Abstract
The corona virus disease 2019 (COVID‐19) is a highly contagious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). More than 18 million people were infected with a total of 0.7 million deaths in ∼188 countries. Controlling the spread of SARS‐CoV‐2 is therefore inherently dependent on identifying and isolating infected individuals, especially since COVID‐19 can result in little to no symptoms. Here, we provide a comprehensive review of the different primary technologies used to test for COVID‐19 infection, discuss the advantages and disadvantages of each technology, and highlight the studies that have employed them. We also describe technologies that have the potential to accelerate SARS‐CoV‐2 detection in the future, including digital PCR, CRISPR, and microarray. Finally, remaining challenges in COVID‐19 diagnostic testing are discussed, including (a) the lack of universal standards for diagnostic testing; (b) the identification of appropriate sample collection site(s); (c) the difficulty in performing large population screening; and (d) the limited understanding of SARS‐COV‐2 viral invasion, replication, and transmission.
Keywords: COVID‐19, diagnostic test, nucleic acid, SARS‐COV‐2, serum
A comprehensive review of the different primary technologies used to test for COVID‐19 infection. The review indicates the advantages and disadvantages of each technology, describes technologies that have the potential to accelerate SARS‐CoV‐2 detection in the future, and discusses the remaining challenges in COVID‐19 diagnostic testing.

1. INTRODUCTION
The corona virus disease 2019 (COVID‐19) is a highly contagious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), and the incubation period is from two to twelve days. 1 , 2 Notably, 17.9% of infected individuals who are asymptomatic or mildly symptomatic are still infectious, which has contributed to the rapid worldwide spread of COVID‐19. 3 , 4 As of August 5, 2020, over 18 million confirmed cases have been reported in ∼188 countries with ∼0.7 million related deaths (Figure 1). 5 A standard epidemiological model has predicted that the reproduction number “R0” of SARS‐CoV‐2, which is 2‐3.5, would be reduced to <1.0 by identifying and isolating the majority of infected individuals, including those who might be asymptomatic or mildly symptomatic. 6 , 7 , 8 Thus, the identification and isolation of infected people is paramount in fighting COVID‐19 since vaccines and effective antiviral drugs have not yet been developed. 6 , 9
FIGURE 1.
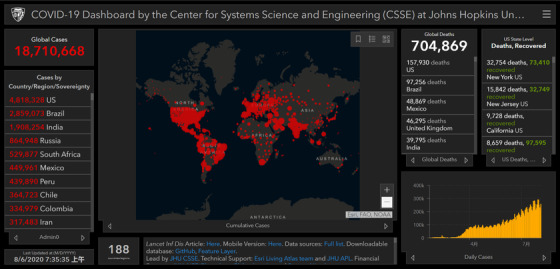
Confirmed COVID‐19 cases as of August 5, 2020. The graph was obtained from an online interactive dashboard developed by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University, Baltimore, MD, USA 5
Tremendous efforts have been employed to develop highly accurate diagnostic testing for COVID‐19 since January 2020. 10 Together, the World Health Organization (WHO) and Foundation for Innovative New Diagnostics (FIND) are collaborating to independently validating tests from different manufactures across the globe. 10 , 11 The U.S. Food and Drug Administration (FDA) initiated fast‐tracked FDA review and approval of COVID‐19 tests through emergency use authorization (EUA) in mid‐March. 11 According to the data from FIND COVID‐19 resource center (https://www.finddx.org/covid-19/) on June 9, 2020, over 400 molecular and serological antibody tests have been approved by different countries’ and regions’ agencies of certification. Among them, 196 molecular tests and 233 serum tests were approved by Conformité Européenne (CE), followed by the US FDA, China National Medical Products Administration (NMPA), and Brazil Brazilian Health Regulatory Agency (ANVISA) (Figure 2A and 2C). Most of the molecular (M) and serum (S) tests for COVID‐19 detection have been produced by China (52 M, 118 S), followed by the USA (20 M, 28 S), Korea (28 M, 19 S), and Germany (13 M, 16 S) (Figure 2B and 2C).
FIGURE 2.
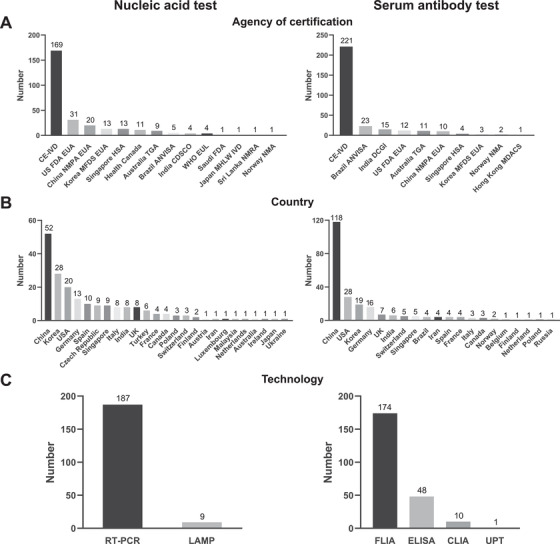
Diagnostic tests that have been approved according by a country's or region's agency of certification. Panels (A‐C) show the nucleic acid and antibody tests that have been approved by different agencies of certification, countries, and technologies, respectively. The data were obtained from the FIND COVID‐19 diagnostics resource center (https://www.finddx.org/covid-19/) on June 9, 2020. Abbreviations: EUA, Emergency Use Authorization; HSA, Health & Safety/Sciences Authority; MFDS, Ministry of Food & Drug Safety; MHRA, Medicines & Health Care Products Regulatory Agency; NRA, National Regulatory Authority; RUO, Research Use Only; TGA, Therapeutic Goods Administration; WHO EUL, World Health Organization Emergency Use Listing Procedure
The type of technologies that a test is based on has a significant impact on the test's diagnostic performance, including sensitivity, specificity, dynamic range, reproducibility, reagent consumption, equipment, cost, throughput, and ease‐of‐use. For example, the successful identification of SARS‐CoV‐2 is attributed to metagenomics next‐generation sequencing (mNGS) technology. 12 SARS‐CoV‐2 nucleic acid testing enables the early COVID‐19 patients detection whereas acute and recovered patients can be detected with antibody tests. 13 , 14
Here, a comprehensive review of the major COVID‐19 diagnostic tests is provided, including their unique advantages and disadvantages. Promising technologies in SARS‐CoV‐2 detection are also discussed. The data were obtained by searching PubMed, Preprint servers (ie, medRxiv, bioRxiv), and related databases for English‐based articles with terms such as “SARS‐CoV‐2,” “COVID‐19,” “nucleic acid testing,” “serum testing,” “diagnostics,” or their lexical variants since January 1, 2020.
2. TECHNOLOGY ON THE COVID‐19 DIAGNOSTIC TESTING
2.1. Molecular assay
A molecular assay tests for the presence of specific genetic material (eg, viral RNA) in a biological specimen. As of June 9, 2020, over ∼200 molecular tests have been approved for in vitro diagnostics of COVID‐19, which include Next‐Generation Sequencing (NGS), 15 reverse transcription polymerase chain reaction (RT‐PCR), 16 , 17 , 18 and loop‐mediated isothermal amplification (LAMP) 19 (Figure 3).
FIGURE 3.
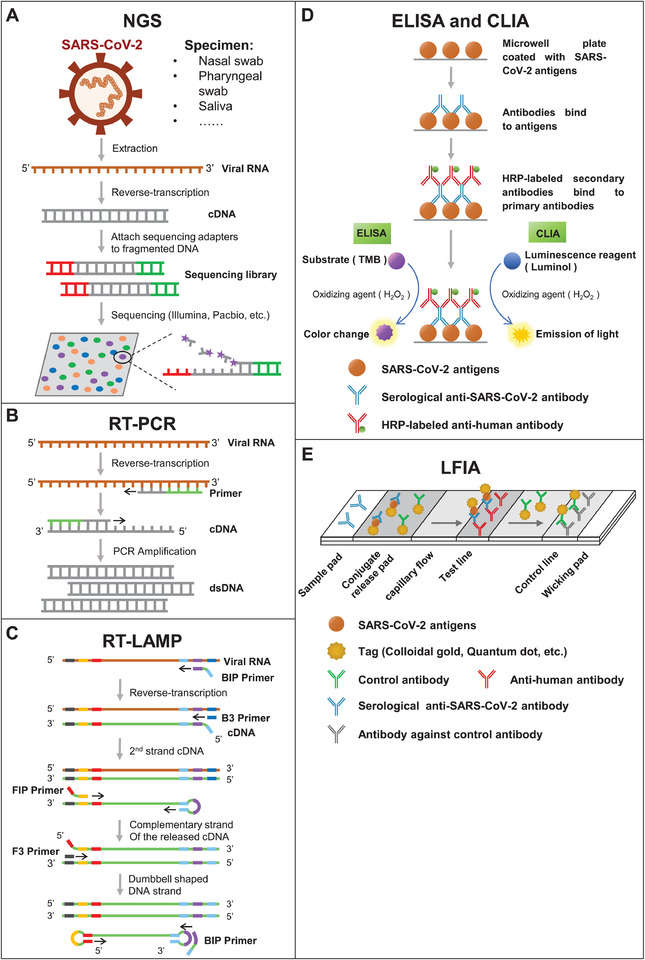
Schematic illustration of nucleic acid and serum testing methods. Panels (A‐E) are the assay principles of NGS, RT‐PCR, RT‐LAMP, ELISA, CLIA, and LFIA, respectively
2.1.1. Next‐generation sequencing
NGS is a genomics technology that enables the simultaneous sequencing of thousands to billions of DNA fragments, and can identify genetic material from entirely different kingdoms of organisms (Figure 3A). 20 Applications of NGS technology include diagnosing infectious diseases, tracking outbreaks, infection control surveillance, and mutation and pathogen discovery 21 with different types of biological specimens (ie, cerebrospinal fluid, blood, respiratory samples, gastrointestinal fluid, and ocular fluid). 22 , 23 Sequencing platforms are available from Illumina (iSeq, MiSeq, MiniSeq, NextSeq, HiSeq, NovaSeq), Thermo Fisher Scientific (Ion Torrent), BGI (BGISEQ platform), and Oxford Nanopore Technologies (MinION, GridION, PromethION). 24
SARS‐CoV‐2 was first isolated as an unknown virus in five COVID‐19 patients, and then identified in two COVID‐19 patients using metagenomics NGS (mNGS). 12 , 15 The sequence of this virus is 79.6% identical to the virus responsible for the 2003 SARS outbreak, SARS‐CoV, and 96% to a bat coronavirus. 12 Phylogenetic analyses of the SARS‐CoV‐2 virus that was extracted from the bronchoalveolar lavage fluid and cultured isolates in nine COVID‐19 patients categorized SARS‐CoV‐2 as subgenus Sarbecovirus of the genus Betacoronavirus, with its closest viral relatives being the bat‐SL‐CoVZC45 and bat‐SL‐CoVZXC21. 25 Notably, the SARS‐CoV spike (S) receptor binding domain (RBD) that interacts with the human host receptor, ACE2, is very different from RBD of SARS‐CoV‐2. 26 , 27 These mutations result in a different binding affinity between the S protein and ACE2. Over 18 000 SARS‐CoV‐2 sequences have thus far been collected and stored in the GISAID database (https://www.gisaid.org/).
However, the analytical sensitivity in pathogen detection is limited with NGS because microbial nucleic acids from resident microbiota often result in high background. In fact, >99% of the reads are often from the human host. 24 Large‐scale population screening is limited by the high cost to perform NGS.
2.1.2. RT‐PCR
Using RT‐PCR, the viral RNA can be reversely transcribed into complementary DNA that is then amplified using directed primers that flank the DNA sequence‐of‐interest (Figure 3B). RT‐PCR is fast, yet also has high specificity and simple quantification; it is considered the gold standard in SARS‐CoV‐2 diagnosis. 28 , 29 , 30 , 31 Briefly, swabs of the back of the throat or nasal cavity are taken, saliva can also be used. RNA is extracted, and then RT‐PCR is performed. Padhye et al evaluated the performance of RT‐PCR with data in 1014 patients, including 601 COVID‐19 positive and 413 healthy controls. The sensitivity and specificity of the RT‐PCR detection was estimated to be 0.777 (95% confidence interval (CI): 0.715‐0.849) and 0.988 (95% CI: 0.933‐1.000), respectively. 32
False negatives can occur with RT‐PCR testing if the viral copies are too low to be amplified. Viral copy varies across individuals, specimen type (ie, nasal, throat or sputum) and days post infection. 16 , 33 , 34 , 35 , 36 Another limitation of RT‐PCR is that the assay must be performed in a Clinical Laboratory Improvement Amendments of 1988 (CLIA) accredited laboratory with appropriate equipment and experienced operators. 11 The process, from RNA extraction to RT‐PCR to data analysis, can be completed in ∼1 day.
2.1.3. RT‐LAMP
Comparing to RT‐PCR, RT‐LAMP does not require a PCR machine and thus the assay is rapid and inexpensive. Briefly, primers, reverse‐transcriptase, Bst DNA polymerase, and a pH indicator dye are mixed with extracted sample RNA and then heated to 65˚C for 60 min (Figure 3C). 37 As the DNA is amplified, the pH decreases, causing a colorimetric change that is visualized by eye. Also, the primers are specially designed so that they “loop” or anneal onto each other to enable rapid DNA amplification without the need for thermal cycling, thus making the RT‐LAMP method faster than RT‐PCR. Unlike RT‐PCR, RT‐LAMP is suitable for on‐site detection because it has high specificity, is simple to perform, cost effective, and only requires a heating block. 38 , 39
Jiang et al developed a RT‐LAMP method using primers targeting SARS‐CoV‐2 genes encoding for the ORF1ab, envelope (E), and nucleocapsid (N) proteins. The sensitivity of 91.4% and the specificity of 99.50% were obtained from 47 and 213 patients with and without SARS‐CoV‐2 infection. 38 Similar results were obtained in an independent study with 17 and 191 patients who had or did not have COVID‐19, respectively. 40 A major drawback of RT‐LAMP is that it is low throughput because only one sample can be measured in an experiment.
2.2. Immunological assay
The detection of SARS‐CoV‐2 antibodies is helpful in identifying prior infection and immunity, which is of significance to epidemiologic and vaccine studies, ongoing surveillance and to evaluate the risk of health care workers. 11 , 41 , 42 , 43 Notably, COVID‐19 antibody profiles across time and the role of IgG in immunity are still being ascertained. Recent studies indicate that the SARS‐CoV‐2 IgM antibodies start ∼7 days after symptom onset and peak at day 28, while IgG start ∼10 days after infection and peak at day 49. 41 However, antibody profiles vary across individuals. Furthermore, the expression of IgM and IgG antibodies are higher in COVID‐19 patients with severe symptoms than patients with milder symptoms (P < .05). COVID‐19 patients with low antibody responses tend to have a higher viral clearance rate than patients with stronger antibody responses (P = .011). 44 , 45 Over 190 COVID‐19 serology tests have been approved by agencies of certification according to the FIND database. These tests include the enzyme‐linked immune sorbent assay (ELISA), chemiluminescence enzyme immunoassay (CLIA), and lateral flow immunochromatographic assay (LFIA) (Figure 3). 46 , 47
2.2.1. Enzyme‐linked immunosorbent assay
As a qualitative or semi‐quantitative detection method, indirect ELISA immobilizes the antigen‐of‐interest on a solid substrate (eg, 96‐well plate) to capture its specific antibody. Antibody binding is often detected using an enzyme‐labeled, secondary antibody that produces a color change in the presence of an enzyme‐catalyzed substrate, which is subsequently detected using a common laboratory instrument (ie, plate reader) (Figures 3D). The advantage of ELISA assay is easy to operate and low cost. It can also be easily made high throughput with an automated workstation.
Zhao et al screened antibodies in the serum of 173 COVID‐19 patients using an indirect ELISA where the S protein's RBD was immobilized onto the plate. The total, IgM, and IgG antibodies were observed in 93.1% (161/173), 82.7% (143/173), and 64.7% (112/173) of COVID‐19 patients, respectively. The median times for seroconversion of total, IgM, and IgG antibodies is 11, 12, and 14 days, respectively. Furthermore, detecting both viral RNA and antibodies can significantly improve the diagnostic sensitivity for COVID‐19 (P < .001), especially for the early viral infection (<7 days following symptom onset) (P = .007). 44
2.2.2. CLIA
CLIA is similar to ELISA, except that antibody binding to the immobilized substrate is detected via a change in luminescence (Figure 3D). More specifically, a different enzyme catalyzed substrate is used for CLIA (ie, luminol) than ELISA (ie, 3,3′,5,5′‐tetramethylbenzidine). The CLIA assay has higher sensitivity than ELISA and has been fully automated, such that hundreds of samples can be screened easily within a day. 48
Using CLIA, Long et al first performed a multi‐center cross‐sectional analyses of 285 patients to determine the diagnostic value of SARS‐CoV‐2 IgM and lgG antibodies. He then analyzed 63 patients in a single‐center follow‐up study. The positive rate of IgM and IgG antibodies reached 94.1% and 100% on days 20‐22 and 17‐19 after symptom onset, respectively. In addition, four and seven patients displaying COVID‐19 symptoms who obtained false negative results with RT‐PCR were confirmed to have COVID‐19 based on their antibody response using CLIA. 49 Similar results were obtained by Zhang et al who analyzed 736 COVID‐19, non‐COVID‐19, or healthy patients. 50 Here, areas under the receiver operating characteristic (ROC) curves for IgM and IgG of 0.988 and 1.000 were obtained, respectively.
Of note, the total, IgM, IgG, and IgA antibodies to SARS‐CoV‐2 were also detected by Zhang et al in 56 asymptomatic patients, in which 23 patients were keeping asymptomatic in the follow‐up period. Nucleic acid testing indicated that the SARS‐CoV‐2 replication in nasopharyngeal cavity were no difference among asymptomatic carriers, pre‐symptomatic, and symptomatic patients. Only IgM antibodies had an obvious difference in the seroconversion rate among the three groups. Interestingly, the asymptomatic patients had low levels of IgM antibodies and high total IgG and IgA compared to symptomatic patients. These results might be helpful to understand viral clearance and antibody changes in COVID‐19 patients of asymptomatic. 51
2.2.3. LFIA
LFIA is a qualitative point‐of‐care (POC) testing device which can detect SARS‐CoV‐2 antibodies within 5‐30 min in urine, saliva, sweat, serum, plasma, whole blood, and other fluids. 52 First, a SARS‐CoV‐2 antigen(s) is immobilized onto the middle of a strip, which is often a nitrocellulose membrane. Then, samples are applied to one end of a strip where Colloidal gold (CG) or quantum dot (QD)‐labeled detection antibodies are present. Then, the sample and labeled antibodies will migrate along the strip. If SARS‐CoV‐2 antibodies are present, they will be bound by the labeled detection antibody and will bind to the immobilized antigen. The immobilized antibody‐detection antibody complex will result in the appearance of a band on the strip due to the accumulation of CG or QD 53 (Figure 3E).
LFIA is simple to use (like a home pregnancy test), cost effective, can be used for onsite detection, and does not require an instrument. The assay performance of nine different LFIA devices was determined by screening 40 RT‐PCR‐positive COVID‐19 individuals and 142 negative controls collected prior to December 2019 from the Biobank in the United Kingdom. The systematic, meta‐analysis comparison of ELISA, CLIA, and LFIA was performed based on 38 studies with 7848 individuals. Regardless of the antigens whose detail is normally not released by the manufactures, the serum testing using ELISA and CLIA‐based methods had higher sensitivity (90% to 94%) than LFIA (Figure 4). 54 These results demonstrate that the serum testing using ELISA and CLIA are more reliable, while the LFIA results should be considered with caution. 43
FIGURE 4.
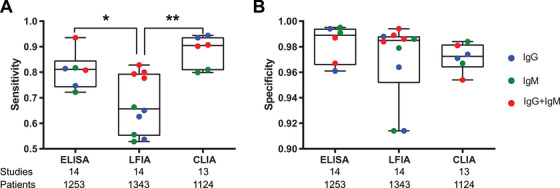
Comparison of different technologies used for serological antibody detection. The data was obtained from the Kontou study by meta‐analysis of antibody testing in 7848 individuals. 54
LFIA is not only used to detect SARS‐CoV‐2 antibodies, but also to detect SARS‐CoV‐2 proteins in clinical specimens. Here, the detection antibody will be specific to the antigen‐of‐interest. However, only one COVID‐19 test that detects SARS‐CoV‐2 proteins (Sofia 2 SARS Antigen FIA) has been approved to date. The point‐of‐care testing must be performed in a certified laboratory under a CLIA Certificate of Waiver. We recently screened the serum of 8 COVID‐19 patients with mild symptoms using a sandwich ELISA developed in our laboratory with a lower detection limit of 200 pg/mL (data not shown). 55 However, none of the samples were positively identified, which is likely because serum has low copies of SARS‐CoV‐2 virus. 47 The levels of SARS‐CoV‐2 proteins in different sampling locations across the stages of COVID‐19 infection should be examined. 56
3. PROMISING TECHNOLOGIES FOR COVID‐19 DETECTION
Asymptomatic and mildly symptomatic individuals are rarely tested for COVID‐19, which is partly because current technologies are limited in throughput. To address this challenge, different technologies other than those described above have either been developed or are in development to detect SARS‐CoV‐2 in high throughput with high sensitivity. 57 , 58 , 59 , 60 Here, we review three such technologies: digital PCR (dPCR), Clustered regularly interspaced short palindromic repeats (CRISPR), and SARS‐CoV‐2 proteome peptide microarray (Figure 5).
FIGURE 5.
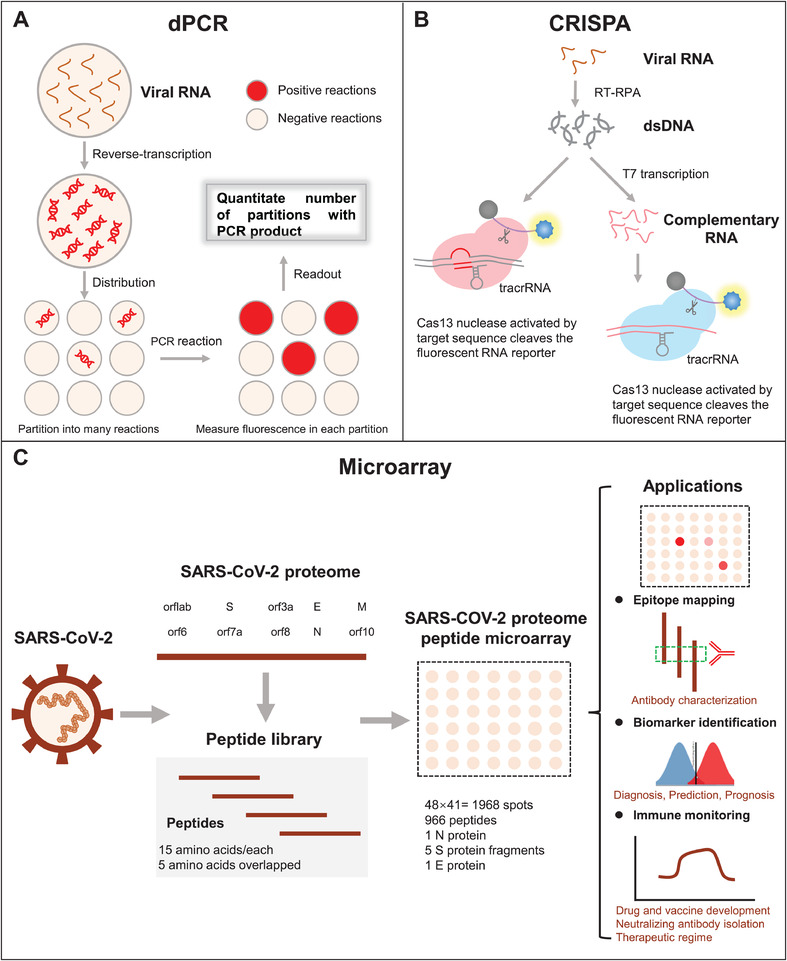
Schematic illustration of promising technologies for SARS‐CoV‐2 detection. (A‐C) are the assay principles of dPCR, CRISPR, and SARS‐CoV‐2 proteome peptide microarray, respectively
3.1. dPCR
dPCR partitions nucleic acid molecules into tens of thousands to hundreds of thousands of reaction units. Each reaction unit carries out PCR amplification independently. Then, the fluorescence signal of each reaction chamber was analyzed, and the copy number of target nucleic acid sequence was calculated and quantified 61 (Figure 5A). According to the type of liquid separation, dPCR can be divided into microfluidic digital PCR (mdPCR), droplet digital PCR (ddPCR), and chip digital PCR (cdPCR). An Important advantage of dPCR is that the quantification is independent of variations in the amplification efficiency. 62 In addition, it does not require internal reference genes, nor does it need a standard curve, which is more tolerant to interference factors such as specific template amplification inhibitors, and can achieve accurate quantification of low concentration nucleic acid samples. dPCR is used to analyze viral load and gene copy number variation. 63 , 64 , 65 , 66 , 67
Using RT‐dPCR, Suo et al detected 57 pharyngeal swab samples for the presence of SARS‐CoV‐2. An overall accuracy of 94.3% was obtained, ∼500 times more sensitive than RT‐PCR. 68 Lu et al compared the RT‐dPCR and RT‐PCR with 108 specimens (pharyngeal swab, stool, and blood) longitudinally collected from 36 COVID‐19 patients. The RT‐dPCR showed the detection limit 10× lower than that of RT‐PCR. Notably, four pharyngeal swab samples showed negative in RT‐PCR testing were positive using RT‐dPCR. 69 Dong et al used RT‐dPCR to measure 194 pharyngeal swab samples from close contacts, suspected, and recovered COVID‐19 patients. The sensitivity of 90% and the specificity of 100% were obtained. Comparing to RT‐PCR sensitivity (28.2%), the RT‐dPCR showed the improved sensitivity of 87.4% in the detection of 103 suspects. 70
3.2. CRISPR
In 2013, CRISPR/Cas technology developed rapidly as a third generation genome editing technology. 71 , 72 Briefly, the CRISPR/Cas system captures and inserts foreign DNA fragments into the bacterial genome, and then guides the Cas endonuclease to remove the foreign nucleic acid (Figure 5B). The CRISPR/Cas technology is characterized by high specificity, high precision, high efficiency, and simple and fast operation. Broughton et al developed an accurate CRISPR‐Cas12‐based lateral flow assay that can easily detect the SARS‐CoV‐2 N and E genes in RNA extracts from respiratory swabs within 40 min. The method was validated using 36 COVID‐19 patients and 42 patients infected with different respiratory viruses, 73 offering a promising poTable POC testing for SARS‐CoV‐2 screening. 13 On May 7, 2020, the US FDA granted its first EUA coronavirus test using CRISPR technology. 74
3.3. Microarray
Microarray technology enables the high‐sensitive detection of hundreds or even thousands of molecules simultaneously using a low sample volume 75 , 76 , 77 (Figure 5C). Our team developed a SARS‐CoV‐2 proteome microarray to detect hundreds of antigen‐antibody interactions in serum at amino acid resolution within 1.5 h. The array has SARS‐COV‐2 N and E full‐length proteins, and five S truncated proteins as well as 966 tiled peptides covering all amino acid sequences of SARS‐CoV‐2 proteins. Using the array, we identified immunogenic regions of the SARS‐CoV‐2 proteins and an epitope for potential neutralizing antibodies to viral entry (ie, the interaction between the S protein's RBD and the human receptor, ACE2). 55 , 78 Similar arrays were developed by Dahlke et al 79 using in situ synthesized peptides and Jiang et al 80 and Assis et al 81 using purified SARS‐CoV‐2 antigens. The comprehensive examination of humoral antibody responses in COVID‐19 patients using protein microarrays would be valuable in understanding immunity, identifying diagnostic targets, and developing COVID‐19 therapies.
4. PROSPECTIVE
Every COVID‐19 diagnostic test has its advantages and limitations (Tables 1 and 2), and there remain several key challenges for all of them. These challenges, which are described in more details below, include the lack of a standard in COVID‐19 diagnostic testing, difficulty in identifying asymptomatic or mildly symptomatic patients as early as possible, and limited understanding of the virus and disease pathology:
TABLE 1.
Advantages and disadvantages of the different technologies used to detect SARS‐CoV‐2 viral RNA
| Technology | NGS | RT‐PCR | RT‐LAMP |
|---|---|---|---|
| Detection time | 0.5‐3 days 38 , 59 | 0.5‐1 h 29 , 111 , 112 | 0.5‐1 h 38 , 40 , 113 |
| Sensitivity | N. A. † | 73%‐86% 32 , 33 , 114 | 81% ‐ 100% 38 , 40 , 115 |
| Specificity | 100% 116 | 86% ‐ 100% 32 , 33 , 114 | 99% ‐ 100% 38 , 40 , 115 |
| Advantages | Unknown virus identification, viral mutation and evolution | High sensitivity and specificity | On‐site detection, simple operation, cost effective |
| Disadvantage | Long turnaround time, expensive | Sophisticated equipment, experienced operators | Low throughput |
| Application | Diagnostics, 117 lineage tracing 15 and mutation discovery, 118 infection control surveillance 119 | Diagnostics 120 , epidemiological surveillance 121 , discharge from hospital 16 | Diagnostics, 39 Epidemiological surveillance 122 |
N. A., not available.
TABLE 2.
Advantages and disadvantages of the different technologies used to detect COVID‐19 antibodies
| Technology | ELISA | CLIA | LFIA |
|---|---|---|---|
| Detection time | 1.5‐2.5 h 123 | 0.5 h 50 | 10‐15 min 57 , 124 |
| Sensitivity | 65‐98% 123 , 125 , 126 , 127 , 128 | 77‐100% 114 , 125 , 127 , 129 | 69‐93% 123 , 124 , 125 , 127 , 130 |
| Specificity | 71‐100% 123 , 125 , 126 , 127 , 128 | 90‐100% 114 , 125 , 127 , 129 | 80‐100% 123 , 124 , 125 , 127 , 130 |
| Advantages | Simple operation, low cost, high‐throughput | Higher sensitivity and specificity, broad linearity, automated | Simple operation, low cost, rapid |
| Disadvantage | Time‐consuming, Vulnerable to contamination | Need Supporting chemiluminescence instruments | Low sensitivity during early SARS‐COV‐2 infection |
| Application | Diagnostics, 131 epidemiological surveillance, 132 discharge from hospital 133 | Diagnostics 49 , epidemiological surveillance 50 , discharge from hospital | Diagnostics, 124 Epidemiological surveillance 134 |
1) No universal standard for diagnostic testing. A universal standard is critical in generating consistent tests by different manufacturers and enables laboratories worldwide to compare results easily. 11 , 56 , 82 For example, there are at least seven versions of PCR primers used in SARS‐CoV‐2 nucleic acid testing (Table 3). 11 , 83 The US Centers for Disease Control (CDC) recommend primers targeting two regions of the N gene. China's CDC recommend primers targeting the ORF1ab and N gene, and France's Institut Pasteur recommends primers targeting the RdRP gene. Similarly, different SARS‐CoV‐2 proteins or protein fragments are employed in immunological testing. 54 To address this issue, WHO, FIND, and other research laboratories are collaborating with each other or working independently to evaluate the performance of different diagnostic tests across the world. For example, Jung et al compared seven primer probe sets for the N gene and the three primer‐probe sets for the Orf1 gene to detect SARS‐CoV‐2 viral RNA. The result showed that the most sensitive primer‐probe sets of N and Orf1 genes are ‘2019‐nCoV_N2, N3’ from the U.S. CDC and the ‘ORF1ab’ from China CDC, respectively. 84
TABLE 3.
Gene targets recommended for nucleic acid testing by different countries and regions‡
| Institute | Gene targets |
|---|---|
| China CDC, China | ORF1ab and N |
| Institut Pasteur, France | Two targets in RdRP |
| US CDC, USA | Three targets in N gene |
| National Institute of Infectious Diseases, Japan | Pancorona and multiple targets, Spike protein |
| Charité, German | RdRP, E, N |
| HKU, Hong Kong SAR | ORF1b‐nsp14 |
| National Institute of Health, Thailan | N |
‡The data were obtained from the Coronavirus disease (COVID‐19) technical guidance: Laboratory testing for 2019‐nCoV in humans in WHO (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance).
We measured the SARS‐CoV‐2 antibodies in early COVID‐19, influenza, and non‐influenza patients using the microarray described above. 55 We found that the N protein might not be an ideal antigen for early SARS‐CoV‐2 testing because the similar IgM antibody profiles was found in COVID‐19 patients and influenza patients. Instead, the RBD and extracellular domain (ECD) from SARS‐CoV‐2 S protein could be a better option for IgM antibody detection, whereas the ECD is more suitable for IgG detection. 85 The results can be also supported by a meta‐analysis of 7848 individuals, in which the S antigen displayed superior sensitivity (81.4% IgG, 81.7% IgM) compared to the N antigen (74.7% IgG, 72.2% IgM) using ELISA. 54 , 77 We speculate that the reason why more antibodies are developed to the S protein is because the S protein is on the SAR‐CoV‐2 surface and is more easily recognized by the immune system. 80 , 85
2) Standardization of sample collection. The specimen and collection time from infected patient may have great influence on the detection result. 30 , 86 , 87 Currently nasal swabs and pharyngeal swabs are the major clinical samples used for nucleic acid testing. However, collection of nasopharyngeal specimens is uncomfortable for the patients and increases the risk of infection to health‐care workers. Wang et al used RT‐PCR to detect the SARS‐CoV‐2 virus in different types of clinical samples. The positive rate of broncho alveolar lavage fluid, sputum, nasal swabs, fibro bronchoscope brush biopsy, pharyngeal swabs, feces, and blood are 93% (14 of 15), 72% (72 of 104), 63% (5 of 8), 46% (6 of 13), 32% (126 of 398), 29% (44 of 153), and 1%, respectively. Whereas, none positive was detected in 72 urine specimens. 86 Using RT‐PCR, To et al detected the SARS‐CoV‐2 S gene in the saliva of 11 out of 12 COVID‐19 patients. 88 In another work, the same group found that the viral load in saliva reached the peak at 7 days of symptom onset and then decreased, indicating the potential of using saliva for diagnosis and viral load monitoring. 89 , 90 , 91 On April 14, 2020, The US FDA authorized the first saliva test for emergency use in COVID‐19 diagnosis, which was developed by Rutgers’ RUCDR Infinite Biologics in partnership with Spectrum Solutions and Accurate Diagnostic Labs (ADL).
Exosomes are small membrane vesicles (40‐100 nm in size) with the potential to transfer complex RNAs and proteins between cells, 92 which had been demonstrated to be a promising specimen for the detection of lung cancer, breast cancer, bladder cancer, and other critical illness. 93 , 94 , 95 , 96 , 97 Recently, it was found that exosomes can transfer ACE2 to receipt cells supporting the SARS‐CoV‐2 internalization and infection, 98 indicating the relationship between exosomes and COVID‐19 pathogenesis. 99 However, the diagnostic value of exosome in COVID‐19 has yet to be determined.
3) Identification of asymptomatic and mildly symptomatic patients require large population screening. Identifying and quarantining infected people – including those who are asymptomatic and mildly symptomatic – at home or shelter is important in stopping the spread of COVID‐19. 100 To do so, large population screening is necessary; thus the assay must be high throughput, rapid, and cheap. 6 , 10 , 51 , 101 , 102 For example, Bendavid et al used an LIFA to test the prevalence of SARS‐CoV‐2 antibodies in 3330 people from Santa Clara County, California. The results indicated that 2.49% (95% CI: 1.80‐3.17%) to 4.16% (95% CI: 2.58‐5.70%) people in Santa Clara might be SARS‐CoV‐2 infected. 103 In another study, Stringhini et al detected IgG antibodies in 1335 participants from Geneva, Switzerland using ELISA. The results show that the seroprevalence increased from 3.1% (95% CI: 0.2‐5.99, n = 343) in the first week to 6.1% (95% CI: 2.6‐9.33, n = 416) and 9.7% (95% CI: 6.1‐13.11, n = 576) in the second and third weeks, respectively. 104
4) Limited understanding of viral invasion, replication, and evolution 105 , 106 , 107 . Accurate COVID‐19 diagnosis and effective treatment will likely depend on better understanding the SARS‐CoV‐2 virus and COVID‐19 pathology. Unfortunately, there are still many unanswered questions. For example, Yao et al recently found that the SARS‐CoV‐2 virus was still present in pneumocytes during a pathological examination of a recovered COVID‐19 patient who passed away due to sudden cardiovascular incident. 108 These results might explain why some recovered COVID‐19 patients end up having positive PCR results after being discharged from the hospital. 16
Here, we discussed the numerous types of technologies available to test for COVID‐19, each with their own advantages and disadvantages. It is clear that a global collaboration is urgently needed. 100 , 109 , 110 There are no standard RT‐PCR primers nor universal SARS‐CoV‐2 antigens for antibody detection. Consequently, diagnostic accuracies vary, and test results cannot be easily compared with each other for meta‐analyses that can help shed light on SARS‐CoV‐2 infection and the host immune response (eg, antibody profiles across time). Finally, individuals displaying COVID‐19 symptoms are tested for COVID‐19. However, large‐scale population screening is necessary since infected individuals who are asymptomatic or mildly symptomatic are still contagious. The global exchange of information about patient data, testing data, and molecular studies will be the most rapid approach to identify the most accurate RT‐PCR approach, immunoassay test, and effective therapeutic regimes to stop the spread of COVID‐19.
AUTHOR CONTRIBUTIONS
M. X., D. W., H. W., X. Z., T. L., J. D., M. L., J. Z., and K. Z., searched the literature. M. X., D. X., and X. Y. prepared the manuscript.
COONFLICT OF INTEREST
The authors have declared no conflict of interest.
ACKNOWLEDGMENT
This work was supported by the National Key R&D Program of China (2020YFE0202200), State Key Laboratory of Proteomics (SKLP‐C202001, SKLP‐O201703 and SKLP‐K201505), the Beijing Municipal Education Commission, National Natural Science Foundation of China (81673040, 31870823), the National Program on Key Basic Research Project (2018YFA0507503, 2017YFC0906703 and 2018ZX09733003). We also thank Dr. Brianne Petritis for her critical review and editing of this manuscript.
Xu M, Wang D, Wang H, et al. COVID‐19 diagnostic testing: Technology perspective. Clin Transl Med. 2020;10:e158 10.1002/ctm2.158
Contributor Information
Danke Xu, Email: xudanke@nju.edu.cn.
Xiaobo Yu, Email: yuxiaobo@mail.ncpsb.org.
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. The Lancet. 2020;395:470‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan JF‐W, Yuan S, Kok K‐H, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. The Lancet. 2020;395:514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID‐19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020:25;2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dong E, Du H, Gardner L. An interactive web‐based dashboard to track COVID‐19 in real time. Lancet Infect Dis. 2020;20:533‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taipale J, Romer P, Linnarsson S. Population‐scale testing can suppress the spread of COVID‐19. medRxiv. 2020.
- 7. Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS‐CoV‐2). Science. 2020;368:489‐493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang C, Liu L, Hao X, et al. Evolving epidemiology and impact of non‐pharmaceutical interventions on the outbreak of coronavirus disease 2019 in Wuhan. China medRxiv. 2020.
- 9. Chowell D, Castillo‐Chavez C, Krishna S, Qiu X, Anderson KS. Modelling the effect of early detection of Ebola. Lancet Infect Dis. 2015;15:148‐149. [DOI] [PubMed] [Google Scholar]
- 10. Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol. 2020;38:515‐518. [DOI] [PubMed] [Google Scholar]
- 11. Cheng MP, Papenburg J, Desjardins M, et al. Diagnostic testing for severe acute respiratory syndrome‐related coronavirus 2: a narrative review. Ann Intern Med. 2020;172:726‐734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmid‐Burgk J, Li D, Feldman D, et al. LAMP‐Seq: population‐Scale COVID‐19 diagnostics using a compressed barcode space. bioRxiv. 2020.
- 14. Sharfstein JM, Becker SJ, Mello MM. Diagnostic testing for the novel coronavirus. JAMA. 2020323(15):1437–1438. [DOI] [PubMed] [Google Scholar]
- 15. Chen L, Liu W, Zhang Q, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9:313‐319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lan L, Xu D, Ye G, et al. Positive RT‐PCR test results in patients recovered from COVID‐19. JAMA. 2020;323:1502‐1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y, Kang H, Liu X, Tong Z. Combination of RT‐qPCR testing and clinical features for diagnosis of COVID‐19 facilitates management of SARS‐CoV‐2 outbreak. J Med Virol. 2020;92:538‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu R, Han H, Liu F, et al. Positive rate of RT‐PCR detection of SARS‐CoV‐2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang M, Pan W, Arasthfer A, et al. Development and validation of a rapid, single‐step reverse transcriptase loop‐mediated isothermal amplification (RT‐LAMP) system potentially to be used for reliable and high‐throughput screening of COVID‐19. Front Cell Infect Microbiol. 2020;10:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levy SE, Myers RM. Advancements in next‐generation sequencing. Annu Rev Genomics Hum Genet. 2016;17:95‐115. [DOI] [PubMed] [Google Scholar]
- 21. Maljkovic Berry I, Melendrez MC, Bishop‐Lilly KA, et al. Next generation sequencing and bioinformatics methodologies for infectious disease research and public health: approaches, applications, and considerations for development of laboratory capacity. J Infect Dis. 2020;221:S292‐S307. [DOI] [PubMed] [Google Scholar]
- 22. Market Trends for Biomarker‐Based IVT Tests (2003‐2014). Amplion Inc. (www.amplion.com) 2015.
- 23. Burger JT, Maree HJ. Metagenomic next‐generation sequencing of viruses infecting grapevines. Methods Mol Biol. 2015;1302:315‐330. [DOI] [PubMed] [Google Scholar]
- 24. Gu W, Miller S, Chiu CY. Clinical metagenomic next‐generation sequencing for pathogen detection. Annu Rev Pathol. 2019;14:319‐338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS‐CoV‐2. Nat Med. 2020;26:450‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Q, Zhang Y, Wu L, et al. Structural and functional basis of SARS‐CoV‐2 entry by using human ACE2. Cell. 2020;181:894‐904. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu R, Fu A, Deng Z, Li Y, Liu T. Promising methods for detection of novel coronavirus SARS‐CoV‐2. View. 2020:1(1):e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020:25;2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chu DKW, Pan Y, Cheng SMS, et al. Molecular diagnosis of a novel coronavirus (2019‐nCoV) causing an outbreak of pneumonia. Clin Chem. 2020;66:549‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Binnicker MJ. Emergence of a novel coronavirus disease (COVID‐19) and the importance of diagnostic testing: why partnership between clinical laboratories, public health agencies, and industry is essential to control the outbreak. Clin Chem. 2020;66:664‐666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Padhye NS. Reconstructed diagnostic sensitivity and specificity of the RT‐PCR test for COVID‐19. medRxiv. 2020.
- 33. Li C, Debruyne DN, Spencer J, et al. High sensitivity detection of coronavirus SARS‐CoV‐2 using multiplex PCR and a multiplex‐PCR‐based metagenomic method. BioRxiv. 2020.
- 34. Bhat TA, Kalathil SG, Bogner PN, Blount BC, Goniewicz ML, Thanavala YM. An animal model of inhaled vitamin E acetate and EVALI‐like lung injury. N Engl J Med. 2020;382:1175‐1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Han Y, Yang H. The transmission and diagnosis of 2019 novel coronavirus infection disease (COVID‐19): a Chinese perspective. J Med Virol. 2020;92:639‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xie C, Jiang L, Huang G, et al. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int J Infect Dis. 2020;93:264‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu X, Wang X, Han L, et al. Reverse transcription loop‐mediated isothermal amplification combined with nanoparticles‐based biosensor for diagnosis of COVID‐19. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 38. Jiang M, Fang W, Aratehfar A, et al. Development and validation of a rapid single‐step reverse transcriptase loop‐mediated isothermal amplification (RT‐LAMP) system potentially to be used for reliable and high‐throughput screening of COVID‐19. MedRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 39. Yu L, Wu S, Hao X, et al. Rapid detection of COVID‐19 coronavirus using a reverse transcriptional loop‐mediated isothermal amplification (RT‐LAMP) diagnostic platform. Clin Chem. 2020;66:975‐977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang W, Dang X, Wang Q, et al. Rapid detection of SARS‐CoV‐2 using reverse transcription RT‐LAMP method. MedRxiv. 2020.
- 41. Huang A, Garcia‐Carreras B, Hitchings M, Yang B, Katzelnick L. A systematic review of antibody mediated immunity to coronaviruses. MedRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 42. Okba NMA, Muller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2‐specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26:1478‐1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Krammer F, Simon V. Serology assays to manage COVID‐19. Science. 2020;368:1060‐1061. [DOI] [PubMed] [Google Scholar]
- 44. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan W, Lu Y, Zhang J, et al. Viral kinetics and antibody responses in patients with COVID‐19. medRxiv. 2020.
- 46. Adams ER, Anand R, Andersson M, et al. Evaluation of antibody testing for SARS‐Cov‐2 using ELISA and lateral flow immunoassays. medRxiv. 2020.
- 47. Amanat F, Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS‐CoV‐2 seroconversion in humans. Nat Med. 2020;26:1033‐1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wan Y, Li Z, Wang K, Li T, Liao P. Performance verification of detecting COVID‐19 specific antibody by using four chemiluminescence immunoassay systems. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 49. Long Q‐x, Deng H‐j, Chen J, et al. Antibody responses to SARS‐CoV‐2 in COVID‐19 patients: the perspective application of serological tests in clinical practice. MedRxiv. 2020.
- 50. Zhang J, Liu J, Li N, et al. Serological detection of 2019‐nCoV respond to the epidemic: a useful complement to nucleic acid testing. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 51. Zhang Z, Xiao T, Wang Y, et al. Early viral clearance and antibody kinetics of COVID‐19 among asymptomatic carriers. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 52. Koczula KM, Gallotta A. Lateral flow assays. Essays Biochem. 2016;60:111‐120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Carter LJ, Garner LV, Smoot JW, et al. Assay techniques and test development for COVID‐19 diagnosis. ACS Cent Sci. 2020;6:591‐605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kontou PI, Braliou GG, Dimou NL, Nikolopoulos G, Bagos PG. Antibody tests in detecting SARS‐CoV‐2 infection: a meta‐analysis. Diagnostics (Basel). 2020:10;319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang H, Hou X, Wu X, et al. SARS‐CoV‐2 proteome microarray for mapping COVID‐19 antibody interactions at amino acid resolution. BioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 56. Liu Y, Yan LM, Wan L, et al. Viral dynamics in mild and severe cases of COVID‐19. Lancet Infect Dis. 2020;20:656‐657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Diao B, Wen K, Chen J, et al. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv. 2020.
- 58. Weiss S, Klingler J, Hioe C, et al. A high through‐put assay for circulating antibodies directed against the s protein of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). MedRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 59. Wang M, Fu A, Hu B, et al. Nanopore targeted sequencing for the accurate and comprehensive detection of SARS‐CoV‐2 and other respiratory viruses. Small. 2020;. 16(32):2002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Park GS, Ku K, Baek SH, et al. Development of reverse transcription loop‐mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). J Mol Diagn. 2020;22:729‐735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Salipante SJ, Jerome KR. Digital PCR‐an emerging technology with broad applications in microbiology. Clin Chem. 2020;66:117‐123. [DOI] [PubMed] [Google Scholar]
- 62. White RA, 3rd , Blainey PC, Fan HC, Quake SR. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci. 1999;96:9236‐9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hindson BJ, Ness KD, Masquelier DA, et al. High‐throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83:8604‐8610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real‐time PCR. Nat Methods. 2013;10:1003‐1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brunetto GS, Massoud R, Leibovitch EC, et al. Digital droplet PCR (ddPCR) for the precise quantification of human T‐lymphotropic virus 1 proviral loads in peripheral blood and cerebrospinal fluid of HAM/TSP patients and identification of viral mutations. J Neurovirol. 2014;20:341‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Miyaoka Y, Mayerl SJ, Chan AH, Conklin BR. Detection and quantification of HDR and NHEJ induced by genome editing at endogenous gene loci using droplet digital PCR. Methods Mol Biol. 2018;1768:349‐362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Suo T, Liu X, Feng J, et al. ddPCR: a more accurate tool for SARS‐CoV‐2 detection in low viral load specimens. Emerg Microbes Infect. 2020;9:1259‐1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lu R, Wang J, Li M, Wang Y, Dong J, Cai W. SARS‐CoV‐2 detection using digital PCR for COVID‐19 diagnosis, treatment monitoring and criteria for discharge. medRxiv. 2020.
- 70. Dong L, Zhou J, Niu C, et al. Highly accurate and sensitive diagnostic detection of SARS‐CoV‐2 by digital PCR. MedRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 71. Marraffini LA. CRISPR‐Cas immunity in prokaryotes. Nature. 2015;526:55‐61. [DOI] [PubMed] [Google Scholar]
- 72. Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA‐programmed genome editing in human cells. Elife. 2013;2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Broughton JP, Deng X, Yu G, et al. CRISPR‐Cas12‐based detection of SARS‐CoV‐2. Nat Biotechnol. 2020;38:870‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guglielmi G. First CRISPR test for the coronavirus approved in the United States. Nature. 2020. [DOI] [PubMed] [Google Scholar]
- 75. Yu X, Schneiderhan‐Marra N, Joos TO. Protein microarrays for personalized medicine. Clin Chem. 2010;56:376‐387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yu X, LaBaer J. High‐throughput identification of proteins with AMPylation using self‐assembled human protein (NAPPA) microarrays. Nat Protoc. 2015;10:756‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Norman M, Gilboa T, Ogata A, et al. Ultra‐sensitive high‐resolution profiling of anti‐SARS‐CoV‐2 antibodies for detecting early seroconversion in COVID‐19 patients. medRxiv. 2020.
- 78. Wu Y, Wang F, Shen C, et al. A noncompeting pair of human neutralizing antibodies block COVID‐19 virus binding to its receptor ACE2. Science. 2020;368:1274‐1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dahlke C, Heidepriem J, Kobbe R, et al. Distinct early IgA profile may determine severity of COVID‐19 symptoms an immunological case series. MedRxiv. 2020.
- 80. Jiang HW, Li Y, Zhang HN, et al. SARS‐CoV‐2 proteome microarray for global profiling of COVID‐19 specific IgG and IgM responses. Nat Commun. 2020;11:3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Assis RRd, Jain A, Nakajima R, et al. Analysis of SARS‐CoV‐2 antibodies in COVID‐19 convalescent plasma using a coronavirus antigen microarray. BioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 82. Bobrovitz N, Arora R, Yan T, et al. Lessons from a rapid systematic review of early SARS‐CoV‐2 serosurveys. medRxiv. 2020.
- 83. Yu L, Wu S, Hao X, et al. Rapid colorimetric detection of COVID‐19 coronavirus using a reverse transcriptional loop‐mediated isothermal amplification (RT‐LAMP) diagnostic platform: iLACO. medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 84. Jung YJ, Park G‐S, Moon JH, et al. Comparative analysis of primer‐probe sets for the laboratory confirmation of SARS‐CoV‐2. bioRxiv. 2020.
- 85. Zhang X, Wu X, Wang D, et al. Proteome‐wide analysis of differentially‐expressed SARS‐CoV‐2 antibodies in early COVID‐19 infection. medRxiv. 2020.
- 86. Wang W, Xu Y, Gao R, et al. Detection of SARS‐CoV‐2 in different types of clinical specimens. JAMA. 2020;323:1843‐1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Moccia F, Gerbino A, Lionetti V, et al. COVID‐19‐associated cardiovascular morbidity in older adults: a position paper from the Italian Society of Cardiovascular Researches. Geroscience. 2020:42;1021‐1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. To KK, Tsang OT, Chik‐Yan Yip C, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71:841‐843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Xu R, Cui B, Duan X, Zhang P, Zhou X, Yuan Q. Saliva: potential diagnostic value and transmission of 2019‐nCoV. Int J Oral Sci. 2020;12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chen Y, Li L. SARS‐CoV‐2: virus dynamics and host response. Lancet Infect Dis. 2020;20:515‐516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis. 2020;20:565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Terrasini N, Lionetti V. Exosomes in critical illness. Crit Care Med. 2017;45:1054‐1060. [DOI] [PubMed] [Google Scholar]
- 93. Zheng R, Du M, Wang X, et al. Exosome‐transmitted long non‐coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. 2018;17:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hannafon BN, Trigoso YD, Calloway CL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sandfeld‐Paulsen B, Jakobsen KR, Baek R, et al. Exosomal proteins as diagnostic biomarkers in lung cancer. J Thorac Oncol. 2016;11:1701‐1710. [DOI] [PubMed] [Google Scholar]
- 96. Goodwin AJ, Guo C, Cook JA, Wolf B, Halushka PV, Fan H. Plasma levels of microRNA are altered with the development of shock in human sepsis: an observational study. Crit Care. 2015;19:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kwon SH, Liu KD, Mostov KE. Intercellular transfer of GPRC5B via exosomes drives HGF‐mediated outward growth. Curr Biol. 2014;24:199‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang J, Chen S, Bihl J. Exosome‐mediated transfer of ACE2 (angiotensin‐converting enzyme 2) from endothelial progenitor cells promotes survival and function of endothelial cell. Oxid Med Cell Longev. 2020;2020:4213541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hassanpour M, Rezaie J, Nouri M, Panahi Y. The role of extracellular vesicles in COVID‐19 virus infection. Infect Genet Evol. 2020;85:104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gates B. Responding to Covid‐19 ‐ A Once‐in‐a‐Century Pandemic?. N Engl J Med. 2020;382:1677‐1679. [DOI] [PubMed] [Google Scholar]
- 101. Virgilio Paradiso A, Summa S, Silvestris N, et al. Rapid serological tests have a role in asymptomatic health workers COVID‐19 screening. medRxiv. 2020.
- 102. Erikstrup C, Hother CE, Pedersen OBV, et al. Estimation of SARS‐CoV‐2 infection fatality rate by real‐time antibody screening of blood donors. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Bendavid E, Mulaney B, Sood N, et al. COVID‐19 antibody seroprevalence in Santa Clara County. California MedRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 104. Stringhini S, Wisniak A, Piumatti G, et al. Repeated seroprevalence of anti‐SARS‐CoV‐2 IgG antibodies in a population‐based sample from Geneva. Switzerland medRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 105. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang B, Wang L, Kong X, et al. Long‐term coexistence of SARS‐CoV‐2 with antibody response in COVID‐19 patients. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Gordon DE, Jang GM, Bouhaddou M, et al. A SARS‐CoV‐2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459‐468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Yao XH, He ZC, Li TY, et al. Pathological evidence for residual SARS‐CoV‐2 in pulmonary tissues of a ready‐for‐discharge patient. Cell Res. 2020;30:541‐543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Benvenuto D, Giovanetti M, Ciccozzi A, Spoto S, Angeletti S, Ciccozzi M. The 2019‐new coronavirus epidemic: evidence for virus evolution. J Med Virol. 2020;92:455‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Holmes EA, O'Connor RC, Perry VH, et al. Multidisciplinary research priorities for the COVID‐19 pandemic: a call for action for mental health science. Lancet Psychiatry. 2020;7:547‐560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lin C, Xiang J, Yan M, Li H, Huang S, Shen C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS‐Cov‐2)‐infected pneumonia (COVID‐19). Clin Chem Lab Med. 2020;58:1089‐1094. [DOI] [PubMed] [Google Scholar]
- 112. Chan JF, Yip CC, To KK, et al. Improved molecular diagnosis of COVID‐19 by the novel, highly sensitive and specific COVID‐19‐RdRp/Hel real‐time reverse transcription‐PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020:58;e00310‐00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lamb LE, Bartolone SN, Ward E, Chancellor MB. Rapid detection of novel coronavirus/severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) by reverse transcription‐loop‐mediated isothermal amplification. PLoS One. 2020;15:e0234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Qian C, Zhou M, Cheng F, et al. Development and multicenter performance evaluation of fully automated SARS‐CoV‐2 IgM and IgG immunoassays. Clin Chem Lab Med. 202058(9):1601–1607. [DOI] [PubMed] [Google Scholar]
- 115. Xiong Y, Li Z‐Z, Zhuang Q‐Z, et al. Comparative performance of four nucleic acid amplification tests for SARS‐CoV‐2 virus. bioRxiv. 2020. [DOI] [PubMed]
- 116. Van Tan L, Thi Thu Hong N, My Ngoc N, et al. SARS‐CoV‐2 and co‐infections detection in nasopharyngeal throat swabs of COVID‐19 patients by metagenomics. J Infect. 2020:81;e175‐e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Quick J, Loman NJ, Duraffour S, et al. Real‐time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Stefanelli P, Faggioni G, Lo Presti A, et al. Whole genome and phylogenetic analysis of two SARS‐CoV‐2 strains isolated in Italy in January and February 2020: additional clues on multiple introductions and further circulation in Europe. Euro Surveill. 2020:25;2000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zhou K, Lokate M, Deurenberg RH, et al. Characterization of a CTX‐M‐15 producing klebsiella pneumoniae outbreak strain assigned to a novel sequence type (1427). Front Microbiol. 2015;6:1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ai T, Yang Z, Hou H, et al. Correlation of Chest CT and RT‐PCR testing for coronavirus disease 2019 (COVID‐19) in China: a report of 1014 cases. Radiology. 2020;296:E32‐E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Chakraborty C, Sharma AR, Sharma G, Bhattacharya M, Lee SS. SARS‐CoV‐2 causing pneumonia‐associated respiratory disorder (COVID‐19): diagnostic and proposed therapeutic options. Eur Rev Med Pharmacol Sci. 2020;24:4016‐4026. [DOI] [PubMed] [Google Scholar]
- 122. Butler DJ, Mozsary C, Meydan C, et al. Shotgun transcriptome and isothermal profiling of SARS‐CoV‐2 infection reveals unique host responses, viral diversification, and drug interactions. bioRxiv. 2020. [DOI] [PMC free article] [PubMed]
- 123. Lassaunière R, Frische A, Harboe ZB, et al. Evaluation of nine commercial SARS‐CoV‐2 immunoassays. medRxiv. 2020.
- 124. Li Z, Yi Y, Luo X, et al. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J Med Virol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Xiang J, Yan M, Li H, et al. Evaluation of enzyme‐linked immunoassay and colloidal gold immunochromatographic assay kit for detection of novel coronavirus(SARS‐Cov‐2) causing an outbreak of pneumonia (COVID‐19). MedRxiv. 2020.
- 126. Zhong L, Chuan J, Gong B, et al. Detection of serum IgM and IgG for COVID‐19 diagnosis. Sci China Life Sci. 2020;63:777‐780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Lou B, Li TD, Zheng SF, et al. Serology characteristics of SARS‐CoV‐2 infection since exposure and post symptom onset. Eur Respir J. 2020;2000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Lin D, Liu L, Zhang M, et al. Evaluations of the serological test in the diagnosis of 2019 novel coronavirus (SARS‐CoV‐2) infections during the COVID‐19 outbreak. Eur J Clin Microbiol Infect Dis. 2020;17:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Jin Y, Wang M, Zuo Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Liu Y, Liu Y, Diao B, et al. Diagnostic indexes of a rapid IgG/IgM combined antibody test for SARS‐CoV‐2. medRxiv. 2020.
- 131. Zhao R, Li M, Song H, et al. Early detection of SARS‐CoV‐2 antibodies in COVID‐19 patients as a serologic marker of infection. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Perera RA, Mok CK, Tsang OT, et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), March 2020. Euro Surveill. 2020:25;2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Zhang L, Pang R, Xue X, et al. Anti‐SARS‐CoV‐2 virus antibody levels in convalescent plasma of six donors who have recovered from COVID‐19. Aging (Albany NY). 2020;12:6536‐6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Montesinos I, Gruson D, Kabamba B, et al. Evaluation of two automated and three rapid lateral flow immunoassays for the detection of anti‐SARS‐CoV‐2 antibodies. J Clin Virol. 2020;128:104413. [DOI] [PMC free article] [PubMed] [Google Scholar]


