Abstract
Background
Previous studies suggest that the nose/throat microbiome may play an important role in shaping host immunity and modifying the risk of respiratory infection. Our aim is to quantify the association between the nose/throat microbiome and susceptibility to influenza virus infection.
Methods
In this household transmission study, index cases with confirmed influenza virus infection and their household contacts were followed for 9–12 days to identify secondary influenza infections. Respiratory swabs were collected at enrollment to identify and quantify bacterial species via high-performance sequencing. Data were analyzed by an individual hazard-based transmission model that was adjusted for age, vaccination, and household size.
Results
We recruited 115 index cases with influenza A(H3N2) or B infection and 436 household contacts. We estimated that a 10-fold increase in the abundance in Streptococcus spp. and Prevotella salivae was associated with 48% (95% credible interval [CrI], 9–69%) and 25% (95% CrI, 0.5–42%) lower susceptibility to influenza A(H3N2) infection, respectively. In contrast, for influenza B infection, a 10-fold increase in the abundance in Streptococcus vestibularis and Prevotella spp. was associated with 63% (95% CrI, 17–83%) lower and 83% (95% CrI, 15–210%) higher susceptibility, respectively.
Conclusions
Susceptibility to influenza infection is associated with the nose/throat microbiome at the time of exposure. The effects of oligotypes on susceptibility differ between influenza A(H3N2) and B viruses. Our results suggest that microbiome may be a useful predictor of susceptibility, with the implication that microbiome could be modulated to reduce influenza infection risk, should these associations be causal.
Keywords: influenza, microbiome, susceptibility, transmission
The upper respiratory microbiome may play a role in susceptibility to influenza virus. This study examines the role of the nose/throat microbiome on influenza infection using a household influenza transmission study.
Influenza causes an estimated 3 to 5 million severe illnesses and 400 000 deaths annually [1, 2]. Vaccination is currently the most effective strategy for controlling influenza transmission, but vaccine effectiveness has been suboptimal in recent seasons mostly due to vaccine–virus mismatch [3]. Moreover, vaccine coverage is typically low in low- and middle-income countries [4]. To develop complementary influenza-prevention strategies, it is important to identify host determinants of influenza susceptibility.
One potential prevention strategy is to manipulate the host nasal/throat microbiome. Both animal models and human studies suggest that the respiratory microbiome in the nose/throat may affect host immunity and modify susceptibility to viral respiratory infections, including influenza [5–9]. However, much remains to be learned, including associations between different microbiota compositions and risk of different viral infections, and the possibility of manipulating the microbiome to prevent infection.
As one of the major venues for influenza transmission, the household is an ideal setting to examine the relationship between biological characteristics of hosts, including the respiratory microbiome, and their risk of influenza infection [10, 11]. In a typical household transmission study, exposed household contacts are closely monitored for influenza infection intensively in the approximately 2 weeks following illness onset in an index case, a period when they are highly infectious [11]. The household secondary attack rate, defined as the risk of infection for household members living with an index case, of seasonal influenza ranges from 10% to 20% [12]. We conducted a household influenza transmission study in Managua, Nicaragua, during 2012–2014 and identified an association between influenza transmission and a microbiome community state type. However, that analysis is based on simple regression methods and the results are not specific to influenza type/subtypes [13]. Another analysis based on the same data found that higher bacterial community diversity prior to infection was associated with longer shedding duration and earlier time to infection [14]. This indicates that the role of the microbiome in influenza transmission could be critical.
Here we present a more thorough analysis of the same data using a transmission model that is capable of quantifying the type/subtype-specific relationship between the nasal/throat microbiota and susceptibility to influenza A(H3N2) and B virus infection. This model accounts for transmission dynamics, including timing of infections, community infection risk, and household transmission chains, providing a more comprehensive picture of the role of the respiratory microbiome on influenza risk.
METHODS
Study Subjects
Detailed methods for the household influenza transmission study have been published [15]. Briefly, index influenza cases were identified at the Health Center Sócrates Flores Vivas, a primary care facility. Index cases were enrolled if (1) they experienced influenza-like illness, defined as fever or feverishness with cough, sore throat, or rhinorrhea, with symptom onset at 48 hours or earlier; (2) they were positive for influenza by rapid antigen test; (3) they had no other household members with influenza symptoms in the previous 2 weeks; and (4) they were living with 1 or more household member. Following enrollment, we conducted a household visit to collect initial respiratory samples and obtain demographic and symptom information. We visited each household up to 4 additional times, every 2–3 days, to collect respiratory samples and daily symptom information. This study was approved by the institutional review boards at the Nicaraguan Ministry of Health and the University of Michigan. Written consent to participate or parental permission was obtained for all participants. Verbal assent was obtained for children aged 6 years or older.
Laboratory Methods
Combined nasal and throat swabs were stored at 4–8°C in viral transport medium and transported to the National Virology Laboratory within 12 hours of collection. Samples were tested for influenza virus types (A and B) and subtypes (H1N1pdm and H3N2) by real-time reverse transcriptase–polymerase chain reaction (RT-PCR) on an Applied Biosystems 7500 Fast PCR platform following validated protocols from the US Centers for Disease Control and Prevention. Sample aliquots were stored at −70°C for microbiota characterization.
Microbiota Characterization
We characterized the microbiota of swabs collected at the first home visit. Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen) and an additional solution consisting of cell lysis solution (Promega), lysozyme, mutanolysin, RNase A, and lysostaphin (Sigma-Aldrich).
The V4 hypervariable region of the 16S rRNA gene was sequenced using Illumina MiSeq V2 chemistry 2x250 (Illumina) and a validated dual-indexing method [13]. Following alignment and quality filtering in mothur v1.38.1 (www.mothur.org/wiki/MiSeq_SOP, accessed 18 November 2016) reads were partitioned into unique taxonomic units (oligotypes) using Minimum Entropy Decomposition with default parameters (-M: 13779.0, -V: 3 nt). Representative sequences of oligotypes were classified using the Human Oral Microbiome Database v14.51 [16] and blastn v2.2.23 [17]. Taxonomic classifications with ≥Taxidentity (≥98% identity) were kept.
After excluding any sample with fewer than 1000 total reads, samples were assigned to 5 bacterial community types using Dirichlet multinomial mixture models [18] and the DirichletMultinomial v1.16.0 R package [19]. The final number of community types was selected based on fit of the negative log models and statistical power in downstream analysis. The community type for an individual was assigned as missing when his/her maximum posterior probabilities for community types were less than 0.9 [18].
Statistical Models
Our analysis focused on RT-PCR–confirmed influenza A(H3N2) and B infection. There were only 16 index cases with influenza A(H1N1)pdm infection, which were insufficient to conduct robust estimation. We used an individual-based hazard household transmission model [20, 21] to characterize transmission in households, involving risks of infection from outside the household (“community infections”) or from infected household members (“secondary or tertiary infections”) to estimate the association between the nose/throat microbiome at enrollment and influenza susceptibility. This relaxed an assumption that all household contacts were infected by their index cases, which is commonly made in regression-type analyses (Supplementary Information, Section 1).
In this model, the infection risk of each contact depended on the infectivity of other infected household members (determined by the time since symptom onset) and a daily infection risk from the community. Serial intervals, defined as the time between 2 consecutive cases, were assumed to follow a discretized Weibull distribution. In the transmission analyses, we adjusted for age (≤18 years vs >18 years), vaccination status (seasonal trivalent inactivated influenza vaccine prior to that influenza season), and household size (<4 people vs ≥5 people).
We used 2 different measures of the nose/throat microbiome in the model. First, we considered the community types identified as described above. Second, we used oligotypes that account for more than 50% of the difference between community types [22]. For the second measure, we conducted the analyses in 2 steps because the number of oligotypes is large. In step 1, we used the model to fit each oligotype separately to explore oligotypes potentially associated with susceptibility, defined as a 90% credible interval of the risk ratio excluding 1. In step 2, we used the transmission model to fit all oligotypes selected by step 1 together.
Inference
Model fitting was conducted in a Bayesian framework. Association is defined as a risk ratio with a 95% credible interval excluding 1. To account for missing data in community type, we constructed a Markov Chain Monte Carlo algorithm [23] that permits the sampling of missing community types. Hence, model parameters and missing community types were estimated jointly (Supplementary Information, Section 2). We conducted simulation to evaluate our model adequacy (Supplementary Information, Section 3). Statistical analyses were conducted using R version 3.2.1. Data and code availability are summarized in Supplementary Information, Section 4.
RESULTS
Study Participants
We recruited 76 individuals with PCR-confirmed influenza A(H3N2) virus and 39 individuals with PCR-confirmed influenza B virus infection together with 286 and 150 household contacts, respectively, between August 2012 and November 2014 (Table 1). The proportion of households with at least 1 secondary case for influenza A(H3N2) index cases was lower than that for influenza B index cases (P = .048). Other characteristics of index cases were similar between the households affected by influenza A(H3N2) and influenza B. The proportion of vaccinated household contacts of influenza A(H3N2) index cases was lower than that of influenza B index cases (P < .0001). Other characteristics for household contacts were similar between influenza A(H3N2) and B households. The observed risks of PCR-confirmed infection in household contacts were 15.0% (43/286; 95% confidence interval [CI], 11.1–19.7%) and 21.3% (32/150; 95% CI, 15.1–28.8%) for influenza A(H3N2) and B, respectively.
Table 1.
Characteristics of Index Case Patients With Influenza A(H3N2) or B Virus Infection and Their Household Contacts
| Characteristics | Influenza A(H3N2) | Influenza B |
|---|---|---|
| Index cases | ||
| No. of index cases | 76 | 39 |
| Age, years | ||
| 0–5 | 47 (62) | 16 (41) |
| 6–17 | 24 (32) | 22 (56) |
| >17 | 5 (7) | 1 (3) |
| Male | 40 (53) | 22 (56) |
| Prior vaccination | 2 (3) | 3 (8) |
| Oseltamivir treatment | 67 (88) | 37 (95) |
| Microbial community type | ||
| 1 | 23 (30) | 8 (21) |
| 2 | 17 (22) | 6 (15) |
| 3 | 10 (13) | 5 (13) |
| 4 | 10 (13) | 8 (21) |
| 5 | 15 (20) | 10 (26) |
| Missing | 1 (1) | 2 (5) |
| Number of household contacts | ||
| 1–3 | 45 (59) | 25 (64) |
| 4–5 | 17 (22) | 6 (15) |
| ≥6 | 14 (18) | 8 (21) |
| Number of secondary cases in household | ||
| 0 | 51 (67) | 18 (46) |
| 1 | 15 (20) | 14 (36) |
| 2 | 5 (7) | 5 (13) |
| ≥3 | 5 (7) | 2 (5) |
| Symptom profile | ||
| Fever | 76 (100) | 39 (100) |
| Rhinorrhea | 74 (97) | 37 (95) |
| Sore throat | 37 (49) | 15 (38) |
| Cough | 72 (95) | 36 (92) |
| ILIa | 72 (95) | 37 (95) |
| Household contacts | ||
| No. of contacts | 286 | 150 |
| Age, years | ||
| 0–5 | 33 (12) | 19 (13) |
| 6–17 | 88 (31) | 45 (30) |
| >17 | 165 (58) | 86 (57) |
| Male | 106 (37) | 58 (39) |
| Prior vaccination | 6 (2) | 18 (12) |
| Bacterial community type | ||
| 1 | 72 (25) | 36 (24) |
| 2 | 57 (20) | 38 (25) |
| 3 | 69 (24) | 30 (20) |
| 4 | 41 (14) | 23 (15) |
| 5 | 36 (13) | 14 (9) |
| Missing | 11 (4) | 9 (6) |
| Household contacts with RT-PCR–confirmed infection | ||
| Overall | 43/286 (15) | 32/150 (21) |
| Age, years | ||
| 0–5 | 13/33 (39) | 8/19 (42) |
| 6–17 | 17/88 (19) | 16/45 (36) |
| >17 | 13/165 (8) | 8/86 (9) |
| Symptom profile | ||
| Fever | 19/43 (44) | 22/32 (69) |
| Rhinorrhea | 24/43 (56) | 20/32 (62) |
| Sore throat | 15/43 (35) | 14/32 (44) |
| Cough | 26/43 (60) | 18/32 (56) |
| ILIa | 17/43 (40) | 15/32 (47) |
Data are presented as n (%) or n/N (%) unless otherwise indicated. Abbreviations: ILI, influenza-like illness; RT-PCR, reverse transcriptase–polymerase chain reaction.
aFever with sore throat or cough.
The characteristics of household contacts stratified by influenza virus type of index cases and baseline bacterial community type are presented in Table 2. The proportion of children among household contacts with community type 4 was lower than those with other community types, regardless of influenza virus type (P < .01 for influenza A(H3N2), P = .046 for influenza B). The observed risk of influenza A(H3N2) virus infection among household contacts with community type 4 was lower (2%; 95% CI, 0–13%) than with other community types (14–22%). Increased and decreased risks of influenza B infection among household contacts were observed for community type 1 (28%; 95% CI, 14–45%) and type 5 (7%; 95% CI, 0–34%), respectively, compared with other community types (17–28%). The distributions of oligotypes in each of the baseline community types among household contacts, stratified by influenza type of the index case, are summarized in Supplementary Table 1. The range of the abundance of those oligotypes is shown in Supplementary Figure 1.
Table 2.
Characteristics of Household Contacts by Bacterial Community Type
| Bacterial Community Type | ||||||
|---|---|---|---|---|---|---|
| Characteristics | 1 | 2 | 3 | 4 | 5 | Missing |
| Influenza A(H3N2) | ||||||
| No. of contacts | 72 | 57 | 69 | 41 | 36 | 11 |
| Age, years | ||||||
| 0–5 | 6/72 (8) | 4/57 (7) | 5/69 (7) | 1/41 (2) | 14/36 (39) | 3/11 (27) |
| 6–17 | 30/72 (42) | 21/57 (37) | 15/69 (22) | 10/41 (24) | 7/36 (19) | 5/11 (45) |
| >17 | 36/72 (50) | 32/57 (56) | 49/69 (71) | 30/41 (73) | 15/36 (42) | 3/11 (27) |
| Male | 29/72 (40) | 17/57 (30) | 21/69 (30) | 18/41 (44) | 16/36 (44) | 5/11 (45) |
| Prior vaccination | 0/72 (0) | 2/57 (4) | 2/69 (3) | 0/41 (0) | 1/36 (3) | 1/11 (9) |
| Infection confirmed | ||||||
| Overall | 11/72 (15) | 8/57 (14) | 13/69 (19) | 1/41 (2) | 8/36 (22) | 2/11 (18) |
| Age 0–5 years | 3/6 (50) | 0/4 (0) | 3/5 (60) | 0/1 (0) | 5/14 (36) | 2/3 (67) |
| Age 6–17 years | 6/30 (20) | 5/21 (24) | 3/15 (20) | 1/10 (10) | 2/7 (29) | 0/5 (0) |
| Age >17 years | 2/36 (6) | 3/32 (9) | 7/49 (14) | 0/30 (0) | 1/15 (7) | 0/3 (0) |
| Symptom profile | ||||||
| Fever | 5/11 (45) | 4/8 (50) | 4/13 (31) | 1/1 (100) | 5/8 (62) | 0/2 (0) |
| Rhinorrhea | 4/11 (36) | 7/8 (88) | 6/13 (46) | 1/1 (100) | 6/8 (75) | 0/2 (0) |
| Sore throat | 4/11 (36) | 4/8 (50) | 5/13 (38) | 0/1 (0) | 2/8 (25) | 0/2 (0) |
| Cough | 5/11 (45) | 7/8 (88) | 8/13 (62) | 1/1 (100) | 5/8 (62) | 0/2 (0) |
| ILIa | 4/11 (36) | 4/8 (50) | 4/13 (31) | 1/1 (100) | 4/8 (50) | 0/2 (0) |
| Influenza B | ||||||
| No. of contacts | 36 | 38 | 30 | 23 | 14 | 9 |
| Age, years | ||||||
| 0–5 | 2/36 (6) | 8/38 (21) | 0/30 (0) | 0/23 (0) | 5/14 (36) | 4/9 (44) |
| 6–17 | 10/36 (28) | 12/38 (32) | 12/30 (40) | 5/23 (22) | 4/14 (29) | 2/9 (22) |
| >17 | 24/36 (67) | 18/38 (47) | 18/30 (60) | 18/23 (78) | 5/14 (36) | 3/9 (33) |
| Male | 13/36 (36) | 14/37 (37) | 9/30 (30) | 12/23 (52) | 6/14 (43) | 4/9 (44) |
| Prior vaccination | 7/36 (19) | 3/37 (8) | 5/30 (17) | 1/23 (4) | 2/14 (14) | 0/9 (0) |
| Infection confirmed | ||||||
| Overall | 10/36 (28) | 8/38 (21) | 5/30 (17) | 4/23 (17) | 1/14 (7) | 4/9 (44) |
| Age 0–5 years | 2/2 (100) | 4/8 (50) | NA | NA | 0/5 (0) | 2/4 (50) |
| Age 6–17 years | 3/10 (30) | 3/12 (25) | 5/12 (42) | 3/5 (60) | 1/4 (25) | 1/2 (50) |
| Age >17 years | 5/24 (21) | 1/18 (6) | 0/18 (0) | 1/18 (6) | 0/5 (0) | 1/3 (33) |
| Symptom profile | ||||||
| Fever | 5/10 (50) | 4/8 (50) | 4/5 (80) | 4/4 (100) | 1/1 (100) | 4/4 (100) |
| Rhinorrhea | 7/10 (70) | 5/8 (62) | 3/5 (60) | 2/4 (50) | 0/1 (0) | 3/4 (75) |
| Sore throat | 3/10 (30) | 4/8 (50) | 3/5 (60) | 3/4 (75) | 1/1 (100) | 0/4 (0) |
| Cough | 5/10 (50) | 6/8 (75) | 3/5 (60) | 2/4 (50) | 1/1 (100) | 1/4 (25) |
| ILIa | 3/10 (30) | 4/8 (50) | 3/5 (60) | 3/4 (75) | 1/1 (100) | 1/4 (25) |
Data are presented as n/N (%) unless otherwise indicated. Abbreviations: ILI, influenza-like illness; NA, not applicable.
aFever with sore throat or cough.
Association Between Community Type and Influenza Virus Infection Susceptibility
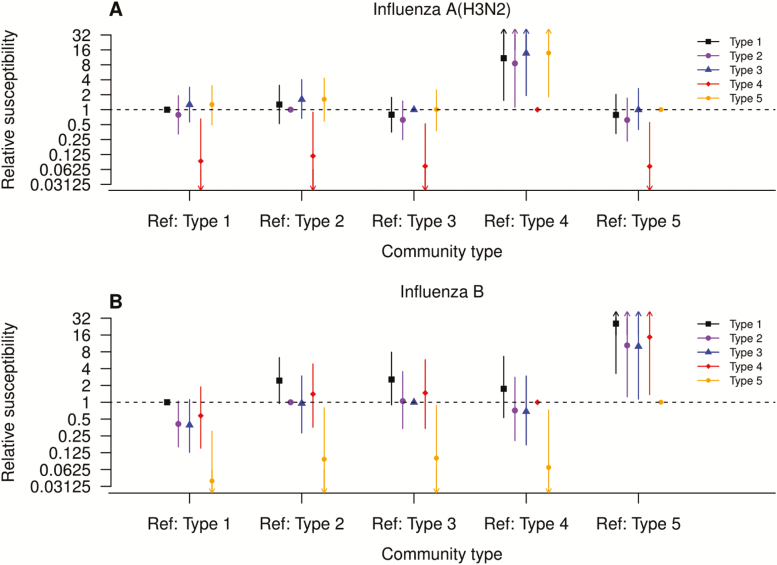
We fitted our transmission model to estimate the association between community type and susceptibility to influenza, while accounting for missing community types (Figure 1A, Supplementary Tables 2 and 3). Simulations suggested that our model provided reasonable fit to the data (Supplementary Figure 2). After adjusting for age groups, vaccination status, and household size, community type 4 was associated with lower susceptibility to influenza A(H3N2) infection, with risk ratios of 0.07 to 0.12 in reference to the other 4 community types. Community type 5 (shown in yellow; Figure 1B) was associated with lower susceptibility to influenza B infection, with risk ratios of 0.04 to 0.10, compared with the other 4 community types. As a sensitivity analysis, we refitted the model with symptomatic influenza infection and the results were similar (Supplementary Table 4).
Figure 1.
A and B, Association between bacterial community types in household contacts and susceptibility to PCR-confirmed influenza infection, by influenza type estimated by the household transmission model. Squares (black), larger circles (purple), triangles (blue), diamonds (red), and smaller circles (orange) represent community types 1 to 5, respectively. Abbreviations: PCR, polymerase chain reaction; Ref, reference group.
Oligotype Contribution to Susceptibility
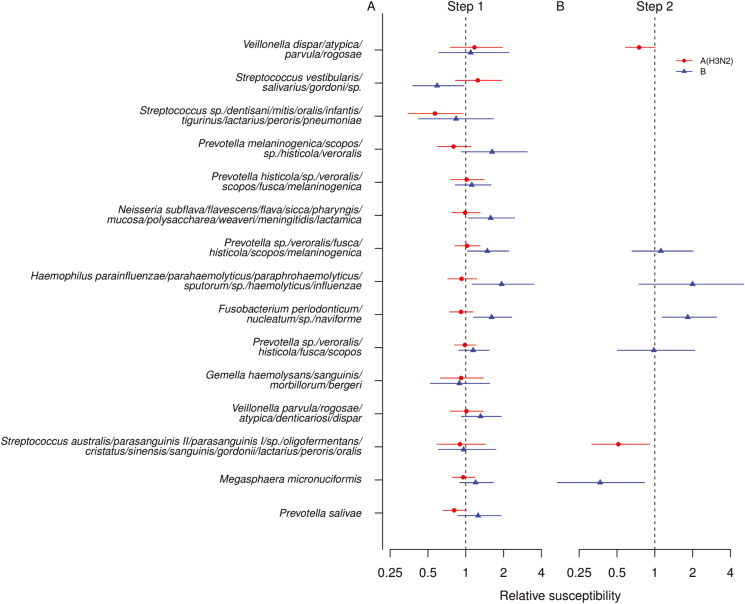
We explored which oligotypes that accounted for more than 50% of the difference between community types contribute to susceptibility. Step 1 transmission analyses were models fitted for each oligotype, adjusted for age, vaccination, and household size (Figure 2A, Supplementary Table 5) We found that Streptococcus species (spp.) (dentisani, mitis, oralis, infantis, tigurinus, lactarius, peroris, pneumoniae) and Prevotella salivae were potentially associated with lower susceptibility to influenza A(H3N2) infection. Step 2 transmission analyses were models fitted with these oligotypes together, adjusted for the same variables in the first step (Figure 2B). We estimated that a 10-fold increase in the abundance in Streptococcus spp. (dentisani, mitis, oralis, infantis, tigurinus, lactarius, peroris, pneumoniae) and Prevotella salivae was associated with 48% (95% credible interval [CrI], 9–69%) and 25% (95% CrI, 0.5–42%) lower susceptibility to influenza A(H3N2) infection. Streptococcus spp. (dentisani, mitis, oralis, infantis, tigurinus, lactarius, peroris, pneumoniae) was the most abundant oligotype in community type 4, suggesting that the lower susceptibility to influenza A(H3N2) infection with community type 4 may be possibly explained by this oligotype.
Figure 2.
Association between bacterial oligotype and susceptibility to PCR-confirmed infection for household contacts estimated by household transmission model. Circles (red) and triangles (blue) indicate PCR-confirmed influenza A(H3N2) and B virus infection, respectively. A, The points and lines represented the point estimate and 90% credible intervals of the association between oligotypes and susceptibility estimated in separate models in step 1. B, The points and lines represented the point estimate and 95% credible intervals of the association between oligotypes and susceptibility estimated in a single model that included those oligotypes with 90% credible intervals did not cover 1 in panel A. Abbreviation: PCR, polymerase chain reaction.
In contrast, we found 5 oligotypes that were potentially associated with susceptibility to influenza B infection in step 1 analyses (Figure 2A, Supplementary Table 3). Based on the multivariate model including these 5 oligotypes, we found that a 10-fold increase in the abundance of Streptococcus vestibularis, salivarius, and gordonii spp. was associated with 63% (95% CrI: 17%, 83%) lower susceptibility, while a 10-fold increase in the abundance of Prevotella spp. (veroralis, fusca, histicola, scopos, melaninogenica) was associated with 83% (95% CrI, 15–210%) higher susceptibility to influenza B infection. Different from community types 1–4, Streptococcus vestibularis, salivarius, and gordonii spp. and Prevotella spp. veroralis, fusca, histicola, scopos, and melaninogenica were the most and least abundant oligotypes in community type 5, suggesting that the lower susceptibility to influenza B infection among individuals with community type 5 may be mostly explained by these 2 oligotypes.
Associations With Age, Household Size, and Vaccination Status
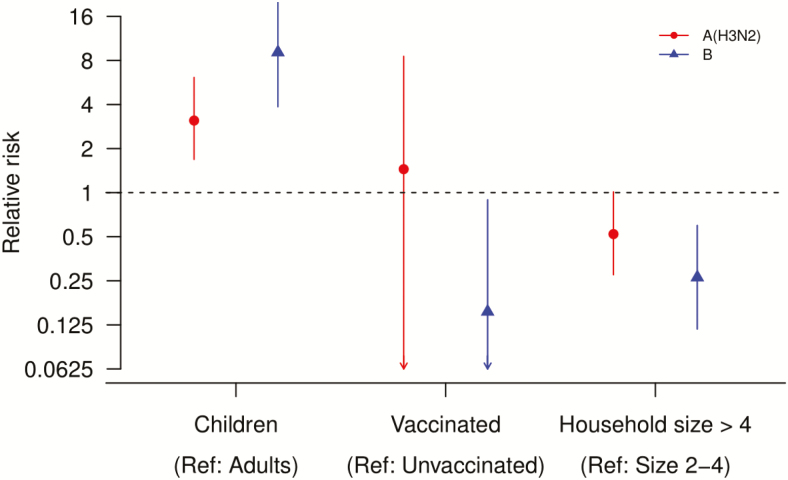
We found that child household contacts (<18 years) were more susceptible to influenza A(H3N2) and B infection than adult contacts (≥18 years), with a risk ratio of 3.1 (95% CrI, 1.7–6.1) and 9.1 (95% CrI, 3.9–23.4), respectively (Figure 3, Supplementary Table 4). We estimated that influenza vaccine protected household contacts against influenza B virus infection, with a risk ratio of 85% (95% CrI, 11–99%), but not for influenza A(H3N2) virus infection (Figure 3, Supplementary Table 4). Residing in a household with 5 or more people was associated with a 48% (95% CrI, −1% to 73%) and 73% (95% CrI, 40–88%) lower risk of influenza A(H3N2) and B virus infection, respectively, compared with those in smaller households (Figure 3, Supplementary Table 4).
Figure 3.
Effect of age group, vaccination status, and household size on transmission estimated under the household transmission model. Circles (red) and triangles (blue) were for household contacts with index cases with PCR-confirmed influenza A(H3N2) and B virus infection, respectively. Abbreviations: PCR, polymerase chain reaction; Ref, reference.
Serial Interval
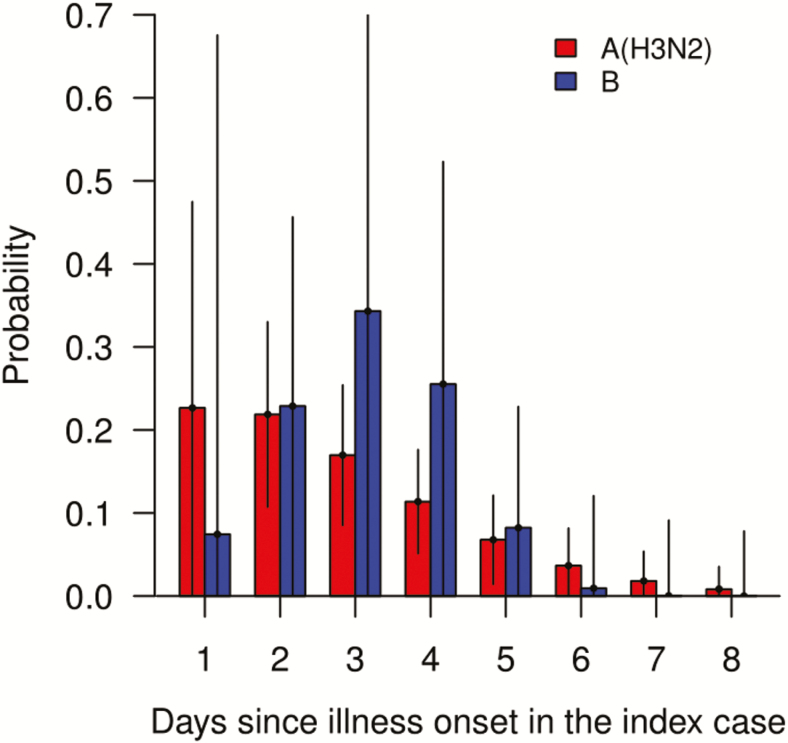
We estimated that the mean serial interval for within-household transmission for influenza A(H3N2) and B virus infection was 2.4 days (95% CrI, 1.1–3.9 days) and 3.1 days (95% CrI, 0.9–6.6 days), respectively (Figure 4).
Figure 4.
Serial interval distribution estimated under the transmission model, by influenza type, accounting for tertiary infections and infections from outside households. The estimated mean serial intervals for influenza A(H3N2) and B virus infection were 2.4 days (95% CrI, 1.1–3.9 days) and 3.1 days (95% CrI, 0.9–6.6 days), respectively. Abbreviation: CrI, credible interval.
DISCUSSION
Compared with the earlier analysis using a mixed-effects regression model [22], the current transmission analyses advance our understanding about the association between the nasal/throat microbiome and the risk of influenza virus infection by providing influenza-type–specific associations in a household transmission setting. We identified bacterial community types and oligotypes associated with the susceptibility to influenza A(H3N2) and B, while adjusting for age, household size and vaccination status. Our framework allows for imputing missing community types to include more households in the analyses to increase the power for detecting associations.
According to the transmission analyses, the community type but also the oligotype of the nasal/throat microbiome were associated with susceptibility to influenza A(H3N2) and B. These associations differ between influenza subtypes. Our findings are consistent with mouse models demonstrating associations between mouse commensal microbiota within the respiratory tract and susceptibility to influenza infection [5, 6].
The abundance of Streptococcus spp. was associated with lower susceptibility for influenza A(H3N2) and B infection in our analyses. This is consistent with previous studies showing that stimulation of the immune system by Streptococcus colonization may inhibit influenza viral replication [24–26]. In a mouse model, prior S. pneumoniae infection protected against severe influenza virus infection [26]. Among healthy young adults inoculated with live attenuated influenza vaccine, Streptococcus infantis was positively associated with influenza H1 immunoglobulin A (IgA) titers [27].
In contrast, the abundance of Prevotella spp. was associated with increased susceptibility to influenza B but not to influenza A(H3N2). Prevotella has previously been associated with increased severity of influenza, tuberculosis, and chronic obstructive pulmonary disease [28–30], but this is the first time its association with susceptibility to influenza has been detected. More in vitro or in vivo studies are needed to verify whether and how Prevotella modify host innate immunity or immune response specific to influenza viruses.
Our dataset has been analyzed without microbiome data [15] based on models without the subtype differences. Here, the microbiome data were added to the model and the estimates for factors affecting transmission were similar, suggesting that the association between microbiome and susceptibility may be independent of those factors. Consistent with the literature [31–33] and our previous analysis [15], we estimated that the mean serial interval for influenza B virus is 3.1 days, longer than the 2.4 days estimated for influenza A(H3N2) virus. Also consistent with the literature and our previous analysis [15], children were more susceptible to influenza A(H3N2) and B virus infection than adults [12, 21, 31, 34] and the differences for influenza B were more extreme than H3N2 [31, 35]. Potential explanations are the lower levels of pre-existing immunity among children, their higher frequency of contact and hence higher risk of exposure to influenza [36], or an inherent difference in the transmissibility between influenza A and B viruses [37]. Also, we found that smaller households were associated with higher secondary attack rate, which was consistent with our previous report [15] and other studies [20, 34].
In our study population, vaccine effectiveness for influenza A(H3N2) was much lower than for influenza B, which is consistent with a meta-analysis published in 2016 [3]. Other studies suggest that this difference may be due to poor vaccine-induced protection in some hosts [38] or mismatch of vaccine strain and circulating A(H3N2) strains due to egg adaptation [39].
Our study has several limitations. Index cases were recruited from health center attendees whose influenza was detected using a rapid test, suggesting they had more severe symptoms and higher transmissibility than general influenza cases. The enrollment of index cases was limited to 2 days or fewer after symptom onset. As adults with illness usually tend to present later to clinics than children, our study likely has an overrepresentation of child index cases. Some tertiary infections may have been missed since the duration of follow-up was from 9–12 days after recruitment. However, the effect of this right-censoring should be minimal since the mean serial interval was around 3 days [15, 21]. Diet information was not collected, and hence the effect of diet on the nose/throat microbiome could not be explored. Infections were defined by PCR positivity, with no culture results. While PCR is the gold standard, it may detect infections with low infectiousness [40]. Finally, although important risk modifiers were adjusted for in our analyses [20, 34], we cannot be sure that detected associations are causal as there could be other important unmeasured confounders. A major strength of our study was that up to 5 respiratory samples were collected from household contacts over a period of 9–12 days after enrollment, regardless of symptoms. Therefore, the likelihood of missing infections due to peak viral shedding occurring between collection of sequential respiratory samples under such an intense sampling should be small.
In conclusion, we found that some bacterial community types and oligotypes of the nose/throat microbiome were associated with susceptibility to influenza. Importantly, these associations were dependent on influenza virus type/subtype. Our results suggest that the microbiome may serve as a useful predictor for susceptibility and have an implication for an alternative approach to the prevention of influenza infection via modulating the microbiome in the upper respiratory tract, should these associations be causal.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the families that participated in the study and our excellent study staff both at the Health Center Sócrates Flores Vives and the Centro Nacional de Diagnóstico y Referencia.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under grant numbers U01AI088654 and R01AI120997 (to A. G.), grant number R21AI119463 (to B. F. and A.G.), and contract number HHSN272201400006C (subcontract to A. G.) and the Center for Statistics and Quantitative Infectious Diseases (grant number U54 GM111274-01) (to T. K. T.).
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Iuliano AD, Roguski KM, Chang HH, et al. ; Global Seasonal Influenza-associated Mortality Collaborator Network Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 2018; 391:1285–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. Influenza. In: Immunization, vaccines and biologicals: influenza. 2017. Available at: http://www.who.int/immunization/topics/influenza/en/. Accessed 17 February 2017. [Google Scholar]
- 3. Belongia EA, Simpson MD, King JP, et al. Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis 2016; 16:942–51. [DOI] [PubMed] [Google Scholar]
- 4. World Health Organization Global Influenza Programme. Seasonal influenza vaccine use in low and middle income countries in the tropics and subtropics: a systematic review [Internet]. Geneva, Switzerland: World Health Organization, 2015. Available at: http://apps.who.int/iris/bitstream/10665/188785/1/9789241565097_eng.pdf. Accessed 19 January 2019. [Google Scholar]
- 5. Abt MC, Osborne LC, Monticelli LA, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012; 37:158–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ichinohe T, Pang IK, Kumamoto Y, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A 2011; 108:5354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol 2017; 35:8–15. [DOI] [PubMed] [Google Scholar]
- 8. Luoto R, Ruuskanen O, Waris M, Kalliomäki M, Salminen S, Isolauri E. Prebiotic and probiotic supplementation prevents rhinovirus infections in preterm infants: a randomized, placebo-controlled trial. J Allergy Clin Immunol 2014; 133:405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Panigrahi P, Parida S, Nanda NC, et al. Corrigendum: a randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 2018; 553:238. [DOI] [PubMed] [Google Scholar]
- 10. Chao DL, Halloran ME, Obenchain VJ, Longini IM Jr. FluTE, a publicly available stochastic influenza epidemic simulation model. PLoS Comput Biol 2010; 6:e1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Monto AS. Studies of the community and family: acute respiratory illness and infection. Epidemiol Rev 1994; 16:351–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsang TK, Lau LLH, Cauchemez S, Cowling BJ. Household transmission of influenza virus. Trends Microbiol 2016; 24:123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013; 79:5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee KH, Foxman B, Kuan G, et al. The respiratory microbiota: associations with influenza symptomatology and viral shedding. Ann Epidemiol 2019; 37:51–56.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gordon A, Tsang TK, Cowling BJ, et al. Influenza transmission dynamics in urban households, Managua, Nicaragua, 2012-2014. Emerg Infect Dis 2018; 24:1882–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microb e taxonomic and genomic information. Database (Oxford) 2010; 2010:baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403–10. [DOI] [PubMed] [Google Scholar]
- 18. Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One 2012; 7:e30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan M. DirichletMultinomial: Dirichlet-multinomial mixture model machine learning for microbiome data. R package version 1.26.0. 2017. Available at: https://bioconductor.org/packages/release/bioc/html/DirichletMultinomial.html . Accessed 19 July 2018. [Google Scholar]
- 20. Cauchemez S, Donnelly CA, Reed C, et al. Household transmission of 2009 pandemic influenza A (H1N1) virus in the United States. N Engl J Med 2009; 361:2619–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsang TK, Cauchemez S, Perera RA, et al. Association between antibody titers and protection against influenza virus infection within households. J Infect Dis 2014; 210:684–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee KH, Gordon A, Shedden K, et al. The respiratory microbiome and susceptibility to influenza virus infection. PLoS One 2019; 14:e0207898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gilks WR, Richardson S, Spiegelhalter D.. Markov Chain Monte Carlo in practice. London: Chapman & Hall, 1996. [Google Scholar]
- 24. Short KR, Habets MN, Hermans PW, Diavatopoulos DA. Interactions between Streptococcus pneumoniae and influenza virus: a mutually beneficial relationship? Future Microbiol 2012; 7:609–24. [DOI] [PubMed] [Google Scholar]
- 25. Diavatopoulos DA, Short KR, Price JT, et al. Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. FASEB J 2010; 24:1789–98. [DOI] [PubMed] [Google Scholar]
- 26. McCullers JA, Rehg JE. Lethal synergism between influenza virus and Streptococcus pneumoniae: characterization of a mouse model and the role of platelet-activating factor receptor. J Infect Dis 2002; 186:341–50. [DOI] [PubMed] [Google Scholar]
- 27. Salk HM, Simon WL, Lambert ND, et al. Taxa of the nasal microbiome are associated with influenza-specific IgA response to live attenuated influenza vaccine. PLoS One 2016; 11:e0162803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langevin S, Pichon M, Smith E, et al. Early nasopharyngeal microbial signature associated with severe influenza in children: a retrospective pilot study. J Gen Virol 2017; 98:2425–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hui AW, Lau HW, Chan TH, Tsui SK. The human microbiota: a new direction in the investigation of thoracic diseases. J Thorac Dis 2013; 5(Suppl 2):S127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheung MK, Lam WY, Fung WY, et al. Sputum microbiota in tuberculosis as revealed by 16S rRNA pyrosequencing. PLoS One 2013; 8:e54574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu C, Chan KH, Tsang TK, et al. Comparative epidemiology of influenza B Yamagata- and Victoria-lineage viruses in households. Am J Epidemiol 2015; 182:705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Levy JW, Cowling BJ, Simmerman JM, et al. The serial intervals of seasonal and pandemic influenza viruses in households in Bangkok, Thailand. Am J Epidemiol 2013; 177:1443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Petrie JG, Ohmit SE, Cowling BJ, et al. Influenza transmission in a cohort of households with children: 2010–2011. PLoS One 2013; 8:e75339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cauchemez S, Ferguson NM, Fox A, et al. Determinants of influenza transmission in South East Asia: insights from a household cohort study in Vietnam. PLoS Pathog 2014; 10:e1004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsang TK, Fang VJ, Ip DKM, et al. Indirect protection from vaccinating children against influenza in households. Nat Commun 2019; 10:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mossong J, Hens N, Jit M, et al. Social contacts and mixing patterns relevant to the spread of infectious diseases. PLoS Med 2008; 5:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen R, Holmes EC. The evolutionary dynamics of human influenza B virus. J Mol Evol 2008; 66:655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cobey S, Gouma S, Parkhouse K, et al. Poor immunogenicity, not vaccine strain egg adaptation, may explain the low H3N 2 influenza vaccine effectiveness in 2012–13. Clin Infect Dis 2018; 67:327–33. doi: 10.1093/cid/ciy097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zost SJ, Parkhouse K, Gumina ME, et al. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci USA 2017; 114:12578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ruest A, Michaud S, Deslandes S, Frost EH. Comparison of the Directigen flu A+B test, the QuickVue influenza test, and clinical case definition to viral culture and reverse transcription-PCR for rapid diagnosis of influenza virus infection. J Clin Microbiol 2003; 41:3487–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.