Abstract
Background
Tuberculosis (TB) control is hindered by absence of rapid tests to identify Mycobacterium tuberculosis (MTB) and detect isoniazid (INH) and rifampin (RIF) resistance. We evaluated the accuracy of the BD MAX multidrug-resistant (MDR)-TB assay (BD MAX) in South Africa, Uganda, India, and Peru.
Methods
Outpatient adults with signs/symptoms of pulmonary TB were prospectively enrolled. Sputum smear microscopy and BD MAX were performed on a single raw sputum, which was then processed for culture and phenotypic drug susceptibility testing (DST), BD MAX, and Xpert MTB/RIF (Xpert).
Results
1053 participants with presumptive TB were enrolled (47% female; 32% with human immunodeficiency virus). In patients with confirmed TB, BD MAX sensitivity was 93% (262/282 [95% CI, 89–95%]); specificity was 97% (593/610 [96–98%]) among participants with negative cultures on raw sputa. BD MAX sensitivity was 100% (175/175 [98–100%]) for smear-positive samples (fluorescence microscopy), and 81% (87/107 [73–88%]) in smear-negative samples. Among participants with both BD MAX and Xpert, sensitivity was 91% (249/274 [87–94%]) for BD MAX and 90% (246/274 [86–93%]) for Xpert on processed sputa. Sensitivity and specificity for RIF resistance compared with phenotypic DST were 90% (9/10 [60–98%]) and 95% (211/222 [91–97%]), respectively. Sensitivity and specificity for detection of INH resistance were 82% (22/27 [63–92%]) and 100% (205/205 [98–100%]), respectively.
Conclusions
The BD MAX MDR-TB assay had high sensitivity and specificity for detection of MTB and RIF and INH drug resistance and may be an important tool for rapid detection of TB and MDR-TB globally.
Keywords: HIV, tuberculosis, Mycobacterium infections, diagnosis, multidrug, resistance
In a prospective, multicenter diagnostic accuracy study, the BD MAX Multidrug-resistant Tuberculosisassay assay had high sensitivity and specificity for detection of Mycobacterium tuberculosis and rifampin and isoniazid drug resistance and allows for rapid detection of tuberculosis and drug resistance.
There were 10 million cases of tuberculosis (TB) in 2017 with 1.6 million deaths; TB is the leading cause of death among persons with human immunodeficiency virus (HIV) and the global burden of multidrug-resistant (MDR) TB is high [1]. Despite current efforts, global case detection of active TB is less than 60% in many endemic settings, with some patients dying before diagnosis [1]. Smear microscopy, the most widely utilized TB diagnostic modality worldwide, has incomplete sensitivity as a screening tool, particularly in cases with presumptive TB and HIV [2]. Sputum culture, the reference standard, is costly, takes weeks to provide results, and remains restricted to higher levels of the health infrastructure because of expertise and equipment requirements.
Two rapid molecular tests, namely the Xpert MTB/RIF assay (Xpert; Cepheid) [3, 4] and the GenoType MTBDRplus (Hain Lifescience) are commercialized and used in some TB-endemic settings, but each has important limitations in addition to attributes. The World Health Organization (WHO)–endorsed Xpert MTB/RIF assay is fully integrated, automated, and appropriate for near-care and requires relatively little training; however, the standard platform for this assay has limited throughput (although larger instruments are now available) and only tests for resistance mutations associated with rifampin (RIF) [3–5]. The GenoType MTBDRplus test can detect mutations associated with isoniazid (INH) and RIF resistance but has suboptimal sensitivity for Mycobacterium tuberculosis (MTB) complex (MTBC) detection and is not fully integrated or automated [6, 7]. There is a need for products that cover a range of performance profiles suitable for the realities of tiered healthcare systems that include testing at point-of-care as well as in more centralized laboratories [8, 9].
The BD MAX MDR-TB assay (BD MAX), performed on the BD MAX System (both from Becton, Dickinson and Company [BD]), is an automated, qualitative in vitro diagnostic test for the direct detection of MTBC DNA in raw induced or expectorated sputum or concentrated sputum sediments from patients for whom there is clinical suspicion of TB and who have not received more than 3 days of anti-TB therapy in the past 6 months [10]. The test utilizes real-time polymerase chain reaction (PCR) for the amplification of specific DNA targets and fluorogenic target-specific hybridization probes to detect MTBC DNA as well as resistance mutations in the rpoB and katG genes and the inhA promoter region associated with MDR-TB. The assay is automated and integrated and requires a stable source of electricity and laboratory technician training; 24 specimens can be tested in 1 run, and turn-around time from the testing start to result is less than 4 hours. Therefore, the BD MAX MDR-TB assay is expected to be most suitable for use in central laboratories in which large numbers of specimens are tested and minimal operator hands-on time is desirable. We sought to assess the diagnostic accuracy of the assay in high-TB-burden low- and middle-income settings.
METHODS
Study Population
We performed a prospective multicenter diagnostic study in which the accuracy of an investigational in vitro molecular diagnostic test (BD MAX) performed on sputum was assessed using the reference standard of liquid culture for mycobacteria growth (BD BACTEC MGIT 960 system [MGIT]; BD) followed by MTBC identification with the BD MGIT TBc Identification Test (TBc ID; BD). Xpert MTB/RIF was performed as a commercially available comparator in secondary analysis. Study sites were located in Kampala, Uganda; Cape Town, South Africa; Pune, India; and Lima, Peru. Participants were recruited consecutively and enrolled between May 2017 and March 2018 into a case-detection group and a drug-resistant detection group in order to enroll individuals with a higher expected prevalence of drug resistance. For both groups, inclusion criteria were age 18 years or older, written informed consent, and symptoms of pulmonary TB (cough ≥2 weeks, and at least 1 other symptom such as fever, night sweats, or weight loss). Individuals receiving more than 2 days or doses of TB treatment within the prior 6 months were excluded from the case-detection group but included in the drug-resistant group if they were suspected or had a history of treatment failure. Non–study-directed HIV testing and CD4 testing was recorded, if available. Study-directed testing included 1 sputum, minimum volume of 3 mL, for acid fast bacilli (AFB) smear microscopy and mycobacterial culture, with MTBC identification. Participants unable to spontaneously expectorate the minimum sputum volume were considered early withdrawals.
The study was approved by review committees of the Faculty of Health Sciences, Human Research Ethics Committee, University of Cape Town, South Africa; Joint Clinical Research Centre, Kampala, Uganda; Uganda National Council for Science and Technology; Universidad Peruana Cayetano Heredia, Comité Institucional de Ética en Investigación; Ethics Committee-B J Medical College and Sassoon General Hospitals, Pune, India; and Johns Hopkins Medical Institutions, Baltimore, Maryland.
Laboratory Testing
All testing was conducted in local laboratories at the study sites (Supplementary Figure 1). The sputum sample was split into 2 components—processed and raw. For the processed component, 1 portion of the specimen was decontaminated with N-acetyl-l-cysteine-sodium hydroxide (BBL MycoPrep; BD). After centrifugation, the pellet was suspended in 2 mL buffer. A concentrated auramine-O smear was examined and graded using a fluorescent microscope (FM). Ziehl-Neelsen (ZN) smear was also examined and graded. A 0.5-mL portion of processed sputum sediment was cultured using MGIT, with species identification using TBc ID [11]. A 0.8-mL portion was tested on BD MAX. All positive cultures had phenotypic drug susceptibility testing (DST) for RIF (1 µg/mL) and INH (0.1 µg/mL) using the MGIT system at standard critical concentrations. A 0.5-mL portion of processed sputum specimen was tested with Xpert MTB/RIF. The raw portion of the sputum specimen was not processed and was also tested using ZN and FM and BD MAX.
BD MAX workflow includes a 30-minute incubation step with sample treatment reagent (STR; STR liquefies specimens and reduces viability of MTBC), transfer to a BD MAX TB sample tube, and then loading onto the instrument with 24 samples per run (run time <4 hours); specimens were batched. The BD MAX assay results for detection of MTB are categorized as MTB Detected (MTBC DNA detected), MTB Not Detected (no MTBC DNA detected and Sample Processing Control detected), MTB Low POS (MTBC DNA detected but resistance metrics not measurable), Indeterminate (due to BD MAX system failure), Incomplete (incomplete run), or Unresolved (no MTBC DNA detected and no sample processing control detected, indicative of an inhibitory sample or reagent failure). The assay additionally reports detection of mutations associated with RIF and INH resistance, as resistance detected (MTBC RIF or INH resistance mutations were detected), not detected (MTBC RIF or INH resistant mutations were not detected), or unreportable (MTBC DNA detected but INH or RIF resistance metrics not measurable).
All index test and reference standard tests were interpreted blinded to any knowledge of clinical information or other tests.
Outcomes Determination
Mycobacterial culture followed by MTBC identification was considered the reference standard. Sputa for which the MGIT culture was still contaminated after a second decontamination, or for which the package inserts were not followed, were excluded from the primary analysis as not assessable. All other sputa were considered to be negative for TB. In secondary analysis, we considered a composite microbiological reference standard using Xpert and mycobacterial culture; for the composite, specimens that were positive by either culture or Xpert were considered positive, were considered negative if results of both assays were negative, and were considered nonevaluable if either method was unevaluable (eg, contaminated) and the other was negative. We evaluated BD MAX results stratified by both ZN and FM status to evaluate the performance among participants with smear-negative TB.
For DST, the reference standard was the results of the phenotypic culture-based DST for INH resistance (0.1 µg/mL) and RIF resistance (1 µg/mL). For all isolates that were discordant by BD MAX assay, Sanger bidirectional sequencing (3500xl Genetic Analyzer and BigDye Terminator v3.1 Cycle sequencing kit; ThermoFisher Scientific) of the rpoB RIF resistance-determining region, katG, and the inhA promoter region was performed on the cultured isolate to help resolve any discordance between molecular and phenotypic results. In secondary analysis, we evaluated performance against a composite reference standard consisting of phenotypic DST and Xpert followed by sequencing. In the composite reference standard, specimens with resistance identified by any of the 2 methods was considered positive for resistance. Specimens were considered negative if results of all assays were negative, or nonevaluable if either method was unevaluable.
Statistical Analyses
Student’s t test was used to compare means. Two-sample proportions were compared by χ 2 tests. McNemar’s test was used to compare BD MAX and Xpert assay sensitivities. A P value of .05 or less was considered statistically significant and 95% confidence intervals (CIs) were used. Confidence intervals for binary outcomes were obtained using Wilson’s score method. Statistical calculations were performed using Stata 14.1 (StataCorp) and the R version 3.5.1 (R Foundation for Statistical Computing).
RESULTS
Characteristics of the Study Population
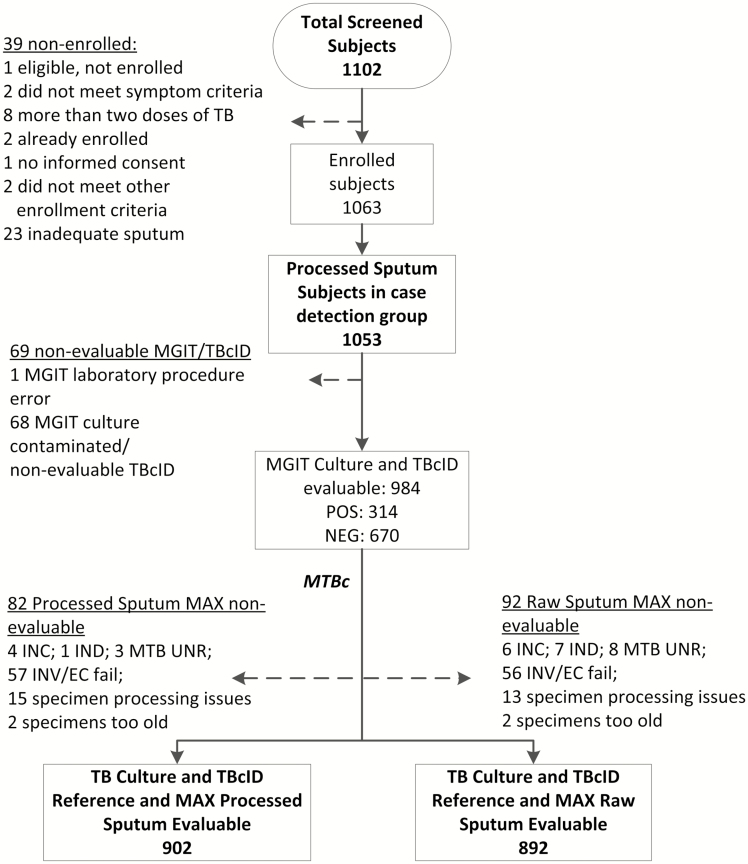
Of the 1102 participants screened, 1053 participants met the enrollment criteria of the case-detection group, had adequate sputa and were enrolled, 69 of whom were excluded due to lack of an interpretable mycobacterial culture for a microbiological reference standard (eg, MGIT culture contamination) and 92 (9%) raw sputa had nonevaluable BD MAX test results (Figure 1, Supplementary Figure 2); 10 additional individuals were enrolled into the drug-resistance detection group and contributed data only to analyses pertaining to drug-resistance determination. Characteristics of the study population are shown in Table 1. Human immunodeficiency virus test results were primarily available in South Africa and Uganda. Among the 712 participants with a known HIV status, 47% (333/712) were positive, with a median CD4 count of 367 (interquartile range [IQR], 228–536). Tuberculosis was microbiologically confirmed on liquid culture in 314 of 984 participants (32%) and differed between study sites (31/320 [10%], South Africa; 81/259 [31%], Uganda; 54/136 [40%], India; 148/269 [55%], Peru; P < .01). A total of 670 of 984 (68%) had no positive cultures and were classified as “not TB.”
Figure 1.
Study population and study flow diagram. Abbreviations: INC, incomplete; IND, indeterminate; INV/EC, invalid/external control; MAX, BD MAX assay; MGIT, BACTEC MGIT 960 system; MTB, Mycobacterium tuberculosis; MTBc, M. tuberculosis complex; NEG, negative; POS, positive; TB, tuberculosis; TBcID, BD MGIT TBc Identification Test; UNR, unresolved.
Table 1.
Study Participant Demographic Characteristics
| Values | |
|---|---|
| Total | 984 |
| Age, years | |
| Median (IQR) | 34 (27–44) |
| Gender | |
| Male | 528 (53.7) |
| Race | |
| Asian | 136 (14) |
| Black | 579 (59) |
| Other/More than one race | 269 (27) |
| Site | |
| India | 136 (14) |
| Peru | 269 (27) |
| South Africa | 320 (33) |
| Uganda | 259 (26) |
| HIV status | |
| Positive | 303 (31) |
| Negative | 347 (35) |
| Unknown | 334 (34) |
| CD4 (among HIV positive)a | |
| Median (IQR) | 365 (231–538) |
Data are n (%) unless otherwise indicated.
Abbreviations: HIV, human immunodeficiency virus; IQR, interquartile range.
aCD4 data available in 113 individuals based on routine testing for clinical purposes at the study sites.
BD MAX MDR-TB Test Performance Among All Participants
Table 2 shows overall BD MAX results for the detection of MTB against the microbiological reference standard on raw sputum (Supplementary Table 1 for processed sputum). BD MAX test sensitivity was 93% (262/282; 95% CI, 89–95%) from raw sputum in participants with confirmed TB. Among individuals categorized as “not TB,” specificity was 97% (593/610; 95% CI, 96–98%). Among these discordant participants (positive BD MAX, negative cultures), all were smear negative (if available), 9 of 17 (53%) were low positive, and 6 of 17 (35%) had a prior history of TB. When using a composite microbiological reference standard of mycobacterial culture and/or Xpert, BD MAX sensitivity was 93% (275/297; 95% CI, 89–95%) from raw sputum. Specificity was 99% (584/592; 95% CI, 97–99%). Specificity estimates were similar against a composite reference standard, comparing those with and without a prior history of TB (147/150 [98%] vs 437/442 [99%], respectively). Positive predictive value (PPV) for confirmed TB and negative predictive value (NPV) were, respectively, 94% (91–96%) and 97% (95–98%) among all participants.
Table 2.
Diagnostic Accuracy of the BD MAX Assay for Detection of Mycobacterium tuberculosis Among Raw Specimens Against a Microbiological Reference Standard of Mycobacterial Culture
| Microbiological Culture | |||
|---|---|---|---|
| Analysis and BD MAX Result | Positive | Negative | Total (95% CI) |
| Overall | |||
| Positive | 262 | 17 | 279 (PPV, 94% [91–96%]) |
| Negative | 20 | 593 | 613 (NPV, 97% [95–98%]) |
| Total | 282 | 610 | 892 |
| Sensitivity | 93% (89–95%)a | ||
| Specificity | 97% (96–98%)a | ||
| Smear stratifiedb | |||
| FM smear positive | |||
| Positive | 175 | 1 | 176 (PPV, 99% [97–100%]) |
| Negative | 0 | 0 | 0 |
| Total | 175 | 1 | 176 |
| Sensitivity | 100% (98–100) | ||
| Specificity | … | ||
| FM smear negative | |||
| Positive | 87 | 15 | 102 (PPV, 85% [77–91%]) |
| Negative | 20 | 591 | 611 (NPV, 97% [95–98%]) |
| Total | 107 | 606 | 713 |
| Sensitivity | 81% (73–88%) | ||
| Specificity | 98% (96–99%) | ||
| ZN smear positive | |||
| Positive | 148 | 0 | 148 (PPV, 100% [98–100%]) |
| Negative | 0 | 0 | 0 (NPV, 100% [21–100%]) |
| Total | 148 | 0 | 148 |
| Sensitivity | 100% (98–100%) | ||
| Specificity | … | ||
| ZN smear negative | |||
| Positive | 114 | 16 | 130 (PPV, 88% [81–92%]) |
| Negative | 20 | 592 | 612 (NPV, 97% [95–98%]) |
| Total | 134 | 608 | 742 |
| Sensitivity | 85% (78–90%) | ||
| Specificity | 97% (96–98%) | ||
Abbreviations: BD MAX, BD MAX multidrug-resistant tuberculosis assay; CI, confidence interval; FM, fluorescence microscopy; NPV, negative predictive value; PPV, positive predictive value; ZN, Ziehl-Neelsen.
aUsing a composite reference standard consisting of Xpert and mycobacterial culture, sensitivity was 93% (275/297 [89–95%]) and specificity was 99% (584/592 [97–99%]).
bSmears performed from raw specimens. Two specimens had a smear status unknown for ZN and 3 specimens had a smear status unknown for FM.
Among ZN and FM smear-positive, culture-positive participants, BD MAX sensitivity was 100% (148/148; 95% CI, 98–100%) and 100% (175/175; 95% CI, 98–100%) from raw sputum specimens, respectively. Among ZN smear-negative specimens, sensitivity was 85% (114/134; 95% CI, 78–90%); among FM smear-negative specimens, sensitivity was 81% (87/107; 95% CI, 73–88%). Overall, 7% (19/282) of participants with confirmed TB were low positive on BD MAX; among these, 100% were smear negative.
Comparison to Xpert MTB/RIF for Detection of MTB
We compared the performance of BD MAX and Xpert MTB/RIF assays for the detection of MTB on processed sputa (Supplementary Table 2). The sensitivity was similar between the 2 assays at 91% (249/274; 95% CI, 87–94%) and 90% (246/274; 95% CI, 86–93%) for the BD MAX and Xpert MTB/RIF assays, respectively. Specificity was 96% (588/615; 95% CI, 94–97%) and 98% (604/615; 95% CI, 97–99%) for BD MAX and Xpert MTB/RIF, respectively. When stratified by smear status, the BD MAX assay sensitivity was 65% (44/68; 95% CI, 53–75%) compared with 59% for Xpert (40/68; 95% CI, 47–70%) among FM smear-negative samples.
Detection of Drug Resistance
Among the 297 cases of TB microbiologically confirmed on liquid culture in both enrollment groups (ie, case-detection and drug-resistance detection groups), resistance results were available by the BD MAX test in 232 (78%) participants, of whom 230 participants had reportable results for both RIF and INH resistance. Overall, 202 (87%) had drug-susceptible TB on phenotypic testing and 29 (13%) had resistance to INH and/or RIF.
Rifampin Resistance Detection
Among 10 patients with microbiologically confirmed TB with RIF resistance on phenotypic DST, sensitivity of BD MAX for the detection of RIF resistance was 90% (9/10; 95% CI, 60–98%). Specificity of BD MAX for RIF susceptibility among 222 participants with TB without detection of RIF resistance by phenotypic DST was 95% (211/222; 95% CI, 91–97%) (Table 3, Supplementary Table 3). Among 11 participants with resistance detected by BD MAX but not phenotypic DST, bidirectional sequencing found 6 with true resistance mutations (2 D435Y, 1 D435F, 2 L430P, 1 L452P) and 2 silent mutations (F433F). Another specimen gave a phenotypic DST error, but Xpert and bidirectional sequencing (H445N) were RIF resistant. Each of these resistance mutations has been previously associated with treatment failure [12–17]. When examined against a composite reference standard inclusive of Xpert and bidirectional sequencing, sensitivity of the BD MAX assay was 94% (16/17; 95% CI, 73–99%) and specificity was 98% (200/205; 95% CI, 94–99%); 2 silent mutations were regarded as susceptible).
Table 3.
Performance of the BD MAX Assay for the Detection of Drug Resistance Among Raw Specimens
| Microbiological Culture–Based DST | |||
|---|---|---|---|
| Analysis and BD MAX Result | Positive | Negative | Total (95% CI) |
| Any drug resistance (INH or RIF)a |
|||
| Positive | 24 | 7 | 31 (PPV, 77%) |
| Negative | 5 | 195 | 200 (NPV, 95%) |
| Total | 29 | 202 | 231 |
| Sensitivity | 83% (66–92%) | ||
| Specificity | 97% (93–98%) | ||
| INH resistance | |||
| Positive | 22b | 0 | 22 (PPV, 100%) |
| Negative | 5c | 205 | 210 (NPV, 98%) |
| Total | 27 | 205 | 232 |
| Sensitivity | 82% (63–92%) | ||
| Specificity | 100% (98–100%) | ||
| RIF resistance | |||
| Positive | 9 | 11d | 20 (PPV, 45%) |
| Negative | 1 | 211 | 212 (NPV, 99.5%) |
| Total | 10 | 222 | 232 |
| Sensitivity | 90% (60–98%) | ||
| Specificity | 95% (91–97%) | ||
Abbreviations: BD MAX, BD MAX multidrug-resistant tuberculosis assay; CI, confidence interval; DST, drug susceptibility testing; INH, isoniazid; NPV, negative predictive value; PPV, positive predictive value; RIF, rifampin.
aCases where BD MAX gave an RIF or INH Not Detected result or an RIF or INH Unreportable result were excluded. A total of 230 samples had reportable results for both RIF and INH resistance; 1 additional sample without INH results available is included, which had RIF resistance detected by BD MAX. Eight samples were resistant both for RIF and INH based on the DST. The BD MAX assay detected the dual resistance for 7 of 8 samples.
bAmong 22 INH-resistant isolates detected by BD MAX assay, mutations were detected in both inhA promoter and katG gene for 2 specimens, in inhA promoter alone for 4 specimens, and in katG gene alone for 16 specimens.
cAmong 5 isolates phenotypically resistant to INH but negative by BD MAX, sequencing of the targeted regions did not find mutations within katG or inhA promotor, suggesting resistance due to mutations outside the targeted regions for 3 isolates. For 1/5, sequencing suggested heteroresistance with a wild-type strain and a strain with a mutation in the inhA promoter may have been present. For 1/5, sequencing suggested heteroresistance with a wild-type strain and a strain with a mutation in the katG gene may have been present.
dEleven samples were resistant with BD MAX assay and sensitive by phenotypic DST. Among these 11, 6 were positive for RIF resistance by Xpert and bidirectional sequencing and 2 were found to have silent mutations. Another specimen gave a phenotypic DST error, but Xpert and bidirectional sequencing were resistant. When examined against a composite reference standard inclusive of Xpert and bidirectional sequencing (222 samples), sensitivity was 94% (16/17 [73–99%]) and specificity was 98% (200/205 [94–99%]).
Isoniazid Resistance Detection
Among 27 patients with microbiologically confirmed TB with INH resistance on phenotypic DST, BD MAX assay sensitivity was 82% (22/27; 95% CI, 63–92%; 4 inhA promoter and 16 katG gene mutations, 2 with both) (Table 3, Supplementary Table 3). Specificity of the assay among 205 participants with TB without detection of INH resistance by phenotypic DST was 100% (205/205; 95% CI, 98–100%).
BD MAX MDR-TB Test Performance Stratified by Human Immunodeficiency Virus Infection
BD MAX test sensitivity and specificity, stratified by HIV-infection status, are shown in Supplementary Tables 4 and 5. Among 273 participants with HIV with evaluable results, assay sensitivity was 86% (44/51; 95% CI, 74–93%) and assay specificity was 98% (217/222; 95% CI, 95–99%). When stratified by ZN smear microscopy status, sensitivity of the BD MAX assay for detection of HIV-associated TB was 100% (22/22; 95% CI, 85–100%) for patients with smear-positive and 76% (22/29; 95% CI, 58–88%) for patients with smear-negative HIV/TB. Among patients with TB with both BD MAX and Xpert test results, sensitivity was 82% (41/50; 95% CI, 69–90%) for both assays.
DISCUSSION
For the diagnosis of active TB in diverse low- and middle-income settings, the BD MAX MDR-TB test had a sensitivity of 93% for confirmed pulmonary TB cases, with accuracy that appeared to be comparable to Xpert MTB/RIF. Of note, Xpert MTB/RIF Ultra was not commercially available at the time of the study. While sample size was limited, BD MAX had high sensitivity and specificity for the detection of both RIF and INH resistance, consistent with current targets for the development of new tools for rapid DSTs for TB [18]. The BD MAX assay may therefore represent a new diagnostic tool in the armamentarium for rapid identification of TB globally.
Globally, the burden of MDR-TB, INH-mono-resistant, and RIF-mono-resistant TB remains high and developing tools that accurately identify both INH and RIF resistance is increasingly important in allowing rapid, individualized therapy. Among the potential benefits of the BD MAX assay is the detection of mutations in inhA promoter, katG, in addition to rpoB, in contrast to other commonly used molecular assays that focus on initial identification of RIF resistance alone. While the distribution of INH mutations has been less well mapped globally, the WHO estimates suggest that nearly 8% of patients with TB worldwide have RIF-susceptible, INH-resistant TB; some parts of the world may have rates of mono-resistance in excess of 10–20%, with poorer treatment outcomes when treated with standard first-line regimens [19–21]. Failure to identify INH mono-resistance may also lead to suboptimal treatment that can select for further acquired resistance [22, 23]. Consequently, in 2018, the WHO issued the first treatment guidelines for Hr-TB (INH resistance with RIF susceptibility), which indicates provisions for a regimen that includes RIF, levofloxacin, pyrazinamide, and ethambutol for 6 months [21, 24]. Empirical treatment of Hr-TB is not currently suggested. Rapid detection of INH resistance is therefore an important consideration for patient treatment and for TB-control programs.
In reference laboratories, the ability to batch or conduct increased numbers of tests simultaneously may improve efficiency in laboratories with very high volumes as countries adopt hub-and-spoke models of sample referral. Given the relatively straightforward workflow of the BD MAX assay, with a short incubation step and run time of less than 4 hours, high-volume laboratories could prepare the next group of samples during the BD MAX system run time. This may offer benefits over alternative platforms with more limited throughput.
Our study has limitations. We conducted the study using a single sputum sample. Consequently, specificity of the BD MAX assay may be underestimated in situations where there was molecular detection of MTB by the BD MAX assay but no detection by culture on a single sputum specimen. Nonetheless, our results show high specificity for the detection of MTB, as well as drug resistance, and our results are strengthened by inclusion of a composite reference standard inclusive of Xpert testing and sequencing. Five individuals had phenotypic INH resistance not detected by BD MAX, reducing sensitivity for INH resistance detection. Sequencing of the targeted regions did not find mutations within katG or inhA promotor for 3 isolates, suggesting mutations outside the targeted regions. For 1 of the 5 cases, sequencing suggested heteroresistance in which a wild-type strain and a strain with an inhA promoter mutation may have been present. For 1 of the 5 cases, sequencing suggested heteroresistance in which a wild-type strain and a strain with katG mutation may have been present. Geographic regions with a higher prevalence of INH resistance mediated by mutations outside katG or inhA promotor regions may consider the inclusion of phenotypic DST within diagnostic algorithms.
On the other hand, our study has several important strengths. We are the first to report on the diagnostic accuracy of the BD MAX assay on a prospective cohort in high-burden, low- and middle-income settings where the test may be most useful. The study was conducted in multiple representative settings that included sites in Asia and South America, in addition to sub-Saharan Africa. We provide results on individuals with HIV as well as those without HIV and included a comparison with Xpert, which is widely used in similar settings. Our results suggest that the BD MAX assay has similar performance to Xpert and offers the added advantage of providing results for INH resistance in addition to RIF.
In conclusion, there is a need for new diagnostic tools to combat the global burden of TB. For many high-burden settings with a high-volume of testing, the BD MAX assay may represent an important automated tool for the rapid detection of both MTB and drug resistance.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. The Johns Hopkins University team (M. S., Y. C. M., J. B., B. K., D. A.) oversaw all data analysis including independent statistical analysis and data verifications, results reporting, and made the decision to publish a manuscript.
Acknowledgments. The authors thank Salma Kodsi from Becton, Dickinson and Company, Becton, Dickinson and Company (BD) Life Sciences–Diagnostic Systems, for her insights. The authors also thank Valentin Parvu, Indrias Berhane, Qing Liang, and Christina Chiao of Becton, Dickinson and Company, BD Life Sciences–Diagnostic Systems, for statistical support. The individuals acknowledged here have no additional funding or additional compensation to disclose.
Financial support. This work was supported by Becton, Dickinson and Company.
Potential conflicts of interest. S. P., D. K., C. E. M., and C. K. C. are employed by Becton, Dickinson and Company (BD), which donated BD MAX instruments and BD MAX multidrug-resistant tuberculosis kits and trained laboratory staff. BD also assisted with the study concept, design, and implementation. M. P. N. reports grants from National Institutes of Health. The other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Global tuberculosis report Available at: http://www.who.int/tb/publications/global_report/en/. Accessed 19 October 2018.
- 2. Kivihya-Ndugga LE, van Cleeff MR, Githui WA, et al. A comprehensive comparison of Ziehl-Neelsen and fluorescence microscopy for the diagnosis of tuberculosis in a resource-poor urban setting. Int J Tuberc Lung Dis 2003; 7:1163–71. [PubMed] [Google Scholar]
- 3. Helb D, Jones M, Story E, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol 2010; 48:229–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boehme CC, Nabeta P, Hillemann D, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med 2010; 363:1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, Dendukuri N. Xpert(R) MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev 2014; 1:CD009593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hillemann D, Rüsch-Gerdes S, Richter E. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol 2007; 45:2635–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ling DI, Zwerling AA, Pai M. Rapid diagnosis of drug-resistant TB using line probe assays: from evidence to policy. Expert Rev Respir Med 2008; 2:583–8. [DOI] [PubMed] [Google Scholar]
- 8. Pai M, Schito M. Tuberculosis diagnostics in 2015: landscape, priorities, needs, and prospects. J Infect Dis 2015; 211 Suppl 2:S21–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Drobniewski F, Cooke M, Jordan J, et al. Systematic review, meta-analysis and economic modelling of molecular diagnostic tests for antibiotic resistance in tuberculosis. Health Technol Assess 2015; 19:1–188, vii–viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. BD MAX™ MDR-TB Assay Package Insert. 2019. BD Life Sciences, Sparks, MD Available at: https://www.bd.com/resource.aspx?IDX=35759. Accessed 21 May 2019.
- 11. Yu MC, Chen HY, Wu MH, et al. Evaluation of the rapid MGIT TBc identification test for culture confirmation of Mycobacterium tuberculosis complex strain detection. J Clin Microbiol 2011; 49:802–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shah NS, Grace Lin SY, Barry PM, Cheng YN, Schecter G, Desmond E. Clinical impact on tuberculosis treatment outcomes of discordance between molecular and growth-based assays for rifampin resistance, California 2003–2013. Open Forum Infect Dis 2016; 3:ofw150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ho J, Jelfs P, Sintchencko V. Phenotypically occult multidrug-resistant Mycobacterium tuberculosis: dilemmas in diagnosis and treatment. J Antimicrob Chemother 2013; 68:2915–20. [DOI] [PubMed] [Google Scholar]
- 14. Williamson DA, Roberts SA, Bower JE, et al. Clinical failures associated with rpoB mutations in phenotypically occult multidrug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis 2012; 16:216–20. [DOI] [PubMed] [Google Scholar]
- 15. van Ingen J, Aarnoutse R, de Vries G, Boeree MJ, van Soolingen D. Low-level rifampicin-resistant Mycobacterium tuberculosis strains raise a new therapeutic challenge. Int J Tuberc Lung Dis 2011; 15:990–2. [DOI] [PubMed] [Google Scholar]
- 16. Tan Y, Hu Z, Zhao Y, et al. The beginning of the rpoB gene in addition to the rifampin resistance determination region might be needed for identifying rifampin/rifabutin cross-resistance in multidrug-resistant Mycobacterium tuberculosis isolates from Southern China. J Clin Microbiol 2012; 50:81–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Van Deun A, Aung KJ, Bola V, et al. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol 2013; 51:2633–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. World Health Organization. High priority target product profiles for new tuberculosis diagnostics Available at: http://www.who.int/tb/publications/tpp_report/en/. Accessed 21 May 2019.
- 19. Stagg HR, Lipman MC, McHugh TD, Jenkins HE. Isoniazid-resistant tuberculosis: a cause for concern? Int J Tuberc Lung Dis 2017; 21:129–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van der Heijden YF, Karim F, Mufamadi G, et al. Isoniazid-monoresistant tuberculosis is associated with poor treatment outcomes in Durban, South Africa. Int J Tuberc Lung Dis 2017; 21:670–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. World Health Organization. WHO treatment guidelines for isoniazid-resistant tuberculosis Available at: http://www.who.int/tb/publications/2018/WHO_guidelines_isoniazid_resistant_TB/en/. Accessed 21 May 2019. [PubMed]
- 22. Gegia M, Winters N, Benedetti A, van Soolingen D, Menzies D. Treatment of isoniazid-resistant tuberculosis with first-line drugs: a systematic review and meta-analysis. Lancet Infect Dis 2017; 17:223–34. [DOI] [PubMed] [Google Scholar]
- 23. Romanowski K, Campbell JR, Oxlade O, Fregonese F, Menzies D, Johnston JC. The impact of improved detection and treatment of isoniazid resistant tuberculosis on prevalence of multi-drug resistant tuberculosis: a modelling study. PLoS One 2019; 14:e0211355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. World Health Organizaiton. WHO consolidated guidelines on drug-resistant tuberculosis Available at: https://www.who.int/tb/publications/2019/consolidated-guidelines-drug-resistant-TB-treatment/en/. Accessed 21 May 2019. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.