Abstract
The numbers of confirmed cases of new coronavirus (Covid-19) are increased daily in different countries. To determine the policies and plans, the study of the relations between the distributions of the spread of this virus in other countries is critical. In this work, the distributions of the spread of Covid-19 in Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran were compared and clustered using fuzzy clustering technique. At first, the time series of Covid-19 datasets in selected countries were considered. Then, the relation between spread of Covid-19 and population's size was studied using Pearson correlation. The effect of the population's size was eliminated by rescaling the Covid-19 datasets based on the population's size of USA. Finally, the rescaled Covid-19 datasets of the countries were clustered using fuzzy clustering. The results of Pearson correlation indicated that there were positive and significant between total confirmed cases, total dead cases and population's size of the countries. The clustering results indicated that the distribution of spreading in Spain and Italy was approximately similar and differed from other countries.
Keywords: Coronaviruses, Covid-19, Statistics, Fuzzy Clustering, Correlation
1. Background
Coronaviruses are a large group of viruses that trace respiratory and neurological systems [1], [2], [3]. In 2003 and 2012 two types of these viruses, called SARS coronavirus (SARS-CoV) and MERS coronavirus (MERS-CoV) were observed in some countries [4]. In last months of 2019, a new type of these viruses, called Covid-19 (2019-nCoV) was reported in Wuhan city in China [5], [6], [7], [8]. The reports show that Covid-19 has been observed in more than 220 countries (up to 18 April 2020). Since January to today 18 April 2020, the spread rate of Covid-19 has increased daily in different countries, specially in Unites States America [9], Spain [10], Italy [11], [12], [13], [14], Germany [15], United Kingdom [16], [17], [18], [19], France [11,[20], [21], [22]], Iran [23] and many others.
The spread rate of the Covid-19 has many dangers and consequently needs strict special policies and plans. Therefore, the study of the relations between the distributions of the spread of this virus in other countries is critical. In this work, the distributions of the spread of Covid-19 in Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran are compared and clustered using fuzzy clustering technique. At first, we consider the time series of Covid-19 datasets in selected countries. Then, the correlations between these time series are computed. Finally, the observed time series are rescaled and categorized using fuzzy clustering technique. The main novelties of the current research can be summarized as following:
-
1
The relation between spread of Covid-19 and population's size is studied.
-
2
The Covid-19 datasets are rescaled based on the population's size of USA.
-
3
The rescaled Covid-19 datasets of the countries with high spread risk are clustered using fuzzy clustering.
2. Material and method
This section discusses various topics such as data collection and data analysis techniques. The first subsection deals with the characteristics of research's dataset. Then the methods used to analyze the dataset are described.
2.1. Dataset
The dataset of this work contained the entire confirmed and dead Covid-19 cases in high risk countries including Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran from 22 February 2020 up to 18 April 2020 based on WHO statistics. Table 1 summarized descriptive statistics about the considered dataset.
Table 1.
Descriptive statistics for confirmed and dead Covid-19 cases in Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran from 22 February 2020 up to 18 April 2020.
| Cases | Country | Number | Minimum | Maximum | Mean | Standard deviation |
|---|---|---|---|---|---|---|
| Confirmed | Unites States America | 59 | 0 | 35527 | 11900.8 | 13327.5 |
| Spain | 59 | 0 | 9222 | 3187.6 | 3016.8 | |
| Italy | 59 | 0 | 6557 | 2922.6 | 2056.1 | |
| Germany | 59 | 0 | 6294 | 2329.2 | 2245.2 | |
| United Kingdom | 59 | 0 | 8719 | 1842.1 | 2207.7 | |
| France | 59 | 0 | 7578 | 1851.5 | 1876.0 | |
| Iran | 59 | 2 | 3186 | 1294.5 | 921.5 | |
| Dead | Unites States America | 59 | 0 | 4928 | 628.0 | 1013.4 |
| Spain | 59 | 0 | 950 | 330.1 | 340.5 | |
| Italy | 59 | 0 | 971 | 385.5 | 309.5 | |
| Germany | 59 | 0 | 315 | 69.7 | 94.9 | |
| United Kingdom | 59 | 0 | 980 | 247.1 | 342.0 | |
| France | 59 | 0 | 2004 | 316.6 | 447.3 | |
| Iran | 59 | 0 | 157 | 80.7 | 55.6 |
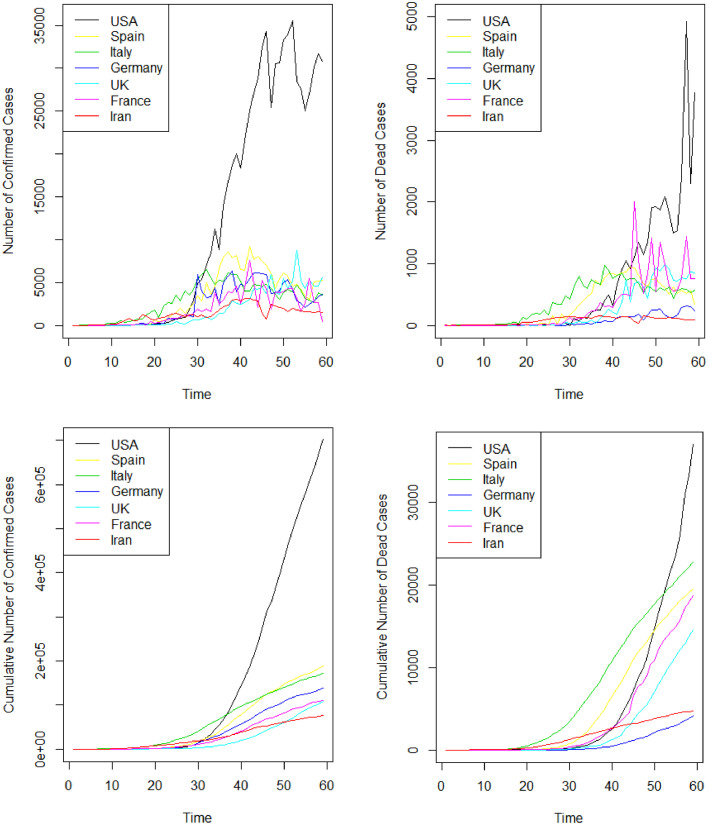
As it can be observed, Unites States America, Spain, Italy, Germany, France, United Kingdom, and Iran have the most means of daily confirmed cases, respectively. Also, Unites States America, Italy, Spain, France, United Kingdom, Iran, and Germany have the most means of daily dead cases, respectively. Fig. 1 also shows the plots of daily confirmed cases, dead cases, cumulative confirmed cases, and cumulative dead cases in in Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran from 22 February 2020 up to 18 April 2020.
Fig. 1.
Daily confirmed cases (Top and Left), dead cases (Top and Right), cumulative confirmed cases (Bottom and Left), and cumulative dead cases (Bottom and Right), in in Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran from 22 February 2020 up to 18 April 2020.
To study the relations between total confirmed cases, total dead cases and population's size of the countries, the Pearson coefficient of correlation is used. The results are reported in Table 2 .
Table 2.
Pearson coefficient of correlation test between confirmed and dead Covid-19 cases and population size up to 18 April 2020.
| Dead cases | Population's size | ||
|---|---|---|---|
| Confirmed cases | Pearson correlation | 0.959 | 0.825 |
| p-value | <0.001 | 0.011 | |
| Dead cases | Pearson correlation | 0.692 | |
| p-value | 0.042 |
The results indicated that there are positive and significant (p-value lower than 0.05) between total confirmed cases, total dead cases and population's size of the countries. Therefore, because the number of cases is dependent to the size of population, the comparison of the countries based on the number of confirmed cases or dead cases are not scientifically true. To solve this problem, the effect of the population's size should be eliminated. We used rescaled data as following:
and
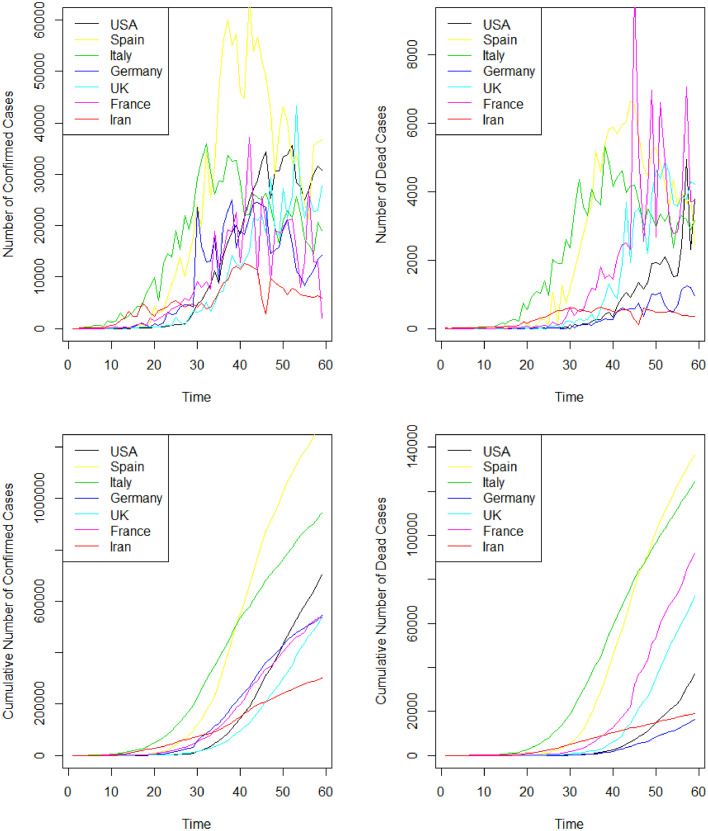
Fig. 2 shows the plots of rescaled data for daily confirmed cases, dead cases, cumulative confirmed cases, and cumulative dead cases in in Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran from 22 February 2020 up to 18 April 2020. Table 3 summarized descriptive statistics about the rescaled dataset. As it can be observed, Spain, Italy, Unites States America, Germany, United Kingdom, France, and Iran have the most mean of rescaled daily confirmed cases, respectively. Also, Spain, Italy, France, United Kingdom, Unites States America, Iran, and Germany have the most mean of daily rescaled dead cases, respectively.
Fig. 2.
Daily rescaled confirmed cases (Top and Left), rescaled dead cases (Top and Right), rescaled cumulative confirmed cases (Bottom and Left), and rescaled cumulative dead cases (Bottom and Right), in in Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran from 22 February 2020 up to 18 April 2020.
Table 3.
Descriptive statistics for rescaled confirmed and dead Covid-19 cases in Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran from 22 February 2020 up to 18 April 2020.
| Cases | Country | Number | Minimum | Maximum | Mean | Standard deviation |
|---|---|---|---|---|---|---|
| Confirmed | Unites States America | 59 | 0.0 | 35527.0 | 11900.8 | 13327.5 |
| Spain | 59 | 0.0 | 64505.5 | 22296.2 | 21101.4 | |
| Italy | 59 | 0.0 | 35861.4 | 15984.0 | 11245.3 | |
| Germany | 59 | 0.0 | 24938.2 | 9228.9 | 8896.1 | |
| United Kingdom | 59 | 0.0 | 43237.7 | 9134.9 | 10948.0 | |
| France | 59 | 0.0 | 37220.6 | 9094.1 | 9214.1 | |
| Iran | 59 | 7.9 | 12589.1 | 5115.0 | 3641.4 | |
| Dead | Unites States America | 59 | 0.0 | 4928.0 | 628.0 | 1013.4 |
| Spain | 59 | 0.0 | 6645.0 | 2309.2 | 2381.4 | |
| Italy | 59 | 0.0 | 5310.6 | 2108.6 | 1692.7 | |
| Germany | 59 | 0.0 | 1248.1 | 276.0 | 376.1 | |
| United Kingdom | 59 | 0.0 | 4859.8 | 1225.1 | 1695.9 | |
| France | 59 | 0.0 | 9843.0 | 1555.1 | 2196.9 | |
| Iran | 59 | 0.0 | 620.4 | 318.7 | 219.8 |
2.2. Fuzzy Clustering
Clustering [24] is a major task in data mining. It has many applications such as image processing, diagnosis systems, classification, missing value management and imputation, optimization, bioinformatics, machine learning [25]. Recently inspiring by classifier ensemble, the clustering ensemble [26] has emerged. But these methods use hard clustering as base clustering algorithm. Recently soft clustering algorithms [27] have been popular and it has been shown that these methods are superior to traditional hard clustering algorithms [28], [29], [30]. We can use soft clustering and fuzzy clustering interchangeably. Each data point belongs to all clusters (although the membership values are different) in soft clustering. It is worthy to be mention that the different membership values of a data point to all clusters should sum up to one. Fuzzy C-means (FCM) clustering algorithm [30] can be arguably considered to be the most popular soft clustering algorithm.
Given a set S of N records , a set of k fuzzy cluster defined by centroids , along with a membership matrix u, a soft clustering algorithm intends to divide S into k partitions , where Sj is achieved according to Eq. (1) (ties are broken randomly).
| (1) |
where cji is ith dimension of jth fuzzy cluster centroid.
All of centers and membership matrix are optimal, if they minimize the error function SSE presented in Eq. (2).
| (2) |
subject to the constraints . Matrix c is of size D × k whose column vectors are denoted by . To solve Eq. (11), we should employ a new set of Lagrange multipliers αx for constraints , and then minimize the final (constraint-free) error function presented in Eq. (3).
| (3) |
For a fix membership matrix u, the optimal can be achieved by setting . Eq. (4) presents .
| (4) |
Solving Eq. (4) with respect to cji gives Eq. (5).
| (5) |
For a fixed cluster center matrix c, we compute the optimal by setting and . Eq. (6) represents .
| (6) |
Eq. (7) presents .
| (7) |
To solve Eq. (6) with respect to ujx we can reach Eq. (8).
| (8) |
Solving Eq. (8) with respect to ujx we obtain Eq. (9).
| (9) |
Substituting this expression in Eq. (7) results in a new equation and solving the resultant equation in terms of αx yields to Eq. (10).
| (10) |
If we substitute Eq. (10) in Eq. (9), we can reach a new based on Eq. (11).
| (11) |
To compare and classify the distributions of the spread of Covid-19 in Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran, the fuzzy clustering technique is applied on rescaled Covid-19 datasets including confirmed cases, dead cases, cumulative confirmed cases, and cumulative dead cases.
3. Results
As it can be seen in Fig. 2 and Table 3, because of the effect of population's size, the rescaled datasets are different from main datasets and are scientifically good choices to compare different countries. In next subsections the results of fuzzy clustering are reported.
3.1. Rescaled number of confirmed cases
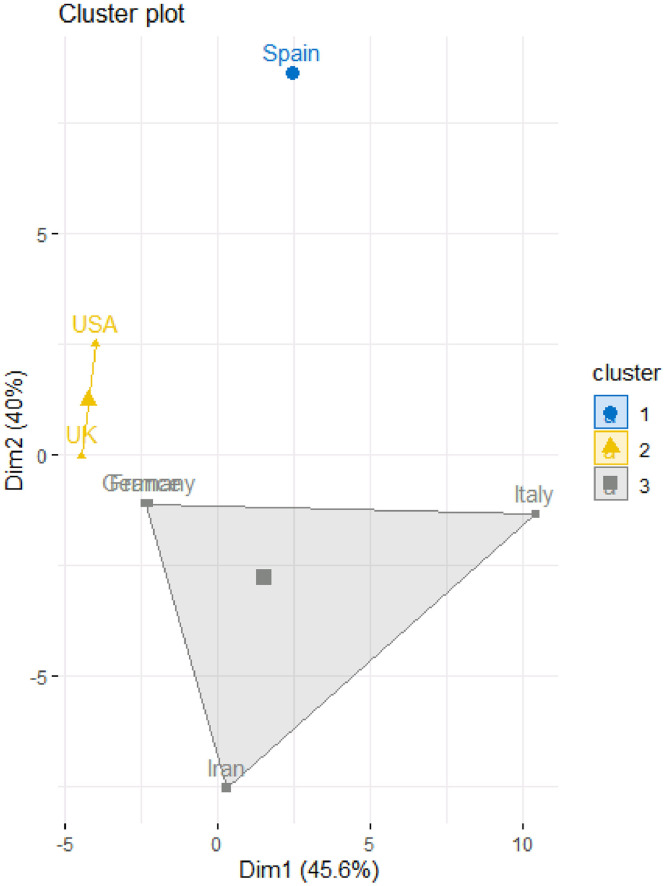
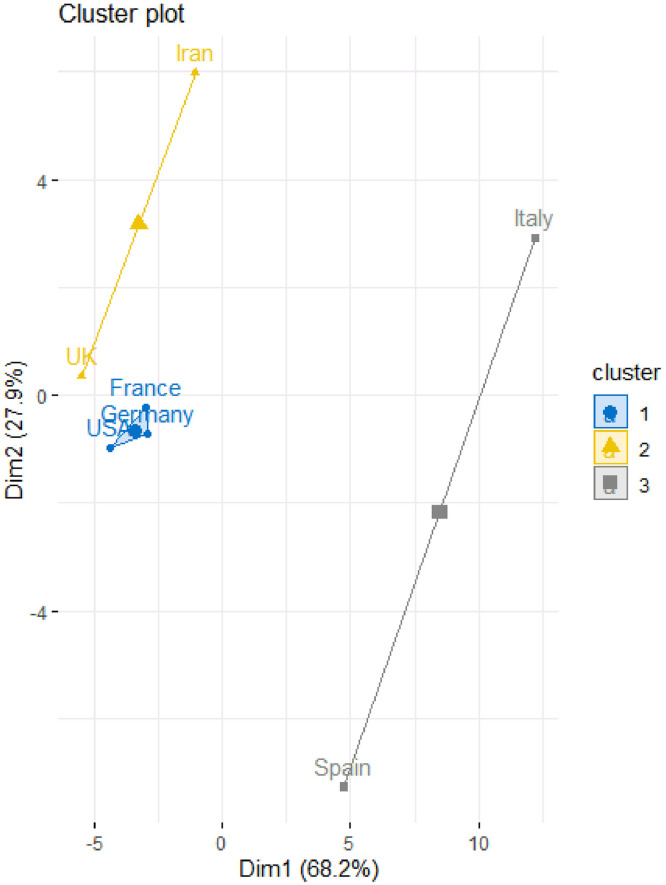
To determine the number of clusters Kaiser Index was used and the number was considered as the number of eigen-values of correlation matrix that are more than 1. Table 4 and Fig. 3, Fig. 4 provide the results of the fuzzy clustering technique. As it can be observed in Table 4 and Figs. 3 and 4, the rescaled numbers of confirmed cases in these considered countries can be divided in three clusters. Table 4 shows the probabilities of the membership of each country in each cluster. For each country, the maximum value of the probabilities of the membership to each cluster has been bolded. Based on these values, the first cluster consists of Spain (with probability 1.00). Also, the second cluster consists of Unites States America and United Kingdom (with probabilities 0.86 and 0.85, respectively). Moreover, the third cluster consists of Italy, Germany, France and Iran (with probabilities 0.46, 0.86, 0.78 and 0.73, respectively). In other words, Unites States America and United Kingdom are statistically similar; Italy, Germany, France and Iran are statistically similar; and Spain are significantly different form them.
Table 4.
The probabilities of membership in different clusters based on rescaled number of confirmed cases.
| Country | Cluster 1 | Cluster 2 | Cluster 3 |
|---|---|---|---|
| Unites States America | 0.03 | 0.86 | 0.11 |
| Spain | 1.00 | 0.00 | 0.00 |
| Italy | 0.21 | 0.33 | 0.46 |
| Germany | 0.02 | 0.12 | 0.86 |
| United Kingdom | 0.02 | 0.85 | 0.13 |
| France | 0.03 | 0.20 | 0.78 |
| Iran | 0.04 | 0.23 | 0.73 |
Fig. 3.
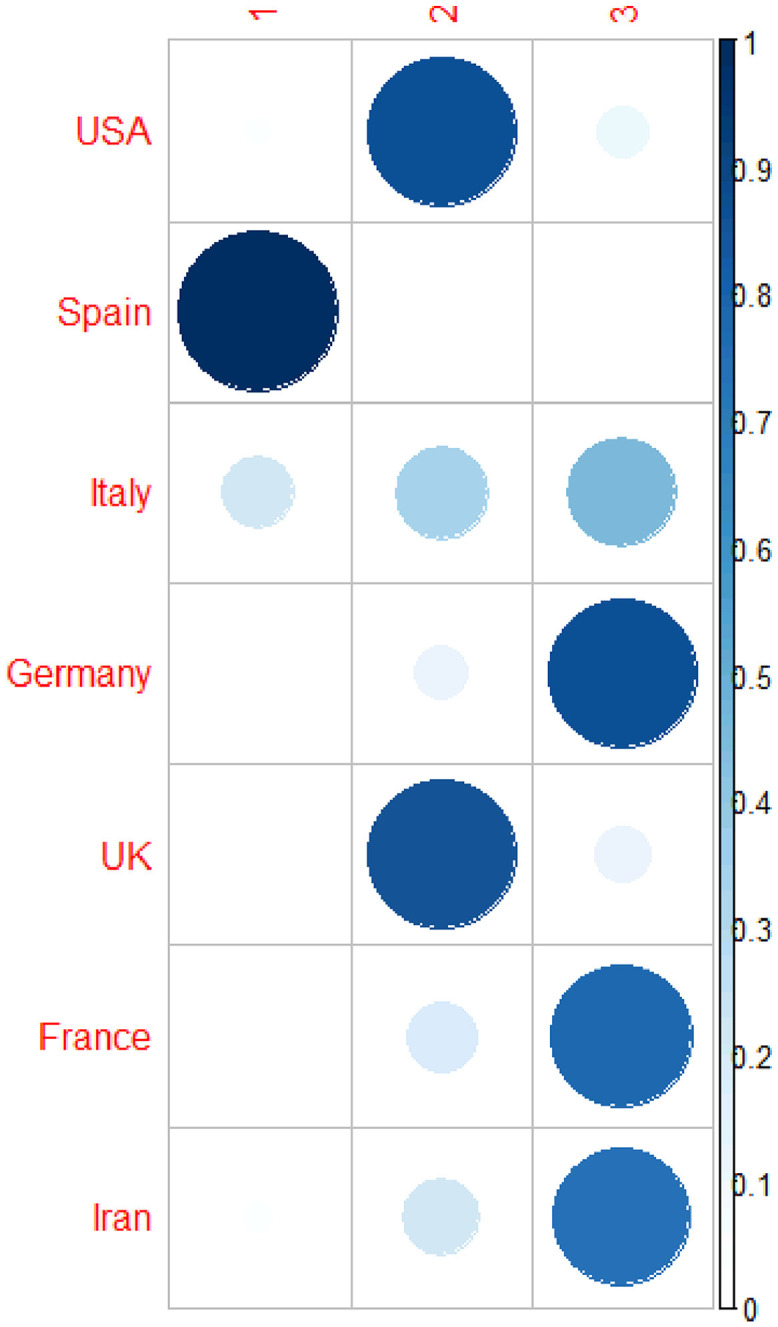
Fuzzy clustering method to classify the countries based on rescaled number of confirmed cases.
Fig. 4.
Fuzzy clustering plot to classify the countries based on rescaled number of confirmed cases.
3.2. Rescaled number of dead cases
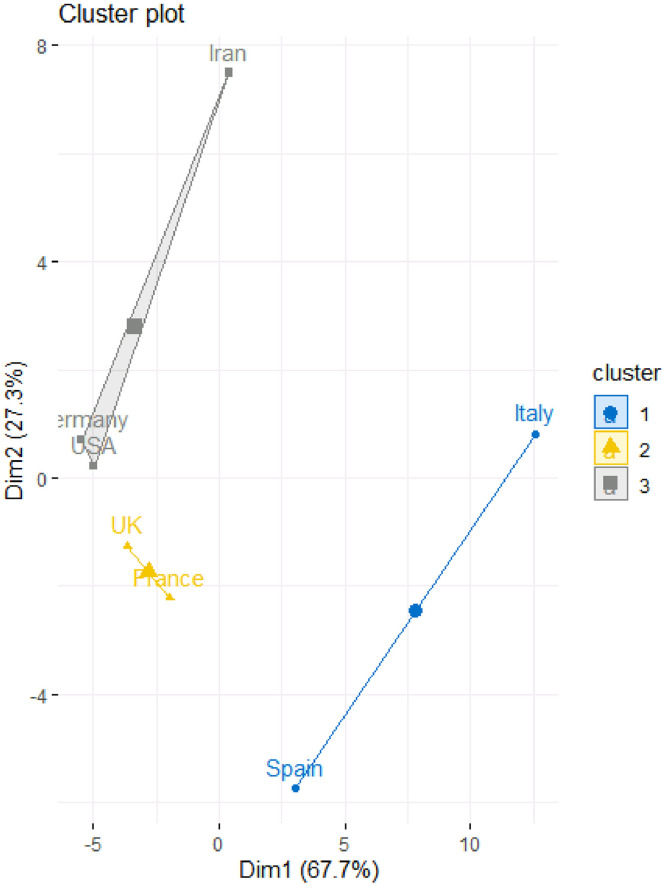
Table 5 and Fig. 5, Fig. 6 provide the results of the fuzzy clustering technique. As it can be observed in Table 5 and Figs. 5 and 6, the rescaled numbers of dead cases in these considered countries can be divided in three clusters. Table 5 shows the probabilities of the membership of each country in each cluster. For each country, the maximum value of the probabilities of the membership to each cluster has been bolded. Based on these values, the first cluster consists of United Kingdom and France (with probabilities 0.75 and 0.86, respectively). Also, the second cluster consists of Unites States America, Germany and Iran (with probabilities 0.77, 0.99 and 0.95, respectively). Moreover, the third cluster consists of Spain and Italy (with probabilities 0.89 and 0.73, respectively). In other words, United Kingdom and France are statistically similar; Unites States America, Germany and Iran are statistically similar; and Spain and Italy are statistically similar.
Table 5.
The probabilities of membership in different clusters based on rescaled number of dead cases.
| Country | Cluster 1 | Cluster 2 | Cluster 3 |
|---|---|---|---|
| Unites States America | 0.17 | 0.77 | 0.06 |
| Spain | 0.08 | 0.03 | 0.89 |
| Italy | 0.10 | 0.06 | 0.84 |
| Germany | 0.01 | 0.99 | 0.00 |
| United Kingdom | 0.75 | 0.14 | 0.11 |
| France | 0.86 | 0.06 | 0.09 |
| Iran | 0.03 | 0.95 | 0.02 |
Fig. 5.
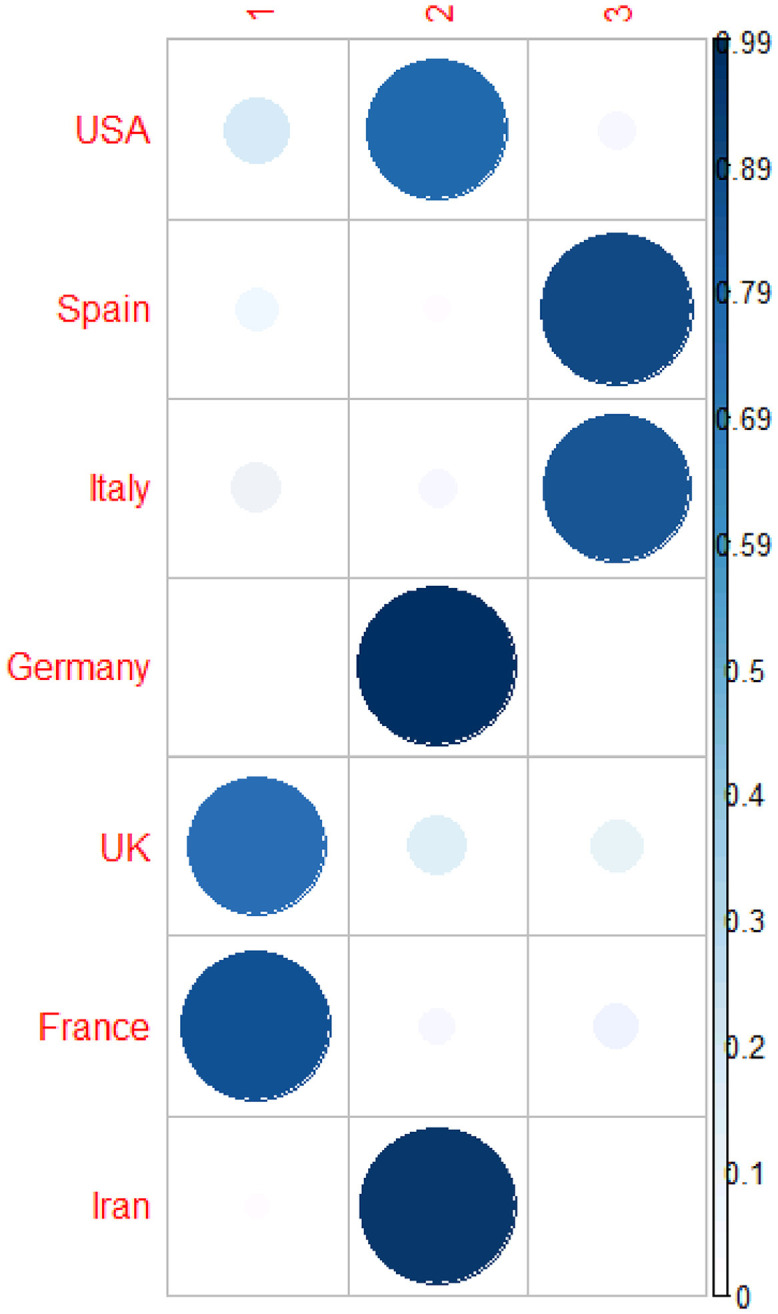
Fuzzy clustering method to classify the countries based on rescaled number of dead cases.
Fig. 6.
Fuzzy clustering plot to classify the countries based on rescaled number of dead cases.
3.3. Rescaled number of cumulative confirmed cases
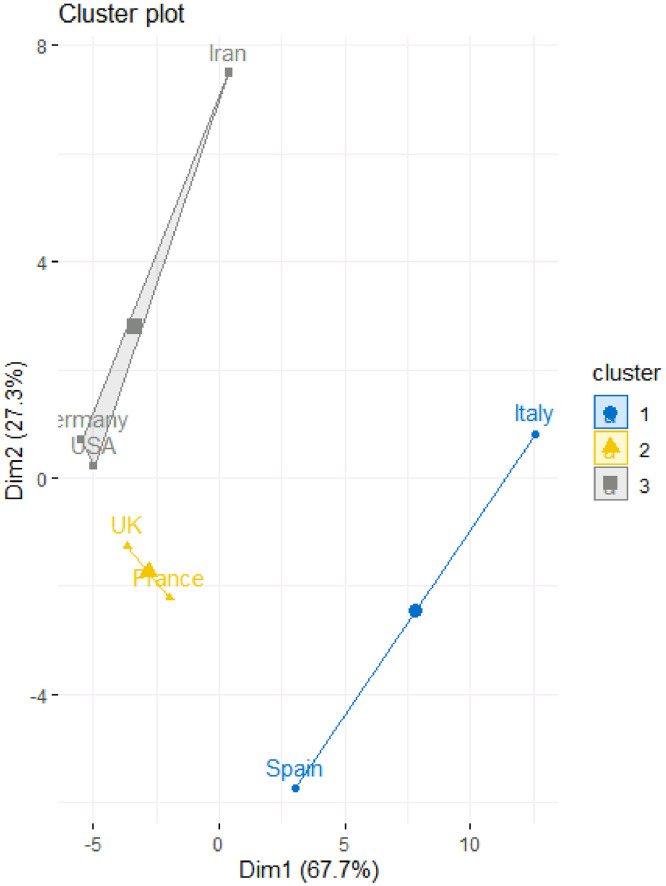
Table 6 and Fig. 7, Fig. 8 provide the results of the fuzzy clustering technique. As it can be observed in Table 6 and Figs. 7 and 8, the rescaled numbers of cumulative confirmed cases in these considered countries can be divided in three clusters. Table 6 shows the probabilities of the membership of each country in each cluster. For each country, the maximum value of the probabilities of the membership to each cluster has been bolded. Based on these values, the first cluster consists of Unites States America, Germany and France (with probabilities 0.90, 0.95 and 0.97, respectively). Also, the second cluster consists of United Kingdom and Iran (with probabilities 0.58 and 0.96, respectively). Moreover, the third cluster consists of Spain and Italy (with probabilities 0.94 and 0.82, respectively). In other words, Unites States America, Germany and France are statistically similar; United Kingdom and Iran are statistically similar; and Spain and Italy are statistically similar.
Table 6.
The probabilities of membership in different clusters based on rescaled number of cumulative confirmed cases.
| Country | Cluster 1 | Cluster 2 | Cluster 3 |
|---|---|---|---|
| Unites States America | 0.90 | 0.09 | 0.01 |
| Spain | 0.03 | 0.02 | 0.94 |
| Italy | 0.11 | 0.06 | 0.82 |
| Germany | 0.95 | 0.05 | 0.00 |
| United Kingdom | 0.41 | 0.58 | 0.01 |
| France | 0.97 | 0.03 | 0.00 |
| Iran | 0.04 | 0.96 | 0.00 |
Fig. 7.

Fuzzy clustering method to classify the countries based on rescaled number of cumulative confirmed cases.
Fig. 8.
Fuzzy clustering plot to classify the countries based on rescaled number of cumulative confirmed cases.
3.4. Rescaled number of cumulative dead cases
Table 7 and Fig. 9, Fig. 10 provide the results of the fuzzy clustering technique. As it can be observed in Table 7 and Figs. 9 and 10, the rescaled numbers of cumulative dead cases in these considered countries can be divided in three clusters. Table 7 shows the probabilities of the membership of each country in each cluster. For each country, the maximum value of the probabilities of the membership to each cluster has been bolded. Based on these values, the first cluster consists of Spain and Italy (with probabilities 0.97 and 0.97, respectively). Also, the second cluster consists of United Kingdom and France (with probabilities 0.89 and 0.94, respectively). Moreover, the third cluster consists of Unites States America, Germany and Iran (with probabilities 0.96, 0.98 and 0.98, respectively). In other words, Spain and Italy are statistically similar; United Kingdom and France are statistically similar; and Unites States America, Germany and Iran are statistically similar.
Table 7.
The probabilities of membership in different clusters based on rescaled number of cumulative dead cases.
| Country | Cluster 1 | Cluster 2 | Cluster 3 |
|---|---|---|---|
| Unites States America | 0.00 | 0.03 | 0.96 |
| Spain | 0.97 | 0.02 | 0.01 |
| Italy | 0.97 | 0.02 | 0.01 |
| Germany | 0.00 | 0.02 | 0.98 |
| United Kingdom | 0.02 | 0.89 | 0.09 |
| France | 0.03 | 0.94 | 0.03 |
| Iran | 0.00 | 0.02 | 0.98 |
Fig. 9.
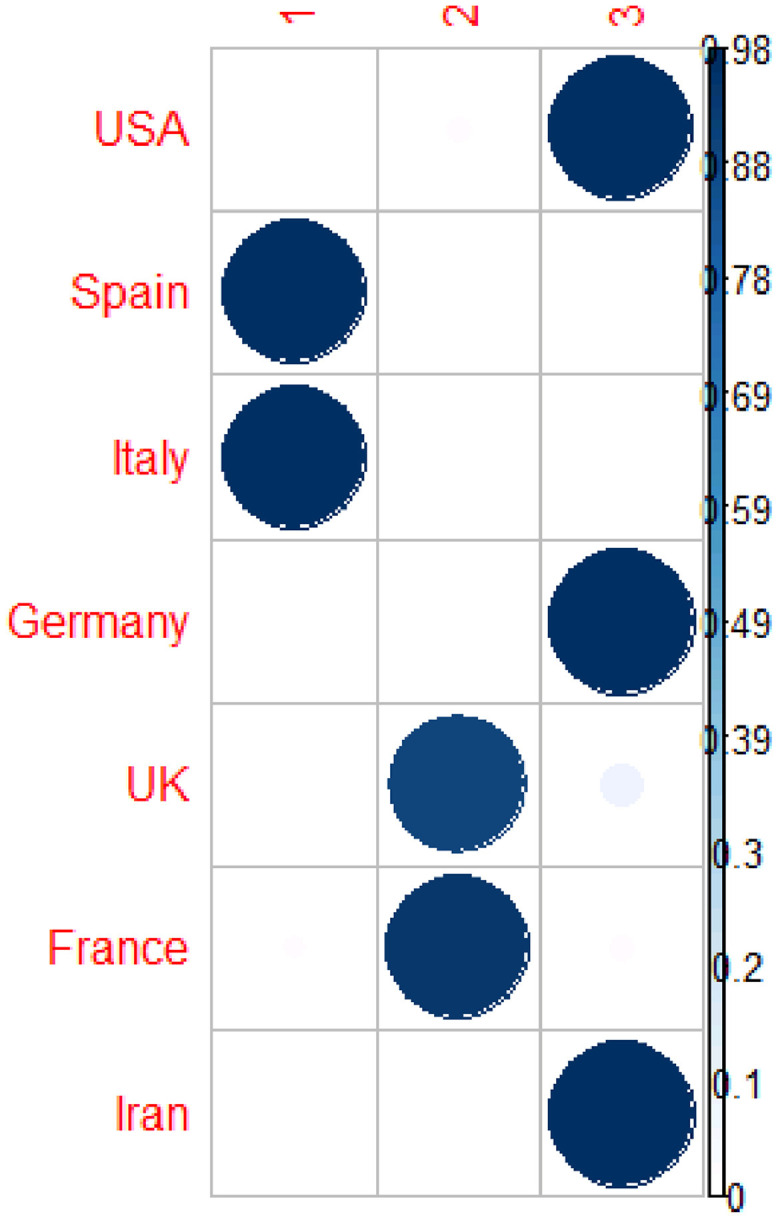
Fuzzy clustering method to classify the countries based on rescaled number of cumulative dead cases
Fig. 10.
Fuzzy clustering plot to classify the countries based on rescaled number of cumulative dead cases.
4. Conclusion
To consider the policies and plans to manage the spread of Covid-19, the study of the relations between the distributions of the spread of this virus in other countries is critical. In this work, the distributions of the spread of Covid-19 in Unites States America, Spain, Italy, Germany, United Kingdom, France, and Iran were compared and clustered using fuzzy clustering technique. In this research, the relation between spread of Covid-19 and population's size was firstly studied. The results indicated that there were positive and significant between total confirmed cases, total dead cases and population's size of the countries. Therefore, because the number of cases was dependent to the size of population, the comparison of the countries based on the number of confirmed cases or dead cases were not scientifically true. To solve this problem, the effect of the population's size has been eliminated by rescaling the Covid-19 datasets based on the population's size of USA. Finally, the rescaled Covid-19 datasets of the countries were clustered using fuzzy clustering. The clustering results indicated that the distribution of spreading in Spain and Italy was approximately similar and differed from other countries. For future works, the authors suggest the researchers categorize the statistical models including regression and time series models and artificial intelligence models that can be fitted on Covid-19 datasets [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61].
Funding
No fund.
CRediT authorship contribution statement
Mohammad Reza Mahmoudi: Data curation, Validation, Writing - original draft. Dumitru Baleanu: Conceptualization, Methodology, Software, Supervision. Zulkefli Mansor: Visualization, Investigation, Writing - review & editing. Bui Anh Tuan: Visualization, Investigation, Writing - review & editing. Kim-Hung Pho: Visualization, Investigation, Writing - review & editing.
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- 1.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y.J. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L.F., Shi Z., Zhang S., Field H., Daszak P., Eaton B.T. Review of bats and SARS. Emerg Infect Dis. 2006;12(12):1834. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cauchemez S., Van Kerkhove M.D., Riley S., Donnelly C.A., Fraser C., Ferguson N.M. Transmission scenarios for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and how to tell them apart. Euro surveillance: bulletin Europeen sur les maladies transmissibles. Eur Commun Dis Bull. 2013;18(24) [PMC free article] [PubMed] [Google Scholar]
- 5.Organization, W.H. Novel Coronavirus (2019-nCoV) 2020, 2020. Available online: https://www.who.int/ (accessed on 27 January 2020).
- 6.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet North Am Ed. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S., Du B. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burke R.M., Midgley C.M., Dratch A. Active monitoring of persons exposed to patients with confirmed COVID-19 — United States, January–February 2020. MMWR Morb Mortal Wkly Rep. 2020;69:245–246. doi: 10.15585/mmwr.mm6909e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legido-Quigley H., Mateos-García J.T., Campos V.R., Gea-Sánchez M., Muntaner C., McKee M. The resilience of the Spanish health system against the COVID-19 pandemic. Lancet Public Health. 2020 doi: 10.1016/S2468-2667(20)30060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fanelli D., Piazza F. Analysis and forecast of COVID-19 spreading in China, Italy and France. Chaos Solitons Fractals. 2020;134 doi: 10.1016/j.chaos.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Remuzzi A., Remuzzi G. COVID-19 and Italy: what next? Lancet North Am Ed. 2020 doi: 10.1016/S0140-6736(20)30627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onder G., Rezza G., Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA. 2020 doi: 10.1001/jama.2020.4683. [DOI] [PubMed] [Google Scholar]
- 14.Lazzerini M., Putoto G. COVID-19 in Italy: momentous decisions and many uncertainties. Lancet Global Health. 2020 doi: 10.1016/S2214-109X(20)30110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahase, E.. Covid-19: UK starts social distancing after new model points to 260 000 potential deaths, 2020. [DOI] [PubMed]
- 17.Lillie P.J., Samson A., Li A., Adams K., Capstick R., Barlow G.D., Easom N., Hamilton E., Moss P.J., Evans A., Ivan M. Novel coronavirus disease (Covid-19): the first two patients in the UK with person to person transmission. J Infect. 2020 doi: 10.1016/j.jinf.2020.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Razai M.S., Doerholt K., Ladhani S., Oakeshott P. Coronavirus disease 2019 (covid-19): a guide for UK GPs. BMJ. 2020;6:368. doi: 10.1136/bmj.m800. [DOI] [PubMed] [Google Scholar]
- 19.Hunter D.J. Covid-19 and the stiff upper lip—the pandemic response in the united kingdom. N Engl J Med. 2020 doi: 10.1056/NEJMp2005755. [DOI] [PubMed] [Google Scholar]
- 20.Gautret P., Lagier J.C., Parola P., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;20 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Stoecklin S.B., Rolland P., Silue Y., Mailles A., Campese C., Simondon A., Mechain M., Meurice L., Nguyen M., Bassi C., Yamani E. First cases of coronavirus disease 2019 (COVID-19) in France: surveillance, investigations and control measures, January 2020. Eurosurveillance. 2020;25(6) doi: 10.2807/1560-7917.ES.2020.25.6.2000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amrane S., Tissot-Dupont H., Doudier B., Eldin C., Hocquart M., Mailhe M., Dudouet P., Ormières E., Ailhaud L., Parola P., Lagier J.C. Rapid viral diagnosis and ambulatory management of suspected COVID-19 cases presenting at the infectious diseases referral hospital in Marseille, France,-January 31st to March 1st, 2020: a respiratory virus snapshot. Travel Med Infect Dis. 2020 doi: 10.1016/j.tmaid.2020.101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takian A., Raoofi A., Kazempour-Ardebili S. COVID-19 battle during the toughest sanctions against Iran. Lancet. 2020;395(10229):1035. doi: 10.1016/S0140-6736(20)30668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nejatian S., Parvin H., Faraji E. Using sub-sampling and ensemble clustering techniques to improve performance of imbalanced classification. Neurocomputing. 2018;276:55–66. [Google Scholar]
- 25.Rashidi F., Nejatian S., Parvin H., Rezaie V. Diversity based cluster weighting in cluster ensemble: an information theory approach. Artif Intell Rev. 2019;52(2):1341–1368. [Google Scholar]
- 26.Abbasi S.O., Nejatian S., Parvin H., Rezaie V., Bagherifard K. Clustering ensemble selection considering quality and diversity. Artif Intell Rev. 2019;52(2):1311–1340. [Google Scholar]
- 27.Niu H., Khozouie N., Parvin H., Alinejad-Rokny H., Beheshti A., Mahmoudi M.R. An ensemble of locally reliable cluster solutions. Appl Sci. 2020;10(5):1891. [Google Scholar]
- 28.Bagherinia A., Minaei-Bidgoli B., Hossinzadeh M., Parvin H. Reliability-based fuzzy clustering ensemble. Fuzzy Sets Syst. 2020 [Google Scholar]
- 29.Bagherinia A., Minaei-Bidgoli B., Hossinzadeh M., Parvin H. Elite fuzzy clustering ensemble based on clustering diversity and quality measures. Appl Intell. 2020;49(5):1724–1747. [Google Scholar]
- 30.Mojarad M., Nejatian S., Parvin H., Mohammadpoor M. A fuzzy clustering ensemble based on cluster clustering and iterative fusion of base clusters. Appl Intell. 2019;49(7):2567–2581. [Google Scholar]
- 31.Haghbin H., Mahmoudi M.R., Shishebor Z. Large sample inference on the ratio of two independent binomial proportions. J Math Ext. 2011;5(1):87–95. [Google Scholar]
- 32.Mahmoudi M.R., Behboodian J., Maleki M. Inference on the ratio of means in two independent populations. J Stat Theory Appl. 2017;16(3):366–374. [Google Scholar]
- 33.Mahmoudi M.R., Mahmoodi M. Inferrence on the ratio of variances of two independent populations. J Math Ext. 2014;7(2):83–91. [Google Scholar]
- 34.Mahmoudi M.R., Mahmoodi M. Inferrence on the ratio of correlations of two independent populations. J Math Ext. 2014;7(4):71–82. [Google Scholar]
- 35.Mahmoudi M.R., Nasirzadeh R., Mohammadi M. On the ratio of two independent skewnesses. Commun Stat-Theor Methods. 2019;48(7):1721–1727. [Google Scholar]
- 36.Mahmoudi M.R., Maleki M., Pak A. Testing the difference between two independent time series models. Iran J Sci Technol A (Sci) 2017;41:665–669. [Google Scholar]
- 37.Mahmoudi M.R., Heydari M.H., Roohi R. A new method to compare the spectral densities of two independent periodically correlated time series. Math Comput Simulat. 2019;160:103–110. [Google Scholar]
- 38.Mahmoudi M.R., Heydari M.H., Avazzadeh Z. Testing the difference between spectral densities of two independent periodically correlated (cyclostationary) time series models. Commun Stat Theory Methods. 2019;48(9):2320–2328. [Google Scholar]
- 39.Mahmoudi M.R., Mahmoudi M., Nahavandi E. Testing the difference between two independent regression models. Commun Stat Theory Methods. 2016;45(21):6284–6289. [Google Scholar]
- 40.Mahmoudi M.R., Maleki M., Pak A. Testing the equality of two independent regression models. Commun Stat Theory Methods. 2018;47(12):2919–2926. [Google Scholar]
- 41.Mahmoudi M.R. On comparing two dependent linear and nonlinear regression models. J Test Eval. 2018;47(1):449–458. [Google Scholar]
- 42.Ji-jun P., Mahmoudi M.R., Baleanu D., Maleki M. On comparing and classifying several independent linear and non-linear regression models with symmetric errors. Symmetry. 2019;11(6):820. [Google Scholar]
- 43.Bahrami M., Amiri M.J., Mahmoudi M.R., Koochaki S. Modeling caffeine adsorption by multi-walled carbon nanotubes using multiple polynomial regression with interaction effects. J Water Health. 2017;15(4):526–535. doi: 10.2166/wh.2017.297. [DOI] [PubMed] [Google Scholar]
- 44.Zarei A.R., Mahmoudi M.R. Evaluation of changes in RDIst index effected by different Potential Evapotranspiration calculation methods. Water Resour Manage. 2017;31(15):4981–4999. [Google Scholar]
- 45.Fisher R.A. On the probable error of a coefficient of correlation deduced from a small sample. Metron. 1921;1:3–32. [Google Scholar]
- 46.Heydari M.H., Avazzadeh Z., Mahmoudi M.R. Chebyshev cardinal wavelets for nonlinear stochastic differential equations driven with variable-order fractional Brownian motion. Chaos Solitons Fractals. 2019;124:105–124. [Google Scholar]
- 47.Mahmoudi M.R., Maleki M. A new method to detect periodically correlated structure. Comput Stat. 2017;32(4):1569–1581. [Google Scholar]
- 48.Nematollahi A.R., Soltani A.R., Mahmoudi M.R. Periodically correlated modeling by means of the periodograms asymptotic distributions. Stat Pap. 2017;58(4):1267–1278. [Google Scholar]
- 49.Mahmoudi M.R., Heydari M.H., Avazzadeh Z. On the asymptotic distribution for the periodograms of almost periodically correlated (cyclostationary) processes. Digit Signal Process. 2018;81:186–197. [Google Scholar]
- 50.Mahmoudi M.R., Heydari M.H., Avazzadeh Z., Pho K.H. Goodness of fit test for almost cyclostationary processes. Digit Signal Process. 2020;96 [Google Scholar]
- 51.Mahmoudi M.R., Maleki M., Borodin K., Pho K.H., Baleanu D. On comparing and clustering the spectral densities of several almost cyclostationary processes. Alexandria Engineering Journal. 2020 [Google Scholar]
- 52.Zhou R., Mahmoudi M.R., Mohammed S.N.Q., Pho K.H. Testing the equality of the spectral densities of several uncorrelated almost cyclostationary processes. Alex Eng J. 2020 [Google Scholar]
- 53.Mahmoudi M.R., Baleanu D., Tuan B.A., Pho K.H. A novel method to detect almost cyclostationary structure. Alex Eng J. 2020 [Google Scholar]; Mahmoudi M.R., Heydari M.H., Pho K.H. Fuzzy clustering to classify several regression models with fractional Brownian motion errors. Alex Eng J. 2020 [Google Scholar]
- 54.Roohi R., Heydari M.H., Aslami M., Mahmoudi M.R. A comprehensive numerical study of space-time fractional bioheat equation using fractional-order Legendre functions. Eur Phys J Plus. 2018;133:412. [Google Scholar]
- 55.Jalali S.M.J., Karimi M., Khosravi A., Nahavandi S. 2019 IEEE international conference on systems, man and cybernetics (SMC) IEEE; 2019. An efficient neuroevolution approach for heart disease detection; pp. 3771–3776. [Google Scholar]
- 56.Jalali S.M.J., Khosravi A., Alizadehsani R., Salaken S.M., Kebria P.M., Puri R., Nahavandi S. ICIT. 2019. Parsimonious evolutionary-based model development for detecting artery disease; pp. 800–805. [Google Scholar]
- 57.Jalali S.M.J., Ahmadian S., Khosravi A., Mirjalili S., Mahmoudi M.R., Nahavandi S. Neuroevolution-based autonomous robot navigation: a comparative study. Cognit Syst Res. 2020 [Google Scholar]
- 58.Mousavirad S.J., Schaefer G., Jalali S.M.J., Korovin I. Proceedings of the 2020 genetic and evolutionary computation conference companion. 2020. A benchmark of recent population-based metaheuristic algorithms for multi-layer neural network training; pp. 1402–1408. [Google Scholar]
- 59.Jalali S.M.J., Ahmadian S., Kebria P.M., Khosravi A., Lim C.P., Nahavandi S. International conference on neural information processing. Springer; Cham: 2019. Evolving artificial neural networks using butterfly optimization algorithm for data classification; pp. 596–607. [Google Scholar]
- 60.Maleki M., Mahmoudi M.R., Wraith D., Pho K.H. Time series modelling to forecast the confirmed and recovered cases of COVID-19. Travel Med Infect Dis. 2020 doi: 10.1016/j.tmaid.2020.101742. [DOI] [PubMed] [Google Scholar]
- 61.Abbasi A.R., Mahmoudi M.R., Avazzadeh Z. Diagnosis and clustering of power transformer winding fault types by cross-correlation and clustering analysis of FRA results. IET Gen Transm Distrib. 2018;12(19):4301–4309. [Google Scholar]