Abstract
Introduction and objectives
Common laboratory parameters are crucial in aiding coronavirus disease 2019 (COVID-19) case detection. This study aimed to determine the differences between laboratory parameters in (1) COVID-19 versus non-COVID-19 pneumonia, and (2) severe versus non-severe COVID-19 cases.
Methods
Studies were collected until March 2020, and retrieved parameters include leukocyte, neutrophil, thrombocyte, and lymphocyte counts in addition to C-reactive protein (CRP), procalcitonin (PCT) and D-dimer levels. In the presence of heterogeneity, the random-effect model (REM) was used instead of the fixed-effect model (FEM).
Results
Seven studies in the first analysis showed significantly lower leukocyte, neutrophil and platelet counts in COVID-19 pneumonia (SMD = −0.42, 95%CI −0.60 to −0.25, p < 0.00001, SMD = −0.23, 95%CI −0.41 to −0.06, p = 0.01, SMD = −0.54, 95%CI −0.91 to −0.16, p = 0.0005) compared to non-COVID-19 pneumonia. Twenty-six studies in the second analysis showed significantly lower lymphocyte and thrombocyte counts (SMD = −0.56, 95%CI −0.71 to −0.40, p < 0.0001, SMD = −0.32, 95%CI −0.49 to −0.15, p = 0.0002) and significantly higher leukocyte, neutrophil, D-dimer, and CRP (SMD = 0.31, 95%CI 0.07–0.56, p = 0.01; SMD = 0.44, 95%CI 0.24–0.64, p < 0.0001; SMD = 0.53, 95%CI 0.31–0.75, p < 0.00001; SMD = 0.97, 95%CI 0.70–1.24, p < 0.00001) in severe COVID-19 compared to non-severe COVID-19.
Conclusions
In conclusion, thrombocyte count is key in both diagnosis and prognosis. Low leukocyte and neutrophil counts are markers of COVID-19 infection, but contrastingly higher counts indicate progressive COVID-19. And although lymphocyte, D-dimer and CRP levels did not demonstrate diagnostic value, all indicate severity of COVID-19. Confirmation of these findings should be performed in future studies.
Keywords: Laboratory parameters, COVID-19, SARS-CoV-2, Diagnosis
Abstract
Introducción y objetivos
Los parámetros comunes de laboratorio son cruciales para ayudar a la detección de casos de enfermedad por coronavirus 2019 (COVID-19). Este estudio tuvo como objetivo determinar las diferencias entre los parámetros de laboratorio en: 1) COVID-19 versus neumonía no COVID-19, y 2) Casos severos versus no severos de COVID-19.
Métodos
Los estudios se recolectaron hasta marzo de 2020, y los parámetros recuperados incluyen recuentos de leucocitos, neutrófilos, trombocitos y linfocitos además de los niveles de proteína C reactiva (PCR), procalcitonina (PCT) y dímero-D. En presencia de heterogeneidad, se utilizó el modelo de efectos aleatorios en lugar del modelo de efectos fijos.
Resultados
Siete estudios en el primer análisis mostraron recuentos de leucocitos, neutrófilos y plaquetas significativamente más bajos en la neumonía por COVID-19 (SMD = −0,42; IC 95%: −0,60 a −0,25; p < 0,00001; SMD = −0,23; IC 95%: −0,41 a −0,06; p = 0,01; SMD = −0,54; IC 95%: −0,91 a −0,16; p = 0,0005) en comparación con la neumonía no COVID-19. Veintiséis estudios en el segundo análisis mostraron recuentos de linfocitos y trombocitos significativamente más bajos (SMD = −0,56; IC 95%: −0,71 a −0,40; p < 0,0001; SMD = −0,32; IC 95%: −0,49 a −0,15; p = 0,0002) y leucocitos, neutrófilos, dímero D y PCR significativamente más altos (SMD = 0,31; IC 95%: 0,07-0,56; p = 0,01; SMD = 0,44; IC 95%: 0,24-0,64; p < 0,0001; SMD = 0,53; IC 95%: 0,31-0,75; p < 0,00001; SMD = 0,97; IC 95%: 0,70-1,24; p < 0,00001) en COVID-19 severo en comparación con COVID-19 no severo.
Conclusiones
En conclusión, el recuento de trombocitos es clave tanto en el diagnóstico como en el pronóstico. Los recuentos bajos de leucocitos y neutrófilos son marcadores de infección por COVID-19, pero los recuentos contrastantemente más altos indican COVID-19 progresivo. Y aunque los niveles de linfocitos, dímero D y PCR no mostraron valor diagnóstico, todos indican la gravedad de COVID-19. La confirmación de estos hallazgos debe realizarse en futuros estudios.
Palabras clave: Parámetros de laboratorio, COVID-19, SARS-CoV-2, Diagnóstico
Introduction
Since the first report1 of Coronavirus Disease 2019 (COVID-19) in Wuhan, the causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has undergone rapid global spread within a matter of months. Due to the alarming rates of transmission, the World Health Organization officially declared the situation as a pandemic, with over 1.051.635 infections and 56,985 deaths worldwide per April 6th 2020.2 The pandemic continues to place a major strain on a country's resources,3 and it is predicted that mortality and morbidity rates will become pronounced with the rising spread amongst lower- and middle-income countries where healthcare is already at capacity.4
Pandemic management emphasizes on prompt identification and containment, achievable through strict surveillance and early diagnosis.5, 6 Gold-standard diagnosis of COVID-19 is achieved through molecular identification of the SARS-CoV-2 using nucleic acid amplification tests such as the reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) or viral gene sequencing.7 In some countries, rapid serological tests are also implemented to complement molecular diagnosis. However, many low resource settings are not equipped with sufficient laboratory and human resource capacity to perform massive molecular identification. The lack of resources in addition to long turnaround time leads to delays between testing and confirmation, during which clinical judgment is crucial. During this period, patient history, hematological and biochemical laboratory parameters, and imaging are necessary for aiding the diagnostic process. Another concerning feature of SARS-CoV-2 infection is the variable case presentation with alarming mortality rates in high-risk groups such as the elderly and immunocompromised.8 Cases of COVID-19 that progress into severe and critical disease are marked with acute respiratory distress (ARDS) and multi-organ failure, requiring highly intensive care. In these scenarios, timely detection of disease progression is crucial for appropriate management and intervention.
In this meta-analysis, we aim to provide an evidence-based summary of the latest studies to (1) determine the differences between laboratory parameters in COVID-19 pneumonia and non-COVID-19 pneumonia, and (2) determine the differences between laboratory parameters in severe and non-severe COVID-19. This study focused on parameters that are routinely tested and readily available even in low-resource settings. The analysis encompasses hematological parameters (including total leukocyte, neutrophil, thrombocyte and lymphocyte counts), D-dimer, procalcitonin (PCT), and C-reactive protein (CRP) levels.
Methods
Literature search and data extraction
This meta-analysis was performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines.9 A literature search was conducted from PubMed, Scopus, the Chinese Medical Journal Network, Google Scholar, and Web of Science. Keywords such as “COVID-19”, “2019-nCOV”, “SARS-COV-2”, “novel coronavirus disease”, “novel coronavirus pneumonia”, non-COVID-19 pneumonia”, and “severe COVID-19” were used singularly and in combination. No date, location, or language restrictions were applied during the literature search.
The literature search was updated until March 2020. The inclusion criteria were as follows: (1) primary research reports of COVID-19 infection with cumulative or raw laboratory values (2) case-control or cohort study, (3) studies with matched controls (COVID-19 versus non-COVID-19 pneumonia and severe versus non-severe COVID-19). Data were extracted as follows: (1) name of the first author, (2) year of publication, (3) country and city of study origin, (4) number of COVID-19 pneumonia (PCR positive) and non-COVID-19 pneumonia (PCR negative), (5) number of severe (i.e., ICU admission, those needing mechanical ventilation, or those who died), and non-severe COVID-19 cases (6) the mean and median of laboratory parameters of the patients.
Data synthesis and statistical analysis
To assess the difference between parameters measured in COVID-19 and non-COVID-19 patients or severe and non-severe cases, pooled standardized mean difference (SMD) with 95% confidence interval (CI) was used. Heterogeneity among studies was evaluated using Q test and I 2 statistic. A significant Q-statistic (p < 0.10) indicated heterogeneity across studies. The I 2 values indicated no (0–24.9%), low (25–49.9%), moderate (50–74.9%) or high (75–100%) heterogeneity. The random-effect model (REM) was used if heterogeneity existed; otherwise, the fixed-effect model (FEM) was used. Begg's funnel plot and Egger's regression test were applied to determine publication bias if the pooled effect size consisted of 10 or more studies.10 A sensitivity test was performed by sequentially omitting one study each time to evaluate the stability of the results. Data that were not expressed as mean and standard deviation were extrapolated according to Hozo et al. and Wan et al.11, 12
Meta-analysis was performed using RevMan ver 5.3. Statistical tests were 2-sided and used a significance threshold of p < 0.05. The mean values of parameters that were found to be significant in the meta-analysis were then used to generate receiver-operating-characteristic (ROC) curves using Graphpad Prism ver 8.1.2 in order to determine the area under the curve (AUC). The optimal cutoff for parameters with a significant p-value (two-sided) were determined using the Youden's index,13, 14 and the corresponding sensitivity and specificity for the cut-off was also calculated.
Results
Laboratory parameters in COVID-19 and non-COVID-19 cases
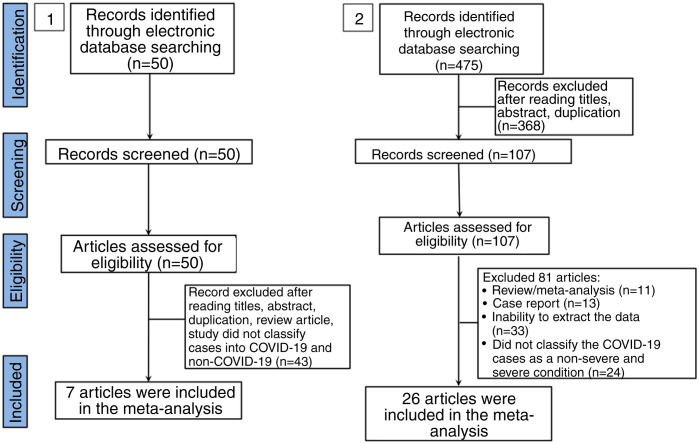
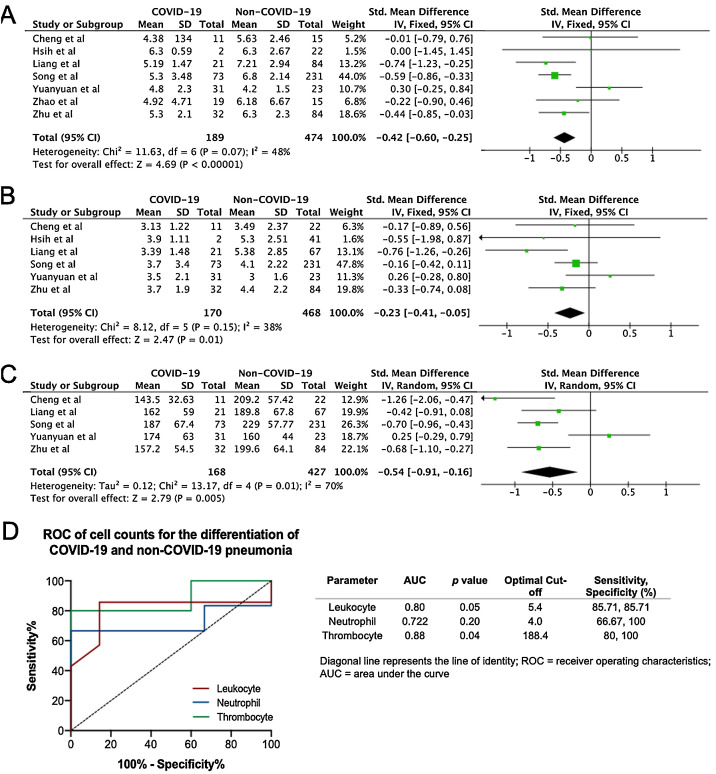
In the literature search, a total of 50 articles were retrieved from databases. After reviewing the title and abstract, 42 articles were excluded on the basis of duplications, being a review article, or if the study did not classify cases into COVID-19 and non-COVID-19 infection. Finally, only 7 studies were included in this meta-analysis (Fig. 1 ).15, 16, 17, 18, 19, 20, 21 The characteristics of included studies are shown in Supplemental Table 1. In general, the pooled studies showed high heterogeneity, except for leukocyte and neutrophil values (Table 1 ). Interestingly, we found that leukocyte, neutrophil, and thrombocyte counts were significantly lower in COVID-19 than in non-COVID-19 cases (SMD = −0.42, 95%CI −0.60 to −0.25, p < 0.00001 (Fig. 2 A); SMD = −0.23, 95%CI −0.41 to −0.06, p = 0.01 (Fig. 2B); SMD = −0.54, 95%CI −0.91 to −0.16, p = 0.0005 (Fig. 2C), respectively). No significant difference in other parameters were observed between the two groups (Table 1). A sensitivity analysis conducted by eliminating one individual study each time (data not shown), showed no change in the pooled analyses, implying the statistical stability of the findings. The ROC curve of crucial parameters is shown in Fig. 2D, demonstrating that thrombocyte count provides good discrimination (AUC = 0.88, p = 0.04) between COVID-19 and non-COVID-19 pneumonia with an optimal cutoff of 188.4 × 109/L, yielding a sensitivity of 80% and specificity of 100%.
Fig. 1.
Flow diagram of the study selection process in this meta-analysis.
Table 1.
Meta-analysis of laboratory findings between COVID-19 and Non-COVID-19 patients.
| Parameters | No. of cohort | Sample size (COVID-19/non-COVID-19) | SMD | 95% CI | I2 (%) | Statistical method | p |
|---|---|---|---|---|---|---|---|
| Leukocyte (×109/L) | 7 | 189/474 | −0.42 | −0.60 to −0.25 | 48 | FEM | <0.00001* |
| Lymphocyte (×109/L) | 7 | 189/474 | −0.37 | −0.81 to −0.07 | 78 | REM | 0.10 |
| Neutrophil (×109/L) | 6 | 170/468 | −0.23 | −0.41 to −0.06 | 38 | FEM | 0.01* |
| Thrombocyte (×109/L) | 5 | 168/427 | −0.54 | −0.91 to −0.16 | 70 | REM | 0.005* |
| D-dimer (μg/mL) | 2 | 63/107 | −0.27 | −0.59 to 0.06 | 0 | FEM | 0.11 |
| CRP (mg/L) | 6 | 168/416 | 0.29 | −0.33 to 0.90 | 87 | REM | 0.35 |
| PCT (ng/mL) | 3 | 136/338 | −0.44 | −1.20 to 0.32 | 91 | REM | 0.26 |
SMD, standard mean difference; CI, confidence interval, FEM, fixed-effect model; REM, random-effect model.
p < 0.05.
Fig. 2.
Forest plot for pooled standardized mean difference (SMD) and 95% confidence interval (CI) of laboratory parameters in COVID-19 and non-COVID-19-infected patients; (A) leukocyte; (B) neutrophil; and (C) thrombocyte. (D) Receiver operating characteristics (ROC) curve of cell counts for the differentiation of COVID-19 and non-COVID-19 pneumonia.
Laboratory parameters in severe and non-severe COVID-19 cases
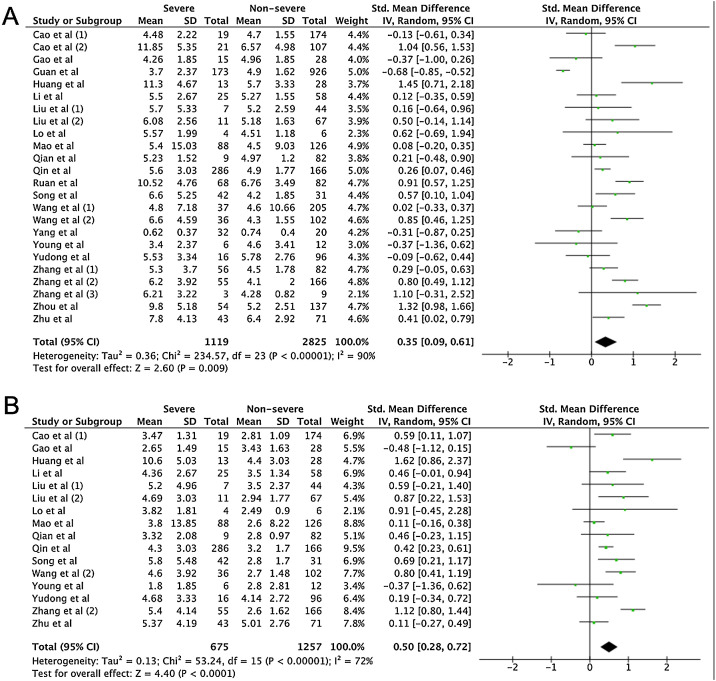
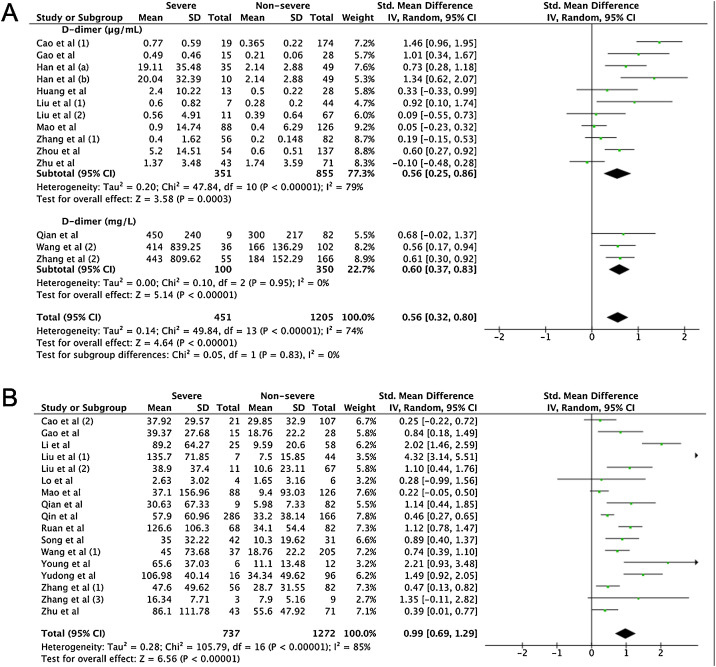
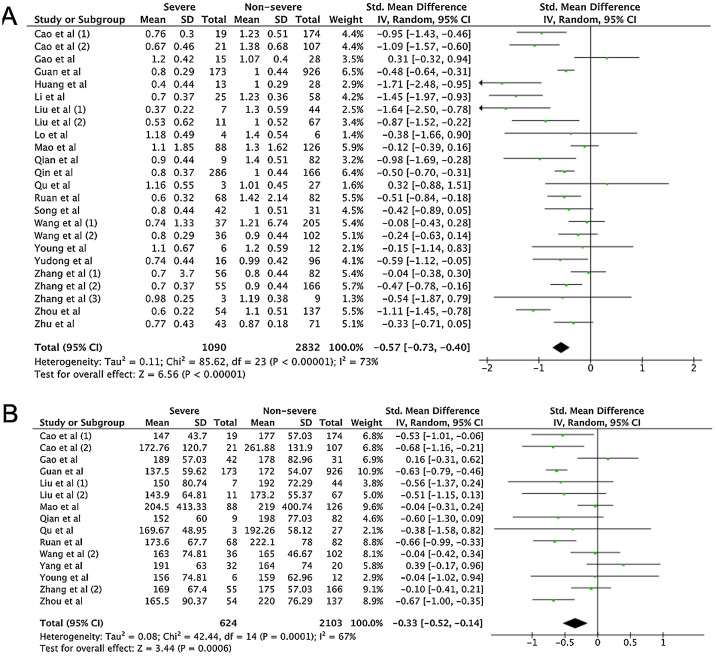
A total of 475 potentially relevant studies were initially detected, 368 of which were excluded after title and abstract review and removing duplications. One hundred and seven studies were eligible for further screening. However, 81 articles were subsequently removed on the basis of (1) being either review articles and meta-analysis (n = 11) or case reports (n = 13) (2) inability to extract the data (n = 33), or (3) did not classify the COVID-19 cases as a non-severe and severe condition (n = 24). Thus, 26 studies were included in this meta-analysis (Fig. 1).3, 18, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 Details of the retrieved studies are shown in Supplemental Table S2. Although the heterogeneity was considerably high in all parameters measured (I 2 = between 67 and 90%, (Table 2 ), we found that leukocytes, neutrophils, D-dimer, and CRP levels were significantly higher (SMD = 0.35, 95%CI 0.09–0.61, p = 0.009 (Fig. 3 A); SMD = 0.50, 95%CI 0.28–0.72, p < 0.0001 (Fig. 3B); SMD = 0.56, 95%CI 0.32–0.80, p < 0.00001 (Fig. 4 A); SMD = 0.99, 95%CI 0.69–1.29, p < 0.00001) (Fig. 4B), respectively) in COVID-19 infected patients with severe condition than in those with non-severe condition. Contrastingly, lymphocyte and thrombocyte counts were significantly lower in severe COVID-19 infected patients (SMD = −0.57, 95%CI −0.73 to −0.40, p < 0.00001; SMD = −0.33, 95%CI −0.52 to −0.14, p = 0.000; Fig. 5 A and B, respectively). No difference was observed in PCT levels (Table 2). Begg's funnel plot (Supplemental Fig. 1A–G) and Egger's test were performed revealing no publication bias (Table 2). A sensitivity analysis was also performed (data not shown), revealing identical results, hence suggesting that the statistical stability and robustness of the findings.
Table 2.
Meta-analysis of laboratory findings between severe and non-severe patients with COVID-19.
| Parameters | No. of cohort | Sample size (Severe/Non-Severe) | SMD | 95% CI | I2 (%) | Statistical method | p | Publication bias (p Egger) |
|---|---|---|---|---|---|---|---|---|
| Leukocyte (×109/L) | 24 | 1119/2825 | 0.35 | 0.09 to 0.61 | 90 | REM | 0.009* | 0.725 |
| Lymphocyte (×109/L) | 24 | 1090/2832 | −0.57 | −0.73 to −0.40 | 73 | REM | <0.00001* | 0.547 |
| Neutrophil (×109/L) | 16 | 675/1257 | 0.50 | 0.28 to 0.72 | 72 | REM | <0.0001* | 0.892 |
| Thrombocyte (×109/L) | 15 | 624/2103 | −0.33 | −0.52 to −0.14 | 67 | REM | 0.0006* | 0.840 |
| D-dimer (μg/mL or mg/L) | 14 | 451/1205 | 0.56 | 0.32 to 0.80 | 74 | REM | <0.00001* | 0.865 |
| CRP (mg/L) | 17 | 737/1272 | 0.99 | 0.69 to 1.29 | 85 | REM | <0.00001* | 0.112 |
| PCT (ng/mL) | 13 | 596/1041 | 0.19 | −0.06 to 0.44 | 75 | REM | 0.14 | 0.930 |
SMD, standard mean difference; CI, confidence interval, FEM, fixed-effect model; REM, random-effect model.
p < 0.05.
Fig. 3.
Forest plot for pooled standardized mean difference (SMD) and 95% confidence interval (CI) of laboratory parameters in severe and non-severe COVID-19-infected patients; (A) leukocyte and (B) neutrophil.
Fig. 4.
Forest plot for pooled standardized mean difference (SMD) and 95% confidence interval (CI) of laboratory parameters in severe and non-severe COVID-19-infected patients; (A) D-dimer and (B) CRP.
Fig. 5.
Forest plot for pooled standardized mean difference (SMD) and 95% confidence interval (CI) of laboratory parameters in severe and non-severe COVID-19-infected patients; (A) lymphocyte and (B) thrombocyte.
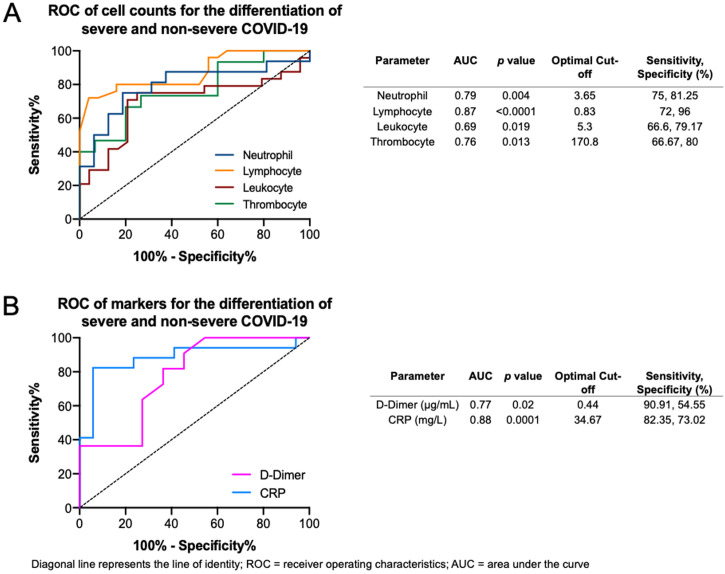
The ROC curves of the crucial blood cell count parameters are shown in Fig. 6 A, demonstrating that fair to good distinction of severe and non-severe COVID-19 was demonstrated by the neutrophil (AUC = 0.79, p = 0.004), lymphocyte (AUC = 0.87, p < 0.0001), leukocyte (AUC = 0.69, p = 0.019), and thrombocyte (AUC = 0.76, p = 0.013) counts. The optimal cut-offs are 3.65 × 109/L for neutrophil count (sensitivity = 75%, specificity = 81.2%), 0.83 × 109/L for lymphocyte count (sensitivity = 72%, specificity = 96%), 5.3 × 109/L for leukocyte count (sensitivity = 66.6%, specificity = 79.1%), and 170.8 × 109/L for thrombocyte count (sensitivity = 66.6%, specificity = 80%). The ROC curve of crucial markers (Fig. 6B), demonstrate that CRP provides better discrimination between severe and non-severe COVID-19 pneumonia relative to D-dimer (AUC = 0.88, p = 0.0001 versus AUC = 0.77, p = 0.02 respectively), with an optimal cutoff of 34.67 mg/L for CRP (sensitivity = 82.3%, specificity = 73%) and 0.44 μg/mL for D-dimer (sensitivity = 90.9%, specificity = 54.55%).
Fig. 6.
Receiver operating characteristics (ROC) curve and optimal thresholds of significant laboratory parameters. (A) Blood cell counts for the differentiation of severe and non-severe COVID-19. (B) Markers for the differentiation of severe and non-severe COVID-19.
Discussion
Molecular identification of SARS-CoV-2 in low-resource settings remains centralized and time-consuming, resulting in a delayed diagnosis of COVID-19. The first analysis performed in this study aimed to summarize current evidence to identify frequently performed laboratory parameters that can assist in COVID-19 case identification. This revealed significantly lower leukocyte, neutrophil, and thrombocyte levels in COVID-19 patients compared to non-COVID-19 patients. Additionally, the pronounced variation in COVID-19 clinical presentation and outcome makes it imperative to predict potentially progressive cases. Hence, the second analysis of this study aimed to identify common laboratory parameters that can act as markers of severe infection, revealing a significant difference in the leukocyte, neutrophil, thrombocyte, and lymphocyte counts as well as D-dimer and CRP levels between severe and non-severe COVID-19 cases.
Relative to non-COVID-19 infections, significantly lower absolute leukocyte or neutrophil counts during early stages of the disease has been broadly demonstrated,15, 18, 21 although the mean leukocyte count of the included studies often do not exceed the lower limit for categorisation as leukopenia or neutropenia. Interestingly, with COVID-19 disease progression, we found that both leukocyte and neutrophil counts were significantly higher in the severe groups in line with previous retrospective studies,41 and other meta-analysis.46 It has been previously established that reduction in circulating leukocytes and neutrophils commonly arise following viral invasion, as a consequence of either bone marrow suppression or peripheral destruction47 during initial stages of infection. This may increase the susceptibility of secondary bacterial infections in severe COVID-19, a consequence that is often marked by leukocytosis. In a dynamic profile of laboratory results presented by Wang et al.,41 non-surviving or severe COVID-19 cases showed steadily increasing levels of the two parameters, resulting in leukocytosis and neutrophilia that was significantly higher compared to the non-severe COVID-19 group, postulated to be a consequence of the secondary infection and infection-induced cytokine storm.
Limited studies have examined the role of the thrombocyte count in COVID-19 identification. In a previous meta-analysis,48 thrombocytopenia has been implicated as a marker for severe SARS-CoV-2 infection. This study augments said findings, revealing that thrombocyte counts can also be used as a marker to differentiate between COVID-19 and non-COVID-19 infections, regardless of severity. In areas with concurrently high dengue infection rates such as South East Asia, the similarities between initial clinical and laboratory presentations of SARS-CoV-2 and dengue infection should be carefully considered during the current pandemic. A report from Singapore49 have shown covert COVID-19 infection in patients originally suspected of dengue infection, with consistent thrombocytopenia observed in the two reported cases. Moreover, false positive dengue serology occurred in the two COVID-19 cases, further complicating the distinction and posing a risk to the intense surveillance required during the current pandemic.
The consistent finding of thrombocytopenia as a marker of severe COVID infection in this study with the aforementioned meta-analysis48 by Lippi et al. further emphasizes that this low-cost and routinely-performed blood parameter can aid both diagnosis and monitoring of progression and should be assessed continually during care of COVID-19 infections. The mechanism behind thrombocytopenia in both early and late stages of COVID-19 infection is likely multifactorial, as a result of both indirect and direct mechanisms that interfere with thrombocyte production, activation, or consumption.50 Although studies are required to confirm, it is highly probable that the mechanisms of SARS-CoV-2 induced thrombocytopenia are analogous to those previously characterized in the SARS-CoV, as both belong to the same family of coronaviruses. Possible direct mechanisms of thrombocytopenia previously characterized in the SARS-CoV is viral bone marrow invasion and infection of progenitor cells, therefore interfering with thrombocyte production from megakaryocytes by inhibition of growth and by inducing apoptosis.51 Other potential mechanisms of thrombocytopenia include excess activation of thrombocytes by virus-triggered immune complexes, and increased consumption due to excessive thrombosis that occur during lung damage.51
Furthermore, extreme thrombocytopenia is often observed in severe and critical conditions of illness due to the development of disseminated intravascular coagulopathy (DIC), reflected excessive coagulation activity marked by significantly higher levels of D-dimers found in our analysis among patients of severe COVID-19 infection. This is in line with previous studies showing D-dimer as a marker for severe COVID-19,30 and even as a marker of mortality at levels higher than 1 μg/L.38, 52
Several reports15, 17, 18, 39, 53, 54 have suggested lymphopenia as a strong indicator of COVID-19 infection. In the first analysis however, despite a trend of lowered lymphocyte count in COVID-19 cases, no statistical significance was observed between the two groups. The possible reason for the insignificant difference between the pooled studies of both groups is that two studies19, 21 included in the analysis showed higher tendency of lymphopenia to occur in the non-COVID group. However, in the second analysis lymphocyte counts were found significantly lower in the severe group, which is in line with previous reports which observed progressive lymphopenia41 as a strong indicator of disease severity.
The results of the first analysis showed that both CRP and procalcitonin were not significant differentiators of COVID-19 and non-COVID-19 cases. However, in the second analysis, a significantly higher level of CRP was observed in the severe COVID-19 group compared to non-COVID-19 cases, confirming previous reports of the clinical utility of CRP levels as an indicator for severe disease and progressive inflammation.25, 31, 40 In contrast to a previous meta-analysis,55 procalcitonin level between severe and non-severe groups were not significantly different in this analysis, which pooled the procalcitonin results of 14 studies. The changes in procalcitonin level among severe COVID patients remain unclear, because although half18, 28, 30, 33, 42, 45, 56 of the 14 analyzed studies showed an insignificant difference between procalcitonin levels between the two groups, the remaining seven analyzed studies reported significantly higher PCT values in the severe group. These reports postulate that raised PCT indicates the presence of secondary bacterial infection.
There are several limitations in our study. First, because of the novelty of the outbreak, the quantity and diversity of the studies are limited, with a bulk originating from various regions in China. Further investigation is required to verify the findings in more diverse geographical settings, and resource conditions. A high heterogeneity was also noted in this study, which may be due to discrepancies between the classification between severe and non-severe COVID-19 across studies, and the lack of data regarding the exact timing of laboratory data collection in the studies. Hence future investigations need to assess the time-sensitive changes in laboratory parameters. And although the follow-up ROC analysis in this study determined the most optimal diagnostic and prognostic markers, future studies should explore the predictive value of marker combinations in order to maximize the discriminatory capacity. Finally, we did not evaluate several other potential parameters such as eosinophil count,16 due to the lack of data associated with such parameter in our collected studies.
Despite the aforementioned limitations, to our knowledge, this is the first analysis that examines laboratory parameters from two perspectives; (1) from a diagnostic perspective to determine parameters that can aid differentiation between COVID-19 and non-COVID-19 pneumonia, and (2) from a prognostic perspective to determine parameters that can aid differentiation between non-severe and severe COVID-19 cases. By inclusion of studies with raw mean data, we could avoid discrepancies that may occur because of varying cut-offs of laboratory parameters across studies.
Conclusion
In conclusion, thrombocyte count was significantly lower in COVID-19 compared to non-COVID-19 infections, and persistently low in severe disease, implying that thrombocyte measurement is a key laboratory parameter for both diagnosis and prognosis. Leukocyte and neutrophil counts were significantly lower in COVID-19 compared to non-COVID-19 infections, but contrastingly higher in severe COVID-19 cases relative to non-severe COVID-19. And although lymphocyte, D-dimer and CRP levels were not significantly different between COVID-19 and non-COVID-19 cases, all three are effective indicators for severe cases of COVID-19. As more studies become available, further analysis is required to confirm these findings.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
None to declare.
Acknowledgements
The author would like to thank to Christian Peinado Garcia for assisting with Spanish translations.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.medcli.2020.05.017.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China. N Engl J Med. 2019:2020. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2020. Coronavirus disease 2019 (COVID-19): situation report, vol. 75. [Google Scholar]
- 3.Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020 doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lloyd-Sherlock P., Ebrahim S., Geffen L., McKee M. Bearing the brunt of covid-19: older people in low and middle income countries. Br Med J Publish Group. 2020 doi: 10.1136/bmj.m1052. [DOI] [PubMed] [Google Scholar]
- 5.Anderson R.M., Heesterbeek H., Klinkenberg D., Hollingsworth T.D. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395:931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowling B.J., Aiello A. Public health measures to slow community spread of COVID-19. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. 2020, World Health Organization.
- 8.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020 doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 9.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Int Med. 2009;151:264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 10.Sterne J.A., Sutton A.J., Ioannidis J.P., Terrin N., Jones D.R., Lau J. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;343:d4002. doi: 10.1136/bmj.d4002. [DOI] [PubMed] [Google Scholar]
- 11.Hozo S.P., Djulbegovic B., Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol. 2005;5:13. doi: 10.1186/1471-2288-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan X., Wang W., Liu J., Tong T. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol. 2014;14:135. doi: 10.1186/1471-2288-14-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fluss R., Faraggi D., Reiser B. Estimation of the Youden Index and its associated cutoff point. Biometr J: J Math Methods Biosci. 2005;47:458–472. doi: 10.1002/bimj.200410135. [DOI] [PubMed] [Google Scholar]
- 14.Youden W.J. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Cheng Z., Lu Y., Cao Q., Qin L., Pan Z., Yan F. Clinical features and chest CT manifestations of coronavirus disease 2019 (COVID-19) in a single-center study in Shanghai, China. Am J Roentgenol. 2020:1–6. doi: 10.2214/AJR.20.22959. [DOI] [PubMed] [Google Scholar]
- 16.Zhao D., Yao F., Wang L., Zheng L., Gao Y., Ye J. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuanyuan L., Weina W., Yu L., Bo Z., Jie Y., Jianwu H. Comparison of the clinical characteristics between RNA positive and negative patients clinically diagnosed with 2019 novel coronavirus pneumonia. Zhonghua jie he he hu xi za zhi = Zhonghua jiehe he huxi zazhi = Chinese J Tuberc Respir Dis. 2020;43:E023. doi: 10.3760/cma.j.cn112147-20200214-00095. [DOI] [PubMed] [Google Scholar]
- 18.Song C.-Y., Xu J., He J.-Q., Lu Y.-Q. COVID-19 early warning score: a multi-parameter screening tool to identify highly suspected patients. medRxiv. 2020 [Google Scholar]
- 19.Hsih W.-H., Cheng M.-Y., Ho M.-W., Chou C.-H., Lin P.-C., Chi C.-Y. Featuring COVID-19 cases via screening symptomatic patients with epidemiologic link during flu season in a medical center of central Taiwan, Journal of Microbiology. Immunol Infect. 2020 doi: 10.1016/j.jmii.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu W., Xie K., Lu H., Xu L., Zhou S., Fang S. Initial clinical features of suspected Coronavirus Disease 2019 in two emergency departments outside of Hubei, China. J Med Virol. 2020 doi: 10.1002/jmv.25763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y., Liang J., Zhou Q., Li X., Lin F., Deng Z. Prevalence and clinical features of 2019 novel coronavirus disease (COVID-19) in the Fever Clinic of a teaching hospital in Beijing: a single-center, retrospective study. medRxiv. 2020 [Google Scholar]
- 22.Han H., Yang L., Liu R., Liu F., Wu K-l, Li J. Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clin Chem Lab Med (CCLM) 2020 doi: 10.1515/cclm-2020-0188. 1 (ahead-of-print) [DOI] [PubMed] [Google Scholar]
- 23.Qu R., Ling Y., Zhang Yh, Wei Ly, Chen X., Li X. Platelet-to-lymphocyte ratio is associated with prognosis in patients with Corona Virus Disease-19. J Med Virol. 2020 doi: 10.1002/jmv.25767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao L., Wang M., Chen S., He Q., Chang J., Hong C. 2020. Neurological Manifestations of Hospitalized Patients with COVID-19 in Wuhan, China: a retrospective case series study. [Google Scholar]
- 25.Lo I.L., Lio C.F., Cheong H.H., Lei C.I., Cheong T.H., Zhong X. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int J Biol Sci. 2020;16:1698–1707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G., Wu C., Zhang Q., Wu F., Yu B., Lv J. 2020. Epidemiological and Clinical Features of Corona Virus Disease 2019 (COVID-19) in Changsha, China. China (3/1/2020) [Google Scholar]
- 27.Young B.E., Ong S.W.X., Kalimuddin S., Low J.G., Tan S.Y., Loh J. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020 doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qian G.-Q., Yang N.-B., Ding F., Ma A.H.Y., Wang Z.-Y., Shen Y.-F. Epidemiologic and clinical characteristics of 91 hospitalized patients with COVID-19 in Zhejiang, China: a retrospective, multi-centre case series. medRxiv. 2020 doi: 10.1093/qjmed/hcaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y., Li T., Han M., Li X., Wu D., Xu Y. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol. 2020 doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020:1–3. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J., Liu Z.-H., Luo X.-H., Xi Y., Ren Z.-Q., Li Y.-N. 2020. Clinical hallmarks of 13 COVID-19 patients revealing SAA biomarker. Available at SSRN 3546066. [Google Scholar]
- 33.Cao M., Zhang D., Wang Y., Lu Y., Zhu X., Li Y. Clinical features of patients infected with the 2019 novel coronavirus (COVID-19) in Shanghai, China. medRxiv. 2020 [Google Scholar]
- 34.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu X., Yuan W., Huang K., Wang Q., Yao S., Lu W. 2020. Clinical features and short-term outcomes of 114 elderly patients with COVID-19 in Wuhan, China: a single-center, retrospective, observational study. SSRN. [Google Scholar]
- 36.Zhang G., Hu C., Luo L., Fang F., Chen Y., Li J. Clinical features and outcomes of 221 patients with COVID-19 in Wuhan, China. MedRxiv. 2020 doi: 10.1016/j.jcv.2020.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao W. Clinical features and laboratory inspection of novel coronavirus pneumonia (COVID-19) in Xiangyang, Hubei. medRxiv. 2020 [Google Scholar]
- 38.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan W-j, Ni Z-y, Hu Y., Liang W-h, Ou C-q, He J-x Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Jj, Dong X., Cao Y.Y., Yuan Yd, Yang Yb, Yan Yq Clinical characteristics of 140 patients infected by SARS-CoV-2 in Wuhan, China. Allergy. 2020 doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 41.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu L., Gao J.-Y., Hu W-m, Zhang X.-X., Guo L., Liu C.-Q. Clinical characteristics of 51 patients discharged from hospital with COVID-19 in Chongqing, China. medRxiv. 2020 [Google Scholar]
- 43.Peng Y., Meng K., Guan H., Leng L., Zhu R., Wang B. Clinical characteristics and outcomes of 112 cardiovascular disease patients infected by 2019-nCoV. Zhonghua xin xue guan bing za zhi. 2020;48:E004. doi: 10.3760/cma.j.cn112148-20200220-00105. [DOI] [PubMed] [Google Scholar]
- 44.Li K., Wu J., Wu F., Guo D., Chen L., Fang Z. The clinical and chest CT features associated with severe and critical COVID-19 pneumonia. Investig Radiol. 2020 doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu W., Tao Z.-W., Lei W., Ming-Li Y., Kui L., Ling Z. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chinese Med J. 2020 doi: 10.1097/CM9.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lippi G., Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med (CCLM) 2020 doi: 10.1515/cclm-2020-0198. [DOI] [PubMed] [Google Scholar]
- 47.Munshi H.G., Montgomery R.B. Evidence-based case review: severe neutropenia: a diagnostic approach. Western J Med. 2000;172:248. doi: 10.1136/ewjm.172.4.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lippi G., Plebani M., Henry B.M. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin Chim Acta. 2020 doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan G., Lee C.K., Lam L.T., Yan B., Chua Y.X., Lim A.Y. Covert COVID-19 and false-positive dengue serology in Singapore. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Assinger A. Platelets and infection – an emerging role of platelets in viral infection. Front Immunol. 2014;5:649. doi: 10.3389/fimmu.2014.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang M., Ng M.H., Li C.K. Thrombocytopenia in patients with severe acute respiratory syndrome. Hematology. 2005;10:101–105. doi: 10.1080/10245330400026170. [DOI] [PubMed] [Google Scholar]
- 52.Weiss P., Murdoch D.R. Clinical course and mortality risk of severe COVID-19. Lancet. 2020 doi: 10.1016/S0140-6736(20)30633-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng Y., Huang Z., Ying G., Zhang X., Ye W., Hu Z. Comparative study of the lymphocyte change between COVID-19 and non-COVID-19 pneumonia cases suggesting uncontrolled inflammation might not be the main reason of tissue injury. medRxiv. 2020 [Google Scholar]
- 54.Rodriguez-Morales A.J., Cardona-Ospina J.A., Gutiérrez-Ocampo E., Villamizar-Peña R., Holguin-Rivera Y., Escalera-Antezana J.P. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis. 2020;1016:23. doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lippi G., Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chim Acta. 2020;505:190–191. doi: 10.1016/j.cca.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mo P., Xing Y., Xiao Y., Deng L., Zhao Q., Wang H. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.