Abstract
Purpose:
Only a minority of patients with advanced non-small-cell lung cancer (NSCLC) truly benefit from single-agent PD-1 checkpoint blockade, and more robust predictive biomarkers are needed.
Experimental design:
We assessed tumor samples from 67 immunotherapy-treated NSCLC cases represented in a tissue microarray, 53 of whom had pre-treatment samples and received monotherapy. Using GeoMx Digital Spatial Profiling system (NanoString), we quantified 39 immune parameters simultaneously in four tissue compartments defined by fluorescence co-localization (tumor [panCK+], leucocytes [CD45+], macrophages [CD68+], and non-immune stroma).
Results:
156 protein variables were generated per case. In the univariate unadjusted analysis, we found 18 markers associated with outcome in spatial context, 5 of which remained significant after multiplicity adjustment. In the multivariate analysis, high levels of CD56 and CD4 measured in the CD45 compartment were the only markers that were predictive for all clinical outcomes, including progression-free survival (PFS) (HR 0.24, p = 0.006; and HR 0.31, p = 0.011, respectively), and overall survival (OS) (HR 0.26, p = 0.014; and HR 0.23, p = 0.007, respectively). Then, using an orthogonal method based on multiplex immunofluorescence and cell counting (inForm), we validated that high CD56+ immune cell counts in the stroma were associated with PFS and OS in the same cohort.
Conclusions:
This pilot scale discovery study shows the potential of the DSP technology in the identification of spatially-informed biomarkers of response to PD-1 checkpoint blockade in NSCLC. We identified a number of relevant candidate immune predictors in spatial context that deserve validation in larger independent cohorts.
Keywords: NSCLC, biomarkers, PD-1 checkpoint blockade, Digital Spatial Profiling, NK and NKT cells, CD4 T cells
Introduction
PD-1 checkpoint blockade is standard of care and a fundamental component in the therapeutic landscape of advanced-stage non-small-cell lung cancer (NSCLC). However, only a minority of NSCLC patients truly benefit from these drugs particularly when given as monotherapies, and more robust predictive biomarkers are needed to optimally deliver these treatments(1).
Several new technologies that facilitate the assessment of multiple markers while preserving the spatial tissue architecture have been developed in recent years(2). These methodologies better characterize the tumor immune microenvironment and are promising tools for immune biomarker discovery. In fact, multiplexed immunohistochemistry (IHC)/immunofluorescence (IF) assays have shown to outperform the accuracy of PD-L1 expression, tumor mutational burden (TMB), and gene expression signatures for predicting response to PD-1 checkpoint blockade across several tumor types(3).
The GeoMx Digital Spatial Profiling (DSP) system (NanoString) is a new platform that enables simultaneous antibody-based detection of multiple proteins from single formalin-fixed paraffin-embedded (FFPE) tissue sections in a quantitative and spatially-resolved manner(4). Due to its high-fold multiplexing capacity from specific regions or marker-selected tissue compartments of interest, it is well suited for the identification of novel spatially-informed tissue biomarkers. In this study, we used DSP technology as a discovery tool to find spatially-resolved protein markers associated with benefit from single-agent PD-1 checkpoint blockade in advanced NSCLC. Then, among the identified candidate predictors, we further assessed CD56 expression by multiplex IF and cell count quantification, with the aim to prove reproducibility in terms of outcome association with an orthogonal quantitative method.
Methods
Patient cohort and tissue microarrays (TMA)
We analyzed retrospectively collected FFPE tumor specimens represented in a TMA format from 81 NSCLC patients treated with PD-1 checkpoint blockade in the advanced setting between 2009-2017 at Yale (YTMA404). All tissue samples were collected and used under the approval from the Yale Human Investigation Committee protocol #9505008219 with an assurance filed with and approved by the US Department of Health and Human Services. The Yale Human Investigation Committee approved the patient inform consent or in some cases waiver of consent all in accordance with the ethical guidelines of the US Common Rule.
For TMA construction, tumors were reviewed by a local pathologist using hematoxylin and eosin–stained preparations to select representative tumor areas. Then, two cores (0.28 mm2 each) were extracted from each tumor block and arrayed in two recipient TMA master-blocks, each TMA block thus containing one non-adjacent 0.28 mm2 tumor core per NSCLC case. Tumor core selection was not based on specific tumor segments or location.
For all the experiments, we assessed two slides derived from two independent YTMA404 blocks, each block containing one non-adjacent tumor core per patient. Sixty-seven cases included in YTMA404 had available or adequate histospots for protein quantification. Of these, 53 had pre-immunotherapy specimens and received single-agent PD-1 checkpoint blockade, constituting our discovery cohort (see consort diagram - supplementary figure S1). Table 1 summarizes the clinicopathologic characteristics of these patients.
Table 1.
Clinical-pathological characteristics of the discovery cohort
| Characteristic | Number of patients (%) |
|---|---|
| Total patients with evaluable tumors | 53 |
| Treatment | |
| Nivolumab | 45 (86) |
| Pembrolizumab | 6 (6) |
| Atezolizumab | 4 (8) |
| Gender | |
| Female | 24 (45) |
| Male | 29 (55) |
| Age | |
| <70yo | 23 (44) |
| ≥70yo | 30 (57) |
| Performance status | |
| 0-1 | 46 (87) |
| >1 | 7 (13) |
| Smoking | |
| Ever smoker | 46 (87) |
| Never smoker | 7 (13) |
| Histology | |
| Adenocarcinoma | 38 (72) |
| Squamous-cell carcinoma | 12 (23) |
| Large-cell carcinoma | 3 (5) |
| Type and site of tumor specimen | |
| Lung primary | 37 (70) |
| Non-lymph node metastasis | 7 (13) |
| Lymph node metastasis | 9 (17) |
| Stage | |
| III | 1 (2) |
| M1a | 13 (24) |
| M1b | 9 (17) |
| M1c | 29 (55) |
| Liver metastasis | |
| Yes | 11 (21) |
| No | 41 (77) |
| Missing | 1 |
| Mutation status | |
| EGFR | 5 (9) |
| KRAS | 15 (28) |
| Others | 5 (9) |
| Wild-type | 28 (53) |
| Derived neutrophil to lymphocyte ratio (dNLR) | |
| ≤3 | 35 (66) |
| >3 | 16 (30) |
| Missing | 2 |
| Lung immune prognostic index (LIPI) score | |
| Good | 22 (41) |
| Intermediate | 19 (36) |
| Poor | 3 (6) |
| Missing | 9 |
| Prior systemic therapies for advanced disease | |
| 0 | 9 (17) |
| 1 | 27 (51) |
| >1 | 17 (32) |
| Best response to immunotherapy | |
| Partial response | 6 (11) |
| Stable disease | 18 (34) |
| Progressive disease | 27 (51) |
| Not evaluable | 2 |
| Benefit from immunotherapy* | |
| Clinical benefit (CB) | 16 (30) |
| Non-clinical benefit (NCB) | 35 (66) |
| Not evaluable | 2 |
We defined clinical benefit (CB) as having experienced partial response or stable disease lasting ≥ 6 months as best response, whereas non-clinical benefit (NCB) was defined as primary progressive disease or stable disease lasting < 6 months.
Digital Spatial Profiling
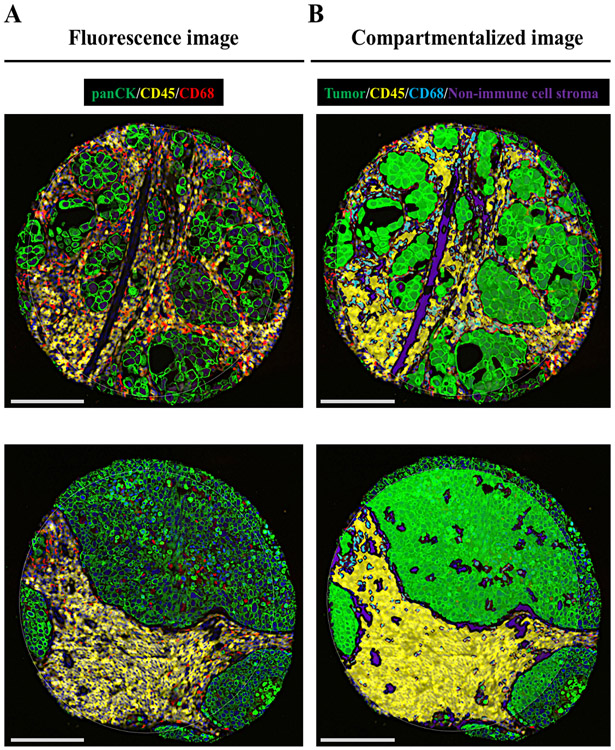
Briefly, once the slides were deparaffined and subjected to antigen retrieval procedures, we co-incubated them overnight with three fluorescent-labeled visualization antibodies to detect tumor cells (pan-cytokeratin [CK]), all immune cells (CD45), and macrophages (CD68), together with a cocktail of 44 unique photocleavable oligonucleotide-labeled primary antibodies targeting immuno-oncology markers (supplementary table S1). Once the staining was completed, we loaded the slides in a prototype Beta version of the GeoMx DSP instrument, where they were scanned to produce a digital fluorescent image of the tissue. Next, we generated individual regions of interest (ROIs) of a maximum of 0.28 mm2 covering the entire TMA core, then each ROI was segmented in four molecularly-defined tissue compartments by fluorescent co-localization: tumor compartment (panCK+), immune cell compartment (CD45+), macrophage compartment (CD68+), and non-immune cell stroma compartment (SYTO13+/panCK−/CD45−/CD68−) (Figure 1). Oligos from these compartments were released upon exposure to UV light in a sequential manner to the macrophage, immune cell, tumor, and finally non-immune cell stromal compartments. Photocleaved oligos were collected via microcapillary aspiration and dispensed into a 96-well plate, then hybridized to 4-color, 6-spot optical barcodes and finally digitally counted in the nCounter system (NanoString). Digital counts from barcodes corresponding to protein probes were first normalized to internal spike-in controls (ERCCs), and then normalized to the area of their compartment. We systematically excluded those compartments with less than 10 nuclei or an area of illumination (AOI) less than 100 μm2. A more detailed description of the protocol can be found in supplementary methods.
Figure 1.
Representative TMA spots showing the fluorescence image (A) and the compartmentalized image created by fluorescence co-localization (B) using GeoMx DSP (scale bar = 100 μm)
Multiplexed IF NK cell panel and cell counting
We performed a multiplexed IF staining protocol for simultaneous detection of CK+ tumor cells, CD3+ lymphocytes, and CD56+ cells. The protocol is detailed in supplementary methods.
We determined cell counts using the inForm Tissue Finder software (Akoya) on multispectral images acquired using a Vectra 3 system (PerkinElmer), as previously described(5) (supplementary methods). First, automated tissue segmentation identified tumor and stroma regions, and two tissue compartments were generated: tumor compartment (DAPI+/CK+) and non-tumor or stromal compartment (DAPI+/CK−). Therefore, in this case, the stromal compartment includes both the CD45+ immune cell compartment and the non-immune cell stromal compartment that were separately generated with DSP. Next, cell segmentation within these regions identified individual cells and respective nuclei, cytoplasm, and membrane components using signal in the nucleus and membrane as internal and external cell borders. Cells were phenotyped based on fluorescent marker intensity, then we calculated the number of individual cell populations as a percentage of the total number of cells in the tumor compartment, the stromal compartment, and the entire TMA core.
Statistical analysis
Pearson’s correlation coefficient was used to analyze the agreement between target counts derived from different tumor regions. For further statistical analysis, we averaged the normalized digital counts or cell counts derived from the two YTMA404 blocks. We stratified each NSCLC case into high and low expression using two exploratory cut-points, median and top tertile. We analyzed the association between target expression and clinical benefit (CB) (supplementary methods and table 1) using binary logistic regression models or the non-parametric Mann-Whitney test. Survival curves were computed with the Kaplan-Meier product-limit method and compared using the log-rank test. We calculated hazard ratios (HR) for progression-free survival (PFS) and overall survival (OS) using the Cox proportional-hazard model. In order to provide more stringent control on false positive results, we used the Benjamini-Hochberg false discovery adjustment method. We applied multiplicity adjustments for PFS and OS associations considering the number of comparisons performed per compartment (tumor, CD45, CD68), and separately for median and top tertile cutpoint comparisons. All hypothesis testing was performed at a two-sided significance level of α=0.05.
Results
We generated 135 ROIs from 67 NSCLC cases, each represented by two TMA cores in two YTMA404 master-blocks. Each ROI was compartmentalized in four tissue compartments, from which 39 protein markers (excluding controls) were separately measured, resulting in 156 quantitative variables per ROI.
First, to assess the performance of the assay, we evaluated the normalized counts of each target relative to non-specific counts (background). To estimate background levels, we averaged the counts from three negative isotype controls for each NSCLC case (supplementary table S1). Most targets showed high signal relative to non-specific counts across all samples (supplementary figure S2). Five markers (PD-1, LAG3, GITR, CD86, and CD40L) showed low signal to background ratios (< 3) across all four tissue compartments in more than 95 % of the TMA spots, and were considered not evaluable for outcome analysis (supplementary figure S3 and S1).
To internally validate the reproducibility of DSP, and also to test the concordance of target count measurements between non-adjacent tumor areas, we compared target counts from each of the two independent cores from each patient, collected in separate DSP runs. In general, abundantly expressed markers in the tumor compartment (e.g. STING or HLA-DR) showed high R2 values (R2 > 0.6), whereas less abundant or heterogenous markers (e.g. CD3 in the tumor compartment or PD-L1 in the CD45 compartment) showed lower R2 values (supplementary figure S4).
Then, we evaluated the association between spatially-informed marker expression and outcome in 53 patients treated with single-agent PD-1 checkpoint blockade. For this analysis, we only included those NSCLC cases with sufficient compartment area for accurate target measurement (≥ 10 nuclei or ≥ 100-μm2 AOI) in the tumor compartment (n = 52), CD68 compartment (n = 47), and CD45 compartment (n = 42) (supplementary figure S1 and S5). Target counts from non-immune cell stroma were considered inadequate for outcome assessment, since immune markers in this compartment were expressed at very low levels (supplementary figure S6).
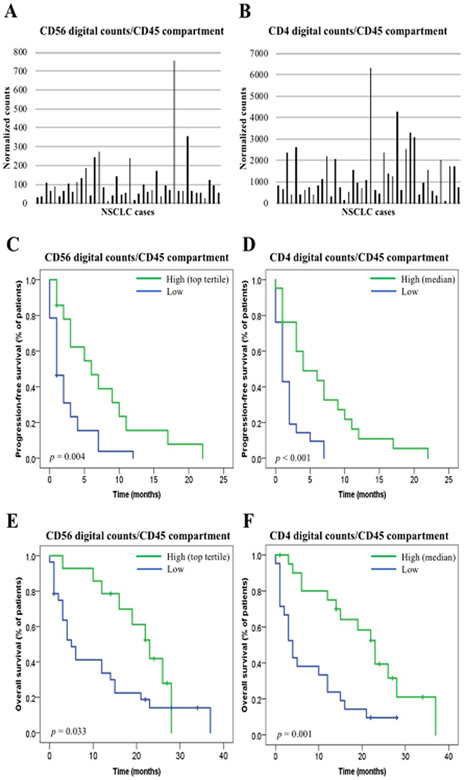
In the univariate unadjusted analysis using two exploratory cutpoints, we found 18 markers associated with PFS and/or OS in spatial context (table 2). After multiplicity adjustment, 5 markers remained significantly associated with outcome: VISTA and CD127 in the tumor compartment, and CD4, Beta-2 microglobulin, and CD3 in the CD45 compartment (table 2). In the multivariate analysis including four clinical prognostic factors (performance status, smoking history, presence of liver metastasis, and LIPI score), high levels of CD56 (top tertile) and CD4 (median) measured in the CD45 compartment were the only markers that were predictive for all clinical outcomes, including durable CB (supplementary table S2), longer PFS (HR 0.24, p = 0.006; and HR 0.31, p = 0.011, respectively), and prolonged OS (HR 0.26, p = 0.014; and HR 0.23, p = 0.007, respectively) (table 2 and figure 2). In contrast, high levels of VISTA (top tertile) and CD127 (top tertile) in the tumor compartment significantly predicted non-clinical benefit (NCB) (supplementary table S2) and shorter PFS (HR 2.49, p = 0.020; and HR 2.39, p = 0.033, respectively), although OS differences did not reach statistical significance (table 2). In this cohort, high PD-L1 expression in the CD45 compartment and the CD68 compartment was associated with longer OS in the univariate analysis (log-rank p = 0.038 and p = 0.035, respectively), although it did not hold significance after adjusting for multiple testing or for clinical prognostic factors in the multivariate model. High PD-L1 expression in the tumor compartment did not show any significant association with outcome (table 2).
Table 2.
Markers significantly associated with PFS and/or OS benefit under PD-1 checkpoint blockade
| Markers associated with PFS benefit | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Compartment | Marker | Cutpoint | Log-rank p |
Adjusted Log-rank p |
Univariate HR (CI 95%) |
p | Adjusted p |
Multivariate HR (CI 95%) |
p |
| Tumor compartment | VISTA | Top tertile | 0.001 | 0.014 | 2.60 (1.37-4.92) | 0.003 | 0.043 | 2.49 (1.15-5.40) | 0.020 |
| CD127 | Top tertile | 0.001 | 0.014 | 2.65 (1.41-4.98) | 0.002 | 0.043 | 2.39 (1.07-5.34) | 0.033 | |
| CD45 compartment | CD56 | Top tertile | 0.004 | 0.124 | 0.38 (0.18-0.80) | 0.011 | 0.341 | 0.24 (0.08-0.66) | 0.006 |
| CD4 | Median | <0.001 | <0.001 | 0.33 (0.16-0.67) | 0.002 | 0.062 | 0.31 (0.12-0.76) | 0.011 | |
| ARG1 | Median | 0.006 | 0.18 | 0.43 (0.21-0.86) | 0.018 | 0.279 | 0.37 (0.16-0.83) | 0.016 | |
| CD68 compartment | CTLA4 | Top tertile | 0.023 | 0.736 | 1.95 (1.01-3.77) | 0.044 | 0.858 | 2.36 (1.06-5.25) | 0.035 |
| Markers associated with OS benefit | |||||||||
| Compartment | Marker | Cutpoint | Log-rank p |
Adjusted Log-rankp |
Univariate HR (CI 95%) |
p | Adjusted p |
Multivariate HR (CI 95%) |
p |
| Tumor compartment | STING | Top tertile | 0.002 | 0.058 | 0.31 (0.14-0.69) | 0.004 | 0.116 | 0.33 (0.12-0.89) | 0.029 |
| CD45 compartment | CD45 | Median | 0.003 | 0.022 | 0.35 (0.16-0.73) | 0.005 | 0.045 | 0.47 (0.15-1.44) | 0.19 |
| CD56 | Top tertile | 0.033 | 0.127 | 0.44 (0.20-0.97) | 0.044 | 0.169 | 0.26 (0.09-0.75) | 0.014 | |
| PD-L1 | Median | 0.038 | 0.167 | 0.48 (0.23-0.99) | 0.049 | 0.20 | 0.43 (0.15-1.23) | 0.11 | |
| CD68 | Top tertile | 0.024 | 0.119 | 0.43 (0.20-0.93) | 0.033 | 0.159 | 0.16 (0.05-0.47) | 0.001 | |
| CD4 | Median | 0.001 | 0.015 | 0.31 (0.15-0.66) | 0.002 | 0.030 | 0.23 (0.08-0.66) | 0.007 | |
| B2M | Median | 0.001 | 0.015 | 0.28 (0.12-0.61) | 0.002 | 0.030 | 0.35 (0.12-0.96) | 0.041 | |
| CD20 | Median | 0.008 | 0.048 | 0.38 (0.18-0.82) | 0.014 | 0.084 | 0.83 (0.32-2.16) | 0.71 | |
| CD3 | Median | <0.001 | <0.001 | 0.24 (0.11-0.53) | <0.001 | <0.001 | 0.24 (0.09-0.64) | 0.005 | |
| CD8 | Top tertile | 0.016 | 0.117 | 0.38 (0.17-0.87) | 0.023 | 0.159 | 0.54 (0.21-1.39) | 0.20 | |
| TIM3 | Median | 0.003 | 0.022 | 0.32 (0.14-0.72) | 0.006 | 0.045 | 0.62 (0.24-1.60) | 0.32 | |
| CD40 | Median | 0.039 | 0.167 | 0.48 (0.24-0.99) | 0.049 | 0.20 | 0.47 (0.17-1.28) | 0.14 | |
| ICOS | Top tertile | 0.006 | 0.093 | 0.35 (0.16-0.78) | 0.010 | 0.155 | 0.26 (0.08-0.79) | 0.018 | |
| CD68 compartment | CD45 | Top tertile | 0.004 | 0.064 | 0.33 (0.15-0.74) | 0.008 | 0.128 | 0.31 (0.11-0.87) | 0.026 |
| PD-L1 | Top tertile | 0.035 | 0.243 | 0.45 (0.21-0.98) | 0.045 | 0.288 | 0.55 (0.22-1.39) | 0.17 | |
| CD20 | Top tertile | 0.004 | 0.064 | 0.33 (0.14-0.74) | 0.007 | 0.128 | 0.56 (0.21-1.45) | 0.23 | |
| GNZB | Top tertile | 0.023 | 0.243 | 0.42 (0.18-0.93) | 0.032 | 0.288 | 0.55 (0.20-1.47) | 0.23 | |
Figure 2.
CD56 and CD4 expression in the CD45 compartment measured by digital counts and their association with outcome. A-B. Histogram showing the distribution of CD56 digital counts (A) and CD4 digital counts (B) in YTMA404 (n = 42); C-D. PFS according to CD56 digital counts in the CD45 compartment (top tertile) (C) and CD4 digital counts in the CD45 compartment (median) (D) (n = 42); E-F. OS according to CD56 digital counts in the CD45 compartment (top tertile) (E) and CD4 digital counts in the CD45 compartment (median) (F) (n = 42)
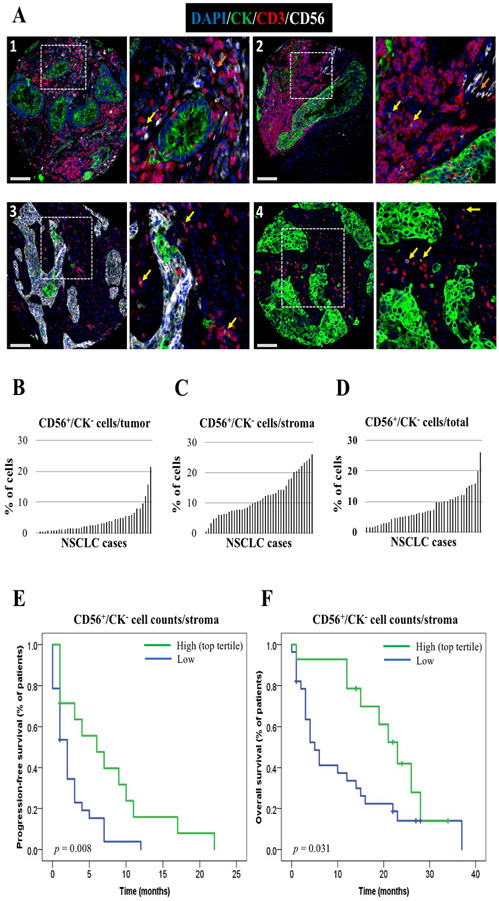
Finally, to be certain that high levels of CD56 in immune-cell stroma were associated with longer PFS and OS in our cohort, we determined its expression using an orthogonal fluorescent-based cell count method in serial YTMA404 sections. For that purpose, we developed a multiplex IF panel to discriminate between CD56+ tumor cells (CD56+/CK+) and CD56+ immune cells (CD56+/CK−, which included CD56+/CD3− NK cells and CD56+/CD3+ NKT cells). Representative images of these cell phenotypes acquired with the Vectra system are shown in figure 3. The median percentage of CD56+/CK− cells from total cells across NSCLC cases was 7%, and were primarily found in the stromal compartment (figure 3b-3d). The distribution of absolute CD56+/CK− cell counts per compartment can be found in supplementary figure S7a-S7c. Using the top tertile cutpoint in the same 42 NSCLC cases, we found that CD56+/CK− cell counts in the stromal compartment were also associated with longer PFS and OS (figure 3e-3f). Patients with CB had a significantly higher median percentage of CD56+/CK− cells in the stroma (20.5%) as compared to those with NCB (9.7%) (p = 0.027). When analyzed as absolute cell counts, patients with CB also had a higher median number of stromal CD56+/CK− cells (85 cells vs. 28 cells), but this difference did not reach statistical significance (p = 0.49) (supplementary figure S7d-S7e). We further explored the predictive significance of CD56+/CD3− NK cells and CD56+/CD3+ NKT cells independently. We observed a stronger trend towards an association with PFS and OS for CD56+/CD3+ cells as compared to CD56+/CD3− cells, but neither of them were significantly associated with outcome when measured separately (supplementary figure S8).
Figure 3.
Orthogonal validation of CD56+/CK− cell counts assessed by Inform as predictors of outcome in YTMA404 cohort. A. Representative images acquired with Vectra Polaris microscope showing CD56 staining pattern in four NSCLC cases (scale bar = 100 μm). CK+ tumor cells are shown in green, CD3+ T cells in red, and CD56+ cells in white. Orange arrows indicate CD56+ NK cells, yellow arrows indicate CD3+/CD56+ NKT cells. Panel 3 illustrates a NSCLC case with strong CD56 positivity in the tumor compartment, and the red asterisk highlights CD56+/CK+ tumor cells; B. Distribution of CD56+/CK− cells in the tumor compartment (B), the stromal compartment, (C) and entire TMA spot (D) (n = 42); E-F. PFS (E) and OS (F) according to CD56+/CK− cell counts (top tertile) in YTMA404 cohort (n = 42)
Discussion
In this pilot scale discovery study, we show the potential of the DSP technology in the identification of spatially-informed biomarkers of clinical benefit to PD-1 checkpoint blockade in NSCLC. By combining high-fold multiplexing with spatial resolution, we identified 12 markers that were associated with PFS and/or OS benefit in spatial context, independently from clinical prognostic factors.
Perhaps one of the most relevant finding in this study is the identification of high levels of CD56 and CD4 in the CD45 compartment as predictors of all favorable clinical outcomes, including CB, longer PFS, and prolonged OS. Although CD56 expression did not hold significant after adjusting for multiple testing, it was significant in the multivariate analysis, and its association with outcome was further validated with an orthogonal method. In this cohort, CD8 levels in the CD45 compartment only predicted longer OS in the univariate analysis, with no differences in terms of CB or PFS. Collectively, these findings support the notion that anti-tumor immune responses following PD-1 checkpoint blockade are likely not exclusively mediated by cytotoxic CD8 T cells, and that NK cells and CD4 T cells also play a role in therapeutic efficacy(6-8).
Our results related to NK cells are concordant with several studies conducted in melanoma cohorts that have shown that NK cell gene signatures correlate with responsiveness from immunotherapy(9,10). In another study, circulating CD56+ cells detected by mass cytometry (CyTOFF) were shown to be upregulated in melanoma patients that responded to PD-1 checkpoint blockade(11). We now extend these findings to NSCLC, having identified an association between CD56 expression in the immune cell stroma and better treatment outcome. By using a multiplex IF panel targeting CD56 and CD3, we quantitatively assessed the abundance of CD56+ NK cells and NKT cells within the tumor microenvironment. Using inForm, we confirmed that these cells were mostly localized in the stroma. Notably, we could reproduce the outcome association by inForm cell count quantification method, with a degree of benefit that compared very similarly to the outcome association obtained when measuring CD56 expression in the CD45+ immune cell compartment (part of the stromal compartment in inForm analysis) using DSP. These results suggest that NK/NKT cells are likely localized mostly in the CD45+ compartment within the stroma, although the absence of CD45 compartmentalization with inForm precludes us to draw definitive conclusions in this regard. We believe that this orthogonal validation, although performed in the same set of patients, strengthens the value of CD56+ immune cells as a candidate predictor of outcome from PD-1 checkpoint blockade in NSCLC.
In line with the findings in our study, it has been suggested that CD4 T cells might play an equally relevant of perhaps more central role than CD8 T cells in mediating efficacy from anticancer immunotherapy(6,12). In a prospective study conducted in patients with NSCLC, functional systemic CD4 immunity was required for deriving significant benefit from PD-1 checkpoint blockade(13). Also, CD4 counts measured in a CD45-defined compartment using DSP was one of the immune parameters associated with longer disease-free survival in melanoma patients treated with neoadjuvant immune checkpoint blockade(14).
The identification of high levels of VISTA and CD127 expression in the tumor compartment as predictors of immunotherapy resistance is also a remarkable finding. Upregulation of compensatory inhibitory checkpoints (including VISTA) has been previously reported as an acquired resistance mechanism to PD-1 checkpoint blockade(15,16). To our knowledge, the role of CD127 (IL-7R) signaling in mediating immunotherapy response in solid tumors has not been previously described. A study conducted in curatively resected NSCLC also found that tumor cell expression of IL-7R was associated with shorter disease-free survival and OS(17), highlighting a potential poor prognostic role of IL-7R signaling in NSCLC.
This study found that PD-L1 expression in immune cells but not tumor cells was associated with OS in the univariate analysis. This reproduces our previous finding with quantitative IF in the same cohort(18). Furthermore, it is also consistent with another study by our group performing DSP in melanoma, where macrophage PD-L1 carried the sensitivity to predict immunotherapy outcomes(19). These findings are in line with previous studies that have shown that targeting PD-1/PD-L1 axis can still be effective regardless of PD-L1 tumor expression(20). Mechanistic studies in mouse models also support macrophages expressing PD-L1 as the key effector cells mediating tumor regression following PD-1 axis blockade(21,22). However, in our study, PD-L1 expression by immune cells and macrophages did not reach significance after controlling for clinical factors or adjusting for multiple testing.
Regarding the potential clinical applicability of the findings in this study, first we need to consider that the recent approval and increasing use of chemo-immunotherapy combinations in the first-line setting has limited the use of single-agent PD-1 axis blockade in unselected NSCLC patients. Monotherapy with PD-1 axis inhibitors is now mostly restricted to patients with high PD-L1 expression, where the response rates is substantially higher (about 45 %)(1) as compared to the response rates observed in our unselected cohort. Therefore, future biomarker discovery studies will preferably need to focus on subgroups of NSCLC patients with high PD-L1 (TPS≥50%) treated with monotherapy, and unselected patients treated with chemo-immunotherapy combinations. If the findings from this study are confirmed in these cohorts, this could suggest that future companion diagnostic tests to predict immunotherapy outcomes may require measuring markers from particular tissue compartments or co-localized with specific cell types (e.g. CD45+ cells). The DSP system is well suited for this aim, utilizing FFPE tumor samples with a relatively simple workflow. Therefore, although still many future efforts are needed to demonstrate the utility of the DSP system to inform therapeutic decisions and impact clinical care, with the appropriate validation it could be potentially scalable as a clinical assay in a CLIA lab in the future.
This study has to be interpreted in the context of a number of limitations. First, it is underpowered to demonstrate the independent predictive value for PD-L1 expression. Our cohort is a retrospective collection of tumors from patients treated in routine practice at a single institution, not a clinical trial. Furthermore, this is a single cohort study in which we assessed multiple hypothesis. Although we applied statistical correction for multiple testing, the false discovery adjustment method that we used is conservative, and does not preclude the need for validation in independent cohorts. As such, the data presented here must be considered hypothesis generating data, and require validation in future external cohorts. It is also a limitation that we used a TMA instead of whole tissue sections. Although we assessed two non-adjacent tumor cores, we recognize that this still represents a small percentage of the area of the standard tissue section. This could potentially under or overrepresent biomarker expression due to the heterogeneity of the tumor immune microenvironment and potentially influence biomarker performance, particularly for those heterogenous immune parameters that are expressed at relatively lower levels. Therefore, future validation efforts should include whole tissue sections assessing a greater number of ROIs. In this same line, the vast majority of the tumor samples included in the TMA and assessed in this study were primary tumors or lymph node biopsies, and mostly from patients that received PD-1 axis inhibition as second or further line of treatment. We tried to perform subgroup analysis and bi-variate Cox proportional hazards models to explore whether the outcome association differed depending on biopsy site (primary/locoregional vs. distant metastasis) or line of therapy (first-line vs. later line), but the subgroups were too small to draw meaningful conclusions in this regard (supplementary tables S3 and S4). Finally, another limitation is perhaps inherent to the DSP protein detection assay. Five markers included in the panel had poor signal to background ratios in nearly all samples, limiting our capacity to assess their predictive value. ARG1 was another marker with low signal relative to non-specific counts in many samples (supplementary figure S9), and therefore its potential favorable predictive value in the CD45 compartment should be cautiously interpreted. These findings could indicate the need for a more rigorous validation of these primary antibodies for future DSP panels. Alternatively, a CD3-restricted compartment could have resulted in an increased signal for some of these markers(23).
In conclusion, this study illustrates the potential of high-plex digital spatial profiling as a research tool to discover biomarkers of response to immunotherapy in NSCLC. We identified a number of relevant candidate immune predictors in spatial context that show promise for future validation in larger independent cohorts.
Supplementary Material
Translational relevance.
The majority of patients with advanced NSCLC do not respond to PD-1 axis blockade, and more robust predictive biomarkers are needed. Using the Digial Spatial Profiling (DSP) system, we identified 12 protein markers independently associated with benefit from single-agent PD-1 checkpoint blockade in spatial context. High expression of CD56 and CD4 in the CD45 compartment were significantly associated with all favorable clinical outcomes, whereas high levels of VISTA and CD127 in the tumor compartment were markers associated with immunotherapy resistance. We also validated the DSP finding that high CD56+ immune cell counts in the stroma were predictive for PFS and OS in the same set of patients using multiplex immunofluorescence, strengthening the relevance of NK/NKT cells as a candidate predictive biomarker for immunotherapy in NSCLC. This work identifies a number of relevant candidate predictors of immunotherapy outcome in spatial context that show promise for future validation in larger independent cohorts.
Acknowledgements
This work was supported by funds from the Yale SPORE in lung cancer P50 CA196530 to Scott Gettinger, Roy Herbst, Kurt A. Schalper, and David L. Rimm. J. Zugazagoitia was supported by a Rio Hortega contract from the Carlos III Research Institute (CM15/00196) and a fellowship from the Spanish Society of Medical Oncology (SEOM). DLR received instrument (GeoMx) support from NanoString Inc.
The authors thank Lori A. Charette and the staff of Yale Pathology tissue services for expert histology services.
Footnotes
Potential conflict of interest statement
J. Zugazagoitia has received consulting honoraria from Guardant Health, speaker fees from Roche, Pfizer, Guardant Health, and NanoString, and travel fees from Roche.
Kit Fuhrman is NanoString employee.
Roy Herbst has served as a consultant for Abbvie Pharmaceuticals, AstraZeneca, Biodesix, Bristol-Myers Squibb, Eli Lilly and Company, EMD Serrano, Genentech/Roche, Heat Biologics, Loxo Oncology, Merck and Company, Nektar, NextCure, Novartis, Pfizer, Sanofi, Seattle Genetics, Shire PLC, Spectrum Pharmaceuticals, Symphogen, and Tesaro. He has received research support from AstraZeneca, Eli Lilly and Company, and Merck and Company
Kurt Schalper has served as a consultant, advisor or served on a Scientific Advisory Board for Clinical Alemana de Santiago, Celgene, Moderna Therapeutics and Shattuck Labs, Agenus, Torque Therapeutics, Pierre-Fabre, Dynamo Therapeutics, EMD Serono, Astra Zeneca. He has received research funding from Genoptix/Navigate (Novartis), Vasculox/Tioma, Tesaro, Onkaido Therapeutics, Takeda Pharmaceuticals, Surface Oncology, Pierre-Fabre Research Institute, Merck, Bristol-Myers Squibb, Astra Zeneca, Eli Lilly.
David Rimm has served as a consultant, advisor or served on a Scientific Advisory Board for Amgen, Astra Zeneca, Agendia, Biocept, BMS, Cell Signaling Technology, Cepheid, Daiichi Sankyo, GSK, Lilly, Merck, NanoString, Perkin Elmer, PAIGE, Ventana and Ultivue. He has received research funding or instrument support from Astra Zeneca, Cepheid, NanoString, Navigate/Novartis, NextCure, Lilly, Ultivue, and Perkin Elmer.
The remaining authors declare no conflicts of interest.
Bibliography
- 1.Doroshow DB, Sanmamed MF, Hastings K, Politi K, Rimm DL, Chen L, et al. Immunotherapy in Non-Small Cell Lung Cancer: Facts and Hopes. Clin Cancer Res. 2019;25(15):4592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods. 2014;70(1):46–58. [DOI] [PubMed] [Google Scholar]
- 3.Lu S, Stein JE, Rimm DL, Wang DW, Bell JM, Johnson DB, et al. Comparison of Biomarker Modalities for Predicting Response to PD-1/PD-L1 Checkpoint Blockade: A Systematic Review and Meta-analysis. JAMA Oncol. 2019. [Epub Ahead of Print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merritt CR, Ong GT, Church S, Barker K, Geiss G, Hoang M, et al. High multiplex, digital spatial profiling of proteins and RNA in fixed tissue using genomic detection methods. BioRxiv 2019. [Epub Ahead of Print]. [DOI] [PubMed] [Google Scholar]
- 5.Huang W, Hennrick K, Drew S. A colorful future of quantitative pathology: validation of Vectra technology using chromogenic multiplexed immunohistochemistry and prostate tissue microarrays. Hum Pathol. 2013;44(1):29–38. [DOI] [PubMed] [Google Scholar]
- 6.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell. 2017. ;168(3):487–502.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böttcher JP, Bonavita E, Chakravarty P, Blees H, Cabeza-Cabrerizo M, Sammicheli S, et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell. 2018;172(5):1022–1037.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu J, Hodgins JJ, Marathe M, Nicolai CJ, Bourgeois-Daigneault M-C, Trevino TN, et al. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J Clin Invest. 2018;128(10):4654–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, et al. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med. 2018;24(8):1178–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zemek RM, De Jong E, Chin WL, Schuster IS, Fear VS, Casey TH, et al. Sensitization to immune checkpoint blockade through activation of a STAT1/NK axis in the tumor microenvironment. Sci Transl Med. 2019;11(501):eaav7816. [DOI] [PubMed] [Google Scholar]
- 11.Krieg C, Nowicka M, Guglietta S, Schindler S, Hartmann FJ, Weber LM, et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med. 2018;24(2):144–53. [DOI] [PubMed] [Google Scholar]
- 12.Binnewies M, Mujal AM, Pollack JL, Combes AJ, Hardison EA, Barry KC, et al. Unleashing Type-2 Dendritic Cells to Drive Protective Antitumor CD4+ T Cell Immunity. Cell. 2019;177(3):556–571.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuazo M, Arasanz H, Fernandez-Hinojal G, García Granda MJ, Gato M, Bocanegra A, et al. Functional systemic CD4 immunity is required for clinical responses to PD-L1/PD-1 blockade therapy. EMBO Mol Med. 2019;11(7):e10293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amaria RN, Reddy SM, Tawbi HA, Davies MA, Ross MI, Glitza IC, et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat Med. 2018;24(11):1649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat Commun. 2016;7:10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Ward JF, Pettaway CA, Shi LZ, Subudhi SK, Vence LM, et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med. 2017;23(5):551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K, Kadota K, Sima CS, Nitadori J-I, Rusch VW, Travis WD, et al. Clinical impact of immune microenvironment in stage I lung adenocarcinoma: tumor interleukin-12 receptor β2 (IL-12Rβ2), IL-7R, and stromal FoxP3/CD3 ratio are independent predictors of recurrence. J Clin Oncol. 2013;31(4):490–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Zugazagoitia J, Ahmed FS, Henick BS, Gettinger S, Herbst RS, et al. Immune cell PD-L1 co-localizes with macrophages and is associated with outcome in PD-1 pathway blockade therapy. Clin Cancer Res. 2019. [Epub Ahead of Print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toki MI, Merritt CR, Wong PF, Smithy JW, Kluger HM, Syrigos KN, et al. High-plex predictive marker discovery for melanoma immunotherapy treated patients using Digital Spatial Profiling. Clin Cancer Res. 2019;25(18):5503–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herbst RS, Soria J-C, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin H, Wei S, Hurt EM, Green MD, Zhao L, Vatan L, et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest. 2018;128(2):805–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang H, Liang Y, Anders RA, Taube JM, Qiu X, Mulgaonkar A, et al. PD-L1 on host cells is essential for PD-L1 blockade-mediated tumor regression. J Clin Invest. 2018;128(2):580–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Datar I, Sanmamed MF, Wang J, Henick BS, Choi J, Badri T, et al. Expression Analysis and Significance of PD-1, LAG-3, and TIM-3 in Human Non-Small Cell Lung Cancer Using Spatially Resolved and Multiparametric Single-Cell Analysis. Clin Cancer Res. 2019;25(15):4663–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.