Abstract
Gram-negative bacteria possess a dual-membrane envelope, which provides defense against environmental assault, as well as formidable resistance against antibiotics. Lipopolysaccharide (LPS) is the primary lipid component in the outermost membrane leaflet of most Gram-negative bacteria, and plays critical roles in cell envelope formation. Newly synthesized LPS at the cytoplasmic side of the inner membrane is flipped across the inner membrane and pushed across the periplasm by two ATP-binding cassette transporters: MsbA and LptB2FGC. Both transporters represent promising targets for developing new classes of antibiotics. In this review, we discuss recent advances in understanding the mechanism of LPS translocation driven by MsbA and LptB2FGC, with a particular focus on the new findings from structural studies.
Keywords: MsbA, LptB2FGC, lipopolysaccharide, ABC transporter, antibiotics
Introduction
Pathogenic Gram-negative bacteria are the major cause of antibiotic-resistant infection[1]. They produce a unique dual-membrane envelope. While the inner membrane (IM) is composed of phospholipids in both membrane leaflets, the outer membrane (OM) is an asymmetric lipid bilayer with the inner leaflet of phospholipids and the outer leaflet predominantly of lipopolysaccharide (LPS). The IM and OM have distinct permeation properties, and together they prevent many antibiotics from entering the cells. LPS is composed of three portions: lipid A, core oligosaccharide, and O-antigen (Fig. 1a). LPS recognition by Toll-like receptor 4 (TLR4) initiates an inflammatory response and in some cases leads to septic shock[2,3]. LPS also stimulates immune cells by binding to a cytoplasmic non-canonical inflammasome complex[4]. Bacteria modify LPS in a variety of ways to alter the OM properties, innate immune stimulation, and pathogenesis[5].
Figure 1 |. Lipopolysaccharide structure and transport pathway.
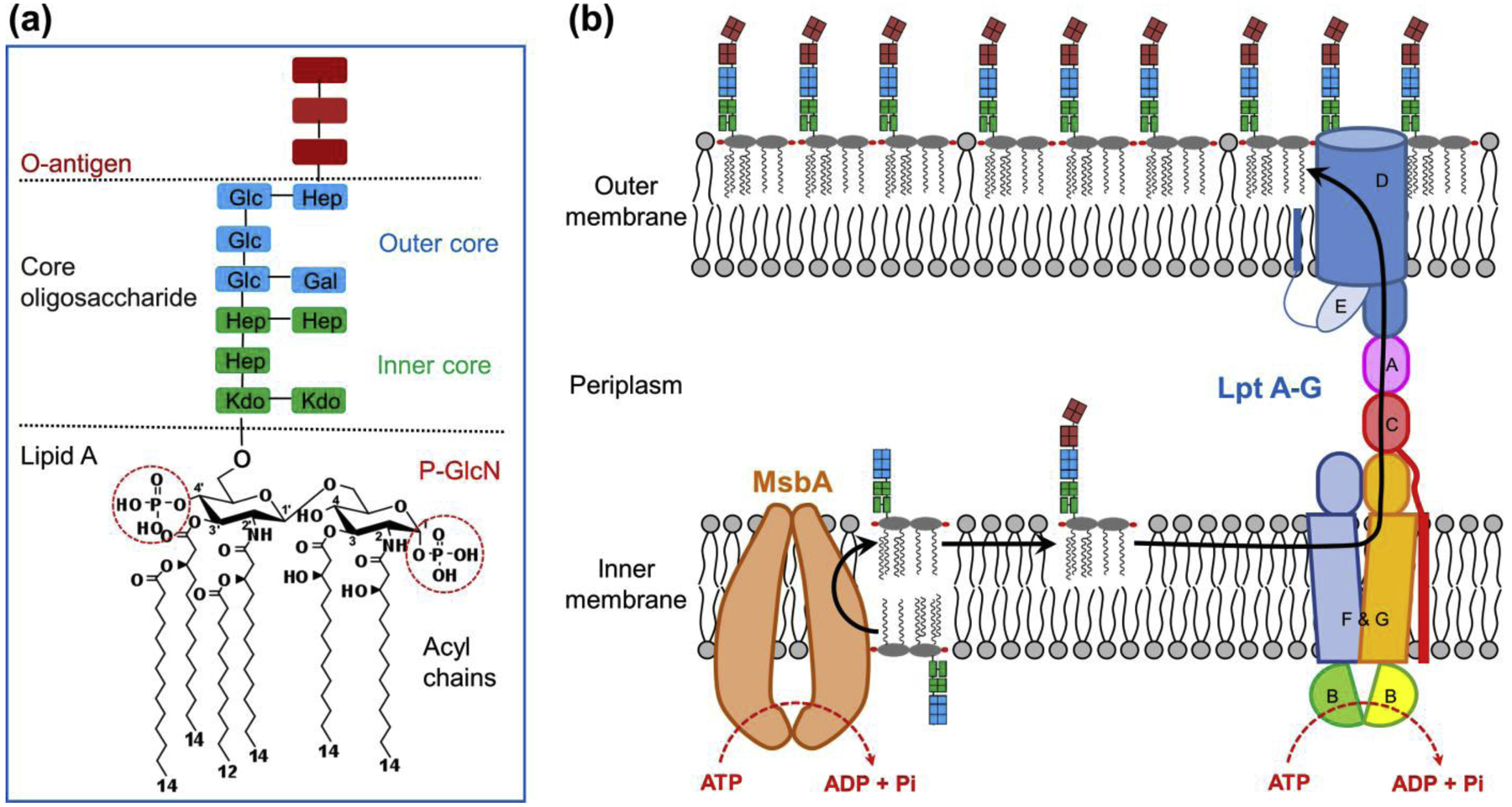
(a) Lipopolysaccharide (LPS) structure in Escherichia coli. LPS is composed of three portions: lipid A, core oligosaccharide, and O-antigen. Lipid A has a glucosamine disaccharide with phosphate groups at the 1- and 4′-positions (phospho-N-acetylglucosamine, or P-GlcN), and primary acyl chains at the 2-, 3-, 2′- and 3′-positions. Two secondary acyl chains are attached to the 2′ and 3′ primary acyl chains. The core oligosaccharide is composed of inner and outer saccharides, including 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo), heptose (Hep), glucose (Glu), and galactose (Gal). The O-antigen consists of many repeats of an oligosaccharide unit. (b) LPS transport pathway in E. coli. Lipid A with core oligosaccharide is synthesized in the cytoplasmic leaflet of the inner membrane (IM). MsbA, a homodimeric ABC transporter, flips the nascent LPS across the IM. The O-antigen is ligated to lipid A-core saccharide on the periplasmic side of the IM. LptB2FGC extracts the mature LPS out of the IM, and subsequently pushes it across the periplasm through LptA and LptDE. LPS is eventually inserted in the outer leaflet of the outer membrane (OM). MsbA and LptB2FGC utilize the energy from ATP hydrolysis to power LPS movement.
During their growth, Gram-negative bacteria assemble millions of LPS molecules in the outermost membrane leaflet, at a speed estimated to be 70,000 molecules per minute[6]. An LPS molecule newly synthesized at the cytoplasmic surface of the IM must cross three cellular compartments (IM, periplasm and OM) before reaching its destination. This remarkable journey of LPS is powered by two ATP-binding cassette (ABC) transporters, MsbA and LptB2FGC[7]. Using the energy of ATP binding and hydrolysis, MsbA flips LPS across the IM and LptB2FGC pushes LPS towards the OM (Fig. 1b).
ABC transporters exist in all kingdoms of life and translocate a multitude of chemically diverse substrates across cell membranes[8,9]. ABC transporters have two transmembrane domains (TMDs), which form the substrate translocation pathway, and two nucleotide binding domains (NBDs), which bind and hydrolyze ATP. Whereas the conformational changes during the functional cycle have been extensively studied for a number of ABC transporters[10–12], much less is known for specific determinants of substrate selectivity and the mechanism underlying tight coupling between substrate translocation and ATP hydrolysis. In this regard, the two ABC transporters (MsbA and LptB2FGC), which work on the same substrate (LPS) but perform completely different tasks, provide an ideal case study for understanding how ABC transporters achieve their extraordinary versatility, specificity and efficiency.
In this review, we summarize recent progress in understanding the mechanistic details of MsbA and LptB2FGC. We highlight the structural analyses and accompanying biochemical data which have revealed the basis of LPS translocation driven by these two ABC transporters, and discuss how these insights may help develop novel antibiotics to combat important bacterial pathogens.
MsbA and LPS flipping: a well-studied protein and an unsolved mystery
MsbA has long been a popular model system for studying ABC transporters, due to its excellent stability and biochemical behavior, as well as to its structural similarity with human multidrug efflux pumps such as P-glycoprotein. As a result, much of the current paradigm of ABC exporter function has been established using results from MsbA. The first set of structures of MsbA in detergent were obtained using X-ray crystallography over a decade ago[13]. These include two different inward-facing open conformations of nucleotide-free MsbA, and one outward-facing open conformation for MsbA bound to AMPPNP (5′-adenylyl-β,γ-imidodiphosphate) or ADP-vanadate. These crystal structures and following spectroscopy studies[14–18] proposed an alternating access transport mechanism, in which LPS from the cytoplasmic leaflet of the IM binds to the inner cavity of MsbA in its inward-facing open conformation, and the subsequent ATP binding shifts MsbA to an outward-facing state in which LPS is released to the periplasmic side. However, because of the absence of LPS in these structures, how MsbA specifically recognizes LPS and drives its 180° rotation remained largely unknown.
MsbA-LPS interaction
By combining single-particle cryo-electron microscopy (cryo-EM)[19–21] and lipid nanodisc technology[22], a breakthrough was made in understanding MsbA-driven LPS flipping[23]. The cryo-EM structure of MsbA in a native-like lipid bilayer clearly resolved a co-purified LPS molecule between the two TMDs of MsbA (Fig. 2a, b). This is the first visualization of LPS in the context of its transport protein machinery, revealing how the inner cavity of MsbA forms highly specific contact with the unique chemical signatures in LPS. Later a crystal structure of an inhibitor-bound MsbA also reveals a similarly bound LPS[24]. Surrounding the negatively charged phosphorylated glucosamines of LPS, a ring of positively charged amino acid residues from both MsbA subunits forms strong electrostatic interactions with LPS (Fig. 2c). Above the glucosamines, the lipid acyl chains are accommodated in a large hydrophobic pocket, which is sealed off from the aqueous periplasm. Notably, the size and shape of the inner cavity of MsbA are complementary to the corresponding portions in LPS such that MsbA would preferentially accommodate the 12–14 carbon chains of LPS acyl tails over a longer phospholipid (i.e. ‘hydrocarbon ruler’)[24]. Below the glucosamines, the inner core of LPS is directly exposed in the cytoplasm, showing no clear contact with MsbA. Taken together, the negatively charged electrostatic surface and the hydrophobic pocket in the inner cavity of MsbA simultaneously interact with two regions of LPS (glucosamines and acyl chains) with distinct surface properties, thus contributing to stable and highly specific LPS binding. These features are highly similar to those seen in the structure of the TLR4–MD-2–LPS complex[25], indicating that proteins with completely different folds use the same strategy to recognize LPS.
Figure 2 |. Cryo-EM structure of MsbA in nanodiscs.
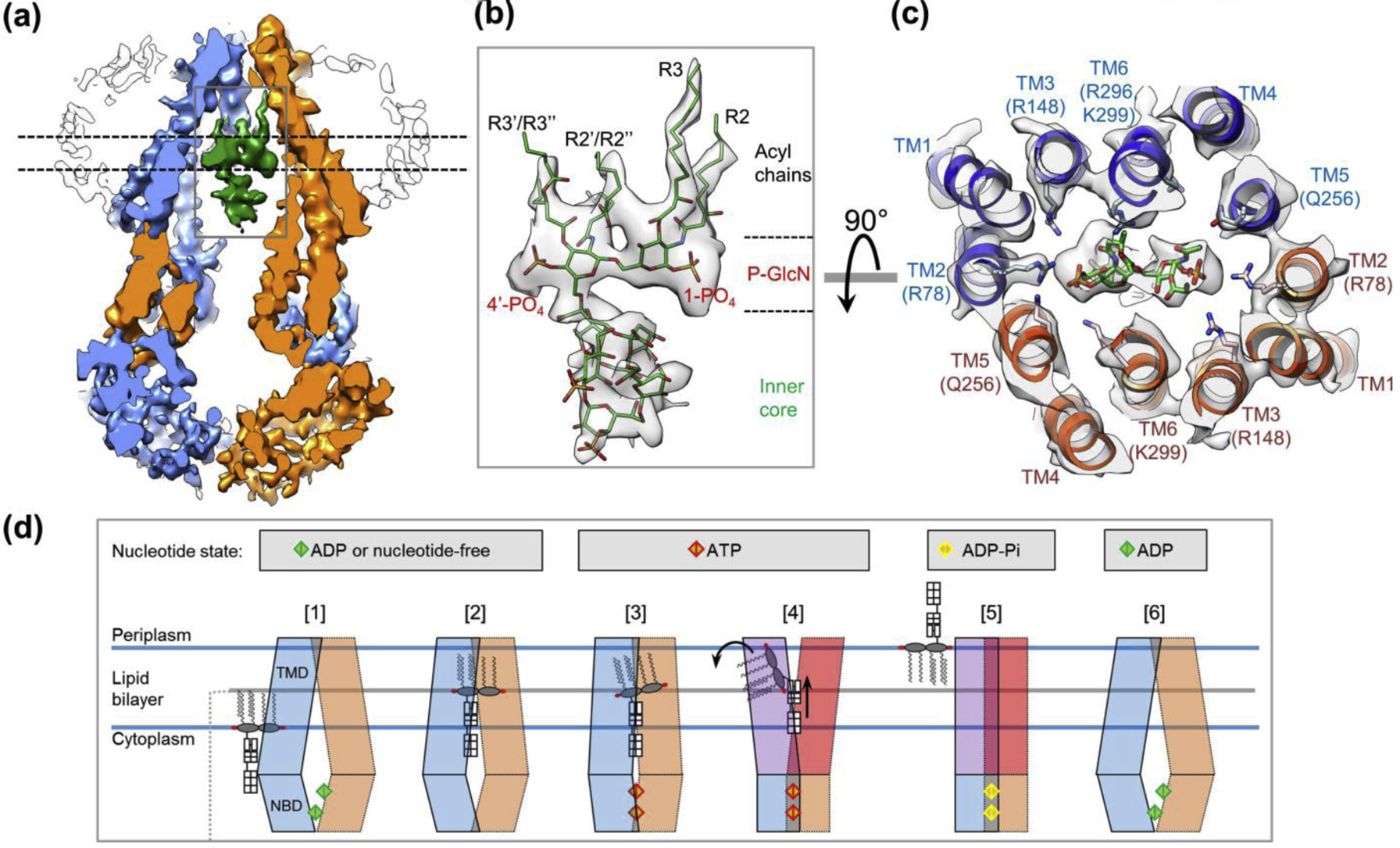
(a) Slice view of the cryo-EM map of MsbA with bound LPS[23]. The two MsbA subunits and LPS are colored in blue, orange and green. Nanodisc is shown as outline. Within the inner cavity of MsbA, LPS is elevated relative to the cytoplasmic membrane leaflet. (b) Close-up view of LPS as highlighted by the rectangle in a. Cryo-EM map (grey) is superimposed with the model of LPS (green). (c) Cross-section of the cryo-EM map of MsbA superimposed with the associated model is viewed from the periplasm in perpendicular to the membrane plane, at the level of the phosphorylated glucosamine (P-GlcN) as indicated by dashed lines in a. (d) Proposed ‘trap and flip’ model for LPS flipping by MsbA. Proposed nucleotide states are indicated at top. 1) LPS in the cytoplasmic leaflet of the IM enter the central cavity of MsbA in its inward-facing open conformation. 2) LPS achieves stable binding through electrostatic and hydrophobic interactions, which restrict the opening of MsbA and align two NBDs for ATP binding. 3) ATP binding induces conformational changes of TMDs to abolish the LPS-MsbA interactions. 4) The outward opening of TMDs exposes the hydrophobic acyl chains of LPS to the aqueous periplasm, and the incompatibility causes the rotation of acyl chains into the periplasmic leaflet of the IM and also upward movement of the core oligosaccharides. 5) After LPS is flipped out, the TM helices move inward and collapse the central cavity, and the ATP is hydrolyzed. 6) MsbA returns to the inward-facing open conformation for the next round LPS flipping.
Inside the MsbA central cavity, LPS acyl chains are located at the level of the periplasmic membrane leaflet (Fig. 2a and step 2 in Fig. 2d). Thus, the cryo-EM structure reveals a previously unrecognized functional state, in which LPS has accomplished much of the translocation across the IM, but without the upside-down rotation. This state is presumably followed by the actual flipping of LPS. We named this process as “trap and flip” (Fig. 2d). This proposed model emphasizes the finding that LPS first undergoes translation within the membrane bilayer without rotation, which had not been observed or speculated as a mechanism for lipid flippases. Interestingly, in a recent cryo-EM structure of a type IV P-type ATPase that translocates phosphatidylserine from the outer to the inner membrane leaflet, which is opposite to the direction of MsbA-mediated LPS flipping, the bound phospholipid has moved towards the target leaflet without flipping its acyl chains[26]. Thus, translation followed by rotation may be a general scheme of ATP-powered lipid flipping.
LPS interacts dynamically and intimately with MsbA throughout its functional cycle, and contributes positively to the high transport efficiency. The LPS binding to MsbA brings two NBDs in proper positions to facilitate ATP hydrolysis[23,24]. Comparison between the nucleotide-free and nucleotide-bound conformations of MsbA further demonstrates that ATP binding-induced movement of TMDs, as well as tight dimerization of NBD and ATP hydrolysis, would not occur unless the bound LPS is moved out of the central cavity (step 3–4 in Fig. 2d). Thus, the translocation of LPS towards the periplasm, conformational changes of MsbA, and ATP hydrolysis must occur in a concerted process. Therefore, rather than a cargo being passively moved, LPS appears more like an essential constituent to maximize the efficiency of the MsbA molecular machine.
MsbA inhibitors
The first MsbA-specific inhibitors were not reported until mid-2018 by two different groups[24,27]. A quinoline class of molecules identified by Genentech selectively inhibits the activity of MsbA and displays bactericidal activity[24,28]. The crystal structures of inhibitor-bound MsbA show that these compounds bind the TMDs and lock MsbA in an inward-facing wide-open conformation. A copurified LPS is resolved inside the central cavity demonstrating similar interactions as those in the cryo-EM structure[23]. The other class of inhibitor was identified from a phenotypic screen, which was designed to target the proteins involved in LPS biogenesis[27]. Despite low bactericidal efficacy, this compound has an interesting property of stimulating the activity of MsbA for ATP hydrolysis while blocking LPS transport, suggesting an action mode distinct from the MsbA inhibitors from Genentech[24].
Lpt machinery with two mysterious pieces: LPS and LptC
After being flipped across the IM by MsbA, LPS in the periplasmic leaflet of the IM travels across the periplasm to the OM, a process mediated by the seven Lpt proteins (LptA through LptG) (Fig. 1b). The structures of individual Lpt proteins or domains have been determined[29–34], and reveal a common β-jellyroll like fold for essentially all the domains that are located in the periplasm, including LptA and the periplasmic domains from LptF, LptG, LptC, and LptD. Combined with this structural knowledge, biochemical experiments have supported what has been termed the “PEZ” model, in which Lpt proteins form a physically connected bridge spanning across the periplasm to shuttle LPS from the IM to the OM[35,36]. LptB2FGC is the sole energy source for LPS extraction out of the IM and LPS movement towards the OM. The concave inner surface of the β-jellyroll domains is hypothesized to protect the hydrophobic acyl chains of LPS during its translocation across the aqueous periplasm. However, the mechanistic details of LPS transport through Lpt machinery remained largely speculative, mainly due to the lack of LPS resolved in the structures of Lpt proteins.
The ABC transporter complex LptB2FGC has several distinct features compared to MsbA. First, LptB2FGC has various domains made of individual proteins. Second, in addition to two TMDs (TM helices from LptF and LptG) and two NBDs (two LptB subunits), it contains a stably associated LptC protein which has a single N-terminal transmembrane segment and a C-terminal β-jellyroll domain in the periplasm[34]. However, the function of LptC has been mysterious. The periplasmic domain of LptC is essential for LPS transport and bacterial growth[37,38]. The TM helix of LptC is dispensable for bacterial survival in laboratory conditions, even though all natural LptC proteins contain the TM helix[37].
LptB2FGC-LPS interaction
The first glimpse of the LptB2FG complex (without LptC) was provided by two crystal structures of LptB2FG from Pseudomonas aeruginosa and Klebsiella pneumoniae[31,32], which display a similar architecture. Between the TMDs formed by LptF and LptG there is a large hydrophobic pocket presumably for LPS binding. However, LPS was not resolved in either structure, and it was unclear what conformation of LptB2FG is competent for LPS binding. Later the cryo-EM structure of nucleotide-free Escherichia coli LptB2FG in nanodiscs resolved a co-purified LPS molecule[23] (Fig. 3a–c). The six lipid acyl chains are tightly packed inside the hydrophobic pocket, and their intimate contact with a number of amino acid residues is directly visualized in the EM density (Fig. 3c). Mutations of many of these residues generate strong inhibitory effects on bacterial growth, indicating their functional importance for LPS binding and transport[31,32,39]. A cluster of positively charged side chains surround the phosphorylated glucosamines of LPS, and most of these residues are from LptG (Fig. 3f). Thus, although LptF and LptG are structurally homologous, their roles in LPS binding and translocation are not equivalent. This is in contrast with MsbA, where the electrostatic interactions around the glucosamines are contributed symmetrically by two identical subunits[23] (Fig. 2c).
Figure 3 |. Cryo-EM structures of LptB2FG and LptB2FGC in nanodiscs.
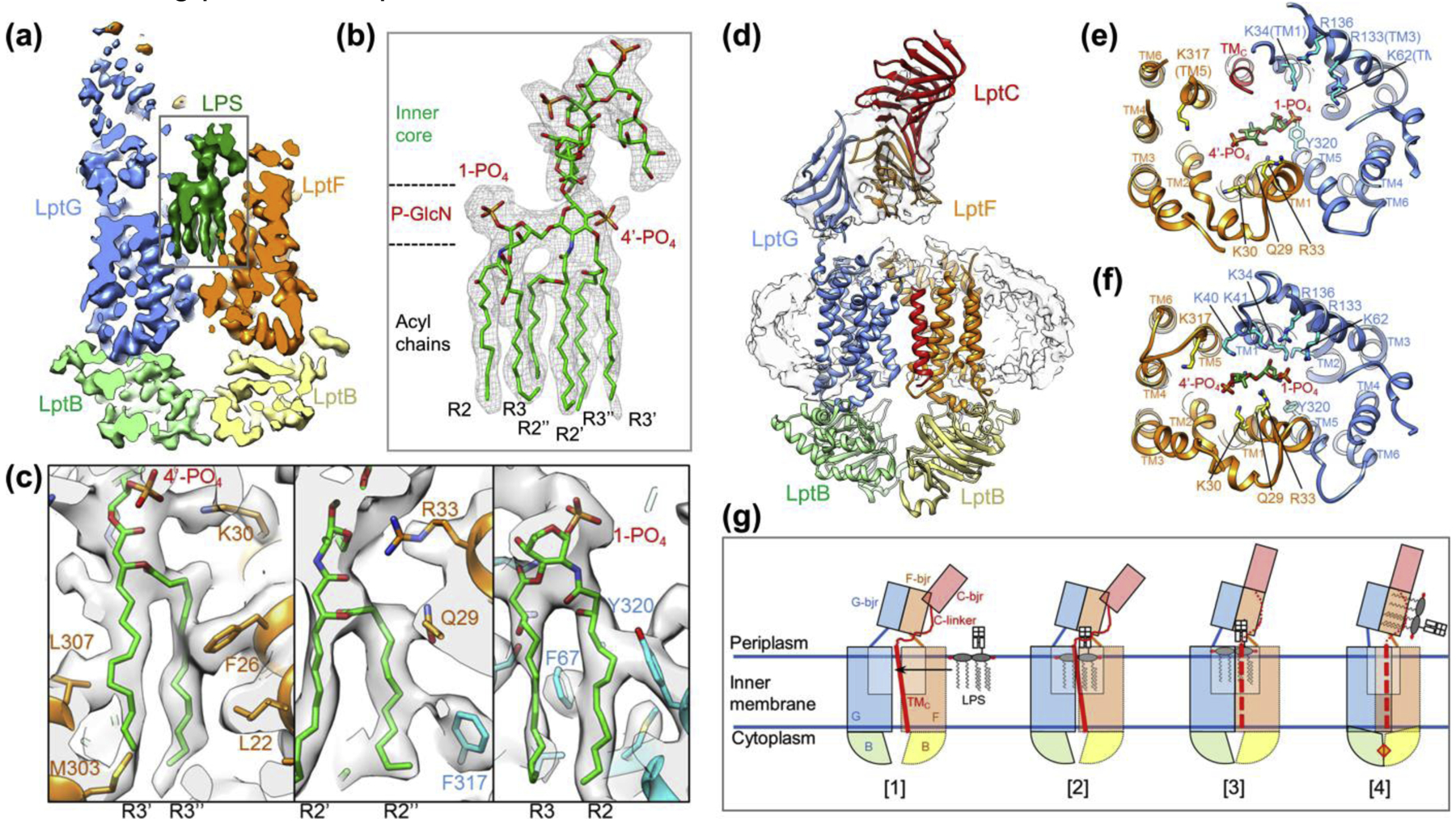
(a) Slice view of the cryo-EM map of LptB2FG with bound LPS[40]. The Lpt proteins and LPS are colored differently. (b) Close-up view of LPS as highlighted by the rectangle in a. The cryo-EM density is of sufficient quality to directly visualize all six acyl chains, phosphorylated glucosamines (P-GlcN), and inner core sugars. (c) Zoomed-in views of LPS binding pocket. (d) Cryo-EM map and associated model of LptB2FGC. The TM helix of LptC (red) binds between the two TMDs formed by LptF and LptG. The β-jellyroll domain of LptC stacks on top of the β-jellyroll domain of LptF (orange), forming a continuous groove path. (e, f) Views from the periplasm of the LPS binding pocket for LptB2FGC (e) and LptB2FG (f). The presence of the LptC TM helix causes the TM helices of LptF and LptG to spread apart (e). In the absence of LptC the two halves of the transporter move closer together, providing for a tight electrostatic lock on the bound LPS (f). (g) Proposed ‘lock and squeeze’ model for LPS extraction by LptB2FGC. 1) LPS traverses laterally from the outer leaflet of the IM into the central cavity of LptB2FGC. 2) Binding of LPS in the central cavity induces the closure of TMDs to lock the bound LPS, and promotes the release of the LptC TM helix. 3) The LptC TM helix (red dashed line) is dissociated from LptB2FG, which facilitates the attachment of the β-jellyroll domain of LptC onto LptF β-jellyroll domain. 4) ATP binding induces tight dimerization of LptB subunits, and causes collapse of the LPS binding pocket to squeeze out the bound LPS.
The cryo-EM structure of E. coli LptB2FGC[40] (Fig. 3d) and two crystal structures of LptB2FGC from Vibrio cholerae and Enterobacter cloacae[41] (Fig. 4e, f) demonstrate a similar architecture of the complex. In these structures, the β-jellyroll domain of LptC is located on top of the β-jellyroll domain of LptF, with essentially no contact with LptG. The concave space in the β-jellyroll domains of LptC and LptF is aligned to form a continuous hydrophobic path, which presumably accommodates the acyl chains of LPS after its extrusion out of the TMDs. Surprisingly, the TM helix of LptC is present between the two TMDs (Fig. 3d, e), mostly interacting with the TM5 of LptF through extensive hydrophobic interactions. The nature of this interaction explains why the length and overall hydrophobicity, but not the exact residues, of the TM helix of LptC are conserved[34]. The presence of an extra TM helix in between two TMDs of an ABC transporter is unprecedented, indicating a novel mechanism for regulation of transporter activity. Indeed, the ATPase activity of LptB2FGC is substantially lower than that of LptB2FG[36,40,41]. Consistent with the notion that an extra TM helix located between the TMDs would interfere with the conformational changes required for ATP hydrolysis, in the cryo-EM structure of LptB2FGC trapped by ADP-vanadate, the TM helix of LptC is dissociated from the TMD interface, allowing the inward movement of TMDs and tight dimerization of NBDs[40].
Figure 4 |. Dynamic β-jellyroll domains.
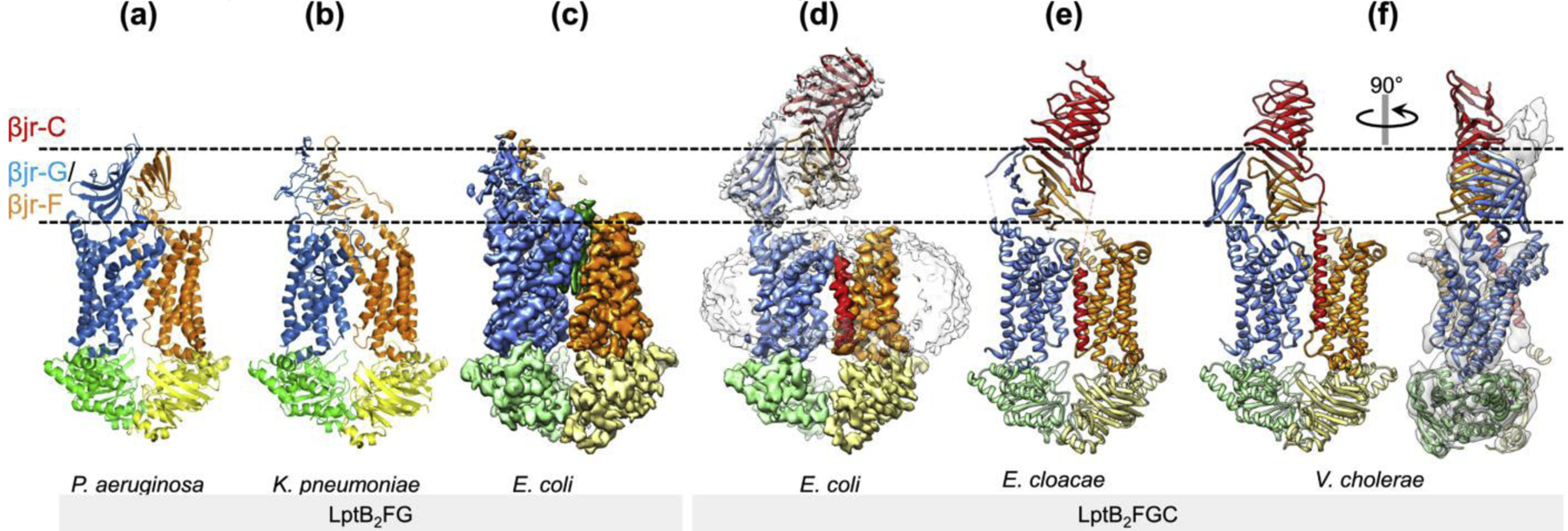
Surface views of the crystal (a, b, e, f) and cryo-EM structures (c, d) of LptB2FG[31,32] and LptB2FGC[40,41] show conformational plasticity of the periplasmic β-jellyroll domains. The regions corresponding to the β-jellyroll domains of LptF/LptG and LptC are delineated by dashed lines. In f, the crystal structure of LptB2FGC from V. cholerae is superimposed onto the cryo-EM map of E. coli LptB2FGC (grey as in d), demonstrating the different orientations of the β-jellyroll domains in these two structures.
Photocrosslinking experiments demonstrate that LPS enters LptB2FGC in the region where the TM helix of LptC is sandwiched by the two TMDs, but not on the opposite side of the transporter[41]. In the cryo-EM structure of LptB2FGC, a relatively weak LPS density is observed in the central pocket, and most of the residues that form electrostatic interactions with LPS in LptB2FG are outside the range for contact (Fig. 3e, f)[40]. This suggests that removal of the TM helix of LptC occurs in concert with the establishment of tight binding to LPS. In the cryo-EM structure of LptB2FGC trapped with ADP-vanadate, the central pocket is completely collapsed without any substrate inside, thus representing a state after LPS exit. Taken together, the results from cryo-EM analysis suggest a “lock and squeeze” model for LptB2FGC-driven LPS extraction from the IM (Fig. 3g). In this model, ATP binding or hydrolysis is not needed for LPS entry but required for LPS translocation from the inner cavity to the β- jellyroll domains, and the TM helix of LptC plays a crucial role in sensing the presence of bound LPS to prevent futile ATP hydrolysis. These notions are supported by the results from photocrosslinking experiments that detect the locations of LPS in the Lpt complex in different ATP states[41].
Lpt β-jellyroll bridge
The β-jellyroll domains of LptF and LptG in the cryo-EM structures of E. coli LptB2FG and LptB2FGC demonstrate two different orientations (Fig. 4c, d), and the structures of LptB2FGC from three different species also demonstrate different titling angles of the β-jellyroll domains (Fig. 4d–f). Interestingly, in the structure of V. cholerae LptB2FGC with the most upright β-jellyroll domains, the LptF β-jellyroll changes its conformation to close its concave path, likely to block backward flow of LPS[41]. Taken together, both the inter-β-jellyroll interactions and the conformation within each β-jellyroll appear to be highly dynamic.
3D classification of cryo-EM particle images of LptB2FGC indicated that the stable attachment of the β-jellyroll domain of LptC on top of LptF is inhibited when the TM helix of LptC is sandwiched between the TMDs; conversely, the interaction between the β-jellyroll domains of LptC and LptF is enhanced when the TM helix of LptC is dissociated from the TMD interface, as observed in the cryo-EM structure of LptB2FGC trapped by ADP-vanadate[40]. These findings indicate that the stable interaction between the β-jellyroll domains of LptC and LptF occurs preferentially when LPS is bound inside the TMDs and the TM helix of LptC is released from the TMD interface. Thus, the β-jellyroll bridge in the periplasm is unlikely to be static or assembled by default. The structural plasticity and dynamic interactions of the β-jellyroll domains likely contribute to high efficiency of LptB2FGC in LPS transport, at least in part, by preferentially connecting the LPS-loaded LptB2FGC to LptA in the periplasm.
Lpt inhibitors
A few compounds have been identified to bind the β-jellyroll domains of Lpt proteins and block LPS transport, including a class of macrocyclic peptidomimetics[42–44], thanatin[45] and IMB-881[46]. Novobiocin, an antibiotic against Gram-positive bacteria by inhibiting DNA gyrase, was co-crystallized with isolated LptB and increases LPS transport instead of inhibiting it[47]. However, when cocrystallized with LptB2FGC, novobicin binds a functionally irrelevant site rather than the interface between LptF/LptG and LptB[41]. Thus, the action mechanism of novobiocin in Gram-negative bacteria has yet to be defined. No small-molecule compounds have been reported to bind the TMDs of LptB2FGC.
Perspectives
A great deal of structural information for the LPS flipping and transport machinery has become available over the past two years. Particularly, single-particle cryo-EM has resolved LPS bound to MsbA and Lpt proteins for the first time[23,40], and demonstrated its power in studying membrane proteins in native-like bilayer membrane, characterizing structural rearrangement of ABC transporters during their functional cycles, and analyzing the conformational ensemble of flexible domains. Despite the great progress, many important questions related to LPS transport remain to be addressed. How does LPS travel into the central cavities of these transporters? Are there unidentified intermediate conformations? What are the structural details of the transporters during LPS exit? What is the architecture of fully assembled Lpt protein bridge? Future structural studies together with biochemical, functional, and spectroscopic approaches will generate deeper insights and probably more unexpected discoveries.
The development of antibiotics to block LPS transport is still in its infancy. It is worth noting that the conformation of MsbA targeted by the compounds developed by Genentech is different from those in all previously published structures[24], and that the binding pocket for these compounds could not be predicted based on available structures. Like other ABC transporters, MsbA and LptB2FGC undergo large conformational transition during their functional cycles, and we know very little about which conformations can be effectively targeted by small molecules. Future structural studies of these essential transporters bound to different classes of inhibitors will be crucial for mapping all druggable conformations. Armed with the wealth of knowledge, the search for new and improved antibiotics that target LPS transport will help solve the worldwide antibiotic crisis.
Acknowledgements
We thank Dr. Wei Mi for critical reading and the members of the Liao laboratory for helpful discussion. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (R01 GM122797) to M.L.. B.J.O. was supported by a Postdoctoral Fellowship, PF-17-212-01 - DMC, from the American Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Nothing declared.
References
- 1.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J: Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 2009, 48:1–12. [DOI] [PubMed] [Google Scholar]
- 2.Raetz CR, Reynolds CM, Trent MS, Bishop RE: Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem 2007, 76:295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitfield C, Trent MS: Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem 2014, 83:99–128. [DOI] [PubMed] [Google Scholar]
- 4.Rathinam VAK, Zhao Y, Shao F: Innate immunity to intracellular LPS. Nat Immunol 2019, 20:527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson BW, Trent MS: Pushing the envelope: LPS modifications and their consequences. Nat Rev Microbiol 2019, 17:403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lima S, Guo MS, Chaba R, Gross CA, Sauer RT: Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 2013, 340:837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruiz N, Kahne D, Silhavy TJ: Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat Rev Microbiol 2009, 7:677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland IB: ABC transporters, mechanisms and biology: an overview. Essays Biochem 2011, 50:1–17. [DOI] [PubMed] [Google Scholar]
- 9.Rees DC, Johnson E, Lewinson O: ABC transporters: the power to change. Nat Rev Mol Cell Biol 2009, 10:218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Locher KP: Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol 2016, 23:487–493. [DOI] [PubMed] [Google Scholar]
- 11.Thomas C, Tampe R: Multifaceted structures and mechanisms of ABC transport systems in health and disease. Curr Opin Struct Biol 2018, 51:116–128. [DOI] [PubMed] [Google Scholar]
- 12.Srikant S, Gaudet R: Mechanics and pharmacology of substrate selection and transport by eukaryotic ABC exporters. Nat Struct Mol Biol 2019, 26:792–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward A, Reyes CL, Yu J, Roth CB, Chang G: Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc Natl Acad Sci U S A 2007, 104:19005–19010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong J, Yang G, McHaourab HS: Structural basis of energy transduction in the transport cycle of MsbA. Science 2005, 308:1023–1028. [DOI] [PubMed] [Google Scholar]
- 15.Borbat PP, Surendhran K, Bortolus M, Zou P, Freed JH, McHaourab HS: Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol 2007, 5:e271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou P, McHaourab HS: Alternating access of the putative substrate-binding chamber in the ABC transporter MsbA. J Mol Biol 2009, 393:574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou P, Bortolus M, McHaourab HS: Conformational cycle of the ABC transporter MsbA in liposomes: detailed analysis using double electron-electron resonance spectroscopy. J Mol Biol 2009, 393:586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collauto A, Mishra S, Litvinov A, McHaourab HS, Goldfarb D: Direct Spectroscopic Detection of ATP Turnover Reveals Mechanistic Divergence of ABC Exporters. Structure 2017, 25:1264–1274 e1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyumkis D: Challenges and opportunities in cryo-EM single-particle analysis. J Biol Chem 2019, 294:5181–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danev R, Yanagisawa H, Kikkawa M: Cryo-Electron Microscopy Methodology: Current Aspects and Future Directions. Trends Biochem Sci 2019. [DOI] [PubMed] [Google Scholar]
- 21.Cheng Y, Grigorieff N, Penczek PA, Walz T: A primer to single-particle cryo-electron microscopy. Cell 2015, 161:438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denisov IG, Sligar SG: Nanodiscs in Membrane Biochemistry and Biophysics. Chem Rev 2017, 117:4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi W, Li Y, Yoon SH, Ernst RK, Walz T, Liao M: Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 2017, 549:233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho H, Miu A, Alexander MK, Garcia NK, Oh A, Zilberleyb I, Reichelt M, Austin CD, Tam C, Shriver S, et al. : Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 2018, 557:196–201. [DOI] [PubMed] [Google Scholar]; Genentech discovered a class of compounds that inhibit the ATPase activity of MsbA and have bactericidal activity. The crystal structure of MsbA bound to one of these inhibitors demonstrates an inward-facing open conformation with an LPS molecule bound in the inner cavity. This is the first time that the mechanism for an MsbA inhibitor is revealed at the structural level. The unprecedented conformation and the compound binding pocket are valuable for further antibiotic development.
- 25.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO: The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 2009, 458:1191–1195. [DOI] [PubMed] [Google Scholar]
- 26.Hiraizumi M, Yamashita K, Nishizawa T, Nureki O: Cryo-EM structures capture the transport cycle of the P4-ATPase flippase. Science 2019, 365:1149–1155. [DOI] [PubMed] [Google Scholar]
- 27.Zhang G, Baidin V, Pahil KS, Moison E, Tomasek D, Ramadoss NS, Chatterjee AK, McNamara CW, Young TS, Schultz PG, et al. : Cell-based screen for discovering lipopolysaccharide biogenesis inhibitors. Proc Natl Acad Sci U S A 2018, 115:6834–6839. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work presents a cell-based screen that identifies inhibitors of LPS biosynthesis and transport. A compound was identified to bind MsbA and inhibit LPS transport, and interestingly the ATPase activity of MsbA is stimulated by this compound.
- 28.Alexander MK, Miu A, Oh A, Reichelt M, Ho H, Chalouni C, Labadie S, Wang L, Liang J, Nickerson NN, et al. : Disrupting Gram-negative bacterial outer membrane biosynthesis through inhibition of the lipopolysaccharide transporter MsbA. Antimicrob Agents Chemother 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Botos I, Majdalani N, Mayclin SJ, McCarthy JG, Lundquist K, Wojtowicz D, Barnard TJ, Gumbart JC, Buchanan SK: Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens. Structure 2016, 24:965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong H, Xiang Q, Gu Y, Wang Z, Paterson NG, Stansfeld PJ, He C, Zhang Y, Wang W, Dong C: Structural basis for outer membrane lipopolysaccharide insertion. Nature 2014, 511:52–56. [DOI] [PubMed] [Google Scholar]
- 31.Dong H, Zhang Z, Tang X, Paterson NG, Dong C: Structural and functional insights into the lipopolysaccharide ABC transporter LptB2FG. Nat Commun 2017, 8:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo Q, Yang X, Yu S, Shi H, Wang K, Xiao L, Zhu G, Sun C, Li T, Li D, et al. : Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat Struct Mol Biol 2017, 24:469–474. [DOI] [PubMed] [Google Scholar]
- 33.Suits MD, Sperandeo P, Deho G, Polissi A, Jia Z: Novel structure of the conserved gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J Mol Biol 2008, 380:476–488. [DOI] [PubMed] [Google Scholar]
- 34.Tran AX, Dong C, Whitfield C: Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J Biol Chem 2010, 285:33529–33539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D: Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 2016, 14:337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman DJ, Xie R, Taylor RJ, George AH, Okuda S, Foster PJ, Needleman DJ, Kahne D: Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 2018, 359:798–801. [DOI] [PMC free article] [PubMed] [Google Scholar]; The Lpt protein complexes in the inner and outer membrane were reconstituted into separate proteoliposomes, and the LPS movement between the stably associated membranes was observed. This is an elegant in vitro assay to support the PEZ dispenser model in which LPS molecules are pushed one after another along a physically connected protein bridge.
- 37.Villa R, Martorana AM, Okuda S, Gourlay LJ, Nardini M, Sperandeo P, Deho G, Bolognesi M, Kahne D, Polissi A: The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J Bacteriol 2013, 195:1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benedet M, Falchi FA, Puccio S, Di Benedetto C, Peano C, Polissi A, Deho G: The Lack of the Essential LptC Protein in the Trans-Envelope Lipopolysaccharide Transport Machine Is Circumvented by Suppressor Mutations in LptF, an Inner Membrane Component of the Escherichia coli Transporter. PLoS One 2016, 11:e0161354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertani BR, Taylor RJ, Nagy E, Kahne D, Ruiz N: A cluster of residues in the lipopolysaccharide exporter that selects substrate variants for transport to the outer membrane. Mol Microbiol 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Orlando BJ, Liao M: Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 2019, 567:486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work presents a series of cryo-EM structures of nanodisc-embedded LptB2FG and LptB2FGC in nucleotide-free and vanadate-trapped conformations. The cryo-EM map of nucleotide-free LptB2FG contains a high-quality EM density of the bound lipid A-inner core molecule, demonstrating unprecedented details of LPS-transporter interaction. This study also reveals the unexpected role of LptC in regulating the action of LptB2FGC. The conformational dynamics of β-jellyroll domains has also been characterized using three-dimensional classification of cryo-EM particle images.
- 41.Owens TW, Taylor RJ, Pahil KS, Bertani BR, Ruiz N, Kruse AC, Kahne D: Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 2019, 567:550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]; Two crystal structures of LptB2FGC from Vibrio cholerae and Enterobacter cloacae show a similar overall architecture as the cryo-EM structure of Escherichia coli LptB2FGC (reference 40), but the orientations and conformations of the β-jellyroll domains in the three structures are different. Elegant photo-crosslinking experiments were also performed to determine the LPS entry gate and the requirement of ATP in specific steps of LPS transport.
- 42.Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, Van der Meijden B, Bernardini F, Lederer A, Dias RL, et al. : Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 2010, 327:1010–1013. [DOI] [PubMed] [Google Scholar]
- 43.Vetterli SU, Moehle K, Robinson JA: Synthesis and antimicrobial activity against Pseudomonas aeruginosa of macrocyclic beta-hairpin peptidomimetic antibiotics containing N-methylated amino acids. Bioorg Med Chem 2016, 24:6332–6339. [DOI] [PubMed] [Google Scholar]
- 44.Werneburg M, Zerbe K, Juhas M, Bigler L, Stalder U, Kaech A, Ziegler U, Obrecht D, Eberl L, Robinson JA: Inhibition of lipopolysaccharide transport to the outer membrane in Pseudomonas aeruginosa by peptidomimetic antibiotics. Chembiochem 2012, 13:1767–1775. [DOI] [PubMed] [Google Scholar]
- 45.Vetterli SU, Zerbe K, Muller M, Urfer M, Mondal M, Wang SY, Moehle K, Zerbe O, Vitale A, Pessi G, et al. : Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Sci Adv 2018, 4:eaau2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X, Li Y, Wang W, Zhang J, Lin Y, Hong B, You X, Song D, Wang Y, Jiang J, et al. : Identification of an anti-Gram-negative bacteria agent disrupting the interaction between lipopolysaccharide transporters LptA and LptC. Int J Antimicrob Agents 2019, 53:442–448. [DOI] [PubMed] [Google Scholar]
- 47.May JM, Owens TW, Mandler MD, Simpson BW, Lazarus MB, Sherman DJ, Davis RM, Okuda S, Massefski W, Ruiz N, et al. : The Antibiotic Novobiocin Binds and Activates the ATPase That Powers Lipopolysaccharide Transport. J Am Chem Soc 2017, 139:17221–17224. [DOI] [PMC free article] [PubMed] [Google Scholar]


