Highlights
-
•
We consider possible repurposed-drugs candidates against SARS-CoV-2.
-
•
10 different HIV protease inhibitors were investigated.
-
•
In silico simulations were used to study protease inhibitors for SARS-CoV-2.
Keywords: Molecular dynamics, SARS-CoV-2, HIV protease inhibitors, Molecular docking, COVID-19
Abstract
In this paper we investigate 10 different HIV protease inhibitors (HPIs) as possible repurposed-drugs candidates against SARS-CoV-2. To this end, we execute molecular docking and molecular dynamics simulations. The in silico data demonstrated that, despite their molecular differences, all HPIs presented a similar behavior for the parameters analyzed, with the exception of Nelfinavir that showed better results for most of the molecular dynamics parameters in comparison with the N3 inhibitor.
1. Introduction
Human coronavirus (HCoV) strains, SARS-CoV-1 (2003), MERS-Cov (2012) as well as the newly emerged SARS-CoV-2 (2019), are responsible for respiratory syndromes that can cause serious infections resulting from a combination of viral replication in the lower respiratory tract with an anomalous host immune response [1]. Specially for SARS-CoV-2, the pandemic outbreak is having severe consequences from both public health and socioeconomic point of view, reaching nowadays over 23 million confirmed cases with about 800 thousand deaths [2]. In addition, SARS-CoV-2 is presenting major challenges to clinical management, since there are still no specific antiviral drugs available or even vaccines proven effective in randomized controlled trials [3]. Therefore, since the emergence of SARS-CoV-1 in 2003, different protocols were adopted to search for possible anti-HCoV treatment options [4]. Part of these protocols involve identifying new uses for approved or investigational antiviral drugs that have been designed to treat other viral infections, the so-called drug repurposing [5], [6], [7], [8].
Drug repurposing emerged primarily as a response of the pharmaceutical industries to the increased time and money spent to bring new drugs to the market in normal conditions [5], [6], [8]. Some successful repurposed drugs appear by the discovering of beneficial side effects associated with their administration during development trials, such as Minoxidil, that was originally designed for hypertension, but through the identification of hair growth in a retrospective clinical analysis is now indicated to hair loss treatments [6]. Another example is the Sildenafil, a drug that was first tested for angina treatment and that became the leading product in the erectile dysfunction (marketed as Viagra) in 2012 [6]. However, despite the identification of beneficial side effects has been proved to be a valid methodology, coordinated efforts are arising to convert the drug repositioning in a deliberately systematic approach [8]. In addition, due to most of the repurposed drugs have already been found to be sufficiently safe in preclinical models, the time frame of clinical trials and drug development is generally drastically reduced. So, this approach presents itself as an excellent option for the search for suitable drugs for emergency treatment in public health crises such as the current SARS-CoV-2 pandemic.
Nevertheless, the drug repurposing approach is highly dependent on identifying suitable drugs for a given target [5], [6], [9]. In this way, the molecular docking, a computational strategy that allows one to predict binding site complementarity between a drug and its therapeutic target, has been massively used to assist drug repositioning for several diseases [6], [9]. For example, in silico simulations were used to evaluate non-nucleoside reverse transcriptase inhibitors for HIV treatment [10], as well as currently used drugs against HIV-1 protease of subtype D [11]; to study the substrate recognition processes for influenza drug targets [12], [13]; to investigate and screening inhibitors against Ebola virus [14], [15], [16]; to explore potential binding pockets and inhibitors for Zika [17], [18], Chikungunya [19] and Dengue virus [20], [21]; to perform structure-based virtual screening studies of potential drug target of Leishmania donovani [22]; to evaluate the anticancer activity of chloro and bromo-pyrazole curcumin knoevenagel condensates and phytoestrogens [23], [24]; to investigate the efficacy of direct acting antivirals (DAAs) to the treatment of different Hepatitis C virus [25] and to design modified peptidomimetic cellulose derivatives Hepatitis C virus protease inhibitors [26]. In addition, in silico simulationshave have also been used to evaluate potential inhibitors of the interaction between ACE2 and SARS-CoV-2 region binding domain [27], to help in the drug repositioning of anti-Hepatitis C virus drugs [28] and anti-polymerase drugs [29] against SARS-CoV-2.
Attending to the drug repurposing strategy, some antiretrovirals (ARV) generally used to the treatment of HIV have been tested against acute respiratory syndromes. These ARV drugs are designed to block different stages of the virus reproduction cycle. There are currently eight classes of ARV drugs, each one classified by the stage of the replication cycle they inhibit: nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs), protease inhibitors (PIs), integrase strand transfer inhibitors (INSTIs), fusion inhibitors (FIs), entry inhibitors (EIs), pharmacokinetic enhancers (PEs) and fixed-dose combinations (FDCs) [30]. In special, PIs act on the proteolytic cleavage of viral polyproteins inhibiting the subsequent replication of individual viral proteins and consequently viral particles into their infectious form [31].
Lopinavir and Ritonavir are HIV protease inhibitors (HPIs) that presented in vitro activity against SARS [32] and MERS [33], [34] coronavirus. This combination of antiretrovirals were also used to treat marmosets with severe disease resembling MERS in humans and results pointed to improved clinical, radiological and pathological conditions [35]. However, an open-label RCT study involving hospitalized adult patients infected with SARS-CoV-2 reported that no benefit was observed with Lopinavir/Ritonavir treatment beyond standard care [36]. In this work, we performed in silico simulations for 10 FDA-approved HPIs drugs in the presence of the main SARS-CoV-2 protease (Mpro) including Lopinavir/Ritonavir (as a reference) in order to have insights about new strategies for the development of anti-SARS therapeutics. The main goal of the present study is to evaluate through molecular dynamics simulation those studies on docking processes recently presented in the literature [37], [38]. Specifically, the compounds used here were based on those discussed in Ref. [37].
2. Materials and methods
The crystal structure under investigation here can be obtained from the RCSB Protein Data Bank [39] (PDB ID 6LU7) consisting of a crystal structure of SARS-CoV-2 main protease in complex with an inhibitor N3, obtained via X-ray diffraction of a crystallized sample prepared by evaporation method at ph 6.0 and temperature of 293 K with resolution of 2.16 Ê. The result related to this structure of SARS-CoV-2 virus main protease in complex with an inhibitor N3 was recently reported in Ref. [40]. The amino acids found in the active site pockets of 6LU7 are THR24, THR26, PHE140, ASN142, GLY143, CYS145, HIS163, HIS164, GLU166, and HIS172 [38].
In our simulations, we fixed the value for the NaCl concentration in the simulation box with normal saline 0.9% (0.154 mol/L) and the temperature as 310 K. These values are very close to those for human blood.
The potential therapeutic agents used in simulations were obtained in PubChem Database [41].
2.1. Molecular docking
The binding mechanism of potential HIV protease inhibitors with SARS-CoV-2 was executed by using Autodock Vina. Our goal was to recover the recent results displayed in Ref. [37]. Indeed, in Ref. [37] the authors investigated 10 different types of HIV protease inhibitors as potential therapeutic agents against SARS-CoV-2 based on the analysis of protease docking. Here, we go beyond the studies implemented in Ref. [37] in view to try to obtain best candidates as potential therapeutic agents by using molecular dynamics simulations. To this end, we first prepare the target protein by excluding any heterogeneous molecules including water (which includes the N3 ligand). Then, Chimera software [42] was used to add polar hydrogen atoms and prepare the main protease as receptor.
Next, based in Ref. [37], we selected ten approved drugs of HIV protease inhibitors [43], namely, Amprenavir, Atazanavir, Darunavir, Fosamprenavir, Indinavir, Lopinavir, Nelfinavir, Ritonavir, Saquinavir, and Tipranavir. The ligand structure was obtained via PubChem Database [41] (see the respective IDs numbers in Table 1). Then, we used the AutoDock Vina, with box center (−10, 11, 69) and size (28, 31, 27), to select the best position for ligand structure, based on the best docking score. We stress that, for ligand structures without 3D representation in PubChem Database we got the 2D one and converted it by using the openbabel.
Table 1.
Molecular docking analysis of several compounds against 6LU7 and properties of SARS-CoV-2 Mpro potential inhibitor candidates.
| PubChem CID |
Name | Affinity (kcal/mol) |
Molecular weight ( ≤ 500 g/mol) |
Log P ( ≤ 5) |
H-Bond donor ( ≤ 5) |
H-bond acceptor ( ≤ 10) |
Bioavailability Score |
|---|---|---|---|---|---|---|---|
| - | N3 | -7.3 | 680.79 | 2.69 | 5 | 9 | 0.17 |
| 65016 | Amprenavir | -7.7 | 505.63 | 2.50 | 3 | 7 | 0.55 |
| 148192 | Atazanavir | -8.8 | 704.86 | 4.02 | 5 | 9 | 0.17 |
| 213039 | Darunavir | -8.0 | 547.66 | 2.47 | 3 | 8 | 0.55 |
| 131536 | Fosamprenavir | -7.7 | 585.61 | 1.69 | 4 | 10 | 0.11 |
| 5362440 | Indinavir | -8.1 | 613.79 | 2.78 | 4 | 7 | 0.55 |
| 92727 | Lopinavir | -8.4 | 628.80 | 4.53 | 4 | 5 | 0.55 |
| 64143 | Nelfinavir | -8.3 | 567.78 | 4.41 | 4 | 5 | 0.55 |
| 392622 | Ritonavir | -7.8 | 720.94 | 5.03 | 4 | 7 | 0.17 |
| 441243 | Saquinavir | -8.8 | 670.84 | 2.87 | 5 | 7 | 0.17 |
| 54682461 | Tipranavir | -7.8 | 602.66 | 6.06 | 2 | 9 | 0.56 |
2.2. Drug-likeness
Additionally, the drug candidate must reach its target in the body in sufficient concentration and stay there in a bioactive form long enough for the expected biologic events to occur. To check some properties related to this context, we take advantage of the SwissADME, that is a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules [44]. The Lipinski rule was used to distinguish between drug like and nondrug like molecules [45], [46]. This rule of 5 predicts high probability of success when there are less than 5 H-bond donors, 10 H-bond acceptors, the molecular weight (MWT) is less than 500 and the calculated LogP is less than 5 [45].
2.3. Molecular dynamics simulations
In view to verify the interactions and stability between the SARS-CoV-2 main protease and its potential inhibitors, i.e., the HIV protease inhibitors used here, explicit solvent molecular dynamics simulations were carried out with GROMACS package [47]. We used the CHARMM36 force field [48] and the tip3p water model to construct the topology. During the production of the topology file for the SARS-CoV-2 main protease we have ignored all hydrogens since they will be added after. The main protease has 306 residues with 4682 total number of atoms. Also, the protease presents total mass 33792.690 a.m.u. and total charge . The simulations were performed under periodic boundary conditions at temperature 310 K and pressure 1 atm.
To prepare the potential inhibitors of the SARS-CoV-2 main protease (listed in Table 1) we take advantage of SwissParam web server [49] in view to generate a new file that contains all of the topology information on the ligand, i.e., atom types, charges, and bonded connectivity. To this end, we use Chimera software [42] to add hydrogen atoms to the ligand. Next, after getting the topology information of the ligand we are prone to building the complex. Note that, the ligands considered here were those selected by high binding affinity in the molecular docking process.
The system size is 5.113 nm × 6.674 nm × 5.991 nm. Then, we generate a rectangular box with size 7.113 nm × 8.674 nm × 7.991 nm, corresponding to a box volume of 493.03 nm3. Then, the system is solvated with 14,639 solvent and sodium atoms are used to neutralize the total charge of the system. Also, additional atoms of NaCl are used to control the salinity of the system (normal saline).
Once the system is prepared we start a steepest descent energy minimization by using the Verlet cutoff-scheme. The Van der Waals forces were assigned a cut-off distance of 12.0 Ê with a smoothening function between 10.0 and 12.0 Ê. The Particle Mesh Ewald method was used to calculate the electrostatic interactions [50]. Indeed, the energy minimization ensures that we have a reasonable starting structure, in terms of geometry and solvent orientation.
Next, we evaluated the equilibration of the system, which is often conducted in two phases, i.e., a NVT ensemble (constant number of particles, volume, and temperature) followed by a NPT ensemble (constant number of particles, pressure, and temperature). In the first case, we conducted a 100-ps NVT equilibration with a position restrain to the protein and to the ligand. The algorithm is based on the leap-frog integrator with a time step of 2 fs and employing temperature coupling by using a modified Berendsen thermostat [51] under 100 ps. Following, a NPT equilibration was executed, using a protocol similar to that used in NVT equilibration, but now employing the Berendsen pressure coupling at 1 atm under 100 ps [51].
Once the system is well-equilibrated at the desired temperature and pressure, we are prone to release the position restraints and run production molecular dynamics simulation for data collection. The final NPT simulations of 30 ns were carried out with a 1 fs time step.
2.4. Ramachandran plot
A Ramachandran plot is a plot of the torsional angles - Phi (Φ) and Psi (Ψ) - of the residues contained in a protein. Here, we used the PROCHECK web software to build the Ramachandran plot [52], [53]. A detailed check on the stereochemistry of a protein structure is obtained by the PROCHECK suite of programs. Also, the results allow us to get an assessment of the overall quality of the structure (as compared with well refined structures of the same resolution) and also highlight regions that may need further investigation.
3. Results
3.1. Molecular docking
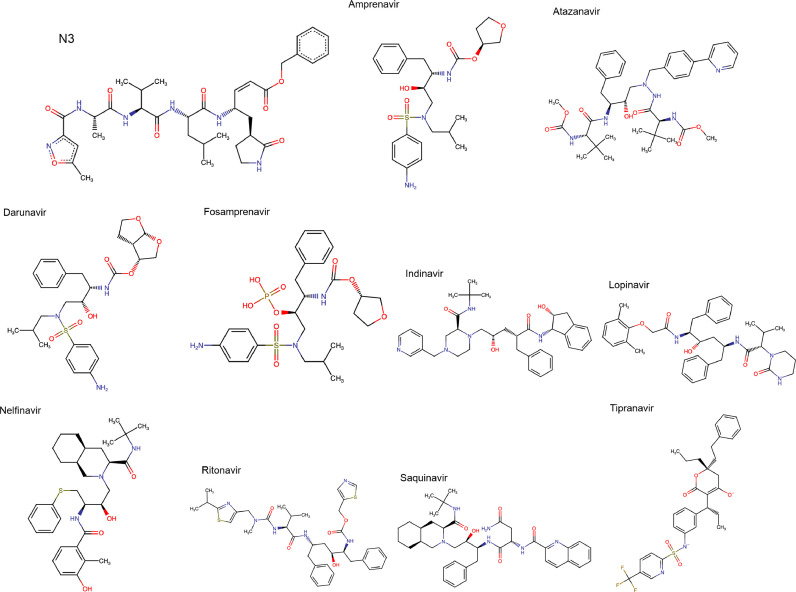
The ligands, obtained via AutoDock Vina, with best docking scores were selected to the sequence in our study. In this sense, in a first round of autodock procedure we get 10 different conformations for the main structure, which were further examined via a redocking process, with a total of 110 samples. The affinity values for the best ranked ligands of each type molecules are listed in Table 1. Also, we display in Fig. 1 the chemical structure depiction in 2D format of all ligands under investigation. Note that the inhibitor N3, present in the complexed structure with Mpro from SARS-CoV-2 virus (PDB: 6LU7), was also shown in Fig. 1 and used as reference for all results. The docking affinity for the N3 compound is also displayed in Table 1. We stress that in the molecular dynamic simulation we use the original crystal structure of SARS-CoV-2 main protease in complex with an inhibitor N3 [40]. Note that we get different values for the relative binding affinity obtained in Ref[37]. However, this value is only an indication of whether or not the ligand added well to the main protein.
Fig. 1.
Chemical structure depiction in 2D format of the compounds described in Table 1.
3.2. Drug-likeness
Next, we analyze the physicochemical descriptors and druglike nature of small molecules to support drug discovery. The corresponding results are listed in Table 1. Also, we presented the bioavailability score for all compounds obtained via SwissADME web tool [44]. The bioavailability score is based on other filters then one by Lipinski. Indeed, it uses the Lipinski, Ghose [54], Veber [55], Egan [56], and Muegge [57] filters. Definitively, some of the compounds with high binding affinity presents low bioavailability score. Then, we have not found a direct correlation between these estimations.
3.3. Molecular dynamics
3.3.1. System check
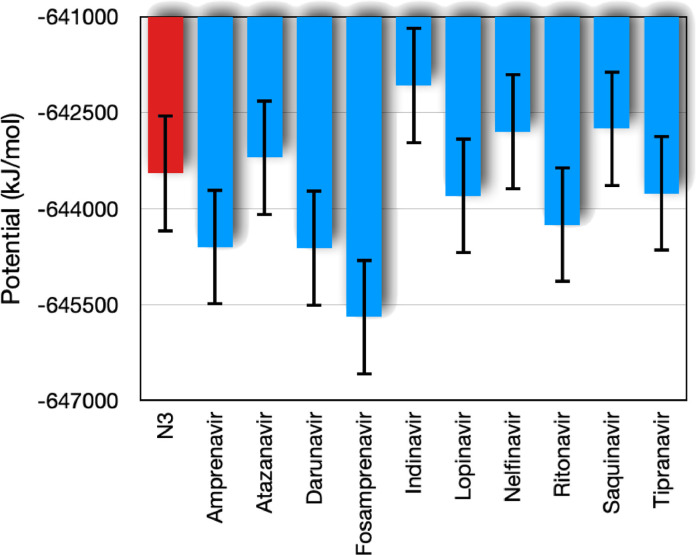
First, we analyze the energy of the system to check the convergence of simulations. To this end, we check the evolution of potential, kinetic and total energy for all complexes studied here. We verify that the kinetic energies of all complexes are very similar. On the other hand, we observe that the potential energy presents different values and some distinct cases. In Fig. 2 we display the mean value (in time) of the potential energy for the complexes with each ligand. This plot shows two compounds with distinct energy values, i.e., the Indinavir ( kJ/mol) and the Fosamprenavir ( kJ/mol). Visually, one can check the statistical compatibility of the Indinavir with N3 compound, considering 1σ interval. However, considering the same interval we verify the incompatibility between the potential energy of the Fosamprenavir and the N3 compound.
Fig. 2.
Time average of the potential energy (kJ/mol) of the complex formed by the SARS-CoV-2 main protease and HIV protease inhibitors (depicted in Table 1). The bars indicate the fluctuations provided by the root-mean-square deviation (σ) of the temporal average.
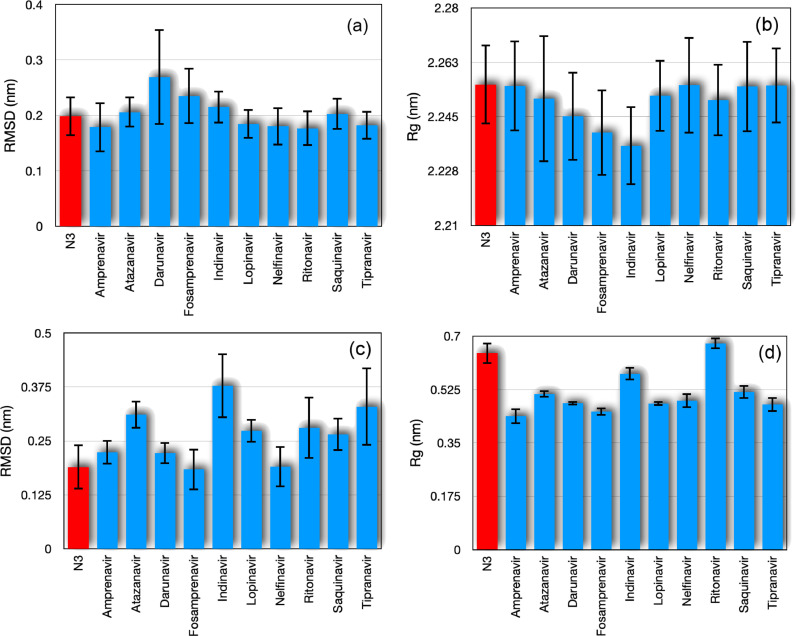
Following, we consider the root-mean-square deviation of atomic positions (RMSD), which is the measure of the average distance between the atoms of the system. This quantity can be used in the study of system conformations by measuring the similarity in three-dimensional structure by the RMSD of the Cα atomic coordinates between the input (at ) and the new atomic positions at some time t. In this sense, in Fig. 3 (a) we show a time average of the RMSD of the Cα atoms of SARS-CoV-2 main protease in complex with HIV protease inhibitors depicted in Table 1. However, we observed that the complexes containing Darunavir and the one with Fosamprenavir showed a higher RMSD value (also greater fluctuations). On the other hand, we can see that the results for the complexes with Amprenavir, Lopinavir, Nelfinavir, Ritonavir and Tipranavir showed RMSD lower than that obtained for the N3 inhibitor.
Fig. 3.
(a) Time average of root-mean-square deviation of atomic positions (RMSD) of the Cα atomic coordinates of protein and (b) radius of gyration (Rg) of all protein atoms. In panels (c) and (d) we show the same as displayed in panels (a) and (b), but considering the ligand atoms. The bars indicate the fluctuations provided by root-mean-square deviation (σ) of the temporal average.
Next, we analyze the radius of gyration (Rg) of the complex, which is a measure for the compactness of a structure defined as the root-mean-square distance of parts of the complex from either its center of mass (here evaluated for the SARS-CoV-2 main protease). We present the time average of the radius of gyration for the SARS-CoV-2 main protease in Fig. 3(b), for which the Atazanavir presents the highest fluctuation value. For this measure, we found that the complex with Indinavir has the lowest radius of gyration. Again, we stress that all observed values are compatible with that for the complex with N3 inhibitor.
In the results shown in 3(a) and 3(b), only the main protease Cα atoms were considered. We also investigated the same measures exclusively for binders. The results of the time average of the RMSD and Rg are displayed in Figs. 3(c) and 3(d), respectively. From these plots we can see that, although Indinavir and Nelfinavir have a lower Rg value than that presented by the N3 inhibitor, the RMSD value of Indinavir is very large and incompatible with that of N3, while Nelfinavir is still compatible with N3. This may be related to a large deformation of the structure of Indinavir ligand found by the autodock tool or that this compound does not bind as well as Nelfinavir to the SARS-CoV-2 main protease. RMSD values much higher than that obtained for the N3 inhibitor were also observed for Atazanavir, Lopinavir, Ritonavir, Saquinavir and Tipranavir.
A more detailed result is shown in Fig. 4 (a) in which we plot the evolution of the RMSD of the three different ligands, namely, N3 inhibitor, Indinavir and Nelfinavir. We focus specifically on the Indinavir and Nelfinavir based on the recent findings reported in Refs. [37], [38], in which they showed these two compounds as probable inhibitors of SARS-CoV-2 main protease in comparison with several other compounds. Here, we found an important feature of the complex with Indinavir, which is the strong fluctuation in RMSD of the ligand. This can imply an effective structural change in the ligand, compromising its complex with the SARS-CoV-2 main protease providing it with a disadvantage during the binding process. In the opposite direction, we observed that Nelfinavir aggregates with a RMSD value close to that of the N3 inhibitor, which suggests that it can keep the structure quite stable. Also, we display the time evolution of Rg in Fig. 4(c), which shows Nelfinavir with the lowest value of this measure.
Fig. 4.
(a) Root mean square deviation (RMSD) vs time, (b) root mean square fluctuation (RMSF) of each residue from SARS-CoV-2 main protease, and (c) Radius of gyration (Rg) vs time. The N3 inhibitor, Indinavir and Nelfinavir were considered, where the corresponding results are shown in black, red and green lines, respectively.
In Fig. 4(b) we display the root mean square fluctuation (RMSF), which is a measure of the deviation between the position of particle i and some reference position over time. Here we considered the Cα atoms as reference. We emphasize that for all potential inhibitors shown in Table 1, the RMSF values do not change considerably when compared to that of the N3 inhibitor.
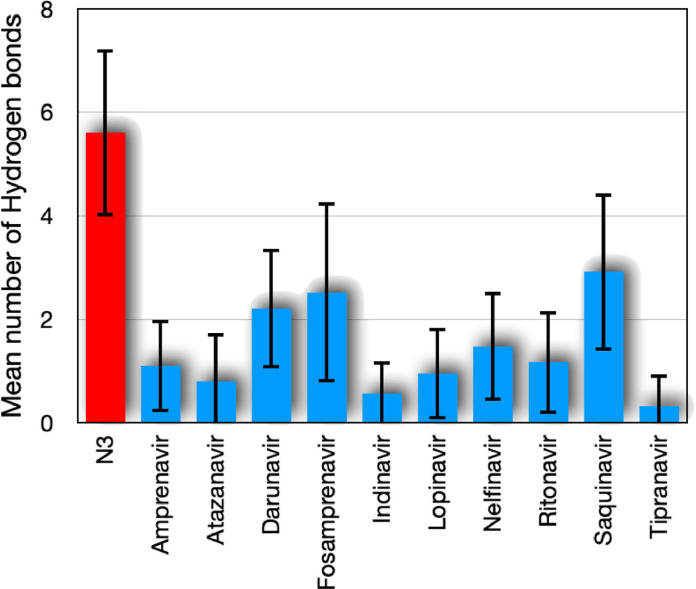
3.3.2. Hydrogen bonds
The interaction of a hydrogen atom covalently attached to an electronegative atom (donor D) with another electronegative atom (acceptor A) is known as the H bond. Protein folding, molecular recognition and protein structure are mostly supported by directional interactions from hydrogen bonds. Secondary structures, such as the α helix and the β sheet, are stabilized by the H bonds between the carbonyl oxygen of the main chain and the amide nitrogen. The structural rigidity of the protein is also associated with hydrogen bonds. The accepted geometry for an H-bond is a distance of less than 3.5 Ê between hydrogen D and A and a D-H-A angle of 180∘ ± 30∘. The time average of the number of hydrogen bonds between the SARS-CoV-2 main protease and the corresponding ligand are shown in Fig. 5 . We observed a higher value of the hydrogen bonds for the N3 inhibitor compared to the other ligands. This can indicate greater stability between the N3 inhibitor and the SARS-CoV-2 main protease. On the other hand, it can be seen that the results of the average number of hydrogen bonds for Darunavir, Fosamprenavir and Saquinavir were the highest among all the HIV inhibitors tested here. Specifically, by using the docked structures, the Darunavir presented a greater tendency to make bounds with the protease sites THR25, GLY143 and GLU166. In the case of the Fosamprenavir, the hydrogen bounds occurred more generally with the protease sites GLU166, GLY143, THR24, and CYS145. Finally, by analyzing the hydrogen bonds made by Saquinavir and the main protease, we observe the presence of the sites GLU166, HIS163, ANS142, GLY143, and SER144.
Fig. 5.
Time average of the number of hydrogen bonds between the SARS-CoV-2 main protease and its corresponding ligand.
3.3.3. Free energy surfaces (FES)
In a next step, we study the free energy surfaces (FES), which provides a pictorial representation for biomolecular processes, such as folding or aggregation. We define the FES as:
| (1) |
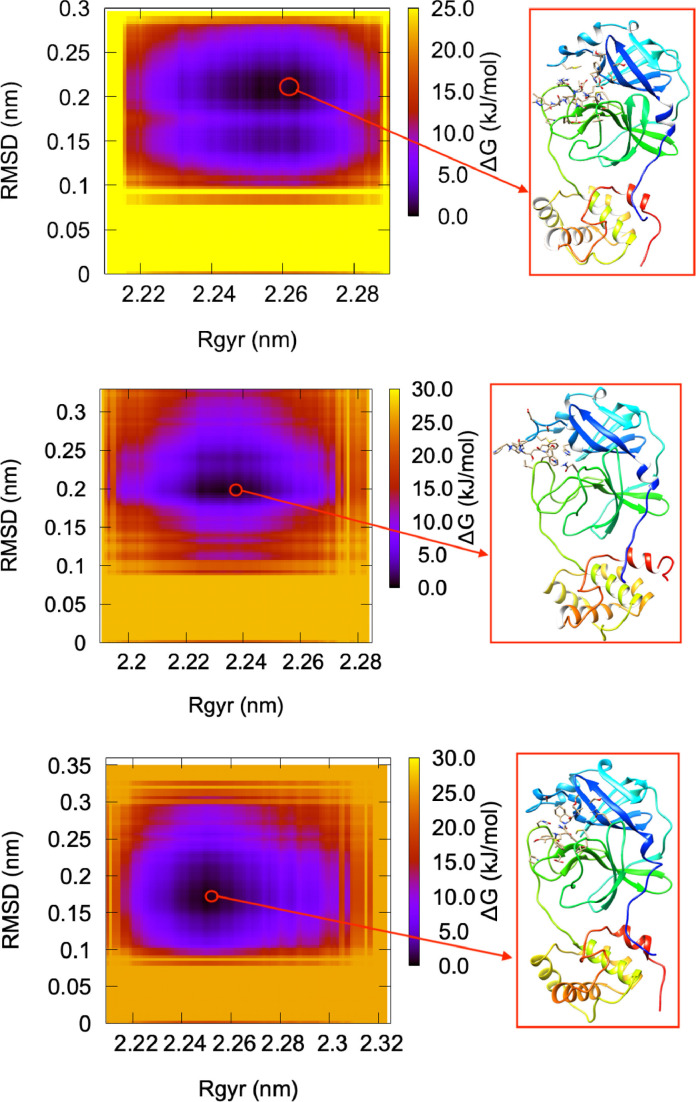
where kB is the Boltzmann constant, P is the probability distribution of the system along some coordinate (so-called order parameters), and denotes the maximum of probability distribution. Note that the rescale in the probability by is to ensure that for the lowest free energy minimum. The free energy is typically plotted along two such order parameters, giving rise to a (reduced) free energy surface (FES). Here, we use as coordinates the RMSD and Rg. As an example, we show in Fig. 6 the FES relating the RMSD and Rg of SARS-CoV-2 main protease in complex with N3 inhibitor, Indinavir, and Nelfinavir. In fact, we note a visible distortion in the structure of the SARS-CoV-2 main protease in complex with Indinavir. This can be an indication of instability in the structure caused by the inhibitor.
Fig. 6.
Free energy surface relating the RMSD and Rg of SARS-CoV-2 main protease in complex with N3 inhibitor (top), Indinavir (center), and Nelfinavir (bottom). The inset shows the minimum energy configuration obtained at ns, ns, and ns, respectively.
3.3.4. Ramachandran plot
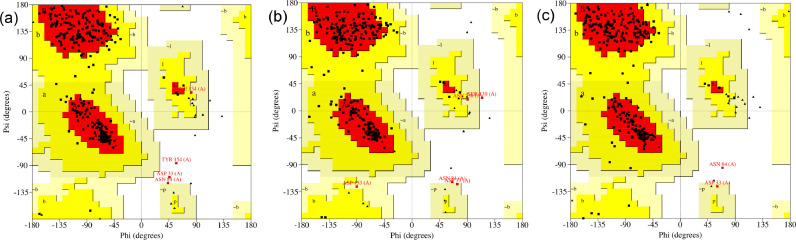
Finally, we analyze the complexes shown in Fig. 6 via the Ramachandran plot. This plot display of the torsional angles - Phi (Φ) and Psi (Ψ) - of the residues contained in a protein. The torsional angles of each residue in a protein define its conformation. The corresponding results are summarized in Fig. 7 . Considering the SARS-CoV-2 main protease in complex with N3 inhibitor, we observed that 89.8% of the residues are in most favored region, 8.7% in additional allowed regions, 0.4% in generously allowed regions, and 1.1% in disallowed regions. These values change to 86.8%, 11.3%, 1.1%, and 0.8%, respectively, to the SARS-CoV-2 main protease in complex with Indinavir. Also, we obtained the values 88.3%, 10.9%, 0.4%, and 0.4%, respectively, to the SARS-CoV-2 main protease in complex with Nelfinavir.
Fig. 7.
Ramachandran Plot. Based on an analysis of 118 structures of resolution of at least 2.0 Angstroms and R-factor no greater than 20%, a good quality model would be expected to have over 90% in the most favored regions [52], [53].
4. Conclusions
Attending to the global efforts responding in a timely fashion to fight against the SARS-CoV-2 pandemic outbreak we provided additional in silico data to 10 HIV HPIs that are supposed to be potential repurposed drugs for the treatment of this HCoV, and that were previously investigated by Refs. [37], [38]. Our molecular dynamics simulations showed that, despite their molecular differences, all HPIs presented a similar behavior for the parameters analyzed. However, in comparison with the N3 inhibitor, Nelfinavir presented better results, mainly for the RMSD and Rg dynamical parameters, and consequently for the free energy associated with structural biomolecular processes. Nevertheless, since our in silico results for Lopinavir and Ritonavir were found to be comparable with those of N3 and also with the other HPIs studied, and due to the recent experimental data that pointed the Lopinavir/Ritonavir combination as ineffective in the treatment of SARS-CoV-2 hospitalized patients [36], it is clear that the computational data must be used only as a guide and that in vitro and in vivo results must be taking in account before the selection of one of these HPIs as a repositioned drug for anti-SARS treatments.
CRediT authorship contribution statement
Wesley B. Cardoso: Conceptualization, Software, Formal analysis, Writing - original draft, Project administration. Sebastião A. Mendanha: Conceptualization, Software, Formal analysis, Writing - original draft, Project administration.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest.
Acknowledgements
The authors acknowledge the financial support of the Brazilian agencies CNPq (#306065/2019-3 & #425718/2018-2), CAPES, and FAPEG (PRONEM #201710267000540, PRONEX #201710267000503). This work was also performed as part of the Brazilian National Institute of Science and Technology (INCT) for Quantum Information (#465469/2014-0).
References
- 1.De Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) Coronavirus Disease (COVID-19) Dashboard, 2020.
- 3.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses – drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashburn T.T., Thor K.B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3(8):673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 6.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., Norris A., Sanseau P., Cavalla D., Pirmohamed M. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 7.Sadati S., Gheibi N., Ranjbar S., Hashemzadeh M. Docking study of flavonoid derivatives as potent inhibitors of influenza H1N1 virus neuraminidase. Biomed. Reports. 2018;10(1):33–38. doi: 10.3892/br.2018.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534(7607):314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- 9.Talevi A., Bellera C.L. Challenges and opportunities with drug repurposing: finding strategies to find alternative uses of therapeutics. Expert Opin. Drug Discov. 2020;15(4):397–401. doi: 10.1080/17460441.2020.1704729. [DOI] [PubMed] [Google Scholar]
- 10.Singh V.K., Srivastava R., Gupta P.S.S., Naaz F., Chaurasia H., Mishra R., Rana M.K., Singh R.K. Anti-HIV potential of diarylpyrimidine derivatives as non-nucleoside reverse transcriptase inhibitors: design, synthesis, docking, TOPKAT analysis and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2020:1–17. doi: 10.1080/07391102.2020.1748111. [DOI] [PubMed] [Google Scholar]
- 11.Sk M.F., Roy R., Kar P. Exploring the potency of currently used drugs against HIV-1 protease of subtype D variant by using multiscale simulations. J. Biomol. Struct. Dyn. 2020;0(0):1–16. doi: 10.1080/07391102.2020.1724196. [DOI] [PubMed] [Google Scholar]
- 12.Durrant J.D., Kochanek S.E., Casalino L., Ieong P.U., Dommer A.C., Amaro R.E. Mesoscale all-Atom influenza virus simulations suggest new substrate binding mechanism. ACS Cent. Sci. 2020;6(2):189–196. doi: 10.1021/acscentsci.9b01071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L., Ai H.-X., Li S.-M., Qi M.-Y., Zhao J., Zhao Q., Liu H.-S. Virtual screening approach to identifying influenza virus neuraminidase inhibitors using molecular docking combined with machine-learning-based scoring function. Oncotarget. 2017;8(47):83142–83154. doi: 10.18632/oncotarget.20915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sankar M., K L., Jeyachandran S., Pandi B. Screening of inhibitors as potential remedial against Ebolavirus infection: pharmacophore-based approach. J. Biomol. Struct. Dyn. 2020;0(0):1–14. doi: 10.1080/07391102.2020.1715260. [DOI] [PubMed] [Google Scholar]
- 15.Pappalardo M., Collu F., Macpherson J., Michaelis M., Fraternali F., Wass M.N. Investigating Ebola virus pathogenicity using molecular dynamics. BMC Genomics. 2017;18(S5):566. doi: 10.1186/s12864-017-3912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou Q., Zhang L. Biomimetic design of peptide neutralizer of Ebola Vvirus with molecular simulation. Langmuir. 2020;36(7):1813–1821. doi: 10.1021/acs.langmuir.9b03832. [DOI] [PubMed] [Google Scholar]
- 17.Choudhry H., Alzahrani F.A., Hassan M.A., Alghamdi A., Abdulaal W.H., Bakhrebah M.A., Zamzami M.A., Helmi N., Bokhari F.F., Zeyadi M., Baothman O.A., Kamal M.A., Warsi M.K., Ali A., Jarullah B., Jamal M.S. Zika virus targeting by screening inhibitors against NS2B/NS3 protease. Biomed Res. Int. 2019;2019:1–11. doi: 10.1155/2019/3947245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elfiky A.A., Ismail A.M. Molecular docking revealed the binding of nucleotide/side inhibitors to Zika viral polymerase solved structures. SAR QSAR Environ. Res. 2018;29(5):409–418. doi: 10.1080/1062936X.2018.1454981. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen P.T.V., Yu H., Keller P.A. Molecular docking studies to explore potential binding pockets and inhibitors for Chikungunya Vvirus envelope glycoproteins. Interdiscip. Sci. Comput. Life Sci. 2018;10(3):515–524. doi: 10.1007/s12539-016-0209-0. [DOI] [PubMed] [Google Scholar]
- 20.Isa D.M., Chin S.P., Chong W.L., Zain S.M., Rahman N.A., Lee V.S. Dynamics and binding interactions of peptide inhibitors of dengue virus entry. J. Biol. Phys. 2019;45(1):63–76. doi: 10.1007/s10867-018-9515-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhowmick S., Alissa S.A., Wabaidur S.M., Chikhale R.V., Islam M.A. Structure-guided screening of chemical database to identify NS3-NS2B inhibitors for effective therapeutic application in dengue infection. J. Mol. Recognit. 2020;33(7):1–18. doi: 10.1002/jmr.2838. [DOI] [PubMed] [Google Scholar]
- 22.Singh S., Vijaya Prabhu S., Suryanarayanan V., Bhardwaj R., Singh S.K., Dubey V.K. Molecular docking and structure-based virtual screening studies of potential drug target, CAAX prenyl proteases, of Leishmania donovani. J. Biomol. Struct. Dyn. 2016;34(11):2367–2386. doi: 10.1080/07391102.2015.1116411. [DOI] [PubMed] [Google Scholar]
- 23.Chaudhary M., Kumar N., Baldi A., Chandra R., Arockia Babu M., Madan J. Chloro and bromo-pyrazole curcumin Knoevenagel condensates augmented anticancer activity against human cervical cancer cells: design, synthesis, in silico docking and in vitro cytotoxicity analysis. J. Biomol. Struct. Dyn. 2020;38(1):200–218. doi: 10.1080/07391102.2019.1578264. [DOI] [PubMed] [Google Scholar]
- 24.Dutta S., Kharkar P.S., Sahu N.U., Khanna A. Molecular docking prediction and in vitro studies elucidate anti-cancer activity of phytoestrogens. Life Sci. 2017;185:73–84. doi: 10.1016/j.lfs.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 25.Ezat A.A., Elshemey W.M. A comparative study of the efficiency of HCV NS3/4A protease drugs against different HCV genotypes using in silico approaches. Life Sci. 2019;217:176–184. doi: 10.1016/j.lfs.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Saleh N.A., Elshemey W.M. Structure-based drug design of novel peptidomimetic cellulose derivatives as HCV-NS3 protease inhibitors. Life Sci. 2017;187:58–63. doi: 10.1016/j.lfs.2017.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Benítez-Cardoza C.G., Vique-Sánchez J.L. Potential inhibitors of the interaction between ACE2 and SARS-CoV-2 (RBD), to develop a drug. Life Sci. 2020;256:117970. doi: 10.1016/j.lfs.2020.117970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248(January):117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elfiky A.A. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X. Anti-retroviral drugs: current state and development in the next decade. Acta Pharm. Sin. B. 2018;8(2):131–136. doi: 10.1016/j.apsb.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Badley A.D. In vitro and in vivo effects of HIV protease inhibitors on apoptosis. Cell Death Differ. 2005;12:924–931. doi: 10.1038/sj.cdd.4401580. [DOI] [PubMed] [Google Scholar]
- 32.Chu C.M. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59(3):252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of middle east respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58(8):4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan J.F.-W., Yao Y., Yeung M.-L., Deng W., Bao L., Jia L., Li F., Xiao C., Gao H., Yu P., Cai J.-P., Chu H., Zhou J., Chen H., Qin C., Yuen K.-Y. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212(12):1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y.-c., Tung Y.-a., Lee K.-h., Chen T.-f., Hsiao Y.-c., Chang C., Hsieh T.-t., Su C.-h., Wang S.-s., Yu J.-y., Lin Y.-h., Lin Y.-h., Tu Y.-c.E., Tung C.-w. Potential therapeutic agents for COVID-19 based on the analysis of protease and RNA polymerase docking. Preprints. 2020;(February):2020020242. doi: 10.20944/preprints202002.0242.v1. [DOI] [Google Scholar]
- 38.Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S. Potential inhibitor of COVID-19 main protease (M pro) from several medicinal plant compounds by molecular docking study. Preprints. 2020;(March):1–14. doi: 10.20944/preprints202003.0226.v1. [DOI] [Google Scholar]
- 39.Berman H.M. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X.X.X., You T., Liu X.X., Yang X.X.X., Bai F., Liu H., Liu X.X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020 doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 41.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., Zaslavsky L., Zhang J., Bolton E.E. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y., Lv Z., Chu Y. HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV/AIDS - Res. Palliat. Care. 2015:95. doi: 10.2147/HIV.S79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7(1):42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46(1–3):3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 46.Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Berendsen H., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91(1–3):43–56. doi: 10.1016/0010-4655(95)00042-E. [DOI] [Google Scholar]
- 48.Huang J., MacKerell A.D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34(25):2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem. 2011;32(11):2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 50.Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103(19):8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 51.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81(8):3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 52.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26(2):283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 53.Laskowski R., Rullmann J., MacArthur M., Kaptein R., Thornton J. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8(4) doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 54.Ghose A.K., Viswanadhan V.N., Wendoloski J.J. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999;1(1):55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 55.Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002;45(12):2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 56.Egan W.J., Merz K.M., Baldwin J.J. Prediction of drug absorption using multivariate statistics. J. Med. Chem. 2000;43(21):3867–3877. doi: 10.1021/jm000292e. [DOI] [PubMed] [Google Scholar]
- 57.Muegge I., Heald S.L., Brittelli D. Simple selection criteria for drug-like chemical matter. J. Med. Chem. 2001;44(12):1841–1846. doi: 10.1021/jm015507e. [DOI] [PubMed] [Google Scholar]