonance Imaging for Characterization of the Neuroligin-3 Knock-in Mouse Model Associated with Autism Spectrum Disorder. PLoS One. 9: e109872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jaramillo TC, Liu S, Pettersen A, Birnbaum SG, Powell CM (2014): Autism-Related Neuroligin-3 Mutation Alters Social Behavior and Spatial Learning. Autism Res. 7: 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaramillo TC, Escamilla CO, Liu S, Peca L, Birnbaum SG, Powell CM (2018): Genetic background effects in Neuroligin-3 mutant mice: Minimal behavioral abnormalities on C57 background. Autism Res. 11: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao W, Lin S, Xia Q qiang, Du Y lan, Yang Q, Zhang M ying, et al. (2018): Gamma Oscillation Dysfunction in mPFC Leads to Social Deficits in Neuroligin 3 R451C Knockin Mice. Neuron. 98: 670. [DOI] [PubMed] [Google Scholar]
- 55.Burrows EL, Eastwood AF, May C, Kolbe SC, Hill T, McLachlan NM, et al. (2017): Social isolation alters social and mating behavior in the R451C neuroligin mouse model of autism. NeuralPlast. 2017. doi: 10.1155/2017/8361290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chadman KK, Gong S, Scattoni ML, Boltuck SE, Gandhy U, Heintz N, Crawley JN (2009): Minimal aberrant behavioral phenotypes of neuroligin-3 R451C knockin mice. Autism. 1: 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Etherton M, Földy C, Sharma M, Tabuchi K, Liu X, Shamloo M (2011): Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. PNAS. 108: 13764–13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hamilton SM, Green JR, Veeraragavan S, Yuva L, McCoy A, Wu Y, et al. (2014): Fmr1 and Nlgn3 knockout rats: Novel tools for investigating autism spectrum disorders. Behav Neurosci. 128: 103–109. [DOI] [PubMed] [Google Scholar]
- 59.Radyushkin K, Hammerschmidt K, Boretius S, Varoqueaux F, El-Kordi A, Ronnenberg A, et al. (2009): Neuroligin-3-deficient mice: Model of a monogenic heritable form of autism with an olfactory deficit. Genes, Brain Behav. 8: 416–425. [DOI] [PubMed] [Google Scholar]
- 60.Bariselli S, Hörnberg H, Prévost-Solié C, Musardo S, Hatstatt-Burklé L, Scheiffele P, Bellone C (2018): Role of VTA dopamine neurons and neuroligin 3 in sociability traits related to nonfamiliar conspecific interaction. Nat Commun. 9. doi: 10.1038/s41467-018-05382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoon M, Soykan T, Falkenburger B, Hammer M, Patrizi A, Schmidt K-F, et al. (2011):Neuroligin-4 is localized to glycinergic postsynapses and regulates inhibition in the retina. Proc Natl Acad Sci U S A. 108: 3053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jamain S, Radyushkin K, Hammerschmidt K, Granon S, Boretius S, Varoqueaux F, et al. (2008): Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc Natl Acad Sci. 105: 1710–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salinas PC, Price SR (2005): Cadherins and catenins in synapse development. Curr Opin Neurobiol. 15: 73–80. [DOI] [PubMed] [Google Scholar]
- 64.Okamura K, Tanaka H, Yagita Y, Saeki Y, Taguchi A, Hiraoka Y, et al. (2004): Cadherin activity is required for activity-induced spine remodeling. J Cell Biol. 167: 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hintsch G, Zurlinden A, Meskenaite V, Steuble M, Fink-Widmer K, Kinter J, Sonderegger P (2002): The calsyntenins - A family of postsynaptic membrane proteins with distinct neuronal expression patterns. Mol Cell Neurosci. 21: 393–409. [DOI] [PubMed] [Google Scholar]
- 66.Ranneva SV, Pavlov KS, Gromova AV, Amstislavskaya TG, Lipina TV (2017): Features of emotional and social behavioral phenotypes of calsyntenin2 knockout mice. Behav Brain Res. 332: 343–354. [DOI] [PubMed] [Google Scholar]
- 67.Hirano S, Yan Q, Suzuki ST (2018): Expression of a Novel Protocadherin, OL-Protocadherin, in a Subset of Functional Systems of the Developing Mouse Brain. J Neurosci. 19: 995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakao S, Platek A, Hirano S, Takeichi M (2008): Contact-dependent promotion of cell migration by the OL-protocadherin-Nap1 interaction. J Cell Biol. 182: 395–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai NP, Wilkerson JR, Guo W, Maksimova MA, Demartino GN, Cowan CW, Huber KM (2012): Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 151: 1581–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luckner R, Obst-Pernberg K, Hirano S, Suzuki ST, Redies C (2001): Granule cell raphes in the developing mouse cerebellum. Cell Tissue Res. 303: 159–172. [DOI] [PubMed] [Google Scholar]
- 71.Aoki E, Kimura R, Suzuki ST, Hirano S (2003): Distribution of OL-protocadherin protein in correlation with specific neural compartments and local circuits in the postnatal mouse brain. Neuroscience. 117: 593–614. [DOI] [PubMed] [Google Scholar]
- 72.Schoch H, Kreibich AS, Ferri SL, White RS, Bohorquez D, Banerjee A, et al. (2017): Sociability Deficits and Altered Amygdala Circuits in Mice Lacking Pcdh10, an Autism Associated Gene. Biol Psychiatry. 81: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peek SL, Mah KM, Weiner JA (2017): Regulation of neural circuit formation by protocadherins. Cell Mol Life Sci. 74: 4133–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsai NP, Huber KM (2017): Protocadherins and the Social Brain. Biol Psychiatry. 81: 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sytnyk V, Leshchyns’ka I, Schachner M (2017): Neural Cell Adhesion Molecules of the Immunoglobulin Superfamily Regulate Synapse Formation, Maintenance, and Function. Trends Neurosci. 40: 295–308. [DOI] [PubMed] [Google Scholar]
- 76.Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC (2002): SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science (80-). 297: 1525–1531. [DOI] [PubMed] [Google Scholar]
- 77.Fogel AI, Akins MR, Krupp AJ, Stagi M, Stein V, Biederer T (2007): SynCAMs Organize Synapses through Heterophilic Adhesion. J Neurosci. 27: 12516–12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Holm J, Hillenbrand R, Steuber V, Bartsch U, Moos M, Hermann L, et al. (1996): Structural features of a close homologue of L1 (CHL1)in the mouse: A new member of the L1 family of neural recognition molecules. Eur J Neurosci. 8: 1613–1629. [DOI] [PubMed] [Google Scholar]
- 79.Robbins EM, Krupp AJ, Perez de Arce K, Ghosh AK, Fogel AI, Boucard A, et al. (2010): SynCAM 1 Adhesion Dynamically Regulates Synapse Number and Impacts Plasticity and Learning. Neuron. 68: 894–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hillenbrand R, Molthagen M, Montag D, Schachner M (1999): The close homologue of the neural adhesion molecule L1 (CHL1): Patterns of expression and promotion of neurite outgrowth by heterophilic interactions. Eur J Neurosci. 11: 813–826. [DOI] [PubMed] [Google Scholar]
- 81.Hoy JL, Constable JR, Vicini S, Fu Z, Washbourne P (2009): SynCAM1 recruits NMDA receptors via Protein 4.1B. Mol Cell Neurosci. 42: 466–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Buhusi M, Midkiff BR, Gates AM, Richter M, Schachner M, Maness PF (2003): Close Homolog of L1 Is an Enhancer of Integrin-mediated Cell Migration. J Biol Chem. 278: 25024–25031. [DOI] [PubMed] [Google Scholar]
- 83.Thomas LA, Akins MR, Biederer T (2008): Expression and Adhesion Profiles of SynCAM Adhesion Molecules Indicate Distinct Neuronal Functions. J Comp Neurol. 510: 47–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takayanagi Y, Fujita E, Yu Z, Yamagata T, Momoi MY, Momoi T, Onaka T (2010): Impairment of social and emotional behaviors in Cadm1-knockout mice. Biochem Biophys Res Commun. 396: 703–708. [DOI] [PubMed] [Google Scholar]
- 85.Andreyeva A, Leshchyns’ka I, Knepper M, Betzel C, Redecke L, Sytnyk V, Schachner M (2010): CHL1 is a selective organizer of the presynaptic machinery chaperoning the SNARE complex. PLoS One. 5. doi: 10.1371/journal.pone.0012018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Leshchyns’ka I, Sytnyk V, Richter M, Andreyeva A, Puchkov D, Schachner M (2006): The Adhesion Molecule CHL1 Regulates Uncoating of Clathrin-Coated Synaptic Vesicles. Neuron. 52: 1011–1025. [DOI] [PubMed] [Google Scholar]
- 87.Morellini F, Lepsveridze E, Kähler B, Dityatev A, Schachner M (2007): Reduced reactivity to novelty, impaired social behavior, and enhanced basal synaptic excitatory activity in perforant path projections to the dentate gyrus in young adult mice deficient in the neural cell adhesion molecule CHL1. Mol Cell Neurosci. 34: 121–136. [DOI] [PubMed] [Google Scholar]
- 88.Hashimoto T, Yamada M, Maekawa S, Nakashima T, Miyata S (2008): IgLON cell adhesion molecule Kilon is a crucial modulator for synapse number in hippocampal neurons. Brain Res. 1224: 1–11. [DOI] [PubMed] [Google Scholar]
- 89.Funatsu N, Miyata S, Kumanogoh H, Shigeta M, Hamada K, Endo Y, et al. (1999): Characterization of a novel rat brain glycosylphosphatidylinositol- anchored protein (Kilon), a member of the IgLON cell adhesion molecule family. J Biol Chem. 274: 8224–8230. [DOI] [PubMed] [Google Scholar]
- 90.Miyata S, Matsumoto N, Taguchi K, Akagi A, Iino T, Funatsu N, Maekawa S (2003): Biochemical and ultrastructural analyses of IgLON cell adhesion molecules, Kilon and OBCAM in the rat brain. Neuroscience. 117: 645–658. [DOI] [PubMed] [Google Scholar]
- 91.Singh K, Loreth D, Pöttker B, Hefti K, Innos J, Schwald K, et al. (2018): Neuronal growth and behavioral alterations in mice deficient for the psychiatric disease-associated negr1 gene. Front Mol Neurosci. 11: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singh K, Jayaram M, Kaare M, Leidmaa E, Jagomäe T, Heinla I, et al. (2019): Neural cell adhesion molecule Negr1 deficiency in mouse results in structural brain endophenotypes and behavioral deviations related to psychiatric disorders. Sci Rep. 9: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Szczurkowska J, Pischedda F, Pinto B, Manago F, Haas CA, Summa M, et al. (2018): NEGR1 and FGFR2 cooperatively regulate cortical development and core behaviours related to autism disorders in mice. Brain. 141: 2772–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de Wit J, Ghosh A (2014): Control of neural circuit formation by leucine-rich repeat proteins. Trends Neurosci. 37: 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ko J, Kim E (2007): Leucine-rich repeat proteins of synapses. J Neurosci Res. 85: 2824–2832. [DOI] [PubMed] [Google Scholar]
- 96.Zhang Q, Wang J, Fan S, Wang L, Cao L, Tang K, et al. (2005): Expression and functional characterization of LRRC4, a novel brain-specific member of the LRR superfamily. FEBS Lett. 579: 3674–3682. [DOI] [PubMed] [Google Scholar]
- 97.Kim S, Burette A, Chung HS, Kwon SK, Woo J, Lee HW, et al. (2006): NGL family PSD- 95-interacting adhesion molecules regulate excitatory synapse formation. Nat Neurosci. 9: 1294–1301. [DOI] [PubMed] [Google Scholar]
- 98.Um SM, Ha S, Lee H, Kim J, Kim K, Shin W, et al. (2018): NGL-2 Deletion Leads to Autistic-like Behaviors Responsive to NMDAR Modulation. Cell Rep. 23: 3839–3851. [DOI] [PubMed] [Google Scholar]
- 99.Wang C-Y, Chang K, Petralia RS, Wang Y-X, Seabold GK, Wenthold RJ (2006): A Novel Family of Adhesion-Like Molecules That Interacts with the NMDA Receptor. J Neurosci. 26: 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ko J, Kim S, Chung HS, Kim K, Han K, Kim H, et al. (2006): SALM Synaptic Cell Adhesion-like Molecules Regulate the Differentiation of Excitatory Synapses. Neuron. 50: 233–245. [DOI] [PubMed] [Google Scholar]
- 101.Li Y, Kim R, Cho YS, Song WS, Kim D, Kim K, et al. (2018): Lrfn2-Mutant Mice Display Suppressed Synaptic Plasticity and Inhibitory Synapse Development and Abnormal Social Communication and Startle Response. J Neurosci. 38: 5872–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morimura N, Yasuda H, Yamaguchi K, Katayama KI, Hatayama M, Tomioka NH, et al. (2017): Autism-like behaviours and enhanced memory formation and synaptic plasticity in Lrfn2/SALM1-deficient mice. Nat Commun. 8. doi: 10.1038/ncomms15800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, Craig AM (2010): LRRTMs and Neuroligins Bind Neurexins with a Differential Code to Cooperate in Glutamate Synapse Development. J Neurosci. 30: 7495–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Linhoff MW, Laurén J, Cassidy RM, Dobie FA, Takahashi H, Nygaard HB, et al. (2009): An Unbiased Expression Screen for Synaptogenic Proteins Identifies the LRRTM Protein Family as Synaptic Organizers. Neuron. 61: 734–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Laurén J, Airaksinen MS, Saarma M, Timmusk T (2003): A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 81: 411–421. [DOI] [PubMed] [Google Scholar]
- 106.Takashima N, Odaka YS, Sakoori K, Akagi T, Hashikawa T, Morimura N, et al. (2011): Impaired cognitive function and altered hippocampal synapse morphology in mice lacking LRRTM1, a gene associated with schizophrenia. PLoS One. 6. doi: 10.1371/journal.pone.0022716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guo B, Chen J, Chen Q, Ren K, Feng D, Mao H, et al. (2019): Anterior cingulate cortex dysfunction underlies social deficits in Shank3 mutant mice. Nat Neurosci. 22. doi: 10.1038/s41593-019-0445-9. [DOI] [PubMed] [Google Scholar]
- 108.Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. (2011): Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 472: 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Duffney LJ, Zhong P, Wei J, Matas E, Cheng J, Qin L, et al. (2015): Autism-like Deficits in Shank3-Deficient Mice Are Rescued by Targeting Actin Regulators. Cell Rep. 11: 1400–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, et al. (2012): Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 486: 256–260. [DOI] [PubMed] [Google Scholar]
- 111.Zhou Y, Kaiser T, Monteiro P, Zhang X, Van Der MS , Wang D, et al. (2016): Mice with Shank3 mutations associated with ASD and schizophrenia display both shared and distinct defects. Neuron. 89: 147–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang N, Zhong P, Shin SM, Metallo J, Danielson E, Olsen CM, et al. (2015): S-SCAM, A Rare Copy Number Variation Gene, Induces Schizophrenia-Related Endophenotypes in Transgenic Mouse Model. J Neurosci. 35: 1892–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang-Xie LF, Liao HM, Chen CH, Chen YT, Ho SY, Lu DH, et al. (2014): Autismassociated gene Dlgap2 mutant mice demonstrate exacerbated aggressive behaviors and orbitofrontal cortex deficits. Mol Autism. 5: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee E, Lee J, Kim E (2017): Excitation/Inhibition Imbalance in Animal Models of Autism Spectrum Disorders. Biol Psychiatry. 81: 838–847. [DOI] [PubMed] [Google Scholar]
- 115.Rubenstein E, Chawla D (2019): Broader autism phenotype in parents of children with autism: a systematic review of percentage estimates. J Child Fam Stud. 27: 1705–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nelson SB, Valakh V (2015): Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron. 87: 684–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Antoine MW, Langberg T, Schnepel P, Feldman DE (2019): Increased Excitation-Inhibition Ratio Stabilizes Synapse and Circuit Excitability in Four Autism Mouse Models. Neuron. 101: 648–661.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fatemi SH, Halt AR, Stary JM, Kanodia R, Schulz SC, Realmuto GR (2002): Glutamic acid decarboxylase 65 and 67 kDa proteins are reduced in autistic parietal and cerebellar cortices. Biol Psychiatry. 52: 805–810. [DOI] [PubMed] [Google Scholar]
- 119.Kim R, Kim J, Chung C, Ha S, Lee S, Lee E, et al. (2018): Cell type-specific Shank2 deletion in mice leads to differential synaptic and behavioral phenotypes. J Neurosci. 38: 2684–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee J, Chung C, Ha S, Lee D, Kim D-Y, Kim H, Kim E (2015): Shank3-mutant mice lacking exon 9 show altered excitation/inhibition balance, enhanced rearing, and spatial memory deficit. Front Cell Neurosci. 9: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, et al. (2012): Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 486: 261–265. [DOI] [PubMed] [Google Scholar]
- 122.Lee EJ, Lee H, Huang TN, Chung C, Shin W, Kim K, et al. (2015): Trans-synaptic zinc mobilization improves social interaction in two mouse models of autism through NMDAR activation. Nat Commun. 6: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jaramillo TC, Speed HE, Reimers JM, Powell CM (2016): Altered striatal synaptic function and abnormal behavior in Shank3 exon 4-9 deletion mouse model of autism. Autism Res. 9: 350–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jaramillo TC, Speed HE, Xuan Z, Reimers JM, Ochoa C, Weaver TP, et al. (2017): Novel Shank3 mutant exhibits behaviors with face validity for autism and altered striatal and hippocampal function. Autism Res. 10: 42–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ma K, Qin L, Matas E, Duffney LJ, Liu A, Yan Z (2018): Histone deacetylase inhibitor MS-275 restores social and synaptic function in a Shank3-deficient mouse model of autism. Neuropsychopharmacology. 43: 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Qin L, Ma K, Yan Z (2019): Chemogenetic Activation of Prefrontal Cortex in Shank3-Deficient Mice Ameliorates Social Deficits, NMDAR Hypofunction, and Sgk2 Downregulation. iScience. 17: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chung C, Ha S, Kang H, Lee J, Um SM, Yan H, et al. (2019): Early Correction of N-Methyl-D-Aspartate Receptor Function Improves Autistic-like Social Behaviors in Adult Shank2 −/− Mice. Biol Psychiatry. 85: 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, et al. (2011): Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 20: 3093–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bariselli S, Tzanoulinou S, Glangetas C, Prévost-Solié C, Pucci L, Viguié J, et al. (2016): SHANK3 controls maturation of social reward circuits in the VTA. Nat Neurosci. 19: 926–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.