Abstract
Background
Intervertebral disc degeneration is related to tissue fibrosis. ADAMTS can degrade the important components of the ECM during the process of intervertebral disc degeneration, ultimately resulting in the loss of intervertebral disc function. sIL-13Rα2-Fc can inhibit fibrosis and slow down the degeneration process, but the mechanism involved remains unclear.
Objective
To determine the mechanism by which sIL-13Rα2-Fc inhibits ECM degradation and reduces intervertebral disc tissue fibrosis using a transcriptomics analysis.
Methods
A rat model of caudal intervertebral disc degeneration was established, and Sirius red staining was used to observe the pathological changes in the caudal intervertebral disc. Transcriptome sequencing was employed to assess the gene expression profiles of the intervertebral disc tissues in the model group and the sIL-13Rα2-Fc-treated group. Differentially expressed genes were identified and analyzed using GO annotation and KEGG pathway analyses. Real-time fluorescence quantitative PCR was used to verify the expression levels of candidate genes. The levels of GAG and HA were quantitatively assessed by ELISA, and the levels of collagen I and collagen II were analyzed by western blotting.
Results
Sirius red staining showed that in the model group, the annulus fibrosus was disordered, the number of breaks increased, and the type I collagen protein levels increased, whereas in the sIL-13Rα2-Fc group, the annulus fibrosus was ordered, the number of breaks decreased, and the type II collagen protein levels increased. In comparison with the model group, we identified 58 differentially expressed genes in the sIL-13Rα2-Fc group, and these were involved in 35 signaling pathways. Compared with those in the model group, the mRNA expression levels of Rnux1, Sod2, and Tnfaip6 in the IL-13Rα2-Fc group were upregulated, and the mRNA expression levels of Aldh3a1, Galnt3, Fgf1, Celsr1, and Adamts8 were downregulated; these results were verified by real-time fluorescence quantitative PCR. TIMP-1 (an ADAMTS inhibitor) and TIMP-1 combined with the sIL-13Rα2-Fc intervention increased the levels of GAG and HA, inhibited the expression of type I collagen, and promoted the expression of type II collagen.
Conclusion
Adamts8 may participate in the degradation of ECM components such as GAG and HA and lead to an imbalance in the ECM of the intervertebral disc, resulting in intervertebral disc degeneration. sIL-13Rα2-Fc promoted anabolism of the ECM and increased the levels of ECM components by inhibiting the expression of Adamts8, thus maintaining the dynamic equilibrium of the ECM and ultimately delaying intervertebral disc degeneration.
1. Introduction
Intervertebral disc degeneration is the underlying basic pathological process of a series of spinal degenerative diseases (such as disc herniation, spinal canal stenosis, spondylolisthesis, spinal instability, and neuropathy) [1]; however, the specific pathophysiological mechanism involved remains unclear. Most scholars believe that the main mechanisms of intervertebral disc degeneration include tissue fibrosis, intervertebral disc cartilage endplate degeneration, a decrease in the number of bone marrow cells (either through increased apoptosis and autophagy or by decreased proliferation), local effects (such as oxidative stress and vascular hyperplasia), decomposition of the extracellular matrix, and the effects of inflammatory cytokines [2–4]. Because of the changes in the rates of the synthesis and decomposition of the extracellular matrix in degenerative discs, there are increased levels of type I collagen and decreased levels of type II collagen, and proteoglycan synthesis is reduced. The degree of nucleus pulposus cell apoptosis, which is considered to be the general pathophysiological process that occurs in intervertebral disc degeneration, is also increased [5]. The extracellular matrix provides mechanical support and physical strength for the integrity of tissues and organs as well as the entire body, thereby playing a prominent role in wound repair and fibrosis [6, 7]. An imbalance between ECM synthesis and degradation can lead to excessive ECM deposition, resulting in fibrosis [8]. Fibrosis is a common underlying pathological change that occurs in many chronic noninfectious diseases, and it is the main cause of disability and death resulting from many chronic diseases and involves almost all organs and systems in the human body [9].
The occurrence and development of fibrosis are closely related to many factors, particularly cytokines [10]. IL-13 and its receptor IL-13Rα2 have become “hot” molecules in asthma research as well as in studies of other fibrotic diseases in recent years[11]. IL-13, a crucial regulator of the fibrotic extracellular matrix, is produced by activated Th2 cells and has been proven to play a role in many inflammatory and fibrotic diseases, such as idiopathic pulmonary fibrosis and systemic sclerosis [12, 13]. IL-13 increases TGF-β levels by acting through its receptor IL-13Rα2, thereby inducing fibrosis [14]. It has been found that overexpression of the high-affinity antagonist IL-13Rα2, which is related to IL-13Rα1 and can bind IL-13 but lacks any signaling capability, inhibits the fibrotic markers induced by IL-13 in vitro and in bleomycin-induced pulmonary fibrosis, indicating that IL-13Rα2 has antifibrotic properties [15]. A recombinant soluble interleukin-13 receptor α2 fusion protein (sIL-13Rα2-Fc) has been generated and shown to also have a high affinity for IL-13, and accordingly, it acts as an IL-13 antagonist to block the profibrotic effects of IL-13 and reduce the deposition of abnormal ECM in injured tissues [16, 17]. Thus, some scholars have proposed that IL-13Rα2 may delay the progression of intervertebral disc degeneration, which has been confirmed by our previous research. We found that intervertebral disc degeneration involves tissue fibrosis. The recombinant sIL-13Rα2-Fc fusion protein was found to regulate the expression levels of types I and II collagen by increasing the levels of GAG, CS, KS, and HA, thereby slowing down the process of intervertebral disc degeneration [18]. Nevertheless, the mechanism by which sIL-13Rα2-Fc delays intervertebral disc degeneration remains unclear. To clarify this issue, we used transcriptomics technology to investigate the molecular mechanism by which sIL-13Rα2-Fc delays intervertebral disc degeneration at the gene transcriptome level.
2. Materials and Methods
2.1. Establishment of a Rat Model with Caudal Intervertebral Disc Degeneration
In this study, all the animal experiments were approved by the Animal Experiment Ethics Committee of the First Hospital of Lanzhou University, Gansu, China. The rats used in the study were SD rats (36 rats, aged 8–10 weeks, weighing 250–300 g) purchased from Lanzhou Veterinary Research Institute of Chinese Academy of Sciences (Lanzhou, Gansu, China). The rat model with caudal intervertebral disc degeneration was established following the experimental method of Chia-Hsian et al. [19] The rats were anesthetized with sodium pentobarbital (1%, 6 mL/kg), the tails of the rats were cleaned, and the limbs of the rats were fixed in the prone position. A 20 G puncture needle was used to puncture the intervertebral space between C7/8 and C8/9 vertically; it was gently rotated 360° and pulled out after 30 seconds, with the puncture depth being 3 mm. One week after successful puncture, the SD rats were randomly divided into five groups: a blank control group, a sham group, a model group and a 2 mg/kg sIL-13Rα2-Fc group, a 1 mg/kg TIMP-1 (ADAMTS inhibitor) group, and a TIMP-1+2 mg/kg sIL-13Rα2-Fc group, with six rats in each group. The model group was given an equivalent volume of normal saline.
2.2. Sirius Red Staining
After 8 weeks of sIL-13Rα2-Fc intervention, three rats were randomly selected from every group for sacrifice. The caudal intervertebral disc tissues of the rats were stripped and fixed with 4% paraformaldehyde, and 6 μm-thick paraffin-embedded sections were generated. After staining with picric acid-Sirius red (Zhongshan Reagent Co., Ltd.) for 30 min, the sections were dehydrated using a stepwise gradient of increasing ethanol concentrations, made transparent with xylene, and then sealed with neutral gum. A polarized light microscope (Olympus BX51) was used to observe the distributions of type I and type II collagen.
2.3. RNA Extraction and Transcriptome Sequencing
After 8 weeks of sIL-13Rα2-Fc intervention, the intervertebral disc tissues of three rats were taken from both the model group and the sIL-13Rα2-Fc group, and total RNA was extracted using RANiso Plus (Takara, Japan) reagent. The concentration of RNA was measured with a NanoDrop 2000 device (Thermo), and the integrity of the RNA was assessed using an RNA Nano 6000 analysis kit (Agilent Technologies, CA, USA). Qualified RNA samples were submitted to Shanghai Baipu Biotechnology Co., Ltd. for transcriptome sequencing.
2.4. Differential Expression Analysis
Fragments per kilobase of transcript per million (FPKM) was used to standardize the gene expression levels, and the differential fold change (FC) in expression was calculated based on the FPKM value. DEseq was employed to analyze the difference in gene expression among different samples and to adjust the P value to strictly control the false discovery rate (FDR). Differentially expressed genes were screened using the adjusted P < 0.01 and log2FC ≥ 2.
2.5. Enrichment Analysis of Differentially Expressed Genes
The DAVID 6.8 online analysis tool (https://david.ncifcrf.gov/home.jsp) was adopted to analyze the enrichment of differentially expressed genes, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). With P < 0.05 indicating significant enrichment, the number of differential genes in each GO term or pathway was counted. The functions and main pathways of differentially expressed genes were determined by GO and KEGG significant enrichment analyses.
2.6. Verification of Candidate Genes
Based on the results of the transcriptome bioinformatics analysis, differentially expressed genes that may be involved in the mechanism by which sIL-13Rα2-Fc protects the intervertebral disc from degeneration were identified. Two of these, namely Fgf1 and Adamts8, were confirmed by real-time quantitative PCR with GAPDH as the housekeeping gene. The primers used are listed in Table 1. The cDNA samples from the intervertebral disc tissues were used as templates, and the PCR verification was carried out using the Bio-Rad CFX 1000 Real-Time PCR system. Each sample was assessed in triplicate.
Table 1.
Primer information.
| Gene | Prime |
|---|---|
| Runx1 | Forward: 5′-TCCAGTGCGGTGTATGACTAC-3′ |
| Reverse:5′-CTTGGAAGCTGCCAGAATGCT-3′ | |
| Sod2 | Forward: 5′-TGG ACA AAC CTG AGC CCT AA-3′ |
| Reverse: 5′-GAC CCA AAG TCA CGC TTG ATA-3′ | |
| Aldh3a1 | Forward: 5′-ACTACATCCTCTGTGACCCC-3′ |
| Reverse: 5′-GCAAGGTGATGTGGACGATGAC-3′ | |
| Galnt3 | Forward: 5′-GTTGCTAGGAGCAACAGTCGCA-3′ |
| Reverse: 5′-AGTTCACCGTGGTAGTATTGTAGT-3′ | |
| Fgf1 | Forward: 5′-GTTCACTTTTCCGCTGCGCC-3′ |
| Reverse: 5′-ACCCCTCCAGATGCTACACA-3′ | |
| Tnfaip6 | Forward: 5′-GTCGTCTCGCAACCTACAAGCAG-3′ |
| Reverse: 5′-CTGACCGTACTTGAGCCGAATGTG-3′ | |
| Celsr1 | Forward: 5′-CTCTTATTCTTGCCACCACT-3′ |
| Reverse: 5′-GATTTCTACATTGAGCCCAC-3′ | |
| Adamts8 | Forward: 5′-TCTGTGACCCCAACAAGAGC-3′' |
| Reverse: 5′-CAAACAGTCTCACGCATGGC-3′ | |
| GAPDH | Forward: 5′-AGGGCTGCCTTCTCTTGTG-3′ |
| Reverse: 5′-CCTGGCATTGCCGACA-3′ |
2.7. ELISA Analysis
According to the aforementioned modeling method, the rat model of intervertebral disc degeneration was reestablished, and the rats were regrouped: a blank control group, a sham group, a model group, a 2 mg/kg sIL-13Rα2-Fc group, a 1 mg/kg TIMP-1 (ADAMTS inhibitor) group, and a TIMP-1+2 mg/kg sIL-13Rα2-Fc group, with six rats in each group. The blank group and model group were given an equivalent volume of normal saline. After 8 weeks of drug intervention, the intervertebral disc tissues of the rats were homogenized in PBS buffer, and the homogenate was centrifuged at 5000 × g for 5 min to generate a supernatant for analysis. Following the instructions for the rat GAG ELISA Kit (mlbio, ml059570) and the HA ELISA Kit (mlbio, ml059167), each antibody was diluted with 0.05 M pH 9 osmium carbonate coating buffer, and blank, standard, and sample wells were prepared, after which samples were added to the appropriate wells. Then, 0.1 mL of enzyme-labeled antibody was added to each reaction well followed by incubation at 37°C for 0.5 h. After the substrate reaction solution (TMB) was added to each well and allowed to incubate for 10–30 min, the terminating reaction solution was added to terminate the reaction. The OD value was measured at 490 nm with a microplate reader (iMark 19718, Bio-Rad, Hercules, CA, USA).
2.8. Western Blot
After 8 weeks of drug intervention, 20 mg of intervertebral disc tissue was taken from the blank control group, the model group, the TIMP-1 group, and the TIMP-1+2 sIL-13Rα2-Fc group and lysed with RIPA (Applygen, YZ-C1053) buffer. The total protein extracted from the rat intervertebral disc tissues was then quantified by the bicinchoninic acid (BCA) method. The tissue lysate samples were loaded onto an SDS-PAGE gel and separated by electrophoresis. Following electrophoretic transfer to a polyvinylidene fluoride (PVDF) membrane, the membrane was blocked with 5% skimmed milk for 1 h and then incubated overnight at 4°C with specific primary antibodies, including an anti-collagen I antibody (ab34710, 1 : 1000, Abcam), an anti-collagen II antibody (ab34712, 1 : 1000, Abcam), and an anti-β-actin antibody (ab8227, 1 : 2000, Abcam). The membrane was then incubated with a horseradish peroxidase- (HRP-) conjugated secondary antibody (E030110, 1 : 5000, Earthhox) for 1.5 h at room temperature. Protein signals were detected using ECL chemiluminescent agents and then quantified by densitometry using Image-Pro plus 6.0 software.
2.9. Statistical Analysis
SPSS19.0 was employed for statistical analysis of the experimental data following the univariate analysis method, and the results of pairwise comparisons between groups are presented as ; P < 0.05 was used to indicate that a difference was statistically significant.
3. Results
3.1. Sirius Red Staining of the Caudal Intervertebral Disc in Rats
We performed a Sirius red staining analysis of the intervertebral disc tissue of rats after the 8th week of the sIL-13Rα2-Fc intervention, and the results are shown in Figure 1. The intervertebral disc tissues in the blank group and sham group mainly consisted of type II collagen, followed by type I collagen, with a neatly arranged annulus fibrosus. Compared with that in the sham group, the intervertebral disc tissue in the model group was mainly composed of type I collagen, with a disordered annulus fibrosus and an increased number of breaks and obvious signs of degeneration. Treatment with sIL-13Rα2-Fc significantly decreased the extent of intervertebral disc degeneration in rats, which was characterized by the dominant expression of type II collagen, an evenly arranged annulus fibrosus, and a decreased number of breaks. These data indicate that sIL-13Rα2-Fc can slow down the process of disc degeneration.
Figure 1.
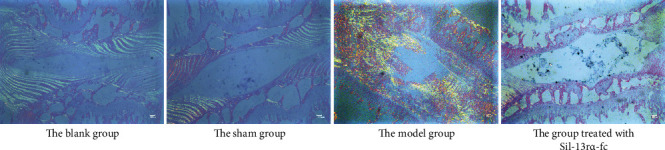
Sirius red staining showing the effect of sIL-13Rα2-Fc on the intervertebral disc tissue in rats.
3.2. Differential Expression Analysis
The FPKM method was used to determine the gene expression patterns in the model group and the sIL-13Rα2-Fc group, after which DEseq was used to identify differentially expressed genes in the caudal intervertebral disc of SD rats. A total of 58 differentially expressed genes were identified between the model group and the sIL-13Rα2-Fc group, of which 22 genes were upregulated and 36 genes were downregulated (Table 2). The volcano plot in Figure 2 presents the distribution of the upregulated and downregulated genes.
Table 2.
Statistical analysis of the number of differentially expressed genes.
| Gene ID | Gene symbol | P value | log2FC | Description | Regulated |
|---|---|---|---|---|---|
| ENSRNOG00000000906 | Medag | 0.00919 | 1.040161 | Predicted: mesenteric estrogen-dependent adipogenesis protein | Up |
| ENSRNOG00000001704 | Runx1 | 0 | 1.091159 | Predicted: runt-related transcription factor 1 isoform X1 | Up |
| ENSRNOG00000003923 | Rab21 | 0 | 1.395875 | Ras-related protein Rab-21 | Up |
| ENSRNOG00000007596 | Rffl | 0 | 1.141026 | Predicted: E3 ubiquitin-protein ligase rififylin isoform X5 | Up |
| ENSRNOG00000011063 | Dennd1b | 0.000568 | 1.636745 | Predicted: DENN domain-containing protein 1B isoform X2 | Up |
| ENSRNOG00000013618 | Ankrd10 | 0 | 1.449423 | Ankyrin repeat domain-containing protein 10 | Up |
| ENSRNOG00000018450 | Slc25a22 | 0 | 5.092963 | Predicted: mitochondrial glutamate carrier 1 isoform X1 | Up |
| ENSRNOG00000019048 | Sod2 | 0 | 1.234773 | Superoxide dismutase (Mn), mitochondrial precursor | Up |
| ENSRNOG00000021119 | Pdcd2l | 0.001631 | 1.483934 | Programmed cell death protein 2-like | Up |
| ENSRNOG00000015002 | Abhd15 | 0.003322 | 1.851272 | Protein ABHD15 precursor | Up |
| ENSRNOG00000025670 | Shisa3 | 0.0014 | 1.954814 | Protein shisa-3 homolog precursor | Up |
| ENSRNOG00000027837 | Gm14569 | 0.005741 | 1.751623 | Similar to novel protein (predicted), partial | Up |
| ENSRNOG00000036662 | Wdr45b | 0 | 1.102958 | WD repeat domain phosphoinositide-interacting protein 3 | Up |
| ENSRNOG00000045941 | Susd6 | 0.000112 | 1.030092 | Predicted: sushi domain-containing protein 6 isoform X1 | Up |
| ENSRNOG00000049185 | LOC100911032 | 0.006145 | 1.808194 | Immunoglobulin light chain, partial | Up |
| ENSRNOG00000050792 | Tnfaip6 | 0.001631 | 1.561718 | Tumor necrosis factor-inducible gene 6 protein precursor | Up |
| ENSRNOG00000052100 | Eddm3b | 0 | 2.071116 | rCG61219 | Up |
| Rattus_norvegicus_newGene_23468 | — | 0.008698 | 1.747348 | mCG61979, isoform CRA_b, partial | Up |
| Rattus_norvegicus_newGene_26511 | — | 0.00812 | 1.748733 | Predicted: uncharacterized protein LOC108348366 isoform X1 | Up |
| Rattus_norvegicus_newGene_26728 | — | 0.009034 | 1.689907 | — | Up |
| Rattus_norvegicus_newGene_37488 | — | 0.008019 | 1.609688 | Predicted: neuron navigator 3 isoform X2 (Microtus ochrogaster) | Up |
| Rattus_norvegicus_newGene_47258 | — | 0 | 2.665634 | rCG63686 | Up |
| ENSRNOG00000010107 | Palld | 0 | -1.176578 | Predicted: palladin isoform X2 | Down |
| ENSRNOG00000011096 | LOC100912369 | 0.000218 | -2.164710 | High-mobility group protein B3-like isoform X1 | Down |
| ENSRNOG00000011151 | Tenm4 | 0.005755 | -1.357832 | Teneurin-4 | Down |
| ENSRNOG00000011595 | Senp8 | 0.001631 | -1.611544 | Sentrin-specific protease 8 | Down |
| ENSRNOG00000013867 | Fgf1 | 0.008019 | -1.262288 | Predicted: fibroblast growth factor 1 isoform X3 | Down |
| ENSRNOG00000014590 | Stk33 | 0.004748 | -1.806869 | Predicted: serine/threonine-protein kinase 33 isoform X1 | Down |
| ENSRNOG00000015550 | Ptgds | 0 | -2.222049 | Prostaglandin-7H2 D-isomerase precursor | Down |
| ENSRNOG00000015636 | Wdr34 | 0 | -1.482727 | WD repeat-containing protein 34 | Down |
| ENSRNOG00000016769 | Rab38 | 0.007215 | -1.475357 | Ras-related protein Rab-38 | Down |
| ENSRNOG00000017233 | Mmachc | 0.006781 | -1.017858 | Methylmalonic aciduria and homocystinuria type C protein | Down |
| ENSRNOG00000002331 | Aldh3a1 | 0.008355 | -1.687169 | Aldehyde dehydrogenase, dimeric NADP-preferring | Down |
| ENSRNOG00000002579 | Parm1 | 0 | -1.803845 | Castration-induced prostatic apoptosis-related protein 1 | Down |
| ENSRNOG00000003031 | Atp2b4 | 0.000242 | -1.261295 | Predicted: plasma membrane calcium-transporting ATPase 4 isoform X1 | Down |
| ENSRNOG00000005535 | Ikzf4 | 0.004819 | -1.501689 | Predicted: zinc finger protein Eos isoform X1 | Down |
| ENSRNOG00000005574 | Adamts8 | 0.004807 | -1.664895 | Predicted: A disintegrin and metalloproteinase with thrombospondin motifs 8 | Down |
| ENSRNOG00000005727 | Galnt3 | 0 | -1.437957 | Polypeptide N-acetylgalactosaminyltransferase 3 | Down |
| ENSRNOG00000009965 | Pih1d2 | 0 | -2.237542 | PIH1 domain-containing protein 2 | Down |
| ENSRNOG00000021285 | Celsr1 | 0.008019 | -1.757083 | Predicted: cadherin EGF LAG seven-pass G-type receptor 1 isoform X4 | Down |
| ENSRNOG00000024237 | Scel | 0.000154 | -2.031804 | Predicted: sciellin isoform X1 | Down |
| ENSRNOG00000024793 | Kctd21 | 0.000107 | -1.933313 | Predicted: BTB/POZ domain-containing protein KCTD21 | Down |
| ENSRNOG00000026211 | Mri1 | 0.001893 | -1.404079 | Methylthioribose-1-phosphate isomerase | Down |
| ENSRNOG00000027590 | Jakmip3 | 0.000471 | -2.080541 | Janus kinase and microtubule-interacting protein 3 | Down |
| ENSRNOG00000029022 | Zfp112 | 0.002058 | -1.570479 | Predicted: zinc finger protein 112 isoform X1 | Down |
| ENSRNOG00000036673 | Sectm1b | 0.009235 | -1.586613 | Secreted and transmembrane protein 1 precursor | Down |
| ENSRNOG00000039969 | Dsc2 | 0.006781 | -1.657599 | Desmocollin-2 precursor | Down |
| ENSRNOG00000043267 | Mrm2 | 0.000159 | -1.505554 | rRNA methyltransferase 2, mitochondrial | Down |
| ENSRNOG00000051238 | Mphosph8 | 0.006353 | -1.709101 | M-phase phosphoprotein 8 | Down |
| ENSRNOG00000057827 | — | 0.001631 | -1.744961 | Predicted: uncharacterized protein RGD1560492 isoform X2 | Down |
| Rattus_norvegicus_newGene_5312 | — | 0.0014 | -1.983142 | mCG1045525, partial | Down |
| Rattus_norvegicus_newGene_7480 | — | 0 | -3.134018 | mCG145399, partial | Down |
| SRattus_norvegicus_newGene_7720 | — | 0.001595 | -1.398793 | rCG35828 | Down |
| Rattus_norvegicus_newGene_14927 | — | 0 | -2.505698 | Ac1071 | Down |
| Rattus_norvegicus_newGene_28112 | — | 0.000159 | -1.719767 | — | Down |
| Rattus_norvegicus_newGene_39936 | — | 0.00156 | -1.964885 | Predicted: protein turtle homolog B isoform X1 | Down |
| Rattus_norvegicus_newGene_40776 | — | 0.001055 | -2.016483 | — | Down |
| Rattus_norvegicus_newGene_40778 | — | 0.000216 | -2.174655 | mCG1028420, partial | Down |
Figure 2.
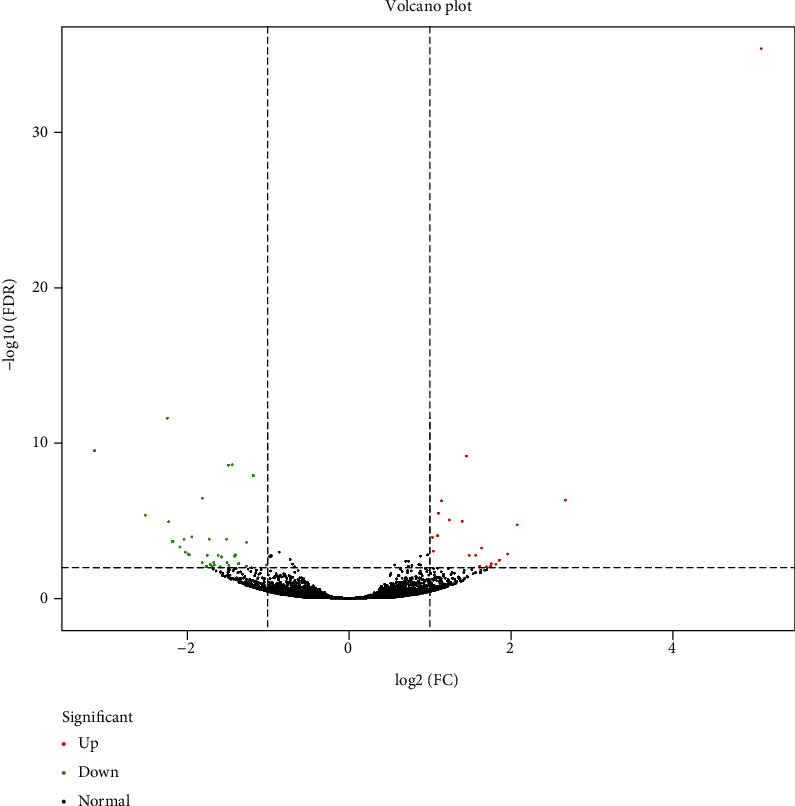
Volcano plot of differentially expressed genes. Notes: each point in the differential expression volcano plot represents a gene. The abscissa represents the absolute value of the fold change in gene expression in the two samples, and the ordinate represents the statistically significant negative logarithm value of the gene expression change. In the plot, green dots represent downregulated differentially expressed genes, red dots represent upregulated differentially expressed genes, and black dots represent nondifferentially expressed genes.
3.3. GO Classification of Differentially Expressed Genes
The differentially expressed genes were functionally annotated according to the GO classification criteria. This analysis revealed that 42 of the differentially expressed genes in the sIL-13Rα2-Fc group could be classified into the three GO branches, namely biological process, molecular function, and cellular components. The 42 differentially expressed genes were further annotated into 61 different subsets of functions under the three major GO branches, including 24 biological process functions, 18 molecular functions, and 19 cellular component functions (Figure 3). The differentially expressed genes were primarily associated with the processes of cell regulation, metabolism, behavior, biological regulation, and biological genes.
Figure 3.
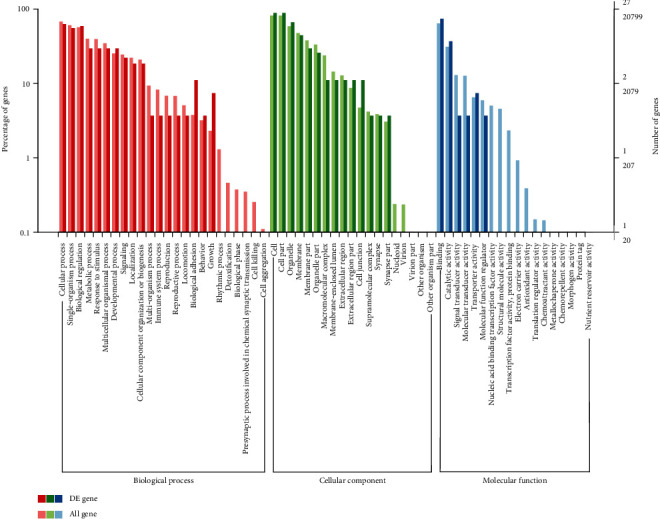
Statistical analysis of the GO annotation classification of differentially expressed genes. Notes: the abscissa represents the GO classification; the left side of the ordinate is the percentage of the number of genes, and the right side is the number of genes.
3.4. KEGG Pathway Enrichment Analysis of Differentially Expressed Genes
In the KEGG pathway analysis, compared with the model group, 26 genes in the sIL-13Rα2-Fc group were upregulated or downregulated, involving 35 metabolic pathways (Figure 4(a)). Among them, the PI3K/Akt, Ras/MEK/ERK, and MAPK signaling pathways were involved in the prevention and treatment of intervertebral disc degeneration by sIL-13Rα2-Fc; among these three, pathways involved in cancer (2 genes) contained the most abundant genes. As shown in the enrichment figure from the KEGG pathway analysis, the differentially expressed genes in the intervertebral disc tissue of SD rats in the sIL-13Rα2-Fc group were mainly enriched in the following pathways: histidine metabolism, vitamin digestion and absorption, mucin type O-glycan biosynthesis, and phenylalanine metabolism (Figure 4(b)), representing the top 20 pathways with the least significant Q values. Table 3 in the pathways with a higher differential gene enrichment rate in the disc tissue of the sIL-13Rα2-Fc group than in the model group. After the intervention with sIL-13Rα2-Fc, the expression levels of Runx1, and Sod2, were upregulated, whereas those of Aldh3a1, Galnt3, Fgf1, Atp2b4, Mri1, Dsc2, Mmachc, Ptgds and Adamts8 were downregulated.
Figure 4.
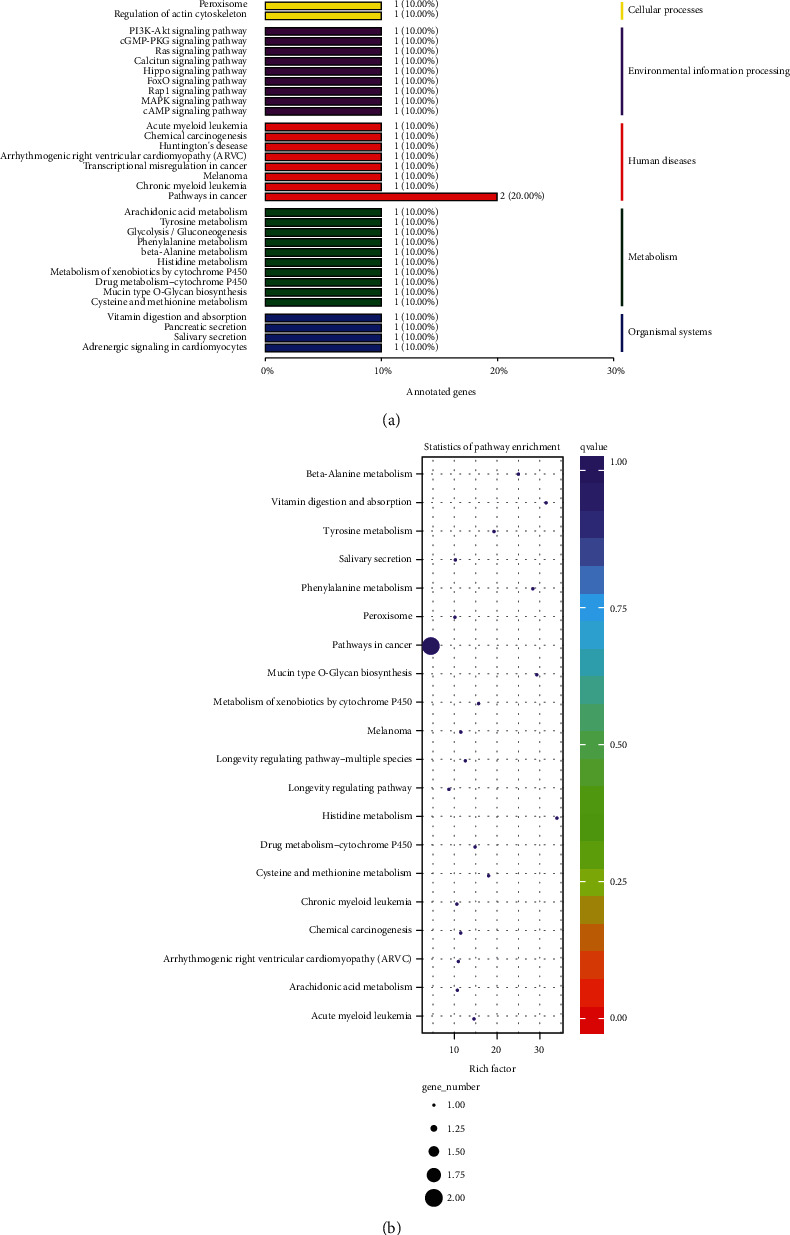
Scatter plot of the KEGG pathway enrichment for differentially expressed genes. Notes: each circle in the figure represents a KEGG pathway; the ordinate is the name of the pathway, and the abscissa is the enrichment factor, which represents the ratio of the proportion of genes annotated to a pathway for differentially expressed genes to the proportion of genes annotated to that pathway for all genes.
Table 3.
Statistical table of differentially expressed genes in significantly enriched KEGG pathways.
| Gene name | Gene symbol | Entry | Log2FC | Regulated |
|---|---|---|---|---|
| ENSRNOG00000001704 | Runx1 | K08367 | 1.091159 | Up |
| ENSRNOG00000019048 | Sod2 | K04564 | 1.234773 | Up |
| ENSRNOG00000002331 | Aldh3a1 | K00129 | -1.687169 | Down |
| ENSRNOG00000003031 | Atp2b4 | K05850 | -1.261295 | Down |
| ENSRNOG00000005727 | Galnt3 | K00710 | -1.437957 | Down |
| ENSRNOG00000013867 | Fgf1 | K18496 | -1.262288 | Down |
| ENSRNOG00000026211 | Mri1 | K08963 | -1.404079 | Down |
| ENSRNOG00000039969 | Dsc2 | K07601 | -1.657599 | Down |
| ENSRNOG00000017233 | Mmachc | K14618 | -1.017858 | Down |
| ENSRNOG00000005574 | Adamts8 | K08623 | -1.664895 | Down |
| ENSRNOG00000015550 | Ptgds | K01830 | -2.222049 | Down |
3.5. Real-Time Fluorescence Quantitative PCR Verification
To verify the transcriptional levels of the genes identified in the transcriptome analysis to be important in the mechanism by which sIL-13Rα2-Fc protects against rat intervertebral disc degeneration and to confirm the accuracy of the bioinformatics analysis, we used real-time fluorescence quantitative PCR to verify the expression of the candidate genes Runx1, Sod2, Tnfaip6, Aldh3a1, Galnt3, Fgf1, Celsr1, and Adamts8. The results are shown in Figure 5. Compared with those in the sham group, the expression levels of Runx1, Sod2, and Tnfaip6 in the model group decreased significantly (P < 0.05), whereas the expression levels of Aldh3a1, Galnt3, Fgf1, Celsr1, and Adamts8 increased significantly (P < 0.05). Compared with those in the model group, the expression levels of Runx1, Sod2, and Tnfaip6 in the sIL-13Rα2-Fc group increased significantly (P < 0.05), whereas the expression levels of Aldh3a1, Galnt3, Fgf1, Celsr1, and Adamts8 in the sIL-13Rα2-Fc group decreased significantly (P < 0.05). The relative expression results were consistent with the sequencing results.
Figure 5.
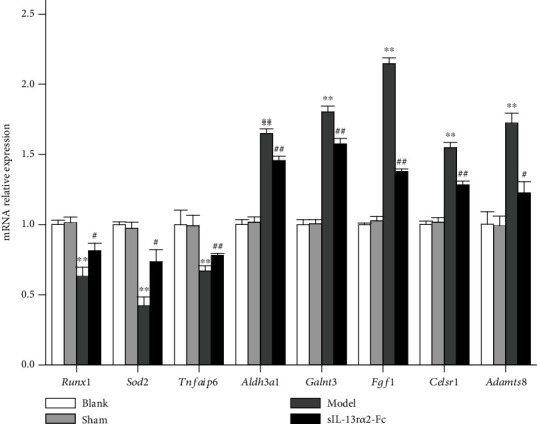
Real-time fluorescence quantitative PCR verification of genes. vs. the sham group, ∗∗P < 0.01; vs. the model group, #P < 0.05, ##P < 0.01.
3.6. Effect of sIL-13Rα2-fc on GAG and HA Levels in the Degenerative Intervertebral Disc Tissue of Rats
We quantitatively measured the GAG and HA levels in degenerative intervertebral disc tissue of rats using an ELISA method, and the results are shown in Figure 6. After 8 weeks of drug intervention, the levels of GAG and HA in the model group were significantly lower than those in the blank control group (P < 0.05). The levels of GAG and HA in the TIMP-1 group or the TIMP-1+2 sIL-13Rα2-Fc group were significantly higher than those in the model group (P < 0.05), and those in the TIMP-1+sIL-13Rα2-Fc group were significantly higher than those in the TIMP-1 group (P < 0.05).
Figure 6.
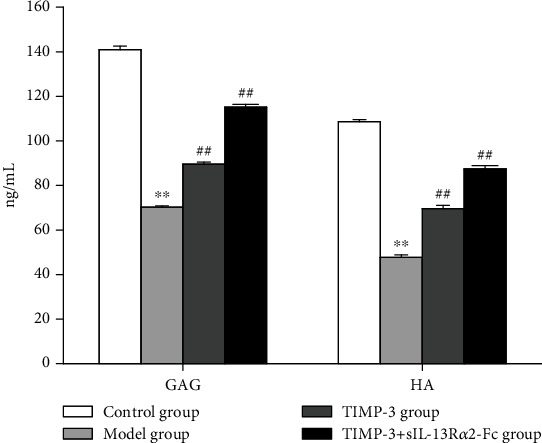
Effect of sIL-13Rα2-Fc on GAG and HA in the degenerative intervertebral disc tissue of rats determined by ELISA. vs. control group, ∗∗P < 0.01; vs. model group, #P < 0.05, ##P < 0.01.
3.7. Effect of sIL-13Rα2-fc on the Expression of Type I Collagen and Type II Collagen in the Degenerative Intervertebral Disc Tissue of Rats
To assess the change in collagen type that occurred during the process of intervertebral disc degeneration, we employed western blotting to measure the expression levels of type I collagen and type II collagen, and the results are shown in Figure 7. After 8 weeks of drug intervention, compared with those in the blank control group, the type I collagen level was significantly higher (P < 0.05), whereas the type II collagen level was significantly lower (P < 0.05) in the model group. Compared with those in the model group, the type I collagen level in the TIMP-1 group or the TIMP-1+2 sIL-13Rα2-Fc group was significantly lower (P < 0.05), whereas the type II collagen level was significantly higher (P < 0.05).
Figure 7.
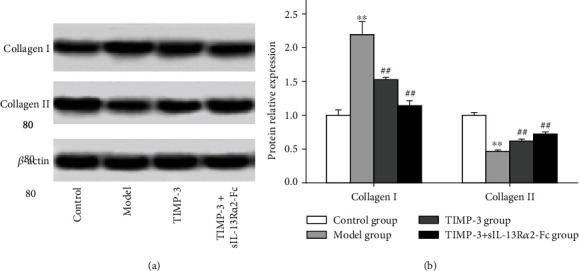
Western blot analysis of the expression of type I collagen and type II collagen in degenerative intervertebral disc tissue. vs. control group, ∗∗P < 0.01; vs. model group, ##P < 0.01.
4. Discussion
Intervertebral disc degeneration can cause intervertebral disc herniation, neuropathy, and spondylolisthesis, resulting in neck, waist, and leg pain and other symptoms [1]. One of the characteristics of intervertebral disc degeneration is the injury of and formation of lesions in the peripheral tissue, which are difficult to heal. When pathological changes occur in the annulus fibrosus or cartilage endplate, it also means that local tissues have been damaged or have undergone an inflammatory response [2, 4]. The structural change in the intervertebral disc tissue is a sign of impaired intervertebral disc function, and such a structural change is permanent [20]. These structural changes can easily be assessed by both physical and biological means [21, 22]. In this study, we used Sirius red staining to observe the pathological changes in the intervertebral disc tissue of rats. In the model group, the annulus fibrosus of the intervertebral disc tissue was disordered, there was an increased number of breaks, and there was obvious degeneration; however, treatment with sIL-13Rα2-Fc clearly improved this condition, as evidenced by an evenly arranged annulus fibrosus, a decreased number of breaks, and a lower degree of degeneration. This suggests that sIL-13Rα2-Fc can reduce the pathological changes in degenerative intervertebral disc tissue.
In this study, we used transcriptome sequencing technology to measure gene expression in the intervertebral disc tissues of the model group and sIL-13Rα2-Fc treatment group, and we identified 58 differentially expressed genes, of which 22 genes were upregulated and 36 genes were downregulated. In addition, the expression levels of 8 differentially expressed genes in the rat intervertebral disc tissues were verified by RT-PCR. In this study, the RNA-seq method was used to study the effect of sIL-13Rα2-Fc on the intervertebral disc tissue of rats, and it was found that the most significant differences in the gene expression were among the biological process-related genes, although there were also varying degrees of influence on genes related to cell composition and molecular function. Pathway enrichment analysis revealed that a variety of pathway-related genes were affected. Amino acid, vitamin, and exogenous substance metabolism pathway-related genes showed a downward trend, and polysaccharide biosynthesis pathway genes showed an upward trend. The degeneration of the intervertebral disc begins when the balance between cell generation and apoptosis is disrupted. sIL-13Rα2-Fc can downregulate metabolism-related genes and upregulate polysaccharide synthesis, which may be one of its important mechanisms of action.
ADAMTS, a depolymerized protein-like metalloproteinase containing a type I platelet binding protein sequence (TSP), is a type of Zn2+-dependent secreted metalloproteinase that was discovered after the matrix metalloproteinases (MMPs). ADAMTS is widely expressed in both mammals and invertebrates and can bind to the cell surface and the extracellular matrix through specific protein motifs, thus playing an influential role in pathophysiological processes such as degenerative aging, inflammatory response, tumor growth, and metastasis. ADAMTS is closely involved in a variety of human diseases [23]. Studies have reported the mRNA and protein expression of ADAMTS1, ADAMTS4, ADAMTS5, ADAMTS9, and ADAMTS15 in human intervertebral disc tissue, suggesting that these ADAMTS play a specific role in the normal physiological processes in the intervertebral disc [24]. In a static pressure-induced rat model with caudal intervertebral disc degeneration, the mRNA levels of ADAMTS4, ADAMTS5, ADAMTS7, and ADAMTS12 were found to be increased significantly [25]. ADAMTS degrades proteoglycans during the degeneration of the intervertebral disc, ultimately resulting in the final loss of function of the intervertebral disc [26]. In this study, Adamts1, Adamts3, Adamts4, Adamts5, Adamts7, Adamts9, and Adamts15 were not found to be significantly differentially expressed genes in the rat intervertebral disc tissue, Adamts8 was found to be significantly downregulated. In addition, the comprehensive GO annotation and KEGG pathway analyses of differentially expressed genes revealed that Adamts8 is involved in the extracellular matrix signaling pathway, and thus, we further speculate that Adamts8 may play a role in the process by which sIL-13Rα2-Fc attenuates intervertebral disc degeneration. The secondary structure of Adamts8 contains a metal peptidase structure and a zinc ion binding site, showing that it has matrix metalloproteinase activity and the ability to bind metal zinc ions [27]. Studies have shown that the Adamts8 could have the ability of exhibiting aggrecanase activity and antiangiogenic properties [28, 29]. In this study, the specific mechanism of Adamts8 remains to be further studied.
The imbalance between the synthesis and degradation of the extracellular matrix, resulting in changes in the EMC composition and content, is one of the important characteristics of the intervertebral disc after intervertebral disc degeneration [30, 31]. The ECM in the intervertebral disc is mainly composed of collagen and proteoglycans [32]. The type of collagen in the degenerative intervertebral disc is significantly different from that in a nondegenerative disc, which may reflect the tissue abnormalities that occur during intervertebral disc degeneration and the related repair processes [33]. The changes in the proteoglycan content and composition are considered to be one of the earliest biochemical changes in intervertebral disc degeneration, and they can directly lead to changes in the biomechanical properties of the intervertebral disc [34]. The ADAMTS family is a class of enzymes that play crucial roles in the process of extracellular matrix degradation, and they can degrade almost all the components of the extracellular matrix; accordingly, these enzymes are the most important factors that regulate the dynamic equilibrium of the ECM [35]. Studies have shown that ADAMTS1, ADAMTS4, ADAMTS5, ADAMTS8, ADAMTS9, ADAMTS15, and ADAMTS18 can degrade polyproteoglycans (important components of the nucleus pulposus ECM), and ADAMTS4 and ADAMTS5 are classified as the major polyproteoglycases [36]. Zhang et al. [37] found that the expression levels of ADAMTS7 and ADAMTS12 were upregulated and that the levels of type II collagen were significantly decreased in the lumbar endplate tissue of patients with intervertebral disc degeneration. In our previous study, we found that the ECM content in the degenerative intervertebral disc tissue of rats decreased to varying degrees, the synthesis of type I collagen increased, and the expression of type II collagen decreased, all of these effects were reversed by intervention with sIL-13Rα2-Fc [18]. In this study, we used the ADAMTS inhibitor TIMP-1 to treat the model rats with intervertebral disc degeneration, and we found that the levels of the ECM components GAG and HA increased, type I collagen expression decreased, and type II collagen expression increased. The levels of the ECM components GAG and HA and the collagen levels in the TIMP-1+2 sIL-13Rα2-Fc intervention group were significantly higher than those in the TIMP-1 intervention group, indicating that TIMP-1 and sIL-13Rα2-Fc had a synergistic effect. Incorporating the transcriptome data analysis results, we hypothesize that sIL-13Rα2-Fc can maintain a dynamic balance in the ECM and delay intervertebral disc degeneration by inhibiting Adamts8-mediated ECM degradation.
In summary, the results of this study indicate that sIL-13Rα2-Fc can inhibit further degradation of the ECM by inhibiting the expression of Adamts8, restoring ECM synthesis and degradation to a dynamic equilibrium, and eventually partially slowing the process of intervertebral disc degeneration in rats.
Acknowledgments
This work was supported by the China Postdoctoral Science Foundation (No. 2017M613342), Lanzhou Chengguan Science and Technology Bureau (No. 2017SHFZ0036), Gansu Natural Science Foundation (No. 18JR3RA353), and the first Hospital of Lanzhou University Foundation (No. ldyyyn2017-26).
Abbreviations
- sIL-13Rα2-Fc:
Soluble interleukin-13 R alpha2 receptor fusion protein
- ECM:
Extracellular matrix
- ADAMTS:
A depolymerized protein-like metalloproteinase containing a type I platelet binding protein sequence
- IL-13:
Interleukin-13
- GAG:
Glycosaminoglycan
- CS:
Chondroitin sulfate
- KS:
Keratan sulfate
- HA:
Hyaluronic
- OD:
Optical density
- RT-PCR:
Reverse transcription-polymerase chain reaction.
Contributor Information
Xin Wang, Email: wangxinldyy@126.com.
Yang Liu, Email: liuyangspine1@hotmail.com.
Data Availability
The datasets supporting the conclusions of this article are included within the article.
Ethical Approval
The research protocol was approved by the Institutional Animal Research Committee Approval of the First Hospital of Lanzhou University (permission number (LDYYLL2019-137)).
Conflicts of Interest
The authors declare that they have no competing interests. All authors are orthopedic doctors and researchers: Xin Wang, Jianshi Tan, Junhao Sun, Pengzhong Fang, and Jinlei Chen from the First Clinical Medical College of Lanzhou University, The First Hospital of Lanzhou University and Wen Yuan, Huajiang Chen and Yang Liu from the Changzheng Orthopedics Hospital, Second Military Medical University.
Authors' Contributions
Xin Wang and Yang Liu conceived the study. The animal model was prepared by Junhao Sun, Jianshi Tan, Pengzhong Fang, and Jinlei Chen. The remaining experiments were performed by Junhao Sun and Jianshi Tan. Statistical analysis was conducted by Wen Yuan and Huajiang Chen. Xin Wang was responsible for writing the manuscript.
References
- 1.Vergroesen P. P. A., Kingma I., Emanuel K. S., et al. Mechanics and biology in intervertebral disc degeneration: a vicious circle. Osteoarthritis and Cartilage. 2015;23(7):1057–1070. doi: 10.1016/j.joca.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 2.Mohanty S., Dahia C. L. Defects in intervertebral disc and spine during development, degeneration, and pain: new research directions for disc regeneration and therapy. Wiley Interdisciplinary Reviews: Developmental Biology. 2019;8(4):p. e343. doi: 10.1002/wdev.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakai D., Schol J. Cell therapy for intervertebral disc repair: clinical perspective. Journal of Orthopaedic Translation. 2017;9(C):8–18. doi: 10.1016/j.jot.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iwahashi M., Matsuzaki H., Tokuhashi Y., Wakabayashi K., Uematsu Y. Mechanism of intervertebral disc degeneration caused by nicotine in rabbits to explicate intervertebral disc disorders caused by smoking. Spine. 2002;27(13):1396–1401. doi: 10.1097/00007632-200207010-00005. [DOI] [PubMed] [Google Scholar]
- 5.Freemont A. J. The cellular pathobiology of the degenerate intervertebral disc and discogenic back pain. Rheumatology. 2008;48(1):5–10. doi: 10.1093/rheumatology/ken396. [DOI] [PubMed] [Google Scholar]
- 6.Kim S. H., Turnbull J., Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. Journal of Endocrinology. 2011;209(2):139–151. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- 7.Herrera J., Henke C. A., Bitterman P. B. Extracellular matrix as a driver of progressive fibrosis. Journal of Clinical Investigation. 2018;128(1):45–53. doi: 10.1172/JCI93557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng L., Liao A., Qu L., Tang J., Li X. Modulation of aberrant extracellular matrix degradation systems by astragali radix and angelicae sinensis radix decoction (A&A) in interstitial fibrotic kidney. Zhongguo Zhong Yao Za Zhi. 2012;37(1):79–84. [PubMed] [Google Scholar]
- 9.Wynn T. A., Ramalingam T. R. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nature Medicine. 2012;18(7):1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bersani-Amado L. E., Dantas J. A., Damião M. J., et al. Involvement of cytokines in the modulation and progression of renal fibrosis induced by unilateral ureteral obstruction in C57BL/6 mice: effects of thalidomide and dexamethasone. Fundamental & Clinical Pharmacology. 2016;30(1):35–46. doi: 10.1111/fcp.12162. [DOI] [PubMed] [Google Scholar]
- 11.Wang W., Shen Y. X., Li J., et al. Enhanced expression of the decoy receptor IL-13Rα2 in macrophages of Schistosoma japonicum-infected mice. Chinese Medical Journal. 2009;122(14):1650–1654. [PubMed] [Google Scholar]
- 12.Hancock A., Armstrong L., Gama R., Millar A. Production of interleukin 13 by alveolar macrophages from normal and fibrotic lung. American Journal of Respiratory Cell and Molecular Biology. 1998;18(1):60–65. doi: 10.1165/ajrcmb.18.1.2627. [DOI] [PubMed] [Google Scholar]
- 13.Wynn T. A. IL-13 effector functions[J] Annual Review of Immunology. 2003;21(1):425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 14.Fichtner-Feigl S., Strober W., Kawakami K., Puri R. K., Kitani A. IL-13 signaling through the IL-13α2 receptor is involved in induction of TGF-β1 production and fibrosis. Nature Medicine. 2006;12(1):99–106. doi: 10.1038/nm1332. [DOI] [PubMed] [Google Scholar]
- 15.Lumsden R. V., Worrell J. C., Boylan D., et al. Modulation of pulmonary fibrosis by IL-13Rα2. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2015;308(7):L710–L718. doi: 10.1152/ajplung.00120.2014. [DOI] [PubMed] [Google Scholar]
- 16.Wilson M. S., Elnekave E., Mentink-Kane M. M., et al. IL-13Rα2 and IL-10 coordinately suppress airway inflammation, airway-hyperreactivity, and fibrosis in mice. Journal of Clinical Investigation. 2007;117(10):2941–2951. doi: 10.1172/JCI31546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Y., Zhou C., Chu D. The effect of recombinant sTGFβ1RII and sIL13Rα2 receptor proteins on schistosomiasis japonica, hepatic fibrosis and signal transduction in a mouse model of schistosome disease. Experimental Parasitology. 2014;142:17–26. doi: 10.1016/j.exppara.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Wang X., Sun J., Tan J., et al. Effect of sIL-13Rα2-Fc on the progression of rat tail intervertebral disc degeneration. Journal of Orthopaedic Surgery and Research. 2019;14(1):p. 386. doi: 10.1186/s13018-019-1361-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C.-H., Chiang C.-J., Wu L.-C., et al. Time course investigation of intervertebral disc degeneration in a rat-tail puncture model. Life Sciences. 2016;156:15–20. doi: 10.1016/j.lfs.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 20.Harris R. I., Macnab I. Structural changes in the lumbar intervertebral DISCS. The Journal of Bone and Joint Surgery. British volume. 1954;36-B(2):304–322. doi: 10.1302/0301-620X.36B2.304. [DOI] [PubMed] [Google Scholar]
- 21.Pfirrmann C. W. A., Metzdorf A., Zanetti M., Hodler J., Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 2001;26(17):1873–1878. doi: 10.1097/00007632-200109010-00011. [DOI] [PubMed] [Google Scholar]
- 22.Fernandez-Moure J., Moore C. A., Kim K., et al. Novel therapeutic strategies for degenerative disc disease: review of cell biology and intervertebral disc cell therapy. SAGE Open Medicine. 2018;6(4):p. 205031211876167. doi: 10.1177/2050312118761674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuno K., Kanada N., Nakashima E., Fujiki F., Ichimura F., Matsushima K. Molecular cloning of a gene encoding a new type of metalloproteinase-disintegrin family protein with thrombospondin motifs as an inflammation associated gene. Journal of Biological Chemistry. 1997;272(1):556–562. doi: 10.1074/jbc.272.1.556. [DOI] [PubMed] [Google Scholar]
- 24.Pockert A. J., Richardson S. M., Le Maitre C. L., et al. Modified expression of the ADAMTS enzymes and tissue inhibitor of metalloproteinases 3 during human intervertebral disc degeneration. Arthritis & Rheumatism. 2009;60(2):482–491. doi: 10.1002/art.24291. [DOI] [PubMed] [Google Scholar]
- 25.Hirata H., Yurube T., Kakutani K., et al. A rat tail temporary static compression model reproduces different stages of intervertebral disc degeneration with decreased notochordal cell phenotype. Journal of Orthopaedic Research. 2014;32(3):455–463. doi: 10.1002/jor.22533. [DOI] [PubMed] [Google Scholar]
- 26.Wang W.-J., Yu X.-H., Wang C., et al. MMPs and ADAMTSs in intervertebral disc degeneration. Clinica Chimica Acta. 2015;448:238–246. doi: 10.1016/j.cca.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 27.Brocker C. N., Vasiliou V., Nebert D. W. Evolutionary divergence and functions of the ADAM and ADAMTS gene families. Human Genomics. 2009;4(1):43–55. doi: 10.1186/1479-7364-4-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins-Racie L. A., Flannery C. R., Zeng W., et al. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biology. 2004;23(4):219–230. doi: 10.1016/j.matbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Choi G. C. G., Li J., Wang Y., et al. The metalloprotease ADAMTS8 displays antitumor properties through antagonizing EGFR–MEK–ERK signaling and is silenced in carcinomas by CpG methylation. Molecular Cancer Research. 2014;12(2):228–238. doi: 10.1158/1541-7786.mcr-13-0195. [DOI] [PubMed] [Google Scholar]
- 30.Wu X., Song Y., Liu W., et al. IAPP modulates cellular autophagy, apoptosis, and extracellular matrix metabolism in human intervertebral disc cells. Cell Death Discovery. 2017;3(1, article 16107) doi: 10.1038/cddiscovery.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bridgen D. T., Gilchrist C. L., Richardson W. J., et al. Integrin-mediated interactions with extracellular matrix proteins for nucleus pulposus cells of the human intervertebral disc. Journal of Orthopaedic Research. 2013;31(10):1661–1667. doi: 10.1002/jor.22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber K. T., Jacobsen T. D., Maidhof R., et al. Developments in intervertebral disc disease research: pathophysiology, mechanobiology, and therapeutics. Current Reviews in Musculoskeletal Medicine. 2015;8(1):18–31. doi: 10.1007/s12178-014-9253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rawlings N. D. MEROPS: the protease database. Nucleic Acids Research. 2002;30(1):343–346. doi: 10.1093/nar/30.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugimoto K., Takahashi M., Yamamoto Y., Shimada E., Tanzawa K. Identification of aggrecanase activity in medium of cartilage culture. Journal of Biochemistry. 1999;126(2):449–455. doi: 10.1093/oxfordjournals.jbchem.a022471. [DOI] [PubMed] [Google Scholar]
- 35.Glasson S. S., Askew R., Sheppard B., et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature (London) 2005;434(7033):644–648. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 36.Gendron C., Kashiwagi M., Lim N. H., et al. Proteolytic activities of human ADAMTS-5. Journal of Biological Chemistry. 2007;282(25):18294–18306. doi: 10.1074/jbc.M701523200. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Q., Huang M., Wang X., Xu X., Ni M., Wang Y. Negative effects of ADAMTS-7 and ADAMTS-12 on endplate cartilage differentiation. Journal of Orthopaedic Research. 2012;30(8):1238–1243. doi: 10.1002/jor.22069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.


