Abstract
The entry of SARS-CoV-2 into host cells proceeds by a proteolysis process, which involves the lysosomal peptidase cathepsin L. Inhibition of cathepsin L is therefore considered an effective method to decrease the virus internalization. Analysis from the perspective of structure-functionality elucidates that cathepsin L inhibitory proteins/peptides found in food share specific features: multiple disulfide crosslinks (buried in protein core), lack or low contents of (small) α-helices, and high surface hydrophobicity.
Lactoferrin can inhibit cathepsin L, but not cathepsins B and H. This selective inhibition might be useful in fine targeting of cathepsin L. Molecular docking indicated that only the carboxyl-terminal lobe of lactoferrin interacts with cathepsin L and that the active site cleft of cathepsin L is heavily superposed by lactoferrin. A controlled proteolysis process might yield lactoferrin-derived peptides that strongly inhibit cathepsin L.
Keywords: COVID-19, Infection, Virus, Food, Protein, Lactoferrin
1. Introduction
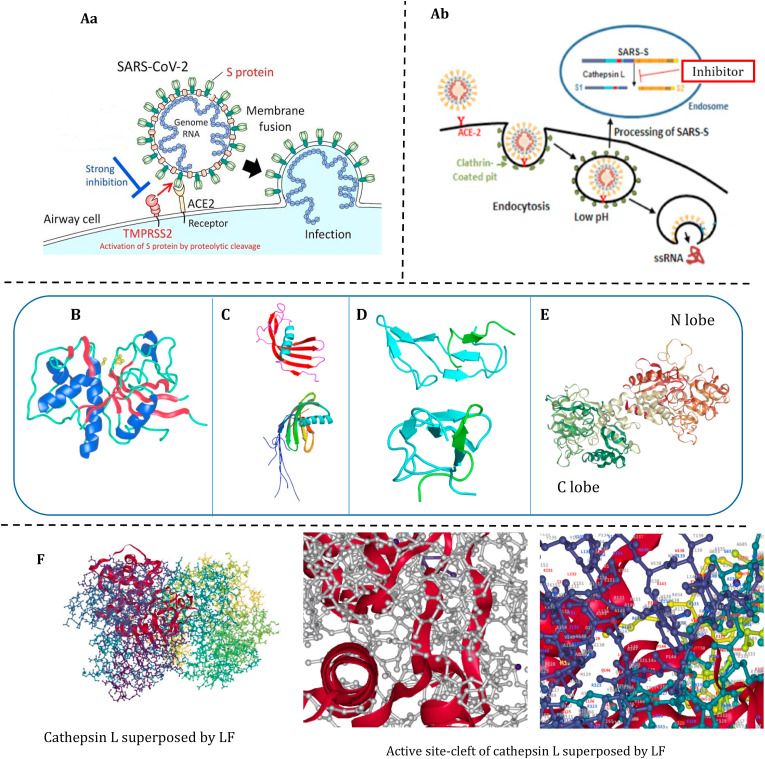
The ongoing coronavirus disease 2019 (COVID-19) is caused by Severe Acute Respiratory Syndrome Corona Virus-2 (SARS-CoV-2). Coronavirus binds though its spike glycoprotein (S) to human angiotensin converting enzyme 2 (hACE2) on the host cell membrane. Proteolysis of the protein S to its subunits S1 and S2 by the transmembrane protease serine 2 (TMPRSS2) enables fusion of the virus envelope with the host cell membrane to gain entry into the cell (Fig. 1 A) (Hoffmann et al., 2020). However, it is now known that SARS-CoV-2 as well (mainly) enters cells through endocytosis. Phosphatidylinositol 3-phosphate 5-kinase and cathepsin L are critical for the endocytosis (Ou et al., 2020). Similar to SARS-CoV, after endocytosis of SARS-CoV-2, the protein S is cleaved (Fig. 1A) by cathepsin L, which allows fusion of the viral membrane with the endosomal membrane, and then the viral genome is released into the host cell. Therefore, cathepsin L inhibition might be helpful in reducing infection by SARS-CoV (Adedeji et al., 2013) and SARS-CoV-2 (Ou et al., 2020). Actually, application of a clinically proven drug that inhibits TMPRSS2 was shown to incompletely inhibit SARS-CoV-2 infection. Full inhibition was achieved by co-employment of two drugs, one for inhibition of TMPRSS2 and the other for cathepsin L/B (Hoffmann et al., 2020).
Fig. 1.
A) Fusion of the coronavirus envelop to the (Aa) host cell membrane (Hoffmann et al., 2020), or (Ab) endo-lysosomal membrane (Adedeji et al., 2013); B) Ribbon drawing structure of cathepsin L. Green ribbon: the fold of the two-chain form of native cathepsin L; Blue: α-helices; Red: β-sheets; Ball and stick: the side chains of the reactive-site cysteine (Cys25) and histidine (His163) (Turk et al., 2012); Ribbon drawing structures of C) cystatin and oryzacystatin, D) bromelain inhibitor VI and E) diferric bovine lactoferrin, which show cathepsin L inhibitory activity. Bromelain inhibitor VI and lactoferrin specifically inhibit cathepsin L (no other cathepsins); F) Structural drawings of human cathpesin L suprposed by bovine lactoferrin. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Cathepsin L (220 amino acid residues) (Fujishima et al., 1997) is a lysosomal cysteine peptidase and has a two-chain form (L and R). The L domain contains three α-helices (one is the longest central helix) and the R domain is a β-barrel, which is closed at the bottom by an α-helix (Fig. 1B). The reactive site-cleft composes of histidine (His163) located at the top of the barrel and cysteine (Cys25) located at the N-terminus of the L domain central helix (Turk et al., 2012). Cathepsin L contributes to protein turnover, and apoptosis in cells. The overexpression of cathepsin L and H in cancer cells, in contrast to cathepsins B, C, S, and X/Z has made cathepsin L a target for anticancer strategies (Lankelma et al., 2010). The enzyme is also implicated in other pathologies such as osteoporosis, and periodontal diseases (Fujishima et al., 1997). Consequently, a plethora of cathepsin L inhibitory drugs are present for assessment. Some of the drugs e.g. E−64 (Fujishima et al., 1997) inhibit several cathepsins including cathepsin L, whereas, a few such as CLIK-148 (an epoxysuccinyl derivate) and iCL (an aldehyde derivative) exclusively inhibit cathepsin L (Lankelma et al., 2010). Many of the cathepsin L inhibitory drugs, like dipeptide epoxyketones, which are used for SARS-CoV suppression are essentially peptidomimetic (Adedeji et al., 2013).
2. Food protein and cathepsin inhibition
Given that food contains naturally occurring bioactive peptides and proteins, recently we outlined how food protein-derived peptides (small fragments, usually <3 kDa) that influence the renin-angiotensin system might have implications on SARS-CoV-2 endocytosis and pulmonary function in COVID-19 patients. The presumed mechanisms of action were over-expression of hACE2, and the receptor Mas, as well as blockage of the angiotensin II receptor type I on cell surface (Goudarzi et al., 2020). However, food proteins/peptides afford further opportunities for inhibition of SARS-CoV-2. Certain foods contain protein/peptide-based cathepsin L inhibitory compounds. This characteristic confers a new vision to exploration and implementation of biomolecules that can inhibit SARS-CoV-2 entry into host cells.
Whereas some naturally occurring bioactive peptides in food such as carnosine (226 Da, β-alanyl-L-histidine, in meat and chicken) do not affect cathepsin activity (Bonner et al., 1995), several cathepsin inhibitory molecules have been identified in plant and animal foods. Oryzacystatin-I (11.4 kDa, residue count 102) in rice is known to inhibit cathepsin L (inhibition constant, Ki: 7.3 × 10−10 M), but also cathepsin B (Ki: 7.9 × 10−8 M) and cathepsin H (Ki: 1.0 × 10−6) (Hellinger and Gruber, 2019). A hydrophobic cluster between the α-helix and the five-stranded antiparallel β-sheet structures (Fig. 1C) stabilizes the helix architecture of oryzacystatin-I (Nagata et al., 2000) and the N-terminal 21 amino acid residues are most probably not essential for cathepsin inhibitory activity of oryzacystatin-I (Abe et al., 1988). It is known that hydrophobic interactions are crucial for the inhibition of cathepsins, likely associated with the hydrophobic wedge-shaped structure of their active-site cleft (Turk et al., 2012). Corn cystatin-I as well inhibits cathepsin L (Ki: 1.7 × 10−8 M) and cathepsin H (Ki: 5.7 × 10−9 M) (Hellinger and Gruber, 2019).
In contrast to cystatin, bromelain inhibitor VI [BI-VI; 5.89 kDa; heavy chain (H, 41 amino acid residues) and light chain (L, 11 amino acid residues)] (Fig. 1D) which is a peptide present in pineapple stem selectively inhibits cathepsin L (Ki 0.2 × 10−6 M) and at a lesser extent trypsin (Polya, 2003). The primary and secondary structures of BI-VI are remarkably distinct from those of cystatin. Hen egg white cystatin consists of two α-helices and a five-stranded antiparallel β-sheet, but the secondary structure of BI-VI lacks α-helix structure. It is composed of two domains A and B, each of which formed by a three-stranded antiparallel β-sheet (Hatano et al., 1995). BI-VI contains ten cysteine residues, and five intra/inter-chain disulfide bonds between Cys3L-Cys7H, Cys6L-Cys39H, Cys8L-Cys5H, Cys14H-Cys21H, and Cys18H-Cys30H crosslink the protein. The disulfide bonds form the protein core, which is not common as a protein core is commonly occupied with some bulky hydrophobic side chains. This arrangement is homologous with Bowman−Birk trypsin/chymotrypsin inhibitor from soybean (BBI-I), a typical serine protease inhibitor (Hatano et al., 1996). The heavily S-S crosslinked double chain conformation might be important for the inhibition selectivity towards cathepsin L. This feature is shared with lactoferrin, which is discussed later in the current communication. It is noteworthy that similar to cathepsin L but not essentially required, trypsin could efficaciously activate SARS-CoV-2 S protein, enabling formation of syncytium (Ou et al., 2020). Therefore, the co-inhibition of trypsin and cathepsin L by BI-VI is advantageous.
Comparable to plant resources, foods of animal origin contain cathepsin L inhibitors. Though the proteins and peptides with cathepsin L inhibitory property in foods of animal origin have been scarcely investigated, some pieces of evidence are available in the literature. Mammalian milk has significant contents of cysteine protease inhibitors, such as lactoferrin, β-casein and β-lactoglobulin. The inhibitory effect of β-lactoglobulin B, also at a lower extent that of β-lactoglobulin A, on cathepsins K and L have been observed (Ogawa et al., 2009). Beta-lactoglobulin is a member of lipocalin family and binds to hydrophobic molecules. In addition to a single cysteine residue buried in protein and protected by α-helix, β-lactoglobulin has 2 internal disulfide linkages that stabilize the protein structure. At physiological pH, β-lactoglobulin exists as a supramolecular dimer. The secondary structure of β-lactoglobulin consists of nine antiparallel β-sheets, three helical turns and only one short α-helix (Ragona et al., 2000). Beta-lactoglobulin A has an additional negative charge compared to β-lactoglobulin B. A shared feature between BI-VI and β-lactoglobulin B is the existence of internal disulfide bonds.
For β-casein, an allosteric-type inhibition mechanism is believed to cause cathepsin inhibitory action (Sano et al., 2005). Beta-casein (23.98 kDa) is a single chain polypeptide and the most hydrophobic casein. Essentially all of β-casein net charge and α-helix structures are positioned at the N-terminal portion of the molecule (Kumosinski et al., 1993). It lacks cysteine. Considerable hydrophobicity and low contents of α-helix structure (<20%) are the shared features between β-casein and β-lactoglobulin B. Taking hydrophobicity into account, I speculate that comparable to oryzacystatin-I (Abe et al., 1988), the hydrophilic N-terminal portion of β-casein may not be essential for cathepsin inhibitory activity. Rather, the C-terminal hydrophobic portion which is also poor in α-helix structure probably causes the inhibition. In fact, inhibition of cathepsin L may occur as a consequence of the interaction between the carbonyl (carboxyl) group of an inhibitor with the electrophilic oxyanion hole of cathepsin L, which consists of side chains of Gln16, Trp189, His163 and the main chain of Cys25 (Fujishima et al., 1997).
Lactoferrin (~80 kDa) is present at much higher contents in human milk than cow's milk. It strongly inhibits cathepsin L. The IC50 of lactoferrin against cathepsin L was 10−7 M, while, that of a synthetic peptide which targets cathepsin L active site was 10−5 M. Notably, lactoferrin does not inhibit cathepsin B and cathepsin H (Sano et al., 2005). This may enable fine targeting of cathepsin L for obstacle SARS-CoV-2 internalization, while avoiding possible jeopardizes to cells. Bovine lactoferrin is a single polypeptide consisting of 689 residues. It is folded into two lobes (N-terminal and C-terminal halves) joined by a small helix (3-turn) (Moore et al., 1997) (Fig. 1E). The crystal structures of lactoferrin and cathepsin L were taken from the RCSB protein data bank. Molecular docking using MDsrv (Tiemann et al., 2017) indicates that the C-lobe of lactoferrin interacts with cathepsin L (Fig. 1F). It also shows that the C-lobe superposes the active-site cleft of cathepsin L (Fig. 1F). The α-helix content of bovine lactoferrin varies between 7% and 16% depending on the pH (Sreedhara et al., 2010). This protein contains 17 disulfide bonds (Wang et al., 2019) and the interactions between the two lobes are mostly hydrophobic caused by packing of nonpolar surfaces on the lobes (Moore et al., 1997). Low α-helix content, small helices, and hydrophobic surfaces are the shared features between lactoferrin, β-lactoglobulin and β-casein. It is worthy to note that both lactoferrin and BI-VI, which show high selectivity towards cathepsin L, include several disulfide bonds, positioned in protein core. As mentioned in an earlier place, cathepsin L inhibition proceeds by thiol chemistry.
In addition to the naturally present protein and peptides with cathepsin L inhibitory activity in foods, some peptides encrypted in food proteins might be able to inhibit cathepsin L and hence assist COVID-19 prevention. Food proteins release diverse biologically active peptides once they are hydrolyzed by the enzymes present in the gastrointestinal tract, and from microbes. Presently some pieces of evidence showing cathepsin L inhibitory activity by food protein-derived peptides are scarce. Two cathepsin B inhibitory peptides derived from β-casein were identified in a pancreatic digest of casein (Lee and Lee, 2000), as well as peptides from the digestion of β-lactoglobulin B by an endopeptidase could inhibit cathepsin K. It remains to be explored whether food protein-derived peptides can inhibit cathepsin L. Isolation of peptides, for instance from lactoferrin that exclusively (or much more preferentially) inhibit cathepsin L rather than other cathepsins can be highly advantageous.
The structural and biofunctional characteristics of many food proteins are extensively characterized. This is especially true for milk proteins, but also egg white proteins. For example, it is known that hen egg white riboflavin-binding protein has a high degree of crosslinking by nine disulfide bonds. Besides, food proteins are generally recognized as safe (GRAS) and inexpensive. Hundreds of biologically active peptide sequences derived from food proteins are known in the literature. The hydrolysis conditions, separation and purification procedures of these peptides are established. In silico examination of the cathepsin L inhibitory property of the known peptide sequences can accelerate the drug discovery.
CRediT authorship contribution statement
Ashkan Madadlou: Conceptualization, Investigation, Writing - original draft, Writing - review & editing.
Declaration of competing interest
The author declares no conflict of interest.
References
- Abe K., Emori Y., Kondo H., Arai S., Suzuki K. The NH2-terminal 21 amino acid residues are not essential for the papain-inhibitory activity of oryzacystatin, a member of the cystatin superfamily. J. Biol. Chem. 1988;263:7655–7659. [PubMed] [Google Scholar]
- Adedeji A.O., Severson W., Jonsson C., Singh K., Weiss S.R., Sarafianos S.G. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 2013;87:8017–8028. doi: 10.1128/JVI.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner A.B., Swann M.E., Marway J.S., Heap L.C., Preedy V.R. Lysosomal and nonlysosomal protease activities of the brain in response to ethanol feeding. Alcohol. 1995;12:505–509. doi: 10.1016/0741-8329(95)00035-6. [DOI] [PubMed] [Google Scholar]
- Fujishima A., Imai Y., Nomura T., Fujisawa Y., Yamamoto Y., Sugawara T. The crystal structure of human cathepsin L complexed with E-64. FEBS Lett. 1997;407:47–50. doi: 10.1016/S0014-5793(97)00216-0. [DOI] [PubMed] [Google Scholar]
- Goudarzi M., Garavand F., Madadlou A., Fogliano V. Food protein-derived antihypertensive peptides in the COVID-19 pandemic: friends of foes? J. Hypertens. 2020;38:1614–1616. doi: 10.1097/HJH.0000000000002534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano K. ‐i, Kojima M., Tanokura M., Takahashi K. Primary structure, sequence‐specific 1H‐NMR assignments and secondary structure in solution of bromelain inhibitor VI from pineapple stem. Eur. J. Biochem. 1995;232:335–343. doi: 10.1111/j.1432-1033.1995.335zz.x. [DOI] [PubMed] [Google Scholar]
- Hatano K.I., Kojima M., Tanokura M., Takahashi K. Solution structure of bromelain inhibitor VI from pineapple stem: structural similarity with Bowman - Birk trypsin/chymotrypsin inhibitor from soybean. Biochemistry. 1996;35:5379–5384. doi: 10.1021/bi952754+. [DOI] [PubMed] [Google Scholar]
- Hellinger R., Gruber C.W. Peptide-based protease inhibitors from plants. Drug Discov. Today. 2019;24:1877–1889. doi: 10.1016/j.drudis.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumosinski T.F., Brown E.M., Farrell H.M. Three-dimensional molecular modeling of bovine caseins: an energy-minimized β-casein structure. J. Dairy Sci. 1993;76:931–945. doi: 10.3168/jds.S0022-0302(93)77420-2. [DOI] [PubMed] [Google Scholar]
- Lankelma J.M., Voorend D.M., Barwari T., Koetsveld J., Van der Spek A.H., De Porto A.P.N.A., Van Rooijen G., Van Noorden C.J.F. Cathepsin L, target in cancer treatment? Life Sci. 2010;86:225–233. doi: 10.1016/j.lfs.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Lee H.S., Lee K.J. Cathepsin B inhibitory peptides derived from β-casein. Peptides. 2000;21:807–809. doi: 10.1016/S0196-9781(00)00212-6. [DOI] [PubMed] [Google Scholar]
- Moore S.A., Anderson B.F., Groom C.R., Haridas M., Baker E.N. Three-dimensional structure of diferric bovine lactoferrin at 2.8 Å resolution. J. Mol. Biol. 1997;274:222–236. doi: 10.1006/jmbi.1997.1386. [DOI] [PubMed] [Google Scholar]
- Nagata K., Kudo N., Abe K., Arai S., Tanokura M. Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica. Biochemistry. 2000;39:14753–14760. doi: 10.1021/bi0006971. [DOI] [PubMed] [Google Scholar]
- Ogawa N., Takahashi M., Ishidoh K., Katunuma N. Inhibition of collagenolytic cathepsins by β-lactoglobulin in milk and its suppressive effect on bone Rresorption. J. Nutr. Sci. Vitaminol. (Tokyo). 2009;55:264–270. doi: 10.3177/jnsv.55.264. [DOI] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polya G.M. 2003. Protein and Non-protein Protease Inhibitors from Plants; pp. 567–641. [DOI] [Google Scholar]
- Ragona L., Fogolari F., Zetta L., Pérez D.M., Puyol P., De Kruif K., Löhr F., Rüterjans H., Molinari H. Bovine β-lactoglobulin: interaction studies with palmitic acid. Protein Sci. 2000;9 doi: 10.1110/ps.9.7.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano E., Miyauchi R., Takakura N., Yamauchi K., Murata E., Le Q.T., Katunuma N. Cysteine protease inhibitors in various milk preparations and its importance as a food. Food Res. Int. 2005;38:427–433. doi: 10.1016/j.foodres.2004.10.011. [DOI] [Google Scholar]
- Sreedhara A., Flengsrud R., Prakash V., Krowarsch D., Langsrud T., Kaul P., Devold T.G., Vegarud G.E. A comparison of effects of pH on the thermal stability and conformation of caprine and bovine lactoferrin. Int. Dairy J. 2010;20:487–494. doi: 10.1016/j.idairyj.2010.02.003. [DOI] [Google Scholar]
- Tiemann J.K.S., Guixà-González R., Hildebrand P.W., Rose A.S. MDsrv: viewing and sharing molecular dynamics simulations on the web. Nat. Methods. 2017;14:1123–1124. doi: 10.1038/nmeth.4497. [DOI] [PubMed] [Google Scholar]
- Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., Turk D. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta Protein Proteonomics. 2012;1824:68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Timilsena Y.P., Blanch E., Adhikari B. Lactoferrin: structure, function, denaturation and digestion. Crit. Rev. Food Sci. Nutr. 2019;59:580–596. doi: 10.1080/10408398.2017.1381583. [DOI] [PubMed] [Google Scholar]