Abstract
Background
Enterovirus (EV) and parechovirus type A3 (PeV-A3) cause infections ranging from asymptomatic to life-threatening. Host immune responses in children, particularly innate responses to PeV-A3, remain largely unknown. The aim of this study was to determine aspects of the cytokine/chemokine responses to EV and PeV-A3 in cerebrospinal fluid (CSF) and plasma obtained from children with systemic/central nervous system infection.
Methods
A total of 74 salvaged CSF samples (27 with EV, 23 with PeV-A3, and 24 with neither EV nor PeV-A3) and 35 paired blood samples (10 with EV, 14 with PeV-A3, and 11 with neither) were studied. Concentrations of cytokines and chemokines were measured using a customized 21-plex MILLIPLEX MAP Human Cytokine/Chemokine Magnetic Bead Panel. Additionally, clinical characteristics data for all the patients were collected from electronic medical records to evaluate the potential association between the immune response and presentations.
Results
We demonstrate that EV and PeV-A3 infections induce different cytokine/chemokine immune responses in children. EV induces more robust responses in CSF with significantly elevated levels of fractalkine, interferon (IFN)-α2, IFN-γ, interleukin (IL)-1Rα, IL-4, IL-8, and tumor necrosis factor α; PeV-A3 induces less robust or absent responses in CSF but robust responses in plasma, with significantly higher concentrations of IFN-α2, IL-15, IL-1Rα, interferon-γ-inducible protein–10, and monocyte chemoattractant protein–1.
Conclusions
High cytokine/chemokine concentrations in the plasma of PeV-A3 patients compared with EV patients could explain higher/more prolonged fever in PeV-A3 patients, whereas relatively low cytokine/chemokine concentrations in PeV-A3 CSF might explain the absence of CSF pleocytosis.
Keywords: cerebrospinal fluid, enterovirus, immune response, parechovirus, plasma
Viral infections of the central nervous system (CNS) remain a public health issue in developing and developed countries [1]. Enterovirus (EV) and parechovirus (PeV-A), particularly PeV-A3, are reported to be the leading causes of viral CNS infections in much of the world, including the United Kingdom, Europe, China, Japan, and the United States [2–9].
EV and PeV-A members of the family Picornaviridae are nonenveloped viruses with single-stranded positive sense RNA genomes, known or suspected to cause a wide spectrum of clinical manifestations in humans [10]. Infections caused by these agents can be asymptomatic but can also be severe, for example, myocarditis, encephalitis, sepsis-like syndrome, and CNS infections [5, 11, 12]. Neonates and infants are at a higher risk for severe clinical illness and sequelae than are older children and adults [10].
The clinical features of both EV and PeV-A3 can be similar when symptoms do occur. Therefore, distinguishing between EV and PeV-A3 based purely on clinical signs or symptoms is difficult at initial presentation [13, 14]. As the infection evolves, young infants with systemic PeV-A3 infections have higher fever, longer duration of fever, higher heart rate, absence of cerebrospinal fluid (CSF) pleocytosis, plus somewhat higher CSF concentrations of glucose and protein compared with EV-infected infants [5, 15].
The innate arm of the immune system produces important initial responses during viral infections, resulting in both local and systemic release of inflammatory and antiviral molecules, for example, cytokines. A subgroup of cytokines categorized as chemokines also influence leukocyte migration and trafficking of immune cells [16]. In particular, early production of cytokines induces an antiviral state and triggers the activation of immune cells [17]. Increased levels of cytokines and chemokines have been observed during systemic viral infections, including dengue fever and influenza [16, 18], and are thought to be associated with aspects of clinical presentations, severity of illness, and degree of abnormalities in laboratory test results.
Clinical characteristics and laboratory findings and their differences in EV- and PeV-A3-infected infants have been documented reasonably well. However, the data for host innate immune responses to EV and PeV-A3 infections in relation to the severity of clinical aspects and abnormalities in laboratory test results are still limited [13, 14, 19]. Therefore, the objective of our study was to evaluate cytokine and chemokine responses in the CSF and plasma of EV- vs PeV-A3-infected patients vs control subjects with neither EV nor PeV-A3, evaluating for possible differences in innate response between PeV-A3, EV, and controls. Differential results for innate responses could be valuable in further defining pathogenesis and possible outcome markers for EV and PeV-A3 CNS infections.
METHODS
Sample Collection
This retrospective study was conducted at Children’s Mercy, Kansas City (CMKC), and was approved by the CMKC Institutional Review Board. De-identified clinical specimens (CSF and plasma) were salvaged from neonates and infants ≤3 months of age who underwent sepsis/meningitis evaluation as part of standard of care during January–December 2018. All samples were collected on the day of admission, which was within 2 days of symptom onset. CSF and blood samples were collected within a 1-hour interval. All CSF samples and residual plasma samples from each group that had enough sample volume were screened for EV and PeV-A by a 2-step reverse transcription polymerase chain reaction (RT-PCR) targeting the 5’untranslated region, as described previously [15]. Positive CSF samples also underwent bidirectional sequencing targeting the VP1 region for determining the EV and PeV-A genotypes [20, 21]. Samples from a total of 71 patients were available for study. Analyzed CSF samples were from 23 patients with EV-positive CSF, 27 patients with PeV-A3-positive CSF, and 24 control patients with CSF that was negative for both EV and PeV-A3. Overall, 35 CSF samples were paired with a plasma sample obtained from the same patient (10 EV, 14 PeV-A3, and 11 samples from the same patient who had neither EV nor PeV-A3). Clinical and laboratory data were obtained from the patients’ electronic medical records (age, gender, maximum temperature [Tmax], length of hospitalization, peripheral blood white blood cell [WBC] count, CSF WBC, CSF glucose, and CSF protein concentration).
Cytokine and Chemokine Analysis
The CSF samples were stored at –70ºC before analysis. Plasma was separated from blood samples by centrifugation (2000g for 10 minutes) at 4ºC and stored at –70ºC until the cytokine analysis. The concentrations of cytokines/chemokines were measured in both the CSF and plasma of the EV, PeV-A3, and control groups using the MILLIPLEX MAP Human Cytokine/Chemokine Magnetic Bead Panel from Millipore Sigma (Burlington, MA, USA). The customized 21-plex cytokine/chemokine panel consisted of the following analytes: granulocyte macrophage–colony-stimulating factor (GM-CSF), fractalkine, interferon (IFN)-α2, IFN-γ, interleukin (IL)-10, IL-12p40, IL-13, IL-15, IL-17A, IL-1Rα, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-8, interferon-γ-inducible protein (IP)–10, chemokine ligand 2 (CCL2)/monocyte chemoattractant protein–1 (MCP-1), CCL3/MIP-1α, chemokine ligand 5 (CCL5)/regulated upon activation, normal T cell expressed and secreted (RANTES), and tumor necrosis factor (TNF)–α. Samples were measured in duplicate, and the concentration was calculated by reference to the standard curve for each cytokine. The average coefficient of variation was measured between the duplicates, and values <30 were considered acceptable. All samples were measured undiluted. The lower detection limit for each cytokine was as follows: 7.5 pg/mL for GM-CSF, 22.7 pg/mL for fractalkine, 2.9 pg/mL for IFN-α2, 0.8 pg/mL for IFN-γ, 1.1 pg/mL for IL-10, 7.4 pg/mL for IL-12p40, 1.3 pg/mL for IL-13, 1.2 pg/mL for IL-15, 0.7 pg/mL for IL-17A, 8.3 pg/mL for IL-1Rα, 0.8 pg/mL for IL-1β, 1.0 pg/mL for IL-2, 4.5 pg/mL for IL-4, 0.5 pg/mL for IL-5, 0.9 pg/mL for IL-6, 0.4 pg/mL for IL-8, 8.6 pg/mL for IP-10, 1.9 pg/mL for MCP-1, 2.9 pg/mL for MIP-1α, 1.2 pg/mL for RANTES, and 0.7 pg/mL for TNF-α. Cytokines that were below the detection limit were categorized as undetectable.
Statistical Analysis
The clinical characteristics of EV, PeV-A3, and control groups are presented as median and interquartile range (IQR). Statistical differences between 2 or multiple groups were analyzed by nonparametric Kruskal-Wallis testing and pairwise post hoc testing in Microsoft Excel 365 with the Real Statistics add-in (http://www.real-statistics.com/). P values <.05 were considered statistically significant.
Discriminant Analysis
Discriminant analysis (DA) was done using SPSS, version 24. The discriminant function aims to create a predictive model of group membership based on a linear combination of variables. Variance–covariance homogeneity was tested using the nonparametric Levene’s test, which is both more robust and suited to non-normal distributed data [22]. Stepwise DA was used so that our predictive model would be based on the most discriminating biomarkers for each data set.
Wilks’ Lambda test was used to evaluate the performance of each biomarker in the model. The values of the Wilks’ Lambda statistic range from 0 to 1, with 0 indicating perfect group discrimination and 1 indicating no discrimination at all. The significance of Wilks’ Lambda was tested using the F test (P values < .05 were considered statistically significant).
Principal Component Analysis
Principal component analysis (PCA) was performed using either BioNumerics, version 6.6 (Applied Maths NV, Austin, TX, USA; www.applied-maths.com), or IBM SPSS Statistics for Windows, version 24.0. (IBM Corp., Armonk, NY, USA). PCA was computed from covariance matrices. To avoid dominance of variables with large values, we normalized the data for each variable by dividing by appropriate variance before principal components computation. Sample adequacy and existence of correlated variables were evaluated by Kaiser-Meyer-Olkin (KMO) measure of sampling adequacy [23, 24] and Bartlett’s test of sphericity [25], respectively; both tests were performed using SPSS. Bartlett’s test of sphericity yields a P value that represents the probability that correlated variables do not exist. For PCA to be meaningful, this null hypothesis must be rejected. The significance of principal components was estimated by Monte Carlo simulation (parallel analysis) using the syntax written for SPSS by Brian O’Connor [26].
Biomarker Evaluation Using the Receiver Operating Characteristic Curve Method
The receiver operating characteristic (ROC) curve was generated by plotting specificity minus 1 and sensitivity on the x- and y-axes, respectively. To simplify the evaluation of any given biomarker, we evaluated the biomarker’s usefulness by the area under the ROC curve (AUC). On such plots, a biomarker with perfect sensitivity and specificity (ie, both sensitivity and specificity are equal to 100%, and “1-specificity” is equal to 0) has an AUC equal to 1, while a useless biomarker with sensitivity and specificity of 50% (ie, using the biomarker is equivalent to flipping a coin) has an AUC equal to .5 [27]. Asymptotic significance is measured by calculating a P value that tests the null hypothesis that the true AUC of the relevant biomarker is equal to .5. The null hypothesis is rejected at P values <.05.
To combine biomarkers into the best possible profiles, we used DA canonical functions 1 and 2 as these profiles. These functions were computed to maximize group separation and included only the best-performing biomarkers. We used the AUC as a measure of biomarker performance.
RESULTS
Demographics and Clinical and Laboratory Findings of Patients From Whom Salvaged Samples Were Obtained
Demographics and laboratory values for overall patient groups (EV, PeV-A3, and controls) are summarized in Supplementary Table 1. There was no statistical difference in length of stay, age, or gender between the EV, PeV-A3, and control groups. The median age of EV patients was 4 weeks with 61% male, 3 weeks for PeV-A3 patients with 63% male, and 4 weeks for the control group with 67% male. Maximum temperature was different among groups overall (P < .001), with Kruskal-Wallis comparison tests showing that PeV-A3-infected patients had higher Tmax than EV (P < .001) and control groups (P < .0001). There was no significant difference in Tmax between EV-infected patients and controls.
Peripheral blood WBC was different among groups overall (P = .001), with lower counts in PeV-A3-infected groups compared with EV (P < .001) and control groups (P < .001). CSF WBC counts were also significantly different among groups overall (P = .0006), with EV patients having higher counts than control (P < .001) and PeV-A3 groups (P < .0001). CSF pleocytosis was present less with PeV-A3 (0/27, 0%) compared with controls (3/24, 12.5%) and EV (8/23, 34.8%; P = .002). CSF protein concentrations were not significantly different among groups. CSF glucose concentrations were significantly different among groups (P = .0003), with concentrations for the EV group being lower than those for both PeV-A3 and controls (P < .0001 for both); PeV-A3 and controls were not significantly different.
Cytokines and Chemokines in the Plasma and CSF of EV- and PeV-Positive Samples Compared With Control Samples
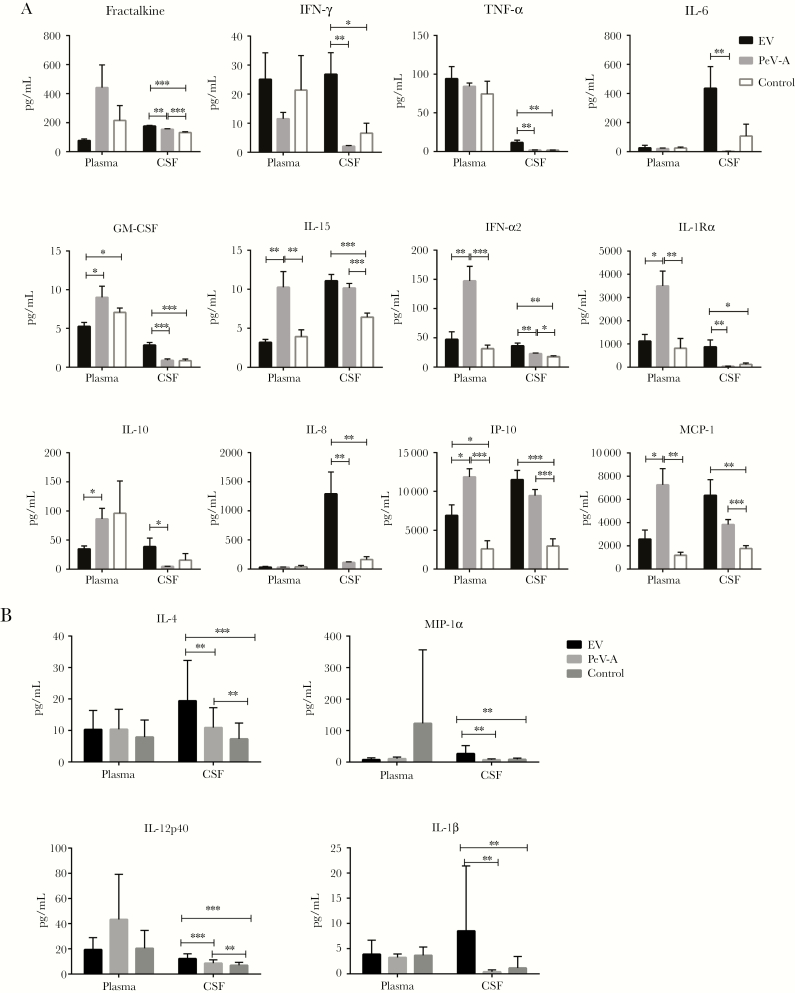
Among the 21 cytokines/chemokines analyzed, the cytokine/chemokine concentrations of 5 (GM-CSF, IL-13, IL-17A, IL-2, and IL-5) were below the detection limit in CSF and close to the limit in plasma. The coefficient of variation between the duplicates was acceptable for all duplicate assays on the same sample. No significant differences between groups were detected for RANTES and IL-5, and hence they were not included in the additional statistical analysis (Figure 1A and B). Several examples of the analytes that had a statistically significant difference between groups and sample types are shown in Figure 1A and B.
Figure 1.
A and B, Bar graphs representing cytokine/chemokine concentrations in the CSF and plasma of the EV, PeV-A3, and control groups. Six analytes demonstrated significantly elevated values in the CSF of EV patients when compared with both the PeV and control groups; fractalkine, IFN-α2, IFN-γ, IL-1Rα, IL-4, IL-8, and TNF-α. In contrast, the values for 5 analytes were significantly higher in the plasma of PeV-A3-infected patients when compared with both the EV and control groups; IFN-α2, IL-15, IL-1Rα, IP-10, and MCP-1. *P < .05; **P < .005; ***P < .0005. Abbreviations: CSF, cerebrospinal fluid; EV, enterovirus; IFN, interferon; IL, interleukin; IP, interferon-γ-inducible protein; MCP, monocyte chemoattractant protein; PeV, parechovirus type A3; TNF, tumor necrosis factor.
The EV group had increased CSF fractalkine, IFN-α2, IFN-γ, IL-1Rα, IL-4, IL-8, and TNF-α when compared with both the PeV-A3 and control groups (Figure 1A and B). Levels of IL-6, IL-10, and TNF-α were high in the EV group but undetectable in the PeV-A3 group. Likewise, the PeV-A3 group’s CSF concentrations for fractalkine, IFN-α2, IL-15, IL-4, IP-10, and MCP-1 were significantly higher compared with controls (Figure 1A and B). The PeV-A3 group’s CSF IFN-γ, IL-6, MIP-1α, and RANTES concentrations, while lower than controls, were not significantly different than controls.
The innate immune response in plasma of the EV group as determined by plasma concentrations of cytokines/chemokines was less intense than the innate immune responses identified in the PeV-A3 and control groups. Compared with PeV-A3 plasma concentrations, EV plasma concentrations were increased only for IFN-γ and TNF-α, but neither increase was significant. Compared with the control plasma concentrations, EV plasma concentrations were significantly increased only for IP-10.
In contrast, the PeV-A3 group had a more robust response in plasma, with significantly higher IFN-α2, IL-15, IL-1Rα, IP-10, and MCP-1 concentrations compared with the EV and control groups (Figure 1A and B).
Determination of the Most Discriminating Biomarkers
We tested CSF and plasma biomarkers separately and in combination. The number of biomarkers that contributed significantly to the model varied between data sets. Six, 2, and 8 biomarkers were significant using CSF, plasma, and both, respectively. Discrimination between EV-infected, PeV-A3-infected, and control subjects was substantially better when using combined CSF–plasma biomarkers.
Better discrimination was evident by visually inspecting canonical function plots (Figure 2), as well as by comparing the rates of correct classification (RCCs) (Table 1). Canonical functions 1 and 2 were used to create a coordinate system in which individual participants were plotted and group centroids were marked (squares in Figure 2). The greatest spread of group centroids and least intergroup overlap were seen using a CSF–plasma combined biomarker profile. Furthermore, the smallest (best) Wilks’ Lambda values were obtained with this combined profile, indicating that the combination provided the best performance within the model as well as the best model performance (Figure 2). Consistent with these data, we saw the highest RCC fidelity using the combined CSF–plasma biomarker profile, with group RCCs ranging between 91% and 100% (Table 1).
Figure 2.
Discriminant analysis predictive model. CSF (control: n = 24; EV: n = 23; PeV: n = 27) and plasma (control: n = 11; EV: n = 10; PeV: n = 14) biomarkers were analyzed separately and in combination. Canonical functions (DSC) 1 and 2 are plotted on the x- and y-axes, respectively. P values shown in the plots indicate model significance. Wilks’ Lambda values for individual biomarkers and the model as a whole are shown in the tables below the plots. Abbreviations: b, plasma biomarker; c, cerebrospinal fluid biomarker; CSF, cerebrospinal fluid; EV, enterovirus; GM-CSF, granulocyte macrophage–colony-stimulating factor; IFN, interferon; IL, interleukin; IP, interferon-γ-inducible protein; MCP, monocyte chemoattractant protein; PeV, parechovirus type A3; TNF, tumor necrosis factor.
Table 1.
Rate of Correct Classification Based on Discriminant Analysis Models
| CSF—Overall RCC: 77.0% | ||||
|---|---|---|---|---|
| Control | Predicted Classification EV |
PeV | ||
| Original classification | Control | 87.5 (21) | 4.2 (1) | 8.3 (2) |
| EV | 8.7 (2) | 60.9 (14) | 30.4 (7) | |
| PeV | 14.8 (4) | 3.7 (1) | 81.5 (22) | |
| Plasma—overall RCC: 71.4% | ||||
| Predicted Classification | ||||
| Control | EV | PeV | ||
| Original classification | Control | 72.7 (8) | 27.3 (3) | 0.0 (0) |
| EV | 30.0 (3) | 50.0 (5) | 20.0 (2) | |
| PeV | 0.0 (0) | 14.3 (2) | 85.7 (12) | |
| CSF & plasma—overall RCC: 97.1% | ||||
| Predicted Classification | ||||
| Control | EV | PeV | ||
| Original classification | Control | 90.9 (10) | 0.0 (0) | 9.1 (1) |
| EV | 0.0 (0) | 100.0 (10) | 0.0 (0) | |
| PeV | 0.0 (0) | 0.0 (0) | 100.0 (14) | |
Rates of correct and incorrect classification are expressed as percentage of group totals. RCCs are in bold, and incorrect classification rates are in roman font. The number of patients per category is in parentheses.
Abbreviations: CSF, cerebrospinal fluid; EV, enterovirus; PeV, parechovirus type A3; RCC, rate of correct classification.
Exploring Natural Partitioning Using PCA
We performed PCA using either all biomarkers or only the significant biomarkers identified in our DA studies. Clearer group separation again occurred with the combination of CSF with plasma biomarkers and also restriction of analysis to the DA-identified biomarkers (Supplementary Figure 1).
Evaluation of Biomarkers’ Performance Using ROC Curves
High CSF cytokine AUC values were more frequent for EV-infected patients, while high plasma AUC values were more frequent for PeV-A3-infected patients (Table 2), that is, blue shaded values in the first 4 columns. Generally, multivariate biomarker profiles performed better than individual biomarkers, as indicated by the higher AUC values in Table 2, Disc1 values. Multivariate profiles had the highest and fourth highest AUC values for PeV-A3 and EV, respectively. The very highest AUC values for both EV- and PeV-A3-infected subjects were obtained using CSF–plasma combined profiles (Table 2 columns 5 and 6), highlighting the potential advantage of using combined profiles.
Table 2.
Evaluation of Biomarkers and Biomarker Profiles Using the Area Under the ROC Curve Method
| CSF | Plasma | CSF & Plasma | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EV | PeV-A3 | EV | PeV-A3 | EV | PeV-A3 | ||||||
| TNF-α | 0.88 | Disc2 | 0.866 | Fractalkine | 0.854 | Disc1 | 0.952 | Disc2 | 0.924 | Disc1 | 1 |
| MIP-1α | 0.861 | IL-1β | 0.726 | GM-CSF | 0.774 | IFN-α2 | 0.901 | Disc1 | 0.912 | Disc2 | 0.558 |
| IFNg | 0.86 | MIP-1α | 0.725 | IL-15 | 0.74 | IL-15 | 0.898 | ||||
| Disc1 | 0.85 | IL-6 | 0.717 | Disc2 | 0.72 | MCP-1 | 0.898 | ||||
| IL-6 | 0.844 | IFNg | 0.686 | IL-17A | 0.718 | IP -10 | 0.891 | ||||
| IL-10 | 0.842 | TNF-α | 0.674 | IL-10 | 0.716 | IL-1Ra | 0.881 | ||||
| IL-1Ra | 0.841 | IL-15 | 0.659 | IFN-α2 | 0.712 | Fractalkine | 0.874 | ||||
| IL-1β | 0.839 | GM-CSF | 0.639 | IL-2 | 0.676 | IL-8 | 0.755 | ||||
| IL-12p40 | 0.835 | IP-10 | 0.632 | MIP-1α | 0.666 | IL-12p40 | 0.753 | ||||
| GM-CSF | 0.826 | MCP-1 | 0.623 | IL-6 | 0.66 | IL-10 | 0.728 | ||||
| IL-8 | 0.812 | IL-10 | 0.61 | IL-12p40 | 0.65 | RANTES | 0.663 | ||||
| Fractalkine | 0.809 | IL-8 | 0.604 | Disc1 | 0.64 | GM-CSF | 0.655 | ||||
| IFN-α2 | 0.8 | Disc1 | 0.601 | MCP1 | 0.632 | TNF-α | 0.609 | ||||
| IL-4 | 0.774 | RANTES | 0.593 | IL-8 | 0.612 | IL-13 | 0.595 | ||||
| IL-13 | 0.765 | IL-17A | 0.548 | TNF-α | 0.604 | IL-6 | 0.595 | ||||
| IP-10 | 0.749 | IL-13 | 0.543 | IL-1Ra | 0.6 | IL-5 | 0.571 | ||||
| Disc2 | 0.702 | IL-12p40 | 0.541 | IP-10 | 0.564 | IL-4 | 0.568 | ||||
| IL-2 | 0.69 | IL-1Ra | 0.54 | IL-4 | 0.548 | MIP-1α | 0.556 | ||||
| MCP-1 | 0.682 | IL-5 | 0.531 | IL-13 | 0.544 | IL-17A | 0.524 | ||||
| IL-15 | 0.679 | IL-2 | 0.527 | IL-5 | 0.54 | IL-2 | 0.519 | ||||
| IL-17A | 0.673 | Fractalkine | 0.525 | IFNg | 0.528 | IL-1β | 0.51 | ||||
| RANTES | 0.649 | IFN-α2 | 0.516 | IL-1β | 0.526 | Disc2 | 0.507 | ||||
| IL-5 | 0.556 | IL-4 | 0.516 | RANTES | 0.524 | IFNg | 0.503 | ||||
Analogous to principal components in PCA, discriminant analysis computes discriminants. Both principal components and discriminant functions are eigenvectors that can be viewed as artificial variables comprised of contributions from observed variables. In a multivariate problem, data points are plotted in multidimensional space with as many axes as variables. To transform this into a simpler 2- or 3-dimensional presentation, variables are combined into eigenvectors ranked by the amount of variance they explain. In PCA, the direction of the eigenvector that explains the most variance (ie, first component) is selected so that it explains the maximum amount of variance that can be explained by 1 vector (ie, maximizing the amount of variance explained). In discriminant analysis, the eigenvector that explains the most variance (first discriminant function) is selected to maximize group separation. As these eigenvectors are linear combinations of observed variables, they may be used as a way to combine variables and test them using the AUC method. Here, we selected discriminant functions (Disc1 and Disc2) as they are likely to be superior to principal components given the way they were computed. The heat map corresponds to a range of AUC values from the lowest (dark red) to highest (dark blue).
Abbreviations: AUC, area under the curve; CSF, cerebrospinal fluid; Disc1/Disc2, first/second discriminant function; EV, enterovirus; GM-CSF, granulocyte macrophage–colony-stimulating factor; IFN, interferon; IL, interleukin; IP, interferon-γ-inducible protein; MCP, monocyte chemoattractant protein; PCA, principal component analysis; PeV, parechovirus type A3; TNF, tumor necrosis factor.
DISCUSSION
The immune system is a complex and coordinated network. Upon invasion of the host by microorganisms, inflammatory mediators are generated in “signaling cascades” through a series of pathways that result in subsequent release of pro-inflammatory and anti-inflammatory cytokines/chemokines intended to inhibit pathogen replication and promote adaptive immunity [14, 28]. While immune responses to EV have been studied in some detail, knowledge gaps persist to a greater extent about PeV-A3, particularly innate responses [1, 14, 29–34].
This is the first US study to describe diverse immune profiles in paired CSF–plasma of EV- vs PeV-A3-infected patients and how differential production of selected molecules might contribute to the differences in certain clinical or laboratory findings of EV vs PeV-A3 in young infants. We confirmed that our current patient groups from which CSF and plasma were obtained shared demographic and clinical characteristics with previously reported patients infected with PeV-A3 and EV infections [5, 6]. Such characteristics include PeV-A3-infected infants having higher Tmax, longer duration of fever, and less frequent CSF pleocytosis compared with EV-infected infants.
We analyzed concentrations of 21 cytokines in plasma and CSF by using DA, PCA, RCC, and AUC methodology. We demonstrate that innate immune responses in CSF and in plasma differ in CNS EV infections compared with PeV-A3. The differential concentrations in CSF vs plasma could explain differences in the rates of CSF pleocytosis for PeV-A3 (rare) vs EV (common) CNS infections, and higher, more prolonged fever in young PeV-A3-infected infants.
Our use of DA or PCA analyses condensed multiple variables into a smaller number of linear combinations or functions. The key difference between the 2 techniques is that DA computes these functions to optimize group prediction; therefore, DA optimizes discrimination between predefined groups, for example, infection by a certain pathogen. PCA, however, ignores investigator-defined groups and aims to maximize the amount of variance accounted for by the smallest number of principal components. Therefore, plotting participants using the PCs obtained from a PCA enabled examination of the unbiased natural partitioning of groups in a data set.
Using multiple analytic tools allowed exploration of distinctions between pathogens and among compartments using data (multiple cytokines/chemokines from differing compartments) from relatively small numbers of patients.
Many cytokines/chemokines (IL-6, IL-8, IL-10, IP-10, MCP-1, IL-1Rα, fractalkine, and GM-CSF) were elevated in the CSF of EV patients but were present in lower concentrations or undetectable in plasma. These cytokines appear to play an important role in early control of both viral replication and also potential tissue injury due to infection [28]. Our data in part support the hypothesis that pro-inflammatory cytokine production in the CSF during EV infection attracts WBCs into the CNS but anti-inflammatory cytokine production terminates the pleocytosis after elimination of virus [30].
Chemokines such as IL-8 and IP-10 that are involved in the regulation of leukocyte migration and inflammation were also more elevated in EV-infected CSF than in plasma. One prior study showed that IL-8 and IP-10 in CSF are principally produced by the CNS microglia or astrocytes, rather than immune cells circulating in the blood in EV-71 infections [35]. IP-10 is known to be a common response in CSF and may be a nonspecific response to any CNS viral infection [13]. That said, these chemokines enhance the production of downstream anti-inflammatory cytokines such as IL-10, IL-1Rα, IL-4, and IL-13, which are important in modulating the inflammatory process in viral meningitis [30].
In contrast to EV infections, innate responses in plasma from PeV-A3-infected patients were more robust than in CSF. In PeV-A3-infected infants, plasma concentrations of pro-inflammatory cytokines (IL-15, fractalkine, and IL-12p40) and chemokines (MCP-1 and IP-10), as well as anti-inflammatory cytokines (IFN-α2, IL-1Rα, and IL-10), were elevated. Yet CSF concentrations of the same cytokines/chemokines were relatively low or were undetectable. The relatively high concentrations of pro-inflammatory cytokines in the plasma of PeV-A3 patients vs EV or control patients could explain the intensity of some clinical manifestations such as higher and longer duration of fever. On the other hand, relatively low chemoattractant cytokine/chemokine concentrations in PeV-A3 CSF could help explain the absence of CSF pleocytosis, a classic finding in PeV-A3-infected children.
Our new data suggest that combined compartment (plasma combined with CSF) biomarker profiles more than individual compartment biomarkers are distinctive and characteristic of EV vs PeV-A3 infections in young infants. This notion is supported by other studies [7, 8] but needs to be confirmed in studies with larger numbers of patients.
Overall, prior studies lend credence to our data. A recent study by Habukha et al. from Japan investigated expression of 22 cytokines and chemokines in the CSF and plasma of infants infected with EV and PeV-A3 [19]. Sixteen biomarkers overlapped between their study panel (n = 22) and ours (n = 21). Seven of 16 biomarkers evaluated in both studies (IL-6, IL-10, IL-1Rα, IFN-α2, IL-8, TNF-α, and IL-15) were elevated in EV CSF, while our study identified an additional 6 biomarkers (fractalkine, IFN-γ, GM-CSF, MIP-1α, IL-12p40, and IL-1β) in EV-CSF samples (Figure 1A and B). Similarly, in PeV-A3 plasma, 5/16 overlapping biomarkers (IFN- α2, fractalkine, IL-15, IL-10, IL-1Rα) were elevated in both studies, and 2 biomarkers (MCP-1 and IP-10) were additionally identified by our study. In studies using only CSF samples, pro-inflammatory CSF cytokines (TNF-α, IL-1, and IL-6) and anti-inflammatory cytokines (IL-4 and IL-10) were significantly lower in PeV-A than in EV CNS infections [14]. Another study evaluating only CSF cytokines in infants showed that some cytokines (IL-1β, IL-5, IL-6, IL-12, and IL-17) were higher in EV infection, while some cytokines (IL-2, IL-4, IL-7, and IL-13) were detected in higher concentrations in PeV-A3 infection [36].
Yet another study evaluated only serum cytokines from 12 patients with PeV-A3-induced sepsis-like syndrome vs 28 healthy children. Serum IL-6, neopterin, sTNFR-I, sTNFR-II, and IL-18 concentrations increased rapidly in early infection but decreased to the levels of healthy children during recovery [37]. There is also a case report of very high serum IL-6 concentrations in an adult with severe epidemic myalgia due to a PeV-A strain that was not typed [38].
Our study has limitations. We collected clinical data retrospectively, so some clinical data may not be completely accurate. We did not have a paired plasma sample for every CSF sample, so our numbers were relatively small for paired analysis. We did not have sufficient paired plasma from all the negative CSF patients to perform EV and PeV-A RT-PCR. Despite small patient numbers, multiple analyses and modeling, including DA, PCA, and RCCs, provided separation of profiles and discrimination between groups. There were multiple detected EV serotypes, but all PeV-A were PeV-A3. EV types can produce different degrees of illness severity, so systemically less virulent and/or more neurotropic EV strains may have contributed to differences in innate responses compared with PeV-A3 infections. We evaluated data for a single year, so years in which differing EVs circulate may produce fewer differences in cytokine profiles. Also, we do not know the pathogen that caused the infection leading to a sepsis workup in the control group, so this group may have a mix of pathogens with varying virulence.
Despite these limitations, our data confirm that an infant <3 months old presenting with high fever, leukopenia, and lack of CSF pleocytosis is likely to have a PeV-A3 systemic/CNS infection and suggest the likelihood of higher plasma but lower CSF concentrations of IP-10, MCP-1, IL-15, IL-1Rα, and IFN-α2 compared with EV-infected infants.
In summary, we found that innate immune responses in different compartments (CSF vs plasma) vary depending on the viral agent. Variations in cytokine and chemokine profiles in patients with EV and PeV-A3 infection may contribute to the pathogenesis, variations in clinical presentation, and disease outcomes of each virus. Analysis of innate immune profiles could be potentially useful to design interventions involving immune-prophylaxis and immunotherapy.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Prior presentation. The study was presented as a poster at the Infectious Diseases Society of America conference 2019; Poster No. 2782.
References
- 1. Bastos MS, Coelho-Dos-Reis JG, Zauli DA, et al. Divergent cerebrospinal fluid cytokine network induced by non-viral and different viral infections on the central nervous system. BMC Infect Dis 2015; 15:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harvala H, McLeish N, Kondracka J, et al. Comparison of human parechovirus and enterovirus detection frequencies in cerebrospinal fluid samples collected over a 5-year period in edinburgh: HPeV type 3 identified as the most common picornavirus type. J Med Virol 2011; 83:889–96. [DOI] [PubMed] [Google Scholar]
- 3. Zhong H, Lin Y, Su L, et al. Prevalence of human parechoviruses in central nervous system infections in children: a retrospective study in Shanghai, China. J Med Virol 2013; 85:320–6. [DOI] [PubMed] [Google Scholar]
- 4. Seo JH, Yeom JS, Youn HS, et al. Prevalence of human parechovirus and enterovirus in cerebrospinal fluid samples in children in Jinju, Korea. Korean J Pediatr 2015; 58:102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Selvarangan R, Nzabi M, Selvaraju SB, et al. Human parechovirus 3 causing sepsis-like illness in children from Midwestern United States. Pediatr Infect Dis J 2011; 30:238–42. [DOI] [PubMed] [Google Scholar]
- 6. Midgley CM, Jackson MA, Selvarangan R, et al. Severe parechovirus 3 infections in young infants—Kansas and Missouri, 2014. J Pediatric Infect Dis Soc 2018; 7(2):104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abedi GR, Watson JT, Pham H, et al. Enterovirus and human parechovirus surveillance - United States, 2009-2013. MMWR Morb Mortal Wkly Rep 2015; 64:940–3. [DOI] [PubMed] [Google Scholar]
- 8. Cilla A, Megias G, Suarez J, et al. Human parechovius and enterovirus in neonates: distinct infections with overlapping features. Early Hum Dev 2015; 91:475–8. [DOI] [PubMed] [Google Scholar]
- 9. Janes VA, Minnaar R, Koen G, et al. Presence of human non-polio enterovirus and parechovirus genotypes in an Amsterdam hospital in 2007 to 2011 compared to national and international published surveillance data: a comprehensive review. Euro Surveill 2014; 19:20964. [DOI] [PubMed] [Google Scholar]
- 10. de Crom SC, Rossen JW, van Furth AM, Obihara CC. Enterovirus and parechovirus infection in children: a brief overview. Eur J Pediatr 2016; 175:1023–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vollbach S, Müller A, Drexler JF, et al. Prevalence, type and concentration of human enterovirus and parechovirus in cerebrospinal fluid samples of pediatric patients over a 10-year period: a retrospective study. Virol J 2015; 12: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Centers for Disease Control and Prevention. Nonpolio enterovirus and human parechovirus surveillance—United States, 2006–2008. MMWR Morb Mortal Wkly Rep 2010; 59:1577–80. [PubMed] [Google Scholar]
- 13. Fortuna D, Hooper DC, Roberts AL, et al. Potential role of CSF cytokine profiles in discriminating infectious from non-infectious CNS disorders. PLoS One 2018; 13:e0205501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fortuna D, Cárdenas AM, Graf EH, et al. Human parechovirus and enterovirus initiate distinct CNS innate immune responses: pathogenic and diagnostic implications. J Clin Virol 2017; 86:39–45. [DOI] [PubMed] [Google Scholar]
- 15. Sharp J, Harrison CJ, Puckett K, et al. Characteristics of young infants in whom human parechovirus, enterovirus or neither were detected in cerebrospinal fluid during sepsis evaluations. Pediatr Infect Dis J 2013; 32:213–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Patro ARK, Mohanty S, Prusty BK, et al. Cytokine signature associated with disease severity in dengue. Viruses 2019; 11:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang PZ, Li ZD, Yu HT, et al. Elevated serum concentrations of inflammatory cytokines and chemokines in patients with haemorrhagic fever with renal syndrome. J Int Med Res 2012; 40:648–56. [DOI] [PubMed] [Google Scholar]
- 18. Cheung CY, Poon LL, Lau AS, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet 2002; 360:1831–7. [DOI] [PubMed] [Google Scholar]
- 19. Habuka R, Aizawa Y, Izumita R, et al. Innate immune responses in serum and cerebrospinal fluid from neonates and infants infected with parechovirus-A3 or enteroviruses. J Infect Dis. In press. [DOI] [PubMed] [Google Scholar]
- 20. Harvala H, Robertson I, Chieochansin T, et al. Specific association of human parechovirus type 3 with sepsis and fever in young infants, as identified by direct typing of cerebrospinal fluid samples. J Infect Dis 2009; 199:1753–60. [DOI] [PubMed] [Google Scholar]
- 21. Nix WA, Maher K, Pallansch MA, Oberste MS. Parechovirus typing in clinical specimens by nested or semi-nested PCR coupled with sequencing. J Clin Virol 2010; 48:202–7. [DOI] [PubMed] [Google Scholar]
- 22. Nordstokke DW, Zumbo BDJP. A new nonparametric Levene test for equal variances. Psicológica 2010; 31:401–30. [Google Scholar]
- 23. Kaiser HF. A note on the equamax criterion. Multivariate Behav Res 1974; 9:501–3. [DOI] [PubMed] [Google Scholar]
- 24. Tomlinson A, Hair M, McFadyen A. Statistical approaches to assessing single and multiple outcome measures in dry eye therapy and diagnosis. Ocul Surf 2013; 11:267–84. [DOI] [PubMed] [Google Scholar]
- 25. Bartlett MS. Properties of sufficiency and statistical tests. Proc R Soc A 1937; 160:268–82. [Google Scholar]
- 26. O’Connor BP. SPSS and SAS programs for determining the number of components using parallel analysis and velicer’s MAP test. Behav Res Methods Instrum Comput 2000; 32:396–402. [DOI] [PubMed] [Google Scholar]
- 27. Perlis RH. Translating biomarkers to clinical practice. Mol Psychiatry 2011; 16:1076–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shang W, Qian S, Fang L, et al. Association study of inflammatory cytokine and chemokine expression in hand foot and mouth disease. Oncotarget 2017; 8:79425–32. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Wiley CA, Bhardwaj N, Ross TM, Bissel SJ. Emerging infections of CNS: avian influenza A virus, Rift Valley fever virus, and human parechovirus. Brain Pathol 2015; 25:634–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sato M, Hosoya M, Honzumi K, et al. Cytokine and cellular inflammatory sequence in enteroviral meningitis. Pediatrics 2003; 112:1103–7. [DOI] [PubMed] [Google Scholar]
- 31. Zhang SY, Xu MY, Xu HM, et al. Immunologic characterization of cytokine responses to enterovirus 71 and coxsackievirus A16 infection in children. Medicine (Baltimore) 2015; 94:e1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beppu M, Sawai S, Misawa S, et al. Serum cytokine and chemokine profiles in patients with juvenile muscular atrophy of distal upper extremity (Hirayama disease). J Neuroimmunol 2017; 302:20–2. [DOI] [PubMed] [Google Scholar]
- 33. Zeng M, Zheng X, Wei R, et al. The cytokine and chemokine profiles in patients with hand, foot and mouth disease of different severities in Shanghai, China, 2010. PLoS Negl Trop Dis 2013; 7:e2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sulik A, Kroten A, Wojtkowska M, Oldak E. Increased levels of cytokines in cerebrospinal fluid of children with aseptic meningitis caused by mumps virus and echovirus 30. Scand J Immunol 2014; 79:68–72. [DOI] [PubMed] [Google Scholar]
- 35. Ye N, Gong X, Pang LL, et al. Cytokine responses and correlations thereof with clinical profiles in children with enterovirus 71 infections. BMC Infect Dis 2015; 15:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Park SE, Song D, Shin K, et al. Prospective research of human parechovirus and cytokines in cerebrospinal fluid of young children less than one year with sepsis-like illness: comparison with enterovirus. J Clin Virol 2019; 119:11–6. [DOI] [PubMed] [Google Scholar]
- 37. Shimizu M, Shimizu H, Jinkawa A, et al. Cytokine profiles in human parechovirus type 3-induced sepsis-like syndrome. Pediatr Infect Dis J 2020; 39:137–9. [DOI] [PubMed] [Google Scholar]
- 38. Nakamura K, Saito K, Hara Y, et al. Severe epidemic myalgia with an elevated level of serum interleukin-6 caused by human parechovirus type 3: a case report and brief review of the literature. BMC Infect Dis 2018; 18:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.